Abstract
Centromeric DNA sequences in multicellular eukaryotes are often highly repetitive and are not unique to a specific centromere or to centromeres at all. Thus, it is a major challenge to study the fine structure of individual plant centromeres. We used a DNA fiber-fluorescence in situ hybridization approach to study individual maize (Zea mays) centromeres using oat (Avena sativa)-maize chromosome addition lines. The maize centromere-specific satellite repeat CentC in the addition lines allowed us to delineate the size and organization of centromeric DNA of individual maize chromosomes. We demonstrate that the cores of maize centromeres contain mainly CentC arrays and clusters of a centromere-specific retrotransposon, CRM. CentC and CRM sequences are highly intermingled. The amount of CentC/CRM sequence varies from ∼300 to >2800 kb among different centromeres. The association of CentC and CRM with centromeric histone H3 (CENH3) was visualized by a sequential detection procedure on stretched centromeres. The analysis revealed that CENH3 is always associated with CentC and CRM but that not all CentC or CRM sequences are associated with CENH3. We further demonstrate that in the chromosomal addition lines in which two CenH3 genes were present, one from oat and one from maize, the oat CENH3 was consistently incorporated by the maize centromeres.
INTRODUCTION
The centromere is the chromosomal domain responsible for kinetochore formation and sister chromatid cohesion. Thus, centromeres play the key role in faithful transmission of chromosomes during mitosis and meiosis. In multicellular eukaryotes, centromeres are embedded within cytologically distinctive heterochromatin and are associated with long tracts of repetitive DNA. Satellite DNA and retrotransposons are the most common DNA elements found in plant centromeres (Jiang et al., 2003). The centromere-specific satellite repeats reported in several plant species show a structure and organization that is similar to the α satellite in human centromeres (Round et al., 1997; Gindullis et al., 2001; Cheng et al., 2002b). Data from several species suggest that satellite DNA has a functional role in chromosome segregation (Kaszas and Birchler, 1996, 1998; Cheng et al., 2002b; Zhong et al., 2002; Nagaki et al., 2003b, 2004). Among the retroelements known at plant centromeres, the centromeric retrotransposon (CR) family in Gramineae species is most intriguing. Unlike other retrotransposon families that diverge rapidly during evolution, the CR family has been found in the centromeres of all Gramineae species studied (Aragon-Alcaide et al., 1996; Jiang et al., 1996; Miller et al., 1998; Presting et al., 1998; Langdon et al., 2000). Highly conserved motifs were found in the long terminal repeats of the CR elements from rice (Oryza sativa), maize (Zea mays), and Hordeum vulgare (barley) (Nagaki et al., 2003a). Recent data from maize showed that the CR elements may play a critical role in centromere function (Zhong et al., 2002).
Whereas there are no universally conserved DNA sequences associated with centromere function, several proteins specific to the centromere/kinetochore complex are highly conserved (Dobie et al., 1999; Yu et al., 2000; Houben and Schubert, 2003). A centromere-specific histone H3–like protein, referred to as CENH3, has been found to underlie the kinetochore (see reviews in Henikoff et al., 2001; Sullivan et al., 2001). The first CENH3, CENP-A (centromeric protein A), was identified in humans (Palmer et al., 1987, 1991). Since then, CENH3 has been found in all model eukaryotes (Henikoff et al., 2001), including Arabidopsis (Arabidopsis thaliana) (Talbert et al., 2002), maize (Zhong et al., 2002), and rice (Nagaki et al., 2004). CENH3s replace the regular histone H3 in centromeric chromatin (Yoda et al., 2000). There are numerous lines of evidence that CENH3 plays the key role in the establishment and function of kinetochores in various organisms. CENP-A is present only in the functional centromeres of cytologically stable dicentric chromosomes in humans (Warburton et al., 1997). CENH3 also is present in significantly rearranged centromeres and human neocentromeres (Sullivan et al., 2001). Inactivation of CENH3 proteins in yeasts, worms, flies, and mammals severely disrupts mitosis and cell-cycle progression and causes mislocation of many other centromere/kinetochore proteins (Sullivan et al., 2001). All of these data indicate that CENH3 is a central component in kinetochore formation and centromere function.
Maize has become an important model for plant centromere research. Maize centromeres contain a 156-bp satellite repeat, CentC, and the centromere-specific retrotransposon CRM (the maize subfamily of CR) (Ananiev et al., 1998; Chen et al., 2000; Zhong et al., 2002; Nagaki et al., 2003a). Both CentC and CRM sequences interact with CENH3, indicating that they are located within the functional maize centromeres (Zhong et al., 2002). Two BAC clones derived from maize centromeres have recently been sequenced (Nagaki et al., 2003a). These two BACs contain primarily CentC and retrotransposon sequences. Alfenito and Birchler (1993) also isolated a repeat specific to the centromeres of maize B chromosomes. The B-specific repeat is thought to reside in the functional portion of the B centromere based on the fact that centromere misdivision derivatives, resulting from a break on the short arm and subsequently on the long arm side of the centromere, still retain substantial B repeat copies. The B centromere repeat is homologous to yet another maize repeat, Cent4, which is only found on chromosome 4 (Page et al., 2001).
Despite the extensive work on maize centromeres, the large-scale DNA structure and organization of individual maize centromeres is not known. In this study, we used the recently developed oat (Avena sativa)-maize addition lines (Kynast et al., 2001) to study individual maize centromeres. We demonstrate that the cores of seven maize centromeres consist almost exclusively of intermingled CentC and CRM sequences. The quantities of CentC and CRM and their organization relative to each other vary significantly among different maize centromeres. The structural relationship between CENH3 and CentC/CRM was visualized by sequential detection of the protein and centromeric DNA on stretched centromeres. We also demonstrate that maize centromeres consistently adopt the oat CENH3 after being transferred into the oat genetic background.
RESULTS
CentC and CRM Are Located in the Cytologically Defined Maize Centromeres
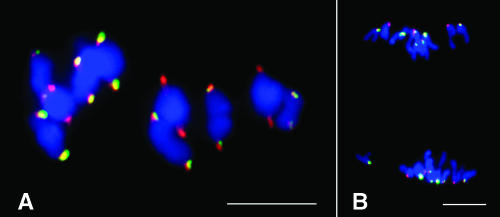
We conducted fluorescence in situ hybridization (FISH) mapping of CentC and CRM sequences on maize chromosomes at different meiotic stages. On the highly extended pachytene chromosomes, both CentC and CRM sequences were restricted to the primary constriction regions. The FISH signals were located at the most poleward positions on chromosomes at metaphase I and anaphase I stages (Figures 1A and 1B). These results confirm that CentC and CRM are located at the cytologically defined maize centromeres (Ananiev et al., 1998; Zhong et al., 2002).
Figure 1.
Cytological Localization of CentC and CRM Sequences on Meiotic Chromosomes of Maize Inbred B73.
(A) Metaphase I. Both CentC and CRM are located on the most poleward position on each bivalent chromosome.
(B) Anaphase I.
CentC, green; CRM, red. Bars = 10 μm.
DNA Organization of Individual Maize Centromeres
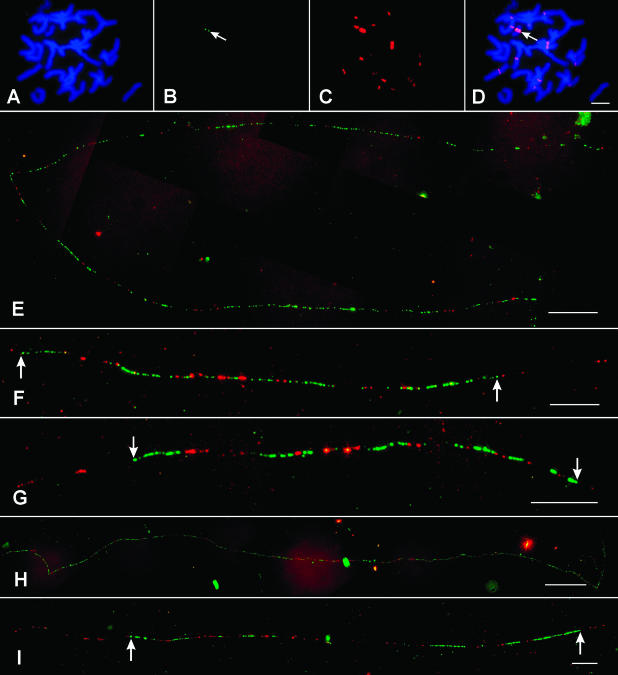
Oat-maize chromosome addition lines (Kynast et al., 2001) were used to study the DNA organization of individual maize centromeres. FISH analysis using CentC and CRM probes confirmed the presence of a single (monosomic addition) or a pair (disomic addition) of maize chromosomes in the seven oat-maize addition lines used in this study (Figures 2A to 2D). Cross-hybridization of the CRM probe to oat centromeres was observed (Figure 2C). However, the CentC probe was highly specific to the maize chromosome (Figure 2B), which facilitates unambiguous identification of fiber-FISH signals derived from the CentC and CRM probes.
Figure 2.
Distribution of CentC and CRM Sequences in Individual Maize Centromeres.
(A) to (D) FISH mapping of CentC (green) and CRM (red) sequences on somatic metaphase chromosomes of oat-maize addition line 6.
(A) 4′6-diamidino-2-phenylindole staining of a somatic metaphase cell.
(B) FISH signals derived from CentC (arrow).
(C) FISH signals derived from CRM.
(D) A merged image of the chromosomes and the FISH signals. Arrow points to the CentC signal.
(E) to (I) Fiber-FISH signals of CentC (green) and CRM (red) from the centromeres of maize chromosomes 1, 2, 3, 7, and 9, respectively. Arrows point to the borders of the central domains of the fiber-FISH signals.
Bars = 10 μm in (D), (F), (G), and (I), 20 μm in (E), and 50 μm in (H).
The distributions of CentC and CRM sequences in the centromeres of maize chromosomes 1, 2, 3, 4, 6, 7, and 9 were analyzed using fiber-FISH. A minimum of eight digital images of the fiber-FISH signals were collected from each centromere and measured using IPLab software. The lengths of the fiber-FISH signals (in micrometers) were converted to kilobases using a 3.21-kb/μm conversion rate (Cheng et al., 2002a) (Table 1). In general, the fiber-FISH signals derived from the CentC and CRM probes were contiguous with few unambiguous gaps, suggesting that these two sequences are the dominant DNA components of maize centromeres (Figures 2E to 2I and 3). Interspersion of the two sequences was observed in all seven maize centromeres (Figures 2E to 2I and 3).
Table 1.
Distribution of the CentC Satellite and CRM Elements among Different Centromeres in Maize
| Chromosome | n | Left Flanking CRM (kb)a | Central Domain (kb)b | Right Flanking CRM (kb)a | Full Size (μm) | Full Size (kb)c | CentC (%) | CRM (%)d |
|---|---|---|---|---|---|---|---|---|
| 1 | 8 | ND | ≥1720 | ND | ≥535 | ≥1720 | 70.4 ± 3.9 | 29.6 ± 3.9 |
| 2 | 18 | ≥34 ± 20 | 282 ± 15 | ≥69 ± 29 | ≥121 ± 11 | ≥388 ± 34 | 70.6 ± 3.7 | 29.4 ± 3.7 |
| 3 | 25 | ≥52 ± 9 | 241 ± 24 | ≥6 ± 3 | ≥93 ± 9 | ≥299 ± 29 | 60.7 ± 5.4 | 39.3 ± 5.4 |
| 4 | 10 | ≥114 ± 38 | 265 ± 27 | ≥164 ± 45 | ≥166 ± 23 | ≥533 ± 74 | 40.0 ± 7.1 | 60.0 ± 7.1 |
| 6 | 12 | ≥76 ± 28 | 351 ± 41 | ≥52 ± 25 | ≥147 ± 17 | ≥471 ± 81 | 42.8 ± 6.4 | 57.2 ± 6.4 |
| 7 | 8 | ND | ≥2830 | ND | ≥882 | ≥2830 | 73.6 ± 4.9 | 26.4 ± 4.9 |
| 9 | 9 | ≥34 ± 8 | 515 ± 82 | ≥160 ± 29 | ≥221 ± 15 | ≥709 ± 47 | 48.4 ± 4.7 | 51.6 ± 4.7 |
The end of the flanking CRM domains cannot be determined unambiguously. We used the most characteristic CRM signals at each end for the measurements that represent the minimum sizes of these flanking domains. ND, no data.
The size of the core domain of centromeres 1 and 7 cannot be determined. The longest fiber-FISH signals were used as the minimum sizes of these domains.
The lengths of the fiber-FISH signals (μm) were converted to kilobases using a 3.21-kb/μm conversion rate (Cheng et al., 2002a).
The percentage of the CentC and CRM sequences are relative to each other without including the unknown sequences in the centromeres.
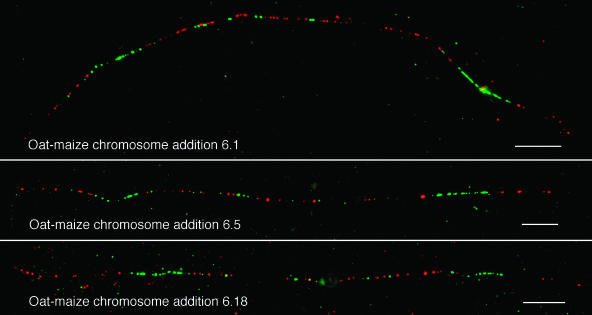
Figure 3.
Distribution Patterns of CentC and CRM Sequences in the Centromeres of Maize Chromosome 6 from Three Independently Isolated Oat-Maize Chromosome 6 Addition Lines.
CentC, green; CRM, red. Bars = 10 μm.
We divided the fiber-FISH signals into a central domain and two flanking domains. The two ends of each central domain are marked by distinct CentC signals (Figures 2F, 2G, and 2I). These end CentC signals are usually flanked by additional CRM signals. The sizes of the fiber-FISH measurements, which include both central and flanking domains, varied from 299 kb ± 29 kb of centromere 3 to >2.8 Mb of centromere 7 (Table 1). Technically, it is difficult to obtain DNA fibers >1 Mb. Thus, most of the fiber-FISH signals of centromeres 1 and 7 may be derived from broken DNA fibers. We used the longest fiber-FISH signals to represent the minimum size of these two centromeres (Table 1).
The relative amount of CentC and CRM sequences within each centromere was calculated by counting the numbers of green and red fluorescence spots in each digital image. The CRM sequences account for 26 to 60% of the fiber-FISH signals in the seven centromeres (Table 1). The CRM signals were as short as one to three consecutive fluorescence spots, which may represent a single CRM element, to loose clusters as long as 350 μm (>1000 kb) (data not shown). Although the signals derived from CentC and CRM in the central domains were largely contiguous, unambiguous gaps can be identified in centromeres 2, 4, and 9. These gaps likely represent DNA sequences other than CentC and CRM.
To investigate whether the organization of the CentC and CRM sequences were rearranged during and/or after the maize chromosomes were transferred into the oat background, we conducted fiber-FISH analysis on three independently isolated oat-maize chromosome 6 addition lines (lines 6.1, 6.5, and 6.18). The fiber-FISH signal patterns derived from the three centromeres were nearly identical (Figure 3). These data suggest that the transfer of a maize chromosome into oat does not cause substantial changes in the content or organization of the centromere.
Centromere 4 Is a CRM-Rich Domain That Does Not Include the Cent4 Repeat
In the original description of CentC, the authors noted that chromosome 4 has an unusually small amount of CentC (Ananiev et al., 1998). Page et al. (2001) later identified a repeat localized to the primary constriction of chromosome 4, called Cent4. Here, we tested the hypothesis that Cent4 may substitute for CentC and extend the size of the functional centromere.
We found that the fiber-FISH pattern of CentC and CRM on chromosome 4 is similar to those from other maize centromeres. The intermingled CentC/CRM sequences span ∼532 kb ± 74 kb and consist of ∼60% CRM and ∼40% CentC (Figure 4A, Table 1). The total amount of CentC in centromere 4 was estimated at ∼100 kb, less than any of the other six chromosomes analyzed.
Figure 4.
Distribution of CentC, CRM, and Cent4 Sequences in the Centromere of Maize Chromosome 4.
(A) Fiber-FISH signal of CentC (green) and CRM (red) from the centromere of maize chromosome 4.
(B) A fiber-FISH signal of Cent4 consists of three contiguous arrays separated by two gaps.
(C) Locations of CentC (red) and Cent4 (green, arrow) on a pachytene chromosome 4 of B73. The signals derived from the two probes are clearly separated from each other.
(D) Locations of CentC (red) and Cent4 (green) on metaphase I chromosome 4 of B73. The Cent4 signals (arrows) are located behind the CentC signals that are at the most poleward positions.
Bars = 10 μm.
Fiber-FISH signals derived from Cent4 were divided into three arrays with different sizes (Figure 4B). The Cent4 locus spans 311 kb ± 25 kb DNA (including the two gaps) based on 14 fiber-FISH measurements. We were not able to find an unambiguous connection between signals derived from Cent4 and CentC on extended DNA fibers, suggesting that these two sequences are separated by >500 kb, the detection limit of fiber-FISH (Jackson et al., 1998). Supporting this view is the fact that the Cent4 and CentC signals were clearly separated from each other on pachytene chromosomes (Figure 4C) and that the signals from Cent4 always lagged behind CentC on centromeres drawn poleward on the spindle (Figure 4D).
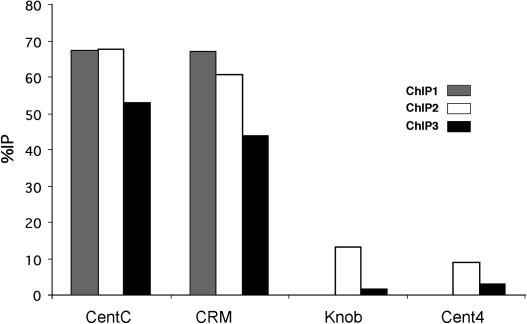
We also conducted chromatin immunoprecipitation (ChIP) analysis to test whether the Cent4 sequence interacts with maize CENH3. Data from three independent experiments showed that whereas CentC and CRM were coimmunoprecipitated at high levels, Cent4 was not (Figure 5). Taken together, the available data indicate that the major cluster of Cent4 does not associate with CENH3.
Figure 5.
ChIP Analysis of Cent4 in Maize.
CentC and CRM sequences were coimmunoprecipitated with CENH3 antibodies at high levels in all three experiments. The percentage of immunoprecipitation (%IP) of Cent4 did not differ significantly from that of the negative control (knob).
Nearly All CENH3 Is Associated with CentC and CRM, but Not All CentC and CRM Sequences Are Associated with CENH3
Data from human neocentromeres showed that CENP-A–associated chromatin domains contain ∼130 to 460 kb DNA (Lo et al., 2001a, 2001b; Alonso et al., 2003). This is roughly consistent with the lengths of the CentC/CRM domains on chromosomes 2, 3, 4, 6, and 9 but substantially less than the length of apparent centromeric DNA on chromosomes 1 and 7 (≥1.7 and 2.8 Mb, respectively). Only a portion of the unusually long centromeric domains of chromosomes 1 and 7 may interact with CENH3. To test this hypothesis, we developed a technique to sequentially visualize CENH3 and CentC/CRM sequences on stretched chromatin fibers.
Interphase nuclei isolated from freshly grown maize callus were used for chromatin fiber preparation (see Methods). Chromatin from purified nuclei was gently stretched, fixed, and treated with the maize antibody to CENH3. Immunostaining signals derived from well-stretched centromeres were recorded. The same slides then were probed with CentC and CRM for fiber-FISH analysis. We frequently observed fiber-FISH signals that spanned regions not covered by anti-CENH3 signals (Figure 6). However, we rarely observed the opposite of this: chromatin fibers that were stained by CENH3 antibodies nearly always coincided with CentC/CRM sequences. Three types of colocalization of CENH3 and CentC/CRM were observed among 25 centromeres analyzed: (1) a nearly perfect overlap of CENH3 and CentC/CRM sequences (13 cases); (2) CENH3 staining located in the middle of the fiber-FISH signals (2 cases); and (3) CENH3 staining located to one end of the fiber-FISH signals (10 cases; Figure 6).
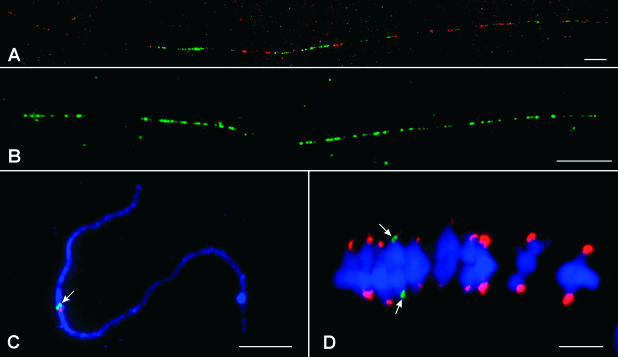
Figure 6.
Association between CENH3 and CentC/CRM Sequences in Maize.
(A1) and (A2) An immunostaining signal derived from the maize CENH3 antibody (A1). CENH3 was detected by rhodamine-conjugated anti-rabbit antibodies, and the signals were pseudocolored in yellow. The same chromatin fiber was probed with CentC (green) and CRM (red) (A2). Arrows point to the regions associated with CENH3. The fiber-FISH signal extends beyond the CENH3-associated region.
(B1) and (B2) Two independent immunostaining signals derived from the maize CENH3 antibody (B1). The signals were pseudocolored in yellow. The same region was probed with CentC (green) and CRM (red) (B2). Arrows and arrowheads point to the two regions associated with CENH3. The fiber-FISH signals extend beyond the two CENH3-associated regions and also are visible on other chromatin fibers that are not associated with CENH3.
Bars = 5 μm.
Functional Adaptation of Maize Centromeres in the Genetic Background of Oat
We previously identified a full-length cDNA from the maize CenH3 gene (Zhong et al., 2002). Physical mapping data indicate that this single maize CenH3 gene (marker CL12047-1) lies close to umc1762 on chromosome 6 (http://genome.arizona.edu/fpc/maize/). To verify this map location, we probed DNA gel blots from the 10 oat-maize addition lines with the CenH3 cDNA. As expected, strong hybridization was only observed in the chromosome 6 addition line (data not shown). Thus, this strain contains both the oat and maize CenH3 genes. We wondered whether maize CENH3 might preferentially associate with the maize centromere, as might be expected if CENH3 has sequence-specific binding properties (Malik and Henikoff, 2002).
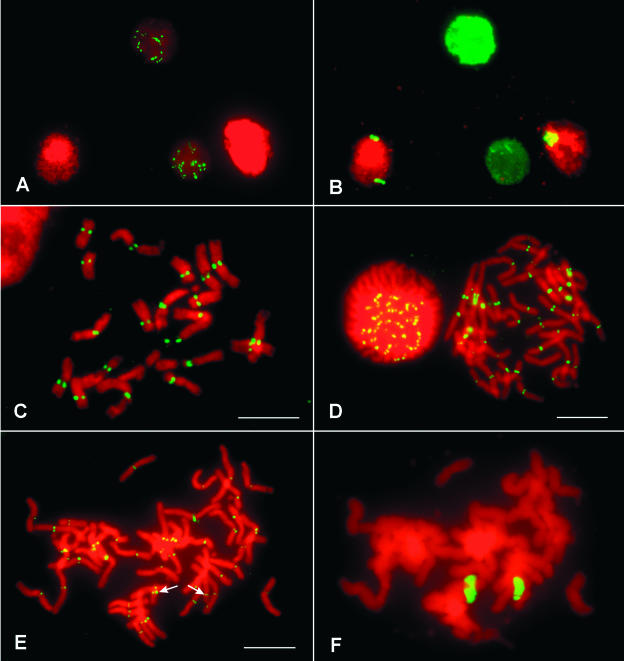
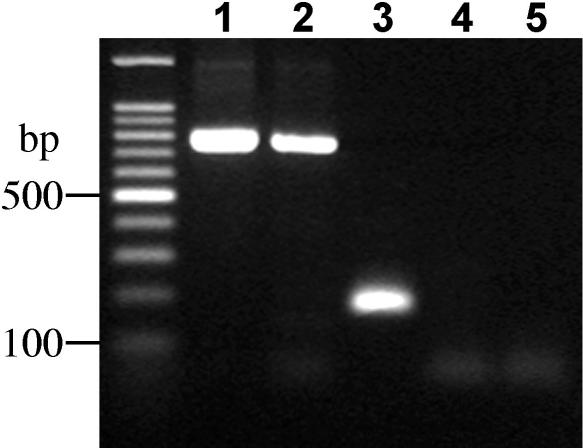
Three independently isolated oat-maize chromosome 6 addition lines (lines 6.1, 6.5, and 6.18) were immunoassayed using the maize-specific antibody to CENH3. Root tip cells from maize and oat-maize chromosome 6 addition lines were mixed in the preparations. After the preparations were stained with the CENH3 antibody, the same slides then were used for genomic in situ hybridization using maize genomic DNA as a probe to confirm the presence of the maize chromosomes. No CENH3 staining was apparent in all three chromosome 6 addition lines (Figures 7A and 7B). The immunostaining data raised the possibility that maize CenH3 is not expressed in the oat background. Reverse transcriptase (RT) PCR was used to test this hypothesis. As shown in Figure 8, CenH3 transcripts were readily detected in the maize Seneca 60 inbred line, but they could not be detected in the oat-maize chromosome 6 addition line that carries the same CenH3 gene from Seneca 60. This result suggests that maize CenH3 is silenced in oat.
Figure 7.
Incorporation of Oat CENH3 in Maize Centromeres.
(A) and (B) Maize CENH3 is not detected in the genetic background of oat. A cytological preparation containing root tip cells from both maize and oat-maize addition line 6 was immunostained by the maize anti-CENH3 antibody (A). Immunostaining signals only were observed on the two maize nuclei. The same cells were hybridized to a maize genomic DNA probe, confirming the presence of the two maize chromosomes 6 in the two nuclei derived from oat-maize addition line 6 (B).
(C) Immunostaining of maize chromosomes by the anti-CENH3 antibody developed in rice.
(D) Immunostaining of oat chromosomes by the anti-CENH3 antibody developed in rice.
(E) and (F) Incorporation of oat CENH3 by the centromere of maize chromosome 6. Immunostaining of chromosomes from an oat-maize chromosome addition 6 by the anti-CENH3 antibody developed in rice (E). Signals were visible on all oat chromosomes and the two maize chromosomes (arrows). The same cell was hybridized to a maize genomic DNA probe to facilitate the identification of the two maize chromosomes (F).
Bars = 10 μm.
Figure 8.
RT-PCR Analysis of the Maize CenH3 Gene.
PCR products were amplified from lane 1, genomic DNA from maize (Seneca 60); lane 2, genomic DNA from oat-maize chromosome 6 addition line 6.1; lane 3, cDNA from maize (Seneca 60); lane 4, cDNA from oat-maize chromosome 6 addition line 6.1; and lane 5, cDNA from oat (Starter-1). A positive RT-PCR product only was observed in lane 3.
We also produced antibodies to rice CENH3 (Nagaki et al., 2004). The rice antibodies stain the centromeres of maize, oat, and several other Gramineae species (Figures 7C and 7D). Maize chromosome 6 and maize chromosomes in the other six addition lines showed clear immunostaining signals derived from the rice antibody (Figures 7E and 7F). These results confirm that the maize centromeres have incorporated the oat CENH3 in the addition lines.
DISCUSSION
Organization, Abundance, and Incorporation of CentC/CRM Sequences in CENH3-Associated Chromatin in Maize Chromosomes
Maize centromeres contain CentC satellite arrays and CRM elements that interact with maize CENH3 (Zhong et al., 2002). Other retroelements are rare within maize centromeres (Mroczek and Dawe, 2003) and do not interact with CENH3 (Zhong et al., 2002). Likewise, CENH3 does not associate with Cent4, which is present at or near the primary constriction of chromosome 4 (Page et al., 2001) (Figures 4 and 5). Here, we confirm the previous interpretations by showing that nearly all CENH3 is associated with CentC and CRM. We also show that fiber-FISH signals derived from CentC and CRM probes are largely contiguous (Figures 2E to 2I and 3), and the CentC and CRM sequences are highly intermingled throughout all seven maize centromeres analyzed (Figures 2 and 3). Notably, some maize centromeres contain more CRM sequences than the CentC satellite in the core domain (Table 1).
It is interesting to note that the centromeres of maize chromosomes 2, 3, 4, 6, and 9 contain similar amounts of the CentC/CRM sequences, ranging from ∼300 to ∼700 kb (Table 1). This size range is consistent with the sizes of reported functional centromeres in a variety of other species. The centromere of a Drosophila melanogaster minichromosome is contained within a 420-kb region of centric repetitive DNA (Murphy and Karpen, 1995; Sun et al., 1997). Similarly, the CENP-A–associated regions in a sampling of human neocentromeres span ∼130 to 460 kb DNA (Lo et al., 2001a, 2001b; Alonso et al., 2003), and a centromere misdivision study in maize suggested that 500 kb of centromeric repeats is the minimum for fully functional B centromeres (Kaszas and Birchler, 1998). The CentC/CRM cores of maize centromeres 2, 3, 4, 6, and 9 may be large enough for formation of full-sized kinetochores.
Sequential detection of CENH3 and the CentC/CRM sequences on stretched chromatin fibers confirmed the interaction that was previously revealed by ChIP analysis (Zhong et al., 2002). We rarely observed chromatin fibers that were stained by CENH3 antibodies but were not associated with CentC/CRM sequences, suggesting that the intermingled CentC/CRM sequence domains account for the majority of the CENH3-associated centromeric chromatin in maize. We also demonstrate that not all of the CentC/CRM sequences are associated with CENH3 (Figure 6), supporting the view that the recognition of centromeric DNA by CENH3 is an epigenetic event. We noticed that the CENH3-associated domains are often located near one end of the CentC/CRM sequences. Similarly, the functional domain of the centromere of human X chromosome is located near the short arm edge of the α satellite array DXZ1 (Spence et al., 2002).
Oat CENH3 Functions as a Kinetochore Protein on Maize Centromeres
CENH3s share high homology with the regular histone H3 in the core domain, but they diverge significantly in the N-terminal tail and the Loop1 region of the histone core (Henikoff et al., 2001). The diverged regions in CENH3s interact with DNA in the nucleosome and show evidence of adaptive evolution (Malik and Henikoff, 2001; Talbert et al., 2002). The anti-CENH3 antibody of A. thaliana does not stain the centromeres in the closely related species A. arenosa but recognizes all centromeres in both synthetic and natural allopolyploids derived from these two species. These results showed that the CENH3 encoded by A. thaliana was successfully incorporated into the centromeric nucleosomes of both A. thaliana and A. arenosa genomes in the allopolyploids, although the centromeric satellite repeats of these two species share only 58 to 80% sequence identity (Kamm et al., 1995). Talbert et al. (2002) suggested that this may be a consequence of the formation of nucleosomes that include CENH3 monomers from both parental genomes and therefore are able to interact with both sets of centromeric repeats. This idea has yet to be tested because the authors did not have the antisera to A. arenosa CENH3 that would have allowed them to determine whether both forms of CENH3 are present at a single centromere.
In this study, we used antibodies developed to both maize CENH3 and to rice CENH3. These two antibodies were developed against peptides located in the different regions of the N-tails (Zhong et al., 2002; Nagaki et al., 2004). The maize CENH3 antibody is specific for maize, but the rice antibody stains the centromeres of rice as well as several other cereal species, including oat and maize (Figure 7). We demonstrated that the CenH3 gene in maize is located on chromosome 6 and that maize CENH3 was not detected in three independently isolated oat-maize chromosome 6 addition lines. However, the maize centromeres in different oat-maize addition lines were recognized by the antibody to rice CENH3. Thus, the centromeric chromatin in both oat and maize chromosomes incorporated only the CENH3 encoded by the oat CenH3 gene. These data and the fact that the maize chromosomes are stable in the genetic background of oat clearly indicate that oat CENH3 is itself sufficient to organize a kinetochore on a maize chromosome.
Rapid elimination of chromosomes from one parental genome is a common phenomenon in somatic and sexual hybrids derived from distantly related species. Elimination of maize chromosomes occurred in sexual hybrids between maize and several cereal species, including Triticum aestivum (wheat) (Laurie and Bennett, 1986; O'Donoughue and Bennett, 1994), H. vulgare (Laurie and Bennett, 1988; Chen et al., 1991), and oat (Rines and Dahleen, 1990). The chromosome elimination occurred in the first three cell-division cycles in the zygotes of T. aestivum × maize hybrids (Laurie and Bennett, 1989). It was noted that the maize chromosomes had small or indistinguishable centromeres and tended to lie away from the plane of the metaphase plate. Laurie and Bennett (1989) suggested that the elimination of maize chromosomes may be caused by impaired centromere function. Laurie and Bennett (1988) also noticed that the maize chromosomes in crosses between H. vulgare and maize had well-differentiated centromeres in zygotes and were retained for more cell cycles than in T. aestivum.
Riera-Lizarazu et al. (1996) reported that 30 out of 90 plants derived from oat × maize crosses retained one to four maize chromosomes, resulting from incomplete maize chromosome elimination in the hybrids. Plants with chromosomal chimeras, in which some tissues of the plants contain maize chromosomes and other tissues do not, also were recovered, suggesting that maize chromosome elimination in oat × maize crosses is more gradual in comparison to T. aestivum × maize crosses (Riera-Lizarazu et al., 1996). By contrast, retention of maize chromosomes was not reported in numerous attempts in the T. aestivum × maize crosses conducted by several different laboratories. It is not known if the centromeric repeats of oat and maize share more sequence similarity than, for instance, the centromeric repeats of T. aestivum and maize. Alternatively, oat and maize CENH3s may share critical amino acid identities in regions that contact DNA that are absent in T. aestivum CENH3. In either case, the data would support the view that CENH3 has evolved to recognize specific centromere repeats (Malik and Henikoff, 2002). It would be interesting to conduct immunostaining experiments on chromosomes in the zygotes derived from the T. aestivum × maize and oat × maize crosses. Such experiments may reveal if the failure/success of incorporation of the T. aestivum/oat CENH3s in maize centromeres play a role in the elimination of maize chromosomes in the hybrids.
METHODS
Materials
Oat-maize chromosome addition lines 1, 2, 3, 4, 6 (including lines 6.1, 6.5, and 6.18), 7, and 9 were provided by Howard Rines and Ronald Phillips of the University of Minnesota. Maize inbred lines B73, W23, and Seneca 60 and oat cultivar Starter-1 were used for molecular and cytological analyses. The centromeric DNA probes, including CentC, CRM, and Cent4, all were described previously (Page et al., 2001; Nagaki et al., 2003a).
FISH and Fiber-FISH
The FISH and fiber-FISH procedures were essentially the same as the published protocols (Jiang et al., 1995; Jackson et al., 1998). DNA probes were labeled with either biotin-dUTP or digoxigenin-dUTP (Roche, Indianapolis, IN). Chromosomes were counterstained by 4′,6-diamidino-2-phenylindole in the antifade solution Vectashield (Vector Laboratories, Burlingame, CA). All of the images were captured digitally using a SenSys CCD camera (Roper Scientific, Tucson, AZ) attached to an Olympus BX60 epifluorescence microscope (Tokyo, Japan). The camera control and measurements of fiber-FISH signals were performed using IPLab Spectrum software version 3.1 (Signal Analytics, Vienna, VA).
ChIP
ChIP was performed as previously (Zhong et al., 2002) except that nuclei from young kernels of the inbred W23 were treated with RNase-free DNase (Promega, Madison, WI) for 10 min at 37°C, using a concentration of 4 units/mg of DNA. Fragments in the size range of 300 to 800 bp were used for immunoprecipitation.
Immunostaining on Somatic Metaphase Chromosomes
Root tips were harvested from actively growing plants and fixed immediately using 4% paraformaldehyde in PHEM (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, 2 mM MgCl2, and 0.3 mM sorbitol, pH 6.8) for 20 min. After washing with PBS (10 mM sodium phosphate, pH 7.0, and 140 mM NaCl), the root tips were treated with 2% cellulase and 1% pectinase (Sigma, St. Louis, MO) in PHEM for ∼2 h and then squashed on slides coated with 3-aminopropyltriethoxysilane. The rabbit antibodies to CENH3 (Biosource International, Camarillo, CA) were diluted to 1:2000 in TNB buffer (0.1 M Tris-HCl, pH 7.5, 0.15 M NaCl, and 0.5% blocking reagent) (Upstate Biochemicals, Lake Placid, NY). Approximately 100 μL of the diluted antibodies was added to each slide, and the slides were incubated in a humid chamber at 37°C for 3 h. After three washes in PBS, 100 μL of rhodamine-conjugated goat anti-rabbit antibody (Jackson ImmunoResearch, West Crove, PA) (1:50 in TNB buffer) was added to the slides. Incubation and washes were the same as for the primary antibody. The slides were counterstained with 4′,6-diamidino-2-phenylindole before being checked with a microscope.
Sequential Detection of CENH3 and CentC/CRM Sequences on Stretched Centromeres
Preparation of stretched chromatin fibers was performed according to Blower et al. (2002) with some modifications. Actively growing maize callus (2 weeks old) was ground with liquid N2, and the powder was resuspended in Tris-buffered saline containing 0.5% Tween 40. The nuclei were purified in a sucrose gradient. Extended chromatin fibers were prepared by centrifuging the nuclei (in 75 mM KCl) onto slides at 1000 rpm for 5 min using Cytospin 4 centrifuge (Shandon, Pittsburgh, PA). The nuclei on the slide were treated with 100 μL of lysis buffer (25 mM Tris, pH 7.5, 500 mM NaCl, 1% Triton X-100, and 0.2 M urea) covered with parafilm for 10 min. The parafilm was slowly dragged down the slide. The slides then were fixed in cold methanol for 10 min and were incubated in TNB buffer for 30 min at room temperature before immunostaining. The immunostaining procedure on stretched chromatin fibers was the same as that used on somatic metaphase chromosomes. After recording the immunostaining signals, the slides were washed in PBS buffer three times (5 min each) and dehydrated in an ethanol series. These slides were probed with CentC and CRM for FISH analysis.
RT-PCR
DNA and total RNA was extracted from leaf tissue of 10-d-old plants using the DNeasy and RNeasy plant mini kits (Qiagen, Valencia, CA). RNA samples were treated with RNase-free DNase (Ambion, Austin, TX). cDNA was synthesized using the RNA and Superscript III RT (Invitrogen, Carlsbad, CA). The primer pairs used for PCR and RT-PCR were CenH3-1 (5′-GGGAGATCAGGAAGTACCAGAAG-3′) and CenH3-2 (5′-GATTCGCCATTTCAAACAGTTC-3′), which were designed from two different exons within the maize CenH3 gene.
Acknowledgments
We thank Ronald Phillips, Howard Rines, and Ralf Kynast (University of Minnesota) for providing the oat-maize chromosome addition lines. We appreciate the contributions from Cathy Zhong and Christopher Topp, who conducted the initial steps of the ChIP studies shown in Figure 5, and from Huihuang Yan and Robert Stupar, who assisted with RT-PCR analysis. We thank William Haun, Peiyu Zeng, and Shawn Kaeppler for providing the meiotic samples and callus of maize. This work was supported by Grant 9975827 from the National Science Foundation to R.K.D. and J.J.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jiming Jiang (jjiang1@wisc.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.018937.
References
- Alfenito, M.R., and Birchler, J.A. (1993). Molecular characterization of a maize B chromosome centric sequence. Genetics 135, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, A., Mahmood, R., Li, S., Cheung, F., Yoda, K., and Warburton, P.E. (2003). Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum. Mol. Genet. 12, 2711–2721. [DOI] [PubMed] [Google Scholar]
- Ananiev, E.V., Phillips, R.L., and Rines, H.W. (1998). Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA 95, 13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon-Alcaide, L., Miller, T., Schwarzacher, T., Reader, S., and Moore, G. (1996). A cereal centromeric sequence. Chromosoma 105, 261–268. [DOI] [PubMed] [Google Scholar]
- Blower, M.D., Sullivan, B.A., and Karpen, G.H. (2002). Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2, 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.C., Chen, C.M., Hsu, F.C., Wang, C.J., Yang, J.T., and Kao, Y.Y. (2000). The pachytene chromosomes of maize as revealed by fluorescence in situ hybridization with repetitive DNA sequences. Theor. Appl. Genet. 101, 30–36. [Google Scholar]
- Chen, F.Q., Hayes, P.M., and Rivin, C.J. (1991). Wide hybridization of Hordeum vulgare x Zea mays. Genome 34, 603–605. [Google Scholar]
- Cheng, Z.K., Buell, C.R., Wing, R.A., and Jiang, J. (2002. a). Resolution of fluorescence in-situ hybridization mapping on rice mitotic prometaphase chromosomes, meiotic pachytene chromosomes and extended DNA fibers. Chromosome Res. 10, 379–387. [DOI] [PubMed] [Google Scholar]
- Cheng, Z.K., Dong, F., Langdon, T., Ouyang, S., Buell, C.B., Gu, M.H., Blattner, F.R., and Jiang, J. (2002. b). Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14, 1691–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobie, K.W., Hari, K.L., Maggert, K.A., and Karpen, G.H. (1999). Centromere proteins and chromosome inheritance: A complex affair. Curr. Opin. Genet. Dev. 9, 206–217. [DOI] [PubMed] [Google Scholar]
- Gindullis, F., Desel, C., Galasso, I., and Schmidt, T. (2001). The large-scale organization of the centromeric region in Beta species. Genome Res. 11, 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., and Malik, H.S. (2001). The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293, 1098–1102. [DOI] [PubMed] [Google Scholar]
- Houben, A., and Schubert, I. (2003). DNA and proteins of plant centromeres. Curr. Opin. Plant Biol. 6, 554–560. [DOI] [PubMed] [Google Scholar]
- Jackson, S.A., Wang, M.L., Goodman, H.M., and Jiang, J. (1998). Application of Fiber-FISH in physical mapping of Arabidopsis thaliana. Genome 41, 566–572. [PubMed] [Google Scholar]
- Jiang, J., Birchler, J.B., Parrott, W.A., and Dawe, R.K. (2003). A molecular view of plant centromeres. Trends Plant Sci. 8, 570–575. [DOI] [PubMed] [Google Scholar]
- Jiang, J., Gill, B.S., Wang, G.L., Ronald, P.C., and Ward, D.C. (1995). Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc. Natl. Acad. Sci. USA 92, 4487–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J., Nasuda, S., Dong, F., Scherrer, C.W., Woo, S., Wing, R.A., Gill, B.S., and Ward, D.C. (1996). A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc. Natl. Acad. Sci. USA 93, 14210–14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamm, A., Galasso, I., Schmidt, T., and Heslop-Harrison, J.S. (1995). Analysis of a repetitive DNA family from Arabidopsis arensa and relationships between Arabidopsis species. Plant Mol. Biol. 27, 853–862. [DOI] [PubMed] [Google Scholar]
- Kaszas, E., and Birchler, J.A. (1996). Misdivision analysis of centromere structure in maize. EMBO J. 15, 5246–5255. [PMC free article] [PubMed] [Google Scholar]
- Kaszas, E., and Birchler, J.A. (1998). Meiotic transmission rates correlate with physical features of rearranged centromeres in maize. Genetics 150, 1683–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kynast, R.G., et al. (2001). A complete set of maize individual chromosome additions to the oat genome. Plant Physiol. 125, 1216–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon, T., Seago, C., Mende, M., Leggett, M., Thomas, H., Forster, J.W., Thomas, H., Jones, R.N., and Jenkins, G. (2000). Retrotransposon evolution in diverse plant genomes. Genetics 156, 313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie, D.A., and Bennett, M.D. (1986). Wheat x maize hybridization. Can. J. Genet. Cytol. 28, 313–316. [Google Scholar]
- Laurie, D.A., and Bennett, M.D. (1988). Chromosome behaviour in wheat x maize, wheat x sorghum and barley x maize crosses. In Kew Chromosome Conference Proceedings III, P.E. Brandham, ed (Norwich, UK: The Stationery Office Books), pp. 167–177.
- Laurie, D.A., and Bennett, M.D. (1989). The timing of chromosome elimination in hexaploid wheat x maize crosses. Genome 32, 953–961. [Google Scholar]
- Lo, A.W., Craig, J.M., Saffery, R., Kalitsis, P., Irvine, D.V., Earle, E., Magliano, D.J., and Choo, K.H. (2001. a). A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J. 20, 2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, A.W.I., Magliano, D.J., Sibson, M.C., Kalitsis, P., Craig, J.M., and Choo, K.H.A. (2001. b). A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res. 11, 448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, H.S., and Henikoff, S. (2001). Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157, 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, H.S., and Henikoff, S. (2002). Conflict begets complexity: The evolution of centromeres. Curr. Opin. Genet. Dev. 12, 711–718. [DOI] [PubMed] [Google Scholar]
- Miller, J.T., Dong, F., Jackson, S.A., Song, J., and Jiang, J. (1998). Retrotransposon-related DNA sequences in the centromeres of grass chromosomes. Genetics 150, 1615–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroczek, R.J., and Dawe, R.K. (2003). Distribution of retroelements in centromeres and neocentromeres of maize. Genetics 165, 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, T.D., and Karpen, G.H. (1995). Localization of centromere function in a Drosophila minichromosome. Cell 82, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki, K., Cheng, Z., Ouyang, S., Talbert, P.B., Kim, M., Jones, K.M., Henikoff, S., Buell, C.R., and Jiang, J. (2004). Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36, 138–145. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., Song, J., Stupar, S.M., Parokonny, A.S., Yuan, Q., Ouyang, S., Liu, J., Hsiao, J., Jones, K.M., Dawe, R.K., Buell, C.R., and Jiang, J. (2003. a). Molecular and cytological analyses of large tracks of centromeric DNA reveal the structure and evolutionary dynamics of maize centromeres. Genetics 163, 759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki, K., Talbert, P.B., Zhong, C.X., Dawe, R.K., Henikoff, S., and Jiang, J.M. (2003. b). Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163, 1221–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donoughue, L.S., and Bennett, M.D. (1994). Comparative responses of tetraploid wheats pollinated with Zea mays L. and Hordeum bulbosum L. Theor. Appl. Genet. 87, 673–680. [DOI] [PubMed] [Google Scholar]
- Page, B.T., Wanous, M.K., and Birchler, J.A. (2001). Characterization of a maize chromosome 4 centromeric sequence: Evidence for an evolutionary relationship with the B chromosome centromere. Genetics 159, 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, D.K., O'Day, K., Trong, H.L., Charbonneau, H., and Margolis, R.L. (1991). Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl. Acad. Sci. USA 88, 3734–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, D.K., O'Day, K., Wener, M.H., Andrews, B.S., and Margolis, R.L. (1987). A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104, 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presting, G.G., Malysheva, L., Fuchs, J., and Schubert, I. (1998). A Ty3/gypsy retrotransposon-like sequence localizes to the centromeric regions of cereal chromosomes. Plant J. 16, 721–728. [DOI] [PubMed] [Google Scholar]
- Riera-Lizarazu, O., Rines, H.W., and Phillips, R.L. (1996). Cytological and molecular characterization of oat x maize partial hybrids. Theor. Appl. Genet. 93, 123–135. [DOI] [PubMed] [Google Scholar]
- Rines, H.W., and Dahleen, L.S. (1990). Haploid oat plants produced by application of maize pollen to emasculated oat florets. Crop Sci. 30, 1073–1078. [Google Scholar]
- Round, E.K., Flowers, S.K., and Richards, E.J. (1997). Arabidopsis thaliana centromere regions: Genetic map positions and repetitive DNA structure. Genome Res. 7, 1045–1053. [DOI] [PubMed] [Google Scholar]
- Spence, J.M., Critcher, R., Ebersole, T.A., Valdivia, M.M., Earnshaw, W.C., Fukagawa, T., and Farr, C.J. (2002). Co-localization of centromere activity, proteins and topoisomerase II within a subdomain of the major human X alpha satellite array. EMBO J. 21, 5269–5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, B.A., Blower, M.D., and Karpen, G.H. (2001). Determining centromere identity: Cyclical stories and forking paths. Nat. Rev. Genet. 2, 584–596. [DOI] [PubMed] [Google Scholar]
- Sun, X.P., Wahlstrom, J., and Karpen, G. (1997). Molecular structure of a functional Drosophila centromere. Cell 91, 1007–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert, P.B., Masuelli, R., Tyagi, A.P., Comai, L., and Henikoff, S. (2002). Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14, 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton, P.E., Cooke, C.A., Bourassa, S., Vafa, O., Sullivan, B.A., Stetten, G., Gimelli, G., Warburton, D., Tyler-Smith, C., Sullivan, K.F., Poirier, G.G., and Earnshaw, W.C. (1997). Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 7, 901–904. [DOI] [PubMed] [Google Scholar]
- Yoda, K., Ando, S., Morishita, S., Houmura, K., Hashimoto, K., Takeyasu, K., and Okazaki, T. (2000). Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl. Acad. Sci. USA 97, 7266–7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H.G., Hiatt, E.N., and Dawe, R.K. (2000). The plant kinetochore. Trends Plant Sci. 5, 543–547. [DOI] [PubMed] [Google Scholar]
- Zhong, C.X., Marshall, J.B., Topp, C., Mroczek, R., Kato, A., Nagaki, K., Birchler, J.A., Jiang, J.M., and Dawe, R.K. (2002). Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14, 2825–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]