Abstract
The Archaea represent the so-called Third Domain of life, which has evolved in parallel with the Bacteria and which is implicated to have played a pivotal role in the emergence of the eukaryotic domain of life. Recent progress in genomic sequencing technologies and cultivation-independent methods has started to unearth a plethora of data of novel, uncultivated archaeal lineages. Here, we review how the availability of such genomic data has revealed several important insights into the diversity, ecological relevance, metabolic capacity, and the origin and evolution of the archaeal domain of life.
1. Introduction
The description of the three (cellular) domains of life—Eukarya, Bacteria, and Archaea—by Carl Woese and George Fox [1] represents a milestone in the modern era of microbiology. In particular, using phylogenetic reconstructions of the small-subunit (16S or 18S) ribosomal RNA gene, Woese discovered that microscopically indistinguishable prokaryotes are not a homogeneous assemblage but are comprised of two fundamentally different groups of organisms: Eubacteria (later Bacteria) on one side and an additional life form referred to as Archaebacteria (later Archaea) on the other side [1]. Though not immediately accepted by the scientific community, this finding was early on supported by Wolfram Zillig through his studies on DNA-dependent RNA polymerases, as well as by Otto Kandler investigating “bacterial” cell walls [2]. Indeed, a subset of prokaryotic organisms subsequently assigned to Archaea was found to harbor DNA-dependent RNA polymerases that bore more similarity to those of eukaryotes, and to contain proteinaceous cell walls that lack peptidoglycan as well as cell membranes composed of L-glycerol ether lipids with isoprenoid chains instead of D-glycerol ester lipids with fatty acid chains [3–6]. Since then, further investigation of cellular characteristics of archaea has revealed that this domain of life contains eukaryotic-like information-processing machineries [7–14]. These findings were later supported by genome sequences and comparative analyses of genes coding for replication, transcription, and translation machineries as well as by protein crystal structures [15–21]. Additionally, some archaeal lineages were shown to contain homologs of eukaryotic cell division and cytoskeleton genes as well as histones and seem to express a chromatin architecture similar to eukaryotes [22–28]. In contrast to information-processing and cell division genes, archaeal operational systems (energy metabolism, biosynthesis pathways, and regulation) often appear to be more closely related to bacteria [29].
Based on phylogenetic reconstructions of the evolutionary history of 16S rRNA genes, the domain Archaea was originally divided into two major phyla: the Euryarchaeota and Crenarchaeota [30], which were separated by a deep split and thought to comprise only extremophilic (thermophilic, halophilic, and acidophilic) as well as methanogenic organisms. However, novel culture-independent and high-throughput sequencing techniques have recently uncovered a huge diversity of so far uncharacterized microorganisms on Earth as well as the ubiquitous occurrence of archaeal species [31–33]. Many of these novel archaeal groups are responsible for important ecological processes and are only distantly related to established lineages within Cren- and Euryarchaeota [31, 32, 34–39]. For example, the acquisition of genome sequences from novel archaeal representatives has led to the proposal of several additional archaeal phyla (including Nanoarchaeota, Korarchaeota, Thaumarchaeota, Aigarchaeota, and Geoarchaeota) [40–46] and the investigation of uncultivated archaea using single cell genomics has already started to add new insights into the phylogenetic diversity of the Third Domain of life and necessitates the definition of additional lineages of high taxonomic rank including novel potential phyla and superphyla [33, 39] (see also below). Furthermore, the investigation of the metabolic potential of these novel organisms has provided fundamentally new insights into major biogeochemical nutrient cycles. Indeed, archaea are now recognized as key players in various biogeochemical processes [47]. For example, the perception of the global nitrogen cycle has been deeply altered by discovering that the ability to gain energy solely from ammonia was not limited to a few bacteria but also included the ammonia-oxidizing Thaumarchaeota [48, 49]. Archaea also appear to play a significant role in the carbon cycle, since, in addition to all known methanogenic organisms on Earth, they also encompass anaerobic methane oxidizing archaea (ANME lineages 1–3) [50].
The study of archaeal genomes and diversity is also of considerable importance for a better understanding of eukaryotic evolution. Indeed, the discovery of eukaryotic features in archaea [10] has initiated a new basis for addressing the origin of eukaryotes [51–54]. Interestingly, recent phylogenetic analyses of universal proteins have suggested that eukaryotes might have evolved from a bona fide archaeal lineage that forms a sister-lineage of or a lineage emerging from within the TACK-superphylum comprised of Thaum-, Aig-, Cren-, and Korarchaeota [55–58].
Below we give a contemporary overview of how recent developments in archaeal genomic research have contributed to revealing new insights into the diversity, ecological relevance, metabolic capacity, and the origin and evolution of the archaeal domain of life.
2. The Methanogenic Nature of Archaea
The scientific community that addressed questions about prokaryotic energy metabolism on the early Earth or in hydrothermal vent systems [59] has proposed that methanogenesis and/or acetogenesis most likely represent ancient metabolic pathways [60–62]. Evidence for the biological production of methane as early as 3.46 Gyr ago supports these scenarios [63]. However, phylogenetic evidence placing methanogens at the base of the archaeal tree is limited and disputed. Depending on the outgroup and phylogenetic methods used, many recent analyses find either members of the Nanohaloarchaea, Nanoarchaeota, ARMAN-lineages, and/or Thermococcales as earliest (eury-)archaeal branches [55, 64, 65]. The latter observation is consistent with results from a base and amino acid composition analysis, which indicated that last archaeal common ancestor (LACA) was a hyperthermophilic organism [66]. The placement of Methanopyrus kandleri as the most basal branch of archaea in some of the earliest phylogenetic analyses can most likely be attributed to long-branch attraction (LBA) artifacts [67]. Notably, in recent phylogenetic analyses that include novel archaeal single cell genomes, Euryarchaeota form a sister group to other archaeal phyla rather than representing an early diverging lineage (Figure 1) [33]. Furthermore, gene content comparisons of extant archaeal lineages and reconstruction of the putative genetic repertoire of the LACA do not support methanogenesis as the earliest archaeal metabolism [57, 68]. In contrast, only one study has so far placed the root of archaea within a methanogenic order [69] and thus favors a methanogenic origin of the Third Domain of life. Gene content comparisons and network analyses that include novel archaeal single-cell amplified genomes (SAGs) Could potentially help to further investigate the metabolic gene repertoire of the archaeal ancestor.
Figure 1.
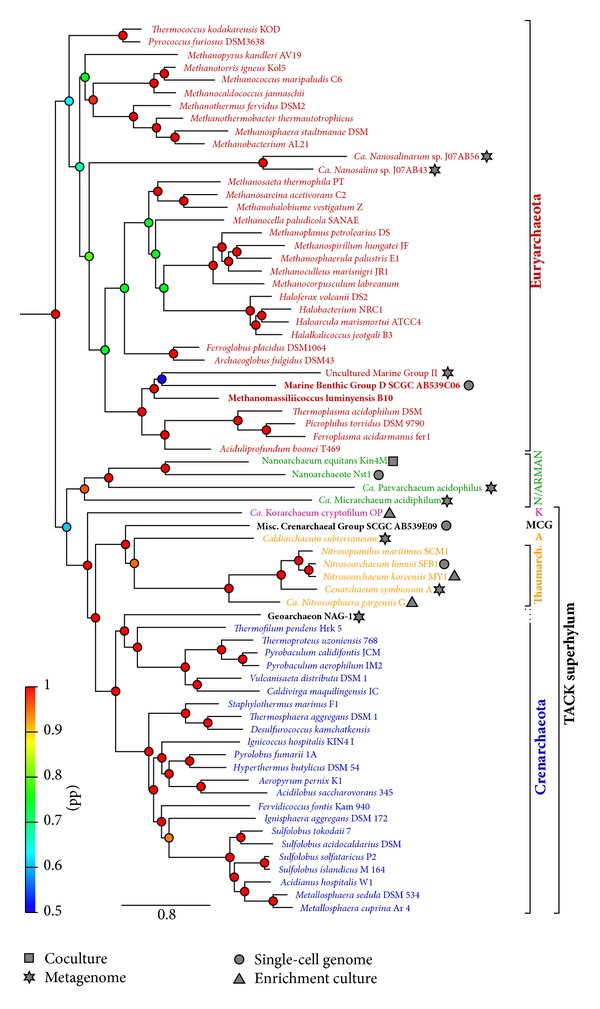
Bayesian phylogeny of 80 representative archaeal species. BLAST databases containing the proteome of 6 new archaeal genomes were retrieved from NCBI (in bold font on the tree): Methanomassiliicoccus luminyensis B10 (acc. no. CAJE01), MCG SCGC AB539E09 (acc. no. ALXK01), Marine Benthic Group D (MBGD) SCGC AB539N05, AB539C06, and AB540F20 (acc. no. ALXL01, AOSH01, and AOSI01, resp.). Protein sequence alignment from the 57 clusters in the discFilter 15 p dataset from [70] for which eukaryotes were removed were used as an input to psi-blast, with the six new proteomes as a database. Orthologs were retrieved as in [70]. For the three MBGD strains, one composite set of orthologs was constituted by using the most complete one (AB539C06) whenever possible and complementing with sequences from the other two if available. Orthologous genes selection, alignment, trimming, and concatenation were performed as in [70] resulting in a 15,069 amino-acid alignment. Four chains of Bayesian phylogenies were run with Phylobayes [71], under the CAT-Poisson model, running for approximately 10000 generations and discarding half as a burn-in. The tree was rooted with bacteria. Posterior probabilities (pp) are represented by colored dots on the nodes, with support values coloured according to the depicted heat-map colour scheme. The scale represents the number of substitutions per site. Species are colored according to the following: red, Euryarchaeota; green, Nanoarchaeota (N) and ARMAN; pink, Korarchaeota (K); black, Misc. Crenarchaeal Group (MCG); orange, Thaumarchaeota and Aigarchaeota (A); blue, Crenarchaeota. The DNA collection method, if different from pure culture, is indicated by a symbol next to the organism name: square, coculture; star, metagenome; circle, single-cell genome; triangle, enrichment culture.
Whereas the origin of methanogenic pathways that include a multitude of specific genes and cofactors is not fully resolved yet [72], it appears that several later emerging euryarchaeal lineages have lost their methanogenic lifestyles. Thus, as already noted more than a decade ago, methanogens comprise a paraphyletic group separated by nonmethanogenic euryarchaeal lineages such as the Thermoplasmatales, Haloarchaeota, and Archaeoglobales [73]. Interestingly, a novel methanogenic archaeal lineage has been described recently that is distantly affiliated with cultivated Thermoplasmatales including Aciduliprofundum sp. [74, 75]. This suggests that the last common ancestor of Thermoplasmatales was a methanogen and the capability to reduce methane has been independently lost several times along some branches within this group [76] or, albeit less likely, that some lineages within the Thermoplasmatales have regained genes for methane production.
A single acquisition of a plethora of genes (>1000) from a bacterial donor has recently been put forward as explanation for the transition from a methanogenic ancestor to aerobic heterotrophic Haloarchaeota [77]. A possible driving force for this massive gene transfer might have been a syntrophic relationship between a methanogenic recipient and a bacterial donor. However, the exact donor lineage could not be determined: the acquired genes bear conflicting phylogenetic signals, supposedly due to prevalent gene transfers between different bacterial species. So far, the origin of alternative energy metabolisms in other non-methanogenic euryarchaeal lineages that evolved from methanogenic ancestors has not been addressed properly. However, comparative genomics suggests that several of these lineages have retained specific genes that trace back to the methanogenic nature of their ancestor (e.g., Archaeoglobus) [78] and might point to a rather transient transition.
3. Phylogeny of New Archaeal Phyla and Lineages
In recent years, several new archaeal lineages have been identified and subjected to whole genome or metagenomic sequencing. Based on phylogenetic analyses of available genomic data, some of these lineages have been proposed to represent novel archaeal phyla. Yet, some of these claims have been challenged or falsified in follow-up studies. Below, we give an overview of several such examples.
The candidate phylum Nanoarchaeota has initially been proposed on basis of the extremely divergent 16S rRNA sequence of the small parasitic cells of Nanoarchaeum equitans growing attached to the cell surface of Ignicococcus hospitalis [41]. Several subsequent and more comprehensive phylogenetic analyses as well as the finding of potentially ancestral genomic features (e.g., split tRNA genes) have provided support for the initial assignment of this tiny archaeal cells to a separate ancient archaeal phylum [65, 79, 80]. Yet, in contrast, other phylogenetic and comparative analyses testing the taxonomic position of N. equitans have suggested that Nanoarchaeota might rather represent a fast-evolving euryarchaeal lineage related to Thermococcales [81]. Genomic data from additional “nanosized” archaea (Ca. Parvarchaeum acidophilus ARMAN-4 and Ca. Micrarchaeum acidiphilum ARMAN-2) [82] as well as of a novel deep-branching member of Nanoarchaeota (Nst1) [83] have enabled a revision of phylogenetic reconstructions and genome comparisons. Although some of these analyses suggest that Nanoarchaeota and Ca. Parvarchaeum acidophilus are monophyletic, the placement of these groups in the archaeal tree remains unclear and is strongly dependent on dataset and phylogenetic methods used [64, 83]. For example, in our phylogenetic reconstructions Nanoarchaeota (including ARMAN-lineages) represent a sister clade of the TACK superphylum (Figure 1), although the support for this clade is low. In a recent study by Rinke et al. [33], the Nanoarchaeota (including all ARMAN strains) were grouped together in the newly proposed superphylum DPANN with two novel groups, DSEG and pMC2A384 (designated “Aenigmarchaeota” and “Diapherotrites”, resp.), as well as the Nanohaloarchaea (see also below). Given that the phylogenetic methods employed by Rinke and coworkers do not accommodate rate heterogeneity across taxa, the proposed grouping of Nanoarchaeota with these archaeal clades has to be taken with care and the exact position of Nanoarchaeota still remains an unresolved question.
The Nanohaloarchaea represent yet another archaeal lineage comprised of small cells and with unresolved phylogenetic position. Based on both 16S rRNA gene and concatenated ribosomal protein phylogenies, this group was suggested to comprise a deep lineage of Haloarchaeota [84]. However, only euryarchaeal sequences were included in these maximum-likelihood (ML) analyses. Depending on the phylogenetic method and evolutionary model used, we obtained contradictory results for the phylogenetic position of this group. Whereas ML analyses tend to recover Nanohaloarchaea as earliest branching archaeal lineage (e.g., see above), a phylogenetic reconstruction using Bayesian methods (and the CAT model [71]) place this lineage within Euryarchaeota, but the exact position could not be resolved with high confidence (Figure 1). Results obtained with Bayesian methods using the CAT model might provide a better approximation of the position of Nanohaloarchaea, as this model accounts for rate variations across sites. As such, the early divergence of Nanohaloarchaea that is observed in ML-based methods is likely caused by LBA artifacts. However, novel phylogenetic analyses including the improved archaeal taxon sampling of Rinke et al. suggest that Nanohaloarchaea form a distinct lineage within the proposed superphylum DPANN and are not closely related to Euryarchaeota [33].
It will be interesting to further address the position of these organisms in the archaeal tree to be able to elucidate whether the adaptation to halophily has evolved only once in archaea or is due to convergence in Halo- and Nanohaloarchaea. The latter has received initial support from comparative genome analyses, which have revealed that each of these two archaeal groups seems to harbor diverse unique features including distinctive amino acid compositions to accommodate high salt conditions [84]. It might also be of value to address the effect of these novel genome sequences on the results obtained in the analysis of Nelson-Sathi et al. studying the origin of Haloarchaeota from a methanogenic ancestor [77].
Another novel archaeal phylum comprises the abundant and ecologically important ammonia-oxidizing archaea (AOA). On the basis of comparative genomics and phylogenetic analyses based on concatenated ribosomal proteins that were rooted with eukaryotes, Brochier-Armanet and coworkers proposed that “mesophilic crenarchaeota” constitute the novel deep branching archaeal phylum Thaumarchaeota [42, 85]. Additional comprehensive phylogenetic analyses including additional members of this group, as well as the discovery of a distinctive set of informational processing genes involved in replication, transcription, and translation as well as DNA repair and cell division machineries, have provided further support for the independent status of the Thaumarchaeota [44]. For example, in contrast to Crenarchaeota, Thaumarchaeota share several characteristics with Euryarchaeota and Korarchaeota including the presence of DNA polymerase D, histones, and cell division protein FtsZ. Furthermore, they contain putative “ancestral” features absent from Cren- or Euryarchaeota but common in Bacteria and eukaryotes (e.g., presence of an unsplit gene encoding DNA polymerase subunit A, and toposimerase IB as well as the absence of ribosomal protein LXa) [20, 44, 85]. The distinct nature of Thaumarchaeota has been accepted by many authors [45, 46, 86, 87] although the taxonomic borders of this phylum are still difficult to delineate and might only be resolved when genomes of uncultivated early branching lineages are made available. The early emergence of Thaumarchaeota in these phylogenetic reconstructions using eukaryotes as outgroup was initially assumed to indicate the ancient nature of this phylum [42, 44]. However, several recent phylogenetic analyses have recovered a monophyletic group of Thaum-, Aig-, Cren, Korarchaeota, and eukaryotes (with varying relationships in between these groups) to the exclusion of Euryarchaeota, which indicates that eukaryotes emerge from within the Archaea [55, 56, 88]. Thus, eukaryotes cannot be used as valid outgroup for the rooting of archaeal phylogenies [54].
Another lineage that emerges as a separate branch in the archaeal tree is comprised of the so-called Hot Water Crenarchaeotic Group I (HWCG I), members of which have been detected in diverse hydrothermal environments but have not yet been cultivated [89, 90]. Until recently, the sole representative with a sequenced genome in this group was Ca. Caldiarchaeum subterraneum, whose composite genome has been obtained from a metagenomic library of a microbial mat in a subsurface geothermal water stream [45]. The investigation of its genome sequence has revealed the presence of components of the eukaryotic ubiquitin-like protein modifier system previously not detected in archaea or bacteria. This unique trait, as well as comparative genomics and phylogenetic analyses of concatenated protein sequences, suggested that this organism and other members of HWCG I might constitute a novel phylum (Aigarchaeota), distinct from both Thaum- and Crenarchaeota [45]. However, due to the presence of a set of informational processing genes most similar to Thaumarchaeota [45] and the highly supported monophyletic grouping of these two lineages in diverse phylogenetic analyses (e.g., see Figure 1), the separation of Thaum- and Aigarchaeota into two distinct phyla is still debated [45, 55, 56, 64, 91, 92].
Uncultivated archaea belonging to the so-called Miscellaneous Crenarchaeotal Group (MCG) (e.g., [39]) have been suggested to represent additional members of Aigarchaeota [55]. Recently, the first single-cell genome of a member of this group has been obtained and phylogenetic analyses of concatenated conserved single copy genes placed the MCG-archaeon as a lineage in between Thaum- and Aigarchaeota [97]. However, our analyses rather suggest that MCG emerges prior to the Thaum/Aigarchaeota (Figure 1). The availability of additional genome sequences of members of this group as well as the comparison of informational processing marker genes [44] of MCG-archaea with other available archaeal genomes might help both to resolve their phylogenetic placement and to determine whether MCG-archaea comprise a separate archaeal phylum [39].
Geoarchaeota represents yet another recently proposed archaeal phylum, which is proposed to emerge as a basal lineage of Crenarchaeota and includes the so-called novel archaeal group I (NAG-1) detected in acidic ferric iron mats from Yellowstone National Park [46, 98]. NAG-1 organisms thrive in hot (60–78°C) acidic mats rich in iron and are suggested to grow heterotrophically from simple carbon compounds. Though not yet enriched in culture, nearly full-length genome sequences of members of this group have been obtained from a de novo metagenome assembly. The description of this lineage as a separate phylum was based on phylogenetic analyses of concatenated ribosomal proteins and 16S/23S rRNA genes as well as on its specific set of informational processing genes with features in common with either Crenarchaeota or Thaum- and Aigarchaeota [46]. However, our analyses, based on a larger dataset, place Geoarchaeota as an early branching lineage of the crenarchaeal order Thermoproteales (Figure 1). This observation is confirmed by Rinke et al., who sequenced six additional NAG-1-related strains [33]. Indeed, detailed phylogenetic analyses, as well as comparative assessment of the NAG-1 composite genome, seem to refute the phylum-level status of NAG-1 (Guy, L., Spang, A., Saw, J.H. and Ettema, T.J.G., unpublished observation).
4. Archaea and the Origin of Eukaryotes
The origin of the eukaryotes remains one of the major unanswered questions in modern biology, and archaea have recently reclaimed the spotlights in heated discussions entailing this enigmatic event. A central issue in this discussion entails the placement of the root within the Tree of Life, as it has a fundamental effect on any hypothesis on the origin of eukaryotes. Whereas diverse competing hypotheses have been put forward in the past, no consensus has been reached on this topic so far. For instance, several studies, including a recent network analysis, place the root between Archaea and Bacteria [99–104]. This view is in agreement with both the observed fundamental differences distinguishing the bacterial and archaeal domains as well as with the geological record. In contrast, studies that were based on transmission analyses or the distribution of indels in protein sequences suggested a rooting within the bacterial domain [105–107], whereas a root in the archaeal domain has been proposed based on analyses of protein folds or the evolution of the tRNA molecules [108, 109]. Yet other hypotheses state that LUCA was a eukaryotic-like organism [110, 111]. Certainly, in order to reach a consensus on this controversial discussion, additional data and analyses are needed. Bearing the uncertainty of the placement of the root in the Tree of Life in mind, we will present current hypotheses on the origin of eukaryotes below, by providing a short review on the most commonly proposed scenarios.
Even though a wide variety of incompatible theories have been suggested regarding the origin of the eukaryotic cell, three aspects are now largely accepted: (i) the last eukaryotic common ancestor (LECA) contained mitochondria, (ii) eukaryotic genomes are chimeric; whereas informational genes are of archaeal descent, many metabolic genes are derived from Bacteria, and (iii) eukaryotes complement a set of proteins not found in either Archaea or Bacteria, the eukaryotic signature proteins (ESPs). Beyond this, the picture becomes blurry. Currently, two major questions are of interest. What was the nature of the cell that was host in the mitochondrial endosymbiosis and when did cellular complexity evolve, before (complexity-first) or after (mitochondria-first) mitochondrial endosymbiosis? From this perspective, theories on eukaryogenesis can be divided into two categories. In the first scenario, the host was a protoeukaryote and complexity evolved first. This theory, often referred to as the “archezoa hypothesis” [112, 113], fits with the three domains tree of life model in which eukaryotes vertically evolved from the archaea-eukaryote common ancestor (Figure 2(a)). In the second scenario, the host was a prokaryote and the acquisition of the mitochondria likely triggered the evolution of cellular complexity. The latter are often referred to as “fusion” hypotheses [51, 58, 93, 95, 96] and these are generally incompatible with the classical three domains model. Rather, in these models, Bacteria and Archaea are the primary domains of life and eukaryotes a secondary, or derived, domain of life (Figure 2(b)). Theories that fit neither of these categories exist as well. These include the neomuran hypothesis [114] (Figure 2(c)), the PVC hypothesis [115–118], virus-assisted eukaryogenesis [119–122], and a hypothesis suggesting a eukaryote-like universal common ancestor [110] (Figure 2(d)). In order to choose the correct category with high confidence, evidence is needed in the form of protoeukaryote intermediate lineage's descendants (“missing links”). Unfortunately, for either category, none has been found so far. Whereas the archezoa theory has lost much support ever since remnants of mitochondria were found in the previously thought archezoa (for review, see [123]), the fusion theory has slowly been gaining favor. Initially lightly supported by ribosomal structural features [124] and an 11-amino acid insertion in EF-1α/EF-Tu [125, 126] shared between eocytes (Crenarchaeota) and eukaryotes to the exclusion of other prokaryotes, it has now received strong support from phylogenomic [55, 56, 70, 88, 127, 128] and gene similarity network analyses [129]. In addition, a large number of ESPs has been found in Archaea, in particular within the recently proposed TACK superphylum [55]. Examples include actin [53, 130], tubulin [28], H3/H4-type histones [55], ESCRT-III [24, 25, 131], and components of the ubiquitin modifier system [45]. Fusion models can be subdivided based upon the nature of the end-product of the “fusion”. In amitochondriate models the symbiosis results in a eukaryotic progenitor lacking mitochondria. They are similar to the archezoa theory in the sense that the origin of eukaryotes and the origin of mitochondria are separate events. These include the serial endosymbiosis theory (SET) [93], the original syntrophy hypothesis [94], and the eocyte hypothesis [96]. In mitochondriate models, the end product is a eukaryotic progenitor containing mitochondria. Here, the origin of eukaryotes and mitochondria are one and the same. These include the hydrogen hypothesis [95], the alternative syntrophy hypothesis [51] and the recently proposed phagocytosing archaeon theory [53, 58]. With exception of the eocyte hypothesis, all fusion theories suggest an archaeal host. Based on extensive, in-depth phylogenomic studies, the archaeal host most likely emerged from within the TACK superphylum [55, 56, 70]. Interestingly, out of all TACK phyla, a sister relationship between the Korarchaeota and eukaryotes was retrieved with significant phylogenetic support [56, 70]. Even though this placement could be a taxon sampling artifact (Korarchaeota are represented by a single, deep rooting taxon), it could also indicate that eukaryotes are affiliated with an unidentified lineage distantly related to Korarchaeota. Genomically unexplored lineages such as DSAG (Deep Sea Archaea Group), MHVG (Marine Hydrothermal Vent Group), and AAG (Ancient Archaea Group) are likely candidates [55, 70].
Figure 2.
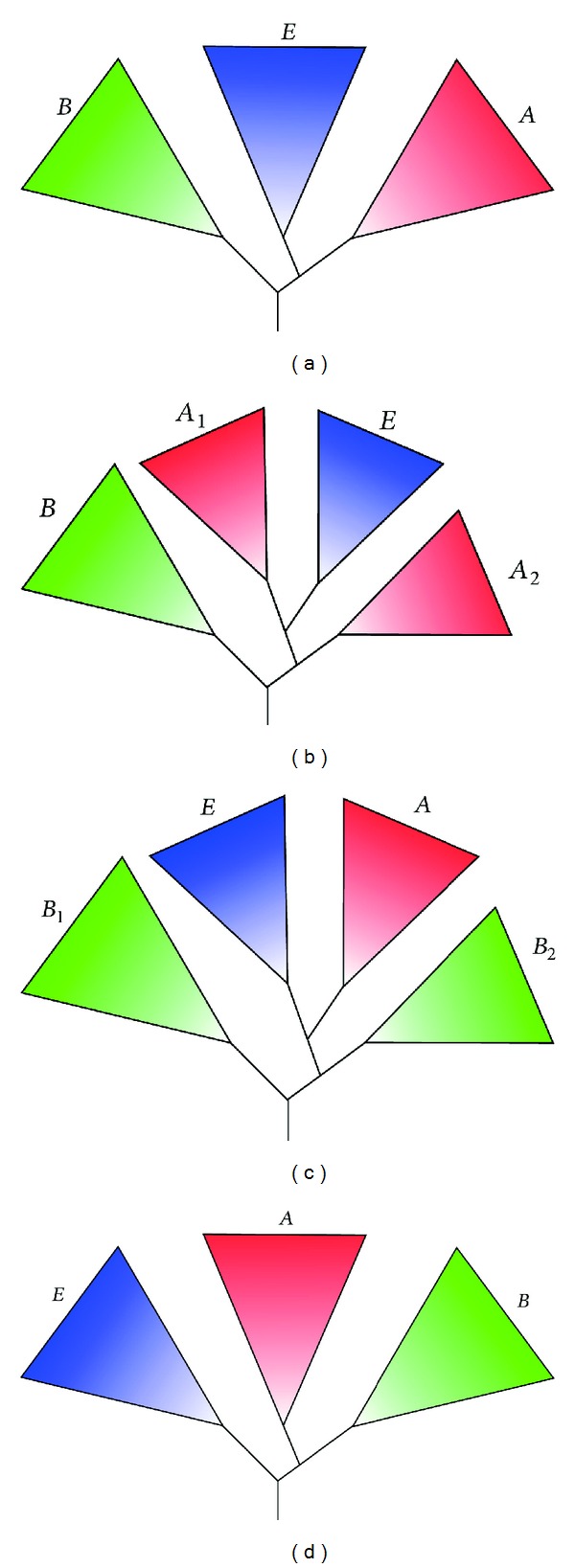
Overview of theories regarding the origin of the eukaryotic nuclear lineage. (a) The classical, Woesean three domains of life tree in which the nuclear lineage vertically evolved from the archaea-eukaryote common ancestor. (b) The fusion tree in which the nuclear lineage originated from the archaeal partner in the fusion. Depending on which fusion model, the archaeal parent's lineage (A 1) was either part of the euryarchaeota [SET [93], original syntrophy hypothesis [94], hydrogen hypothesis [95] or alternative syntrophy hypothesis [51]], the Crenarchaeota (eocyte hypothesis) [96], or the TACK superphylum (PhAT) [58]. “A 2” represents all archaea not directly affiliated with “A 1.” (c) The neomuran tree in which the eukaryotic and archaeal lineage (combined referred to as “neomurans”), evolved vertically from ancestor shared with actinobacteria (B 2) as a result of the loss of bacterial-type cell wall (the neomuran revolution). B 1 represents all bacteria not directly affiliated with B 2. (d) The eukaryote-early tree, which suggests that the last common universal ancestor was more eukaryote-like than prokaryote-like.
5. Genomic Assessment and Taxonomic Classification of Archaeal Diversity
Recent progress in genomic sequencing technologies and cultivation-independent methods has started to unearth a plethora of novel, uncultivated archaeal lineages. The availability of such genomic data has revealed several important insights into the diversity, ecological relevance, metabolic capacity, and the origin and evolution of the archaeal domain of life. Several new archaeal lineages have been obtained by means of metagenomics approaches, such as sequencing of enrichment cultures or environmental samples. Examples of the former include the first korarchaeal genome [43] and several of the available thaumarchaeal genomes (e.g. [132, 133]). Archaeal genomes that have been retrieved from metagenomic datasets include the first thaumarchaeal genome (Ca. Cenarchaeum symbiosum [134]), the genome of the proposed Aigarchaeon Ca. C. subterraneum [45], the proposed Geoarchaeon NAG-1 [46], representatives of the Nanohaloarchaea [84], several ARMAN lineages that were part of an acid mine drainage microbial community [82], and a genome derived from a representative of the uncultivated marine group II euryarchaeota [135] (Figure 1). More recently, a number of studies have employed single cell genomic approaches to probe the genetic diversity of uncultivated archaea. For example, Lloyd and coworkers have reported the first genomic data of a representative of the Miscellaneous Crenarchaeal Group (MCG) and of members of the Marine Benthic Group D that were isolated from marine sediments and speculate that these lineages are involved in the degradation of detrital proteins [97] (Figure 1). Another large scale study that aimed at uncovering the coding potential of so-called “microbial dark matter” using single cell genomics approaches reported several genome sequences of cells that potentially represented novel phylum-level archaeal lineages, including the members of the uncultured DSEG and pMC2A384 clades, designated Aenigmarchaeota and Diapherotrites, respectively [33]. A combination of single cell genomics and metagenomics has been used to sequence the genome of the thaumarchaeon Ca. Nitrosoarchaeum limnia SFB1 [136].
Obviously, single cell and metagenomics-oriented projects will continue to probe the existing archaeal diversity during the coming years, and most likely, the availability of genomic data will reveal interesting insights into novel characteristics and the diversity within the Third Domain of life. In addition, the availability of such genomic data is likely to trigger discussions regarding the higher-order taxonomic classification of the major archaeal lineages. To many (micro-)biologists, it would appear that the archaeal domain is far less diverse than the bacterial domain. A reason for this could be, for instance, the discrepancy in assigned or proposed phyla, which ranges from a handful in Archaea, to well over a hundred in Bacteria. But is it really fair to say that the bacterial domain of life is more diverse than that of the Archaea? Whereas bacterial phyla generally have been assigned based on the diversity of the 16S rDNA gene sequence, archaeal taxonomy is largely founded on historic grounds, that is, adhering to the classical Cren-Euryarchaeota dichotomy (sensu Woese [30]). Only during the past decade, a handful of additional archaeal phyla have been proposed based on genome sequencing, such as the Nano-, Kor-, and Thaumarchaeota and a few other lineages that may or may not represent phylum-level archaeal clades (also see above). Yet, the majority of archaeal species that have been sequenced in recent years have been assigned to the phyla Cren- or Euryarchaeota, each of which now comprise genetically distinct groups, which differ in terms of metabolic capacity, lifestyle, and environmental distribution. In light of this and of the abovementioned “superficial” imbalance in bacterial versus archaeal diversity, one could argue that a revision of archaeal higher-order taxonomy is in place. The suggestion to bring order into archaeal systematics was recently put forward [92], but thus far, a framework as to how novel phyla and/or superphyla should be defined is debated. Nevertheless, to be able to fully appreciate the overall archaeal diversity and compare it to the diversity observed within the bacterial domain of life, a reappraisal of the archaeal taxonomy, whether it will be at the level of rRNA genes, large datasets of concatenated protein sequences, genome content, or gene networks analyses, seems to be a conditio sine qua non.
Conflicts of Interests
The authors declare that they do not have a direct financial relation with the trademarks mentioned in the paper that might lead to a conflict of interests for the authors.
Acknowledgments
The work in Ettema laboratory is supported by the Swedish Research Council (Grant no. 621-2009-4813), by the European Research Council (ERC) (Grant no. 310039-PUZZLE_CELL), by a Marie Curie European Reintegration Grant (ERG) (Grant no. 268259-RICKOCHET), and by the Carl Tryggers Stiftelse (Grant no. CTS11:127).
Abbreviations
- ANME:
Anaerobic methanotrophic archaea
- LACA:
Last archaeal common ancestor
- LBA:
Long-branch attraction
- MCG:
Miscellaneous Crenarchaeotal Group
- ML:
Maximum-likelihood
- SAGs:
Single-cell amplified genomes.
References
- 1.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proceedings of the National Academy of Sciences of the United States of America. 1977;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woese CR. The birth of the Archaea: a personal retrospective. In: Garrett RA, Klenk H-P, editors. Archaea. Blackwell Publishing; 2007. [Google Scholar]
- 3.Langworthy TA, Smith PF, Mayberry WR. Lipids of Thermoplasma acidophilum. Journal of Bacteriology. 1972;112(3):1193–1200. doi: 10.1128/jb.112.3.1193-1200.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langworthy TA, Mayberry WR, Smith PF. Long chain glycerol diether and polyol dialkyl glycerol triether lipids of Sulfolobus acidocaldarius. Journal of Bacteriology. 1974;119(1):106–116. doi: 10.1128/jb.119.1.106-116.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kandler O, Hippe H. Lack of peptidoglycan in the cell walls of Methanosarcina barkeri. Archives of Microbiology. 1977;113(1-2):57–60. doi: 10.1007/BF00428580. [DOI] [PubMed] [Google Scholar]
- 6.Zillig W, Stetter KO, Janekovic D. DNA-dependent RNA polymerase from the archaebacterium Sulfolobus acidocaldarius . European Journal of Biochemistry. 1979;96(3):597–604. doi: 10.1111/j.1432-1033.1979.tb13074.x. [DOI] [PubMed] [Google Scholar]
- 7.Huet J, Schnabel R, Sentenac A, Zillig W. Archaebacteria and eukaryotes possess DNA-dependent RNA polymerases of a common type. EMBO Journal. 1983;2(8):1291–1294. doi: 10.1002/j.1460-2075.1983.tb01583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lake JA. Ribosome evolution: the structural bases of protein synthesis in archaebacteria, eubacteria, and eukaryotes. Progress in Nucleic Acid Research and Molecular Biology. 1983;30:163–194. doi: 10.1016/s0079-6603(08)60686-8. [DOI] [PubMed] [Google Scholar]
- 9.Henderson E, Oakes M, Clark MW, Lake JA, Matheson AT, Zillig W. A new ribosome structure. Science. 1984;225(4661):510–512. doi: 10.1126/science.6429855. [DOI] [PubMed] [Google Scholar]
- 10.Zillig W, Schnabel R, Stetter KO. Archaebacteria and the origin of the eukaryotic cytoplasm. Current Topics in Microbiology and Immunology. 1985;114:1–18. doi: 10.1007/978-3-642-70227-3_1. [DOI] [PubMed] [Google Scholar]
- 11.Puhler G, Leffers H, Gropp F, et al. Archaebacterial DNA-dependent RNA polymerase testify to the evolution of the eukaryotic nuclear genome. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(12):4569–4573. doi: 10.1073/pnas.86.12.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandman K, Krzycki JA, Dobrinski B, Lurz B, Reeve JN. HMf, a DNA-binding protein isolated from the hyperthermophilic archaeon Methanothermus fervidus, is most closely related to histones. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(15):5788–5791. doi: 10.1073/pnas.87.15.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langer D, Hain J, Thuriaux P, Zillig W. Transcription in archaea: similarity to that in eucarya. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(13):5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kandler O, König H. Cell wall polymers in Archaea (Archaebacteria) Cellular and Molecular Life Sciences. 1998;54(4):305–308. doi: 10.1007/s000180050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olsen GJ, Woese CR. Lessons from an Archaeal genome: what are we learning from Methanococcus jannaschii? Trends in Genetics. 1996;12(10):377–379. doi: 10.1016/0168-9525(96)30092-9. [DOI] [PubMed] [Google Scholar]
- 16.Lecompte O, Ripp R, Thierry J-C, Moras D, Poch O. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Research. 2002;30(24):5382–5390. doi: 10.1093/nar/gkf693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barry ER, Bell SD. DNA replication in the archaea. Microbiology and Molecular Biology Reviews. 2006;70(4):876–887. doi: 10.1128/MMBR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gribaldo S, Brochier-Armanet C. The origin and evolution of Archaea: a state of the art. Philosophical Transactions of the Royal Society B. 2006;361(1470):1007–1022. doi: 10.1098/rstb.2006.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grochowski LL, Xu H, White RH. Methanocaldococcus jannaschii uses a modified mevalonate pathway for biosynthesis of isopentenyl diphosphate. Journal of Bacteriology. 2006;188(9):3192–3198. doi: 10.1128/JB.188.9.3192-3198.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwapisz M, Beckouët F, Thuriaux P. Early evolution of eukaryotic DNA-dependent RNA polymerases. Trends in Genetics. 2008;24(5):211–215. doi: 10.1016/j.tig.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nature Reviews Microbiology. 2011;9(2):85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- 22.Pereira SL, Reeve JN. Histones and nucleosomes in Archaea and Eukarya: a comparative analysis. Extremophiles. 1998;2(3):141–148. doi: 10.1007/s007920050053. [DOI] [PubMed] [Google Scholar]
- 23.Reeve JN, Bailey KA, Li W-T, Marc F, Sandman K, Soares DJ. Archaeal histones: structures, stability and DNA binding. Biochemical Society Transactions. 2004;32(2):227–230. doi: 10.1042/bst0320227. [DOI] [PubMed] [Google Scholar]
- 24.Lindås A-C, Karlsson EA, Lindgren MT, Ettema TJG, Bernander R. A unique cell division machinery in the Archaea. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(48):18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322(5908):1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernander R, Lind AE, Ettema TJG. An archaeal origin for the actin cytoskeleton. Communicative & Integrative Biology. 2011;4:664–667. doi: 10.4161/cib.16974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ammar R, Torti D, Tsui K, et al. Chromatin is an ancient innovation conserved between Archaea and Eukarya. Elife. 2012;1 doi: 10.7554/eLife.00078.e00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yutin N, Koonin EV. Archaeal origin of tubulin. Biology Direct. 2012;7, article 10 doi: 10.1186/1745-6150-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rivera MC, Jain R, Moore JE, Lake JA. Genomic evidence for two functionally distinct gene classes. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(11):6239–6244. doi: 10.1073/pnas.95.11.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schleper C, Jurgens G, Jonuscheit M. Genomic studies of uncultivated archaea. Nature Reviews Microbiology. 2005;3(6):479–488. doi: 10.1038/nrmicro1159. [DOI] [PubMed] [Google Scholar]
- 32.Teske A, Sørensen KB. Uncultured archaea in deep marine subsurface sediments: have we caught them all? The ISME Journal. 2008;2(1):3–18. doi: 10.1038/ismej.2007.90. [DOI] [PubMed] [Google Scholar]
- 33.Rinke C, Schwientek P, Sczyrba A, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013 doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 34.McInerney JO, Wilkinson M, Patching JW, Embley TM, Powell R. Recovery and phylogenetic analysis of novel archaeal rRNA sequences from a deep-sea deposit feeder. Applied and Environmental Microbiology. 1995;61(4):1646–1648. doi: 10.1128/aem.61.4.1646-1648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delong EF. Everything in moderation: Archaea as ‘non-extremophiles’. Current Opinion in Genetics & Development. 1998;8(6):649–654. doi: 10.1016/s0959-437x(98)80032-4. [DOI] [PubMed] [Google Scholar]
- 36.Takai K, Horikoshi K. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics. 1999;152(4):1285–1297. doi: 10.1093/genetics/152.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker BJ, Tyson GW, Webb RI, et al. Lineages of acidophilic archaea revealed by community genomic analysis. Science. 2006;314(5807):1933–1935. doi: 10.1126/science.1132690. [DOI] [PubMed] [Google Scholar]
- 38.Chaban B, Ng SYM, Jarrell KF. Archaeal habitats—from the extreme to the ordinary. Canadian Journal of Microbiology. 2006;52(2):73–116. doi: 10.1139/w05-147. [DOI] [PubMed] [Google Scholar]
- 39.Kubo K, Lloyd KG, F Biddle J, Amann R, Teske A, Knittel K. Archaea of the Miscellaneous Crenarchaeotal Group are abundant, diverse and widespread in marine sediments. The ISME Journal. 2012;6:1949–1965. doi: 10.1038/ismej.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barns SM, Delwiche CF, Palmer JD, Pace NR. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(17):9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, Stetter KO. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417(6884):63–67. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- 42.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nature Reviews Microbiology. 2008;6(3):245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 43.Elkins JG, Podar M, Graham DE, et al. A korarchaeal genome reveals insights into the evolution of the Archaea. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(23):8102–8107. doi: 10.1073/pnas.0801980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spang A, Hatzenpichler R, Brochier-Armanet C, et al. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends in Microbiology. 2010;18(8):331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 45.Nunoura T, Takaki Y, Kakuta J, et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Research. 2011;39(8):3204–3223. doi: 10.1093/nar/gkq1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kozubal MA, Romine M, Jennings R, et al. Geoarchaeota: a new candidate phylum in the Archaea from high-temperature acidic iron mats in Yellowstone National Park. The ISME Journal. 2013;7:622–634. doi: 10.1038/ismej.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Offre P, Spang A, Schleper C. Archaea in biogeochemical cycles. Annual Review of Microbiology. 2013;67:437–457. doi: 10.1146/annurev-micro-092412-155614. [DOI] [PubMed] [Google Scholar]
- 48.Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437(7058):543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 49.Treusch AH, Leininger S, Kietzin A, Schuster SC, Klenk H-P, Schleper C. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environmental Microbiology. 2005;7(12):1985–1995. doi: 10.1111/j.1462-2920.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- 50.Knittel K, Boetius A. Anaerobic oxidation of methane: progress with an unknown process. Annual Review of Microbiology. 2009;63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- 51.López-Garćia P, Moreira D. Metabolic symbiosis at the origin of eukaryotes. Trends in Biochemical Sciences. 1999;24:88–93. doi: 10.1016/s0968-0004(98)01342-5. [DOI] [PubMed] [Google Scholar]
- 52.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440(7084):623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 53.Yutin N, Wolf MY, Wolf YI, Koonin EV. The origins of phagocytosis and eukaryogenesis. Biology Direct. 2009;4, article 9 doi: 10.1186/1745-6150-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gribaldo S, Poole AM, Daubin V, Forterre P, Brochier-Armanet C. The origin of eukaryotes and their relationship with the Archaea: are we at a phylogenomic impasse? Nature Reviews Microbiology. 2010;8(10):743–752. doi: 10.1038/nrmicro2426. [DOI] [PubMed] [Google Scholar]
- 55.Guy L, Ettema TJG. The archaeal “TACK” superphylum and the origin of eukaryotes. Trends in Microbiology. 2011;19(12):580–587. doi: 10.1016/j.tim.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 56.Williams TA, Foster PG, Nye TMW, Cox CJ, Embley TM. A congruent phylogenomic signal places eukaryotes within the Archaea. Proceedings of the Royal Society B. 2012;279(1749):4870–4879. doi: 10.1098/rspb.2012.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolf YI, Makarova KS, Yutin N, Koonin EV. Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer. Biology Direct. 2012;7, article 46 doi: 10.1186/1745-6150-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martijn J, Ettema Thijs JG. From archaeon to eukaryote: the evolutionary dark ages of the eukaryotic cell. Biochemical Society Transactions. 2013;41:451–457. doi: 10.1042/BST20120292. [DOI] [PubMed] [Google Scholar]
- 59.Martin W, Baross J, Kelley D, Russell MJ. Hydrothermal vents and the origin of life. Nature Reviews Microbiology. 2008;6(11):805–814. doi: 10.1038/nrmicro1991. [DOI] [PubMed] [Google Scholar]
- 60.Lane N, Martin WF. The origin of membrane bioenergetics. Cell. 2012;151:1406–1416. doi: 10.1016/j.cell.2012.11.050. [DOI] [PubMed] [Google Scholar]
- 61.Liu Y, Beer LL, Whitman WB. Methanogens: a window into ancient sulfur metabolism. Trends in Microbiology. 2012;20(5):251–258. doi: 10.1016/j.tim.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 62.Poehlein A, Schmidt S, Kaster A-K, et al. An ancient pathway combining carbon dioxide fixation with the generation and utilization of a sodium ion gradient for ATP synthesis. PLoS ONE. 2012;7(3) doi: 10.1371/journal.pone.0033439.e33439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ueno Y, Yamada K, Yoshida N, Maruyama S, Isozaki Y. Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era. Nature. 2006;440(7083):516–519. doi: 10.1038/nature04584. [DOI] [PubMed] [Google Scholar]
- 64.Brochier-Armanet C, Forterre P, Gribaldo S. Phylogeny and evolution of the Archaea: one hundred genomes later. Current Opinion in Microbiology. 2011;14(3):274–281. doi: 10.1016/j.mib.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 65.Yutin N, Puigbò P, Koonin EV, Wolf YI. Phylogenomics of prokaryotic ribosomal proteins. PLoS ONE. 2012;7(5) doi: 10.1371/journal.pone.0036972.e36972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Groussin M, Gouy M. Adaptation to environmental temperature is a major determinant of molecular evolutionary rates in archaea. Molecular Biology and Evolution. 2011;28(9):2661–2674. doi: 10.1093/molbev/msr098. [DOI] [PubMed] [Google Scholar]
- 67.Brochier C, Forterre P, Gribaldo S. Archaeal phylogeny based on proteins of the transcription and translation machineries: tackling the Methanopyrus kandleri paradox. Genome Biology. 2004;5(3):p. R17. doi: 10.1186/gb-2004-5-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Makarova KS, Sorokin AV, Novichkov PS, Wolf YI, Koonin EV. Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of archaea. Biology Direct. 2007;2, article 33 doi: 10.1186/1745-6150-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kelly S, Wickstead B, Gull K. Archaeal phylogenomics provides evidence in support of a methanogenic origin of the Archaea and a thaumarchaeal origin for the eukaryotes. Proceedings of the Royal Society B. 2011;278(1708):1009–1018. doi: 10.1098/rspb.2010.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guy L, Saw JH, Ettema TJG. The archaeal legacy of eukaryotes: a phylogenomic perspective. doi: 10.1101/cshperspect.a016022. Cold Spring Harbor Perspectives in Biology. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25(17):2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- 72.Ferry JG. How to make a living by exhaling methane. Annual Review of Microbiology. 2010;64:453–473. doi: 10.1146/annurev.micro.112408.134051. [DOI] [PubMed] [Google Scholar]
- 73.Matte-Tailliez O, Brochier C, Forterre P, Philippe H. Archaeal phylogeny based on ribosomal proteins. Molecular Biology and Evolution. 2002;19(5):631–639. doi: 10.1093/oxfordjournals.molbev.a004122. [DOI] [PubMed] [Google Scholar]
- 74.Dridi B, Fardeau M-L, Ollivier B, Raoult D, Drancourt M. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. International Journal of Systematic and Evolutionary Microbiology. 2012;62:1902–1907. doi: 10.1099/ijs.0.033712-0. [DOI] [PubMed] [Google Scholar]
- 75.Paul K, Nonoh JO, Mikulski L, Brune A. ‘Methanoplasmatales,’ Thermoplasmatales-related archaea in termite guts and other environments, are the seventh order of methanogens. Applied and Environmental Microbiology. 2012;78:8245–8253. doi: 10.1128/AEM.02193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Borrel G, O'Toole PW, Harris HMB, Peyret P, Brugère J-F, Gribaldo S. Phylogenomic data support a seventh order of methylotrophic methanogens and provide insights into the evolution of methanogenesis. Genome Biology and Evolution. 2013 doi: 10.1093/gbe/evt128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nelson-Sathi S, Dagan T, Landan G, et al. Acquisition of 1, 000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proceedings of the National Academy of Sciences. 2012;109:20537–20542. doi: 10.1073/pnas.1209119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schworer B, Breitung J, Klein AR, Stetter KO, Thauer RK. Formylmethanofuran: tetrahydromethanopterin formyltransferase and N5,N10-methylenetetrahydromethanopterin dehydrogenase from the sulfate-reducing Archaeoglobus fulgidus: similarities with the enzymes from methanogenic Archaea. Archives of Microbiology. 1993;159(3):225–232. doi: 10.1007/BF00248476. [DOI] [PubMed] [Google Scholar]
- 79.Waters E, Hohn MJ, Ahel I, et al. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(22):12984–12988. doi: 10.1073/pnas.1735403100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Di Giulio M. Formal proof that the split genes of tRNAs of nanoarchaeum equitans are an ancestral character. Journal of Molecular Evolution. 2009;69(5):505–511. doi: 10.1007/s00239-009-9280-z. [DOI] [PubMed] [Google Scholar]
- 81.Brochier C, Gribaldo S, Zivanovic Y, Confalonieri F, Forterre P. Nanoarchaea: representatives of a novel archaeal phylum or a fast-evolving euryarchaeal lineage related to Thermococcales? Genome Biology. 2005;6(5):p. R42. doi: 10.1186/gb-2005-6-5-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baker BJ, Comolli LR, Dick GJ, et al. Enigmatic, ultrasmall, uncultivated Archaea. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(19):8806–8811. doi: 10.1073/pnas.0914470107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Podar M, Makarova KS, Graham DE, Wolf YI, Koonin EV, Reysenbach A-L. Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park. Biology Direct. 2013;8, article 9 doi: 10.1186/1745-6150-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Narasingarao P, Podell S, Ugalde JA, et al. De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. The ISME Journal. 2012;6(1):81–93. doi: 10.1038/ismej.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brochier-Armanet C, Gribaldo S, Forterre P. A DNA topoisomerase IB in Thaumarchaeota testifies for the presence of this enzyme in the last common ancestor of Archaea and Eucarya. Biology Direct. 2008;3, article 54 doi: 10.1186/1745-6150-3-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gupta RS, Shami A. Molecular signatures for the Crenarchaeota and the Thaumarchaeota. Antonie van Leeuwenhoek. 2011;99(2):133–157. doi: 10.1007/s10482-010-9488-3. [DOI] [PubMed] [Google Scholar]
- 87.Pester M, Schleper C, Wagner M. The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Current Opinion in Microbiology. 2011;14(3):300–306. doi: 10.1016/j.mib.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM. The archaebacterial origin of eukaryotes. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(51):20356–20361. doi: 10.1073/pnas.0810647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nunoura T, Hirayama H, Takami H, et al. Genetic and functional properties of uncultivated thermophilic crenarchaeotes from a subsurface gold mine as revealed by analysis of genome fragments. Environmental Microbiology. 2005;7(12):1967–1984. doi: 10.1111/j.1462-2920.2005.00881.x. [DOI] [PubMed] [Google Scholar]
- 90.Nunoura T, Oida H, Nakaseama M, et al. Archaeal diversity and distribution along thermal and geochemical gradients in hydrothermal sediments at the yonaguni knoll IV hydrothermal field in the Southern Okinawa Trough. Applied and Environmental Microbiology. 2010;76(4):1198–1211. doi: 10.1128/AEM.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brochier-Armanet C, Gribaldo S, Forterre P. Spotlight on the Thaumarchaeota. The ISME Journal. 2012;6(2):227–230. doi: 10.1038/ismej.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gribaldo S, Brochier-Armanet C. Time for order in microbial systematics. Trends in Microbiology. 2012;20(5):209–210. doi: 10.1016/j.tim.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 93.Margulis L, Chapman M, Guerrero R, Hall J. The last eukaryotic common ancestor (LECA): acquisition of cytoskeletal motility from aerotolerant spirochetes in the Proterozoic Eon. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(35):13080–13085. doi: 10.1073/pnas.0604985103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Moreira D, López-García P. Symbiosis between methanogenic archaea and δ-proteobacteria as the origin of eukaryotes: the syntrophic hypothesis. Journal of Molecular Evolution. 1998;47(5):517–530. doi: 10.1007/pl00006408. [DOI] [PubMed] [Google Scholar]
- 95.Martin W, Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392(6671):37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 96.Lake JA. Origin of the eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences. Nature. 1988;331(6152):184–186. doi: 10.1038/331184a0. [DOI] [PubMed] [Google Scholar]
- 97.Lloyd KG, Schreiber L, Petersen DG, et al. Predominant archaea in marine sediments degrade detrital proteins. Nature. 2013;496:215–218. doi: 10.1038/nature12033. [DOI] [PubMed] [Google Scholar]
- 98.Kozubal MA, Macur RE, Jay ZJ, et al. Microbial iron cycling in acidic geothermal springs of yellowstone national park: integrating molecular surveys, geochemical processes, and isolation of novel fe-active microorganisms. Frontiers in Microbiology. 2012;3, article 109 doi: 10.3389/fmicb.2012.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gogarten JP, Kibak H, Dittrich P, et al. Evolution of the vacuolar H+-ATPase: implications for the origin of eukaryotes. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(17):6661–6665. doi: 10.1073/pnas.86.17.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Iwabe N, Kuma K, Hasegawa M, Osawa S, Miyata T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(23):9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gribaldo S, Cammarano P. The root of the universal tree of life inferred from anciently duplicated genes encoding components of the protein-targeting machinery. Journal of Molecular Evolution. 1998;47(5):508–516. doi: 10.1007/pl00006407. [DOI] [PubMed] [Google Scholar]
- 102.Zhaxybayeva O, Lapierre P, Gogarten JP. Ancient gene duplications and the root(s) of the tree of life. Protoplasma. 2005;227(1):53–64. doi: 10.1007/s00709-005-0135-1. [DOI] [PubMed] [Google Scholar]
- 103.Boussau B, Blanquart S, Necsulea A, Lartillot N, Gouy M. Parallel adaptations to high temperatures in the Archaean eon. Nature. 2008;456(7224):942–945. doi: 10.1038/nature07393. [DOI] [PubMed] [Google Scholar]
- 104.Dagan T, Roettger M, Bryant D, Martin W. Genome networks root the tree of life between prokaryotic domains. Genome Biology and Evolution. 2010;2(1):379–392. doi: 10.1093/gbe/evq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cavalier-Smith T. Rooting the tree of life by transition analyses. Biology Direct. 2006;1, article 19 doi: 10.1186/1745-6150-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lake JA, Skophammer RG, Herbold CW, Servin JA. Genome beginnings: rooting the tree of life. Philosophical Transactions of the Royal Society B. 2009;364(1527):2177–2185. doi: 10.1098/rstb.2009.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cavalier-Smith T. Deep phylogeny, ancestral groups and the four ages of life. Philosophical Transactions of the Royal Society B. 2010;365:111–132. doi: 10.1098/rstb.2009.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Di Giulio M. The tree of life might be rooted in the branch leading to Nanoarchaeota. Gene. 2007;401(1-2):108–113. doi: 10.1016/j.gene.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 109.Kim KM, Caetano-Anollés G. The evolutionary history of protein fold families and proteomes confirms that the archaeal ancestor is more ancient than the ancestors of other superkingdoms. BMC Evolutionary Biology. 2012;12(1, article 13) doi: 10.1186/1471-2148-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kurland CG, Collins LJ, Penny D. Genomics and the irreducible nature of eukaryote cells. Science. 2006;312(5776):1011–1014. doi: 10.1126/science.1121674. [DOI] [PubMed] [Google Scholar]
- 111.Glansdorff N, Xu Y, Labedan B. The Last Universal Common Ancestor: emergence, constitution and genetic legacy of an elusive forerunner. Biology Direct. 2008;3, article 29 doi: 10.1186/1745-6150-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cavalier-Smith T. Molecular phylogeny. Archaebacteria and Archezoa. Nature. 1989;339(6220):l00–01. doi: 10.1038/339100a0. [DOI] [PubMed] [Google Scholar]
- 113.Poole AM, Penny D. Evaluating hypotheses for the origin of eukaryotes. BioEssays. 2007;29(1):74–84. doi: 10.1002/bies.20516. [DOI] [PubMed] [Google Scholar]
- 114.Cavalier-Smith T. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. International Journal of Systematic and Evolutionary Microbiology. 2002;52(1):7–76. doi: 10.1099/00207713-52-1-7. [DOI] [PubMed] [Google Scholar]
- 115.Devos DP, Reynaud EG. Evolution. Intermediate steps. Science. 2010;330(6008):1187–1188. doi: 10.1126/science.1196720. [DOI] [PubMed] [Google Scholar]
- 116.Forterre P, Gribaldo S. Bacteria with a eukaryotic touch: a glimpse of ancient evolution? Proceedings of the National Academy of Sciences of the United States of America. 2010;107(29):12739–12740. doi: 10.1073/pnas.1007720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Forterre P. A new fusion hypothesis for the origin of Eukarya: better than previous ones, but probably also wrong. Research in Microbiology. 2011;162(1):77–91. doi: 10.1016/j.resmic.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 118.Reynaud EG, Devos DP. Transitional forms between the three domains of life and evolutionary implications. Proceedings of the Royal Society B. 2011;278(1723):3321–3328. doi: 10.1098/rspb.2011.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Livingstone Bell PJ. Viral eukaryogenesis: was the ancestor of the nucleus a complex DNA virus? Journal of Molecular Evolution. 2001;53(3):251–256. doi: 10.1007/s002390010215. [DOI] [PubMed] [Google Scholar]
- 120.Takemura M. Poxviruses and the origin of the eukaryotic nucleus. Journal of Molecular Evolution. 2001;52(5):419–425. doi: 10.1007/s002390010171. [DOI] [PubMed] [Google Scholar]
- 121.Forterre P. The two ages of the RNA world, and the transition to the DNA world: a story of viruses and cells. Biochimie. 2005;87(9-10):793–803. doi: 10.1016/j.biochi.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 122.Forterre P. Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: a hypothesis for the origin of cellular domain. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(10):3669–3674. doi: 10.1073/pnas.0510333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Embley TM. Multiple secondary origins of the anaerobic lifestyle in eukaryotes. Philosophical Transactions of the Royal Society B. 2006;361(1470):1055–1067. doi: 10.1098/rstb.2006.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lake JA, Henderson E, Oakes M, Clark MW. Eocytes: a new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proceedings of the National Academy of Sciences of the United States of America. 1984;81(12):3786–3790. doi: 10.1073/pnas.81.12.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Baldauf SL, Palmer JD, Doolittle WF. The root of the universal tree and the origin of eukaryotes based on elongation factor phylogeny. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(15):7749–7754. doi: 10.1073/pnas.93.15.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hashimoto T, Hasegawa M. Origin and early evolution of eukaryotes inferred from the amino acid sequences of translation elongation factors 1α/Tu and 2/G. Advances in Biophysics. 1996;32:73–120. doi: 10.1016/0065-227x(96)84742-3. [DOI] [PubMed] [Google Scholar]
- 127.Lasek-Nesselquist E, Gogarten JP. The effects of model choice and mitigating bias on the ribosomal tree of life. Molecular Phylogenetics and Evolution. 2013;69:17–38. doi: 10.1016/j.ympev.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 128.Foster PG, Cox CJ, Martin Embley T. The primary divisions of life: a phylogenomic approach employing composition-heterogeneous methods. Philosophical Transactions of the Royal Society B. 2009;364(1527):2197–2207. doi: 10.1098/rstb.2009.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Alvarez-Ponce D, Lopez P, Bapteste E, McInerney JO. Gene similarity networks provide tools for understanding eukaryote origins and evolution. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1594–1603. doi: 10.1073/pnas.1211371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ettema TJG, Lindås A-C, Bernander R. An actin-based cytoskeleton in archaea. Molecular Microbiology. 2011;80(4):1052–1061. doi: 10.1111/j.1365-2958.2011.07635.x. [DOI] [PubMed] [Google Scholar]
- 131.Ettema TJG, Bernander R. Cell division and the ESCRT complex: a surprise from the archaea. Communicative and Integrative Biology. 2009;2(2):86–88. doi: 10.4161/cib.7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kim BK, Jung M-Y, Yu DS, et al. Genome sequence of an ammonia-oxidizing soil archaeon, “Candidatus Nitrosoarchaeum koreensis” MY1. Journal of Bacteriology. 2011;193(19):5539–5540. doi: 10.1128/JB.05717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Spang A, Poehlein A, Offre P, et al. The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environmental Microbiology. 2012;14:3122–3145. doi: 10.1111/j.1462-2920.2012.02893.x. [DOI] [PubMed] [Google Scholar]
- 134.Hallam SJ, Konstantinidis KT, Putnam N, et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(48):18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Iverson V, Morris RM, Frazar CD, Berthiaume CT, Morales RL, Armbrust EV. Untangling genomes from metagenomes: revealing an uncultured class of marine euryarchaeota. Science. 2012;335(6068):587–590. doi: 10.1126/science.1212665. [DOI] [PubMed] [Google Scholar]
- 136.Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS ONE. 2011;6(2) doi: 10.1371/journal.pone.0016626.e16626 [DOI] [PMC free article] [PubMed] [Google Scholar]


