Abstract
We demonstrate the hybridization-induced fluorescence detection of DNA on an origami-based paper analytical device (oPAD). The paper substrate was patterned by wax printing and controlled heating to construct hydrophilic channels and hydrophobic barriers in a three-dimensional fashion. A competitive assay was developed where the analyte, a single-stranded DNA (ssDNA), and a quencher-labeled ssDNA competed for hybridization with a fluorophore-labeled ssDNA probe. Upon hybridization of the analyte with the fluorophore-labeled ssDNA, a linear response of fluorescence vs. analyte concentration was observed with an extrapolated limit of detection < 5 nM and a sensitivity relative standard deviation as low as 3%. The oPAD setup was also tested against OR/AND logic gates, proving to be successful in both detection systems.
Keywords: Paper-based microfluidics, origami, DNA, fluorescence, logic gate, strand-displacement
The design and implementation of paper-based sensors pose a number of challenges. Among the most difficult are achieving quantitation,1-5 low limits of detection (LODs) for bioassays,6-8 minimal nonspecific adsorption (NSA),6,9 timing of reactions and washing steps,10-14 and the integration of detection systems that are consistent with the low-cost philosophy of using paper as the platform.15 In the present article we address the first two of these issues by introducing a three-dimensional (3D) origami-based paper analytical device (oPAD)16 that detects target DNA using toehold-induced strand displacement.17 The key finding is that single-stranded DNA (ssDNA) can be quantitatively detected at nanomolar concentrations via fluorescence using a simple device made of wax-patterned, folded paper.
Paper is an attractive substrate for the development of low-cost, point-of-care (POC) sensors because it is biocompatible, flexible, easily remediated, and has built-in filtering functionality.18,19 The first paper-based sensor was reported in 1883,20 and since that time the field has evolved from lateral flow assays (1D)6,21-23 to 3D devices.16,24-29 Historically, most paper-based assays are implemented using nitrocellulose,30,31 but the characteristics of this material are best suited for lateral flow sensing rather than multidimensional sensor systems. Alternatively, various forms of cellulose have been preferred for the latter application because of their inherent flexibility, ability to be easily patterned, lower cost, and lower susceptibility to NSA.15,16,24-27 For example, our group developed a 3D paper analytical device fabricated using the principles of origami (Japanese art of paper folding). As mentioned earlier, we call this family of devices oPADs, and we have used them for detection of bovine serum albumin (BSA), glucose,16 and adenosine.29 The origami method allows the user to pattern the paper substrate in only one step, easily fold and align all layers, avoid the use of cellulose powder between layers, and unfold the device so that the results of analysis can be read on any of the layers.
A number of different detection methods have been used on paper devices.15,32 These include chemiluminescence,33,34 electrogenerated chemiluminescence,35 electrochemistry,4,7,36,37 thermochromic detection,38 and colorimetry.1-3,5,39-42 In the present work we have focused on fluorescence. Fluorescence is well-known for its high sensitivity and low LODs.6,43 However, its implementation on paper matrices has been limited due to high background fluorescence (particularly those caused by paper whitening additives) present in some cellulose papers.18 Nonetheless, a few groups have successfully developed paper-based sensors that utilize fluorescence detection16,25 and yield low LODs.23,44
The field of DNA detection is quite extensive,45 but only a few such studies have been carried out using paper devices and most of these are lateral-flow assays.46-48 The limited number of paper-based DNA assays is likely a consequence of the low detection levels required for most DNA analyses,45 and the difficulty of attaining those levels using paper substrates.46 Nevertheless, there are some success stories. For example, Yu and co-workers developed a 3D paper platform for the electrochemical detection of DNA with an LOD of 20 pM.49 They used screen-printed electrodes modified with graphene and gold nanoparticles for the immobilization of capture DNA. When the target ssDNA was present, it was retained by the capture probe and induced hybridization of an electro-active amplification label. Araújo et al. immobilized single-strand probe DNA on a previously activated paper strip.46 The paper was then dipped in a solution containing a fluorophore-labeled target DNA and, upon capillary flow, fluorescence was observed (with LODs of 0.2 pM) as the target hybridized with the probe. Similarly, also in a lateral flow-type assay, Ellington and co-workers implemented catalyzed hairpin assemblies for the fluorescence detection of ssDNA with LODs as low as 0.3 μM.47 This detection system is based on the strand displacement reaction of a quencher-labeled ssDNA from a fluorophore-labeled ssDNA in the presence of the target. The catalytic reaction takes place only when the target is present, as it triggers the hybridization reaction between two hairpins that later displace the quencher-labeled ssDNA from the fluorophore-labeled ssDNA.
In the present manuscript we describe the hybridization-induced fluorescent detection of ssDNA using an oPAD platform. Detection is based on the strand displacement of a quencher-labeled ssDNA from a fluorophore-labeled ssDNA upon addition of the analyte. This displacement produces a fluorescence signal proportional to the amount of analyte present and the overall detection is reproducible with a slope relative standard deviation (RSD) as low as 3% from device-to-device. Moreover, LODs < 5 nM have been achieved with linear dose-response curves. In addition, OR and AND logic gates were executed, further demonstrating the versatility of the oPAD approach.
EXPERIMENTAL SECTION
Chemicals and Materials
All oligonucleotides used in this work were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). The DNA sequences from 5′ to 3′ are as follows (toehold is underlined): AACCTAGC CCTTGTCA TAGAGCAC (A1), CGAAGATGCT CCTGATGTG GGCTAAAG (A2), Cy5-GTGCTCTA TGACAAGG GCTAGGTT (F1), Cy5-CAT CTTTAGCC CACATCAG G AGCATCTTCG (F2), Cy5-CGA GTGCTCTA TGACAAGG GCTAGGT CTTTAGCC CACATCAGG AGCATCTTCG (F3), CCTTGTCA TAGAGCAC-Iowa Black® RQ (Q1), C CTGATGTG GGCTAAAG ATG-Iowa Black® RQ (Q2), CCTTGTCA TAGAGCAC TCG-Iowa Black RQ (Q3), CCTGATGTG GGCTAAAG ACCTAGC (M), TTTTTTTTTTTTTTTTTTTTT ((dT)21).
The fluorophore (Cy5) has excitation and emission wavelengths of 648 and 668 nm, respectively. The quencher, Iowa Black RQ, has an absorption wavelength of 656 nm. TNak buffer (20.0 mM Tris-HCl, 140.0 mM NaCl, and 5.0 mM KCl) was used to prepare all DNA solutions. TNak buffer chemicals were purchased from Sigma Aldrich (St. Louis, MO). A 0.5% w/v bovine serum albumin (BSA, Acros, NJ) solution was prepared in 10.0 mM phosphate buffer saline (PBS, Sigma, St. Louis, MO) as a NSA blocker. 10.0 mM PBS (138.0 mM NaCl and 2.7 mM KCl, pH 7.4) was prepared by dissolving a package of dry PBS powder (in foil pouches) in 1 L of deionized water (18 MΩ•cm, Milli-Q Gradient System, Millipore, Bedford, MA). Parafilm paper was purchased from Pechiney Plastic Packaging (IL). Whatman Grade 1 chromatography paper (180 μm thick, linear flow rate (water) of 13 cm/30 min) was purchased from Fisher Scientific (Pittsburg, PA).
DNA preparation
All oligonucleotides were received as powders and centrifuged (16,100 rpm) so that they would reside at the bottom of the containers. The powder was then dissolved with deionized water (18 MΩ•cm, Milli-Q Gradient System, Millipore, Bedford, MA) and quantified using a nanodrop UV-Vis spectrophotometer (Thermo Scientific, Inc. Wilmington, DE) before use (peak absorbance reading at 260 nm). These stock solutions were further diluted in TNak buffer (pH 7.5) to the necessary final concentrations.
Data acquisition and instrumentation
Fluorescence images were obtained using a Typhoon Trio fluorescence scanner (Variable Mode Imager, GE Healthcare, Piscataway, NJ) operated using Typhoon Scanner Control v5.0 software (Build 5.0.0409.0700, Amersham Biosciences 2004). The excitation and emission wavelengths used for all experiments were 633 and 670 nm, respectively (PMT = 400). The pixel size for all scans was 100 μm and the focal plane was set to Platen. Fluorescence images were processed with ImageQuant v5.2 to obtain fluorescence intensity (volume) values for each reservoir of the third oPAD layer. All fluorescence intensities reported were background-corrected by subtracting the fluorescence intensity of the selected reservoir from a section of the same device where the paper was pristine. Note that the fluorescence intensity of F1 was higher after drying than when it was wet (Figure S1a, purple and orange dashed circles, respectively). This high fluorescence saturated the detector and so there was no correlation between F1 concentration and the fluorescence intensity. Therefore, unless otherwise indicated, all fluorescence images were obtained while the paper was wet.
oPAD fabrication and patterning
6 cm × 1.5 cm patterns were designed using CorelDRAW Graphics Suite 12 (Figure S2) and printed on a 20 cm × 20 cm paper sheet using a Xerox ColorQube 8570DN inkjet printer that deposited wax-based solid ink on the paper (a total of 24 devices were printed per paper sheet).50,51 Next, each individual oPAD was cut from the paper sheet and placed wax-side up on a hot plate at 120 °C for 10 s and then cooled to 25 °C. This causes the wax to melt through the paper, creating a hydrophobic barrier that defines the 3D hydrophilic channels and reservoirs (white sections of Figure S2).50 The reagents were then dispensed onto the appropriate reservoirs of the unfolded oPAD. These reagents were left at room temperature until completely dry (5-30 min depending on the volume added). Then the paper strip could be folded along the “fold lines” (Figure S2) to form the oPAD. All devices were immediately used following drying. Note that layers 1-3 of the oPAD each contain four reservoirs that allow four independent reactions to take place simultaneously with no cross-talk between fluidic pathways.16 These four different channels are color coded on Figures S1b, S3a, S5a, S7a, and S8a but they also apply to Figures 1-4 in the main text. The fourth layer of the oPAD shown in Figure S2 prevents the reagents from contacting the aluminium clamp during incubation, but this layer has no direct effect on assay performance. For clarity, therefore, the fourth oPAD layer is not shown in the other figures.
Figure 1.
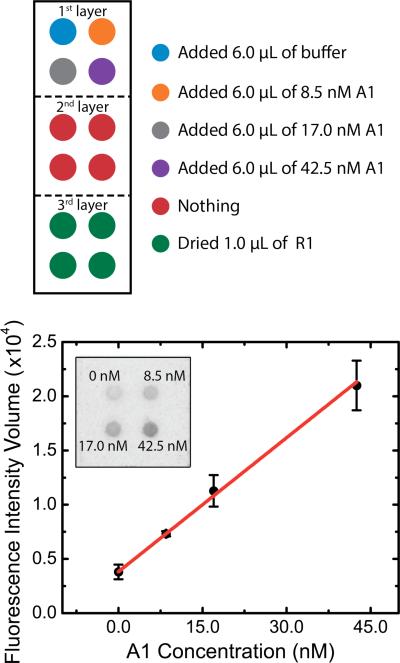
Strand displacement-induced fluorescence detection of ssDNA for Case 1. Top: Color-coded schematic representation of reagent placement on the oPAD. Bottom: Plot of fluorescence intensity volume (FIV) as a function of the concentration of A1. The inset is the fluorescence image of the third oPAD layer showing, qualitatively, the dependence of the FIV on A1 concentration. Note that the error bars for the data points correspond to standard deviations obtained by performing experiments on three different oPADs on the same day.
Figure 4.
Strand displacement-induced fluorescence detection of DNA coupled to oPAD-based AND gate. The device was passivated with BSA and all solutions added to the first oPAD layer contained 1.0 μM (dT)21 as an additional blocker. Top left: Color-coded schematic representation of the reagent placement on the oPAD. Top right: Truth table. Bottom left: Fluorescence image of the third oPAD layer showing the resulting fluorescence response upon addition of different inputs. Bottom right: Histogram of the fluorescence intensity volumes (FIV) measured at the third oPAD layer as a function of the input type. Note that the error bars for the histogram correspond to standard deviations obtained by performing experiments on three different oPADs on the same day.
Blocking
The hydrophilic sections of all the devices were blocked to prevent NSA of the reagents. This was done by drying 5.0 μL of 0.5% w/v BSA in 10.0 mM PBS at 25 °C in each reservoir prior to addition of reagents. Additionally, in some cases 1.0 μM of non-complementary ssDNA ((dT)21) was used to prepare the solutions added to the first oPAD layer once the device was assembled (see Supporting Information section for more details).
RESULTS AND DISCUSSION
Principles of the DNA detection system
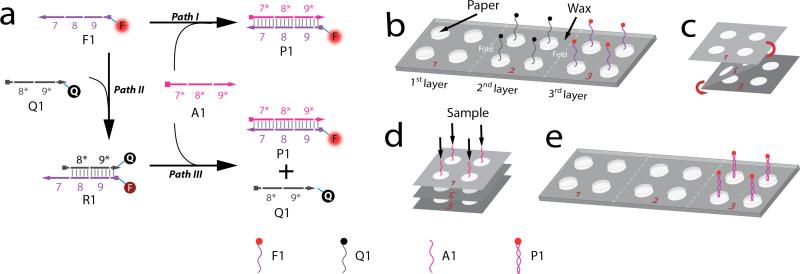
As shown in Scheme 1, the DNA detection system reported here consists of a fluorophore-labeled ssDNA probe, F1; a quencher-labeled ssDNA, Q1; and the analyte (target), A1 (a ssDNA fully complementary to F1). If F1 is dried in the reservoir of a paper substrate and A1 is added (Scheme 1a, Path I), hybridization occurs resulting in fluorescent hybridization product P1. However, if ssDNA quencher Q1 is added to the reservoir containing dried F1 (Scheme 1a, Path II), then the resulting hybridized molecule (R1) does not fluoresce. The assay is established when both the target, A1, and the quencher, Q1, compete simultaneously to hybridize with F1. In this case, A1 and Q1 will hybridize with F1 at similar reaction rates (Scheme 1a, Paths I and II), forming both P1 and R1 hybrids. However, A1 contains 8 more bases complementary to F1 (domain 7* in Scheme 1a) compared to Q1. These 8 bases serve as a toe-hold to initiate a strand displacement reaction that removes Q1 from R1 (Scheme 1a, Path III). According to previous kinetic studies reported by Winfree and Zhang,17 an 8-base toe-hold is sufficient to provide fast (kf = 106 M−1s−1) and irreversible (kb ≈ 1 M−1s−1) reaction displacement rate constant.
Scheme 1.
Operation and performance of the device
We performed two distinct experiments to demonstrate the potential of this paper-based DNA detection system. In Case 1, F1 and Q1 are each dried on different oPAD layers. In Case 2, F1 and Q1 are hybridized in solution first, and the resulting complex, R1, is dried on one of the oPAD layers. In both cases, the paper reservoirs were passivated with BSA as indicated in the Experimental Section.52,53 This step results in lower LODs while improving the sensitivity (~3-fold, Figure S3).
It is important to mention that F1 is specific for the target analyte, A1. This is demonstrated by the results shown in the black dashed circle of Figure S1c. In this experiment, F1, Q1, and a non-complementary ssDNA target (e.g. (dT)21) are mixed together. As a consequence, Path II in Scheme 1a is followed, resulting in R1. This last control experiment proves the hybridization selectivity of the system for A1. Note that the results described here, using a paper substrate, mirror those observed in homogenous solutions (Figure S4).
Case 1
The experimental arrangement for Case 1 is displayed in Scheme 1b. The oPAD sensor was constructed by drying 6.0 μL of 475.0 nM Q1 and 1.0 μL of 40.0 nM F1 within the reservoirs of the second and third layers of the oPAD, respectively. As discussed in the Supporting Information (Figure S5), the experimental parameters for depositing F1 on the oPAD were optimized for maximum signal response upon addition of A1. The oPAD was then folded (Scheme 1c) and compressed (Scheme 1d) via an aluminum clamp (Figure S6) so that the layers would be in direct contact with each other. Next, 6.0 μL of the TNak buffer solution containing 0, 8.5, 17.0, and 42.5 nM A1 was introduced to the reservoirs of the first layer of the oPAD (Scheme 1d) through the holes present in the aluminum clamp. The device was immediately sealed with Parafilm (to avoid water evaporation) and incubated at 37 °C.
The solution containing A1 flows through the second layer of the device, where it picks up pre-dried Q1, and then both of these components move to the third layer where they encounter pre-dried F1. Because A1, F1, and Q1 are all present, all three pathways shown in Scheme 1a are possible. However, on the basis of results from solution-phase experiments and thermodynamics, all A1 should be present within the fluorescent P1 complex, and excess F1 should be quenched by Q1. Hence, the concentration of A1 should be related to the total measured fluorescence. After 40 min of incubation time, the oPAD was unfolded (Scheme 1e) and the third layer of the oPAD was read with a fluorescence scanner.
Figure 1 shows the results of the just-described assay for different concentrations of A1. The plot shown at the bottom of the figure indicates a linear dependence on A1 concentration (sensitivity RSD = 14°%) and a LOD of 3.1 ± 0.4 nM. This LOD is defined as three times the standard deviation of the blank signal, divided by the slope of the linear fit from Figure 1. In addition, the sensitivity RSD was calculated by dividing the standard deviation of the slope over the slope's mean value. From these results, we conclude that the assay summarized by Scheme 1 proceeds as indicated in the oPAD.
Case 2
In this experiment, 1.0 μL of R1, prepared from a solution containing 40.0 nM F1 and 475.0 nM Q1, was dried on the third layer of the oPAD. This situation is in contrast to Case 1, in which Q1 and F1 were dried on separate layers of the device. After folding the oPAD, 6.0 μL of the same concentrations of A1 used for Case 1 were added to the top oPAD layer. After incubating for 40 min, the displacement reaction represented by Path III of Scheme 1a is predicted to take place. The results shown at the bottom of Figure 2 indicate a better linear relationship between fluorescence intensity and the concentration of A1 than for Case 1 (sensitivity RSD = 3%). The LOD was determined to be 4.9 ± 0.1 nM, which is a little higher than Case 1.
Figure 2.
Strand displacement-induced fluorescence detection of ssDNA for Case 2. Top: Color-coded schematic representation of reagent placement on the oPAD. Bottom: Plot of fluorescence intensity volume (FIV) as a function of the concentration of A1. The inset is the fluorescence image of the third oPAD layer showing, qualitatively, the dependence of the FIV on A1 concentration. Note that the error bars for the data points correspond to standard deviations obtained by performing experiments on three different oPADs on the same day.
The difference in sensitivity RSD between Cases 1 and 2 (14% and 3%, respectively) could be due to a number of factors. First, despite using BSA as a blocker, the reagents still exhibit considerable NSA to the paper (Figure S7). For Case 1, this means that a significant fraction of F1 and Q1 remain trapped within the cellulose matrix even after the initiation of flow. Therefore, upon addition of sample A1, only a fraction of A1 and Q1 reach the third oPAD layer where F1 is present and that only the accessible fraction of F1 can take part in the reaction. The inaccessible fraction of F1 contributes to the overall high background fluorescence observed at the reporting layer of the device in Case 1.
In contrast, Case 2 suffers less from NSA because the F1-Q1 hybridized complex (R1), rather than the individual ssDNA components, is dried on the paper. Consequently, the background fluorescence in Case 2 is lower than that observed in Case 1 and, therefore, the linearity of the fit is better for Case 2 (Figure 1b). Nevertheless, we believe Case 1 is better for future POC devices than Case 2, because no pre-hybridization of reagents is required for the experiments to be successful, while still achieving similar LODs (3.1 ± 0.4 nM vs. 4.9 ± 0.1 nM).
Smart DNA detection systems
Boolean logic gates are the fundamental building blocks of many electronic devices. Biological analogs of these devices, which are based on enzyme-substrate, aptamer-target, and DNA-anti-sense DNA interactions, are of particular scientific interest as they could lead to interesting new functions.54 In the next two sections, we present logic gates, built on the oPAD platform, that exhibit the OR and AND operations. We carried out these experiments to expand the functional scope of paper-based fluidics, generally, and the multi-level oPAD format, specifically. Note that in this section, NSA was minimized by preparing all solutions added to the first layer of the folded device with 1.0 μM (dT)21 in addition to drying BSA on the paper reservoirs (Figure S8).
OR logic gate
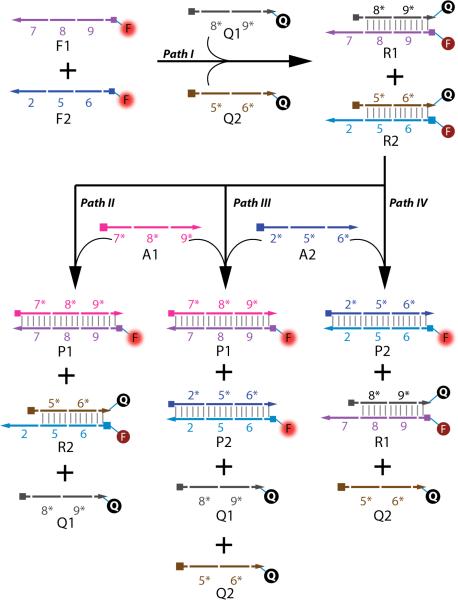
OR gates yield an output of 1 when either of the two inputs is positive.54 In the present case, the OR gate inputs are two analytes, A1 and A2 (Scheme 2), and the output will be 1 when either A1 or A2 is present. In the opposite case, when neither A1 nor A2 is present, the output will be 0. In analogy to Case 1 and Case 2, discussed earlier, A1 is complementary to F1, and Q1 quenches the fluorescence of F1 in the absence of A1. Likewise, A2 is complementary to F2, and Q2 quenches the fluorescence of F2 in the absence of A2. Moreover, A1 can only hybridize with F1 and A2 can only hybridize with F2. Therefore, there is no cross-talk between the parallel response circuits.
Scheme 2.
The OR function was demonstrated as follows. First, the paper was passivated with BSA. Second, 1.0 μL of a mixture containing TNka buffer plus 3.0 μM Q1 and 3.0 μM Q2, and 1.0 μL of a mixture containing TNka buffer plus 50.0 nM F1 and 50.0 nM F2, was dried on the reservoirs of the second and third oPAD layers, respectively (Figure 3). Third, the oPAD was folded and compressed with the aluminum clamp. Finally, 6.0 μL of solutions containing different concentrations of A1 and A2 (plus 1.0 μL of (dT)21 as a secondary blocker (Figure S8)) were introduced to the reservoirs of the first oPAD layer. These combinations of A1 and A2 are designated as inputs 0,0; 0,1; 1,0; and 1,1. Input 0,0 contained only TNka buffer. Input 0,1 was a solution containing TNka buffer plus 200.0 nM A1. Input 1,0 consisted on a solution containing TNka buffer plus 200.0 nM A2. And input 1,1 consisted on a solution containing TNka buffer plus 200.0 nM A1 and 200.0 nM A2.
Figure 3.
Strand displacement-induced fluorescence detection of DNA coupled to an oPAD-based OR gate. The device was passivated with BSA and all solutions added to the first oPAD layer contained 1.0 μM (dT)21 as an additional blocker. Top left: Color-coded schematic representation of the reagent placement on the oPAD. Top right: Truth table. Bottom left: Fluorescence image of the third oPAD layer showing the resulting fluorescence response upon addition of different inputs. Bottom right: Histogram of the fluorescence intensity volumes (FIV) measured at the third oPAD layer as a function of the input type. Note that the error bars for the histogram correspond to standard deviations obtained by performing experiments on three different oPADs on the same day.
The fluorescence image at the bottom left of Figure 3 shows the signal obtained at the third oPAD layer at the end of the experiment. These results are quantitatively represented by the FIV histograms at the bottom right of Figure 3. Here, the black dashed line is the threshold, and its magnitude is defined as twice the FIV of the blank. Inputs resulting in a signal above the threshold lead to an output of 1, while lower FIVs result in an output of 0.
The results in Figure 3 can be rationalized in terms of Scheme 2. When 0,0 is input to the first oPAD layer, the reaction is predicted to follow Path I in Scheme 2. In this case, both F1 and F2 are quenched, and the output of the OR gate is 0. When 0,1 is input, Path II is predicted. In this case, A1 hybridizes with F1 and displaces Q1 from hybrid R1. The resulting product, P1, is fluorescent, and therefore the FIV exceeds the threshold and the gate output is 1 (Figure 3). The same result is obtained for input 1,0, although in this case it arises from Path IV (Scheme 2). Lastly, when both A1 and A2 are present (input 1,1), the reaction follows Path III. This leads to a FIV that also exceeds the threshold, and again the OR gate output is 1 due to the presence of both P1 and P2. It is clear that the transfer function of this paper-based device is consistent with the truth table for an OR gate (Figure 3, top right corner).
AND logic gate
The AND gate is analogous to the just-described OR gate, except that both inputs must be positive for the output to be 1.54 Scheme 3 shows the functional pathways of the oPAD-based AND gate.
Scheme 3.
After passivating the paper with BSA, 1.0 μL of a solution containing TNka buffer plus 100.0 nM R3 (100.0 nM F3, 1.0 μM M, and 1.0 μM Q3) was dried on the third layer of the oPAD while nothing was dried on the second oPAD layer. Once the oPAD was folded and compressed, 6.0 μL of TNka buffer containing 1.0 μL of (dT)21 and the different inputs ((0,0), (0,1), (1,0), and (1,1)) were introduced to the first oPAD layer. These input designations correspond to the same A1 and A2 concentrations described for the OR gate. A FIV lower than the threshold (twice the blank signal, dashed black line at the bottom of Figure 4) corresponds to an output of 0.
The fluorescence image at the bottom left of Figure 4 shows the signal obtained from the third oPAD layer at the end of the experiment. These results are quantitatively represented at the bottom right of Figure 4 with a histogram of FIV as a function of input type. When 0,0 (A1 and A2 absent) is input to the first oPAD layer, R3 is predicted to be unchanged (Scheme 3), and therefore the FIV is below the threshold and the output of the AND gate is 0 (Figure 4). For input 0,1 (only A1 present), no fluorescence response is observed. This is because A1 cannot hybridize with F3 until A2 displaces M from R3 (Scheme 3). A 0 output is also observed for input 1,0 (only A2 present). In this case, A2 displaces M from R3; however, Q3 still quenches the F3 fluorescence in the absence of A1. Only for an input of 1,1 (both A1 and A2 present) is the FIV above the threshold for an output response of 1 (Figure 4). In this case A2 hybridizes with F3 and displaces M. In the absence of M, the bases in F3 designated by the number 7 (Scheme 3) are free to start hybridizing with A1. Consequently, A1 displaces Q3 and the resulting complex, P3, is fluorescent. Hence, consistent with the truth table for an AND gate (Figure 4, top right corner), the output of this oPAD is only 1 when both A1 and A2 are present.
By demonstrating that we can pattern and program both flow (via wax printing and folding) and molecules (such as the OR and AND gates demonstrated herein), many possibilities arise for carrying out and evaluating multiplexed reactions within the same device. Reactions can be distinguished either by flow or by molecular programming or by combinations thereof. This should make the oPAD configurations coupled to nucleic acid circuitry especially compelling into the future. For example, an effective OR gate oPAD could report on whether a patient has been infected by any of the different types of common influenza viruses or drug resistant tuberculosis. An AND gate oPAD could provide more accurate and confident diagnosis for a target or a reaction having multiple markers, such as long amplified products from polymerase chain reactions or isothermal amplification.
SUMMARY AND CONCLUSIONS
We have reported three significant findings in this manuscript. First, implementation of strand-displacement fluorescence detection of ssDNA at concentrations as low as 3.1 ± 0.4 nM using 3D paper analytical devices. Second, a multiplexed version of strand displacement that results in logical OR and AND operations. Third, the multilayer device, which is prepared by simple wax printing and paper folding, permits reagent storage on different layers of the oPAD before experiments are initiated. This avoids cross-reaction between the reagents until they are triggered to mix by capillary flow of the sample solution. These findings provide an important demonstration of functional integration, without resorting to increased complexity.
The simple (but important) DNA reactions presented in this work are the building blocks for understanding and developing more complicated assays on paper (e.g. both enzymatic and non-enzymatic DNA amplification systems widely used to detect pathogens and viruses). In the future, we seek to further improve LODs via reduced NSA and on-chip amplification. Accordingly, we are currently studying NSA of DNA on paper substrates to better understand and improve the results obtained in this work. On-chip amplification could be introduced using our recently described SlipPAD approach, which enables timed reaction and washing steps.10 Additionally, we are currently working to replace fluorescence with electrochemical detection, thereby simplifying read-out without sacrificing quantitation.
Supplementary Material
ACKNOWLEDGMENTS
This project is sponsored by the Department of the Defense, Defense Threat Reduction Agency (contract number HDTRA-1-13-1-0031). The content of the information does not necessarily reflect the position or the policy of the federal government, and no official endorsement should be inferred. We also thank the Robert A. Welch Foundation (Grants F-0032 and F-1654). ADE also acknowledges support from the NIH (U54EB015403-01).
Footnotes
SUPPORTING INFORMATION
Control experiments for the proposed reaction pathways of Cases 1 and 2, a schematic diagram showing the oPAD design and dimensions, a discussion of partial blocking of NSA by BSA, a plot of fluorescence intensity vs. time for the strand displacement reactions in homogeneous solution, an explanation of how the placement of F1 in the device was optimized, a representation of the experimental protocol for assembly of the oPAD, and a semi-quantitative analysis of NSA on paper. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Dungchai W, Chailapakul O, Henry CS. Anal. Chim. Acta. 2010;674:227–233. doi: 10.1016/j.aca.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 2.Ellerbee AK, Phillips ST, Siegel AC, Mirica KA, Martinez AW, Striehl P, Jain N, Prentiss M, Whitesides GM. Anal. Chem. 2009;81:8447–8452. doi: 10.1021/ac901307q. [DOI] [PubMed] [Google Scholar]
- 3.Shen L, Hagen JA, Papautsky I. Lab Chip. 2012;12:4240–4243. doi: 10.1039/c2lc40741h. [DOI] [PubMed] [Google Scholar]
- 4.Dungchai W, Chailapakul O, Henry CS. Anal. Chem. 2009;81:5821–5826. doi: 10.1021/ac9007573. [DOI] [PubMed] [Google Scholar]
- 5.Vella SJ, Beattie P, Cademartiri R, Laromaine A, Martinez AW, Phillips ST, Mirica KA, Whitesides GM. Anal. Chem. 2012;84:2883–2891. doi: 10.1021/ac203434x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gubala V, Harris LF, Ricco AJ, Tan MX, Williams DE. Anal. Chem. 2011;84:487–515. doi: 10.1021/ac2030199. [DOI] [PubMed] [Google Scholar]
- 7.Nie Z, Deiss F, Liu X, Akbulut O, Whitesides GM. Lab Chip. 2010;10:3163–3169. doi: 10.1039/c0lc00237b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H, Crooks RM. Anal. Chem. 2012;84:2528–2532. doi: 10.1021/ac203457h. [DOI] [PubMed] [Google Scholar]
- 9.Locke IC, Cox SF, Carpenter BG. J. Chromatog. A. 1997;788:75–80. [Google Scholar]
- 10.Liu H, Li X, Crooks RM. Anal. Chem. 2013;85:4263–4267. doi: 10.1021/ac4008623. [DOI] [PubMed] [Google Scholar]
- 11.Jahanshahi-Anbuhi S, Chavan P, Sicard C, Leung V, Hossain SMZ, Pelton R, Brennan JD, Filipe CD. M. Lab Chip. 2012;12:5079–5085. doi: 10.1039/c2lc41005b. [DOI] [PubMed] [Google Scholar]
- 12.Cate D, Dungchai W, Cunningham J, Volckens J, Henry C. Lab Chip. 2013;13:2397–2404. doi: 10.1039/c3lc50072a. [DOI] [PubMed] [Google Scholar]
- 13.Fridley GE, Le HQ, Fu E, Yager P. Lab Chip. 2012;12:4321–4327. doi: 10.1039/c2lc40785j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Zwanenburg P, Liu X. Lab Chip. 2013;13:2609–2614. doi: 10.1039/c3lc00006k. [DOI] [PubMed] [Google Scholar]
- 15.Nery EW, Kubota LT. Anal. Bioanal. Chem. 2013:1–23. doi: 10.1007/s00216-013-6911-4. [DOI] [PubMed] [Google Scholar]
- 16.Liu H, Crooks RM. J. Am. Chem. Soc. 2011;133:17564–17566. doi: 10.1021/ja2071779. [DOI] [PubMed] [Google Scholar]
- 17.Zhang DY, Winfree E. J. Am. Chem. Soc. 2009;131:17303–17314. doi: 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]
- 18.Pelton R. TrAC-Trend. Anal. Chem. 2009;28:925–942. doi: 10.1016/j.trac.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yetisen AK, Akram MS, Lowe CR. Lab Chip. 2013;13:2210–2251. doi: 10.1039/c3lc50169h. [DOI] [PubMed] [Google Scholar]
- 20.Rocco RM. Landmark papers in analytical chemistry. San Francisco State University; Elsevier: 2006. [Google Scholar]
- 21.Rohrman BA, Leautaud V, Molyneux E, Richards-Kortum RR. PLoS ONE. 2012;7:e45611. doi: 10.1371/journal.pone.0045611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasooly AH, K. E. Biosensors and Biodetection. Methods and Protocols: Electrochemical and Mechanical Detections, Lateral Flow and Ligands for Biosensors. Vol. 504. Springer; 2009. [PubMed] [Google Scholar]
- 23.Aragay G, Monton H, Pons J, Font-Bardia M, Merkoci A. J. Mater. Chem. 2012;22:5978–5983. [Google Scholar]
- 24.Martinez AW, Phillips ST, Whitesides GM. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19606–19611. doi: 10.1073/pnas.0810903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thom NK, Yeung K, Pillion MB, Phillips ST. Lab Chip. 2012;12:1768–1770. doi: 10.1039/c2lc40126f. [DOI] [PubMed] [Google Scholar]
- 26.Lewis GG, DiTucci MJ, Baker MS, Phillips ST. Lab Chip. 2012;12:2630–2633. doi: 10.1039/c2lc40331e. [DOI] [PubMed] [Google Scholar]
- 27.Schilling KM, Jauregui D, Martinez AW. Lab on a Chip. 2013;13:628–631. doi: 10.1039/c2lc40984d. [DOI] [PubMed] [Google Scholar]
- 28.Govindarajan AV, Ramachandran S, Vigil GD, Yager P, Bohringer KF. Lab Chip. 2012;12:174–181. doi: 10.1039/c1lc20622b. [DOI] [PubMed] [Google Scholar]
- 29.Liu H, Xiang Y, Lu Y, Crooks RM. Angew. Chem. Int. Ed. 2012;51:6925–6928. doi: 10.1002/anie.201202929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mansfield MA. The Use of Nitrocellulose Membranes in Lateral-Flow Assays. In: R.C.T. Wong HY, editor. Drug Abuse: Body Fluid Testing. Humana Press; 2005. pp. 71–85. [Google Scholar]
- 31.Wong RCT, H. Y. Lateral Flow Immunoassay. Humana Press; New York, NY: 2009. [Google Scholar]
- 32.Coltro WKT, de Jesus DP, da Silva JAF, do Lago CL, Carrilho E. Electrophoresis. 2010;31:2487–2498. doi: 10.1002/elps.201000063. [DOI] [PubMed] [Google Scholar]
- 33.Wang S, Ge L, Song X, Yu J, Ge S, Huang J, Zeng F. Biosens. Bioelectron. 2012;31:212–218. doi: 10.1016/j.bios.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 34.Yu J, Wang S, Ge L, Ge S. Biosens. Bioelectron. 2011;26:3284–3289. doi: 10.1016/j.bios.2010.12.044. [DOI] [PubMed] [Google Scholar]
- 35.Delaney JL, Hogan CF, Tian J, Shen W. Anal. Chem. 2011;83:1300–1306. doi: 10.1021/ac102392t. [DOI] [PubMed] [Google Scholar]
- 36.Lankelma J, Nie Z, Carrilho E, Whitesides GM. Anal. Chem. 2012;84:4147–4152. doi: 10.1021/ac3003648. [DOI] [PubMed] [Google Scholar]
- 37.Nie Z, Nijhuis CA, Gong J, Chen X, Kumachev A, Martinez AW, Narovlyansky M, Whitesides GM. Lab Chip. 2010;10:477–483. doi: 10.1039/b917150a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siegel AC, Phillips ST, Wiley BJ, Whitesides GM. Lab Chip. 2009;9:2775–2781. doi: 10.1039/b905832j. [DOI] [PubMed] [Google Scholar]
- 39.Zhao W, Ali MM, Aguirre SD, Brook MA, Li Y. Anal. Chem. 2008;80:8431–8437. doi: 10.1021/ac801008q. [DOI] [PubMed] [Google Scholar]
- 40.Mentele MM, Cunningham J, Koehler K, Volckens J, Henry CS. Anal. Chem. 2012;84:4474–4480. doi: 10.1021/ac300309c. [DOI] [PubMed] [Google Scholar]
- 41.Klasner S, Price A, Hoeman K, Wilson R, Bell K, Culbertson C. Anal. Bioanal. Chem. 2010;397:1821–1829. doi: 10.1007/s00216-010-3718-4. [DOI] [PubMed] [Google Scholar]
- 42.Jokerst JC, Adkins JA, Bisha B, Mentele MM, Goodridge LD, Henry CS. Anal. Chem. 2012;84:2900–2907. doi: 10.1021/ac203466y. [DOI] [PubMed] [Google Scholar]
- 43.Shera EB, Seitzinger NK, Davis LM, Keller RA, Soper SA. Chem. Phys. Let. 1990;174:553–557. [Google Scholar]
- 44.Yu A, Shang J, Cheng F, Paik BA, Kaplan JM, Andrade RB, Ratner DM. Langmuir. 2012;28:11265–11273. doi: 10.1021/la301661x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sassolas A, Leca-Bouvier BD, Blum LJ. J. Chem. Rev. 2007;108:109–139. doi: 10.1021/cr0684467. [DOI] [PubMed] [Google Scholar]
- 46.Araújo AC, Song Y, Lundeberg J, Ståhl PL, Brumer H. Anal. Chem. 2012;84:3311–3317. doi: 10.1021/ac300025v. [DOI] [PubMed] [Google Scholar]
- 47.Allen PB, Arshad SA, Li B, Chen X, Ellington AD. Lab Chip. 2012;12:2951–2958. doi: 10.1039/c2lc40373k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ali MM, Aguirre SD, Xu Y, Filipe CDM, Pelton R, Li Y. Chem. Commun. 2009:6640–6642. doi: 10.1039/b911559e. [DOI] [PubMed] [Google Scholar]
- 49.Lu J, Ge S, Ge L, Yan M, Yu J. Electrochim. Acta. 2012;80:334–341. [Google Scholar]
- 50.Carrilho E, Martinez AW, Whitesides GM. Anal. Chem. 2009;81:7091–7095. doi: 10.1021/ac901071p. [DOI] [PubMed] [Google Scholar]
- 51.Lu Y, Shi W, Jiang L, Qin J, Lin B. Electrophoresis. 2009;30:1497–1500. doi: 10.1002/elps.200800563. [DOI] [PubMed] [Google Scholar]
- 52.Le NCH, Gubala V, Gandhiraman RP, Daniels S, Williams DE. Langmuir. 2011;27:9043–9051. doi: 10.1021/la2011502. [DOI] [PubMed] [Google Scholar]
- 53.Miller-Jaster K, Petrie Aronin C, Guilford W. Cell. Mol. Bioeng. 2012;5:44–51. [Google Scholar]
- 54.Szacitowski K. Chem. Rev. 2008;108:3481–3548. doi: 10.1021/cr068403q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.