Abstract
Long-chain acyl-CoA synthetase (LACS) activities are encoded by a family of at least nine genes in Arabidopsis (Arabidopsis thaliana). These enzymes have roles in lipid synthesis, fatty acid catabolism, and the transport of fatty acids between subcellular compartments. Here, we show that the LACS2 gene (At1g49430) is expressed in young, rapidly expanding tissues, and in leaves expression is limited to cells of the adaxial and abaxial epidermal layers, suggesting that the LACS2 enzyme may act in the synthesis of cutin or cuticular waxes. A lacs2 null mutant was isolated by reverse genetics. Leaves of mutant plants supported pollen germination and released chlorophyll faster than wild-type leaves when immersed in 80% ethanol, indicating a defect in the cuticular barrier. The composition of surface waxes extracted from lacs2 leaves was similar to the wild type, and the total wax load was higher than the wild type (111.4 μg/dm2 versus 76.4 μg/dm2, respectively). However, the thickness of the cutin layer on the abaxial surface of lacs2 leaves was only 22.3 ± 1.7 nm compared with 33.0 ± 2.0 nm for the wild type. In vitro assays showed that 16-hydroxypalmitate was an excellent substrate for recombinant LACS2 enzyme. We conclude that the LACS2 isozyme catalyzes the synthesis of ω-hydroxy fatty acyl-CoA intermediates in the pathway to cutin synthesis. The lacs2 phenotype, like the phenotypes of some other cutin mutants, is very pleiotropic, causing reduced leaf size and plant growth, reduced seed production, and lower rates of seedling germination and establishment. The LACS2 gene and the corresponding lacs2 mutant will help in future studies of the cutin synthesis pathway and in understanding the consequences of reduced cutin production on many aspects of plant biology.
INTRODUCTION
Long-chain acyl-CoA synthetases (LACSs) catalyze the conversion of free fatty acids to fatty acyl-CoAs (Kornberg and Pricer, 1953). LACS activity has been localized to several subcellular locations, including the outer envelope of chloroplasts, microbodies, oilbodies of germinating seeds, and the endoplasmic reticulum (ER) (Andrews and Keegstra, 1983; Block et al., 1983; Gerbling and Gerhardt, 1987; Fulda et al., 2002; Schnurr et al., 2002; Shockey, et al., 2002). Chloroplast (plastid) LACS activity is essential to lipid metabolism because chloroplasts are the principal site of de novo fatty acid synthesis (Browse and Somerville, 1991). Fatty acids are synthesized via the addition of two-carbon units to a growing acyl chain attached to acyl carrier protein (ACP) (Ohlrogge and Jaworski, 1997). The final products of fatty acid synthesis are 16:0-ACP and 18:0-ACP, but most of the 18:0-ACP is desaturated to 18:1-ACP. These fatty acid products can enter one of two pathways for the synthesis of membrane glycerolipids. In mesophyll cells of Arabidopsis (Arabidopsis thaliana), 38% of newly synthesized fatty acids remain in the chloroplast and are incorporated into lipids via the prokaryotic pathway. The remaining 62% of newly synthesized fatty acids are cleaved from ACP by thioesterases, which target the fatty acids for export to the eukaryotic pathway in the ER. Before export from the chloroplast, fatty acids must be converted to fatty acyl-CoAs by an outer envelope LACS activity. Acyl-CoAs produced by de novo fatty acid synthesis can then be used in several metabolic pathways besides those leading to the synthesis of membrane glycerolipids. These pathways include those leading to storage lipid (triacylglycerol) synthesis in developing seeds (Browse and Somerville, 1991), to cutin and suberin production (Kolattukudy, 2001; Nawrath, 2002), and to the synthesis of fatty acid–derived components of the cuticular wax layer (Post-Beittenmiller, 1996; Jenks et al., 2002).
In epidermal cells of aerial plant tissues, LACS activity is required for the activation of fatty acids that are used for the synthesis of cutin and wax, two main components of the cuticle. The main functions of the cuticle include reduction in transpirational water loss and protection against insects, pathogens, UV light, and frost (Becraft, 1999). Cutin and wax are synthesized exclusively in the epidermis, and they are the dominant sink of de novo synthesized fatty acids in epidermal cells (Post-Beittenmiller, 1996; Kolattukudy, 2001; Nawrath, 2002). A simplified scheme of these pathways is shown in Figure 1. The major structural component of the cuticle is cutin, which consists of interesterified hydroxy and epoxy-hydroxy fatty acids with chain lengths of 16 or 18 carbons. These derivatized fatty acids thus form a strong, three-dimensional polyester coating over all aerial organs of the plant (Kolattukudy, 2001; Nawrath, 2002). The cutin layer is embedded with intracuticular waxes and covered by epicuticular waxes. The principal components of these waxes are very-long-chain fatty acids (VLCFAs) with chain lengths of 24 to 36 carbons, along with hydrocarbons, alcohols, aldehydes, ketones, and wax esters derived from VLCFAs (Post-Beittenmiller, 1996; Jenks et al., 2002). The pathways for the synthesis of these compounds begin with the extraplastidial elongation of de novo synthesized fatty acids (primarily 18:0) to VLCFAs by microsomal enzymes. The VLCFAs produced then give rise to aldehydes, alkanes, secondary alcohols, and ketones by a pathway involving fatty acyl decarbonylation (Cheesbrough and Kolattukudy, 1984) and to primary alcohols, aldehydes, and wax esters by acyl reduction reactions and esterification (Vioque and Kolattukudy, 1997).
Figure 1.
A Simplified Outline of the Pathways of Fatty Acid Modification That Result in the Synthesis of Cutin and Cuticular Waxes in Epidermal Cells.
FA, fatty acid.
The fatty acid precursor of epicuticular wax is predominantly 18:0, whereas the monomers of cutin are synthesized from 16:0 and 18:1 fatty acyl-CoAs. After de novo synthesis of fatty acids, a partitioning occurs that delivers 18:0 to wax biosynthesis and 16:0 and 18:1 to cutin biosynthesis. Although there is a moderately good understanding of the biochemistry of fatty acid derivatization, there is considerably less information about how the pathways are organized within the cell and, in particular, how the cutin monomers are transported and assembled into the polyester layer that is formed at the outer surface of the cell wall. We can anticipate that the enzymes catalyzing cutin polymerization exist at an extracellular location in close association with the polymer (Kolattukudy, 1981, 2001). In Vicia faba, 10,16-dihydroxypalmitate is the predominant cutin monomer accounting for 78% of the structure (Kolattukudy and Walton, 1972). A particulate fraction prepared from V. faba epidermis incorporated 10,16-dihydroxypalmitate into cutin in a reaction sequence that required ATP and CoA (Croteau and Kolattukudy, 1974). These results indicate that epidermal cells possess a LACS isozyme capable of synthesizing 10,16-dihydroxypalmitoyl-CoA and that this CoA ester is a substrate for a transacylase that incorporates the hydroxy fatty acid into the insoluble cutin layer of the extracellular matrix. In vivo, LACS may act at an earlier step in the pathway because ω-hydroxylation of 16:0 by a cytochrome P450 monooxygenase (CYP86A8 encoded in Arabidopsis by the LACERATA [LCR] gene; Wellesen et al., 2001) can occur on the free acid, whereas 16-hydroxypalmitoyl-CoA is required as the substrate for the second hydroxylation at the C10 position (Walton and Kolattukudy, 1972).
Mutant analysis and molecular-genetic approaches have been used to investigate both cuticular wax and cutin synthesis. Because epicuticular waxes influence light refraction, mutations in genes involved in accumulation of wax can be detected visually. In this way, 22 eceriferum (cer) loci have been identified (Koornneef et al., 1989; McNevin et al., 1993). All of the cer mutants, in addition to the reduced glaucous appearance, have alterations in wax load and composition. Most of the cer mutants are relatively normal in growth and appearance. However, mutations at four of the cer loci produce plants of smaller stature that have reduced fertility under low humidity (Koornneef et al., 1989). One of these, cer10, also exhibits a weak organ fusion phenotype in floral organs, and leaf surfaces of cer10 plants support hydration of wild-type pollen, evidently as a result of a reduced water barrier (Lolle et al., 1998). In addition, cer10 tissues leach chlorophyll more rapidly than the wild type when immersed in ethanol (Lolle et al., 1998). Other mutants with defects in wax synthesis include wax1 and fiddlehead (fdh). Like cer10, wax1 and fdh plants are smaller than the wild type (Lolle et al., 1992; Jenks et al., 1996). In the case of the fdh mutant, it has been shown that leaves will support the germination of pollen on their surfaces and will rapidly leach chlorophyll when immersed in 80% ethanol (Lolle et al., 1997). Cloning of the FDH locus (Yephremov et al., 1999; Pruitt et al., 2000) revealed that it encodes a homolog of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) protein, which is a 3-ketoacyl-CoA synthase required for elongation of 18:1 in developing seeds (James et al., 1995). This suggests that the FDH enzyme is involved in the synthesis of VLCFAs required for cuticular wax synthesis; however, detailed characterization of wax components from fdh tissues has not been made (Lolle et al., 1997).
Genetic studies of cutin synthesis are less well developed, but the two Arabidopsis mutants that have been characterized to date both exhibit phenotypes that are similar to those of the most severe wax synthesis mutants described above. To investigate the role of cutin in plant–environment interactions and plant development, Nawrath and colleagues expressed a fungal cutinase in transgenic Arabidopsis (Sieber et al., 2000). Characterization of three transgenic lines demonstrated that the rate of chlorophyll release from leaves immersed in 80% ethanol was 2 to 10 times that of the wild type and was correlated to the level of cutinase expression in the three lines. Leaf surfaces of the transgenics supported pollen germination. Finally, the cutinase-expressing transgenics were reduced in size and also distorted because the reduction in cutin allowed fusion of both leaves and floral organs as seen in the fdh mutants (Lolle et al., 1997; Sieber et al., 2000). The Arabidopsis LCR gene encodes a cytochrome P450 monooxygenase, CYP86A8, that catalyzes ω-hydroxylation of fatty acids and is suggested to be a key enzyme for the production of ω-hydroxy fatty acids that are substrates for cutin synthesis (Wellesen et al., 2001). lcr mutant plants are smaller than the wild type and support pollen germination on their leaf surfaces. Experiments to measure chlorophyll leaching have not been reported for lcr mutants. Typically, lcr mutants show a weak organ fusion phenotype. Homozygous lcr plants are fertile (unlike fdh mutants) but have reduced fertility (Wellesen et al., 2001).
The Arabidopsis wax2 mutants (Chen et al., 2003) are deficient in both cutin and cuticular wax synthesis. wax2 mutant plants are approximately wild type in size with only a slight reduction in leaf size. Chlorophyll leaching and water loss are both increased relative to the wild type, and wax2 mutants exhibit a weak organ fusion phenotype (Chen et al., 2003).
Here, we report the characterization of the LACS2 gene of Arabidopsis that encodes one of nine LACS isozymes (Shockey et al., 2002). In leaves, LACS2 expression is limited to the epidermis. A lacs2 null mutant is phenotypically similar to mutants deficient in cutin and wax synthesis, except that it does not exhibit organ fusion of either leaves or flowers. The total wax load on mutant leaves is similar to the wild type, but the thickness of the cutin layer covering the abaxial (lower) epidermis is reduced by ∼40%. Taken together, our results suggest that the LACS2 enzyme is required for synthesis of cutin but not wax components in the epidermal cells of Arabidopsis.
RESULTS
LACS2 Transcript Is Abundant in Young, Expanding Tissues
Genomics analysis identified LACS2 as one of nine genes that encode LACS isozymes in Arabidopsis. Preliminary results showed that LACS2 was expressed in stems, leaves, roots, and flowers (Shockey et al., 2002). To extend these results, we investigated tissue-specific expression patterns by RNA gel blot analysis. The expression pattern of LACS2 is shown in Figure 2. Total RNA from wild-type plants probed with the LACS2 cDNA sequence revealed that expression was high in leaves from 14-d-old rosettes (Figure 2, lane 1). LACS2 transcript was present at considerably lower levels in RNA from mature leaves of 42-d-old rosettes (Figure 2, lane 2), and from roots (Figure 2, lane 3). The LACS2 transcript was most abundant in buds and flowers (Figure 2, lane 4) and also in young siliques (Figure 2, lane 5), whereas older siliques showed much lower levels of expression (Figure 2, lanes 6 and 7). The presence of greater amounts of LACS2 transcript in RNA from younger versus older leaves and younger versus older siliques suggested it was an isoform that might be involved in rapid cell growth.
Figure 2.
Expression Pattern of LACS2 in Arabidopsis Wild Type.
Total RNA (15 μg) from wild-type tissues was probed with a full-length LACS2 cDNA. Lane 1, leaves from 14-d-old rosettes; lane 2, mature leaves (2 to 4 cm long) from 42-d-old rosettes; lane 3, roots; lane 4, buds and flowers; lane 5, siliques 1 to 5 d after flowering (DAF); lane 6, siliques 6 to 11 DAF; and lane 7, siliques 12 to 20 DAF. An ethidium bromide stain of the gel is shown to confirm equal RNA loading.
Isolation of the lacs2-1 Mutant by Reverse Genetics
Pooled DNA stocks available through the ABRC at Ohio State University were used as templates in a PCR screen designed to amplify a product specific for the DNA sequence flanking aT-DNA insertion in the LACS2 coding region. A screen using the right border primer (XR2) and the 5′ gene-specific primer (2F) yielded a PCR product consistent with a T-DNA insertion in the 3′ portion of the LACS2 coding region. Sequence analysis of the PCR product indicated that the right border of the T-DNA was inserted in exon 15 of the LACS2 gene (Figure 3A), at 3040 bp from the ATG start codon in the genomic sequence (1446 bp in the cDNA sequence). A 3′ gene-specific primer (2R) was also synthesized. Screening of subpools of the population, using 2F-XR2 and 2F-2R primer combinations for PCR, allowed identification of one plant, designated 11-4, that was homozygous for the mutation at lacs2. Significantly, this plant was smaller than other plants in the experiment and had fewer, smaller leaves with a wrinkled appearance (Figure 2B). However, when plant 11-4 was backcrossed (BC) to the wild type, analysis of the BC2 population indicated the presence of at least three independently segregating T-DNA insertions. One BC2 line, A1-3, produced BC3 seed that segregated 3:1 for the kanamycin marker present in the T-DNA (19 of 75 kanamycin sensitive; χ2 = 0.0044, P > 0.95). The kanamycin-resistant plants were screened by PCR to identify an individual homozygous for the lacs2 mutation, and this plant formed the basis of the line designated lacs2-1. In the process of scoring BC3 individuals for phenotypes, 35 individuals were identified with a dwarfed phenotype. PCR analysis of genomic DNA from these 35 individuals yielded bands with the 2F and XR2 primers but never the 2F and 2R primers. This was a strong indication that the phenotype observed in this mutant line was the result of a recessive mutation at the lacs2 locus.
Figure 3.
The lacs2-1 T-DNA Knockout Mutant.
(A) Diagram of the genomic sequence showing the location of the T-DNA insertion. The mutant was identified in a PCR screen of pooled genomic DNA using the 5′ gene-specific primer (2F) in combination with the T-DNA right border primer (XR2).
(B) Photograph of 37-d-old wild-type (left) and lacs2-1 mutant plants. The lacs2-1 mutant has smaller, wrinkled leaves and displays overall reduced vigor.
Complementation of the lacs2-1 Phenotype
The only intact (neomycin phosphotransferase II–expressing) T-DNA insert in the lacs2-1 line is in the LACS2 gene, and its presence cosegregated with the reduced-growth phenotype (Figure 3B). However, T-DNA insertion can cause secondary mutations at linked or unlinked sites. To demonstrate that the T-DNA insertion in the LACS2 gene is the basis of the phenotype observed in lacs2-1 plants, we transformed the mutant with a construct containing a LACS2 cDNA under control of the constitutive 35S promoter of Cauliflower mosaic virus (CaMV) together with the Basta-resistance marker. Initial attempts to transform homozygous lacs2-1 plants failed to produce Basta-resistant T1 individuals (data not shown), so heterozygous plants were used for transformation. Resultant T1 seeds were collected and sown on Basta-treated soil to identify transformed T1 individuals. Basta-resistant T1 plants were grown to maturity, and resultant T2 seeds from individual plants were sown on soil treated with Basta to determine segregation ratios. One line, 3B-19, showed a segregation ratio that was a good fit to the expected 3:1 ratio for a single insert (9 of 47 were Basta sensitive; χ2 = 0.854; P > 0.5), and this line was chosen for further analysis. T2 seeds were 100% kanamycin resistant (n = 137), showing that only T-DNA-interrupted lacs2-1 alleles were present.
In a second cosegregation experiment, seeds were germinated on soil without selection. Nineteen plants were wild type in appearance, whereas five exhibited the dwarf phenotype. When these plants were 3 weeks old, they were sprayed with Basta. All the normal-sized plants survived this treatment, indicating that they contained the LACS2 transgene construct, whereas the dwarf plants all died. This result indicates that the LACS2 transgene successfully complements the dwarf phenotype. Subsequent investigation of the complemented plants indicated that seed yield, seed color, and chlorophyll leaching from leaves (all aspects of the lacs2 phenotype, as described below) were also restored to wild type. Genomic DNA isolated from the phenotypically wild-type progeny of line 3B-19 (Figure 4A) was analyzed by PCR amplification using the intronF and 2R primers (Figure 3A). No wild-type LACS2 alleles were detected (n = 10), providing further evidence that the lacs2-1 phenotype is complemented by the presence of the LACS2 transgene.
Figure 4.
The lacs2-1 Phenotype Is Complemented by Expression of the LACS2 cDNA.
(A) Photograph of the wild type (WT), lacs2-1 mutant, and lacs2-1 plant expressing a LACS2 cDNA under the control of the CaMV 35S promoter. Plants shown here are 24 d old.
(B) Gel blot analysis of RNA from the wild type (lane 1), lacs2-1 heterozygote (lane 2), lacs2-1 homozygote (lane 3), and a complemented line (lane 4). Total RNA (15 μg) was probed with the LACS2 cDNA sequence. Ethidium bromide is shown as a loading control.
(C) PCR analysis of genomic DNA from the wild type (1), lacs2-1 (2), and a complemented line (3). The complemented line did not contain a wild-type LACS2 genomic fragment, although the presence of the LACS2 transgene is indicated. Primer locations within the genomic sequence are shown in Figure 3.
To analyze the expression profile in both the lacs2-1 mutant and the complemented line 3B-19-4, total RNA isolated from leaves of the wild type, heterozygous lacs2-1, homozygous lacs2-1, and the 3B-19-4 line was probed with the LACS2 cDNA sequence. Gel blot analysis (Figure 4B) shows that wild-type RNA contained a full-length LACS2 transcript. Comparatively, plants heterozygous for the lacs2-1 allele contained lesser amounts of LACS2 transcript, probably because of the presence of only one wild-type allele. The homozygous lacs2-1 plants did not contain any detectable LACS2 transcript, suggesting that any truncated mRNA transcribed from the T-DNA–disrupted lacs2-1 locus is degraded. RNA from the complemented line showed high levels of expression of LACS2 transcript.
The RNA gel blot analysis in Figure 4B did not distinguish the wild-type LACS2 transcript from the LACS2 cDNA transcribed from the CaMV 35S promoter. To reconfirm that the complemented line was indeed homozygous for the lacs2-1 allele and contained the 35S:LACS2 transgene, we performed PCR analysis on genomic DNA isolated from the wild type, lacs2-1 (homozygote), and pooled plants from the segregating 3B-19 line. The intronF and 2R primers (Figure 3A) were used to screen for the presence of the wild-type LACS2 allele, and the 2F and 2R primers were used to distinguish between the 4-kb genomic copy of LACS2 and the smaller (2 kb) cDNA present in the 35S:LACS2 transgene. Lastly, the 2F and XR2 primers were used to detect the presence of the mutant, T-DNA-interrupted lacs2-1 allele. Figure 4C summarizes the results of the PCR analysis. The intronF and 2R primer combination, which was designed to amplify only the wild-type LACS2 allele, produced a 1.5-kb band in the wild type but not in lacs2-1 or in the complemented line. The 2F and 2R primer combination yielded a band in the wild type corresponding to the endogenous LACS2 gene (4 kb), whereas a 2.0-kb band was present in the complemented line. The 2F and XR2 primer pair detected the mutant lacs2-1 allele in both lacs2-1 and the complemented line. Taken together, the results of the gel blot and PCR experiments confirmed that the complemented line was indeed homozygous for the lacs2-1 mutant allele and contained the LACS2 transgene.
Initial Characterization of the lacs2-1 Phenotype
To obtain further evidence about the precise biochemical role of LACS2 in lipid metabolism, we first compared the total lipid content and lipid composition of lacs2-1 with wild-type leaf tissue. However, we did not identify any significant difference in total lipid content (on a fresh weight or dry weight basis), the proportion of different glycerolipid classes, or the fatty acid compositions of the major membrane lipids in a series of analyses (data not shown).
The smaller lacs2-1 rosettes were slower to flower than wild-type plants (34.1 ± 0.5 d versus 30.2 ± 0.6 d under a 16-h photoperiod). lacs2-1 plants produced smaller bolts, but flower morphology and fertility were normal, and siliques were similar in size to those on wild-type plants. The smaller bolts of the mutant yielded only 62 ± 20 mg seed/plant compared with 177 ± 17 mg seed/plant from wild-type controls. Harvested lacs2-1 seeds appeared smaller and darker than seeds from wild-type plants that had been grown alongside the mutants. Examination by light microscopy revealed that ∼50% of lacs2-1 seeds contained green embryos that could be distinguished through the testa. When the green and normal brown seeds from a lacs2 plant were germinated separately, the green seeds showed 79% germination compared with 100% germination for the brown seeds. To determine if lacs2-1 seeds were indeed smaller, five replicates of 250 seeds each for the wild type and lacs2-1 were counted and subsequently weighed. The wild-type samples gave a weight of 20.1 ± 0.6 μg/seed, whereas the lacs2-1 samples weighed 18.6 ± 0.3 μg/seed. We also measured the total fatty acid content of the seeds by quantitative gas chromatography (GC) of fatty acid methyl esters (FAMES) using 17:0 as an internal standard. lacs2-1 seeds averaged only 2.23 ± 0.09 μg FAMES/seed compared with 3.03 ± 0.10 μg FAMES/seed for the wild type.
To quantify differences in germination between the wild type and lacs2-1, seeds were sown on soil and covered with plastic wrap to maintain high humidity. At 6 d, the plastic wrap was removed, and seedlings were scored. Of the wild-type seeds, 99.3% had emerged and produced cotyledons (134 of 135). By contrast, only 80.0% of lacs2-1 seedlings emerged (108 of 135). It was apparent at 6 d that a percentage of the emerged lacs2-1 seedlings would not grow and develop into viable plants. These seedlings had white cotyledons, and they eventually died. At 19 d, 91.0% (122 of 134) of the wild-type seeds that germinated had produced viable rosettes with four to five pairs of true leaves. For lacs2-1 mutants, only 72.2% of seeds that had germinated (58% of those sown) produced rosettes (78 of 108). At this stage, the mutant had only two to three pairs of true leaves.
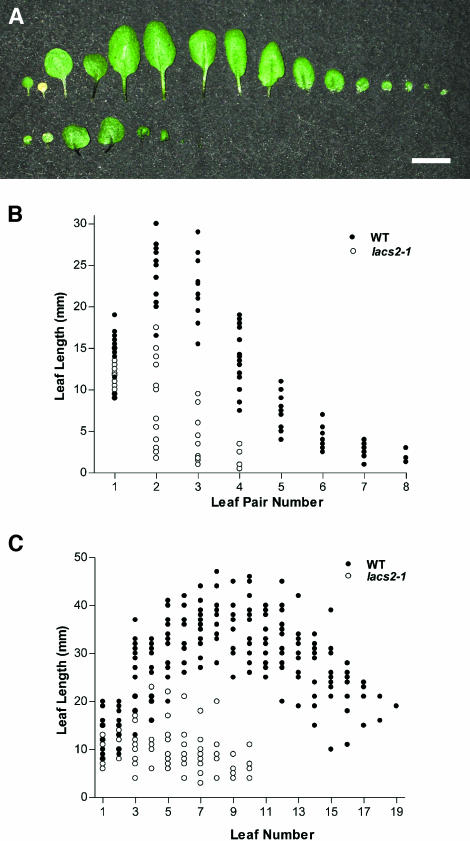
The lacs2-1 seedlings that developed into viable rosettes were significantly smaller than the wild type under standard growth conditions (Figure 3B). To quantitatively evaluate differences in rosette size and development, we measured leaf length and number in 22-d-old rosettes. As shown in Figure 5A, the lacs2-1 mutant had fewer leaves, and the leaves that were present were shorter and misshapen. Figure 5B summarizes the results of leaf measurements. Although the wild type had up to 16 leaves, lacs2-1 had no more than eight leaves (n = 15) at 22 d. To determine if the mutant was developmentally delayed and ultimately approached wild-type numbers or length, 35-d-old plants were analyzed, at which point both the wild type and mutant had produced inflorescences. As shown in Figure 5C, lacs2-1 did not approach the wild type either in the number of leaves or leaf length. At 35 d, wild-type rosettes had up to 19 leaves, whereas lacs2-1 rosettes had no more than 10.
Figure 5.
Leaf Phenotype of the lacs2-1 Mutant.
(A) Leaves were removed from 22-d-old rosettes and measured. Scale bar = 1 cm.
(B) Summary of leaf length versus leaf number at 22 d (n = 15).
(C) Summary of leaf length versus leaf number at 35 d (n = 12).
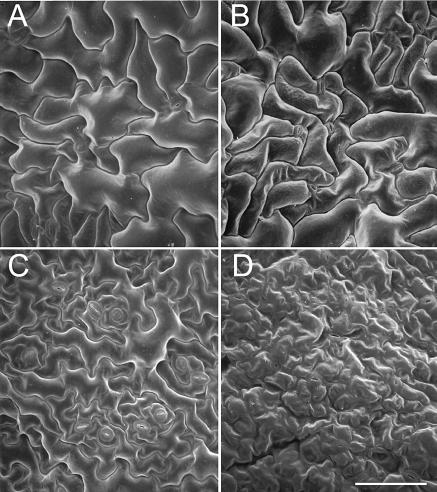
As shown in Figure 5A, lacs2-1 leaves were small and irregular in shape compared with the wild type. To determine if there were differences at the microscopic level, we analyzed surfaces of wild-type and lacs2-1 leaves from 49-d-old rosettes by scanning electron microscopy. Both surfaces of lacs2-1 leaves differed from the wild type (Figure 6). Adaxial surfaces of the wild type (Figure 6A) showed the characteristic jigsaw puzzle shape of pavement cells (Telfer and Poethig, 1994). In lacs2-1, the cells contained fewer, larger lobes (Figure 6B). Epidermal cells on the abaxial surface of lacs2-1 were collapsed (Figure 6D). This collapsed appearance was consistently seen in views of the abaxial leaf surface of lacs2-1 plants but was never observed on the abaxial surface of wild-type leaves (Figure 6C) or on the adaxial surfaces of either the wild-type or mutant plants.
Figure 6.
Scanning Electron Microscopy of Wild-Type and lacs2-1 Leaf Surfaces.
(A) Wild-type leaf, adaxial side.
(B) lacs2-1 leaf, adaxial side.
(C) Wild-type leaf, abaxial side.
(D) lacs2-1 leaf, abaxial side. Scale bar represents 75 μm.
LACS2 Is Expressed in the Epidermis
Many of the phenotypic characteristics of lacs2-1 plants are consistent with a defect in the supply of fatty acids from the chloroplast for membrane and storage lipid synthesis. For example, in Arabidopsis, the mosaic death1 (mod1) mutation causes a 95 to 98% reduction in activity of enoyl-ACP reductase, a key enzyme of the plastid fatty acid synthase (Mou et al., 2000). Like lacs2-1 plants, mod1 plants are reduced in size and produce fewer, smaller seeds that exhibit poor germination and seedling establishment. Although highly deficient in enoyl-ACP reductase activity, mod1 plants show only a small (<10%) decrease in lipid content per gram fresh weight compared with the wild type, suggesting that plant growth is reduced to match the lower availability of fatty acids for lipid synthesis and membrane biogenesis. To test the possibility that LACS2 encoded a chloroplast acyl-CoA synthetase, we performed in vitro chloroplast import assays (Froehlich et al., 2001), and transient expression experiments with a LACS2:GFP construct (Schnurr et al., 2002), but these did not provide convincing evidence that LACS2 is localized to the chloroplast (data not shown). Moreover, in parallel with our studies on LACS2, we also investigated the LACS9 gene. Our characterization of LACS9 indicated that it encodes the major chloroplast LACS isozyme, which is responsible for ∼90% of the LACS activity in mesophyll chloroplasts (Schnurr et al., 2002).
In principle, it was possible that the chloroplast activity attributable to LACS9 is not the major contributor to cellular lipid metabolism and that the isozyme encoded by LACS2 is primarily responsible for export of fatty acids through the chloroplast envelope. To help resolve this issue, we generated a construct containing 1.8 kb of DNA upstream of the LACS2 start codon fused to a cDNA encoding the β-glucuronidase (GUS) reporter (Jefferson et al., 1987). Wild-type Arabidopsis plants were transformed with the LACS2 promoter:GUS construct, and resultant T1 seeds were sown on selective medium to screen for transformed individuals. Kanamycin-resistant T1 seedlings were transferred to soil after 10 d on selection and grown to maturity. Tissue samples were removed at various ages and stained for GUS activity.
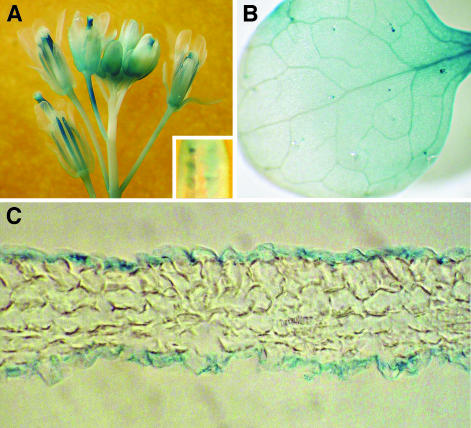
The RNA gel blot analysis shown in Figure 2 indicated that highest LACS2 expression was found in buds and flowers. In these tissues of the lines expressing the LACS2 promoter:GUS fusion, GUS expression was found in the stigma, style, filaments, sepals, and pedicels (Figure 7A). GUS staining was also observed in the developing seed (Figure 7A, inset). As shown in Figure 2, LACS2 transcript was also abundant in young leaves, and leaves from 15-d-old rosettes also showed GUS activity. This activity was diffusely spread across the leaf lamina, with strongest staining in the expanding basal portion of the leaf (Figure 7B). To determine if LACS2 expression was localized in specific tissues of the leaf, histochemically stained leaves were fixed, embedded in paraffin, and sectioned. Inspection of the sections by light microscopy clearly demonstrated that GUS activity was entirely confined to the epidermal cells (Figure 7C). This result strongly indicates that LACS2 is not involved in general membrane lipid synthesis, which is highest in the leaf mesophyll, and instead suggests a role in cutin or cuticular wax synthesis, which are major functions of the epidermal cells.
Figure 7.
Histochemical Staining of Transgenic Lines Containing the LACS2 Promoter Fused to the GUS Reporter Gene.
(A) Flower cluster showing GUS staining in the stigma, style, filament, sepals, and pedicel tissues. Staining can also be seen in developing seeds of a dissected developing silique (inset).
(B) Overview of a leaf from a 15-d-old plant.
(C) Cross section of a leaf from a 15-d-old plant showing expression only in epidermal cells.
The lacs2-1 Mutation Results in a Defective Cutin Layer
In principle, the lacs2-1 lesion might cause a deficiency in synthesis of cutin, synthesis of cuticular wax, or both because many components of the cuticle are derivatized fatty acids produced in the epidermal cells (Kolattukudy, 2001; Jenks et al., 2002). A reliable method for the quantitative analysis of cutin composition in Arabidopsis has not yet been established because the cuticle is thin and fragile (Nawrath, 2002). However, preliminary results suggest that 9,16- and 10,16-dihydroxypalmitate are major components of Arabidopsis cutin (C. Nawrath, personal communication). To determine if the cutin layer in lacs2-1 was affected, we analyzed cross sections of epidermal cells from leaves of the wild type and lacs2-1 by transmission electron microscopy (TEM). Sample electron micrographs showing the adaxial and abaxial cutin layers of wild-type and mutant leaves are shown in Figure 8. We measured the thickness of the cutin layer from TEM cross sections in five independent samples of wild-type and mutant leaves. The cutin layer on the adaxial surface of lacs2-1 leaves was not significantly different in cross-sectional thickness from that of control wild-type leaves (31.6 ± 1.7 nm in the wild type versus 33.0 ± 2.0 nm in lacs2-1). However, the abaxial cutin layer on lacs2-1 leaves was substantially thinner than the wild type (22.3 ± 1.7 nm compared with 36.5 ± 2.1 nm in the wild type). Thus, substantial reduction in thickness of the abaxial cutin layer correlates with the scanning electron microscopy results showing collapse of abaxial leaf surfaces in the mutant (Figure 6D).
Figure 8.
Electron Micrographs of the Cuticle Layer on Wild-Type and lacs2-1 Leaves.
(A) Wild type, adaxial surface.
(B) lacs2-1, adaxial surface.
(C) Wild type, abaxial surface.
(D) lacs2-1, abaxial surface. Scale bar = 100 nm.
We also investigated whether the wax load and composition were altered in lacs2-1. The lacs2-1 plants do not display an eceriferum phenotype (Koornneef et al., 1989). For quantification of wax load and composition, we analyzed total epicuticular wax from leaves from 38-d-old lacs2-1 plants. Tissue was immersed in chloroform for 30 s to extract epicuticular wax components, and the extracts were analyzed by GC–mass spectrometry after derivatization. Results from one set of triplicate analyses are summarized in Table 1. The total wax load on lacs2-1 leaves was 111 μg/dm2 compared with 76 μg/dm2 for wild-type controls. There were small changes in the proportions of some components but no indication that lacs2-1 plants had any substantial defect in wax composition.
Table 1.
Analysis of Leaf Wax Components from Leaves of 38-d-old Wild-Type and lacs2-1 Plants
| Wax Component | Wild Type | lacs2-1 |
|---|---|---|
| Alkanes | ||
| C27 | 0.7 (0.1) | 1.2 (0.1) |
| C29 | 17.2 (0.5) | 29.2 (0.9) |
| C31 | 37.6 (0.3) | 49.7 (1.6) |
| C33 | 13.1 (0.8) | 12.5 (0.3) |
| Primary alcohols | ||
| C26 | 2.4 (0.3) | 3.2 (0.3) |
| C28 | 1.6 (0.6) | 7.9 (0.3) |
| C30 | 0.9 (0.1) | 1.2 (0.2) |
| Fatty acids | ||
| C24 | 0.1 (0.01) | 0.3 (0.01) |
| C26 | 0.5 (0.07) | 3.9 (0.25) |
| C28 | 2.0 (0.03) | 1.7 (0.15) |
| C30 | 0.3 (0.01) | 0.6 (0.13) |
| Total | 76.4 | 111.4 |
Compounds were separated and quantified by gas chromatography as described in Methods. Data are μg/dm2 of leaf surface and the average of three replicate analyses, with SE shown in parentheses. This experiment was repeated with similar results.
Both the lcr mutant, which is defective in cutin synthesis (Wellesen et al., 2001), and transgenic plants expressing a fungal cutinase (Sieber et al., 2000) lose chlorophyll more rapidly than the wild type when tissue is immersed in ethanol. This effect can be attributed to more rapid penetration of the solvent into the leaf through the defective cuticle, and some mutants in cuticular wax synthesis, such as fdh and wax2 mutants, also show accelerated chlorophyll leaching into ethanol (Yephremov et al., 1999; Chen et al., 2003). To determine if the lacs2-1 mutant has a similar defect, we immersed intact rosettes from 36-d-old wild-type and lacs2-1 plants into 80% ethanol and monitored the accumulation of chlorophyll in the external solution by spectrophotometry. Data from a typical experiment are shown in Figure 9 and indicate that the rate of chlorophyll leaching from lacs2-1 plants was threefold higher than the rate from wild-type controls. The total chlorophyll content of lacs2-1 plants was 778 ± 36 μg/g fresh weight compared with 815 ± 109 μg/g fresh weight in wild-type plants.
Figure 9.
Chlorophyll Leaching from Wild-Type and lacs2-1 Leaves.
Rosettes were placed in 80% EtOH, and aliquots were removed at the indicated time points. Results shown here are representative of three independent experiments.
The leaf surfaces of lcr, fdh, and the cutinase-expressing transgenic Arabidopsis were all shown to support the germination of wild-type pollen (Lolle and Cheung, 1993; Sieber et al., 2000; Wellesen et al., 2001). Evidently, these mutants have cuticular defects that alleviate the barrier that inhibits pollen hydration and germination on wild-type plants. To test whether lacs2-1 leaf surfaces supported pollen germination, wild-type pollen was applied to both surfaces of attached rosette leaves, and plants were returned to the growth chamber. After 24 h, leaves were removed and prepared for scanning electron microscopy. Wild-type surfaces (Figures 10A and 10B) did not support the germination of pollen. The fdh-3 mutant leaves served as a positive control, and pollen was found to germinate on both surfaces (not shown). The surfaces of lacs2-1 (both adaxial and abaxial) also supported the germination of pollen (Figures 10C and 10D). Although pollen tubes often grew in random directions along the leaf surface, tubes also appeared to penetrate the cell wall (data not shown), similar to pollen grains observed on leaves of fdh (Lolle and Cheung, 1993). Quantitative data for pollen germination on wild-type, fdh-3, and lacs2-1 leaves are shown in Table 2. No germinating pollen grains were found on wild-type surfaces. In fdh-3, 55.6% and 54.7% of pollen grains germinated on adaxial and abaxial surfaces, respectively. Germination on lacs2-1 abaxial surfaces was higher than adaxial surfaces (52.5% compared with 20.3%, respectively). Thus, lacs2 mutant plants exhibit phenotypes that are characteristic of mutants deficient in cuticle formation.
Figure 10.
Pollen Germination on lacs2-1 Leaf Surfaces.
Wild-type pollen was applied to the surfaces of wild-type or lacs2-1 leaves on 42-d-old plants. After 24 h, leaves were fixed for scanning electron microscopy analysis.
(A) Wild-type adaxial surface.
(B) Wild-type abaxial surface.
(C) lacs2-1 adaxial surface.
(D) lacs2-1 abaxial surface. The scale bar represents 75 μm.
ω-Hydroxypalmitic Acid Is a Substrate for the LACS2 Isozyme
Recombinant (and purified) acyl-CoA synthetase enzymes typically show broad substrate specificities (Shockey et al., 2002). This makes it difficult to deduce the physiological substrate(s) and, thus, the likely biological role of a particular enzyme using biochemical approaches. Thus, the recombinant LACS2 protein showed activity against a broad range of long-chain fatty acids, as did the eight other Arabidopsis LACS proteins that were recombinantly expressed in both Saccharomyces cerevisiae (yeast) and Escherichia coli cells (Shockey et al., 2002).
Given the results described above, we wished to directly test the ability of LACS2 enzyme to activate 16-hydroxypalmitate. The product of this reaction, 16-hydroxypalmitoyl-CoA, serves as the substrate for a second hydroxylation reaction, which produces the cutin monomer (Walton and Kolattukudy, 1972). We first assayed the kinetics of LACS2 activity against 16:0 and 16-hydroxypalmitate. With 16:0 as substrate, LACS2 showed a Vmax of 7.5 nmol/min/mg protein and a Km of 81.4 μM. By comparison, Vmax for 16-hydroxypalmitate was 11.8 nmol/min/mg protein with a Km of 31.4 μM. These data indicate that 16-hydroxypalmitate is a better substrate than 16:0 for the LACS2 enzyme. To compare this substrate specificity of LACS2 with other LACS isoforms, we measured the activities of LACS2, LACS4, LACS8, and LACS9 enzymes against 16:0 and 16-hydroxypalmitate at saturating substrate concentrations (500 μM). The results in Table 3 indicate that although LACS2 exhibited a twofold preference for 16-hydroxypalmitate over 16:0, the remaining three enzymes had a twofold preference for the unhydroxylated fatty acid. These results lend further support to the hypothesis that LACS2 contributes to the synthesis of cutin monomers in epidermal cells of Arabidopsis.
Table 3.
Substrate Specificities of Acyl-CoA Synthetase Isozymes
| Isozyme | Substrate | Synthetase Activitynmol/min/mg Protein | Ratio of Activities |
|---|---|---|---|
| LACS2 | Palmitate | 4.6 | 1.98 |
| 16-hydroxypalmitate | 9.1 | ||
| LACS4 | Palmitate | 9.4 | 0.45 |
| 16-hydroxypalmitate | 4.2 | ||
| LACS8 | Palmitate | 11.2 | 0.44 |
| 16-hydroxypalmitate | 4.9 | ||
| LACS9 | Palmitate | 2.6 | 0.50 |
| 16-hydroxypalmitate | 1.3 |
Recombinant proteins for Arabidopsis LACS isozymes were produced in E. coli and assayed for activity against palmitate (16:0) and 16-hydroxypalmitate as described in Methods.
DISCUSSION
Our results indicate that the LACS2 gene encodes an acyl-CoA synthetase involved in cutin synthesis and required for the correct assembly of the cuticular barrier that covers leaf surfaces and the surfaces of other plant organs. The expression pattern of LACS2, determined by both RNA gel blot analysis and the use of LACS2 promoter:GUS fusions, is similar to the expression patterns reported for other genes involved in production of cuticle components. For example, RNA gel blot data for LCR (Wellesen et al., 2001) show high expression in young leaf tissue of seedlings, with much lower levels in older rosette and cauline leaves; inflorescence and silique tissues have the highest levels of transcript, as we found for LACS2 (Figure 2). Both LCR and LACS2 are expressed at low levels in root tissues. In roots, the LCR and LACS2 gene products may be involved in the synthesis and activation of hydroxy fatty acids, which are also used for suberin synthesis (Kolattukudy, 2001; Nawrath, 2002). Predominant expression in the epidermal cells of leaves was observed for a GUS reporter driven by the Brassica oleracea (broccoli) wax9D promoter (Pyee and Kolattukudy, 1995). The wax9D gene encodes the major protein of B. oleracea cuticular wax. wax9D and other genes, such as FDH, are also highly expressed in flower buds and flowers (Pyee and Kolattukudy, 1995; Yephremov et al., 1999). In our LACS2 promoter:GUS transgenic plants, GUS staining is darkest in expanding tissues of the floral organs—the base of the sepals on the young flower buds, the newly elongated pedicel (but not in mature, fully elongated pedicels), and the anther filaments as they elongate just before and during flower opening. The style also shows strong GUS expression (Figure 7A). The very specific labeling of epidermal cells in our leaf cross sections (Figure 7C) suggests a role for the LACS2 enzyme in the synthesis of cuticle components. This suggestion is supported by our observation in lacs2 mutant plants of two well-recognized phenotypes associated with defects in cuticle production—germination of pollen on leaf surfaces (Figure 10, Table 2) and accelerated leaching of chlorophyll from leaves immersed in 80% ethanol (Figure 9).
Table 2.
Quantitation of Pollen Germination on Leaf Surfaces
| Genotype | Leaf Surface | No. of Pollen Grains Counted | No. of Pollen Grains Germinated | % Germination |
|---|---|---|---|---|
| Wild type | Adaxial | 478 | 0 | 0.0 |
| Wild type | Abaxial | 495 | 0 | 0.0 |
| fdh-3 | Adaxial | 142 | 79 | 55.6 |
| fdh-3 | Abaxial | 139 | 76 | 54.7 |
| lacs2-1 | Adaxial | 691 | 140 | 20.3 |
| lacs2-1 | Abaxial | 472 | 248 | 52.5 |
Wild-type pollen was applied to either abaxial or adaxial surfaces of leaves on 42-d-old plants, and the plants returned to the growth chamber for 24 h before leaves were removed and fixed for scanning electron microscopy.
It is unlikely that LACS2 encodes a plastid envelope activity involved in fatty acid export from the organelle. We obtained no results to support this notion from either in vitro chloroplast import assays or transient expression of LACS2:GFP translational fusions in Allium cepa (onion) epidermal cells (data not shown). Furthermore, LACS9 promoter:GUS fusions revealed that the LACS9 gene, which does encode a plastid LACS isozyme (Schnurr et al., 2002), is strongly expressed in leaf epidermal cells as well as in the mesophyll cells (data not shown). Both LACS9 and LACS2 are active with a range of 16- and 18-carbon fatty acids, including 16:0 and 18:1, which are the main products of plastid fatty acid synthesis (Shockey et al., 2002). However, LACS2 is considerably more active with 16-hydroxypalmitate than with palmitate, whereas LACS9 shows low activity on 16-hydroxypalmitate (Table 3).
Leaf wax load is increased rather than decreased in lacs2 mutant plants, whereas there is a 40% decrease in thickness of the abaxial cutin layer of lacs2 leaves relative to the wild type. Based on these observations, we hypothesize that LACS2 encodes an acyl-CoA synthetase isozyme required for cutin synthesis but not for synthesis of cuticular wax components. The thickness of the adaxial cutin layer appears unchanged, but results of the pollen germination tests imply that there may be changes in the density or composition of the cutin that also compromise the cuticular barrier on the adaxial leaf surface.
Many details of the pathways of fatty acid modification and interesterification that lead to cutin synthesis remain unclear (Kolattukudy, 2001; Nawrath, 2002). In particular, there is no demonstrated role for an acyl-CoA synthetase reaction beyond the initial activation that occurs at the plastid envelope (Andrews and Keegstra, 1983; Block et al., 1983; Schnurr et al., 2002). However, we can envisage two possible roles for the LACS2 enzyme (Figure 1). The first step in 16:0 (and 18:1) modification is a ω-hydroxylation catalyzed by a P450 monooxygenase, encoded in Arabidopsis by the LCR gene, that recognizes free fatty acids as substrates (Wellesen et al., 2001). It is not clear if LCR (CYP86A8) can also accept 16:0-CoA as a substrate, but biochemical studies of a 16:0 ω-hydroxylase activity from V. faba provided indirect evidence suggesting that 16:0-CoA may not be an efficient substrate (Soliday and Kolattukudy, 1977). Thus, it is possible that 16:0-CoA is hydrolyzed to the free acid before being acted on by the ω-hydroxylase. In this scenario, LACS2 may be required to synthesize 16-hydroxypalmitoyl-CoA before further modification of the fatty acid can take place. In V. faba, 16-hydroxypalmitoyl-CoA, not the free acid, is the substrate for synthesis of 10,16-dihydroxypalmitate (Walton and Kolattukudy, 1972). The strong activity of recombinant LACS2 against 16-hydroxypalmitate supports the hypothesis that this reaction is the physiological function for the LACS2 gene product. Our complementation experiment used the CaMV 35S promoter to drive expression of a LACS2 cDNA, and this would have led to production of the LACS2 enzyme in mesophyll (and other) cells as well as the epidermis. However, we did not detect any phenotypic consequences of this ectopic expression, probably because the proposed hydroxyacyl fatty acid substrates of the enzyme are only synthesized in the epidermal cells.
The second possibility is that LACS2 is required for membrane transfer of cutin monomers. For example, it is generally assumed that 10,16-dihydroxypalmitate and other cutin precursors are transferred through the plasma membrane (or an endomembrane) to the cell exterior. In many cases, cross-membrane transport of fatty acids requires an acyl-CoA synthetase activity on the distal side of the membrane (Schaffer and Lodish, 1994; Gargiulo et al., 1999; Zou et al., 2002), and acyl-CoAs have been shown to be substrates for cutin synthesis in vitro (Croteau and Kolattukudy, 1974). Thus, LACS2 may act either at the external face of the plasma membrane or the lumenal face of the ER or Golgi to synthesize hydroxyacyl-CoA substrates for cutin synthesis. However, LACS2 does not contain a recognizableN-terminal signal peptide sequence expected in a protein targeted to the secretory pathway.
Whatever the role of LACS2, our characterization of the lacs2-1 null mutant indicates that some cutin synthesis occurs in the absence of this acyl-CoA synthetase isozyme. The LACS gene family in Arabidopsis contains at least nine members (Shockey et al., 2002). The leakiness of the lacs2-1 phenotype with respect to cutin production may result from overlapping functions among this family of enzymes as previously described for other LACS isozymes (Fulda et al., 2002; Schnurr et al., 2002). Alternatively, the leaky phenotype may reflect a low activity of the LCR ω-hydroxylase on 16:0-CoA.
The lacs2-1 mutants exhibit a range of defects relative to wild-type plants, and many of these are very similar to the pleiotropic effects reported for lcr mutants (Wellesen et al., 2001) and for transgenic Arabidopsis expressing a cutinase enzyme (Sieber et al., 2000). The reduced size and altered shape of epidermal cells, together with the associated distortion of the leaf lamina, are symptoms shared with the other cutin mutants, although these do not appear to be features of the wax2 mutant (Chen et al., 2003). The cutinase transgenics show highest expression (Sieber et al., 2000), and most lcr individuals exhibit fusion of leaves and floral organs, but we never observed any organ fusion in lacs2-1plants. In the absence of the LACS2 isozyme, cutin production is apparently still sufficient to preclude organ fusion. Consistent with this conclusion, Wellesen et al. (2001) reported some lcr individuals that lacked organ fusions but displayed other features (such as reduced plant size) of the cutin deficiency phenotype. Similarly, transgenic plants expressing low levels of cutinase do not exhibit organ fusion (C. Nawrath, personal communication).
The 40% reduction in thickness of the cutin layer on the abaxial epidermis of lacs2-1 leaves is accompanied by a 40% reduction in the thickness of the primary cell wall of the epidermal cells. Very probably it is this reduction in cell wall that causes the collapse of the epidermal cells observed in SEM of the lower epidermis (Figure 6D). There is no evidence to suggest that an acyl-CoA synthase activity is involved directly in primary cell wall synthesis, and we hypothesize that the reduced cell wall thickness is a pleiotropic consequence of the defect in cutin production.
The reduced plant size, reduced seed set, and lower germination/seedling establishment percentages for lacs2-1 all closely parallel results reported for lcr and cutinase-expressing mutants (Sieber et al., 2000; Wellesen et al., 2001). In addition, we noted that ∼50% of lacs2-1 seeds had green cotyledons when harvested from fully mature and senescent plants. The LACS2 gene is strongly expressed in both developing seeds (Figure 7A, inset) and young, expanding siliques of wild-type plants. This suggests that inadequate cutin synthesis in lacs2 plants may cause incomplete maturation of seeds, perhaps as a result of excessive water loss from the siliques and/or seeds.
At present, we have a very poor understanding of the cellular (and extracellular) organization and precise biochemical reactions of the pathway leading to cutin synthesis (Kolattukudy, 2001). Furthermore, the recent isolation of cutin mutants (Sieber et al., 2000; Wellesen et al., 2001; Chen et al., 2003) has revealed the wide range of phenotypes associated with defects in cutin synthesis, but we lack a mechanistic understanding of exactly how these phenotypes arise. The LACS2 gene and the corresponding lacs2-1 mutant provide new tools for the investigation of the cutin biosynthesis pathway and the many roles that cutin and related polymers play in the cell biology, development, and physiology of plants.
METHODS
Plant Material and Growth Conditions
The wild-type ecotype Columbia (Col) of Arabidopsis was used in this study. The lacs2-1 knockout mutant lines were identified from the population of T-DNA–tagged plants (Campisi et al., 1999) available through ABRC (seed stock CS19786; ecotype gl-1 background). Plants were germinated and grown on a commercial potting mixture at 22°C under illumination of fluorescent lights (175 μmol·m−2·s−1) under 16-h-light/8-h-dark photoperiod unless otherwise noted. For germination and establishment experiment, seeds grown and harvested at the same dates were dried on silica chips for 6 weeks. Randomized seeds were sown on wetted potting mixture, covered with plastic wrap, and placed at 4°C. After 2 d, the covered trays were moved to the growth chamber. At 6 d after sowing, the plastic wrap was removed and seedlings were scored. At 19 d, plants were scored again.
RNA Gel Blot Analysis
Plants were germinated and grown under standard growth conditions as described above. Total RNA was isolated from leaf, flower, and root tissues according to the Trizol protocol (Sigma, St. Louis, MO). Total RNA from developing siliques and mature seeds was isolated using the protocol of Vicient and Delseny (1999). RNA was separated on a 1% agarose gel in formaldehyde containing 0.3 μg·mL−1 ethidium bromide. The RNA was transferred to a nylon membrane overnight in 10× SSC (1.5 M NaCl and 0.15 M sodium citrate, pH 7.0). Radioactive-labeled probes were synthesized with the DecaPrime II kit (Ambion, Austin, TX), and blots were hybridized and washed at high stringency to avoidcross-hybridization.
T-DNA Mutant Identification, Isolation, and Complementation
The Arabidopsis T-DNA–tagged lines available through ABRC (Feldmann, 1991) were screened for the presence of a mutant containing a T-DNA insertion in the LACS2 gene. PCR was performed on pooled DNA using the T-DNA right border primer (XR2, 5′-TGGGAAAACCTGGCGTTACCCAACTTAAT-3′) in combination with the 5′ gene-specific primer (5′F, 5′-GATAATGTGTTGTTGGTGGAAGAAGGAAG-3′) or the 3′ gene specific primer (3′R, 5′-CTATGCCCTTGTAATGTTGGAGGAGCTGT-3′). The DNA pools were screened using Takara ExTaq DNA polymerase (PanVera, Madison, WI) according to the protocol at the Arabidopsis Knockout Facility at the University of Wisconsin Biotechnology Center. The PCR products were analyzed on an agarose gel and blotted to nylon membrane in transfer buffer (1.5 M NaCl and 0.25 M NaOH). The membranes were probed with 32P-labeled fragments representing the full-length LACS2 genomic sequence synthesized with the DECAprime II kit (Ambion).
Seeds corresponding to the appropriate pool were surface-sterilized in 20% bleach + 0.1% SDS for 20 min and rinsed three times in sterile water. Sterilized seeds suspended in 0.1% agarose were germinated on medium containing 4.3 g·L−1 MS salts (Sigma), pH 5.8, 1% w/v sucrose, 0.35% w/v Phytagel (Sigma), and 100 mg·L−1 kanamycin. After 10 d, kanamycin-resistant plants were removed and transferred to soil. Genomic DNA was isolated from tissue (Asemota, 1995) and screened with primer combinations of 2F, 2R, XR2, and intronF (5′-CTTCTCCTAGCCTTCTCAAATGGAAGAT-3′).
To isolate the lacs2 mutant, genomic DNA samples isolated from 12 pools of six plants each were screened using the 2F and XR2 primers. Two pools yielded PCR products, and genomic DNA was isolated from individual plants in each pool to screen for the lacs2 mutant allele. Genomic DNA from two individuals, 3-1 and 11-4, yielded PCR products with the 2F and XR2 primers. DNA from these two lines was used as template in PCR reactions with the 2F and 2R primers. Only one line, 3-1, yielded a PCR product consistent with the presence of a wild-type LACS2 allele, indicating that 3-1 was heterozygous for the insert in LACS2, whereas 11-4 was homozygous. We used the kanamycin resistance marker present in the T-DNA construct to determine the number of T-DNA inserts present in the lacs2 mutants. Seed was germinated on media containing kanamycin and scored for resistance or sensitivity. Seed from line 11-4 was 100% kanamycin resistant (n = 95), which was consistent with PCR tests showing this line was homozygous for the T-DNA insertional allele of lacs2. However, >99% of seeds from line 3-1 (450 of 454) were also resistant to kanamycin, indicating that there were several unlinked T-DNA insertions in this line.
To obtain a lacs2 mutant with a single T-DNA insertion, progeny from plant 11-4 were backcrossed to the wild type. The resulting BC1 progeny were grown to maturity, and BC2 seeds were collected from individual plants. The BC2 seeds were plated on kanamycin to determine segregation of the T-DNA inserts. Of 19 BC2 lines, none showed a 3:1 segregation ratio on kanamycin. In one line, designated A1, four of 95 BC2 seeds were kanamycin sensitive, providing a good approximation to the 15:1 hypothesis (χ2 = 0.674; P > 0.5) and indicating that two inserts were likely present. Kanamycin-resistant plants from line A1 (n = 85) were transferred to soil and scored for the reduced growth phenotype observed in line 11-14. Of the 85 kanamycin-resistant plants, 19 displayed the 11-4 phenotype (χ2 = 0.807; P > 0.5). The fact that the kanamycin segregation showed a 15:1 ratio while the phenotype was present in 25% of the plants is in agreement with two unlinked insertional events. BC3 seeds were collected from several individuals of line A1 and subsequently plated on kanamycin to determine the segregation ratios. Nineteen of 75 plants from line A1-3 were kanamycin sensitive, which is a good fit to the 3:1 hypothesis for a single insertion (χ2 = 0.0044; P > 0.95).
All of the BC4 progeny of 1-1-10D displayed the lacs2-1 phenotype (Figure 4B). The 1-1-10D line was grown in large quantity to bulk up seed for experiments. For all experiments characterizing the lacs2-1 phenotype, the BC5 seeds of 1-1-10D were used. For the complementation of lacs2-1, the LACS2 cDNA sequence was subcloned into pBART, a derivative of pART27 (Gleave, 1992), which contains the bar gene in place of the neomycin phosphotransferase II gene. The resultant plasmid, pJAS44, was electroporated into Agrobacterium tumefaciens strain GV3101 (Holsters et al., 1980) and used to transform the lacs2-1 heterozygous plants by the floral dip method (Clough and Bent, 1998). Transformants were selected on soil containing 25 mg·L−1 glufosinate ammonium (Finale; AgrEvo, Montvale, NJ).
Leaf Measurements and Chlorophyll Measurements
To quantify differences in leaf length, wild-type and lacs2-1 plants were grown under standard growing conditions. At 22 d, rosette leaves were removed in sequential order, starting with cotyledons, and measured. For chlorophyll leaching experiments, the aerial portions of rosettes (36 d) were removed from the soil, weighed, and immersed in 80% EtOH. At the indicated time points, aliquots were removed, and absorbance was measured at 664 nm and 647 nm. Total chlorophyll was calculated according to Wintermans and de Mots (1965). Chlorophyll content of lacs2-1 leaves was not significantly different from the wild type on a fresh weight basis (data not shown).
Electron Microscopic Techniques
Leaf surfaces were visualized with standard scanning electron microscopy preparative techniques. At 49 d, wild-type and lacs2-1 leaves were removed at the petiole and fixed overnight in 2% paraformaldehyde + 2% glutaraldehyde in 0.05 M Pipes buffer, pH 7.2. After an overnight postfix in 1% osmium tetroxide, leaf samples were dehydrated through a graded ethanol series. After being processed through a critical point dryer, leaves were sputter-coated with gold (150 to 200 Å). Surfaces were viewed with a scanning electron microscope (Model S570; Hitachi, Tokyo, Japan). For the pollen germination assay, wild-type Col pollen was applied to abaxial and adaxial surfaces of wild-type Col, lacs2-1, and fdh-3 leaves on 42-d-old rosettes. The fdh-3 seeds were a kind gift from Susan Lolle (Purdue University, West Lafayette, IN). The plants remained in the growth chamber for 24 h, at which point the leaves were removed from the plants and fixed according to protocol above. The apparent hydration of pollen on the wild type is likely an artifact of fixation and processing for scanning electron microscopy (Lolle and Cheung, 1993).
TEM visualization of epidermal cell cuticle was done according to Sieber et al. (2000), with modifications. Briefly, 34-d-old leaves were fixed overnight in 2% paraformaldehyde + 2% glutaraldehyde in 0.05 M Pipes, pH 7.2. After an overnight postfix in 1% osmium tetroxide, leaf pieces were dehydrated through a graded acetone series into Spurr's epoxy resin. Ultrathin sections (80 to 100 nm) were stained in 4% uranyl acetate for 20 min, followed by alkaline lead citrate for 10 min (Reynolds, 1963). Sections were visualized on a TEM (JEM 1200EX; JEOL, Tokyo, Japan) for quantification of cell wall and cutin thickness. Leaf samples were positioned perpendicular to the knife before sectioning. A minimum of four negatives each for the wild type and lacs2-1 (from at least two grids, each grid representing a different leaf sample) were scanned and imported into Scion Image for measurements (http://www.scioncorp.com/). On each negative, triplicate measurements were made.
Fatty Acid and Wax Analysis
FAMES analysis of leaf and seed tissues was done by placing tissue in 1.5 mL of 2.5% H2SO4 (v/v) in methanol. After a 1-h incubation at 80°C, 1.5 mL water and 0.3 mL hexane were added. The hexane phases were then transferred to vials for GC analysis. One-microliter samples were analyzed by GC on a 15 m × 0.53 mm Supelcowax column (Supelco, Bellefonte, PA) and quantified with flame ionization detection. The chromatograph was programmed for an initial temperature of 150°C for 1 min, followed by a 15°C · min−1 ramp to 240°C, and then held for 2 min. For quantification of FAMES, a 17:0 free fatty acid standard was added before derivatization.
For analysis of wax components, leaves (38 d) and stems (43 d) were immersed in chloroform for 30 s. An internal standard (n-octacosane, Sigma) was added before derivatization using N,O-bis(trimethylsilyl) fluoroacetamide (BSTFA) containing 1% (v/v) trimethylchlorosilane (TMCS). Chloroform extracts were dried down under a gentle stream of argon. After addition of 100 μL pyridine and 100 μL BSTFA + 1% TMCS (Regis Technologies, Morton Grove, IL), sealed tubes were incubated at 110°C for 10 min. After samples were dried down, the extracts were dissolved in heptane:toluene (1:1; v/v). Samples (1 μL) were injected (splitless) into an HP-5 column. The 60-min program consisted of an initial temperature of 90°C for 3 min, followed by a 5°C min−1 ramp to 325°C, and then held for 10 min. Peak areas were determined from background-subtracted spectra.
Histochemical GUS Analysis
A 1785-bp portion of the LACS2 promoter region was amplified by PCR using primer 2PROM5 (5′-ATAGGATCCTGAGAAACCTGTTCTGACCAAT-3′) and 2PROM3 (5′-CTAGGATCCTTGTGTAATATATAAAGTTGAGG-3′), which included sequence immediately before the ATG. The primers contain BamHI sites to facilitate insertion into the transformation vector pBI101.1 (Sigma). The resultant plasmid, pJAS45, was sequenced and did not contain any errors when compared with the BAC sequence (F13F21; GenBank accession number AC007504). The resultant plasmids were electroporated into A. tumefaciens and used to transform wild-type Arabidopsis (Col) as described above. T2 seeds were harvested and selected on kanamycin medium and transferred to soil. Fifteen days after sowing, leaves were removed and placed in the GUS substrate buffer (50 mM sodium phosphate buffer, pH 7.0, 0.1% Triton X-100, 3 mM potassium ferricyanide, 3 mM potassium ferrocyanide, and 1 mM X-Gluc). After a 5-min vacuum infiltration, samples were placed in a 37°C incubator. The next day, tissue was fixed in FAA (50% EtOH, 5% acetic acid, and 3.7% formaldehyde) for 60 min to overnight and dehydrated through an ethanol series. Samples were processed through ethanol to CitriSolv and then into paraffin. The paraffin-embedded samples sat at 62°C for 3 d, after which blocks were cast. Sections (15 μm) were deparaffinized in CitriSolv and hydrated in oil (under coverslip) for viewing.
LACS Enzyme Expression and Activity Assays
LACS enzymes were overproduced in the K27 mutant of E. coli, which is mutated in its sole LACS gene, fadD (Ray and Cronan, 1976), as described previously (Shockey et al., 2002). Protein concentrations of membrane fractions were determined using the BCA kit (Pierce, Rockford, IL). The potassium salts of palmitic acid (Nu-Chek Prep, Elysian, MN) or 16-hydroxypalmitic acid (Sigma) were prepared by neutralization of methanolic solutions of the free fatty acids with methanolic potassium hydroxide. The fatty acid soaps were then dissolved in 20 mM Triton X-100. Enzyme activity levels were measured using the colorimetric, enzyme-coupled assay described by Ichihara and Shibasaki (1991).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AC007504.
Acknowledgments
We thank the staff at the Electron Microscopy Center of Washington State University, including Valerie Lynch-Holm, Vince Franceschi, and Chris Davitt. We also thank Christiane Nawrath, Irene Benveniste, Pappachan Kolattukudy, Susan Lolle, Penny von Wettstein-Knowles, and Mechthild Tegeder for helpful discussions. Special thanks to Gustavo Bonaventure (Michigan State University; Ohlrogge Lab) for detailed wax analysis protocols. This work was supported by Dow Chemical, Dow AgroSciences, the National Science Foundation (Grant IBN-0084329), and the Agricultural Research Center, Washington State University.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: John Browse (jab@wsu.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.017608.
References
- Andrews, J., and Keegstra, K. (1983). Acyl-CoA synthetase is located in the outer membrane and acyl-CoA thioesterase in the inner membrane of pea chloroplast envelopes. Plant Physiol. 72, 735–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asemota, H.N. (1995). A fast, simple, and efficient miniscale method for the preparation of DNA from tissues of yam (Dioscorea spp.). Plant Mol. Biol. Rep. 13, 214–218. [Google Scholar]
- Becraft, P.W. (1999). Development of the leaf epidermis. Curr. Top. Dev. Biol. 45, 1–40. [DOI] [PubMed] [Google Scholar]
- Block, M., Dorne, A.-J., Joyard, J., and Douce, R. (1983). The acyl-CoA synthetase and acyl-CoA thioesterase are located on the outer and inner membrane of the chloroplast envelope, respectively. FEBS Lett. 153, 377–381. [Google Scholar]
- Browse, J., and Somerville, C.R. (1991). Glycerolipid metabolism, biochemistry and regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42, 467–506. [Google Scholar]
- Campisi, L., Yang, Y., Yi, Y., Heilig, E., Herman, B., Cassista, A.J., Allen, D.W., Xiang, H., and Jack, T. (1999). Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 17, 699–707. [DOI] [PubMed] [Google Scholar]
- Cheesbrough, T.M., and Kolattukudy, P.E. (1984). Alkane biosynthesis by decarbonylation of aldehydes catalyzed by a particulate preparation from Pisum sativum. Proc. Natl. Acad. Sci. USA 81, 6613–6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Goodwin, S.M., Boroff, V.L., Liu, X., and Jenks, M.A. (2003). Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15, 1170–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Croteau, R., and Kolattukudy, P.E. (1974). Biosynthesis of hydroxyfatty acid polymers. Enzymatic synthesis of cutin from monomer acids by cell-free preparations from the epidermis of Vicia faba leaves. Biochemistry 13, 3193–3202. [DOI] [PubMed] [Google Scholar]
- Feldmann, K.A. (1991). T-DNA insertion mutagenesis in Arabidopsis: Mutational spectrum. Plant J. 1, 71–82. [Google Scholar]
- Froehlich, J.E., Itoh, A., and Howe, G.A. (2001). Tomato allene oxide synthase and fatty acid hydroperoxide lyase, two cytochrome p450s involved in oxylipin metabolism, are targeted to different membranes of chloroplast envelope. Plant Physiol. 125, 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda, M., Shockey, J., Werber, M., Wolter, F.P., and Heinz, E. (2002). Two long-chain acyl-CoA synthetases from Arabidopsis thaliana involved in peroxisomal fatty acid beta-oxidation. Plant J. 32, 93–103. [DOI] [PubMed] [Google Scholar]
- Gargiulo, C.E., Stuhlsatz-Krouper, S.M., and Schaffer, J.E. (1999). Localization of adipocyte long-chain fatty acyl-CoA synthetase at the plasma membrane. J. Lipid Res. 40, 881–892. [PubMed] [Google Scholar]
- Gerbling, H., and Gerhardt, B. (1987). Activation of fatty acids by non-glyoxysomal peroxisomes. Planta 171, 386–392. [DOI] [PubMed] [Google Scholar]
- Gleave, A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203–1207. [DOI] [PubMed] [Google Scholar]
- Holsters, M., et al. (1980). The functional organization of the nopaline A. tumefaciens plasmid pTiC58. Plasmid 3, 212–230. [DOI] [PubMed] [Google Scholar]
- Ichihara, K., and Shibasaki, Y. (1991). An enzyme-coupled assay for acyl-CoA synthetase. J. Lipid Res. 32, 1709–1712. [PubMed] [Google Scholar]
- James, D.W., Jr., Lim, E., Keller, J., Plooy, I., Ralston, E., and Dooner, H.K. (1995). Directed tagging of the Arabidopsis FATTY ACID ELONGATION 1 (FAE1) gene with the maize transposon Activator. Plant Cell 7, 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks, M.A., Eigenbrode, S.D., and Lemieux, B. (2002). Cuticular waxes of Arabidopsis. In The Arabidopsis Book, C.R. Somerville and E.M. Myerowitz, eds (Rockville, MD: American Society of Plant Biologists), http://www.aspb.org/publications/arabidopsis. [DOI] [PMC free article] [PubMed]
- Jenks, M.A., Rashotte, A.M., Tuttle, H.A., and Feldmann, K.A. (1996). Mutants in Arabidopsis thaliana altered in epicuticular wax and leaf morphology. Plant Physiol. 110, 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolattukudy, P.E. (1981). Structure, biosynthesis, and biodegradation of cutin and suberin. Annu. Rev. Plant Physiol. 32, 539–567. [Google Scholar]
- Kolattukudy, P.E. (2001). Polyesters in higher plants. Adv. Biochem. Eng. Biotechnol. 71, 1–49. [DOI] [PubMed] [Google Scholar]
- Kolattukudy, P.E., and Walton, T.J. (1972). Structure and biosynthesis of the hydroxy fatty acids of cutin in Vicia faba leaves. Biochemistry 11, 1897–1907. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Hanhart, C.J., and Thiel, F. (1989). A genetic and phenotypic description of eceriferum (cer) mutants in Arabidopsis thaliana. J. Hered. 80, 118–122. [Google Scholar]
- Kornberg, A., and Pricer, W.E., Jr. (1953). Enzymatic synthesis of the coenzymeA derivatives of long chain fatty acids. J. Biol. Chem. 204, 329–343. [PubMed] [Google Scholar]
- Lolle, S.J., Berlyn, G.P., Engstrom, E.M., Krolikowski, K.A., Reiter, W.-D., and Pruitt, R.E. (1997). Developmental regulation of cell interactions in the Arabidopsis fiddlehead-1 mutant: A role for the epidermal cell wall and cuticle. Dev. Biol. 189, 311–321. [DOI] [PubMed] [Google Scholar]
- Lolle, S.J., and Cheung, A.Y. (1993). Promiscuous germination and growth of wild-type pollen from Arabidopsis and related species on the shoot of the Arabidopsis mutant, fiddlehead. Dev. Biol. 155, 250–258. [DOI] [PubMed] [Google Scholar]
- Lolle, S.J., Cheung, A.Y., and Sussex, I.M. (1992). Fiddlehead: An Arabidopsis mutant constitutively expressing an organ fusion program that involves interactions between epidermal cells. Dev. Biol. 152, 383–392. [DOI] [PubMed] [Google Scholar]
- Lolle, S.J., Hsu, W., and Pruitt, R.E. (1998). Genetic analysis of organ fusion in Arabidopsis thaliana. Genetics 149, 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNevin, J.P., Woodward, W., Hannoufa, A., Feldmann, K.A., and Lemieux, B. (1993). Isolation and characterization of eceriferum (cer) mutants induced by T-DNA insertions in Arabidopsis thaliana. Genome 36, 610–618. [DOI] [PubMed] [Google Scholar]
- Mou, Z., He, Y., Dai, Y., Liu, X., and Li, J. (2000). Deficiency in fatty acid synthase leads to premature cell death and dramatic alterations in plant morphology. Plant Cell 12, 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C. (2002). The biopolymers cutin and suberin. In The Arabidopsis Book, C.R. Somerville and E.M. Myerowitz, eds (Rockville, MD: American Society of Plant Biologists), http://www.aspb.org/publications/arabidopsis. [DOI] [PMC free article] [PubMed]
- Ohlrogge, J.B., and Jaworski, J.G. (1997). Regulation of fatty acid synthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 109–136. [DOI] [PubMed] [Google Scholar]
- Post-Beittenmiller, D. (1996). Biochemistry and molecular biology of wax production in plants. Annu. Rev. Plant Physiol. and Plant Mol. Biol. 47, 405–430. [DOI] [PubMed] [Google Scholar]
- Pruitt, R.E., Vielle-Calzada, J.-P., Ploense, S.E., Grossniklaus, U., and Lolle, S.J. (2000). FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc Natl. Acad. Sci. USA 97, 1311–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyee, J., and Kolattukudy, P.E. (1995). The gene for the major cuticular wax-associated protein and three homologous genes from broccoli (Brassica oleracea) and their expression patterns. Plant J. 7, 49–59. [DOI] [PubMed] [Google Scholar]
- Ray, T.K., and Cronan, J.E. (1976). Mechanism of phospholipids biosynthesis in Escherichia coli: acyl-CoA synthetase is not required for the incorporation of intracellular free fatty acids into phospholipids. Biochem. Biophys. Res. Commun. 69, 506–513. [DOI] [PubMed] [Google Scholar]
- Reynolds, E.S. (1963). The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17, 208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer, J.E., and Lodish, H.F. (1994). Expression cloning and characterization of a novel adipocyte long chain fatty acid transport protein. Cell 79, 427–436. [DOI] [PubMed] [Google Scholar]
- Schnurr, J.A., Shockey, J., deBoer, G.-J., and Browse, J. (2002). Fatty acid export from the chloroplast: Molecular characterization of a major plastidial acyl-coenzymeA synthetase from Arabidopsis. Plant Physiol. 129, 1700–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey, J., Fulda, M., and Browse, J. (2002). Arabidopsis contains nine long-chain acyl-coenzymeA synthetases that participate in fatty acid and glycerolipid metabolism. Plant Physiol. 129, 1710–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber, P., Schorderet, M., Ryser, U., Buchala, A., Kolattukudy, P., Métraux, J.-P., and Nawrath, C. (2000). Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell 12, 721–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliday, C.L., and Kolattukudy, P.E. (1977). Biosynthesis of cutin. Plant Physiol. 59, 1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer, A., and Poethig, R.S. (1994). Leaf development in Arabidopsis. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 379–401.
- Vicient, C.M., and Delseny, M. (1999). Isolation of total RNA from Arabidopsis thaliana seeds. Anal. Biochem. 268, 412–413. [DOI] [PubMed] [Google Scholar]
- Vioque, J., and Kolattukudy, P.E. (1997). Resolution and purification of an aldehyde-generating and an alcohol-generating fatty acyl-CoA reductase from pea leaves (Pisum sativum L.). Arch. Biochem. Biophys. 340, 64–72. [DOI] [PubMed] [Google Scholar]
- Walton, T.J., and Kolattukudy, P.E. (1972). Enzymatic conversion of 16-hydroxypalmitic acid into 10,16-dihydroxypalmitic acid in Vicia faba epidermal extracts. Biochem. Biophys. Res. Commun. 46, 16–21. [DOI] [PubMed] [Google Scholar]
- Wellesen, K., Durst, F., Pinot, F., Benveniste, I., Nettesheim, K., Wisman, E., Steiner-Lange, S., Saedler, H., and Yephremov, A. (2001). Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid ω-hydroxylation in development. Proc. Natl. Acad. Sci. USA 98, 9694–9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermans, J.F., and de Mots, A. (1965). Spectrophotometric characteristics of chlorophylls a and b and their pheophytins in ethanol. Biochim. Biophys. Acta 109, 448–453. [DOI] [PubMed] [Google Scholar]
- Yephremov, A., Wisman, E., Huijser, P., Huijser, C., Wellesen, K., and Saedler, H. (1999). Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11, 2187–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, Z., DiRusso, C.C., Ctrnacta, V., and Black, P.N. (2002). Fatty acid transport in Saccharomyces cerevisiae. J. Biol. Chem. 277, 31062–31071. [DOI] [PubMed] [Google Scholar]