Abstract
Unlike adult mammals, adult zebrafish vigorously regenerate lost heart muscle in response to injury. The epicardium, a mesothelial cell layer enveloping the myocardium, is activated to proliferate after cardiac injury and can contribute vascular support cells or provide mitogens to regenerating muscle. Here, we applied proteomics to identify secreted proteins that are associated with heart regeneration. We found that Fibronectin, a main component of the extracellular matrix, is induced and deposited after cardiac damage. In situ hybridization and transgenic reporter analyses indicated that expression of two fibronectin paralogues, fn1 and fn1b, are induced by injury in epicardial cells, while the itgb3 receptor is induced in cardiomyocytes near the injury site. fn1, the more dynamic of these paralogs, is induced chamber-wide within one day of injury before localizing epicardial Fn1 synthesis to the injury site. fn1 loss-of-function mutations disrupted zebrafish heart regeneration, as did induced expression of a dominant-negative Fibronectin cassette, defects that were not attributable to direct inhibition of cardiomyocyte proliferation. These findings reveal a new role for the epicardium in establishing an extracellular environment that supports heart regeneration.
Keywords: zebrafish, heart regeneration, epicardium, extracellular matrix, cardiomyocyte, Fibronectin, Integrin
Introduction
Fetal and neonatal mice can regenerate lost regions of heart muscle through cardiomyocyte proliferation (Drenckhahn et al., 2008; Porrello et al., 2011), whereas the injured adult mammalian heart has a limited regenerative capacity. By contrast, zebrafish do not significantly lose regenerative potential as they mature, and can regenerate large portions of adult myocardium lost from resection, cryoinjury, or genetic ablation (Gonzalez-Rosa et al., 2011; Poss et al., 2002; Wang et al., 2011). In both zebrafish and mice, the epicardium, a mesothelial cell sheet that covers the heart, is activated to induce embryonic markers after cardiac damage (Lepilina et al., 2006). Epicardial cells have been studied as a source of paracrine signals, a supply of perivascular components or other cell types, and a mediator of inflammation during cardiac repair or regeneration (Huang et al., 2012; Kikuchi et al., 2011a; Smart et al., 2011; Zhou et al., 2011).
The extracellular matrix (ECM) has long been recognized as a key influence on organ formation and repair (Martino et al., 2011). During embryonic heart development, the ECM provides cues for assembly, proliferation and maturation of cardiac cell types (George et al., 1997; Ieda et al., 2009; Magnusson and Mosher, 1998; Trinh and Stainier, 2004). For instance, early cardiomyocytes that shape the zebrafish heart require the core ECM component Fibronectin (Fn) as they migrate toward the midline, a function that also explains observations during murine heart development (George et al., 1997; Trinh and Stainier, 2004). Fn deposition following cardiac damage in adult mammals has been previously documented, where it has been associated with adverse effects like fibrosis (Knowlton et al., 1992; Rysa et al., 2005; Willems et al., 1996; Zhong et al., 2010). Here, we find that Fn is dynamically produced by epicardial cells in response to cardiac injury, and that it is essential for heart regeneration.
Materials and methods
Zebrafish and cardiac injuries
Outbred Ekkwill zebrafish strains (4-10 months of age) were used for ventricular resection surgeries (Poss et al., 2002), or for genetic cardiomyocyte ablation (Wang et al., 2011). Animal density was maintained at approximately 4 fish per liter in all experiments. To ablate cardiomyocytes, animals were treated for 16 hours in 0.1 μM 4-hydroxytamoxifen (4-HT) in fish water. Transgenic tcf21:nucEGFP and zebrafish cardiac genetic ablation strains were previously described (Wang et al., 2011). Progeny from heterozygous matings were raised at a water temperature of 22°C to 2 months of age, then maintained at 26°C. fn1 homozygous mutants from these clutches were identified by PCR screening, as described (Trinh and Stainier, 2004). Heat-shock experiments were performed as described (Kikuchi et al., 2011b). Newly constructed strains are described below. All transgenic strains were analyzed as hemizygotes. All animal procedures were performed in accordance with Duke University guidelines.
fn1:mCherry-NTR
The translational start codon of fn1 in the BAC clone CH211-160E15 was replaced with the mCherry-NTR cassette by Red/ET recombineering technology (Gebe Bridges) (Singh et al., 2012). The 5′ and 3′ homologous arms for recombination were a 50-base pair (bp) fragment upstream and downstream of the start codon, and were included in PCR primers to flank the mCherry-NTR cassette. To avoid aberrant recombination between the mCherry-NTR cassette and endogenous loxp site in the BAC vector, we replaced the vector-derived loxp site with an I-SceI site using the same technology. The final BAC was purified with Nucleobond BAC 100 kit (Clontech), and co-injected with I-SceI into one-cell-stage zebrafish embryos. The full name of this transgenic line is Tg(fn1:mCherry-NTR)pd65.
itgb3:EGFP
The translational start codon of itgb3 in the BAC clone DKEYP-287G12 was replaced with the EGFP cassette by Red/ET recombineering technology (GeneBridges). The procedures of the homologous arm design, the loxp replacement strategy and BAC preparation are the same as described above. The full name of this transgenic line is Tg(itgb3:EGFP)pd66.
hsp70:dn-fnl1-9
A gene cassette encoding human fibronectin l1-9 fragment was PCR amplified with primers containing ClaI/ClaI (plasmid kindly provided by Harold Erickson) (Ohashi and Erickson, 2011). PCR products was gel-purified, digested with restriction enzymes, and ligated into an ClaI digested vector containing the 1.5 kb zebrafish hsp70 promoter (Halloran et al., 2000). The plasmid was injected into one-cell zebrafish embryos along with I-SceI to generate transgenic animals. The full name of this transgenic is: Tg(hsp70:fnl1-9)pd67.
Proteomics
Z-CAT zebrafish were treated with EtOH vehicle or 4-HT and ventricles were collected at 7 days post incubation (dpi), 14 dpi, and 30 dpi. Three separate pools of 3 hearts were collected for each time point. Proteomic analysis was performed using a label-free quantitative liquid chromatography – tandem mass spectrometry (LC-MS/MS) approach after tissue solubilization (Geromanos et al., 2009). The Duke Proteomics Core Facility received snap-frozen extracted zebrafish cardiomyocyte tissue. Each tissue sample was solubilized using a MS-compatible surfactant/ burst sonication procedure in which samples were suspended in 50 mM ammonium bicarbonate, pH 8 with 0.25% ALS-1 and subjected to 3X 10s probe sonication bursts at 30% power. Samples were spun at 15,000 rpm for 5 min and insoluble material was discarded. A Bradford assay (mini-Bradford, BioRad, Inc) of all samples was taken after protein isolation to determine protein yield. 25 μg protein from each sample was aliquoted and normalized to 1.0 μg/μL for reduction (10 mM DTT), cysteine alkylation (20 mM iodoacetamide), and trypsin digestion according to a standard protocol (http://www.genome.duke.edu/cores/proteomics/sample-preparation/documents/In-solutionDigestionProtocol_012309.doc). After digestion, all samples were spiked with ADH1_YEAST digest (Massprep standard, Waters Corporation) as a surrogate standard at a concentration of 50 fmol/ug, and acidified to a final concentration of 2% v/v acetonitrile and 1% trifluoroacetic acid. A sample “pool” to be used for column conditioning and QC purposes was generated by removing an equal quantity (5 μg) from each of the samples.
Quantitative LC/MS/MS was performed on 1 μg of protein digest per sample, using a nanoAcquity UPLC system (Waters Corp) coupled to a Synapt G2 HDMS high resolution accurate mass tandem mass spectrometer (Waters Corp.) via a nanoelectrospray ionization source. Briefly the sample was first trapped on a Symmetry C18 300 mm × 180 mm trapping column (5 μl/min at 99.9/0.1 v/v water/acetonitrile), after which the analytical separation was performed using a 1.7 um Acquity BEH130 C18 75 mm × 250 mm column (Waters Corp.) using a 90-min gradient of 5 to 40% acetonitrile with 0.1% formic acid at a flow rate of 300 nanoliters/minute (nL/min) with a column temperature of 45°C. Quantitative data collection for each sample in singlicate and on the sample pool (5x) on the Synapt G2 mass spectrometer was performed in data-independent acquisition mode (MSE) using 0.6 second alternating cycle time between low (6V) and high (15-40V) collision energy (CE) in the trapping region. Additional qualitative analyses were performed using the pooled sample in both ion-mobility assisted data-independent acquisition (HDMSE) mode or data-dependent acquisition (DDA) mode. Scans performed at low CE measure peptide accurate mass and intensity (abundance), while scans at elevated CE allow for qualitative identification of the resulting peptide fragments via database searching. The total analysis cycle time for each sample injection was approximately 2 hours.
The QC pool containing equivalent amounts of all samples was used to condition the UPLC column prior to the study and was run five times throughout the study for quantitative QC and five additional runs for supplementary qualitative identifications (for a total of 34 LC-MS/MS analyses). Treatment groups were evenly distributed across the run queue in a block design, and within each block the sample order was randomized. Following the analyses, data was imported into Rosetta Elucidator v3.3 (Rosetta Biosoftware, Inc), and all LC/LC-MS runs were aligned based on the accurate mass and retention time of detected ions (“features”) using PeakTeller algorithm (Elucidator). The relative peptide abundance was calculated based on area-under-the- curve (AUC) of aligned features across all runs. The overall dataset had 204,872 deisotoped features, and high collision energy (peptide fragment) data was collected in 370,834 spectra for sequencing by database searching. This MS/MS data was searched against an NCBI RefSeq database with danio rerio taxonomy (http://www.ncbi.nlm.nih.gov/protein/), which also contained a reversed-sequence “decoy” database for false positive rate determination. After individual peptide scoring using PeptideProphet algorithm (Elucidator), the data was annotated at a <1% peptide false discovery rate. This analysis yielded identifications for 3897 peptides and 545 proteins across all samples, including 325 proteins with two or more peptides quantified. For quantitative processing, the data was first curated to contain only high quality peptides with appropriate chromatographic peak shape and the dataset was intensity scaled to the robust mean across all samples analyzed; the final quantitative dataset for cardiomyocytes was based on 3744 peptides and contains 521 proteins. Protein expression within a sample was determined by summing the intensity of all peptides to a protein, and this sum was compared between groups using standard statistical tools.
Histological methods
In situ hybridization (ISH) was performed on 10 μm cryosections of paraformaldehyde-fixed hearts using digoxygenin-labeled cRNA probes as described (Poss et al., 2002) with the aid of an InSituPro robot (Intavis). Acid Fuchsin-Orange G staining was performed as described (Poss et al., 2002). Primary antibodies used in this study were: anti-Myosin heavy chain (MHC; F59, mouse; Developmental Studies Hybridoma Bank), anti-Mef2 (rabbit; Santa Cruz), anti-PCNA (mouse; Sigma), anti-DsRed (Rabbit; Clontech) and anti-Fibronectin (Sigma, F3648). Secondary antibodies (Invitrogen) used in this study were: Alexa Fluor 488 goat anti-rabbit; Alexa 594 goat anti-rabbit, goat anti-rat and goat anti-mouse antibodies.
Results
Fibronectin is expressed during heart regeneration
To identify proteins that are upregulated after cardiac injury and during regeneration, we used an unbiased proteomic profiling approach. We used the zebrafish cardiac ablation transgenic (Z-CAT) animals to induce cardiac damage by induced cardiomyocyte ablation, a procedure that ablates ∼60% or more of all cardiomyocytes diffusely throughout both chambers (Wang et al., 2011). These massive injuries provoke cardiomyocyte proliferation throughout the heart, with regeneration typically completing in 3 weeks. We performed LC-MS analysis on cardiac tissue at 7, 14, and 30 days post-injury (dpi). We observed the largest changes in differential protein expression at 7 dpa. Over the course of regeneration, proteomic profiles increasingly became more similar to the uninjured heart (Fig 1A). Proteins significantly induced at 7 dpa include proteins known to be induced during heart regeneration, such as Raldh2, Nppa, and Nppb (Gupta et al., 2013; Kikuchi et al., 2011b; Lepilina et al., 2006). Furthermore, we noted large changes in mitochondrial proteins such as Atp5j, Atp5h, Mt-co2, Ndufs1, Uqcrh, and Cycsb, consistent with ultrastructural changes observed in mitochondria during heart regeneration (Kikuchi et al., 2010). Fibronectin (Fn), a major ECM component, was highly induced at 7 and 14 dpi, showing the 16th largest fold-change at 7 dpi of 141 significantly differentially expressed proteins (Fig. 1A and B; the annotated dataset is provided in Table S1).
Fig. 1.

Fibronectin expression is upregulated during heart regeneration. (A) Heat-map of the 141 differentially regulated proteins during cardiac regeneration. Red indicates increased expression and blue indicates reduced expression. Raw data are shown in accompanying Supplemental dataset. (B) Results of quantitative proteomics show an increase of Fn in ablated hearts at 7 days post 4-HT or vehicle incubation (dpi) and 14 dpi compared to animals treated with vehicle. Student's t-test, *p < 0.05.
fibronectin paralogs are dynamically expressed during heart regeneration
To confirm this induction, we assessed ventricles injured locally by partial ventricular resection, using an antibody that recognizes gene products of the two zebrafish Fn paralogs, Fn1 and Fn1b, (Sun et al., 2005), and identified strong Fn immunoreactivity in injury sites at 7 days post-amputation (7 dpa) (Fig. 2A and B).
Fig. 2.
Fibronectin is dynamically expressed during heart regeneration. (A, B) Fibronectin expression by immunostaining in uninjured (A) and 7 dpa (B) ventricles, localizing to the injury site. MHC, Myosin heavy chain. (C-J) In situ hybridization for fn1 and fn1b in uninjured, 1, 7 and 30 dpa ventricles. In each section, violet indicates a positive signal. Dashed line indicates approximate resection plane. Scale bars: 50 μm.
To examine the expression patterns of individual Fn paralogs during heart regeneration, we performed in situ hybridization using riboprobes that are specific for either fn1 or fn1b. fn1 was undetectable in the uninjured heart (Fig. 2C), strongly induced on the periphery of the ventricular wall both near and away from the injury site by 1 dpa (Fig. 2D), and localized to the injured ventricular apex by 7 and 14 dpa (Fig. 2E and data not shown). By 30 dpa, a time when the myocardial wall has typically been replaced, we detected little or no fn1 (Fig. 2F). The fn1b paralog was expressed in cells all along the periphery of uninjured ventricles (Fig. 2G), was induced strongly at 1, 7, and 14 dpa in a similar manner as fn1 (Fig. 2H, I and data not shown), and retained expression in the ventricle at 30 dpa (Fig. 2J). These results revealed dynamic, organ-wide induction by both fn paralogs, possibly in the same cell type(s) within the ventricular wall and injured apex. Notably, the fn1 paralog displayed an injury profile that suggested a role specific to regeneration.
fn1 is induced predominantly in epicardial cells during heart regeneration
The immunofluoresence and in situ hybridization results suggested expression of one or both fn paralogs in epicardial cells, but were unable to definitively specify the source of Fn. Therefore, we generated a BAC transgenic reporter strain that would mark cells activating fn1 gene expression Tg(fn1:mCherry-NTR)pd65 (referred to hereafter as fn1:mCherry-NTR). This cassette includes a bacterial nitroreductase coding sequence that can in practice be used for ablation of expressing cells (Curado et al., 2007). Fluorescent protein expression in fn1:mCherry-NTR embryos largely recapitulated endogenous fn1 expression (Fig. 3A) (Rauch, 2003). In adults, fn1:mCherry-NTR expression was undetectable in the uninjured heart (Fig. 3B, C and D), but appeared to reproduce endogenous expression of fn1 in the injured ventricular apex by 7 dpa (Fig. 3H-J and Fig. 2C, E). In double transgenic animals, fn1:mCherry-NTR fluorescence co-localized with that of the epicardial marker tcf21:nucEGFP, demonstrating epicardial expression at 7 dpa during regeneration (Fig. 3K, L and M). We found that fn1:mCherry-NTR fluorescence colocalized with tcf21:nucEGFP at 1 dpa in areas lateral to the injury site (Fig. 3E, F and G). Although we had observed some fn1-expressing cells in the 1 dpa injury site by in situ hybridization (Fig. 2D), we typically did not see cells in the wound positive for fn1:mCherry-NTR and/or tcf21:nucEGFP fluorescence. Therefore, we suspect that the reporter may have a lag time for strong fluorescence versus mRNA induction, or is simply not as sensitive. However, we cannot rule out that there is another cell type(s) that expresses fn1 and/or fn1b at 1 dpa. Additionally, we did not observe colocalization with cardiomyocyte-specific or endothelial/endocardial cell markers. These results revealed that Fn1 is induced and secreted predominantly by epicardial cells in response to cardiac injury, and suggested a role for Fn1 in facilitating heart regeneration.
Fig. 3.
Fibronectin is synthesized predominantly by epicardial cells during heart regeneration. (A) Visualization of fn1:mCherry-NTR-expressing cells at 2 dpf. White arrows show fn1:mCherry-NTR+ cells. (B-M) Visualization of fn1:mCherry-NTR+ cells (red) and epicardial cells (tcf21:nucEGFP+; green) in uninjured, 1 and 7 dpa ventricles. Boxed area indicates an enlarged view. Arrows indicate double-positive cells. Dashed line indicates approximate resection plane. Scale bars: 50 μm.
The Fibronectin receptor itgb3 is induced in cardiomyocytes near the injury site
Fn interacts with various Integrin partners that mediate signal transduction (Labat-Robert, 2012). We performed in situ hybridization to screen for expression of several integrin genes, and identified expression of integrin b3 (itgb3) and αV (itgαV) during heart regeneration (Fig. 4A, B and data not shown). These integrin receptors, which are also present during heart development (Ablooglu et al., 2007), were upregulated in the injured ventricular apex at 7 dpa, with itgb3 showing particularly strong expression (Fig. 4A and B). To determine the cell type that expresses itgb3 during heart regeneration, we generated a BAC transgenic reporter strain using itgb3 regulatory sequences, Tg(itgb3:EGFP)pd66 (hereafter referred to as itgb3:EGFP). EGFP fluorescence in itgb3:EGFP embryos recapitulated endogenous itgb3 expression domains (Ablooglu et al., 2007) (Fig. 4C). In the uninjured heart, itgb3:EGFP fluoresence was occasionally present in blood cells (Fig. 4D). However, by 7 dpa, we observed expression in many cardiomyocytes near and within the injury site (Fig. 4E). These results indicated that cardiomyocytes in the area of trauma induce expression of integrins that can transduce signals from the Fn deposited by epicardial cells.
Fig. 4.
The Fibronectin partner integrin β3 is expressed during heart regeneration. (A, B) itgb3 expression by in situ hybridization in uninjured (A) and 7 dpa (B) ventricles. (C) Visualization of embryonic itgb3:EGFP-expressing cells (green). Boxed areas indicates a higher magnification view of the heart (area 1), pectoral fin (area 2) and posterior fin fold (area 3). (D, E) Visualization of itgb3:EGFP+ cells (green) in uninjured (D) and 7 dpa (E) ventricles. Insets: High magnification of the boxed area. Blue arrows indicate EGFP-positive cells. Yellow arrows indicate EGFP and MHC double-positive cells, indicative of cardiomyocytes. Dashed line indicates approximate resection plane. Scale bars: 50 μm.
Fn is required for heart regeneration
Fn has been implicated in multiple cellular processes relevant to tissue repair and regeneration, including cell proliferation, cell migration, cellular dedifferentiation, and fibrosis (Frangogiannis, 2008; Singh et al., 2010; Singh and Schwarzbauer, 2012; Willems et al., 1996). To examine whether Fn is required for heart regeneration, we generated a transgenic strain that enables heat-inducible expression of a dominant-negative human fibronectin fragment Tg(hsp70:fnI1-9)pd67 (Halloran et al., 2000; Ohashi and Erickson, 2011). Following a single heat-shock, transgenic embryos displayed a shortened body axis similar to the reported phenotype upon co-injection of morpholinos against both fn paralogs (Julich et al., 2005) (Fig. 5A, B). Adult transgenic fish and wild- type clutchmates were given partial resection injuries followed by a daily heat-shock from 3 to 30 dpa. This treatment had no detectable effect on the distribution of Fn protein in wild-type and transgenic clutchmates (Fig. S1). Ventricles were collected and analyzed at 30 dpa. This treatment induced fnI1-9 strongly in cardiomyocytes throughout the ventricle (Fig. 5C and D) and caused an increased incidence of regenerative failure (Fig. 5E and F) and fibrosis (Fig. 5G and H), presumably by disupting Fn function in the area of regeneration. While we did not notice gross deleterious consequences of expressing truncated Fn on other aspects of cardiac or animal physiology, such effects are possible in this type of experiment.
Fig. 5.
Fibronectin is required for heart regeneration. (A, B) Wild-type embryos (A), or embryos from a cross between hemizygous hsp70:fnI1-9 and wild-type parents (B), were heat-shocked at 6 hours post-fertilization (hpf) at 38°C for 40 minutes and imaged at 24 hpf. Wild-type embryos appeared largely normal after this treatment (92.4% normal, 7.6% general dysmorphology, n = 303). By contrast 61.4% of embryos from the transgenic cross developed a markedly shortened body axis (n = 264). (C, D) In situ hybridization for fnI1-9 in wild-type (C) and hsp70:fnI1-9 clutchmate (D) ventricles at 1 day post heat-shock (dphs). (E-H) MHC (Myosin heavy chain; green) staining and collagen staining of hsp70:fnI1-9 (E, G) and wild type clutchmate (F, H) ventricles at 30 dpa. Three of 10 ventricles of heat-shocked clutchmates showed obvious areas of missing myocardium, compared to 9 of 10 ventricles in hsp70:fnI1-9 fish. Fisher Irwin exact test, *p < 0.05. Black arrows indicate fibrosis. Dashed line indicates approximate resection plane.Scale bars: 50 μm.
As the fn1 paralog is localized to the injured ventricular apex by 7 and 14 dpa (Fig. 2E), we then analyzed zebrafish homozygous for a null mutation in fn1. While fn1 mutants have defects in cardiac development at temperatures typically used to raise zebrafish, some mutant animals are capable of surviving to adulthood when raised at 22 °C (Trinh and Stainier, 2004). The temperature-sensitive nature of the mutation is likely due to functional compensation by Fn1b, combined with the fact that embryogenesis happens more slowly at lower temperatures. To examine the requirement for Fn1 function during heart regeneration, we raised animals from fn1/+ crosses at 22 °C until 2 months of age, after which we maintained animals at 26 °C. We injured adult fn1/fn1 and wild-type or fn1/+ clutchmates and assessed regeneration after 30 days. In these experiments, similar to hsp70:fnI1-9 animals, fn1/fn1 fish were less frequently able to regenerate a contiguous wall of new muscle (Fig. 6A and B), resulting in fibrosis (Fig. 6C and D). Quantification of the regenerated muscle at 30 dpa indicated fn1/fn1 fish showed significantly less muscle in the injury site than fn1/+ clutchmates (Fig. S2A, B and C). Based on these genetic data, we conclude that Fn is required for normal heart regeneration.
Fig. 6.
fibronectin1 mutants show defective heart regeneration. (A-D) Myosin heavy chain (MHC; green) staining (A, B) and collagen staining (C, D) of wild-type and homozygous fn1 mutant ventricles at 30 dpa. One of 10 wild-type and 7 of 12 fn1 mutant ventricles showed obvious areas of depleted myocardium. Dashed line indicates approximate resection plane. Fisher Irwin exact test, *p < 0.05. Scale bars: 50 μm.
Fibronectin loss-of-function does not directly inhibit cardiomyocyte proliferation
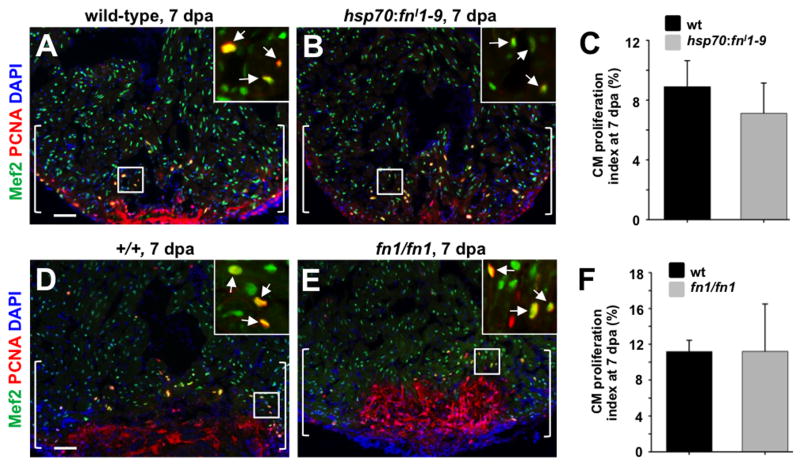
The source of new heart muscle in zebrafish is pre-existing cardiomyocytes, which are stimulated by injury to proliferate and replace lost myocardium (Jopling et al., 2010; Kikuchi et al., 2010). To determine if heart regeneration defects caused by Fn loss-of-function result directly from inhibited cardiomyocyte proliferation, we quantified the percentage of proliferating myocytes in our transgenic and mutant strains. Adult transgenic fish and wild-type clutchmates were given partial resection injuries followed by a daily heat-shock from 3 to 7 dpa. Ventricles were collected and analyzed at 7 dpa. At 7 dpa, cardiomyocyte proliferation indices were comparable between fn1 mutant, fnI1-9-expressing, and their respective control groups (Fig. 7A-F). These results indicate that Fn deposited in the ECM after cardiac injury does not facilitate muscle regeneration via direct regulation of cardiomyocyte proliferation. We then examined the distribution of cardiomyocytes adjacent to the injured area at 30 dpa in fn1/fn1 mutant and fn1/+ clutchmates. Quantification of cardiomyocyte nuclear density in areas flanking the injury at 30 dpa indicated a higher cellular density in fn1/fn1 mutant animals (Fig. S2D, E, and F). These data suggest that Fn is required for cardiomyocyte mobilization and integration into the injury site.
Fig. 7.
Evidence that Fibronectin does not directly regulate cardiomyocyte proliferation during regeneration. (A, B) Assessment of Mef2+PCNA+ cells (arrows) in hsp70:fnI1-9 (A) and clutchmate (B) 7 dpa ventricles. Insets, enlarged view of the rectangle. Brackets indicate injury site. (C) Quantification of cardiomyocyte proliferation in hsp70:fnI1-9 and clutchmate 7 dpa ventricles. For each group, 6 zebrafish were assessed. Student's t-test, p = 0.23. Mean +/- s.e.m. (D, E) Assessment of Mef2+PCNA+ cells (arrows) in wild-type (D) and homozygous fn1 mutant (E) 7 dpa ventricles. Brackets indicate injury site. (F) Quantification of cardiomyocyte proliferation in wild-type and homozygous fn1 mutant 7 dpa ventricles. For each group, 3-4 zebrafish were assessed. Student's t-test, p = 0.99. Mean +/- s.e.m. Scale bars: 50 μm.
Discussion
Here, we have used a series of expression analyses to identify a key response by the epicardium that enriches Fn in the injury site after cardiac damage in zebrafish. Together, our genetic, transgenic, and expression data point to a model for epicardial regulation of the ECM. Cardiac damage boosts Fn1 and Fn1b levels predominantly in epicardial cells throughout the ventricle, a response that localizes to the wound site with similar dynamics as other epicardial markers induced by injury (Kikuchi et al., 2011b; Lepilina et al., 2006). As the injury site accumulates Fn, itgb3 is upregulated in cardiomyocytes where it can facilitate regenerative responses. Our evidence for a pro-regenerative ECM thus suggests a new interaction between the epicardium and the myocardium that is important for regeneration, and that may be relevant to ECM changes during other examples of tissue regeneration (Bentzinger et al., 2013; Calve et al., 2010; Martino et al., 2011; Tonge et al., 2012). These findings add to a growing body of literature identifying diverse roles for the epicardium in promoting heart regeneration.
Fn is well-studied for its influences on cell adhesion and migration, and loss of Fn function in zebrafish, avian or mouse embryos leads to complete or partial failure of fusion by the cardiac primordia (George et al., 1997; Linask and Lash, 1988; Trinh and Stainier, 2004). Although Fn secretion from embryonic cardiac fibroblasts can promote the proliferation of cultured mammalian cardiomyocytes (Ieda et al., 2009), our data do not implicate Fn signaling directly in cardiomyocyte proliferation during heart regeneration. As a recent study suggested that migration of cardiomyocytes is important for zebrafish heart regeneration (Itou et al., 2012), our findings raise the possibility that Fn/Itgb3 interactions are involved in such an event. Other possible functions of Fn in a regenerative ECM include regulation of inflammatory cell function; epicardial cell behaviors like migration, proliferation, and differentiation; and vascularization of the regenerate.
Supplementary Material
Table S1. Annotated dataset of proteomics. Table of proteins differentially expressed during heart regeneration. Column 1 gives the annotated RefSeq protein identifier. Column 2 give the ProteinTeller probability, indicating the likelihood for correctly identifying each protein using the peptide fragments obtained from mass spectroscopy. Column 3 gives the number of unique peptides used to correctly identify a candidate protein. Columns 4-6 provide fold-change information across the time course of regeneration. Only proteins with an FDR corrected p-value less than 0.05 were considered to be differentially expressed.
Highlights.
The epicardium produces the ECM component Fibronectin in response to cardiac injury
Dynamic Fibronectin induction starts organ-wide and then localizes to injury
Fibronectin receptor integrin β3 is induced in cardiomyocytes near the injury site
Loss-of-function approaches indicate Fibronectin is required for heart regeneration
Fibronectin loss-of-function does not directly inhibit cardiomyocyte proliferation
Acknowledgments
We thank Harold Erickson for human fibronectin l1-9 fragment; Didier Stainier for fn1 mutant fish; Duke Proteomics Core Facility for proteomic analysis; J. Holdway for help with surgeries; J. Burris, A. Eastes, N. Blake and P. Williams for fish care; and Poss laboratory members for comments on the manuscript. K.D.P. is an Early Career Scientist of the Howard Hughes Medical Institute. This work was funded by a postdoctoral fellowship from the American Heart Association (AHA; to J.W.); an AHA Fellow-to-Faculty Award (12FTF11660037 to R.K.); a voucher received through Duke's CTSA grant 1UL1 RR024128-01 from NCRR/NIH; and a grant from NHLBI (HL081674 to K.D.P.).
Footnotes
Supporting information: Annotated proteomics dataset, Two Supplemental Figures
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ablooglu AJ, Kang J, Handin RI, Traver D, Shattil SJ. The zebrafish vitronectin receptor: characterization of integrin alphaV and beta3 expression patterns in early vertebrate development. Dev Dyn. 2007;236:2268–76. doi: 10.1002/dvdy.21229. [DOI] [PubMed] [Google Scholar]
- Bentzinger CF, Wang YX, von Maltzahn J, Soleimani VD, Yin H, Rudnicki MA. Fibronectin regulates Wnt7a signaling and satellite cell expansion. Cell Stem Cell. 2013;12:75–87. doi: 10.1016/j.stem.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calve S, Odelberg SJ, Simon HG. A transitional extracellular matrix instructs cell behavior during muscle regeneration. Dev Biol. 2010;344:259–71. doi: 10.1016/j.ydbio.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, Stainier DY. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn. 2007;236:1025–35. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- Drenckhahn JD, Schwarz QP, Gray S, Laskowski A, Kiriazis H, Ming Z, Harvey RP, Du XJ, Thorburn DR, Cox TC. Compensatory growth of healthy cardiac cells in the presence of diseased cells restores tissue homeostasis during heart development. Dev Cell. 2008;15:521–33. doi: 10.1016/j.devcel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Frangogiannis NG. The immune system and cardiac repair. Pharmacol Res. 2008;58:88–111. doi: 10.1016/j.phrs.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George EL, Baldwin HS, Hynes RO. Fibronectins are essential for heart and blood vessel morphogenesis but are dispensable for initial specification of precursor cells. Blood. 1997;90:3073–81. [PubMed] [Google Scholar]
- Geromanos SJ, Vissers JP, Silva JC, Dorschel CA, Li GZ, Gorenstein MV, Bateman RH, Langridge JI. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics. 2009;9:1683–95. doi: 10.1002/pmic.200800562. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rosa JM, Martin V, Peralta M, Torres M, Mercader N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development. 2011;138:1663–74. doi: 10.1242/dev.060897. [DOI] [PubMed] [Google Scholar]
- Gupta V, Gemberling M, Karra R, Rosenfeld GE, Evans T, Poss KD. An injury-responsive Gata4 program shapes the zebrafish cardiac ventricle. Curr Bio. 2013;23:1221–1227. doi: 10.1016/j.cub.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halloran MC, Sato-Maeda M, Warren JT, Su F, Lele Z, Krone PH, Kuwada JY, Shoji W. Laser-induced gene expression in specific cells of transgenic zebrafish. Development. 2000;127:1953–60. doi: 10.1242/dev.127.9.1953. [DOI] [PubMed] [Google Scholar]
- Huang GN, Thatcher JE, McAnally J, Kong Y, Qi X, Tan W, DiMaio JM, Amatruda JF, Gerard RD, Hill JA, Bassel-Duby R, Olson EN. C/EBP transcription factors mediate epicardial activation during heart development and injury. Science. 2012;338:1599–603. doi: 10.1126/science.1229765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Tsuchihashi T, Ivey KN, Ross RS, Hong TT, Shaw RM, Srivastava D. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev Cell. 2009;16:233–44. doi: 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itou J, Oishi I, Kawakami H, Glass TJ, Richter J, Johnson A, Lund TC, Kawakami Y. Migration of cardiomyocytes is essential for heart regeneration in zebrafish. Development. 2012;139:4133–42. doi: 10.1242/dev.079756. [DOI] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Marti M, Raya A, Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–9. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julich D, Geisler R, Holley SA. Integrinalpha5 and delta/notch signaling have complementary spatiotemporal requirements during zebrafish somitogenesis. Dev Cell. 2005;8:575–86. doi: 10.1016/j.devcel.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Gupta V, Wang J, Holdway JE, Wills AA, Fang Y, Poss KD. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development. 2011a;138:2895–902. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Major RJ, Blum N, Dahn RD, Begemann G, Poss KD. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev Cell. 2011b;20:397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–5. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton AA, Connelly CM, Romo GM, Mamuya W, Apstein CS, Brecher P. Rapid expression of fibronectin in the rabbit heart after myocardial infarction with and without reperfusion. J Clin Invest. 1992;89:1060–8. doi: 10.1172/JCI115685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labat-Robert J. Cell-Matrix interactions, the role of fibronectin and integrins. A survey Pathol Biol (Paris) 2012;60:15–9. doi: 10.1016/j.patbio.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–19. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Linask KK, Lash JW. A role for fibronectin in the migration of avian precardiac cells I Dose-dependent effects of fibronectin antibody. Dev Biol. 1988;129:315–23. doi: 10.1016/0012-1606(88)90378-8. [DOI] [PubMed] [Google Scholar]
- Magnusson MK, Mosher DF. Fibronectin: structure, assembly, and cardiovascular implications. Arterioscler Thromb Vasc Biol. 1998;18:1363–70. doi: 10.1161/01.atv.18.9.1363. [DOI] [PubMed] [Google Scholar]
- Martino MM, Tortelli F, Mochizuki M, Traub S, Ben-David D, Kuhn GA, Muller R, Livne E, Eming SA, Hubbell JA. Engineering the growth factor microenvironment with fibronectin domains to promote wound and bone tissue healing. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3002614. 100ra89. [DOI] [PubMed] [Google Scholar]
- Ohashi T, Erickson HP. Fibronectin aggregation and assembly: the unfolding of the second fibronectin type III domain. J Biol Chem. 2011;286:39188–99. doi: 10.1074/jbc.M111.262337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–80. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–90. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- Rauch GJ, Lyons DA, Middendorf I, Friedlander B, Arana N, Reyes T, Talbot WS. Submission and Curation of Gene Expression Data. ZFIN Direct Data Submission. 2003 http://zfin.org.
- Rysa J, Leskinen H, Ilves M, Ruskoaho H. Distinct upregulation of extracellular matrix genes in transition from hypertrophy to hypertensive heart failure. Hypertension. 2005;45:927–33. doi: 10.1161/01.HYP.0000161873.27088.4c. [DOI] [PubMed] [Google Scholar]
- Singh P, Carraher C, Schwarzbauer JE. Assembly of fibronectin extracellular matrix. Annu Rev Cell Dev Biol. 2010;26:397–419. doi: 10.1146/annurev-cellbio-100109-104020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Schwarzbauer JE. Fibronectin and stem cell differentiation - lessons from chondrogenesis. J Cell Sci. 2012;125:3703–12. doi: 10.1242/jcs.095786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SP, Holdway JE, Poss KD. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev Cell. 2012;22:879–86. doi: 10.1016/j.devcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart N, Bollini S, Dube KN, Vieira JM, Zhou B, Davidson S, Yellon D, Riegler J, Price AN, Lythgoe MF, Pu WT, Riley PR. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011;474:640–4. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zou Z, Collodi P, Xu F, Xu X, Zhao Q. Identification and characterization of a second fibronectin gene in zebrafish. Matrix Biol. 2005;24:69–77. doi: 10.1016/j.matbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Tonge DA, de Burgh HT, Docherty R, Humphries MJ, Craig SE, Pizzey J. Fibronectin supports neurite outgrowth and axonal regeneration of adult brain neurons in vitro. Brain Res. 2012;1453:8–16. doi: 10.1016/j.brainres.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh LA, Stainier DY. Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev Cell. 2004;6:371–82. doi: 10.1016/s1534-5807(04)00063-2. [DOI] [PubMed] [Google Scholar]
- Wang J, Panakova D, Kikuchi K, Holdway JE, Gemberling M, Burris JS, Singh SP, Dickson AL, Lin YF, Sabeh MK, Werdich AA, Yelon D, Macrae CA, Poss KD. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138:3421–30. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems IE, Arends JW, Daemen MJ. Tenascin and fibronectin expression in healing human myocardial scars. J Pathol. 1996;179:321–5. doi: 10.1002/(SICI)1096-9896(199607)179:3<321::AID-PATH555>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Zhong J, Basu R, Guo D, Chow FL, Byrns S, Schuster M, Loibner H, Wang XH, Penninger JM, Kassiri Z, Oudit GY. Angiotensin-converting enzyme 2 suppresses pathological hypertrophy, myocardial fibrosis, and cardiac dysfunction. Circulation. 2010;122:717–28. doi: 10.1161/CIRCULATIONAHA.110.955369. [DOI] [PubMed] [Google Scholar]
- Zhou B, Honor LB, He H, Ma Q, Oh JH, Butterfield C, Lin RZ, Melero-Martin JM, Dolmatova E, Duffy HS, Gise A, Zhou P, Hu YW, Wang G, Zhang B, Wang L, Hall JL, Moses MA, McGowan FX, Pu WT. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J Clin Invest. 2011;121:1894–904. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Annotated dataset of proteomics. Table of proteins differentially expressed during heart regeneration. Column 1 gives the annotated RefSeq protein identifier. Column 2 give the ProteinTeller probability, indicating the likelihood for correctly identifying each protein using the peptide fragments obtained from mass spectroscopy. Column 3 gives the number of unique peptides used to correctly identify a candidate protein. Columns 4-6 provide fold-change information across the time course of regeneration. Only proteins with an FDR corrected p-value less than 0.05 were considered to be differentially expressed.