Abstract
Oligomer-based DNA Affymetrix GeneChips representing about one-third of Arabidopsis (Arabidopsis thaliana) genes were used to profile global gene expression in a single cell type, guard cells, identifying 1309 guard cell–expressed genes. Highly pure preparations of guard cells and mesophyll cells were isolated in the presence of transcription inhibitors that prevented induction of stress-inducible genes during cell isolation procedures. Guard cell expression profiles were compared with those of mesophyll cells, resulting in identification of 64 transcripts expressed preferentially in guard cells. Many large gene families and gene duplications are known to exist in the Arabidopsis genome, giving rise to redundancies that greatly hamper conventional genetic and functional genomic analyses. The presented genomic scale analysis identifies redundant expression of specific isoforms belonging to large gene families at the single cell level, which provides a powerful tool for functional genomic characterization of the many signaling pathways that function in guard cells. Reverse transcription–PCR of 29 genes confirmed the reliability of GeneChip results. Statistical analyses of promoter regions of abscisic acid (ABA)–regulated genes reveal an overrepresented ABA responsive motif, which is the known ABA response element. Interestingly, expression profiling reveals ABA modulation of many known guard cell ABA signaling components at the transcript level. We further identified a highly ABA-induced protein phosphatase 2C transcript, AtP2C-HA, in guard cells. A T-DNA disruption mutation in AtP2C-HA confers ABA-hypersensitive regulation of stomatal closing and seed germination. The presented data provide a basis for cell type–specific genomic scale analyses of gene function.
INTRODUCTION
In the leaf epidermis, pairs of guard cells form stomatal pores, which provide CO2 intake conduits for photosynthesis and sites for controlling transpirational water loss in plants. The guard cell system has been adapted and developed for studying the transduction of environmental and endogenous signals in plants because guard cells cell-autonomously respond to stimuli such as blue light, temperature, CO2, drought, abscisic acid (ABA), and other hormones. Moreover, models have been developed allowing individual mechanisms and branches in early signaling to be characterized with mechanistic and temporal resolution (reviewed in MacRobbie, 1998; Schroeder et al., 2001; Finkelstein et al., 2002).
During drought, the phytohormone ABA triggers cellular responses resulting in stomatal closing, thus reducing plant water loss. Characterization of guard cell–expressed genes on a genomic scale would greatly facilitate identification of new ABA and other signal transduction mechanisms because guard cells can be probed at different steps and branches within early signal transduction cascades (Mäser et al., 2003). Furthermore, only recently genomic studies have been reported to examine which genes are expressed in a single plant cell type (Becker et al., 2003; Birnbaum et al., 2003; Honys and Twell, 2003), largely because of the fact that it is not easy to obtain a large amount of a single purified cell type from Arabidopsis (Arabidopsis thaliana). An initial EST sequencing study previously reported ∼500 guard cell–expressed sequence tags in Brassica campestris (Kwak et al., 1997). Expression profiling in Arabidopsis has been used to obtain valuable information on functions of genes under a variety of conditions (Harmer et al., 2000; Wang et al., 2000; Hugouvieux et al., 2001; Chen et al., 2002). However, expression analyses in plants and other systems have thus far focused on whole organs (Schena et al., 1995; Harmer et al., 2000; Perez-Amador et al., 2001; Seki et al., 2001, 2002; Hoth et al., 2002; Yin et al., 2002).
Only a few genes have been cloned from recessive ABA-insensitive mutants (Giraudat et al., 1992; Finkelstein, 1994; Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Wang et al., 2001; Kwak et al., 2002, 2003; Mustilli et al., 2002; Zhu et al., 2002). In addition, several negative regulators of ABA signaling, which produce ABA hypersensitivity when mutated, have been characterized (Cutler et al., 1996; Pei et al., 1998; Lu and Fedoroff, 2000; Hugouvieux et al., 2001; Lee et al., 2001; Merlot et al., 2001; Xiong et al., 2001a, 2001b). However, conventional genetic screens have left large portions of plant signal transduction networks uncharacterized, which is likely because of redundancy within plant gene families. Genetic analysis of redundant pathways requires the study of double or multiple mutants. Unfortunately, this is impractical for large gene families. For example, for the ∼69 protein phosphatase 2C (PP2C) genes expressed in Arabidopsis, 2346 distinct double mutant combinations could be generated for phenotypic analyses. This could partly explain why no recessive PP2C knockout mutant phenotypes have yet been reported in plants, with the exception of intragenic suppressor mutations (Merlot et al., 2001).
Given the substantial number of large gene families found in plants, gaining knowledge of the expression of specific gene family members in individual cell types will provide a powerful single cell–type genomic approach to identifying which members of a gene family are likely to function in a specific pathway. For instance, the RCN1 protein phosphatase 2A subunit and the redundant NADPH oxidase AtrbohD and AtrbohF genes were shown to function in early ABA signal transduction by initially revealing high expression levels of these genes in guard cells (Kwak et al., 2002, 2003). The combined analysis of guard cell–specific expression profiles with phenotypic analysis of Arabidopsis knockout mutants of homologous gene family members will allow the functional identification of redundant and unknown genes that play important roles in plant signal transduction.
In an effort to provide genomic scale information on gene expression in a single cell type, we identify transcripts expressed in Arabidopsis guard cells, compare these with expression in whole leaf mesophyll tissues, and report their regulation by ABA. In addition, our results provide new information on cis-elements conferring gene regulation by ABA. Interestingly, a current working model for early ABA signal transduction is presented, unexpectedly demonstrating that ABA modulates the mRNA levels of many known negatively regulating and positively transducing ABA signal transduction mechanisms in guard cells. Additionally, to demonstrate the utility of these expression profile analyses, one PP2C gene, AtP2C-HA, which is strongly induced by ABA in guard cells, was selected. T-DNA disruption of AtP2C-HA confers ABA-hypersensitive regulation of both stomatal closing and seed germination. These results show that this cell type–oriented genomic scale approach is a powerful tool for identifying new signal transduction mechanisms.
RESULTS
To establish a genomic scale analysis of gene function in guard cells, we performed expression-profiling experiments using Affymetrix GeneChips, representing ∼8100 genes in the Arabidopsis genome. Experiments were performed over the course of 18 months. Four- to five-week-old Arabidopsis (Columbia) plants were either control treated or treated with 100 μM ABA for 4 h, and guard cells and mesophyll cells were extracted at the same time from plants grown in parallel. Total RNA was prepared for individual microarray hybridizations from control and ABA-treated guard cells for each of the three experiments and from control and ABA-treated mesophyll cells for two of the three experiments (one mesophyll hybridization failed), resulting in three pairs of guard cell hybridizations and two pairs of mesophyll cell hybridizations (for a total of 10 microarray hybridizations). In addition, for each individual hybridization, three RNA samples from three independent cell preparations were extracted and pooled (a total of 30 independent RNA extractions) (Excel files reporting expression data for all 10 experiments are available as supplemental data online and at http://www-biology.ucsd.edu/labs/schroeder/guardcellchips.html). For each microarray, overall intensity normalization for the entire probe set was performed as described by Zhu et al. (2001). Then, for each probe set, the signal value, which assigns a relative measure of abundance to the transcript, and the detection P-value, which indicates whether a transcript is reliably detected, were calculated from each independent guard cell and mesophyll cell hybridization (see Methods).
Effect of Transcription Inhibitors on Stress-Inducible Genes
To inhibit the modulation of gene expression in response to stress during physical isolation and enzymatic digestions for guard cell and mesophyll cell protoplast preparations, two transcription inhibitors, actinomycin D and cordycepin, were applied during all steps of protoplast extraction. To test whether these inhibitors suppressed induction of stress-inducible genes, reverse transcription (RT)–PCR was performed using cDNA synthesized from guard cell RNA from protoplasts prepared in the presence or absence of these inhibitors.
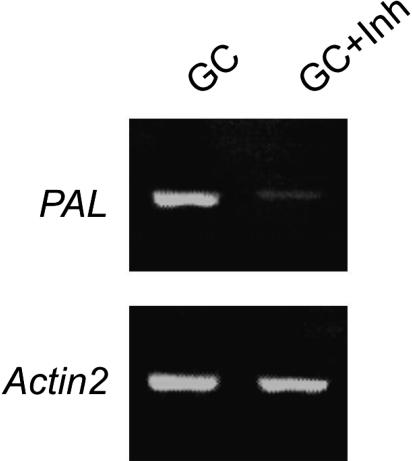
Phe ammonia-lyase (PAL) gene mRNA levels were used to monitor the effect of the inhibitors because PAL gene expression is induced by stress (Wanner et al., 1995). As shown in Figure 1, expression of PAL was highly induced in guard cells prepared in the absence of transcription inhibitors. By contrast, PAL expression was low in guard cells prepared in the presence of transcription inhibitors. Similar results were observed for other stress-inducible genes, including COR47, genes encoding heat shock proteins (At5g04140 and At1g16030), and glutathione S-transferase (At5g02200) (data not shown). Thus, inhibitors of transcription suppressed stress-inducible gene induction during cell isolations.
Figure 1.
Transcription Inhibitors Suppress Stress-Induced PAL Gene Expression during Guard Cell Isolation.
RT-PCR analyses of PAL in guard cells extracted in the absence (GC) or presence (GC + Inh) of the transcription inhibitors actinomycin D (33 mg/L) and cordycepin (100 mg/L). The Actin2 gene was amplified as a control.
Reproducibility of Chip Hybridizations
To test the variability between our chip hybridization experiments, raw signal intensities from all probe sets were plotted for all possible pairs of independent experiments. Scatter plots of the raw data for the control-treated guard cells, ABA-treated guard cells, control-treated mesophyll cells, and ABA-treated mesophyll cells are shown in Figure 2. The diagonal lines in Figure 2 indicate twofold relative intensity differences between two experiments. These scatter plots show that the majority (>90%) of the significantly expressed genes fall between the two lines, showing that most genes exhibit less than twofold variation in signal intensity between the two independent chip hybridizations, even though individual mRNA isolations were performed over a period of 18 months. This level of reproducibility is likely attributed to tightly controlled growth and experimental conditions. However, some genes with low raw signal intensities corresponding to either noise or low abundance mRNAs showed more variability (Figure 2, gray points). The same comparisons were performed with the other samples against each other (see Supplemental Figure 1 online).
Figure 2.
GeneChip Hybridizations Show Genomic Scale Reproducibility.
Scatter plots comparing the raw signal intensities of two independent experiments from guard cells (A), guard cells treated with ABA (B), mesophyll cells (C), and mesophyll cells treated with ABA (D). Each gene is represented by one dot. For each gene, the raw RNA expression level in one experiment is given on the x axis, and the expression level for the same gene in the other experiment is plotted on the y axis. The solid diagonal lines indicate a difference by a factor of 2 between the two hybridizations for visual reference. Significantly expressed genes detected as Present or Marginal in two or three samples (see Methods) are represented by black dots, whereas genes for which expression levels were not significant in two or three samples are shown as gray dots.
Functional Classification of Genes Expressed in Guard Cells and Mesophyll Cells
Among the >8100 genes analyzed, 1309 transcripts were found to be significantly expressed in guard cells and 1479 in mesophyll cells (see Supplemental Tables 1 and 2 online). A previously reported (Ghassemian et al., 2001) and recently further enhanced annotation of the genes represented on the GeneChip was used for analyses (see supplemental data online and http://www-biology.ucsd.edu/labs/schroeder/index.html, click on Genechip). We classified these genes according to their putative functions based on the classification of the Munich Information Center for Protein Sequence (MIPS) database to determine whether these specialized cells allocate gene expression patterns differently among classifications.
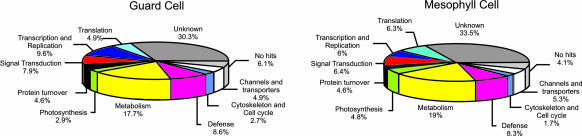
Figure 3 shows a summary of the functional categorization of the transcripts detected in guard cells and mesophyll cells, showing that the majority of the known genes are predicted to function in metabolic pathways, followed by genes implicated in transcription and replication and signal transduction in both guard cells and mesophyll cells. Interestingly, the percentage of genes in each category is similar between the two cell types, with some exceptions, including photosynthesis-related genes with 4.8% in mesophyll cells and 2.9% in guard cells. This correlates with the fact that mesophyll cells possess the major photosynthetic activity in leaves.
Figure 3.
Comparison of Predicted Functional Distribution of Guard Cell– and Mesophyll Cell–Expressed Genes Shows a Higher Relative Portion of Photosynthesis Gene Expression in Mesophyll Cells.
Distribution of guard cell and mesophyll cell profiles among 11 major classes was performed using the MIPS database (http://mips.gsf.de/proj/thal/db/tables/tables_func_frame.html).
Identification of Genes Preferentially Expressed in Guard Cells
To identify genes expressed preferentially in guard cells compared with mesophyll cells, scatter plots of guard cell versus mesophyll cell expression were generated. As shown in Figure 4, many guard cell–expressed genes are also expressed in mesophyll cells. These data provide experimental support for the prediction that ∼60 to 77% of plant genes do not have a strict tissue-specific expression (Okamura and Goldberg, 1989).
Figure 4.
Comparison of Guard Cell–Expressed Genes versus Mesophyll Cell–Expressed Genes.
Scatter plot of the normalized signal intensity values from guard cell versus mesophyll cell comparison (average of two and three replicates for mesophyll cells and guard cells, respectively) shows that many genes are expressed in both guard cells and mesophyll cells, albeit many at substantially different levels.
Interestingly, guard cell versus mesophyll cell analyses revealed that only 64 transcripts were detectable solely in guard cells in all three independent guard cell chip hybridizations compared with the two independent mesophyll chip hybridizations. These genes encode transcription factors, signal transduction proteins such as protein kinases, receptor protein kinases, and metabolic pathway proteins (Table 1). Among these genes, several have been reported previously to be predominantly expressed in guard cells, indicating the reliability of the method. These include ECERIFERUM2 (CER2) that functions in cuticular wax accumulation. CER2 is expressed in guard cells and trichomes but not in mesophyll cells (Xia et al., 1997). The KAT1 gene encoding an inward-rectifying K+ channel (Anderson et al., 1992; Schachtman et al., 1992) has also been reported to be expressed predominantly in guard cells (Nakamura et al., 1995). Importantly, we also identified previously uncharacterized genes that are preferentially expressed in guard cells and thus are strong candidates for contributing to guard cell signal transduction and development. Many of these highly expressed genes encode proteins of unknown function and therefore may be of particular interest for future studies (Table 1).
Table 1.
Genes Preferentially Expressed in Guard Cells
| Experiment 1
|
Experiment 2
|
Experiment 3
|
||||||
|---|---|---|---|---|---|---|---|---|
| Affy Probea | Description | AGI Numberb | Signal Valuec | P-Valued | Signal Value | P-Value | Signal Value | P-Value |
| 12100_at | MYB family transcription factor | At2g21650 | 3,851 | 0.04397 | 7,039 | 0.00653 | 4,685 | 0.00869 |
| 12188_at | Unknown protein | At2g28870 | 6,782 | 0.03937 | 7,601 | 0.02187 | 3,274 | 0.03937 |
| 12196_at | Unknown protein | At2g28410 | 5,143 | 0.03934 | 6,363 | 0.00999 | 3,333 | 0.00114 |
| 12261_at | Receptor kinase | At1g11340 | 18,071 | 0.00039 | 20,116 | 0.00160 | 24,685 | 0.00067 |
| 12284_at | Somatic embryogenesis receptor-like kinase | At1g34210 | 7,252 | 0.00486 | 13,534 | 0.00307 | 10,207 | 0.00056 |
| 12448_at | Acyl-CoA synthetase | At2g47240 | 3,761 | 0.01145 | 4,334 | 0.01145 | 2,148 | 0.01494 |
| 12493_g_ | Chloride channel | At5g49890 | 4,235 | 0.03516 | 6,627 | 0.00189 | 2,519 | 0.01309 |
| 12499_at | Phosphatidylinositol/phosphatidylcholine transfer protein | At2g16380 | 9,010 | 0.00754 | 5,813 | 0.02786 | 1,780 | 0.00114 |
| 12780_s_ | Unknown protein | At4g10840 | 9,242 | 0.00486 | 10,923 | 0.00223 | 21,807 | 0.00114 |
| 12796_s_ | β-Ketoacyl-CoA synthase | At2g26250 | 36,481 | 0.00189 | 40,054 | 0.00067 | 91,193 | 0.00022 |
| 12959_at | Ser/Thr protein kinase | At2g32850 | 8,793 | 0.01930 | 4,356 | 0.04397 | 1,139 | 0.10730 |
| 13110_at | Symbiosis-related protein | At4g04620 | 18,390 | 0.02471 | 11,383 | 0.02471 | 15,203 | 0.00039 |
| 13179_at | Unknown protein | At2g43680 | 6,676 | 0.00754 | 11,499 | 0.00039 | 9,828 | 0.00081 |
| 13482_at | Nodulin-like protein | At2g37450 | 9,952 | 0.00869 | 17,055 | 0.00135 | 11,928 | 0.00869 |
| 13554_at | β-Ketoacyl-CoA synthase | At2g16280 | 41,511 | 0.00094 | 39,345 | 0.01157 | 89,871 | 0.00074 |
| 13577_s_ | CER2 | At4g24510 | 37,080 | 0.00586 | 45,432 | 0.00195 | 80,154 | 0.00586 |
| 13612_at | Ubiquitin-conjugating enzyme | At2g16740 | 6,194 | 0.03516 | 2,892 | 0.02786 | 4,891 | 0.00223 |
| 14048_at | Protein kinase | At2g18890 | 7,378 | 0.04397 | 4,701 | 0.04397 | 4,578 | 0.00869 |
| 14527_at | Unknown protein | At2g30500 | 7,794 | 0.01930 | 4,417 | 0.04397 | 2,668 | 0.00307 |
| 14663_s_ | Glycosyl hydrolase | At4g24040 | 6,918 | 0.01930 | 6,836 | 0.00486 | 4,016 | 0.00564 |
| 14664_i_ | Trehalose-6-phosphate synthase | At1g78580 | 4,641 | 0.03516 | 3,586 | 0.00081 | 3,987 | 0.00032 |
| 14665_r_ | Trehalose-6-phosphate synthase | At1g78580 | 7,050 | 0.01494 | 7,118 | 0.03516 | 4,178 | 0.00262 |
| 14712_s_ | Translation initiation factor | At5g43810 | 5,681 | 0.00653 | 8,440 | 0.03516 | 2,845 | 0.01700 |
| 14870_at | Auxilin-like protein | At1g21660 | 4,453 | 0.03516 | 4,310 | 0.02187 | 2,089 | 0.03516 |
| 15034_at | Unknown protein | At2g39340 | 10,976 | 0.00096 | 9,830 | 0.00999 | 26,291 | 0.00027 |
| 15187_s_ | Fimbrin 2 | At5g48460 | 3,705 | 0.03937 | 3,067 | 0.01494 | 1,538 | 0.03516 |
| 15251_at | No hits | 55,883 | 0.00022 | 66,764 | 0.00022 | 93,068 | 0.00022 | |
| 15368_at | Unknown protein | At4g14830 | 45,332 | 0.00032 | 51,212 | 0.00022 | 43,714 | 0.00027 |
| 15389_at | Unknown protein | At2g22860 | 9,290 | 0.00307 | 10,139 | 0.00359 | 15,106 | 0.00032 |
| 15528_at | Electron transfer flavoprotein-ubiquinone oxidoreductase | At2g43400 | 9,730 | 0.01494 | 6,377 | 0.01145 | 4,914 | 0.00189 |
| 15548_at | Unknown protein | At1g11820 | 6,745 | 0.02187 | 7,496 | 0.00160 | 15,819 | 0.00032 |
| 15550_at | Unknown protein | At1g15200 | 5,624 | 0.01494 | 5,624 | 0.01145 | 4,719 | 0.00307 |
| 15642_at | Unknown protein | At1g68530 | 10,249 | 0.00358 | 7,315 | 0.00223 | 10,099 | 0.00081 |
| 15695_s_ | Histone H1 | At2g18050 | 5,054 | 0.03134 | 5,267 | 0.02187 | 1,873 | 0.00999 |
| 15709_at | Protein kinase | At1g62400 | 18,377 | 0.00135 | 18,227 | 0.00359 | 17,259 | 0.00067 |
| 15835_at | NAM (no apical meristem)-like protein | At2g02450 | 8,368 | 0.00160 | 14,219 | 0.00189 | 6,552 | 0.00096 |
| 16092_s_ | Potassium channel protein KAT1 | At5g46240 | 12,663 | 0.04397 | 17,358 | 0.00114 | 17,941 | 0.00022 |
| 16148_s_ | Kinesin-like protein | At5g54670 | 15,495 | 0.03937 | 12,073 | 0.00032 | 17,155 | 0.00039 |
| 16161_s_ | Gly-rich protein (AtGRP2) | At2g21060 | 56,617 | 0.00027 | 42,066 | 0.00160 | 74,085 | 0.00022 |
| 16403_at | ADP-glucose pyrophosphorylase | At2g21590 | 20,298 | 0.00261 | 12,647 | 0.00564 | 13,381 | 0.00223 |
| 16453_s_ | Histone | At1g06760 | 5,961 | 0.00754 | 4,816 | 0.02471 | 2,732 | 0.00653 |
| 16567_s_ | Unknown protein | At2g47980 | 6,685 | 0.00869 | 8,013 | 0.02187 | 2,120 | 0.03516 |
| 16624_s_ | Transcription factor | At1g08810 | 6,792 | 0.03516 | 4,895 | 0.03516 | 8,108 | 0.00189 |
| 17143_s_ | Unknown protein | At2g38300 | 7,847 | 0.00564 | 6,204 | 0.00999 | 12,622 | 0.00096 |
| 17386_at | Pro-rich protein | At2g21140 | 5,524 | 0.04900 | 4,479 | 0.00262 | 6,674 | 0.00039 |
| 17518_s_ | Transcriptional regulator | At3g26790 | 5,288 | 0.00754 | 3,076 | 0.00359 | 2,588 | 0.00039 |
| 17539_at | Nuclear cap binding protein CBP20 | At5g44200 | 5,628 | 0.03516 | 3,657 | 0.01145 | 4,012 | 0.01700 |
| 17567_at | Unknown protein | At5g03540 | 9,045 | 0.02187 | 8,730 | 0.00067 | 25,037 | 0.00027 |
| 17902_s_ | Unknown protein | At2g35330 | 10,193 | 0.00047 | 9,572 | 0.00160 | 11,924 | 0.00027 |
| 18319_g_ | Transcription factor ZAP1 | At2g04880 | 3,044 | 0.05447 | 7,593 | 0.00223 | 3,729 | 0.00096 |
| 18478_at | Receptor protein kinase | At1g78530 | 12,521 | 0.00564 | 9,739 | 0.01930 | 9,218 | 0.00056 |
| 18522_at | Kinase-like protein | At4g14480 | 12,486 | 0.00114 | 14,055 | 0.00754 | 20,933 | 0.00027 |
| 18625_at | Unknown protein | At1g03290 | 4,489 | 0.05444 | 2,519 | 0.03937 | 6,282 | 0.00486 |
| 18663_s_ | Unknown protein | At4g24130 | 7,330 | 0.05447 | 15,540 | 0.00262 | 13,017 | 0.00067 |
| 18918_at | No hits | 6,232 | 0.01700 | 6,271 | 0.00999 | 6,302 | 0.00135 | |
| 18953_at | Branched-chain alpha-keto acid dehydrogenase | At1g21400 | 6,569 | 0.02786 | 5,540 | 0.04900 | 4,649 | 0.01494 |
| 18975_g_ | Protein kinase | At4g35780 | 6,382 | 0.01700 | 6,307 | 0.00418 | 4,428 | 0.00754 |
| 19008_s_ | Glycosyl hydrolase | At2g28470 | 20,096 | 0.00486 | 28,800 | 0.00418 | 29,361 | 0.00032 |
| 19365_s_ | Cinnamyl-alcohol dehydrogenase | At4g39330 | 7,367 | 0.01930 | 6,090 | 0.00307 | 9,559 | 0.00022 |
| 19439_at | Aldehyde dehydrogenase protein | At4g36250 | 3,851 | 0.04397 | 7,039 | 0.00653 | 4,685 | 0.00869 |
| 19948_at | Unknown protein | At2g31580 | 6,782 | 0.03937 | 7,601 | 0.02187 | 3,274 | 0.03937 |
| 20067_at | Unknown protein | At2g04280 | 5,143 | 0.03934 | 6,363 | 0.00999 | 3,333 | 0.00114 |
| 20129_at | Pyrophosphate–fructose-6-phosphate 1-phosphotransferase | At2g22480 | 18,071 | 0.00039 | 20,116 | 0.00160 | 24,685 | 0.00067 |
| 20518_at | Unknown protein | At1g10060 | 7,252 | 0.00486 | 13,534 | 0.00307 | 10,207 | 0.00056 |
| 20570_at | Trehalose-6-phosphate phosphatase | At4g12430 | 3,761 | 0.01145 | 4,334 | 0.01145 | 2,148 | 0.01494 |
Describes names of probe set on Affymetrix chip.
AGI locus numbers.
Signal value, which assigns a relative measure of abundance of transcript of each gene.
Detection P-value, which indicates whether a transcript is reliably detected.
Identification of Genes Regulated by ABA
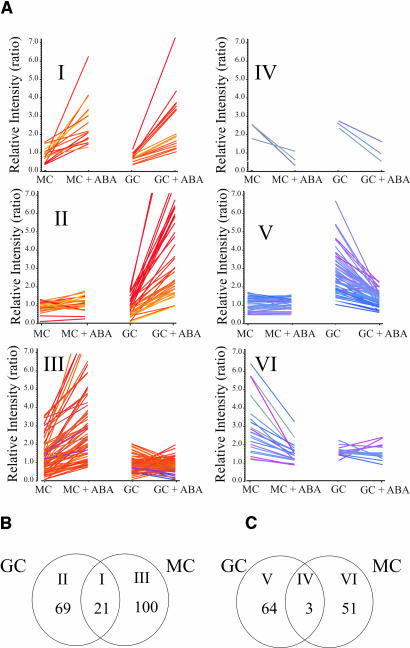
To identify genes regulated by ABA in guard cells and/or in mesophyll cells, six groups with distinct expression profiles were analyzed based on cluster analyses (see Methods) and are shown in Figure 5. The number of genes found in each cluster will depend on the applied statistical algorithms and parameters. Group I contains 21 transcripts that are ABA induced in both guard cells and mesophyll cells (Table 2, Figures 5A and 5B). This group includes several known genes that were shown previously to be induced by ABA in different tissues, including dehydrins ERD14 and COR47 (Gilmour et al., 1992; Yamaguchi-Shinozaki and Shinozaki, 1993; Kiyosue et al., 1994; Nylander et al., 2001; Hoth et al., 2002; Seki et al., 2002), as well as several genes that encode signal transduction components such as the two PP2Cs, AtP2C-HA (At1g72770) and AtPP2CA (At3g11410; Rodriguez et al., 1998; Cherel et al., 2002). In addition, several transcription factors were also found in this group, including Leu zipper DNA binding proteins.
Figure 5.
Cluster Analyses of Six Distinguishable ABA-Dependent Expression Responses of Guard Cell– and Mesophyll Cell–Expressed Genes.
(A) Six clusters showing distinctive ABA gene regulation patterns. Note that in the presented y axis scales, a value of 2 does not refer to a twofold increase in expression level (see Methods for log-derived values). Group I cluster contains ABA-induced genes in both guard cells and mesophyll cells. Group II cluster contains ABA-induced mRNAs only in guard cells. Group III cluster contains ABA-induced mRNAs only in mesophyll cells. Group IV cluster contains ABA-repressed mRNAs both in guard cells and mesophyll cells. Group V cluster contains ABA-repressed mRNAs only in guard cells. Group VI cluster contains ABA-repressed mRNAs only in mesophyll cells. Colors represent relative expression level of a gene after ABA treatment. Red indicates increased expression, and blue indicates reduced expression.
(B) Venn diagram presentation shows that 69 mRNAs are ABA induced only in guard cells (GC), 100 only in mesophyll cells (MC), and 21 in both cell types.
(C) Venn diagram presentation shows that 64 mRNA levels are repressed by ABA only in guard cells (GC), 51 only in mesophyll cells (MC), and three in both cell types.
Table 2.
ABA-Induced Genes in Guard Cells
| Experiment 1
|
Experiment 2
|
Experiment 3
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Affy Probea | Description | AGI Numberb | Clusterc | Ratiod | P-Valuee | Fold Changef | Ratio | P-Value | Fold Change | Ratio | P-Value | Fold Change | Seki et al.g | Hoth et al.h |
| 12046_at | Unknown protein | At1g30690 | II | 1.6 | 0.05391 | 3.0 | 1.4 | 0.00031 | 2.6 | 2.1 | 0.00000 | 4.3 | ||
| 12426_at | GTP binding protein | At2g21880 | II | 2.1 | 0.00001 | 4.3 | 2.8 | 0.00000 | 7.0 | 1.1 | 0.00036 | 2.1 | ||
| 12515_at | Expansin | At2g39700 | II | 1.2 | 0.00679 | 2.3 | 1.5 | 0.00018 | 2.8 | 5.4 | 0.00000 | 42.2 | ||
| 12521_at | Ca2+/H+-exchanging protein | At3g51860 | II | 2 | 0.00004 | 4.0 | 2.3 | 0.00108 | 4.9 | 1.3 | 0.00000 | 2.5 | ||
| 12645_at | Fibrillin precursor | At4g22240 | I | 1.1 | 0.00090 | 2.1 | 1.7 | 0.00011 | 3.2 | 1.2 | 0.00000 | 2.3 | ||
| 12749_at | Cold acclimation protein | At2g15970 | I | 2 | 0.00000 | 4.0 | 1.9 | 0.00000 | 3.7 | 1.4 | 0.00008 | 2.6 | 2.6 | 17.7 |
| 12753_at | Nonspecific lipid-transfer prot. | At2g38540 | I | 3 | 0.00000 | 8.0 | 2.1 | 0.00000 | 4.3 | 1.1 | 0.00000 | 2.1 | ||
| 12754_g_at | Nonspecific lipid-transfer prot. | At2g38540 | II | 2.4 | 0.00001 | 5.3 | 4 | 0.00015 | 16.0 | 3.7 | 0.00000 | 13.0 | ||
| 12781_at | Unknown protein | At1g13930 | II | 1.4 | 0.00000 | 2.6 | 1.7 | 0.00000 | 3.2 | 1.5 | 0.00001 | 2.8 | ||
| 12964_at | Protease | At5g47040 | II | 3.2 | 0.00001 | 9.2 | 3.8 | 0.00130 | 13.9 | 1.7 | 0.00000 | 3.2 | ||
| 13004_at | Senescence-associated protein | At2g17840 | II | 2.6 | 0.00053 | 6.1 | 3.5 | 0.00024 | 11.3 | 3 | 0.00000 | 8.0 | 2.63 | 4.1 |
| 13015_s_at | Zinc finger protein | At5g59820 | II | 2.6 | 0.00157 | 6.1 | 1.5 | 0.00177 | 2.8 | 1.6 | 0.00000 | 3.0 | 4 | 3.4 |
| 13067_s_at | Calcium binding protein | At1g09210 | II | 1.3 | 0.00019 | 2.5 | 1.6 | 0.00007 | 3.0 | 1.1 | 0.00002 | 2.1 | ||
| 13084_at | Unknown protein | At1g76990 | II | 4.1 | 0.00199 | 17.1 | 1.8 | 0.08910 | 3.5 | 1.8 | 0.00012 | 3.5 | ||
| 13128_at | PP2C | At4g31860 | II | 1.8 | 0.00000 | 3.5 | 1.8 | 0.00000 | 3.5 | 2.7 | 0.00000 | 6.5 | ||
| 13158_at | Cinnamoyl-CoA reductase | At2g33590 | I | 2.3 | 0.00000 | 4.9 | 1.6 | 0.00000 | 3.0 | 2.1 | 0.00001 | 4.3 | 2.65 | |
| 13275_f_at | Heat shock protein | At3g46230 | II | 1.6 | 0.00061 | 3.0 | 1 | 0.00044 | 2.0 | 2.1 | 0.00065 | 4.3 | 10.71 | |
| 13279_at | Heat shock protein | At5g12020 | II | 4.8 | 0.00000 | 27.9 | 3.9 | 0.00001 | 14.9 | 1.2 | 0.00000 | 2.3 | ||
| 13370_at | MAP kinase | At1g05100 | II | 3.6 | 0.00130 | 12.1 | 2.9 | 0.06494 | 7.5 | 3.1 | 0.00000 | 8.6 | 191 | |
| 13426_at | Unknown protein | At2g41190 | I | 3.4 | 0.00009 | 10.6 | 1.9 | 0.00044 | 3.7 | 2.4 | 0.00000 | 5.3 | 28.6 | |
| 13595_at | Unknown protein | At4g39730 | II | 1.3 | 0.00211 | 2.5 | 1.9 | 0.00022 | 3.7 | 1.8 | 0.00007 | 3.5 | ||
| 13616_s_at | 60S ribosomal protein | At2g39460 | II | 3.9 | 0.00000 | 14.9 | 3 | 0.00000 | 8.0 | 2.1 | 0.00000 | 4.3 | ||
| 13617_at | Mitoch. dicarboxylate carrier | At2g22500 | II | 2.9 | 0.00177 | 7.5 | 2.0 | 0.34040 | 4.0 | 3.4 | 0.00199 | 10.6 | 1.74 | |
| 13916_at | Unknown protein | At2g19800 | II | 2.3 | 0.00000 | 4.9 | 3.3 | 0.00000 | 9.8 | 1.5 | 0.00000 | 2.8 | ||
| 13918_at | Unknown protein | At1g54710 | II | 2.6 | 0.00715 | 6.1 | 3.2 | 0.00130 | 9.2 | 3.1 | 0.00000 | 8.6 | 5.4 | |
| 13965_s_at | ABREs | At4g34000 | II | 3.7 | 0.00003 | 13.0 | 3.6 | 0.00008 | 12.1 | 5 | 0.00000 | 32.0 | 13.5 | |
| 13966_at | Protein pEARLI 4 | At4g35110 | II | 2.9 | 0.00014 | 7.5 | 1.5 | 0.00375 | 2.8 | 1.6 | 0.00001 | 3.0 | ||
| 13977_at | G-3-P dehydrogenase | At2g41540 | II | 4.1 | 0.00002 | 17.1 | 1.7 | 0.00041 | 3.2 | 3.3 | 0.00000 | 9.8 | ||
| 14052_at | Tat binding protein | At1g10070 | II | 2.3 | 0.00130 | 4.9 | 3.3 | 0.00003 | 9.8 | 3.7 | 0.00001 | 13.0 | 9.21 | |
| 14526_at | Unknown protein | At4g36210 | II | 2.4 | 0.00004 | 5.3 | 1.2 | 0.00074 | 2.3 | 1.6 | 0.00001 | 3.0 | ||
| 14542_i_at | Putative prolylcarboxypeptidase | At2g24280 | II | 3.6 | 0.00020 | 12.1 | 1.2 | 0.00886 | 2.3 | 1.2 | 0.00002 | 2.3 | ||
| 14666_s_at | Trehalose-6-phosphate synthase | At1g78580 | II | 2 | 0.00047 | 4.0 | 1.8 | 0.00000 | 3.5 | 2.5 | 0.00000 | 5.7 | ||
| 14701_s_at | 14-3-3 protein | At5g38480 | II | 1.9 | 0.00002 | 3.7 | 1.4 | 0.00001 | 2.6 | 1.7 | 0.00000 | 3.2 | ||
| 14722_s_at | 14-3-3 protein | At5g10450 | II | 1.2 | 0.00000 | 2.3 | 1 | 0.00000 | 2.0 | 1.4 | 0.00000 | 2.6 | ||
| 14734_s_at | 14-3-3 protein | At5g38480 | II | 1.7 | 0.00002 | 3.2 | 1.2 | 0.00001 | 2.3 | 1.8 | 0.00000 | 3.5 | ||
| 14846_at | Calmodulin binding protein | At1g67310 | II | 3.8 | 0.00878 | 13.9 | 2.3 | 0.00252 | 4.9 | 3.0 | 0.00130 | 8.0 | ||
| 15052_at | RD20 protein | At2g33380 | I | 3.8 | 0.00166 | 13.9 | 3.4 | 0.00000 | 10.6 | 1.7 | 0.00000 | 3.2 | 24.09 | 18 |
| 15053_at | DNA binding protein | At2g41870 | II | 2.3 | 0.00493 | 4.9 | 3.2 | 0.00267 | 9.2 | 3.5 | 0.00000 | 11.3 | ||
| 15110_s_at | Dehydrin | At1g76180 | I | 2.1 | 0.00029 | 4.3 | 1.9 | 0.00000 | 3.7 | 1.7 | 0.00000 | 3.2 | ||
| 15124_s_at | Pro oxidase | At3g30775 | II | 3.4 | 0.00000 | 10.6 | 4.9 | 0.00007 | 29.9 | 6.1 | 0.00000 | 68.6 | 6.78 | |
| 15153_at | Chlorophyll a/b binding proteini | At3g27690 | II | 1.7 | 0.00050 | 3.2 | 2 | 0.00022 | 4.0 | 1.2 | 0.00000 | 2.3 | 1.69 | |
| 15214_s_at | G-box binding transcription factor | At2g46270 | II | 3.7 | 0.00147 | 13.0 | 3.9 | 0.00004 | 14.9 | 3.9 | 0.00000 | 14.9 | 3.15 | |
| 15481_at | Unknown protein | At2g30610 | II | 2.7 | 0.00069 | 6.5 | 2 | 0.00005 | 4.0 | 2.2 | 0.00000 | 4.6 | ||
| 15519_s_at | 3-Methylcrotonyl-CoA carboxylase | At1g03090 | II | 3.3 | 0.00002 | 9.8 | 2.9 | 0.00000 | 7.5 | 2.3 | 0.00000 | 4.9 | 3.58 | |
| 15544_at | Cation transport protein | At4g31290 | II | 1.6 | 0.01183 | 3.0 | 1.8 | 0.00130 | 3.5 | 3.7 | 0.00057 | 13.0 | 3 | |
| 15582_s_at | Alternative oxidase 1a precursor | At3g22370 | II | 2.2 | 0.00000 | 4.6 | 1.7 | 0.00000 | 3.2 | 2.1 | 0.00000 | 4.3 | 3.79 | |
| 15611_s_at | Low-temperature-induced protein | At5g52310 | II | 2.6 | 0.00000 | 6.1 | 3.2 | 0.00002 | 9.2 | 3.7 | 0.00000 | 13.0 | 9.09 | 58.3 |
| 15625_at | Glyoxalase II | At1g53580 | I | 2.4 | 0.00000 | 5.3 | 1.7 | 0.00000 | 3.2 | 1.6 | 0.00000 | 3.0 | 5.63 | |
| 15672_s_at | Arabinogalactan protein (AGP2) | At2g22470 | II | 2.3 | 0.00006 | 4.9 | 3.8 | 0.00000 | 13.9 | 1.9 | 0.00000 | 3.7 | 7.52 | |
| 15863_at | Unknown protein | At2g21820 | II | 2.6 | 0.00000 | 6.1 | 3 | 0.00000 | 8.0 | 2.6 | 0.00000 | 6.1 | ||
| 15997_s_at | Unknown protein | At1g20440 | I | 2.4 | 0.00000 | 5.3 | 2.2 | 0.00000 | 4.6 | 2.7 | 0.00000 | 6.5 | 6.28 | 17 |
| 16031_at | Ferritin1 precursor | At5g01600 | I | 1.1 | 0.00147 | 2.1 | 2.1 | 0.00000 | 4.3 | 1.6 | 0.00000 | 3.0 | 2.77 | |
| 16037_s_at | Unknown protein | At1g20696 | II | 2.1 | 0.00001 | 4.3 | 1.4 | 0.00115 | 2.6 | 3 | 0.00000 | 8.0 | ||
| 16038_s_at | Dehydrin | At5g66400 | I | 3.5 | 0.00000 | 11.3 | 3.4 | 0.00000 | 10.6 | 3.8 | 0.00000 | 13.9 | 6.83 | 77.6 |
| 16062_s_at | DRE CRT binding protein | At4g25470 | II | 2.1 | 0.00025 | 4.3 | 1.5 | 0.00096 | 2.8 | 1.8 | 0.00000 | 3.5 | ||
| 16099_at | MYB family transcription factor | At4g09460 | II | 1.5 | 0.30683 | 2.8 | 2.1 | 0.00238 | 4.3 | 1.5 | 0.00000 | 2.8 | ||
| 16115_at | Homeobox protein ATHB-12 | At3g61890 | I | 2 | 0.00084 | 4.0 | 3.1 | 0.00002 | 8.6 | 1.1 | 0.00002 | 2.1 | 9.11 | 58 |
| 16440_at | Unknown protein | At2g40000 | II | 2.0 | 0.00549 | 4.0 | 2.7 | 0.00018 | 6.5 | 1.0 | 0.00188 | 2.0 | 3.17 | |
| 16510_at | Unknown protein | At4g32480 | II | 2.9 | 0.00079 | 7.5 | 3.6 | 0.00029 | 12.1 | 1.8 | 0.00001 | 3.5 | ||
| 16524_at | Aldehyde dehydrogenase | At1g54100 | I | 1.9 | 0.00002 | 3.7 | 1.3 | 0.00001 | 2.5 | 1.5 | 0.00000 | 2.8 | 6.61 | |
| 16544_s_at | 60S ribosomal protein | At2g39460 | II | 3.4 | 0.00000 | 10.6 | 2.9 | 0.00000 | 7.5 | 2.1 | 0.00000 | 4.3 | ||
| 16953_at | Ribosomal protein | At1g67430 | II | 2.4 | 0.00007 | 5.3 | 2.5 | 0.06734 | 5.7 | 1.9 | 0.00000 | 3.7 | ||
| 17384_at | Unknown protein | At4g23630 | II | 1.0 | 0.00001 | 2.0 | 1.3 | 0.00000 | 2.5 | 1.9 | 0.00000 | 3.7 | ||
| 17407_s_at | Low-temperature-induced protein | At5g52300 | II | 3.4 | 0.00098 | 10.6 | 2.7 | 0.00211 | 6.5 | 5.3 | 0.00000 | 39.4 | 12.82 | |
| 17441_s_at | Unknown protein | At1g78860 | II | 3.4 | 0.00007 | 10.6 | 2.6 | 0.00157 | 6.1 | 3.7 | 0.00000 | 13.0 | ||
| 17442_i_at | Glycoprotein | At1g78850 | II | 3 | 0.00081 | 8.0 | 2.8 | 0.00022 | 7.0 | 1.7 | 0.00022 | 3.2 | 1.99 | |
| 17769_s_at | Trehalose-6-phosphate synthase | At1g78580 | II | 1.6 | 0.00000 | 3.0 | 1.4 | 0.00011 | 2.6 | 3 | 0.00000 | 8.0 | ||
| 17824_s_at | MYB-related protein | At4g21440 | II | 5 | 0.00000 | 32.0 | 4.5 | 0.00000 | 22.6 | 2 | 0.00000 | 4.0 | 33 | |
| 17921_s_at | Shaggy-like protein kinase | At4g18710 | II | 2.1 | 0.00252 | 4.3 | 1.6 | 0.13906 | 3.0 | 1.3 | 0.00000 | 2.5 | ||
| 17978_s_at | MYB96 transcription factor | At5g62470 | II | 1.8 | 0.00024 | 3.5 | 1.8 | 0.00096 | 3.5 | 1.2 | 0.00000 | 2.3 | ||
| 18594_at | Unknown protein | At1g01470 | I | 1.1 | 0.00000 | 2.1 | 1 | 0.00000 | 2.0 | 1.5 | 0.00000 | 2.8 | 4.01 | 4.6 |
| 18624_at | Unknown protein | At2g39570 | II | 4.2 | 0.00013 | 18.4 | 2.7 | 0.00267 | 6.5 | 2.9 | 0.00000 | 7.5 | ||
| 18646_at | Unknown protein | At1g10590 | II | 1.3 | 0.00010 | 2.5 | 1.1 | 0.00004 | 2.1 | 1.3 | 0.00000 | 2.5 | ||
| 18663_s_at | Unknown protein | At4g24130 | II | 2.2 | 0.00000 | 4.6 | 2.1 | 0.00000 | 4.3 | 1.5 | 0.00000 | 2.8 | 6.8 | |
| 18872_at | LEA-like protein | At3g17520 | II | 5.4 | 0.00001 | 42.2 | 6.2 | 0.00000 | 73.5 | 5.2 | 0.00000 | 36.8 | ||
| 18936_at | PP2C AtP2C-HA | At1g72770 | I | 4 | 0.00000 | 16.0 | 5.8 | 0.00001 | 55.7 | 2.2 | 0.00000 | 4.6 | 3.5 | |
| 18949_at | MYB-related protein | At5g67300 | II | 5.1 | 0.00000 | 34.3 | 1.6 | 0.00065 | 3.0 | 2.2 | 0.00000 | 4.6 | ||
| 18955_at | β-Ketoacyl-CoA synthase | At1g04220 | II | 1.6 | 0.01183 | 3.0 | 2.5 | 0.00299 | 5.7 | 2.1 | 0.00000 | 4.3 | 3.2 | |
| 19152_at | LEA | At5g06760 | I | 3.5 | 0.00000 | 11.3 | 6.6 | 0.00000 | 97.0 | 3.4 | 0.00000 | 10.6 | 13.29 | 328 |
| 19186_s_at | Dehydrin Xero2 | At3g50970 | I | 2.9 | 0.00000 | 7.5 | 3.1 | 0.00000 | 8.6 | 4.5 | 0.00000 | 22.6 | ||
| 19441_s_at | G-box binding factor | At4g01120 | II | 4.3 | 0.00139 | 19.7 | 1.7 | 0.00012 | 3.2 | 3 | 0.00004 | 8.0 | ||
| 19638_at | PP2C | At3g11410 | I | 3.6 | 0.00000 | 12.1 | 2.1 | 0.00000 | 4.3 | 2.9 | 0.00000 | 7.5 | 7.58 | 4.6 |
| 19646_at | Homeodomain transcription factor | At2g46680 | I | 3.9 | 0.00000 | 14.9 | 3.4 | 0.00000 | 10.6 | 2.3 | 0.00000 | 4.9 | 8.84 | 27.7 |
| 19688_at | Low-temperature-induced protein | At4g25580 | II | 1.0 | 0.01897 | 2.0 | 3.5 | 0.00166 | 11.3 | 2.6 | 0.00000 | 6.1 | ||
| 19852_s_at | Cytoplasmatic aconitate hydratase | At4g35830 | II | 1.1 | 0.00003 | 2.1 | 1.1 | 0.00001 | 2.1 | 1.8 | 0.00000 | 3.5 | ||
| 19860_at | Transcription factor | At1g56170 | II | 1.6 | 0.00038 | 3.0 | 1.4 | 0.00130 | 2.6 | 1.8 | 0.00000 | 3.5 | 4 | |
| 19982_at | Unknown protein | At1g79270 | I | 3.5 | 0.00753 | 11.3 | 1.3 | 0.00044 | 2.5 | 3.8 | 0.00000 | 13.9 | ||
| 20042_at | No hits | II | 2.7 | 0.00000 | 6.5 | 2.4 | 0.00000 | 5.3 | 1.3 | 0.00000 | 2.5 | |||
| 20060_at | No hits | II | 5 | 0.00001 | 32.0 | 2.8 | 0.00006 | 7.0 | 2.3 | 0.00000 | 4.9 | |||
| 20186_at | Unknown protein | At1g08630 | II | 2.3 | 0.00299 | 4.9 | 1.1 | 0.00009 | 2.1 | 2.1 | 0.00001 | 4.3 | ||
| 20200_at | Unknown protein | At4g25690 | II | 1.4 | 0.02480 | 2.6 | 1.2 | 0.00029 | 2.3 | 1.3 | 0.00000 | 2.5 | 1.89 | |
| 20323_at | Small heat shock protein | At2g29500 | II | 3.4 | 0.00018 | 10.6 | 1.2 | 0.00027 | 2.3 | 2.1 | 0.00001 | 4.3 | ||
| 20635_s_at | MYB96 transcription factor | At5g62470 | II | 1.0 | 0.02705 | 2.0 | 2.2 | 0.00019 | 4.6 | 1.1 | 0.00000 | 2.1 | ||
| 20641_at | LEA | At1g52690 | I | 4.9 | 0.00001 | 29.9 | 6.6 | 0.00000 | 97.0 | 3.6 | 0.00000 | 12.1 | 30.75 | 1799 |
Describes names of probe set on Affymetrix chip.
AGI locus numbers.
Indicates clusters in Figure 5.
Signal log value, which measures the change in expression level for a transcript between two arrays. This change is expressed as the log2 ratio. A log2 ratio of 1 is the same as a fold change of 2.
Change P-value, which measures the probability that the expression levels of a probe in two different arrays are the same. Change P-values of 0.00000 correspond to P-value <0.000005.
Fold change is calculated using the signal log ratio (see Methods).
Obtained from Supplemental Table 1 online (column for 5-h ABA treatment; Seki et al., 2002).
Obtained from Supplemental Table Induced.xls online (Hoth et al., 2002).
Chlorophyll a/b binding protein gene expression is repressed by ABA at 1, 2, 10, and 24 h of ABA treatment but shows ABA induction at 5 h of ABA treatment (see Seki et al., 2002, Supplemental Table 5 online).
Group II contains 69 transcripts that are ABA induced in guard cells but not in mesophyll cells (Figure 5A, Table 2). Most of these genes have not been described at the cellular level. However, several have been shown to play roles in drought tolerance. For example, the trehalose-6-phosphate synthase gene is induced by ABA (Table 2) and is part of the trehalose synthase complex that functions in protection against heat and desiccation stress in microorganisms (Bell et al., 1998). In addition, overexpression of a Saccharomyces cerevisiae (yeast) or bacterial trehalose-6-phosphate synthase gene in Nicotiana tabacum (tobacco) plants was reported to lead to an increase in drought resistance (Romero et al., 1997).
Group III contains 100 transcripts that are ABA induced only in mesophyll cells (Figure 5A, see Supplemental Table 3 online). This group includes several known genes that were previously shown to be induced by ABA, such as cor15b (Wilhelm and Thomashow, 1993). Interestingly, several genes encoding proteins involved in sugar sensing were identified as members of this group, suggesting the existence of cross talk between sugar and ABA signaling pathways as observed in genetic analyses (reviewed in Sheen et al., 1999; Gazzarrini and McCourt, 2001).
In addition, comparison analyses with previously reported results show that among the 190 ABA-induced genes in guard cells, mesophyll cells, or both cells types, 37 genes (∼20%) have been previously described to be induced in whole seedlings after 8-h treatment with 50 μM ABA: 11 only in guard cells (representing 16% of ABA-induced transcripts in guard cells), 14 only in mesophyll cells (representing 14% of ABA-induced transcripts in mesophyll cells), and 12 in both cell types (representing 52% of ABA-induced transcripts in both cell types) (Table 2, see Supplemental Table 3 online; Hoth et al., 2002). Similarly, a comparison of our results with Seki et al. (2002) revealed that 51 transcripts (∼27%) of 190 genes listed in Table 2 and Supplemental Table 3 online were also reported to be induced in whole plants treated hydroponically with 100 μM ABA for 5 h: 16 only in guard cells (representing 23% of ABA-induced transcripts only in guard cells), 21 only in mesophyll cells (representing 21% of ABA-induced transcripts only in mesophyll cells), and 14 in both cell types (representing 67% of ABA-induced transcripts in both cell types) (Table 2, see Supplemental Table 3 online; Seki et al., 2002). Thus, many of the ABA-induced genes detected here differ from ABA-induced genes detected in whole seedlings and whole plants.
ABA-Repressed Gene Clusters
Previous studies have focused mainly on ABA upregulated transcripts. Very little knowledge exists on genes downregulated by ABA. The groups IV to VI contain transcripts that show lower expression in guard cells, mesophyll cells, or in both cell types after ABA treatment (Figures 5A and 5C). The number of genes in these three clusters is small. For example, group IV contains only three transcripts, which are slightly lower in both ABA-treated mesophyll and guard cells (Figure 5A, Table 3).
Table 3.
ABA-Repressed Genes in Guard Cells
| Experiment 1
|
Experiment 2
|
Experiment 3
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Affy Probea | Description | AGI Numberb | Clusterc | Ratiod | P-Valuee | Fold Changef | Ratio | P-Value | Fold Change | Ratio | P-Value | Fold Change | Seki et al.g | Hoth et al.h |
| 12100_at | MYB family transcription factor | At2g21650 | V | −3.8 | 0.99997 | −13.9 | −3.3 | 0.99999 | −9.8 | −3.4 | 0.99997 | −10.6 | ||
| 12185_at | Unknown protein | At1g54860 | V | −1.8 | 0.95007 | −3.5 | −2.1 | 0.99898 | −4.3 | −1.7 | 0.99733 | −3.25 | ||
| 12196_at | Unknown protein | At2g28410 | V | −1.5 | 1.00000 | −2.8 | −1.8 | 1.00000 | −3.5 | −2 | 0.99998 | −4.0 | ||
| 12284_at | Somatic embryogenesis receptor kinase | At1g34210 | V | −0.8 | 0.96060 | −2 | −1.4 | 0.99977 | −2.6 | −2 | 0.99980 | −4.0 | 4.7 | |
| 12338_at | Pectin methylesterase | At3g14310 | V | −4.1 | 0.99994 | −17.1 | −1.7 | 0.99977 | −3.2 | −2.3 | 0.97409 | −4.9 | ||
| 12381_at | No hits | V | −0.6 | 0.94811 | −2 | −0.7 | 0.99733 | −2 | −1.5 | 0.99996 | −2.8 | |||
| 12409_at | Small nuclear ribonucleoprotein U1A | At2g47580 | V | −0.8 | 1.00000 | −2 | −0.9 | 0.99992 | −2 | −2.4 | 1.00000 | −5.3 | ||
| 12768_at | Unknown protein | At2g15890 | V | −0.9 | 0.99999 | −2 | −0.7 | 0.99995 | −2 | −1.6 | 0.99973 | −3.0 | ||
| 12773_at | Membrane channel protein | At2g28900 | V | −0.9 | 1.00000 | −2 | −0.6 | 1.00000 | −2 | −1.2 | 0.99122 | −2.3 | ||
| 12843_s_at | Unknown protein | At2g16590 | V | −1.0 | 0.99994 | −2 | −0.6 | 0.99947 | −2 | −0.8 | 0.50000 | −2 | ||
| 12846_s_at | No hits | V | −2.1 | 1.00000 | −4.3 | −0.8 | 0.99999 | −2.0 | −1.5 | 1.00000 | −2.8 | |||
| 12881_s_at | No hits | V | −1.6 | 0.99995 | −3.0 | −0.7 | 1.00000 | −2 | −2.3 | 1.00000 | −4.9 | |||
| 12893_at | Alcohol dehydrogenase | At5g43940 | V | −1.6 | 0.99989 | −3.0 | −0.6 | 0.99964 | −2 | −1.6 | 0.99998 | −3.0 | ||
| 13080_at | 40S ribosomal protein | At2g21580 | V | −0.7 | 0.99776 | −2 | −0.6 | 0.99905 | −2 | −1.8 | 1.00000 | −3.5 | ||
| 13184_s_at | Thioredoxin | At5g42980 | V | −2.3 | 1.00000 | −4.9 | −0.6 | 1.00000 | −2.0 | −0.9 | 0.86906 | −2 | ||
| 13236_at | Aldolase | At4g39980 | V | −0.6 | 0.97627 | −2 | −1.2 | 0.99843 | −2.3 | −2.1 | 1.00000 | −4.3 | ||
| 13254_at | Farnesyl pyrophosphate synthetase2 | At4g17190 | V | −0.7 | 0.99999 | −2 | −0.7 | 0.99973 | −2 | −1.8 | 1.00000 | −3.5 | ||
| 13554_at | β-Ketoacyl-CoA synthase | At2g16280 | V | −1.1 | 0.99853 | −2.1 | −1.6 | 1.00000 | −3.0 | −1.8 | 0.99776 | −3.5 | ||
| 13672_at | No hits | V | −0.9 | 0.99967 | −2 | −1 | 0.99993 | −2 | −1.4 | 0.99939 | −2.6 | |||
| 14090_i_at | Unknown protein | At2g15830 | V | −0.6 | 0.99820 | −2 | −0.6 | 1.00000 | −2 | −0.7 | 0.76103 | −2 | ||
| 14663_s_at | Glycosyl hydrolase family | At4g24040 | V | −1.0 | 0.99978 | −2 | −0.6 | 0.99910 | −2 | −0.6 | 0.49254 | −2 | ||
| 14667_at | TRP synthase | At5g54810 | V | −0.7 | 0.99977 | −2 | −0.9 | 0.99967 | −2 | −2.3 | 0.99999 | −4.9 | ||
| 14717_s_at | CLC-a chloride channel protein | At5g40890 | V | −0.7 | 0.99993 | −2 | −1.1 | 1.00000 | −2.1 | −2.6 | 1.00000 | −6.1 | ||
| 15105_s_at | Gly-rich RNA binding protein | At2g21660 | V | −1.1 | 0.99992 | −2.1 | −1.3 | 1.00000 | −2.5 | 0.2 | 0.00000 | −2 | 0.8 | |
| 15194_s_at | GASA4 | At5g15230 | IV | −0.8 | 1.00000 | −2 | −0.9 | 1.00000 | −2 | −0.8 | 0.89823 | −2 | ||
| 15213_s_at | Orn carbamoyltransferase | At1g75330 | V | −0.8 | 0.99991 | −2 | −0.9 | 1.00000 | −2 | −2 | 1.00000 | −4.0 | ||
| 15453_at | Unknown protein | At4g14020 | V | −0.9 | 0.99999 | −2 | −0.7 | 0.99990 | −2 | −1.9 | 1.00000 | −3.7 | ||
| 15510_r_at | No hits | V | −1.8 | 0.82023 | −3.5 | −0.8 | 0.99989 | −2 | −2.1 | 1.00000 | −4.3 | |||
| 15548_at | Unknown protein | At1g11820 | V | −0.9 | 0.99971 | −2 | −0.7 | 0.99978 | −2 | −2.3 | 1.00000 | −4.9 | ||
| 15624_s_at | ADPG pyrophosphorylase | At5g48300 | IV | −0.6 | 0.99931 | −2 | −1.2 | 1.00000 | −2.3 | −2.2 | 1.00000 | −4.6 | ||
| 15626_s_at | Vacuolar sorting receptor | At3g52850 | V | −0.6 | 0.98817 | −2 | −0.8 | 0.99998 | −2 | −1.7 | 0.99999 | −3.2 | ||
| 15703_i_at | Putative trypsin inhibitor | At2g43520 | V | −2.6 | 0.99971 | −6.1 | −0.8 | 1.00000 | −2.0 | −1.8 | 0.99934 | −3.5 | ||
| 16014_at | Unknown protein | At1g75750 | V | −0.8 | 1.00000 | −2 | −0.8 | 1.00000 | −2 | −1.9 | 0.99999 | −3.7 | ||
| 16042_s_at | Cytochrome P450i | At5g05690 | V | −0.7 | 0.99910 | −2 | −1.3 | 0.99986 | −2.5 | −0.7 | 0.54467 | −2 | 1.2 | |
| 16051_at | Tubulin β-9 chain | At4g20890 | V | −1.2 | 0.99997 | −2.3 | −0.9 | 0.99988 | −2 | −1.6 | 0.99990 | −3.0 | ||
| 16092_s_at | Potassium channel protein KAT1 | At5g46240 | V | −2.3 | 0.99981 | −4.9 | −1.5 | 0.99999 | −2.8 | −2.3 | 0.99999 | −4.9 | ||
| 16118_s_at | WD-40 repeat protein | At2g19520 | V | −0.7 | 0.50000 | −2 | −0.9 | 0.99905 | −2 | −2.5 | 1.00000 | −5.7 | ||
| 16145_at | Vacuolar ATP synthase | At1g75630 | V | −1.0 | 1.00000 | −2.0 | −0.9 | 0.99996 | −2 | −2.9 | 0.99999 | −7.5 | ||
| 16416_at | Plant defensin protein | At2g02130 | V | −0.8 | 0.99989 | −2 | −1.8 | 1.00000 | −3.5 | −0.9 | 0.92765 | −2 | ||
| 16430_at | Glutathione S-transferasej | At2g30860 | V | −0.8 | 0.99733 | −2 | −1.3 | 1.00000 | −2.5 | −2 | 1.00000 | −4.0 | 1.1 | |
| 16451_at | Ketol-acid reductoisomerase | At3g58610 | V | −1.3 | 0.99420 | −2.5 | −1.5 | 1.00000 | −2.8 | −1.6 | 0.99999 | −3.0 | ||
| 16458_s_at | No hits | V | −0.9 | 0.99993 | −2 | −0.7 | 0.99980 | −2 | −1.6 | 0.70618 | −3.0 | |||
| 16624_s_at | Transcription factor | At1g08810 | V | −0.6 | 0.84813 | −2 | −1.4 | 1.00000 | −2.6 | −1.9 | 0.99997 | −3.7 | ||
| 16897_i_at | Unknown protein | At5g15350 | V | −0.7 | 0.99713 | −2 | −1 | 0.99992 | −2 | −2.3 | 0.99994 | −4.9 | ||
| 16985_s_at | Cytosolic ribosomal protein | At3g48930 | V | −0.7 | 1.00000 | −2 | −0.6 | 0.99999 | −2 | −1.9 | 0.99994 | −3.7 | ||
| 16998_at | dTDP-glucose 4-6-dehydratase | At3g62830 | V | −0.7 | 0.99962 | −2 | −0.6 | 0.99985 | −2 | −1.8 | 0.99998 | −3.5 | ||
| 17033_at | Unknown protein | At5g39410 | V | −1.0 | 0.99910 | −2.0 | −1 | 0.99999 | −2.0 | −1.4 | 0.90472 | −2.64 | ||
| 17126_at | WD-40 repeat protein | At5g58230 | V | −0.9 | 0.82023 | −2 | −0.6 | 0.99992 | −2 | −2.1 | 1.00000 | −4.3 | ||
| 17187_at | Putative arginase | At4g08870 | V | −3.2 | 0.99853 | −9.2 | −1.6 | 0.99763 | −3.0 | −1.4 | 0.99479 | −2.64 | ||
| 17215_at | Trypsin inhibitor | At2g43550 | V | −0.8 | 0.94609 | −2 | −1 | 0.99950 | −2 | −2.1 | 0.99991 | −4.3 | ||
| 17237_at | Unknown protein | At2g22840 | V | −1.7 | 0.77836 | −3.2 | −2.3 | 0.99926 | −4.9 | −2.6 | 0.99973 | −6.06 | ||
| 17865_at | NADPH oxidoreductase | At1g75280 | V | −1.2 | 0.97295 | −2.3 | −2.7 | 0.99939 | −6.5 | −1.8 | 0.99984 | −3.48 | ||
| 17957_at | Acetone-cyanohydrin lyase | At2g23600 | IV | −1.2 | 0.99910 | −2.3 | −3.3 | 1.00000 | −9.8 | −2.9 | 1.00000 | −7.46 | ||
| 18063_i_at | No hits | V | −1.2 | 0.99425 | −2.3 | −1 | 0.99915 | −2.0 | −2.7 | 0.99997 | −6.5 | |||
| 18272_at | Unknown protein | At2g40080 | V | −1.1 | 0.94402 | −2.1 | −3.9 | 1.00000 | −14.9 | −2.7 | 0.99999 | −6.5 | ||
| 18315_s_at | β-Glucosidase | At4g27830 | V | −0.7 | 0.99718 | −2 | −1.1 | 0.98873 | −2.1 | −2.3 | 0.99999 | −4.9 | ||
| 18611_at | Putative pyruvate water dikinase | At5g26570 | V | −1.1 | 0.99823 | −2.1 | −0.7 | 0.99843 | −2 | 0.4 | 0.31342 | −2 | ||
| 18678_at | Peptidylprolyl isomerase ROC4 | At3g62030 | V | −1.8 | 0.99853 | −3.5 | −0.9 | 0.99995 | −2 | −2.5 | 1.00000 | −5.7 | 0.19 | |
| 18687_at | Pyruvate dehydrogenase subunit | At1g59900 | V | −0.7 | 0.99973 | −2 | −0.8 | 1.00000 | −2 | −1.8 | 1.00000 | −3.5 | ||
| 18708_at | Ripening-related protein | At5g62350 | V | −1.4 | 0.99998 | −2.6 | −2.3 | 1.00000 | −4.9 | −2.8 | 1.00000 | −7.0 | 0.66 | |
| 18882_at | No hits | V | −0.7 | 0.99996 | −2 | −0.7 | 0.99943 | −2 | −1.8 | 1.00000 | −3.5 | |||
| 18956_at | Ethylene response sensor | At2g40940 | V | −0.7 | 0.99943 | −2 | −0.6 | 0.99992 | −2 | −1.9 | 1.00000 | −3.7 | ||
| 19171_at | Trypsin inhibitork | At2g43510 | V | −1.5 | 0.99967 | −2.8 | −0.6 | 0.99885 | −2 | −2.5 | 1.00000 | −5.7 | 2.97 | |
| 20282_at | Protein kinase | At2g28930 | V | −1.3 | 0.99989 | −2.5 | −1.3 | 1.00000 | −2.5 | −3.2 | 1.00000 | −9.2 | ||
| 20537_at | Extensin | At4g13340 | V | −1.6 | 0.99892 | −3.0 | −0.9 | 0.99733 | −2 | −1.2 | 0.93019 | −2.3 | ||
| 20585_at | Glutaredoxin | At2g47880 | V | −1.7 | 0.99994 | −3.2 | −1.9 | 0.94811 | −3.7 | −1.6 | 0.99950 | −3.03 | ||
| 20709_s_at | Putative 3-oxoacyl reductase | At1g24360 | V | −0.6 | 0.99998 | −2 | −0.6 | 0.99998 | −2 | −2.2 | 1.00000 | −4.6 | ||
Describes names of probe set on Affymetrix chip.
AGI locus numbers.
Indicates clusters in Figure 5.
Signal Log value which measures the change in expression level for a transcript between two arrays. This change is expressed as the log2 ratio. A log2 ratio of 1 is the same as a Fold change of 2.
Change P-value, which measures the probability that the expression levels of a probe in two different arrays are the same. Change P-values of 1.00000 correspond to P-value >0.99995.
Fold change is calculated using the signal log ratio.
Obtained from Supplemental Table 5 online (column for 5-h ABA treatment; Seki et al., 2002).
Obtained from Supplemental Table Repressed.xls online (Hoth et al., 2002).
Cytochrome p450 gene expression is repressed by ABA at 1, 2, 10, and 24 h of ABA treatment but shows ABA induction at 5 h of ABA treatment.
GST gene expression is slightly induced by ABA at 1, 5, and 10 h of ABA treatment but repressed by ABA at 2 and 24 h of ABA treatment (see Seki et al., 2002, Supplemental Table 5 online).
Trypsin inhibitor gene is induced by ABA (see Seki et al., 2002, Supplemental Table 1 online).
Group V contains 64 transcripts that are lower only in ABA-treated guard cells but not in mesophyll cells (Figure 5A, Table 3). Interestingly, among these genes, the KAT1 mRNA encoding an inward potassium channel was found to be downregulated by ABA. This finding correlates with the ABA inhibition of guard cell inward K+ channel activity that has been previously shown in guard cells from different species (Blatt and Armstrong, 1993; Lemtiri-Chlieh and MacRobbie, 1994; Schwartz et al., 1994; Lemtiri-Chlieh, 1996). In addition, of the 64 ABA-repressed transcripts in guard cells, six transcripts (∼9%) were shown to be repressed by ABA in ABA-treated whole plants (Seki et al., 2002) and only one in ABA-treated whole seedlings (Hoth et al., 2002).
Group VI contains 51 transcripts that are slightly lower in ABA-treated mesophyll cells but not in guard cells (Figure 5A, see Supplemental Table 4 online). This group includes several genes encoding proteins that function in photosynthesis, such as carbonic anhydrase, phosphoenolpyruvate carboxylase, and ribulose-1,5-bisphosphate carboxylase subunit binding protein (see Supplemental Table 4 online). The finding that ABA inhibits the expression of photosynthetic genes correlates with findings in other species (Bartholomew et al., 1991; Chang and Walling, 1991; Reinbothe et al., 1993; Weatherwax et al., 1996). In addition, ABA-repressed transcripts in mesophyll cells encode proteins that are implicated in biosynthetic functions, including amino acid and carbohydrate metabolism. Similar results have been observed in drought-stressed Hordeum vulgare (barley) (Ozturk et al., 2002). Of the 51 ABA-repressed transcripts in mesophyll cells (see Supplemental Table 4 online), only two transcripts were ABA-repressed in whole seedlings (Hoth et al., 2002) and in whole plants (Seki et al., 2002).
RT-PCR Confirms Microarray Data
To confirm the reliability of results from GeneChip expression profile analyses, RT-PCR experiments were performed using cDNAs synthesized from guard cell and mesophyll cell RNA. Twenty-nine genes exhibiting expression changes in response to ABA or showing differential expression in guard cells versus mesophyll cells were selected and tested two or three times. Results from eight of the tested genes are shown in Figure 6 (CER2 [At4g24510], trehalose-6-phosphate synthase [At1g78580], LEA [At5g06760], KAT1 [At5g46240], CDPK [At3g51850], dehydrin [At3g50970], PP2C [At3g11410], and COR47 [At1g20440]). All of the analyzed genes exhibited cell type specificity or ABA regulation in RT-PCR experiments that correlated with our GeneChip results (see Supplemental Figure 2 online), indicating reliability of the microarray data.
Figure 6.
RT-PCR Analyses Independently Confirm Results Obtained from Chip Hybridization Experiments.
RT-PCR was performed using guard cell and mesophyll cell RNA with primers for selected genes from guard cell preferential genes showing no ABA modulation (CER2 and calcium-dependent protein kinase [At3g50530]) and from Figure 5–derived group I (dehydrin [At3g50970], PP2C [At3g11410], and COR47), group II (trehalose-6-phosphate synthase and LEA) and group V (KAT1). Results are from 24 and 27 RT-PCR cycles. Actin2 gene was used as control. Results from 8 of 29 tested genes are illustrated. PCR was repeated at least twice. GC; guard cells; GC + ABA, guard cells treated with ABA; MC, mesophyll cells; MC + ABA, mesophyll cells treated with ABA.
Promoter Element Analyses
To analyze ABA-regulated promoter sequences, statistical analyses of cis-acting elements were pursued. Functional analyses and sequence comparison of ABA-inducible promoters allowed the identification of cis-acting sequences, such as the G-box–containing elements designated ABA-regulated elements (ABREs) and the functionally equivalent coupling element3 (CE3)–like sequences (reviewed in Busk and Pages, 1998). Our gene expression results allow us to statistically explore the correlation between these elements and ABA regulation on a global scale and to test the generality of hypotheses generated from the few experimentally analyzed promoters in different plant species. Table 4 shows results from our analysis of the occurrence of several known ABA-related cis-regulatory motifs in the 500-bp upstream sequences. Pattern match searches were performed by writing custom Python scripts with C extensions (see Methods). The number and percentage of occurrences (hits) of each motif are given for each gene set (Table 4, columns A to E). To evaluate the significance of these numbers and percentages, a P-value was computed giving the probability of obtaining at least as many hits as were observed in the set, if the set were a random sample drawn without replacement from the complete set of Arabidopsis promoter sequences. A small P-value for a given motif and set shows that the observed number of hits is unlikely to be because of chance (Table 4). In further controls, ABA-repressed transcripts were also analyzed, and the analyzed motifs were found not to be significantly overrepresented (Table 4, columns B and D).
Table 4.
Occurrence of Motifs in 500-bp Region Upstream of ABA-Regulated Genes
| A
|
B
|
C
|
D
|
E
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All Sequences
|
Guard Cell, ABA Downregulated
|
Guard Cell, ABA Upregulated
|
Mesophyll Cell, ABA Downregulated
|
Mesophyll Cell, ABA Upregulated
|
|||||||||||||||||||
| Motif | Sequence | Allowed Mismatches | Reference | Hits | Set Size | Pct | Hits | Set Size | Pct | P-Value | Hits | Set Size | Pct | P-Value | Hits | Set Size | Pct | P-Value | Hits | Set Size | Pct | P-Value | |
| 1 | ABRE from AlignACE | (C/G)ACGTG(G/T)(A/C) | 0 | This article | 831 | 7749 | 11 | 3 | 59 | 5.1 | 9.60E−01 | 34 | 92 | 37 | 2.26E−11 | 6 | 53 | 11 | 5.09E−01 | 33 | 120 | 28 | 2.07E−07 |
| 2 | Classical ABRE | (C/G/T)ACGTG(G/T)(A/C) | 0 | Ref 1 | 1121 | 7749 | 14 | 4 | 59 | 6.8 | 9.79E−01 | 43 | 92 | 47 | 9.78E−14 | 10 | 53 | 19 | 2.29E−01 | 42 | 120 | 35 | 1.21E−08 |
| 3 | CE1 | TGCCACCGG | 0 | Ref 2 | 3 | 7749 | 0.0 | 0 | 59 | 0.0 | 1.00E+00 | 0 | 92 | 0.0 | 1.00E+00 | 0 | 53 | 0.0 | 1.00E+00 | 0 | 120 | 0.0 | 1.00E+00 |
| 4 | CE1 | TGCCACCGG | 1 | Ref 2 | 135 | 7749 | 2.0 | 2 | 59 | 3.4 | 2.75E−01 | 2 | 92 | 2.2 | 4.79E−01 | 3 | 53 | 5.7 | 6.45E−02 | 3 | 120 | 2.5 | 3.48E−01 |
| 5 | CE3, Full | ACGCGTGTCCTC | 1 | Ref 2 | 14 | 7749 | 0.0 | 0 | 59 | 0.0 | 1.00E+00 | 1 | 92 | 1.1 | 1.54E−01 | 0 | 53 | 0.0 | 1.00E+00 | 0 | 120 | 0.0 | 1.00E+00 |
| 6 | CE3, Short | ACGCGTGTC | 0 | This article | 11 | 7749 | 0.0 | 0 | 59 | 0.0 | 1.00E+00 | 2 | 92 | 2.2 | 7.15E−03 | 0 | 53 | 0.0 | 1.00E+00 | 0 | 120 | 0.0 | 1.00E+00 |
| 7 | DRE | TACCGACAT | 0 | Ref 2 | 14 | 7749 | 0.0 | 0 | 59 | 0.0 | 1.00E+00 | 2 | 92 | 2.2 | 1.16E−02 | 0 | 53 | 0.0 | 1.00E+00 | 4 | 120 | 3.3 | 4.86E−05 |
| 8 | DRE | TACCGACAT | 1 | Ref 2 | 364 | 7749 | 5.0 | 1 | 59 | 1.7 | 9.42E−01 | 8 | 92 | 8.7 | 6.67E−02 | 5 | 53 | 9.4 | 1.02E−02 | 14 | 120 | 12 | 1.42E−03 |
For each motif, the number (Hits) and percentage (Pct) of promoters in each gene set containing at least one occurrence of the given motif are shown. Occurrence of the motifs was determined by scanning for either exact (Allowed Mismatches = 0) or inexact (Allowed Mismatches = 1) pattern matches on both strands of the 500 nucleotides upstream of the translational start. Significantly overrepresented motifs have P-values ≤0.01, which are in bold. Ref 1, Shinozaki and Yamaguchi-Shinozaki (2000); Ref 2, Busk and Pages (1998).
Our analyses showed that the classical G-box containing ABRE is significantly overrepresented in the set of ABA-induced genes in both guard cells (P < 10−13) and mesophyll cells (P < 10−7) (Table 4, row 2, columns C and E). Moreover, its occurrence in non-ABA-induced gene sets is not more than would be expected by chance (Table 4, rows 1 and 2, columns B and D; data not shown). This result statistically confirms that the classical ABRE is a predictor of ABA induction. A sequence (ACGCGTGTC) was computationally derived with similarity to the coupling element CE3 from H. vulgare. This sequence is also mildly overrepresented in the ABA-induced gene sets (Table 4, row 6, column C). The dehydration-responsive element (DRE), which has been reported in the promoter regions of drought- and cold-inducible genes, is also statistically overrepresented in the ABA-induced genes in mesophyll cells but not in guard cells (Table 4, rows 7 and 8, columns C and E).
In addition to searching for known motifs, we applied the motif-finding program AlignACE 3.0 that uses a Gibbs sampling algorithm (http://atlas.med.harvard.edu/; Hughes et al., 2000) to each of the promoter sequence sets. AlignACE found a strong signal for an ABRE-like motif with the consensus sequence (C/G)ACGTG(G/T)(A/C) in both sets of ABA-induced genes (Table 4, row 1) but not in the non-ABA-induced gene sets. This pattern represents a subset of the classical G-box containing ABRE (C/G/T)ACGTG(G/T)(A/C) (Shinozaki and Yamaguchi-Shinozaki, 2000).
Expression Profiling of ABA Signaling Components in Guard Cells
Expression profiling has been proposed as a method to produce new hypotheses for possible roles of genes and proteins in signal transduction (Hughes et al., 2000; Perez-Amador et al., 2001). To test this hypothesis, we investigated the genes that were described previously to function in early ABA signaling in guard cells and that were present on the chip. To test whether transcript levels of these genes are modulated, we analyzed their regulation by ABA. A simplified model including previously demonstrated positive transducers and negative regulators mediating guard cell ABA signal transduction is shown in Figure 7 (Cutler et al., 1996; Leung et al., 1997; Pei et al., 1997, 1998, 2000; Allen et al., 2001, 2002; Hugouvieux et al., 2001; Murata et al., 2001; Cherel et al., 2002; Kwak et al., 2002, 2003; Mustilli et al., 2002; Yoshida et al., 2002). This figure is color coded to show the relative expression of the components before and after ABA treatment. Interestingly, many mRNAs of genes known to function in ABA signal transduction (see Mäser et al., 2003) were regulated by ABA in guard cells (Figure 7). For example, the two PP2C genes, ABI1 and ABI2, are induced by ABA in both guard cells and mesophyll cells. Interestingly, ABH1 and the dominant abi1-1 and abi2-1 alleles are negative regulators of ABA signaling, and their mRNA levels are increased by ABA, whereas the transcript level of another negative regulator ERA1 is reduced by ABA.
Figure 7.
Schematic Representation of ABA-Regulated Guard Cell–Expressed Genes in Current Working Model for ABA Signal Transduction Shows That the Transcript Levels of ABA Signal Transducers Are Regulated by ABA in Guard Cells.
Negative regulators and effectors are shown in red for clarity. Colors in boxes represent relative expression level of a gene before and after ABA treatment (+ABA). Low message level genes are included for this model, and genes not present on the chip are excluded in the model.
K+ uptake channels, which contribute to K+ uptake during stomatal opening, are negative effectors of ABA-induced ion efflux. The K+ uptake channel transcripts such as KAT1, KAT2, and AKT2 are repressed by ABA. Moreover, AtPP2CA, which has been shown to inhibit the AKT2 inward-rectifying K+ channel activity in heterologous systems (Cherel et al., 2002), is induced by ABA.
From Single Cell Expression Profiling to Gene Function: Identification of a PP2C That Functions in ABA Signaling
To verify that the microarray approach to functional discovery is effective, we selected one PP2C gene, AtP2C-HA, because it is a PP2C transcript that showed a very high level of ABA upregulation in guard cells and because no recessive knockout mutant phenotypes of PP2Cs have yet been reported in plants. AtP2C-HA was identified previously as an ABA-induced transcript with homology to ABI1 and ABI2 (Rodriguez et al., 1998).
We identified an atp2C-HA-1 disruption mutant carrying a single T-DNA insertion from the SALK Institute Genomic Analysis Laboratory (SIGnAL) database of sequenced T-DNA mutagenized Arabidopsis lines. The T-DNA insertion was in the third intron of AtP2C-HA (Figure 8A). RNA gel blot analysis confirmed the ABA induction of AtP2C-HA mRNA in the wild type, which was abolished in homozygous atp2C-HA-1 plants (Figure 8B). To determine whether the atp2C-HA-1 disruption mutation affects stomatal responses to ABA, we performed ABA-induced stomatal closing assays. Figure 8C shows that ABA-induced stomatal closing in atp2C-HA-1 mutant plants was more sensitive to ABA than that in the wild type at 0.01 μM ABA. In addition, we examined whether AtP2C-HA may also function in ABA-mediated inhibition of seed germination. Interestingly, the loss of AtP2C-HA function confers a substantial ABA-hypersensitive regulation of seed germination under the conditions assayed (Figure 8D). ABA hypersensitivity showed a 1:3 segregation in backcrossed F2 lines, indicating that atp2C-HA-1 is a recessive mutant. To test whether the ABA-hypersensitive phenotype of atp2C-HA-1 is the result of AtP2C-HA gene disruption, we tested complementation of the atp2C-HA-1 mutant with the At2PC-HA cDNA, which restored At2PC-HA expression (Figure 8E). Two independent complemented atp2C-HA-1 lines showed wild-type responses in both ABA-induced stomatal closing assays (Figure 8C) and ABA inhibition of seed germination assays (Figure 8D). These results suggest that AtP2C-HA functions as a negative regulator of ABA signal transduction and illustrate that the combination of guard cell expression data and functional genomic approaches can be used to identify gene function.
Figure 8.
atp2C-HA-1 Mutant Shows ABA Hypersensitivity in Stomatal Response and ABA Inhibition of Seed Germination.
(A) Genomic organization of AtP2C-HA gene. Exons are shown as boxes, whereas introns are shown as lines. The insertion site and orientation of the T-DNA in the atp2C-HA-1 mutant are indicated.
(B) RNA gel blot confirms disruption of the AtP2C-HA mRNA in atp2C-HA-1 mutant and ABA induction in the wild-type plants.
(C) Stomatal aperture measurements show that ABA-induced stomatal closing is ABA hypersensitive in the atp2C-HA-1 T-DNA disruption mutant (open bars) compared with the wild type (black bars) and that the AtP2C-HA cDNA complemented this phenotype (hatched bars, line 1). Stomatal apertures were measured 3 h after addition of 0.01 or 0.1 μM ABA.
(D) The atp2C-HA-1 (atp2C-HA) mutation causes ABA hypersensitivity in ABA inhibition of seed germination, which is complemented by the AtP2C-HA cDNA.
(E) RNA gel blots (left) and RT-PCR (right) show that At2PC-HA expression is restored in the atp2C-HA-1 complemented lines. Bottom left, total RNA control; bottom right, actin 2 control. Error bars represent SE of n = 6 independent experiments with 240 stomata each analyzed for the wild type and atp2C-HA-1 (atp2C-HA) and of two independent experiments with 80 stomata each analyzed for a complemented atp2C-HA-1 (line 1) in (C) and >50 seeds at each data point in (D).
DISCUSSION
This study reports cell type–specific characterization of gene expression patterns in guard cells and mesophyll cells and their regulation by ABA. For most of the genes studied here, no previously published data have been available with respect to their expression in guard cells and mesophyll cells. These data provide initial information, which could lead to characterization of gene functions in these cell types. For example, the identification of two ABA-induced guard cell preferential genes encoding trehalose synthase complex proteins can direct future analysis of the phenotype of mutants or transgenic plants altered in the expression of these genes. Moreover, a large number of the guard cell–expressed genes have unknown functions, and, therefore, the tools and signal transduction branches characterized in guard cells can guide future work in functional genomic, cell biological, biophysical, and biochemical characterizations. Analyses of expression data show differences in guard cell and mesophyll cell expression profiles (Figures 5 and 6, Table 1, see Supplemental Tables 1 and 2 online). Importantly, the results indicate that a relatively small fraction of genes are expressed in a strictly cell type–specific manner. Note, however, that the relative expression level of a given gene or protein is an additional important factor and can differ among cell types. This finding supports a model in which combinatorial and temporal permutation in gene expression profiles and transcript processing are important for achieving specialized functions of cells, a model that is also being strongly considered for other organisms, in which relatively small genomes give rise to a high level of sophistication (e.g., The International Human Genome Mapping Consortium, 2001).
Identification of New ABA-Regulated Genes
Only a few ABA-regulated genes had been reported until recently. The present guard cell expression study and recently reported analyses of mRNA extracted from whole seedlings or whole plants (Seki et al., 2001, 2002; Hoth et al., 2002) show that the mRNA levels of many genes are ABA regulated, providing additional marker genes for mutant analyses. Interestingly, the presented data also show differences in ABA regulation of gene expression in these different studies (Tables 2 and 3, see Supplemental Tables 2 and 3 online). This is likely because of the differences in methods used, including differences in plant materials (whole plants versus guard cells), differences in the methods for growing plants and ABA treatments, and differences in microarray technologies (full-length cDNA versus computationally selected oligonucleotide probes). Our results, therefore, together with Seki et al. (2002) and Hoth et al. (2002), provide diverse resources of ABA-regulated genes with different spectrums in genes identified from microarray experiments.
On the basis of microarray and RT-PCR analyses presented here, we found 190 ABA-induced genes and 118 ABA-repressed genes, which have been grouped according to cell type expression (Figure 5). Among ABA-induced genes, 69 were induced preferentially in guard cells (Figures 5A and 5B, Table 2), 100 in mesophyll cells (Figures 5A and 5B, see Supplemental Table 3 online), and 21 in both cell types (Figures 5A and 5B, Table 2, see Supplemental Table 3 online). Differences in ABA-induced genes in guard cells and mesophyll cells may in part be the result of differential accessibility of these cells to exogenous ABA.
The ABA upregulated genes reported here can be classified in two major classes with respect to their putative functions: (1) genes implicated in cell protection and in the production of important metabolic proteins and (2) genes implicated in signal transduction. The first class includes proteins that probably function in stress tolerance, such as enzymes required for the biosynthesis of osmoprotectants (sugars) and proteins that may protect macromolecules and membranes (LEA and Chaperones). The second class contains proteins that may function in signal transduction and stress tolerance, such as protein kinases, protein phosphatases, receptor kinases, transcription factors, 14-3-3 proteins, reactive oxygen turnover enzymes (NADPH oxidases, catalase, and glutathione S-transferase), and protein turnover mechanisms (ubiquitination). The identification and characterization of these putative regulatory proteins will contribute to further analyses of the ABA signaling network in plants. Moreover, the study of uncharacterized transcription factors, which are highly induced in response to ABA, may provide clues toward further characterizing a transcriptional control network of genes that mediate physiological ABA responses.
In addition, we show here that the transcripts of 118 genes are repressed by ABA; 64 of these are preferentially ABA repressed in guard cells (Figures 5A and 5C, Table 3), 51 in mesophyll cells (Figures 5A and 5C, see Supplemental Table 4 online), and three in both cell types (Figures 5A and 5C, Table 3, see Supplemental Table 4 online). Until now, mainly photosynthesis-related genes have been characterized as repressed by ABA (Weatherwax et al., 1996), but our data show that several genes that function in signal transduction and osmoregulation, such as protein kinases and the K+ channels KAT1, KAT2, and AKT2, are also repressed by ABA.
Statistical Analysis of ABA-Responsive cis-Elements
Several cis-regulatory elements have been shown previously to contribute to ABA responses of individual genes such as the G-box–containing ABREs, the coupling elements CE1 and CE3, the RY/Sph elements, and the recognition sequences for the MYB and MYC class of transcription factors (reviewed in Busk and Pages, 1998). In this study, analysis of the promoter regions of ABA-induced genes shows that promoters containing one or more copies of an ABRE-like motif, (C/T)ACGTG(G/T)(A/T), and a motif similar to the H. vulgare coupling element CE3 are significantly overrepresented in the set of ABA induced genes (Table 4, row 1). Interestingly, ABRE-like elements are not found for all ABA-induced genes, and the large number of new ABA-induced or -repressed genes described in this study provides a basis for combinatorial analyses of different elements and a better understanding of the contribution of new cis-acting elements to ABA-dependent gene expression and cell type specificity.
ABA Modulation of Guard Cell ABA Signal Transduction Components
To date, relatively few genes acting in ABA signaling have been identified using genetic screens based on whole-plant or whole-tissue phenotypes (see Introduction). The guard cell system has been adapted and developed to dissect where and how ABA signaling mechanisms function within the network. Unexpectedly, we have found that all of the known ABA signal transducers and negative regulators of ABA signaling represented on the GeneChip are ABA modulated at the transcript level in guard cells (Figure 7). These findings suggest the existence of feedback regulation or parallel ABA regulation of transcript levels, which may occur via transcription and mRNA processing. Recent studies suggest the importance of RNA processing in ABA signal transduction (Lu and Fedoroff, 2000; Hugouvieux et al., 2001; Xiong et al., 2001a; Li et al., 2002). These data indicate that modulation of mRNA levels may be of broad general importance for early ABA signal transduction. Therefore, ABA-mediated expression profiling provides an approach for selection of putative signal transducers that are members of large gene families (Figure 8) or are genes presently of unknown function for biochemical and cell biological analyses.
The large number of multimember gene families found in plant genomes likely causes redundancies and partial redundancies, thus greatly limiting the number of genes that can be identified in conventional genetic mutant screens. Furthermore, knockout mutant analyses based on the many possible combinations of double or triple mutants of homologous genes are limited without knowledge of cell type–specific expression. In one of many possible examples, 10 NADPH oxidase genes are found in the Arabidopsis genome, giving rise to 45 possible double mutant combinations. We identified two ABA-induced NADPH oxidase Atrboh genes expressed at high levels in guard cells from the presented data sets. A double mutant in these two AtrbohD and AtrbohF genes shows ABA insensitivity in ABA activation of plasma membrane calcium permeable ICa channels (Kwak et al., 2003). In another example, assuming ≥69 PP2C genes found in Arabidopsis, ≥2346 distinct double mutant lines could be generated, and no recessive gene deletion mutant phenotypes have been reported previously for plant PP2Cs. Among 69 genes encoding PP2Cs in the whole genome of Arabidopsis, only four of those present on the array are highly expressed in guard cells. From our published findings using the guard cell expression data sets presented here, analyses of present genes (called Present by Affymetrix Microarray Suite 5.0 software) and selection, in particular, of highly expressed genes for functional genomic analyses can lead to identification of new players and new mechanisms that mediate guard cell and/or ABA signal transduction (e.g., Figure 8; Kwak et al., 2003). This approach circumvents the redundancy problem in signal transduction. Moreover, this approach will also be valuable for characterizing the functions of many highly expressed genes that are presently annotated as of unknown function.
We identified the AtP2C-HA gene (Rodriguez et al., 1998) as one of the most strongly ABA-induced PP2C mRNA in guard cells and one of the most highly expressed among the PP2Cs. To determine whether AtP2C-HA may play a role in ABA signaling, we characterized a T-DNA disruption mutant atp2C-HA-1. The atp2C-HA-1 mutant shows ABA-hypersensitive regulation of stomatal closing (Figure 8C) and seed germination (Figure 8D). The ABA hypersensitivity is complemented by the AtP2C-HA cDNA (Figures 8C and 8D). These findings suggest that AtP2C-HA is a component of ABA signaling, acting as a negative regulator of the analyzed ABA responses.
In conclusion, genomic scale cell type–specific expression profiling provides a powerful tool for narrowing down the genome and greatly facilitates the determination of the functions of highly expressed and ABA-regulated genes belonging to large gene families or to genes of unknown function in plants. The identification of an ABA-hypersensitive disruption mutant in the AtP2C-HA gene illustrates the power of this approach. These analyses allow focused cell type–dependent phenotypic and mechanistic characterizations.
METHODS
Plant Growth and ABA Treatment
Wild-type Arabidopsis plants, ecotype Columbia, were grown in soil in a plant growth chamber with a 16-h-light (80 μE light fluence rate) and 8-h-dark cycle at 20°C and 40% humidity. After 5 to 6 weeks, plants were sprayed with 100 μM ABA (+/− cis,trans ABA; Sigma, St. Louis, MO) or with water 4 h before cell isolations at the same time of day, after 1 h light, to eliminate variation as a result of diurnal changes in gene expression. In addition, cell isolations from ABA-treated and control-treated plants were performed simultaneously to eliminate variation because of growth conditions.
Isolation of Guard Cell Protoplasts
Arabidopsis rosette leaves of ∼100 plants were excised and blended for 3 min with a Waring blender at room temperature in water in the presence of actinomycin D (33 mg/L) and cordycepin (100 mg/L). Epidermal fragments were collected and filtered through a nylon mesh (pore size 100 μm). To remove epidermal and mesophyll cells, epidermal peels from ∼1600 leaves were transferred into a flask containing 50 mL of 0.7% Cellulysin (Calbiochem, San Diego, CA), 0.01% polyvinylpyrrolidone 40, 0.25% BSA, 55% basic medium, pH 5.5, and incubated at 27°C in a linear shaking water bath in the dark. The basic medium contained 0.5 mM CaCl2, 0.5 mM MgCl2, 0.01% cordycepin, 0.0033% actinomycin D, 5 mM Mes, pH 5.5, and the osmolarity was adjusted to 500 mmol/kg by addition of d-sorbitol. After 3 h of incubation, epidermal fragments were examined under a microscope. If few mesophyll cells remained, incubation was continued for an additional 30 min. When all epidermal and mesophyll cells were digested, epidermal fragments were collected on a nylon mesh (pore size 100 μm) and rinsed two to three times with the basic medium. After washing, epidermal fragments were transferred to a flask containing 50 mL of basic medium containing 1.5% Cellulase RS (Yakult, Tokyo, Japan), 0.03% Pectolyase Y23 (Seishin Pharmaceutical, Tokyo, Japan), 0.25% BSA (Sigma), 0.01% cordycepin (Sigma), 0.0033% actinomycin D (Sigma), pH 5.5, and incubated at 19°C in a shaking water bath (70 rpm) until guard cell protoplasts were released, usually after ∼2 h. The solution containing guard cell protoplasts was filtered through four layers of a nylon mesh (pore size 10 μm), and guard cell protoplasts were collected by centrifugation at 1000g for 5 min. This last step was repeated three to four times to keep guard cell protoplasts free from other cells or cell fragment contamination. Guard cells and mesophyll cells were collected simultaneously, and RNA was extracted, thus resulting in equal durations from leaf excision to RNA extractions for guard cells and mesophyll cells. Guard cell purity was determined using a hemacytometer. The guard cell protoplasts that showed a purity of ∼99% were selected for further experiments. To prevent changes of gene expression during protoplast isolation, two different transcription inhibitors, actinomycin D (33 mg/L) and cordycepin (100 mg/L), were used in all procedures of the isolation, including the first step of blending. Total RNA from guard cell protoplasts was then extracted using Trizol reagent according to the manufacturer's instructions (Life Technologies, Cleveland, OH). For each of the 18 independent guard cell RNA extractions, approximately 5 μg of total RNA were obtained from 109 guard cell protoplasts, which were from ∼100 plants.
Isolation of Mesophyll Cell Protoplasts
Five grams of rosette leaves were cut from Arabidopsis plants treated with ABA or water and placed in a Petri dish containing 0.5 mM CaCl2, 0.5 mM MgCl2, 5 mM MES, 1.5% Cellulase RS, 0.03% Pectolyase Y23, 0.25% BSA, actinomycin D (33 mg/L), and cordycepin (100 mg/L), pH 5.5. Osmolarity was adjusted to 550 mmol/kg by addition of d-sorbitol. Leaves were chopped with a sterile razor blade into small squares of 5 to 10 mm2 and incubated for 2 h at room temperature in the dark with gentle agitation until mesophyll cell protoplasts were released. The protoplast suspension was filtered through a nylon mesh (30 μm), washed several times, and resuspended with the basic medium under the same shaking conditions (70 rpm) until guard cells were ready in parallel isolations (see above). Cell purity was examined by a hemacytometer, and total RNA was extracted using Trizol reagent.
GeneChip Microarray Experiments and Data Analyses
Ten micrograms of total RNA from three independent protoplast extractions were pooled and used for each DNA chip hybridization, representing a total of 30 independent RNA isolations. Affymetrix GeneChips (Santa Clara, CA) were used, representing ∼8100 Arabidopsis genes. The transcript population of guard cells and mesophyll cells treated by ABA or water was amplified, labeled, and hybridized at the University of California, Irvine, and University of California, San Diego Gene Chip Core facilities. For each condition (with or without ABA), two (mesophyll cell) or three (guard cell) independent hybridizations were performed. Control and ABA-treated plants were grown in parallel for each isolation. The isolations took place over a period of 18 months and showed reproducibility (Figure 2, see Supplemental Figure 1). A set of spiked biotin-labeled bacterial RNAs, all at varying total concentrations, was added to the samples at the time of hybridization and included in the overall intensity normalization. The data were first analyzed with Microarray Suite 5.0 software (Affymetrix). For each microarray, overall intensity normalization for the entire probe sets was performed using Affymetrix Microarray Suite 5.0. Using the GeneChip Suite 5.0 default parameters, the detection P-value and the signal value were calculated for each probe set from each independent guard cell and mesophyll cell hybridization. The detection P-value generated by this analysis allowed us to determine whether a transcript is reliably detected (Present, P-value <0.04), and the signal value assigns a relative measure of abundance to the transcript. The data were then passed through a quality filter using Excel (Microsoft, Redmond, WA), through which transcripts were selected for further analyses according to the following ad hoc criteria: a significantly expressed transcript must have been significantly expressed in at least two samples obtained under the same condition (P for Present, detection P-value <0.04. In the case of guard cells, a third experiment call of M for Marginal was allowed, detection P-value <0.06 in Affymetrix nomenclature). Note that low abundance transcripts may actually encode important cellular proteins even if statistical analyses define these as Absent (A).
For the comparison analyses reported in the tables, in the study of ABA responses and cell type expression, each sample was compared with the corresponding control-treated sample from plants grown at the same times and samples isolated in parallel using Affymetrix Microarray Suite 5.0, which provides values as the log2 of the intensity ratio (signal log2 ratio). The signal log2 ratio corresponding to the change in expression level determined for each probe set was calculated by comparing each probe pair from one array to the corresponding probe pair on the other array. Then, using the signal log2 ratio value, we calculated the fold change for each probe set according to the following formula:
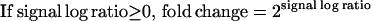 |
 |
In addition, for all comparison analyses that we performed, the change P-value, measuring the probability that the expression levels of each probe set are different between two arrays, was calculated. When the change P-value is close to 0.5, they are likely to be the same. When the change P-value is close to 0, the expression level in the ABA-treated array is higher than that of the control array. When the change P-value is close to 1, the expression level in the ABA-treated array is lower than that of the control array (Affymetrix). Only probe sets that showed significant differences in the two independent comparison analyses for mesophyll cells or in the three independent comparison analyses for guard cells were selected.
To perform cluster analysis (Figure 5), the statistically normalized preselected data of each array were analyzed using GeneSpring 4.2 software (Silicon Genetics, Redwood, CA) as replicate for mesophyll cells and triplicate for guard cells, and a second type of normalization, a per-gene normalization, was applied across all experiments using algorithms implemented in the GeneSpring program. This normalization method, also referred to as normalizing to the median for each gene, is intended to remove the differing intensity scales and binding rates from multiple experimental readings, allowing comparisons of multiple GeneChip hybridizations. It normalizes each gene to itself, so the median of all of the measurements is 1 for each replicate or triplicate experiment. To calculate this, GeneSpring divides each gene's signal by the median of all the signals for that particular gene. This normalization allows comparison of the relative changes in gene expression levels and display of these levels on a similar scale on the same graph. The expression pattern clusters were predefined by subjecting the relative differential expression data set obtained as described above to K-means clustering algorithms (K = 6), as implemented in the GeneSpring program using the standard correlation distance definition.
Affymetrix probe identifiers were matched with Arabidopsis Genome Initiative (AGI) identifiers by Basic Local Alignment Search Tool analysis of the published target sequences against the Institute for Genomic Research (TIGR) ATH1 sequences. For ∼5% of the detected transcripts, we were unable to unambiguously identify a gene in the ATH1 genome sequence corresponding to that probe set. These genes are listed in the tables as no hit. A full description of how we identified the AGI identifiers for the probe sets is available at http://www-biology.ucsd.edu/labs/schroeder/index.html, click on Genechip (Ghassemian et al., 2001). Note that if less stringent matches are allowed for annotation of the Affymetrix probes, some of the no-hit probes listed in the tables may conceivably cross-hybridize with known genes (see supplemental data online, http://www-biology.ucsd.edu/labs/schroeder/index.html, and http://genetics.mgh.harvard.edu/sheenweb.html). In addition to the supplemental data online, the complete set of raw Excel data files from Affymetrix chip experiments have been deposited at http://www-biology.ucsd.edu/labs/schroeder/guardcellchips.html, allowing users to pursue additional analyses and to develop functional genomic applications in guard cells and mesophyll cells.
RT-PCR Analyses
The expression profiles obtained from chip hybridizations were further validated by RT-PCR using first-strand cDNA synthesized from independently isolated RNA samples. Two micrograms of total RNA was converted into cDNA. Each cDNA was diluted 50 times, and 1 μL of cDNA was used for 27 cycle three-step PCR. RT-PCR amplifications were performed with 29 genes that were preferentially expressed in guard cells and showed transcript level induction or repression in response to ABA preferentially in guard cells or in both cell types. The Actin2 gene (forward primer, 5′-ggccgatggtgaggatattcagccacttg-3′; reverse primer, 5′-tcgatggacctgactcatcgtactcactc-3′) was used as a control for RT-PCR experiments. Guard cell preferentially expressed genes include a calcium-dependent protein kinase gene (forward primer, 5′-tggtagtgcatactatgtagc-3′; reverse primer, 5′-gacacaccatgtagcaat-3′) and CER2 (forward primer, 5′-cataccttacattcgctgc-3′; reverse primer, 5′-actacacttacaatccttgc-3′). ABA-induced genes preferentially expressed in guard cells include a trehalose-6-phosphate synthase (forward primer, 5′-ggcttctaatcctgggt-3′; reverse primer, 5′-ggtggtcttcggtctc-3′) and a LEA (forward primer, 5′-gaggaaagtgtacggtttg-3′; reverse primer, 5′-cgtcatatcgctcgcc-3′). The KAT1 gene (forward primer, 5′-ccgatcttctaccatctcttggagccagg-3′; reverse primer, 5′-agttgcagcctccaaacttctcacttgc-3′) is an ABA-repressed gene preferentially expressed in guard cells. ABA-induced genes in guard cells and mesophyll cells include a dehydrin (forward primer, 5′-aagattaaagagcaactgcc-3′; reverse primer, 5′-aacgaaaccagaagtagatatt-3′), the protein phosphatase AtP2C-HA (forward primer, 5′-taccgctttggggcac-3′; reverse primer, 5′-ggacctagacatggcga-3′), and COR47 (forward primer, 5′-ctgaaccggagttagc-3′; reverse primer, 5′-cttggcatgataacctgga-3′). The following AtP2C-HA primers were used to confirm AtP2C-HA cDNA expression in the complemented lines: forward primer, 5′-atggaggagatgactcccgcagtt-3′; reverse primer, 5′-tcaggttctggtcttgaactttctt-3′.
Promoter Analyses
The upstream nucleotide sequences used were obtained from the file At.upstream.1000.20020107 obtained from the FTP site of the Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org). A file of 500-bp sequences was derived from the above file by extracting 500 bp from the 3′ end of each sequence. The association between the Affymetrix probe sets and the AGI identifiers used to identify the sequences was determined by Basic Local Alignment Search Tool analysis of the Affymetrix target sequences against the TIGR nucleotide sequences. Stringent criteria were applied to matches (http://www-biology.ucsd.edu/labs/schroeder/howandwhy.html). We were able to unambiguously identify upstream sequences for 7749 of the genes on the GeneChip. Motif searches were performed by writing custom Python scripts (http://www.python.org) and C extensions, which search for both exact and approximate matches to a pattern. Both forward and reverse strands were searched, and some patterns contained wildcard nucleotides (e.g., S = C or G, Y = pyrimidine). Because the degree of conservation of many of these patterns is unknown, each pattern was searched using criteria of 0 or 1 allowed mismatches (Table 4). The P-values given in the table are based on an upper tail hypergeometric cumulative distribution function and represent the probability of obtaining at least as many hits as were observed in the set, if the set were a random sample drawn without replacement from the complete set of promoter sequences. The equation used was:
 |
where N = population size, R = number of hits in the population, n = sample size, and r = number of hits in the sample.
Stomatal Aperture Measurements
Leaves of 5- to 6-week-old wild type, atp2C-HA, and complemented atp2C-HA-1 plants were used for stomatal aperture measurements. For Figure 8C, epidermal strips were peeled and incubated 2 h in white light in stomatal opening solution containing 10 mM KCl, 7.5 mM iminodiacetic acid, and 10 mM MES/Tris, pH 6.2, at 20°C.
Seed Germination Analyses
Seeds of the wild type and atp2C-HA were plated on Murashige and Skoog medium (Sigma) containing 0, 0.2, 0.4, 0.6, or 1 μM ABA. Seeds were stratified at 4°C for 4 d and then transferred to a growth chamber (24°C under 16-h-light/8-h-dark regime). Seed germination rates were scored after 6 d in the growth chamber. Germination is defined as the emergence of both cotyledons and roots from the seed.
Complementation of the atp2C-HA-1 Mutant
A 1.54-kb fragment corresponding to the AtP2C-HA coding region was amplified by RT-PCR from Arabidopsis cDNA with forward (5′-ATCCATGGAGGAGATGACTCCCGCAGTT-3′) and reverse (5′-TAGCTAGCTCAGGTTCTGGTCTTGAACTTTCTT-3′) primers. The PCR product was digested with NcoI and NheI and cloned into the NcoI and PmlI sites of the binary vector pCAMBIA 1302 (CAMBIA, Canberra, Australia) to express the AtP2C-HA coding sequence under the 35S promoter of the Cauliflower mosaic virus. This construct was verified by DNA sequencing and transformed into the atp2C-HA-1 mutant plants by the floral dip method using the Agrobacterium tumefaciens strain C58. Transgenic lines were selected on Murashige and Skoog medium containing 25 μg/mL hygromycin.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Grant MCB 0077791, National Institutes of Health Grant R01GM060396, and partly by U.S. Department of Energy Grant DE-FG02-03ER15449 to J.I.S. and fellowships to J.M.K. and N.L. from the Human Frontier Science Program Organization.
On-line version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Julian Schroeder (julian@ucsd.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.019000.
References
- Allen, G.J., Chu, S.P., Harrington, C.L., Schumacher, K., Hoffmann, T., Tang, Y.Y., Grill, E., and Schroeder, J.I. (2001). A defined range of guard cell calcium oscillation parameters encodes stomatal movements. Nature 411, 1053–1057. [DOI] [PubMed] [Google Scholar]
- Allen, G.J., Murata, Y., Chu, S.P., Nafisi, M., and Schroeder, J.I. (2002). Hypersensitivity of abscisic acid-induced cytosolic calcium increases in the Arabidopsis farnesyltransferase mutant era1–2. Plant Cell 14, 1649–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.A., Huprikar, S.S., Kochian, L.V., Lucas, W.J., and Gaber, R.F. (1992). Functional expression of a probable Arabidopsis thaliana potassium channel in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 89, 3736–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew, D.M., Bartley, G.E., and Scolnik, P.A. (1991). Abscisic acid control of rbcs and cab transcription in tomato leaves. Plant Physiol. 96, 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, J.D., Boavida, L.C., Carneiro, J., Haury, M., and Feijo, J.A. (2003). Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol. 133, 713–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, W., Sun, W.N., Hohmann, S., Wera, S., Reinders, A., De Virgilio, C., Wiemken, A., and Thevelein, J.M. (1998). Composition and functional analysis of the Saccharomyces cerevisiae trehalose synthase complex. J. Biol. Chem. 273, 33311–33319. [DOI] [PubMed] [Google Scholar]
- Birnbaum, K., Shasha, D.E., Wang, J.Y., Jung, J.W., Lambert, G.M., Galbraith, D.W., and Benfey, P.N. (2003). A gene expression map of the Arabidopsis root. Science 302, 1956–1960. [DOI] [PubMed] [Google Scholar]
- Blatt, M.R., and Armstrong, F. (1993). K+ channels of stomatal guard cells: Abscisic-acid-evoked control of the outward rectifier mediated by cytoplasmic pH. Planta 191, 330–341. [Google Scholar]
- Busk, P.K., and Pages, M. (1998). Regulation of abscisic acid-induced transcription. Plant Mol. Biol. 37, 425–435. [DOI] [PubMed] [Google Scholar]
- Chang, Y.C., and Walling, L.L. (1991). Abscisic acid negatively regulates expression of chlorophyll a/b binding protein genes during soybean embryogeny. Plant Physiol. 97, 1260–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W.Q., et al. (2002). Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14, 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherel, I., Michard, E., Platet, N., Mouline, K., Alcon, C., Sentenac, H., and Thibaud, J.B. (2002). Physical and functional interaction of the Arabidopsis K+ channel AKT2 and phosphatase AtPP2CA. Plant Cell 14, 1133–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler, S., Ghassemian, M., Bonetta, D., Cooney, S., and McCourt, P. (1996). A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science 273, 1239–1241. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R. (1994). Mutations at two new Arabidopsis ABA response loci are similar to abi3 mutations. Plant J. 5, 765–771. [Google Scholar]
- Finkelstein, R.R., Gampala, S.S.L., and Rock, C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14, S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Lynch, T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini, S., and McCourt, P. (2001). Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr. Opin. Plant Biol. 4, 387–391. [DOI] [PubMed] [Google Scholar]
- Ghassemian, M., Waner, D., Tchieu, J., Gribskov, M., and Schroeder, J.I. (2001). An integrated Arabidopsis annotation database for Affymetrix Genechip(R) data analysis, and tools for regulatory motif searches. Trends Plant Sci. 6, 448–449. [DOI] [PubMed] [Google Scholar]
- Gilmour, S.J., Artus, N.N., and Thomashow, M.F. (1992). cDNA sequence analysis and expression of two cold-regulated genes of Arabidopsis thaliana. Plant Mol. Biol. 18, 13–22. [DOI] [PubMed] [Google Scholar]
- Giraudat, J., Hauge, B.M., Valon, C., Smalle, J., Parcy, F., and Goodman, H.M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4, 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer, S.L., Hogenesch, J.B., Straume, M., Chang, H.-S., Han, B., Zhu, T., Wang, X., Kreps, J.A., and Kay, S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113. [DOI] [PubMed] [Google Scholar]
- Honys, D., and Twell, D. (2003). Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 132, 640–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoth, S., Morgante, M., Sanchez, J.P., Hanafey, M., Tingey, S., and Chua, N.-H. (2002). Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1–1 mutant. J. Cell Sci. 115, 4891–4900. [DOI] [PubMed] [Google Scholar]
- Hughes, J.D., Estep, P.W., Tavazoie, S., and Church, G.M. (2000). Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 296, 1205–1214. [DOI] [PubMed] [Google Scholar]
- Hugouvieux, V., Kwak, J.M., and Schroeder, J.I. (2001). An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106, 477–487. [DOI] [PubMed] [Google Scholar]
- The International Human Genome Mapping Consortium. (2001). A physical map of the human genome. Nature 409, 934–941. [DOI] [PubMed] [Google Scholar]
- Kiyosue, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). Characterization of two cDNAs (ERD10 and ERD14) corresponding to genes that respond rapidly to dehydration stress in Arabidopsis thaliana. Plant Cell Physiol. 35, 225–231. [PubMed] [Google Scholar]
- Kwak, J.M., Kim, S.A., Hong, S.W., and Nam, H.G. (1997). Evaluation of 515 expressed sequence tags obtained from guard cells of Brassica campestris. Planta 202, 9–17. [DOI] [PubMed] [Google Scholar]
- Kwak, J.M., Moon, J.-H., Murata, Y., Kuchitsu, K., Leonhardt, N., DeLong, A., and Schroeder, J.I. (2002). Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 14, 2849–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak, J.M., Mori, I.C., Pei, Z.-M., Leonhardt, N., Torres, M.A., Dangl, J.L., Bloom, R.E., Bodde, S., Jones, J.D.G., and Schroeder, J.I. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22, 2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H., Xiong, L., Gong, Z., Ishitani, M., Stevenson, B., and Zhu, J.-K. (2001). The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 15, 912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemtiri-Chlieh, F. (1996). Effects of internal K+ and ABA on the voltage- and time-dependence of the outward K+-rectifier in Vicia guard cells. J. Membr. Biol. 153, 105–116. [DOI] [PubMed] [Google Scholar]
- Lemtiri-Chlieh, F., and MacRobbie, E.A.C. (1994). Role of calcium in the modulation of Vicia guard cell potassium channels by abscisic acid: A patch-clamp study. J. Membr. Biol. 137, 99–107. [DOI] [PubMed] [Google Scholar]
- Leung, J., Merlot, S., and Giraudat, J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Kinoshita, T., Pandey, S., Ng, C.K.-Y., Gygi, S.P., Shimazaki, K.-I., and Assmann, S.M. (2002). Modulation of an RNA-binding protein by abscisic-acid-activated protein kinase. Nature 418, 793–797. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina, L., and Chua, N.-H. (2000). A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 41, 541–547. [DOI] [PubMed] [Google Scholar]
- Lu, C., and Fedoroff, N. (2000). A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12, 2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRobbie, E.A.C. (1998). Signal transduction and ion channels in guard cells. Phil. Trans. Roy. Soc. London 1374, 1475–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäser, P., Leonhardt, N., and Schroeder, J.I. (2003). The clickable guard cell: Electronically linked model of guard cell signal transduction pathways. In The Arabidopsis Book, http://www-biology.ucsd.edu/labs/schroeder/clickablegc.html. [DOI] [PMC free article] [PubMed]
- Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., and Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25, 295–303. [DOI] [PubMed] [Google Scholar]
- Murata, Y., Pei, Z.-M., Mori, I.C., and Schroeder, J.I. (2001). Abscisic acid activation of plasma membrane Ca2+ channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1–1 and abi2–1 protein phosphatase 2C mutants. Plant Cell 13, 2513–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli, A.-C., Merlot, S., Vavasseur, A., Fenzi, F., and Giraudat, J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14, 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, R.L., McKendree, W.L., Hirsch, R.E., Sedbrook, J.C., Gaber, R.F., and Sussman, M.R. (1995). Expression of an Arabidopsis potassium channel gene in guard cells. Plant Physiol. 109, 371–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander, M., Svensson, J., Palva, E.T., and Welin, B.V. (2001). Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol. Biol. 45, 263–279. [DOI] [PubMed] [Google Scholar]
- Okamura, J.K., and Goldberg, R.B. (1989). Regulation of plant gene expression: General principles. In The Biochemistry of Plants, Vol. 115, P.K. Stumpf and E.E. Conn, eds (New York, NY: Academic Press), pp. 1–82.
- Ozturk, Z.N., Talame, V., Deyholos, M., Michalowski, C.B., Galbraith, D.W., Gozukirmizi, N., Tuberosa, R., and Bohnert, H.J. (2002). Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley. Plant Mol. Biol. 48, 551–573. [DOI] [PubMed] [Google Scholar]
- Pei, Z.-M., Ghassemian, M., Kwak, C.M., McCourt, P., and Schroeder, J.I. (1998). Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 282, 287–290. [DOI] [PubMed] [Google Scholar]
- Pei, Z.-M., Kuchitsu, K., Ward, J.M., Schwarz, M., and Schroeder, J.I. (1997). Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell 9, 409–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, Z.-M., Murata, Y., Benning, G., Thomine, S., Klusener, B., Allen, G.J., Grill, E., and Schroeder, J.I. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406, 731–734. [DOI] [PubMed] [Google Scholar]
- Perez-Amador, M.A., Lidder, P., Johnson, M.A., Landgraf, J., Wisman, E., and Green, P.J. (2001). New molecular phenotypes in the dst mutants of Arabidopsis revealed by DNA microarray analysis. Plant Cell 13, 2703–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe, S., Reinbothe, C., and Parthier, B. (1993). Methyl jasmonate represses translation initiation of a specific set of mRNAs in barley. Plant J. 4, 459–467. [Google Scholar]
- Rodriguez, P.L., Benning, G., and Grill, E. (1998). ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett. 421, 185–190. [DOI] [PubMed] [Google Scholar]
- Romero, C., Belles, J.M., Vaya, J.L., Serrano, R., and Culianezmacia, F.A. (1997). Expression of the yeast trehalose-6-phosphate synthase gene in transgenic tobacco plants: Pleiotropic phenotypes include drought tolerance. Planta 201, 293–297. [DOI] [PubMed] [Google Scholar]
- Schachtman, D., Schroeder, J.I., Lucas, W.J., Anderson, J.A., and Gaber, R.F. (1992). Expression of an inward-rectifying potassium channel by the Arabidopsis KAT1 cDNA. Science 258, 1654–1658. [DOI] [PubMed] [Google Scholar]
- Schena, M., Shalon, D., Davis, R.W., and Brown, P.O. (1995). Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470. [DOI] [PubMed] [Google Scholar]
- Schroeder, J.I., Kwak, J.M., and Allen, G.J. (2001). Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410, 327–330. [DOI] [PubMed] [Google Scholar]
- Schwartz, A., Wu, W.-H., Tucker, E.B., and Assmann, S.M. (1994). Inhibition of inward K+ channels and stomatal response by abscisic acid: An intracellular locus of phytohormone action. Proc. Natl. Acad. Sci. USA 91, 4019–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., et al. (2002). Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct. Integr. Genomics 2, 282–291. [DOI] [PubMed] [Google Scholar]
- Seki, M., Narusaka, M., Abe, H., Kasuga, M., Yamaguchi-Shinozaki, K., Carninci, P., Hayashizaki, Y., and Shinozaki, K. (2001). Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen, J., Zhou, L., and Jang, J.C. (1999). Sugars as signaling molecules. Curr. Opin. Plant Biol. 2, 410–418. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. [PubMed] [Google Scholar]
- Wang, R.C., Guegler, K., LaBrie, S.T., and Crawford, N.M. (2000). Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12, 1491–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.-Q., Ullah, H., Jones, A.M., and Assmann, S.M. (2001). G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292, 2070–2072. [DOI] [PubMed] [Google Scholar]
- Wanner, L.A., Li, G.Q., Ware, D., Somssich, I.E., and Davis, K.R. (1995). The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol. Biol. 27, 327–338. [DOI] [PubMed] [Google Scholar]
- Weatherwax, S.C., Ong, M.S., Degenhardt, J., Bray, E.A., and Tobin, E.M. (1996). The interaction of light and abscisic acid in the regulation of plant gene expression. Plant Physiol. 111, 363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm, K.S., and Thomashow, M.F. (1993). Arabidopsis thaliana cor15b, an apparent homologue of cor15a, is strongly responsive to cold and ABA, but not drought. Plant Mol. Biol. 25, 1073–1077. [DOI] [PubMed] [Google Scholar]
- Xia, Y.J., Nikolau, B.J., and Schnable, P.S. (1997). Developmental and hormonal regulation of the Arabidopsis CER2 gene that codes for a nuclear-localized protein required for the normal accumulation of cuticular waxes. Plant Physiol. 115, 925–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., Gong, Z., Rock, C.D., Subramanian, S., Guo, Y., Xu, W., Galbraith, D., and Zhu, J.-K. (2001. a). Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev. Cell 1, 771–781. [DOI] [PubMed] [Google Scholar]
- Xiong, L., Lee, B.-H., Ishitani, M., Lee, H., Zhang, C., and Zhu, J.-K. (2001. b). FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 15, 1971–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1993). Characterization of the expression of a desiccation-response RD29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol. Gen. Genet. 236, 331–340. [DOI] [PubMed] [Google Scholar]
- Yin, Y.H., Wang, Z.Y., Mora-Garcia, S., Li, J.M., Yoshida, S., Asami, T., and Chory, J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191. [DOI] [PubMed] [Google Scholar]
- Yoshida, R., Hobo, T., Ichimura, K., Mizoguchi, T., Takahashi, F., Alonso, J., Ecker, J.R., and Shinozaki, K. (2002). ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 43, 1473–1483. [DOI] [PubMed] [Google Scholar]
- Zhu, T., Budworth, P., Han, B., Brown, D., Chang, H.-S., Zou, G., and Wang, X. (2001). Toward elucidating the global gene expression patterns of developing Arabidopsis: Parallel analysis of 8300 genes by a high-density oligonucleotide probe array. Plant Physiol. Biochem. 39, 221–242. [Google Scholar]
- Zhu, J., Gong, Z., Zhang, C., Song, C.-P., Damsz, B., Inan, G., Koiwa, H., Zhu, J.-K., Hasegawa, P.M., and Bressan, R.A. (2002). OSM1/SPY61: A syntaxin protein in Arabidopsis controls abscisic acid–mediated and non-abscisic acid–mediated responses to abiotic stress. Plant Cell 14, 3009–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.