Abstract
Chemokines are small molecules that when secreted by tissues under pathological conditions such as inflammation are believed to be involved in carcinogenesis. Recent reports have found that the genetic variation in chemokine encoding genes are associated with risk of breast cancer. Methods: Using data from a population-based case-control study of 845 invasive breast cases and 807 controls, we genotyped 34 single nucleotide polymorphisms (SNPs) in 8 chemokine candidate genes (CCL3, CCL4, CCL5, CCL20, CCR5, CCR6, CXCL12 and CXCR4). Associations with breast cancer were computed for individual SNPs, groups of SNPs within genes, and in a gene-set analysis. We also performed a meta-analysis of CXCL12 rs1801157 and a haplotype analysis for two SNPs: CXCR4 rs2228014 and CXCL12 rs1801157. Results: We found no significant associations between the risk of breast cancer and any individual SNPs, single genes, or combined gene sets. Some individual variants were marginally associated with some histologic subtypes, but these associations were not significant after adjustment for multiple comparisons. In the meta-analysis of five studies of European ancestry, CXCL12 rs1801157 was marginally associated with breast cancer risk (OR=1.14, 95% CI: 1.00, 1.30). Conclusions: Our findings suggest that genetic variants in the 8 candidate genes coding for chemotactic cytokines have little influence in the risk of breast cancer in postmenopausal women. Additional examination of the relationship between CXCL12 rs1801157 and breast cancer risk is warranted.
Keywords: Breast cancer, chemokines, genetic variation, epidemiology
Introduction
It is well established that certain inflammatory processes, such as obesity [1] and diabetes [2], increase the risk of breast cancer in postmenopausal women. Chemokines are small chemotactic cytokines that are important for migration of cells, and under pathological conditions, such as inflammation, they contribute to leukocyte migration to the injured tissue [3]. Chemokines may regulate immune function against developing tumors [4]. Although many genome wide association studies (GWAS) have been performed in breast cancer, most platforms have not included single nucleotide polymorphism (SNP) rs1801157 located in the chemokine gene CXCL12 (http://www.broadinstitute.org/mpg/snap/ldsearch.php), which has been associated with several cancers [5]. Therefore, the candidate gene approach provides information not accounted for in most GWAS.
The association between CXCL12 rs1801157 and breast cancer has been inconsistent. While some studies found that CXCL12 rs1801157 was associated with a significant increased risk of breast cancer [6,7], others found no association [8-11]. However, all these studies were small (less than 300 cases each) and conducted in different populations (Greece, Iran, China, Brazil and Poland). A meta-analysis using data from most of the above studies reported a significant association [12]. In addition, Lin et al. found a reduced breast cancer risk with the combination of CXCL12 rs1801157 and other SNPs including CXCR4 rs2228014 [9].
Using data from a population-based case-control study, we performed a comprehensive analysis of the association between the common genetic variation in eight chemokines or chemokine receptors and the risk of breast cancer in post-menopausal women.
Materials and methods
The study population and methods have been described elsewhere [13]. Briefly, a population-based case-control study was conducted in King, Pierce and Snohomish counties in western Washington State. Cases were identified via the Cancer Surveillance System (CSS), a population-based cancer registry that participates in the Surveillance Epidemiology and End Results (SEER) program [14]. Cases were 65-79 years old at diagnosis of a first primary invasive breast cancer between April 1, 1997 and May 31, 1999. Controls were identified from the general population using Health Care Financing Administration records and frequency matched to cases in 5-year age groups. Estrogen receptor (ER) and progesterone receptor (PR) status of the tumor were available from the CSS.
TagSNPs in eight candidate chemokine-related genes (CCL3, CCL4, CCL5, CCL20, CCR5, CCR6, CXCL12 and CXCR4) were selected using the Genome Variation Server (http://gvs.gs.washington.edu/GVS/) if they had minor allele frequency (MAF) greater than 5% and r2≤0.80 in the CEU-HapMap population. In addition, SNPs located in exons or the untranslated region (UTR) that had MAF of at least 2% were selected. The SNAGGER algorithm was used to select a subset of the tagSNPs, which yielded similar coverage, resulting in the attempted genotyping of 48 SNPs [15]. Genotying was performed using the Illumina GoldenGate multiplex platform and was successful for 33 SNPs. One SNP from CXCR4 (rs2228014) that failed on the Illumina platform and one SNP from CXCL12 (rs266093) that was not suited for the Illumina panel were genotyped by a parallel Taqman assay (KASPAR, Kbioscience Inc). One of the resulting 35 SNPs was monomorphic and was not included in the analysis.
Computations were carried out using the R software environment (version 2.15.0 for Macintosh). Analysis was restricted to women of European ancestry, as determined by self-report and principal component analysis as described previously [13]. Linkage disequilibrium (LD) between variants and departures from Hardy-Weinberg equilibrium were computed in controls [16]. Odds ratios (OR) and 95% confidence intervals (CI) were computed using logistic regression adjusting for age as a continuous linear variable and assuming an additive model of inheritance. The analysis of the combined set of genes included in the study was performed using the GRASS algorithm [17]. For single gene tests, we corrected for multiple tagSNPs within a gene with a P-min procedure using 10,000 permutations [18]. Gene-based P-values were computed separately for the main and subgroup analyses. Fixed-effects analysis was done to obtain meta-analytic estimates of the summary OR across studies looking at the association between CXCL12 rs1801157 and breast cancer risk using the rmeta package. Haplotype analysis was performed to explore the association between tagSNPs CXCR4 rs2228014 and CXCL12 rs1801157 with breast of risk cancer using the haplo.stats R library.
Results
We analyzed the association between 34 tagSNPs in 8 chemokine-related genes and case status in our population of 845 cases and 807 controls. No departures from Hardy-Weinberg equilibrium were observed among controls. None of the tagSNPs in the chemokine-related genes studied altered the risk of breast cancer (Table 1). When we combined all the genes in a gene-set analysis, no association was observed with the risk breast cancer (p=0.112). In the meta-analysis of five studies of European ancestry looking at the association between CXCL12 rs1801157 and breast cancer, there was a marginally significant 14% increased risk (OR=1.14, 95% CI: 1.00, 1.30) (Figure 1B).
Table 1.
Association between common sequence variants in chemokine-related genes and breast cancer risk in postmenopausal women
| Controls | Cases | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| Gene | Variant | Function | Genotype | N=807 | (%) | N=845 | (%) | OR* | (95% CI)* | P-value# |
| CXCR4 | 0.22 | |||||||||
| rs2228014 | Synonymous | 0.70 | (0.47-1.06) | |||||||
| CC | 689 | (92.24) | 723 | (94.39) | ||||||
| TC | 56 | (7.50) | 43 | (5.61) | ||||||
| TT | 2 | (0.27) | 0 | (0.00) | ||||||
| rs2680880 | 5’UTR | 1.04 | (0.91-1.19) | |||||||
| TT | 259 | (32.33) | 267 | (31.79) | ||||||
| AT | 388 | (48.44) | 397 | (47.26) | ||||||
| AA | 154 | (19.23) | 176 | (20.95) | ||||||
| rs2471859 | Intron | 0.88 | (0.75-1.03) | |||||||
| AA | 451 | (56.30) | 507 | (60.36) | ||||||
| AG | 300 | (37.45) | 285 | (33.93) | ||||||
| GG | 50 | (6.24) | 48 | (5.71) | ||||||
| CCL20 | 0.32 | |||||||||
| rs6749704 | Flanking 5’UTR | 0.92 | (0.79-1.07) | |||||||
| AA | 385 | (48.06) | 430 | (51.19) | ||||||
| AG | 347 | (43.32) | 339 | (40.36) | ||||||
| GG | 69 | (8.61) | 71 | (8.45) | ||||||
| rs940339 | Flanking 3’UTR | 1.09 | (0.95-1.25) | |||||||
| AA | 203 | (25.37) | 210 | (25.00) | ||||||
| AG | 415 | (51.88) | 405 | (48.21) | ||||||
| GG | 182 | (22.75) | 225 | (26.79) | ||||||
| CCR5 | 0.85 | |||||||||
| rs11575816 | Flanking 3’UTR | 0.99 | (0.86-1.14) | |||||||
| GG | 318 | (39.7) | 340 | (40.48) | ||||||
| AG | 376 | (46.94) | 387 | (46.07) | ||||||
| AA | 107 | (13.36) | 113 | (13.45) | ||||||
| CCR6 | 0.57 | |||||||||
| rs3093027 | Flanking 5’UTR | 1.22 | (0.94-1.59) | |||||||
| AA | 682 | (85.14) | 692 | (82.48) | ||||||
| AC | 112 | (13.98) | 144 | (17.16) | ||||||
| CC | 7 | (0.87) | 3 | (0.36) | ||||||
| rs3093026 | Flanking 5’UTR | 0.92 | (0.8-1.05) | |||||||
| GG | 215 | (26.84) | 235 | (28.01) | ||||||
| AG | 388 | (48.44) | 422 | (50.30) | ||||||
| AA | 198 | (24.72) | 182 | (21.69) | ||||||
| rs3093023 | Flanking 5’UTR | 1.04 | (0.9-1.19) | |||||||
| GG | 274 | (34.21) | 263 | (31.31) | ||||||
| AG | 368 | (45.94) | 420 | (50.00) | ||||||
| AA | 159 | (19.85) | 157 | (18.69) | ||||||
| rs3798315 | Intron | 1.05 | (0.85-1.3) | |||||||
| GG | 639 | (79.78) | 664 | (79.05) | ||||||
| AG | 150 | (18.73) | 162 | (19.29) | ||||||
| AA | 12 | (1.50) | 14 | (1.67) | ||||||
| rs3093012 | Intron | 1.01 | (0.88-1.16) | |||||||
| GG | 271 | (33.83) | 279 | (33.21) | ||||||
| AG | 390 | (48.69) | 415 | (49.40) | ||||||
| AA | 140 | (17.48) | 146 | (17.38) | ||||||
| rs3093010 | Intron | 1.00 | (0.86-1.15) | |||||||
| CC | 365 | (45.57) | 375 | (44.64) | ||||||
| AC | 336 | (41.95) | 369 | (43.93) | ||||||
| AA | 100 | (12.48) | 96 | (11.43) | ||||||
| rs3093007 | Synonymous | 0.93 | (0.78-1.11) | |||||||
| AA | 534 | (66.67) | 577 | (68.69) | ||||||
| AG | 239 | (29.84) | 234 | (27.86) | ||||||
| GG | 28 | (3.50) | 29 | (3.45) | ||||||
| rs3093006 | 3’UTR | 1.06 | (0.87-1.3) | |||||||
| GG | 608 | (75.91) | 625 | (74.40) | ||||||
| AG | 179 | (22.35) | 201 | (23.93) | ||||||
| AA | 14 | (1.75) | 14 | (1.67) | ||||||
| rs3093005 | 3’UTR | 1.04 | (0.64-1.7) | |||||||
| AA | 768 | (96.00) | 805 | (95.83) | ||||||
| AG | 32 | (4.00) | 35 | (4.17) | ||||||
| GG | 0 | (0.00) | 0 | (0.00) | ||||||
| rs11575083 | 3’UTR | 0.99 | (0.58-1.67) | |||||||
| GG | 773 | (96.50) | 811 | (96.55) | ||||||
| AG | 28 | (3.50) | 29 | (3.45) | ||||||
| AA | 0 | (0.00) | 0 | (0.00) | ||||||
| rs3093002 | Flanking 3’UTR | 0.90 | (0.78-1.05) | |||||||
| GG | 355 | (44.38) | 395 | (47.08) | ||||||
| AG | 371 | (46.38) | 378 | (45.05) | ||||||
| AA | 74 | (9.25) | 66 | (7.87) | ||||||
| rs3093001 | Flanking 3’UTR | 1.13 | (0.98-1.29) | |||||||
| AA | 208 | (26.10) | 194 | (23.23) | ||||||
| AC | 399 | (50.06) | 416 | (49.82) | ||||||
| CC | 190 | (23.84) | 225 | (26.95) | ||||||
| rs367523 | Flanking 3’UTR | 0.92 | (0.8-1.06) | |||||||
| CC | 303 | (37.83) | 334 | (39.76) | ||||||
| CG | 392 | (48.94) | 410 | (48.81) | ||||||
| GG | 106 | (13.23) | 96 | (11.43) | ||||||
| rs4710189 | Flanking 3’UTR | 0.92 | (0.8-1.07) | |||||||
| AA | 340 | (42.45) | 377 | (44.88) | ||||||
| AG | 374 | (46.69) | 380 | (45.24) | ||||||
| GG | 87 | (10.86) | 83 | (9.88) | ||||||
| CXCL12 | 0.68 | |||||||||
| rs266093 | 3’UTR | 1.07 | (0.93-1.24) | |||||||
| GG | 299 | (40.46) | 304 | (39.79) | ||||||
| GC | 338 | (45.74) | 333 | (43.59) | ||||||
| CC | 102 | (13.8) | 127 | (16.62) | ||||||
| rs1029153 | 3’UTR | 0.91 | (0.79-1.05) | |||||||
| AA | 391 | (48.81) | 427 | (50.89) | ||||||
| AG | 322 | (40.20) | 337 | (40.17) | ||||||
| GG | 88 | (10.99) | 75 | (8.94) | ||||||
| rs1801157 | 3’UTR | 1.04 | (0.87-1.23) | |||||||
| GG | 515 | (64.29) | 523 | (62.26) | ||||||
| AG | 251 | (31.34) | 288 | (34.29) | ||||||
| AA | 35 | (4.37) | 29 | (3.45) | ||||||
| rs266087 | Intron | 0.93 | (0.81-1.08) | |||||||
| GG | 325 | (40.68) | 360 | (42.91) | ||||||
| AG | 371 | (46.43) | 378 | (45.05) | ||||||
| AA | 103 | (12.89) | 101 | (12.04) | ||||||
| rs2839695 | 3’UTR | 1.07 | (0.91-1.27) | |||||||
| AA | 504 | (62.92) | 528 | (62.86) | ||||||
| AG | 270 | (33.71) | 264 | (31.43) | ||||||
| GG | 27 | (3.37) | 48 | (5.71) | ||||||
| rs2236534 | Intron | 0.98 | (0.83-1.16) | |||||||
| CC | 493 | (61.55) | 530 | (63.10) | ||||||
| AC | 276 | (34.46) | 268 | (31.90) | ||||||
| AA | 32 | (4.00) | 42 | (5.00) | ||||||
| rs3780891 | Intron | 0.90 | (0.71-1.13) | |||||||
| GG | 647 | (80.77) | 692 | (82.38) | ||||||
| AG | 144 | (17.98) | 140 | (16.67) | ||||||
| AA | 10 | (1.25) | 8 | (0.95) | ||||||
| CCL5 | 0.62 | |||||||||
| rs2107538 | Flanking 5’UTR | 0.95 | (0.8-1.14) | |||||||
| GG | 541 | (67.54) | 590 | (70.24) | ||||||
| AG | 240 | (29.96) | 218 | (25.95) | ||||||
| AA | 20 | (2.50) | 32 | (3.81) | ||||||
| CCL3 | 0.93 | |||||||||
| rs9972960 | Flanking 5’UTR | 1.00 | (0.87-1.16) | |||||||
| GG | 307 | (38.33) | 330 | (39.29) | ||||||
| AG | 395 | (49.31) | 398 | (47.38) | ||||||
| AA | 99 | (12.36) | 112 | (13.33) | ||||||
| rs1634502 | Flanking 5’UTR | 0.98 | (0.85-1.12) | |||||||
| AA | 286 | (35.75) | 294 | (35.00) | ||||||
| AT | 372 | (46.50) | 412 | (49.05) | ||||||
| TT | 142 | (17.75) | 134 | (15.95) | ||||||
| CCL4 | 0.92 | |||||||||
| rs10491121 | Flanking 5’UTR | 0.97 | (0.85-1.11) | |||||||
| GG | 288 | (35.96) | 298 | (35.48) | ||||||
| AG | 368 | (45.94) | 406 | (48.33) | ||||||
| AA | 145 | (18.10) | 136 | (16.19) | ||||||
| rs1634517 | Intron | 1.05 | (0.89-1.24) | |||||||
| CC | 472 | (59.97) | 492 | (59.78) | ||||||
| AC | 282 | (35.83) | 284 | (34.51) | ||||||
| AA | 33 | (4.19) | 47 | (5.71) | ||||||
| rs17679451 | Flanking 3’UTR | 1.03 | (0.72-1.47) | |||||||
| GG | 736 | (91.89) | 770 | (91.67) | ||||||
| AG | 63 | (7.87) | 68 | (8.10) | ||||||
| AA | 2 | (0.25) | 2 | (0.24) | ||||||
| rs1619526 | Flanking 3’UTR | 1.05 | (0.89-1.24) | |||||||
| GG | 472 | (58.93) | 492 | (58.57) | ||||||
| AG | 292 | (36.45) | 297 | (35.36) | ||||||
| AA | 37 | (4.62) | 51 | (6.07) | ||||||
Odds ratios (ORs) and 95% confidence intervals (CI) are computed using a multiplicative model (additive on log scale).
If genotype counts were less than 5 for any of the three cells, a dominant model was used. ORs and 95% CI are age adjusted (age as a continuous linear variable).
P-value adjusted for multiple comparisons within a gene using a P-min permutation test.
Figure 1.
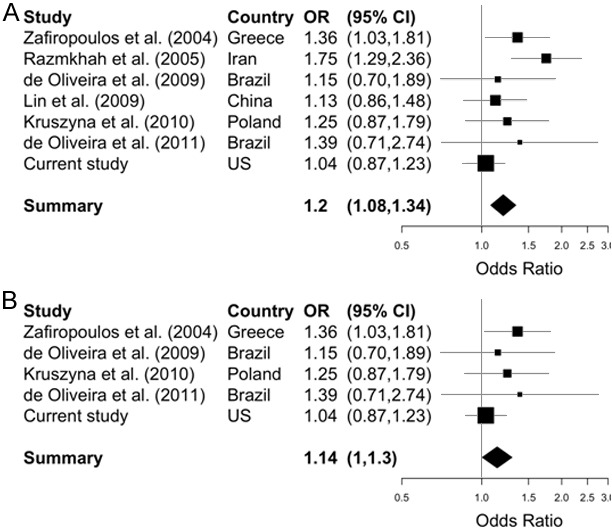
Forest plot summarizing odds ratios (ORs) and 95% confidence intervals (CIs) for the association between tagSNP rs1801157 and breast cancer risk. A. Pooled effect estimated from 7 case-control studies conducted in 6 different countries using fixed-effect meta-analytic models (I2=40.6% [0%, 75%]; Q: 10.09, P=0.121). B. Pooled effect estimated from 5 case-control studies of European ancestry using fixed-effect meta-analytic models (I2=0% [0%, 75%]; Q: 3.32, P=0.505).
None of the SNPs were related to the risk of luminal and non-luminal breast cancers, after adjusting for multiple comparisons (Table 2). Finally, compared to the most common haplotype formed by tagSNPs CXCR4 rs2228014 and CXCL12 rs1801157, other haplotypes did not significant alter the risk of breast cancer (Table 3).
Table 2.
Association between common sequence variants in chemokine-related genes and breast cancer risk in postmenopausal women by tumor subtypes
| Controls (N=807) | Luminal Cases (N=744) | Non-luminal Cases (N=73) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||||
| Gene | Variant | Genotype | n | (%) | n | (%) | OR* | (95% CI)* | P-value# | n | (%) | OR* | (95% CI)* | P-value# |
| CXCR4 | 0.08 | 0.75 | ||||||||||||
| rs2228014 | 0.61 | (0.39-0.95) | 1.28 | (0.56-2.93) | ||||||||||
| CC | 689 | (92.24) | 639 | (95.09) | 62 | (89.86) | ||||||||
| TC | 56 | (7.50) | 33 | (4.91) | 7 | (10.14) | ||||||||
| TT | 2 | (0.27) | 0 | (0.00) | 0 | (0.00) | ||||||||
| rs2680880 | 1.02 | (0.89-1.18) | 1.04 | (0.74-1.47) | ||||||||||
| TT | 259 | (32.33) | 243 | (32.88) | 19 | (26.03) | ||||||||
| AT | 388 | (48.44) | 341 | (46.14) | 43 | (58.90) | ||||||||
| AA | 154 | (19.23) | 155 | (20.97) | 11 | (15.07) | ||||||||
| rs2471859 | 0.91 | (0.77-1.07) | 0.80 | (0.49-1.31) | ||||||||||
| AA | 451 | (56.30) | 440 | (59.54) | 45 | (61.64) | ||||||||
| AG | 300 | (37.45) | 255 | (34.51) | 26 | (35.62) | ||||||||
| GG | 50 | (6.24) | 44 | (5.95) | 2 | (2.74) | ||||||||
| CCL20 | 0.22 | 0.28 | ||||||||||||
| rs6749704 | 0.91 | (0.78-1.06) | 1.11 | (0.77-1.6) | ||||||||||
| AA | 385 | (48.06) | 380 | (51.42) | 35 | (47.95) | ||||||||
| AG | 347 | (43.32) | 299 | (40.46) | 29 | (39.73) | ||||||||
| GG | 69 | (8.61) | 60 | (8.12) | 9 | (12.33) | ||||||||
| rs940339 | 1.12 | (0.97-1.29) | 0.79 | (0.55-1.11) | ||||||||||
| AA | 203 | (25.37) | 181 | (24.49) | 24 | (32.88) | ||||||||
| AG | 415 | (51.88) | 356 | (48.17) | 35 | (47.95) | ||||||||
| GG | 182 | (22.75) | 202 | (27.33) | 14 | (19.18) | ||||||||
| CCR5 | 0.93 | 0.77 | ||||||||||||
| rs11575816 | 0.99 | (0.86-1.15) | 0.95 | (0.67-1.35) | ||||||||||
| GG | 318 | (39.70) | 294 | (39.78) | 32 | (43.84) | ||||||||
| AG | 376 | (46.94) | 348 | (47.09) | 30 | (41.1) | ||||||||
| AA | 107 | (13.36) | 97 | (13.13) | 11 | (15.07) | ||||||||
| CCR6 | 0.51 | 1.00 | ||||||||||||
| rs3093027 | 1.22 | (0.93-1.60) | 1.05 | (0.53-2.05) | ||||||||||
| AA | 682 | (85.14) | 609 | (82.52) | 62 | (84.93) | ||||||||
| AC | 112 | (13.98) | 126 | (17.07) | 11 | (15.07) | ||||||||
| CC | 7 | (0.87) | 3 | (0.41) | 0 | (0) | ||||||||
| rs3093026 | 0.91 | (0.79-1.05) | 0.92 | (0.65-1.28) | ||||||||||
| GG | 215 | (26.84) | 210 | (28.46) | 20 | (27.4) | ||||||||
| AG | 388 | (48.44) | 369 | (50.00) | 37 | (50.68) | ||||||||
| AA | 198 | (24.72) | 159 | (21.54) | 16 | (21.92) | ||||||||
| rs3093023 | 1.05 | (0.92-1.21) | 1.05 | (0.75-1.47) | ||||||||||
| GG | 274 | (34.21) | 228 | (30.85) | 22 | (30.14) | ||||||||
| AG | 368 | (45.94) | 370 | (50.07) | 38 | (52.05) | ||||||||
| AA | 159 | (19.85) | 141 | (19.08) | 13 | (17.81) | ||||||||
| rs3798315 | 1.09 | (0.87-1.35) | 0.86 | (0.46-1.61) | ||||||||||
| GG | 639 | (79.78) | 579 | (78.35) | 60 | (82.19) | ||||||||
| AG | 150 | (18.73) | 147 | (19.89) | 13 | (17.81) | ||||||||
| AA | 12 | (1.50) | 13 | (1.76) | 0 | (0) | ||||||||
| rs3093012 | 1.02 | (0.89-1.18) | 0.95 | (0.67-1.34) | ||||||||||
| GG | 271 | (33.83) | 242 | (32.75) | 26 | (35.62) | ||||||||
| AG | 390 | (48.69) | 368 | (49.80) | 35 | (47.95) | ||||||||
| AA | 140 | (17.48) | 129 | (17.46) | 12 | (16.44) | ||||||||
| rs3093010 | 1.02 | (0.88-1.18) | 0.91 | (0.64-1.31) | ||||||||||
| CC | 365 | (45.57) | 324 | (43.84) | 34 | (46.58) | ||||||||
| AC | 336 | (41.95) | 328 | (44.38) | 32 | (43.84) | ||||||||
| AA | 100 | (12.48) | 87 | (11.77) | 7 | (9.59) | ||||||||
| rs3093007 | 0.92 | (0.77-1.11) | 0.94 | (0.57-1.58) | ||||||||||
| AA | 534 | (66.67) | 510 | (69.01) | 49 | (67.12) | ||||||||
| AG | 239 | (29.84) | 203 | (27.47) | 23 | (31.51) | ||||||||
| GG | 28 | (3.50) | 26 | (3.52) | 1 | (1.37) | ||||||||
| rs3093006 | 1.04 | (0.85-1.29) | 1.17 | (0.68-2.01) | ||||||||||
| GG | 608 | (75.91) | 551 | (74.56) | 53 | (72.6) | ||||||||
| AG | 179 | (22.35) | 178 | (24.09) | 18 | (24.66) | ||||||||
| AA | 14 | (1.75) | 10 | (1.35) | 2 | (2.74) | ||||||||
| rs3093005 | 0.98 | (0.59-1.64) | 1.04 | (0.31-3.51) | ||||||||||
| AA | 768 | (96.00) | 710 | (96.08) | 70 | (95.89) | ||||||||
| AG | 32 | (4.00) | 29 | (3.92) | 3 | (4.11) | ||||||||
| GG | 0 | (0.00) | 0 | (0.00) | 0 | (0) | ||||||||
| rs11575083 | 0.89 | (0.51-1.55) | 1.2 | (0.35-4.05) | ||||||||||
| GG | 773 | (96.50) | 716 | (96.89) | 70 | (95.89) | ||||||||
| AG | 28 | (3.50) | 23 | (3.11) | 3 | (4.11) | ||||||||
| AA | 0 | (0.00) | 0 | (0) | 0 | (0) | ||||||||
| rs3093002 | 0.90 | (0.77-1.05) | 1.07 | (0.74-1.55) | ||||||||||
| GG | 355 | (44.38) | 347 | (47.02) | 33 | (47.02) | ||||||||
| AG | 371 | (46.38) | 335 | (45.39) | 31 | (42.47) | ||||||||
| AA | 74 | (9.25) | 56 | (7.59) | 9 | (12.33) | ||||||||
| rs3093001 | 1.14 | (0.99-1.31) | 1.00 | (0.71-1.4) | ||||||||||
| AA | 208 | (26.1) | 170 | (23.16) | 17 | (23.29) | ||||||||
| AC | 399 | (50.06) | 363 | (49.46) | 41 | (56.16) | ||||||||
| CC | 190 | (23.84) | 201 | (27.38) | 15 | (20.55) | ||||||||
| rs367523 | 0.90 | (0.77-1.05) | 1.15 | (0.81-1.63) | ||||||||||
| CC | 303 | (37.83) | 294 | (39.78) | 28 | (38.36) | ||||||||
| CG | 392 | (48.94) | 367 | (49.66) | 31 | (42.47) | ||||||||
| GG | 106 | (13.23) | 78 | (10.55) | 14 | (19.18) | ||||||||
| rs4710189 | 0.92 | (0.79-1.08) | 1.02 | (0.71-1.47) | ||||||||||
| AA | 340 | (42.45) | 329 | (44.52) | 34 | (46.58) | ||||||||
| AG | 374 | (46.69) | 340 | (46.01) | 28 | (38.36) | ||||||||
| GG | 87 | (10.86) | 70 | (9.47) | 11 | (15.07) | ||||||||
| CXCL12 | 0.69 | 0.45 | ||||||||||||
| rs266093 | 1.06 | (0.91-1.23) | 1.07 | (0.75-1.54) | ||||||||||
| GG | 299 | (40.46) | 272 | (40.54) | 24 | (35.29) | ||||||||
| GC | 338 | (45.74) | 286 | (42.62) | 36 | (52.94) | ||||||||
| CC | 102 | (13.8) | 113 | (16.84) | 8 | (11.76) | ||||||||
| rs1029153 | 0.94 | (0.80-1.09) | 0.81 | (0.56-1.18) | ||||||||||
| AA | 391 | (48.81) | 370 | (50.14) | 39 | (53.42) | ||||||||
| AG | 322 | (40.2) | 299 | (40.51) | 29 | (39.73) | ||||||||
| GG | 88 | (10.99) | 69 | (9.35) | 5 | (6.85) | ||||||||
| rs1801157 | 1.03 | (0.87-1.24) | 1.05 | (0.64-1.73) | ||||||||||
| GG | 515 | (64.29) | 459 | (62.11) | 46 | (63.01) | ||||||||
| AG | 251 | (31.34) | 256 | (34.64) | 25 | (34.25) | ||||||||
| AA | 35 | (4.37) | 24 | (3.25) | 2 | (2.74) | ||||||||
| rs266087 | 0.91 | (0.78-1.06) | 1.14 | (0.8-1.62) | ||||||||||
| GG | 325 | (40.68) | 325 | (44.04) | 24 | (32.88) | ||||||||
| AG | 371 | (46.43) | 325 | (44.04) | 41 | (56.16) | ||||||||
| AA | 103 | (12.89) | 88 | (11.92) | 8 | (10.96) | ||||||||
| rs2839695 | 1.07 | (0.90-1.27) | 1.25 | (0.83-1.88) | ||||||||||
| AA | 504 | (62.92) | 466 | (63.06) | 43 | (58.9) | ||||||||
| AG | 270 | (33.71) | 231 | (31.26) | 25 | (34.25) | ||||||||
| GG | 27 | (3.37) | 42 | (6.68) | 5 | (6.85) | ||||||||
| rs2236534 | 1.04 | (0.88-1.24) | 0.65 | (0.38-1.1) | ||||||||||
| CC | 493 | (61.55) | 456 | (61.71) | 52 | (71.23) | ||||||||
| AC | 276 | (34.46) | 242 | (32.75) | 20 | (27.4) | ||||||||
| AA | 32 | (4) | 41 | (5.55) | 1 | (1.37) | ||||||||
| rs3780891 | 0.86 | (0.68-1.09) | 1.2 | (0.67-2.15) | ||||||||||
| GG | 647 | (80.77) | 615 | (83.22) | 57 | (78.08) | ||||||||
| AG | 144 | (17.98) | 116 | (15.70) | 16 | (21.92) | ||||||||
| AA | 10 | (1.25) | 8 | (1.08) | 0 | (0) | ||||||||
| CCL5 | 0.56 | 0.63 | ||||||||||||
| rs2107538 | 0.94 | (0.78-1.14) | 1.12 | (0.72-1.73) | ||||||||||
| GG | 541 | (67.54) | 520 | (70.37) | 50 | (68.49) | ||||||||
| AG | 240 | (29.96) | 192 | (25.98) | 18 | (24.66) | ||||||||
| AA | 20 | (2.5) | 27 | (3.65) | 5 | (6.85) | ||||||||
| CCL3 | 0.99 | 0.10 | ||||||||||||
| rs9972960 | 0.99 | (0.85-1.15) | 1.03 | (0.72-1.49) | ||||||||||
| GG | 307 | (38.33) | 296 | (40.05) | 25 | (34.25) | ||||||||
| AG | 395 | (49.31) | 342 | (46.28) | 41 | (56.16) | ||||||||
| AA | 99 | (12.36) | 101 | (13.67) | 7 | (9.59) | ||||||||
| rs1634502 | 1.01 | (0.87-1.16) | 0.71 | (0.5-1.01) | ||||||||||
| AA | 286 | (35.75) | 251 | (49.80) | 33 | (45.21) | ||||||||
| AT | 372 | (46.5) | 368 | (16.24) | 32 | (43.84) | ||||||||
| TT | 142 | (17.75) | 120 | (34.51) | 8 | (10.96) | ||||||||
| CCL4 | 0.97 | 0.06 | ||||||||||||
| rs10491121 | 1.00 | (0.87-1.15) | 0.68 | (0.48-0.98) | ||||||||||
| GG | 288 | (35.96) | 255 | (34.51) | 33 | (45.21) | ||||||||
| AG | 368 | (45.94) | 361 | (48.85) | 33 | (45.21) | ||||||||
| AA | 145 | (18.1) | 123 | (16.64) | 7 | (9.59) | ||||||||
| rs1634517 | 1.02 | (0.86-1.22) | 1.57 | (1.07-2.32) | ||||||||||
| CC | 472 | (59.97) | 437 | (60.36) | 35 | (49.3) | ||||||||
| AC | 282 | (35.83) | 248 | (34.25) | 29 | (40.85) | ||||||||
| AA | 33 | (4.19) | 39 | (5.39) | 7 | (9.86) | ||||||||
| rs17679451 | 1.06 | (0.73-1.52) | 0.99 | (0.41-2.36) | ||||||||||
| GG | 736 | (91.89) | 676 | (91.47) | 67 | (91.78) | ||||||||
| AG | 63 | (7.87) | 61 | (8.25) | 6 | (8.22) | ||||||||
| AA | 2 | (0.25) | 2 | (0.27) | 0 | (0) | ||||||||
| rs1619526 | 1.04 | (0.88-1.23) | 1.39 | (0.95-2.05) | ||||||||||
| GG | 472 | (58.93) | 435 | (58.86) | 37 | (50.68) | ||||||||
| AG | 292 | (36.45) | 261 | (35.32) | 30 | (41.1) | ||||||||
| AA | 37 | (4.62) | 43 | (5.82) | 6 | (8.22) | ||||||||
Odds ratios (ORs) and 95% CI intervals (CI) are adjusted for age.
If genotype counts were less than 5 for any of the three cells, a dominant model was used; otherwise, an additive model was used.
P-value adjusted for multiple comparisons within a gene using a P-min permutation test.
Table 3.
Association between haplotype formed by tagSNPs CXCR4 rs2228014 and CXCL12 rs1801157 and breast cancer risk
| CXCR4 rs2228014 | CXCL12 rs1801157 | Controls (%) | Cases (%) | OR* | (95% CI)* |
|---|---|---|---|---|---|
| C | G | (76.35) | (77.15) | 1.00 | (Ref.) |
| C | A | (19.63) | (20.04) | 1.02 | (0.85, 1.21) |
| T | G | (3.61) | (2.25) | 0.63 | (0.39, 1.01) |
| T | A | (0.41) | (0.55) | 1.26 | (0.21, 7.55) |
Odds ratios (ORs) and 95% confidence intervals (CI) are age adjusted (age as a continuous linear variable).
Discussion
No strong evidence of association between chemokine-related genes and breast cancer was observed in our study, and this was true regardless of the receptor subtypes of the tumors.
Our study has a few limitations that could have influenced our results. Study sample size could have prevented us from detecting a small association. However, our sample size (845 cases and 807 controls) gave us better power than either of the two previous studies (233 cases and 210 controls [6]; 278 cases and 181 controls [7]) that did find a significant association for CXCL12 rs1801157. Because of the possible small effect of each single SNP on breast cancer, we also combined all the SNPs into gene regions and all the genes in a gene-set analysis, but still were unable to observe an association.
A previous meta-analysis of five studies looking at CXCL12 rs1801157 and breast caner risk reported a positive association [12]. We performed a meta-analysis to include a Brazilian and our current study and observed a marginal significant increased risk. However, while our study was restricted to postmenopausal women and the risk of invasive breast cancer, other studies included pre- and postmenopausal women as well as all stages, possibly in situ and invasive [6-11].
In summary, our study fails to support the hypothesis that genetic variation in chemokine-related genes is involved in breast cancer etiology in postmenopausal Caucasian women.
Acknowledgements
The study was supported by grants R01 CA116786 and R01 CA72787 from the National Cancer Institute.
Disclosure of conflict of interest
None.
References
- 1.Cleary MP, Grossmann ME. Minireview: Obesity and breast cancer: the estrogen connection. Endocrinology. 2009;150:2537–2542. doi: 10.1210/en.2009-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larsson SC, Mantzoros CS, Wolk A. Diabetes mellitus and risk of breast cancer: a meta-analysis. Int J Cancer. 2007;121:856–862. doi: 10.1002/ijc.22717. [DOI] [PubMed] [Google Scholar]
- 3.Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392:565–568. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Baruch A. The multifaceted roles of chemokines in malignancy. Cancer Metastasis Rev. 2006;25:357–371. doi: 10.1007/s10555-006-9003-5. [DOI] [PubMed] [Google Scholar]
- 5.Gong H, Tan M, Wang Y, Shen B, Liu Z, Zhang F, Liu Y, Qiu J, Bao E, Fan Y. The CXCL12 G801A polymorphism and cancer risk: Evidence from 17 case-control studies. Gene. 2012;509:228–231. doi: 10.1016/j.gene.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 6.Zafiropoulos A, Crikas N, Passam AM, Spandidos DA. Significant involvement of CCR2-64I and CXCL12-3a in the development of sporadic breast cancer. J Med Genet. 2004;41:e59. doi: 10.1136/jmg.2003.013649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Razmkhah M, Talei AR, Doroudchi M, Khalili-Azad T, Ghaderi A. Stromal cell-derived factor-1 (SDF-1) alleles and susceptibility to breast carcinoma. Cancer Lett. 2005;225:261–266. doi: 10.1016/j.canlet.2004.10.039. [DOI] [PubMed] [Google Scholar]
- 8.de Oliveira KB, Oda JM, Voltarelli JC, Nasser TF, Ono MA, Fujita TC, Matsuo T, Watanabe MA. CXCL12 rs1801157 polymorphism in patients with breast cancer, Hodgkin’s lymphoma, and non-Hodgkin’s lymphoma. J Clin Lab Anal. 2009;23:387–393. doi: 10.1002/jcla.20346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin GT, Tseng HF, Yang CH, Hou MF, Chuang LY, Tai HT, Tai MH, Cheng YH, Wen CH, Liu CS, Huang CJ, Wang CL, Chang HW. Combinational Polymorphisms of Seven CXCL12-Related Genes Are Protective against Breast Cancer in Taiwan. OMICS. 2009;13:165–172. doi: 10.1089/omi.2008.0050. [DOI] [PubMed] [Google Scholar]
- 10.Kruszyna L, Lianeri M, Rubis B, Knunla H, Rybczynska M, Grodecka-Gazdecka S, Jagodzinski PP. CXCL12-3’ G801A polymorphism is not a risk factor for breast cancer. DNA Cell Bio. 2010;29:423–427. doi: 10.1089/dna.2010.1030. [DOI] [PubMed] [Google Scholar]
- 11.de Oliveira KB, Guembarovski RL, Oda JM, Mantovani MS, Carrera CM, Reiche EM, Voltarelli JC, da Silva do Amaral Herrera AC, Watanabe MA. CXCL12 rs1801157 polymorphism and expression in peripheral blood from breast cancer patients. Cytokine. 2011;55:260–265. doi: 10.1016/j.cyto.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 12.Shen W, Cao X, Xi L, Deng L. CXCL12 G801A polymorphism and breast cancer risk: a meta-analysis. Mol Biol Rep. 2012;39:2039–2044. doi: 10.1007/s11033-011-0951-7. [DOI] [PubMed] [Google Scholar]
- 13.Resler AJ, Malone KE, Johnson LG, Malkki M, Petersdorf EW, McKnight B, Madeleine MM. Genetic variation in TLR or NFkappaB pathways and the risk of breast cancer: a case-control study. BMC Cancer. 2013;13:219. doi: 10.1186/1471-2407-13-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hankey BF, Ries LA, Edwards BK. The surveillance, epidemiology, and end results program: a national resource. Cancer Epidemiol Biomarkers Prev. 1999;8:1117–1121. [PubMed] [Google Scholar]
- 15.Edlund CK, Lee WH, Li D, Van Den Berg DJ, Conti DV. Snagger: a user-friendly program for incorporating additional information for tagSNP selection. BMC Bioinformatics. 2008;9:174. doi: 10.1186/1471-2105-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen LS, Hutter CM, Potter JD, Liu Y, Prentice RL, Peters U, Hsu L. Insights into Colon Cancer Etiology via a Regularized Approach to Gene Set Analysis of GWAS Data. Am J Hum Genet. 2010;86:860–871. doi: 10.1016/j.ajhg.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Westfall PH, Young SS. Resampling-based multiple testing: Examples and methods for p-value adjustment. New York: Wiley-Interscience; 1993. [Google Scholar]


