Abstract
Type-A Arabidopsis (Arabidopsis thaliana) response regulators (ARRs) are a family of 10 genes that are rapidly induced by cytokinin and are highly similar to bacterial two-component response regulators. We have isolated T-DNA insertions in six of the type-A ARRs and constructed multiple insertional mutants, including the arr3,4,5,6,8,9 hextuple mutant. Single arr mutants were indistinguishable from the wild type in various cytokinin assays; double and higher order arr mutants showed progressively increasing sensitivity to cytokinin, indicating functional overlap among type-A ARRs and that these genes act as negative regulators of cytokinin responses. The induction of cytokinin primary response genes was amplified in arr mutants, indicating that the primary response to cytokinin is affected. Spatial patterns of ARR gene expression were consistent with partially redundant function of these genes in cytokinin signaling. The arr mutants show altered red light sensitivity, suggesting a general involvement of type-A ARRs in light signal transduction. Further, morphological phenotypes of some arr mutants suggest complex regulatory interactions and gene-specific functions among family members.
INTRODUCTION
Cytokinins are N6-substituted adenine derivatives that have been implicated in nearly all aspects of plant growth and development, including cell division, shoot initiation and development, light responses, and leaf senescence (Mok and Mok, 2001b). Lowering endogenous levels of cytokinin inhibits shoot development and increases primary root growth and branching, indicating that cytokinin plays opposite roles in the shoot and root meristems (Werner et al., 2001). Ectopic expression and overexpression of cytokinin biosynthetic genes have also demonstrated that elevated levels of cytokinin can release apical dominance, reduce root development, delay senescence, and enhance shoot regeneration in cultured tissues (Medford et al., 1989; Smigocki, 1991; Li et al., 1992; Gan and Amasino, 1995; Sa et al., 2001; Zubko et al., 2002).
The current model for cytokinin signaling in plants is similar to the two-component phosphorelay system with which bacteria sense and respond to environmental changes. A simple two-component system involves a His sensor kinase and a response regulator (Stock et al., 2000; West and Stock, 2001). The His kinase perceives environmental stimuli via the input domain and autophosphorylates on a conserved His residue within the kinase domain. The phosphoryl group is subsequently transferred to a conserved Asp residue on the receiver domain of a response regulator, which mediates downstream responses via the output domain. Multicomponent phosphorelay systems occur in most eukaryotic and some prokaryotic systems, which employ His kinase signal transduction in a multistep His-Asp-His-Asp phosphotransfer process (Stock et al., 2000; West and Stock, 2001). The Arabidopsis (Arabidopsis thaliana) cytokinin receptors (CRE1, AHK2, and AHK3) are similar to bacterial His sensor hybrid kinases in two-component signaling, containing a receiver domain fused to the His kinase domain (Inoue et al., 2001; Suzuki et al., 2001; Ueguchi et al., 2001a, 2001b; Yamada et al., 2001). The cytokinin receptors are predicted to signal through His phosphotransfer proteins to ultimately alter the phosphorylation state of the Arabidopsis response regulators (ARRs) in a multistep phosphorelay (Hutchison and Kieber, 2002).
ARRs can be broadly classified into two groups (type A and type B) by the similarity of their receiver domain sequences and by their C-terminal characteristics. Like most bacterial response regulators, type-B ARRs have C-terminal domains that contain DNA binding, nuclear localization, and transcription activator domains (Sakai et al., 1998, 2000, 2001). C-terminal sequences of type-A ARRs are short and have yet to be assigned functions. Type-A and type-B ARR homologs are found in other dicotyledonous and monocotyledonous plants, including maize (Zea mays) and rice (Oryza sativa) (Kieber, 2002; Asakura et al., 2003).
There are 10 type-A ARRs that fall into five very similar pairs (Figure 1A). The rates of transcription of most of the type-A ARRs, but not the type-B ARRs, are rapidly and specifically induced in response to exogenous cytokinin, and this induction occurs in the absence of de novo protein synthesis (Taniguchi et al., 1998; D'Agostino et al., 2000). Gene expression differs among various type As, with ARR4, ARR8, and ARR9 displaying relatively high basal levels and ARR5, ARR6, ARR7, and ARR15 showing the greatest fold induction in response to cytokinin (D'Agostino et al., 2000; Rashotte et al., 2003). Transcription of type-A ARRs is regulated in part by type-B ARRs (Hwang and Sheen, 2001; Sakai et al., 2001). Overexpression of some type-A ARRs inhibits expression of an ARR6 promoter-luciferase reporter in cultured Arabidopsis cells, suggesting that type-A ARRs have the ability to negatively regulate their own transcription (Hwang and Sheen, 2001). Consistent with this, overexpression of ARR15 leads to decreased cytokinin sensitivity (Kiba et al., 2003). ARR4 has been shown to interact with and stabilize the far-red active form of phytochrome B (PhyB); overexpression of ARR4 in Arabidopsis also confers hypersensitivity to red light (Sweere et al., 2001), indicating a role in light-regulated development.
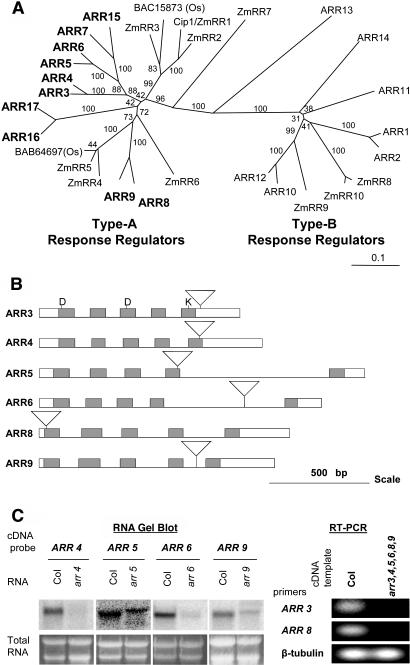
Figure 1.
Type-A ARR Phylogeny and Positions of T-DNA Insertions.
(A) An unrooted phylogenetic tree was made using receiver domain sequences of type-A and type-B response regulators from Arabidopsis (ARR), maize (ZmRR), and rice (Os with accession numbers). Full-length protein sequences of the response regulators were obtained from Entrez Protein Database (National Center for Biotechnology Information [NCBI]), and their receiver domain sequences were identified by searching Conserved Domain Database (version 1.62; NCBI). Receiver domain sequences were aligned using the CLUSTAL W program (version 1.81; University of Nijmegen, http://www.cmbi.kun.nl/bioinf/tools/clustalw.shtml), and the phylogenetic tree was constructed with 1000 bootstrapping replicates. The unrooted tree is presented in TreeView (version 1.6.6, 2001; Page, 1996). The bootstrap values are indicated on the tree. Scale bar represents 0.1 amino acid substitution per site.
(B) Positions of T-DNA insertions in the type-A arr mutants. The insertional mutants were identified by PCR screening, and the site of insertion determined by DNA sequencing of the border fragment. Boxes represent exons, lines represent introns, and inverted triangles indicate T-DNA insertions. Receiver domains are shaded. The DDK residues that are conserved in two-component receiver domains are indicated.
(C) Expression of type-A ARRs in insertional mutants. RNA from 3-d-old seedlings was either blotted to nylon for RNA gel blot analysis (left) or transcribed in vitro to cDNA for use in an RT-PCR reaction (right) as described in Methods. For the RNA gel blot, different cDNA clones were used as hybridization probes, as indicated above the figure, and the ethidium bromide–stained agarose gel is shown below. For RT-PCR, primers were designed to amplify the first three exons of ARR3 or the entire β-tubulin gene as a control. Col, wild-type Columbia.
Using the model plant Arabidopsis, we took a reverse genetic approach to study the function of type-A ARRs. We isolated T-DNA insertions in six of the 10 type-A ARRs (three of the five most similar pairs) and have constructed various combinations of these mutations, including the arr3,4,5,6,8,9 hextuple mutant. Overall, we show that these genes have overlapping functions and act as negative regulators of cytokinin signaling. We show that the mutants are affected in their response to light. In addition, we identify morphological phenotypes in a subset of arr mutants that support some functional specificity within the type-A family of ARRs.
RESULTS
Isolation of Insertions in Response Regulator Loci
To study the function of type-A ARRs, we isolated T-DNA insertions in six of the 10 genes: ARR3 (At1g59940), ARR4 (At1g10470), ARR5 (At3g48100), ARR6 (At5g62920), ARR8 (At2g41310), and ARR9 (At3g57040). These mutations cover three of the five gene pairs, ARR3/ARR4, ARR5/ARR6, and ARR8/ARR9, identified by phylogenetic analysis (Figure 1A; D'Agostino et al., 2000). We identified individual insertions in each gene by PCR screening and located the sites of insertions by DNA sequencing (see supplemental data online). In arr3, the T-DNA inserted in the C-terminal domain, 26 bp downstream of the sequence encoding the receiver domain (Figure 1B). The insertions in arr4, arr5, arr6, arr8, and arr9 are predicted to disrupt the receiver domain of the respective genes. Furthermore, the insertions in arr5, arr6, arr8, and arr9 occur in the coding region before an invariant Lys residue in the receiver domain, and thus are unlikely to produce functional proteins (Figure 1B).
We examined RNA expression of the type-A ARRs to determine if the T-DNA insertions affected the level of RNA in each of the mutant lines. RNA gel blot analysis showed that arr4, arr6, and arr9 mutants had substantially reduced levels of the transcripts corresponding to the mutated genes (Figure 1C). The arr5 mutant displayed a shift in transcript size, as well as a decrease in transcript levels (Figure 1C). Reverse transcription (RT)–PCR analysis showed that the T-DNA insertions in ARR3 and ARR8 abolished expression of the respective transcripts (Figure 1C). We conclude that the T-DNA insertions in arr3 and arr8 result in null alleles, whereas the remaining insertions result in hypomorphic alleles.
Adult Phenotype of arr Mutants
When grown under long-day conditions on soil, the six single arr insertion lines were indistinguishable at all stages of growth when compared with their wild-type counterparts (data not shown). Likewise, arr3, arr6, arr8, and arr9 grown under short days were also indistinguishable from the wild type (Figure 2A). However, arr4 and arr5 displayed subtle alterations in rosette morphology when grown under short-day conditions: arr4 adult plants developed mildly elongated petioles, and the rosette size of the arr5 mutant was reduced (Figure 2A; see supplemental data online).
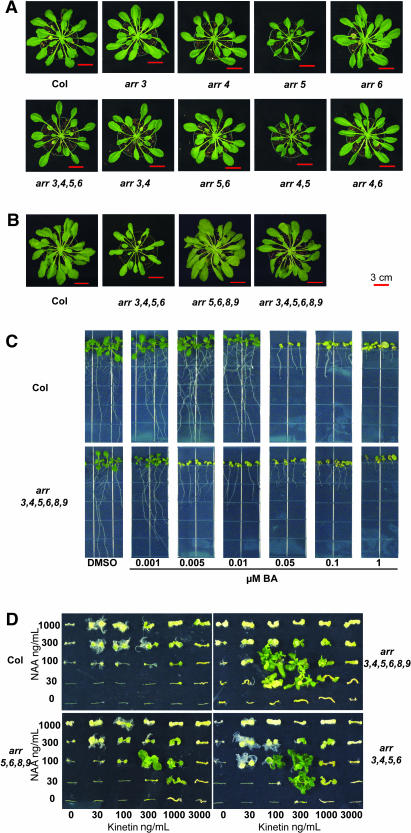
Figure 2.
arr Mutant Phenotypes.
(A) and (B) arr adult plants are affected in short-day conditions. Plants of the genotypes noted were grown in short-day conditions (8-h-light/16-h-dark) for 9 weeks. At least eight plants per genotype were examined, and photographs of representative plants for each line are shown. The experiment was conducted three times with similar results. The red scale bar in each photograph corresponds to 3 cm. Plants in (A) and (B) are from two separate experiments.
(C) arr seedlings are more sensitive to cytokinin. Seedlings were grown vertically on plates supplemented with the specified concentrations of BA or a DMSO vehicle control under constant light conditions at 23°C. Seedlings were photographed at 10 d.
(D) arr mutants form elaborate shoot structures on low cytokinin concentrations and fewer roots on high auxin concentrations in shoot initiation assay. Hypocotyls were excised from seedlings grown for 3 d in the dark, followed by 3 d in dim light, and transferred to media containing various concentrations of auxin (NAA) and cytokinin (kinetin) for 4 weeks under constant light. Five hypocotyls of each genotype were examined at each concentration. One hypocotyl representative of the response at each concentration was selected and arranged to create a composite photograph for each genotype.
To examine the genetic interactions among the six type-A arr mutations, higher order mutants were generated. These include double mutants between each highly similar pair (arr3,4, arr5,6, and arr8,9), double mutants across pairs (arr4,5 and arr4,6), quadruple mutants (arr3,4,5,6, arr3,4,8,9, and arr5,6,8,9), and the arr3,4,5,6,8,9 hextuple mutant. The elongated petioles of the arr4 single mutant were enhanced in the arr3,4 double mutant, indicating functional redundancy between the two members of this gene pair (Figure 2A; see supplemental data online). Surprisingly, the reduced rosette size of arr5 was not enhanced but suppressed by the arr6 mutation, suggesting antagonistic function. The arr4,5 double mutant appeared similar to the arr5 parent, and the arr4,6 double mutant was similar to the arr4 parent. The elongated petioles of arr4 and arr3,4 were further enhanced in arr3,4,5,6, but the overall rosette size was similar to that of the wild-type parent (Figures 2A and 2B; see supplemental data online). The increased petiole elongation in the arr3,4,5,6 quadruple mutant suggests that although ARR5 and ARR6 may act antagonistically to each other in regulating rosette size, as a pair they still function additively with ARR3 and ARR4 in the regulation of petiole elongation.
The arr5,6,8,9 quadruple mutant was indistinguishable from the wild type, as were the arr5,6 and arr8,9 double mutants (Figures 2A and 2B). However, the arr3,4,5,6,8,9 hextuple mutant had intermediate petiole length between arr3,4,5,6 and the wild type (Figure 2B; see supplemental data online), suggesting complex interactions between these genes.
arr Mutant Seedling Root Elongation Is More Sensitive to Cytokinin Inhibition
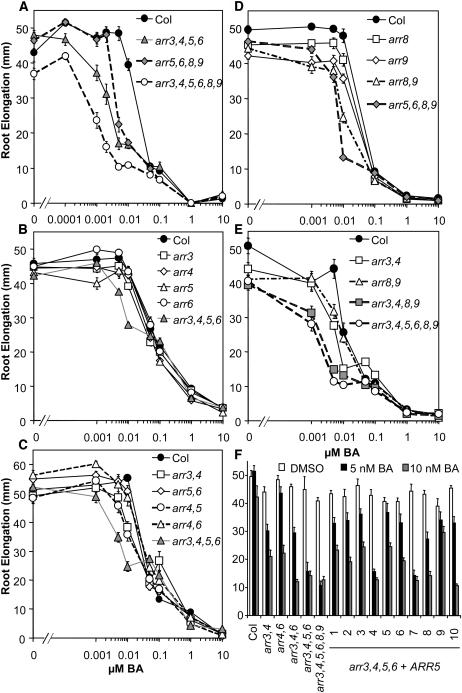
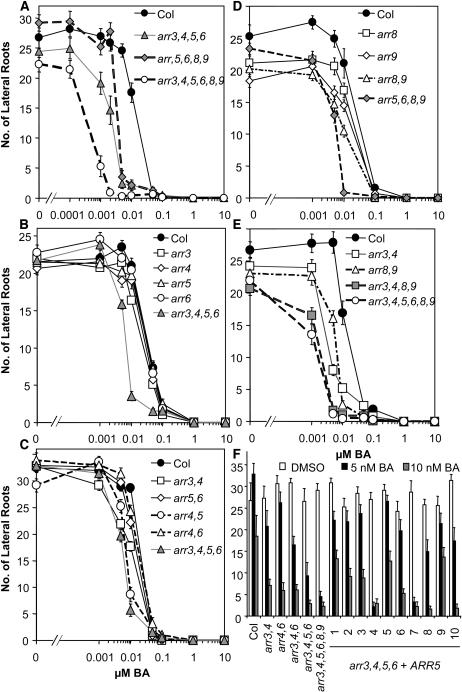
To assess the role of type-A ARRs in the cytokinin response pathway, we examined root elongation in response to exogenous cytokinin. We compared root elongation of wild-type with arr3,4,5,6,8,9 mutant seedlings across a range of cytokinin concentrations between 1 nM and 10 μM benzyladenine (BA) (Figures 2C and 3). Wild-type root elongation was not affected by BA concentrations <5 nM. Upon further increase in BA concentration, primary root elongation decreased sharply, with a half-maximal inhibition at ∼12 nM (Figure 3A). In the absence of exogenous cytokinin, roots of the arr3,4,5,6,8,9 hextuple mutant were shorter than roots of the wild type (P < 10−4 in two-tailed Student's t test). In the presence of low doses (<50 nM) of BA, the arr3,4,5,6,8,9 mutant displayed increased sensitivity to BA, as shown by a greater inhibition of root elongation than wild-type roots at comparable concentrations. The arr3,4,5,6,8,9 hextuple mutant reached half-maximal inhibition at ∼2 nM BA. At higher BA concentrations (≥50 nM), the mutant response was similar to that of the wild type (Figure 3A). This resulted in a change in the overall shape of the dose–response curve from primarily monophasic in the wild type to biphasic in the hextuple mutant. Interestingly, the central part of the response curve in the hextuple mutant showed little or no change in inhibition of root elongation as the concentration of BA was increased from 8 to 100 nM BA. This dramatic shape change in the dose–response curve was very reproducible, consistently observed among three separate experiments (Figures 3A and 3E, and data not shown).
Figure 3.
arr Seedlings Are More Sensitive to Cytokinin Inhibition of Root Elongation.
(A) to (E) Seedlings were grown vertically on plates supplemented with the specified concentrations of BA or a DMSO vehicle control under constant light conditions at 23°C. Root elongation between days 4 and 9 was measured as described in Methods. Results shown were pooled from an experimental set of three independent samples of 10 to 15 individual seedlings. Error bars represent se (n > 30). Each experiment was repeated at least twice with consistent results.
(F) Complementation of arr3,4,5,6 phenotype with ARR5. A construct containing a wild-type ARR5 cDNA driven by the ARR5 promoter was transformed into arr3,4,5,6. Wild-type seedlings, various arr mutant seedlings, and 10 transformed lines were grown as in (A) to (E) in the presence of 5 nM BA (black bars), 10 nM BA (shaded bars), or a DMSO vehicle control (open bars). Ten independent T1 lines are denoted 1 to 10. Error bars represent se (n = 15).
To examine the contributions of individual ARR genes to cytokinin responsiveness and their interactions, inhibition of primary root elongation of single, double, and quadruple mutants in response to increasing concentrations of exogenous BA was examined. Single arr mutants were indistinguishable from the wild type in this cytokinin response (Figures 3B and 3D), which coupled with the cytokinin-hypersensitive phenotype of the higher order mutants indicates genetic redundancy among these genes. The arr5,6 and arr4,6 double mutants showed subtle differences in cytokinin sensitivity compared with the wild type, whereas the arr3,4 and arr4,5 double mutants exhibited a significant increase in cytokinin inhibition of root elongation intermediate between arr3,4,5,6 and the wild type (Figure 3C). arr8,9 also exhibited a significant increase in cytokinin sensitivity intermediate between the wild type and the arr5,6,8,9 or arr3,4,8,9 quadruple mutants (Figures 3D and 3E), indicating that all these mutations additively contribute to this phenotype of arr3,4,5,6,8,9.
The arr3,4,5,6 and arr5,6,8,9 quadruple mutants showed root elongation responses intermediate between arr3,4,5,6,8,9 and the wild type (Figure 3A). The arr3,4,8,9 mutant exhibited the greatest increase in cytokinin sensitivity among the three quadruple mutants examined, almost approaching the hypersensitivity of the arr3,4,5,6,8,9 hextuple mutant (Figure 3E), indicating that the component ARRs play a key role in this cytokinin response. However, ARR5 and ARR6 still contribute to the effect of cytokinin on root elongation because arr3,4,8,9 is significantly less sensitive than arr3,4,5,6,8,9 at 5 and 10 nM BA (t test P < 0.01), whereas arr3,4,5,6 and arr5,6,8,9 are also significantly more sensitive than arr3,4 and arr8,9 (t test P < 10−5 and P < 10−10 at 10 nM BA), respectively (Figures 3A, 3C, and 3D).
arr Mutant Seedling Lateral Root Formation Is More Sensitive to Cytokinin Inhibition
Formation of lateral roots is inhibited by cytokinin in plants (Werner et al., 2001). We examined the number of lateral roots on wild-type and all the arr mutant 10-d-old seedlings across the same concentration range used in the root elongation assay. In wild-type seedlings, the effect of BA on lateral root formation decreased dramatically between 5 and 50 nM BA, reaching half-maximal inhibition at ∼12 nM BA, and essentially no lateral roots were detected at BA concentrations >1 μM (Figures 2C and 5A). In the arr3,4,5,6,8,9 hextuple mutant, significantly fewer lateral roots than the wild type were formed in the absence of BA (t test P < 10−7) (Figures 2C and 4A). The range of inhibition of lateral roots was also markedly shifted to lower BA concentrations in arr3,4,5,6,8,9, with a half-maximal inhibition of ∼1 nM BA (Figure 4A).
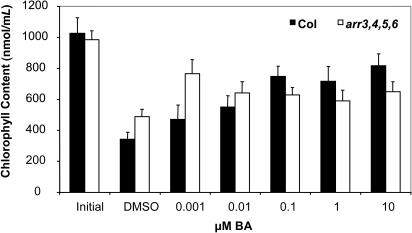
Figure 5.
arr3,4,5,6 Shows Delayed Leaf Senescence.
Fully expanded leaves were excised from 3.5-week-old plants and floated on water supplemented with various concentrations of cytokinin for 10 d in the dark. Chlorophyll content was determined spectrophotometrically as described in Methods. Three independent plates with six leaves per plate were examined at each concentration. Two chlorophyll measurements were taken per plate. Results shown are pooled from three independent experiments ±se (n = 18).
Figure 4.
arr Seedlings Are More Sensitive to Cytokinin Inhibition of Lateral Root Formation.
(A) to (E) Seedlings were grown vertically on plates supplemented with the specified concentrations of BA or a DMSO vehicle control under constant light conditions at 23°C. The total number of lateral roots was quantified at 9 d. Results shown were collected from the same experimental sets as in Figure 2. Error bars represent SE (n > 30).
(F) Complementation of arr3,4,5,6 phenotype with ARR5. A construct containing a wild-type ARR5 cDNA driven by the ARR5 promoter was transformed into arr3,4,5,6. Wild-type seedlings, various arr mutant seedlings, and seven transformed lines were grown as in (A) to (E) in the presence of 5 nM BA (black bars), 10 nM BA (shaded bars), or a DMSO vehicle control (open bars). Ten independent T1 lines are denoted 1 to 10. Error bars represent SE (n = 15).
Overall, the partial genetic redundancy among these type-A ARRs in the lateral root assay was similar to that observed in the root elongation response. In general, the single mutants exhibited near wild-type cytokinin sensitivity (Figures 4B and 4D), whereas the double mutants displayed cytokinin sensitivity that was intermediate between the wild type and the quadruple mutants (Figures 4C to 4E). The arr3,4,5,6, arr5,6,8,9, and arr3,4,8,9 quadruple mutants showed intermediate responses between the wild type and the arr3,4,5,6,8,9 hextuple mutant (Figures 4A and 4E), with the sensitivity of arr3,4,8,9 closest to arr3,4,5,6,8,9.
The arr8 and arr9 single mutants and the arr8,9 double mutant developed slightly fewer lateral roots in the absence of exogenous BA (t test P < 0.01). The difference in lateral root number in the absence of exogenous BA was further enhanced in arr3,4,8,9 and arr3,4,5,6,8,9 but not in arr5,6,8,9 (Figures 4D and 4E). This indicates that ARR5 and ARR6 do not act redundantly with ARR8 and ARR9 in the root without exogenous application of cytokinin and that ARR8 and ARR9 may be key elements in cytokinin inhibition of lateral root formation.
arr Seedlings Develop Pale Rosettes on Lower Concentrations of Cytokinin
When grown in the presence of exogenous BA, rosettes of wild-type seedlings were smaller, and the leaves were progressively paler with increasing concentrations of the hormone. The transition from dark to pale green rosettes occurred at similar doses to those that inhibited root formation in wild-type and mutant seedlings, respectively (Figure 2C). Chlorophyll content was quantified for wild-type Columbia and arr3,4,5,6 seedlings grown in the presence and absence of BA. In the absence of BA, chlorophyll content of the wild type and the arr3,4,5,6 quadruple mutant were not significantly different (1180 nmol/g ± 185 nmol/g and 862 nmol/g ± 161 nmol/g fresh weight, respectively). As observed in the seedling root assays, the most dramatic difference occurred at 10 nM BA. Chlorophyll levels in the wild type decreased to 790 nmol/g ± 220 nmol/g fresh weight in the presence of 10nM BA (∼67% of chlorophyll content in the absence of BA), whereas chlorophyll levels in arr3,4,5,6 decreased further to 234 nmol/g ± 47 nmol/g fresh weight (∼27% of chlorophyll content in the absence of BA). This analysis confirmed that wild-type seedlings contained significantly less chlorophyll (t test P = 0.025) when grown in the presence of BA and that the arr3,4,5,6 mutant was hypersensitive to cytokinin in this assay.
Complementation of arr Seedling Response to Cytokinin
To confirm that the altered cytokinin responses were the result of the disruption of type-A ARRs, a wild-type ARR5 gene (see Methods) was reintroduced into arr3,4,5,6 mutants. T1 transformants were selected on hygromycin, and homozygous T3 progeny from independent T1 lines were analyzed. The selected T3 progeny were assayed for cytokinin responsiveness in the seedling root assay. Eight of 11 selected lines showed strong complementation based on analysis of cytokinin-regulated root elongation, lateral root formation, and shoot chlorophyll content on 10 nM BA (Figures 3F and 4F, and data not shown). Three of the 11 lines did not complement these mutant phenotypes (Figures 3F and 4F, and data not shown). These results indicate that the altered cytokinin sensitivity of the arr3,4,5,6 mutant is the result of disruption of the type-A ARR genes. Reintroducing ARR5 into the arr3,4,5,6 quadruple background restored the cytokinin response to the levels of arr3,4,6 in two of the 11 lines, whereas six of the 11 lines resulted in a cytokinin responsiveness intermediate between the wild type and the arr3,4 mutant, suggesting that reintroduction of an ARR5 construct lacking introns (see Methods), multiple and or tandem T-DNA insertions, or positional effects may have resulted in higher levels of expression.
arr Mutations Affect the Response to Cytokinin:Auxin Ratios in Shoot Initiation Assays
Cytokinins promote cell division and initiate shoots in concert with auxin in cultured plant tissues (Miller et al., 1955, 1956; Mok and Mok, 2001a). We examined the response of excised hypocotyls from wild-type and several type-A arr mutant seedlings in response to various concentrations of the cytokinin kinetin and the auxin naphthaleneacetic acid (NAA).
Wild-type Columbia hypocotyl explants formed green foci only at high cytokinin:auxin ratios. However, no recognizable shoots were formed under these conditions, which is consistent with previous reports indicating that the Columbia ecotype does not efficiently form shoots from undifferentiated tissues in culture (Valvekens et al., 1988). At low cytokinin:auxin ratios, initiation of root primordia was observed, with the most prominent root structures observed at 30 ng/mL kinetin and 1000 ng/mL NAA; at intermediate ratios of these hormones, undifferentiated calli predominated (Figure 1D). The arr mutants formed larger calli on comparable concentrations of hormones that were able to induce wild-type calli (Figure 1D). arr3,4,5,6,8,9, arr3,4,5,6, and arr5,6,8,9 mutants also formed recognizable shoot structures; large leafy and flowering structures were found in the arr3,4,5,6,8,9 hextuple mutant between 100 to 300 ng/mL kinetin and 30 to 100 ng/mL NAA (Figure 1D; see supplemental data online). The range of calli-inducing media was expanded to lower cytokinin:auxin ratios relative to the wild type, and the ability to form shoots on concentrations at which the wild type was only able to form calli indicates an increase in both cytokinin sensitivity and responsiveness. The effect of the arr mutations was additive in this assay. arr3,4, arr5,6, and arr8,9 all formed larger calli than the wild type on comparable concentrations of hormones (data not shown). arr3,4 and arr5,6 generated small leaves at 300 ng/mL kinetin and 100 ng/mL NAA and 1000 ng/mL kinetin and 100 ng/mL NAA, respectively, whereas arr8,9 did not produce obvious shoot structures (data not shown). arr3,4,5,6 was more sensitive than arr5,6,8,9 in this assay and produced prominent shoot structures at a lower range of cytokinin concentrations than arr5,6,8,9 (Figure 1D; see supplemental data online), consistent with the seedling responses of the component double mutants. Further, root formation in the arr3,4,5,6,8,9 hextuple mutant was inhibited by cytokinin, resulting in elimination of root structures in some concentrations, most prominent at 30 ng/mL kinetin and 1000 ng/mL NAA (Figure 1D). Interestingly, in the absence of exogenous hormones, arr3,4,5,6,8,9 hypocotyl explants appeared swollen from disorganized cell divisions, suggesting a shift in the response to endogenous hormone levels (Figure 1D).
The increase in sensitivity and responsiveness of the arr3,4,5,6,8,9, arr3,4,5,6, and arr5,6,8,9 in callus formation and root inhibition and the ability to form recognizable shoots in this assay further indicate that these type-A ARRs act as negative regulators of cytokinin signaling with overlapping function.
Leaf Senescence Is Delayed in arr Mutants
Cytokinins inhibit leaf senescence in a variety of plant species (Gan and Amasino, 1995; Mok and Mok, 2001b). We used chlorophyll loss in a detached leaf assay to determine the effect of arr mutations on senescence. After 10 d of dark-induced senescence, wild-type leaf chlorophyll levels were substantially reduced relative to the initial content (Figure 5). This decrease in chlorophyll levels was inhibited in the presence of cytokinin in wild-type leaves, with maximal inhibition at ∼100 nM BA (Figure 5). arr3,4,5,6 exhibited a higher rate of chlorophyll retention in the absence of exogenous cytokinin (t test P < 10−4), and the maximal response occurred at lower cytokinin concentrations than the wild type (Figure 5). As in the root assays, these results indicate that the arr mutant is hypersensitive to cytokinin in adult leaves.
Expression Patterns of Type-A ARRs
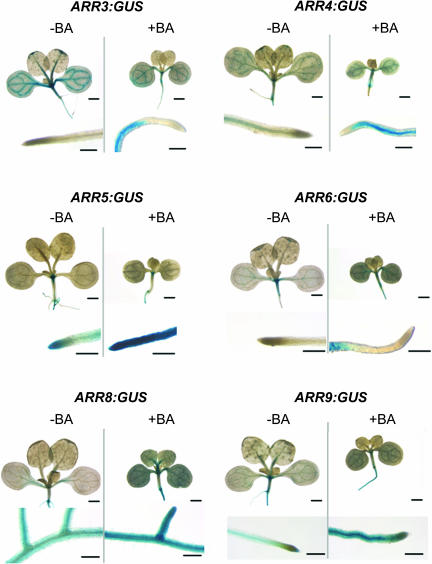
Functional redundancy of the type-A ARRs predicts that the genes would have overlapping patterns of expression. To test this hypothesis, we generated β-glucuronidase (GUS) reporter constructs fused to promoters of these six type-A ARRs. We examined the expression of these genes both in the presence and the absence of 10 nM BA, which is the concentration of BA at which the greatest differences in seedling response was observed. Consistent with RNA gel blot analysis (Taniguchi et al., 1998; D'Agostino et al., 2000), lines harboring the ARR5 and ARR6 promoter fusions displayed the highest level of induction by cytokinin, whereas the ARR3, ARR4, ARR8, and ARR9 promoter fusions only showed a moderate increase in reporter activity in response to cytokinin (Figure 6). Members of the most similar pairs showed similar patterns of expression (Figure 6).
Figure 6.
Expression Analysis of ARR Gene Promoters.
ARR promoter–driven GUS constructs were generated and introduced into wild-type Columbia background. Transgenic seedlings were grown on MS media (−BA) or media supplemented with 10 nM BA (+BA) for 9 d and assayed for GUS activity. Ten transformed lines were examined, and one representative line for each construct was photographed. With the exception of ARR8:GUS, close-up images show the relative GUS activity at the primary root tip. For ARR8:GUS, the close-up images show lateral root junctions on the primary root. Scale bars = 1 mm for aerial tissues, 250 μm for roots.
ARR3:GUS and ARR4:GUS were constitutively expressed in the vasculature of both shoots and roots, with stronger expression in the shoot. When grown on 10 nM BA, the region of expression was expanded to tissues surrounding vasculature in the root but was excluded from the root tip/meristematic region. ARR5 expression was as reported previously (D'Agostino et al., 2000), primarily found in the root and shoot meristems in the absence of exogenous cytokinin. In the presence of 10 nM BA, the ARR5:GUS expression region was enlarged to include tissues around the shoot meristematic region; strong ARR5:GUS expression was induced in all tissues in the root, from the hypocotyl–root junction through the root tip. Basal ARR6:GUS expression was detected in the shoot meristematic region and cotyledon vasculature. Cytokinin treatment resulted in overall higher levels of ARR6:GUS expression, with GUS staining expanded to the hypocotyl and root tissues but excluded from the root tip. ARR8 and ARR9 were expressed strongly throughout the root and weakly in the seedling vasculature, with an overall increase in GUS activity in the same tissues on exogenous cytokinin.
Although basal expression patterns differed among the ARR gene pairs, their expression patterns mostly overlap in the presence of exogenous cytokinin, particularly in the root. This is consistent with the functional redundancy that we observe among type-A ARRs in root assays in the presence of BA.
arr Mutations Affect Cytokinin Primary Response
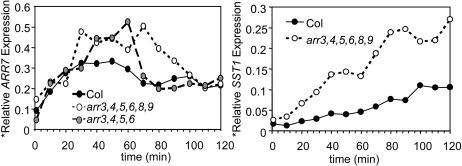
To investigate whether the increase in cytokinin sensitivity of the arr mutants was a result of altered primary response, we examined gene expression in response to cytokinin. Ten-day-old light-grown seedlings were treated with 10 nM BA, and the expression levels of two cytokinin primary response genes, ARR7 and a steroid sulfotransferase (SST1) (D'Agostino et al., 2000; J. To and J. Kieber, unpublished data), were analyzed by RNA gel blot. Two independent full experiments were conducted, and critical time points were further repeated in triplicate, all of which produced consistent results. The results from one of the experiments are shown in Figure 7.
Figure 7.
arr Mutants Are Affected in the Cytokinin Primary Response Pathway.
RNA was extracted from 10-d-old light-grown seedlings treated with 10 nM BA in liquid MS with 1% sucrose for the indicated time. The RNA was analyzed by RNA gel blotting. The blots were probed with either an ARR7, SST1, or β-tubulin radiolabeled probe. The signal obtained for each was quantified using a PhosphorImager, and the ARR7 and SST1 signals were normalized to the β-tubulin signal. The experiment was conducted twice with similar results.
In wild-type seedlings, ARR7 was induced rapidly by cytokinin treatment and reached twofold above basal level after 10 min, after which the signal continued to increase to maximal levels of ∼3.5-fold at 30 min (Figure 7). The arr3,4,5,6 quadruple mutant exhibited a greater amplitude in cytokinin-induced ARR7 expression. The arr3,4,5,6,8,9 hextuple mutant displayed an induction amplitude similar to that seen in arr3,4,5,6 but also showed an extended peak of elevated ARR7 expression. As with the ARR7 genes, the rapid induction of SST1 was magnified in the arr3,4,5,6,8,9 hextuple mutant (Figure 7). The amplified rapid induction of cytokinin response genes in arr3,4,5,6 and arr3,4,5,6,8,9 mutants indicates that type-A ARRs negatively regulate the primary cytokinin signal transduction pathway.
arr Mutants Exhibit Altered Responses to Red Light
ARR4 has previously been implicated in modulating red light responses in Arabidopsis, based on its ability to interact with PhyB and the effects of ARR4 overexpression upon the red light sensitivity of seedlings (Sweere et al., 2001). However, no loss-of-function mutants within the type-A ARR family have been characterized for their red light sensitivity. We therefore investigated the response of arr seedling hypocotyl elongation to red light.
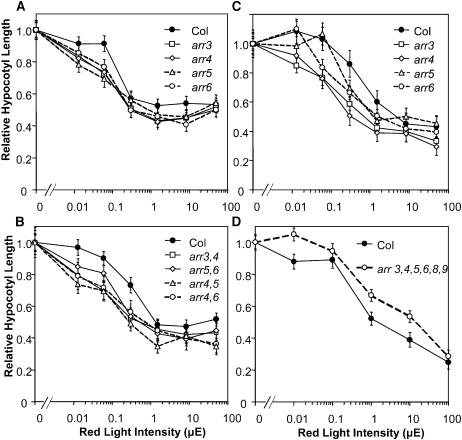
Differences between the single arr3, arr4, arr5, and arr6 mutants and wild-type hypocotyl lengths were observed over the entire red light range from 0.013 to 50 μE (Figure 8A). Among the double mutants, arr3,4, arr4,5, and arr4,6 demonstrated the greatest increase in sensitivity to red light, whereas arr5,6, although more sensitive to red light than the wild type, did not show as dramatic a shift in response as the three double mutants carrying the arr4 mutation (Figure 8B). These results suggest that ARR3 and ARR4 play a more substantial role in the red light response than ARR5 and ARR6. Interestingly, the arr3,4,5,6,8,9 hextuple mutant was less sensitive to red light than the wild type (Figure 8D), suggesting complex interactions among type-A ARRs as observed in the rosette phenotypes.
Figure 8.
arr Seedlings Exhibit Altered Hypocotyl Growth Response to Red Light.
Mutant and wild-type seeds were stratified and pretreated with fluorescent light before incubation under various red light intensities for 3 d ([A], [B], and [D]) or directly irradiated with red light after stratification (C). Mean hypocotyl lengths at various light intensities are normalized to the mean value of the etiolated seedlings of the respective genotypes. Mean etiolated hypocotyl heights (mm) are 9.7, 8.5, 8.7, 9.3, and 10 for arr3, arr4, arr5, arr6, and the wild type in (A); 8.6, 8.8, 8.2, 9.7, and 9.6 for arr3,4, arr4,6, arr4,5, arr5,6, and the wild type in (B); 8.6, 7.8, 6.4, 7.0, and 6.7 for arr3, arr4, arr5, arr6, and the wild type in (C); and 9.4 and 9.2 for arr3,4,5,6,8,9 and the wild type in (D), respectively. Bars represent SE (n > 13). The experiment was conducted twice with consistent results.
Because the initial ratios of active and inactive forms of phytochrome in the seeds may affect the red light sensitivity, we also conducted an experiment without the 15-h light pretreatment. The results showed a similar trend to the experiment with light pretreatment, with arr3 and arr4 showing the most pronounced increase in red light sensitivity (Figure 8C). Thus, the red light hypersensitivity of the mutants is not an artifact of pretreatment with fluorescent light. The higher order mutants were delayed in germination relative to the wild type under these growth conditions, hence their sensitivity to red light could not be assessed.
DISCUSSION
We have described the characterization of six type-A response regulator genes in Arabidopsis. A variety of cytokinin response assays indicate that all six of these type-A ARRs act as negative regulators of cytokinin function. This is observed in both root and shoot tissues in seedlings, in fully expanded adult leaves, and in tissue culture. Furthermore, consistent with their highly similar sequences, our analyses indicate that these genes have at least partially overlapping functions. However, we also detect morphological differences among the mutants that are consistent with gene-specific functions and potential antagonistic functions within this gene family.
arr Mutations Increase Cytokinin Sensitivity
arr mutants display increased cytokinin sensitivity at low concentrations of cytokinin in various responses, including seedling root elongation and lateral root formation, hypocotyl shoot initiation assays, senescence delay, and induction of cytokinin response genes. Intriguingly, in the root elongation assay, mutations in the type-A ARRs only affect the response at lower concentrations of cytokinin (<0.1 μM), thus changing the shape of the dose–response curve from monophasic in the wild type to biphasic in the quadruple and higher order arr mutants. This suggests that the monophasic response in the wild type may be comprised of a more complex response. Alternatively, root inhibition at the higher doses (0.1 to 10 μM BA) could represent a nonphysiological, toxic effect on root elongation. However, cytokinin receptor mutants are insensitive to such concentrations of cytokinin with no observable toxic effects (Inoue et al., 2001; Ueguchi et al., 2001b), and a similar range of concentrations of BA has been shown to induce cytokinin primary response genes (D'Agostino et al., 2000). Together, these results suggest that these higher doses of BA are not simply toxic but rather constitute part of the cytokinin responsive range.
Hwang and Sheen (2001) have shown previously that overexpression of a subset of type-A ARRs in plant protoplasts inhibits the expression of an ARR6 promoter-luciferase reporter. Here, we demonstrate that multiple loss-of-function type-A arr alleles result in an increase in both the amplitude and period of cytokinin induction of cytokinin primary response genes. This effect occurs with kinetics that strongly suggest that type-A ARRs modulate the sensitivity of the cytokinin primary response pathway.
Role of Type-A ARRs in Cytokinin Signaling
Type-A ARRs are generally rapidly upregulated by exogenous cytokinin (D'Agostino et al., 2000) which, in conjunction with our results here, suggests that type-A ARRs mediate a feedback mechanism by which the plant decreases its sensitivity to the hormone. Type-B ARRs have been shown to be transcription factors that positively mediate cytokinin responses (Hwang and Sheen, 2001; Sakai et al., 2001). Type-A ARRs may negatively regulate cytokinin responses by interfering with type-B ARR activity. This could occur via direct protein–protein interactions between type-A and type-B ARRs in a manner similar to the Aux/IAA early auxin response genes and auxin response factors in auxin response (Hutchison and Kieber, 2002; Leyser, 2002), though evidence for direct protein–protein interactions between type-A and type-B ARRs is lacking. A more likely model is that type-A ARRs inhibit type-B ARR activation by competing for phosphotransfer from upstream His phosphotransfer proteins, as has been demonstrated in a few bacterial two-component systems (Rabin and Stewart, 1993; Li et al., 1995; Sourjik and Schmitt, 1998). An additional possibility is that type-A ARRs may act indirectly by increasing the function of a negative regulator of type-B ARRs.
arr Mutants Have Weak Morphological Phenotypes
Cytokinin has been linked to fundamental processes in plant growth and development, including the regulation of cell division, and altering endogenous cytokinin levels can have dramatic consequences on plant development and morphology (Miller et al., 1955, 1956; Medford et al., 1989; Werner et al., 2001). Thus, it is somewhat surprising that a shift in cytokinin sensitivity of >10-fold, as is seen in the arr3,4,5,6,8,9 mutant, does not result in a strong morphological phenotype. Furthermore, it is remarkable that disruption of six out of 10 members of a gene family involved in cytokinin response does not significantly impact basal development. The T-DNA insertions in the type-A ARRs described herein do not all result in transcript nulls, and, thus, the hextuple mutant may still retain partial function in these genes, which may contribute to the lack of a substantial phenotype. However, this would not explain why a 10-fold shift in cytokinin sensitivity does not affect basal development. The plant may compensate for increased cytokinin sensitivity by decreasing active hormone levels. Attempts to increase cytokinin levels by constitutive overexpression of bacterial isopentenyl transferases in whole plants resulted in no striking morphological effects because the plant may compensate for elevated biosynthesis by increasing the conjugation and degradation of the hormone (Medford et al., 1989; Smigocki, 1991; Mok and Mok, 2001a). Consistent with this model, a global analysis of gene expression has revealed that a primary response of Arabidopsis seedlings treated with high levels of exogenous cytokinin is to alter genes whose combined function is to decrease cytokinin levels and responsiveness (Rashotte et al., 2003).
Another explanation for the lack of a phenotype is that, although the type-A arr mutants alter cytokinin sensitivity, this change is not beyond a threshold that dramatically affects basal development under laboratory conditions. These genes may play a role in response to some factor not present in laboratory growth conditions, or they may play a role in environmental transitions, which are minimized under controlled growth conditions. A more dynamic environment that requires intact mechanisms for developmental plasticity (and, thus, fluctuations in hormonal responsiveness) may reveal more pronounced morphological alterations in the arr mutants.
Finally, cytokinin regulation of development may be redundant with other control mechanisms. For example, cell division is controlled by multiple regulatory inputs, some subset of which may compensate for the altered cytokinin function of the type-A arr mutants.
arr Mutants Are Affected in Light Responses
We found that mutations in ARR3, ARR4, ARR5, and ARR6 independently or together result in increased sensitivity to red light, similar to PhyB overexpressers (McCormac et al., 1993; Krall and Reed, 2000), suggesting that these genes function as negative regulators of red light signal transduction. The arr double mutants did not show an obvious increase in red light sensitivity over their component single mutants, which may indicate that type-A ARRs modulate only part of the seedling red light response and/or that there is not substantial redundancy in this function of the type-A ARRs. The elongated petiole phenotypes of the arr3,4,5,6 mutant also suggest an altered shade avoidance response mediated by light and/or ethylene signaling pathways (Finlayson et al., 1999). The long petiole phenotype in arr3,4,5,6 is similar to that observed for phyB mutants, albeit the arr3,4,5,6 petiole phenotype is weaker. However, the arr3,4,5,6,8,9 hextuple mutant exhibited a decrease in red light sensitivity compared with arr3,4,5,6, suggesting that arr8 and arr9 may antagonize the effects of the other four arr mutations or that an overall decrease in the abundance of ARRs beyond a certain threshold may have an opposite effect on the light response.
Sweere et al. (2001) have shown that ARR4 overexpression resulted in increased red light sensitivity in hypocotyls and proposed that this was because of a direct interaction between ARR4 and PhyB, which inhibited the conversion of PhyB from the active to the inactive form. Our data support the involvement of ARR4 as well as other type-A ARRs in red light signal transduction. However, the overexpression data predicts a decrease in red light sensitivity in a loss-of-function arr4 mutant, in contrast with what we observed in our mutant analysis. It is possible that overexpression of ARR4 dramatically changes the stoichiometry between ARR4 and PhyB or other interacting proteins. If interactions with phytochrome play a significant role, it may be that the activity of the ARRs is regulated by phytochromes rather than the ARRs regulating phytochrome activity, as originally proposed (Sweere et al., 2001). Alternatively, the type-A ARRs could be involved in a cytokinin signaling pathway that impinges upon the phytochrome-mediated pathway (Su and Howell, 1995) and, thus, indirectly regulate red light sensitivity. Finally, differences in growth conditions may alter the role of the type-A ARRs in red light responses.
Redundancy and Specificity among Type-A ARRs
Phylogenetic analysis reveals that the 10 type-A ARRs fall into five distinct pairs (Figure 1A), and analysis of the positions of these genes within the genome indicates that these pairs arose from a genome duplication event (Vision et al., 2000). Interestingly, most of the Arabidopsis type-A ARRs generally fall into a clade that is distinct from those formed by the rice and maize type-A ARR genes (Figure 1A), and, thus, the progenitor of monocots and dicots may have had only a relatively small number of type-A ARRs. If this is the case, then it is likely that the expansion of this family occurred in both monocots and dicots. Alternatively, common ancestral genes may have been deleted in each lineage. Evidence for accelerated gene loss in duplicated regions of the Arabidopsis genome (Ku et al., 2000) suggests that there has been pressure for maintenance of all 10 type-A ARRs, despite the partial redundancy found in our analysis. Furthermore, the commonality of a large type-A ARR gene family in both monocots and dicots also suggests some selective advantage.
Although our studies suggest that there is significant functional overlap among members of the type-A ARR gene family, several lines of evidence also support a model for some gene-specific function. Analysis of basal patterns of expression reveal some differences among the type-A ARRs, largely defined by the most similar pairs. ARR3 and ARR4 are expressed mainly in the shoot vasculature, ARR5 and ARR6 are expressed in the shoot meristematic region, and ARR8 and ARR9 are expressed strongly throughout the root. Several of the single and double mutants have subtle but distinct morphological phenotypes, which are in general consistent with their patterns of expression. Disruption of ARR8 and ARR9 loci affect lateral root number in seedlings in the absence of cytokinin application but do not affect shoot development. Under short-day conditions, adult plants of arr5 develop smaller rosettes and arr4 develop longer petioles, but neither mutant is affected in basal root development. Thus, it is likely that these genes have acquired some specificity that may have contributed to their retention.
Interactions between Type-A ARRs
A previous study examined the effect of overexpression of ARR4 and ARR8 on shoot formation from cultured Arabidopsis roots. Interestingly, ARR4 overexpression resulted in a cytokinin hypersensitive phenotype, but overexpression of ARR8 caused cytokinin insensitivity in this assay (Osakabe et al., 2002). The authors concluded that ARR4 and ARR8 have opposing effects on cytokinin responsiveness. Our loss-of-function analysis does not support a positive role for ARR4 in cytokinin signaling, and the discrepancy may reflect complications arising from overexpression in the earlier study.
However, phenotypes of adult arr mutant plants are consistent with some members of these gene pairs having antagonistic effects. For example, the small rosette phenotype of the arr5 mutant is suppressed by the arr6 mutation (its closest homolog) but not by arr4. Additionally, the arr8 and arr9 mutations appear to partially suppress the elongated petiole phenotype of the arr3,4,5,6 mutant and antagonize the red light hypersensitivity of single and double mutants containing mutations in arr3, arr4, arr5, and arr6. These results suggest that there may be interactions among the type-A ARRs involving both additive and antagonistic functions.
Implications in Tissue Culture
The change in the response of type-A ARR hypomorphic mutants in tissue culture is both quantitative (i.e., shoot formation is shifted to lower concentrations of cytokinin) and qualitative (i.e., well-developed shoots form in the mutant, but only green foci form in the wild type). Plant tissue and species vary widely in their regenerative potential, which poses major obstacles for transformation of some species. This conversion of a tissue that is recalcitrant to regeneration (i.e., Columbia hypocotyls) to one that readily forms shoots in culture (i.e., the mutant hypocotyls) is intriguing and implies that the relative level of functional type-A ARRs may be one of the factors underlying the differences in regenerative capacity.
In conclusion, we have shown that type-A ARRs are negative regulators with overlapping function in cytokinin signaling. These genes also affect light-regulated development. Morphological differences among arr mutants predict some specific functions and suggest regulatory interactions among these genes. Additional genetic studies may further dissect the role of type-A ARRs in development and their complex interactions, and biochemical analyses may reveal the mechanism by which these genes inhibit cytokinin signaling.
METHODS
Isolation of arr Mutants
A total of 80,000 Arabidopsis lines from the Salk T-DNA collection in the Columbia ecotype were screened for T-DNA insertions in the type-A ARRs using a PCR-based method as described previously (Alonso et al., 2003). Gene-specific primers used and sites of T-DNA insertions are described in the supplemental data online.
Single mutants arr3 and arr4, arr5 and arr6, and arr8 and arr9 were crossed to generate double mutants arr3,4, arr5,6, and arr8,9, respectively. Double mutants arr3,4, arr5,6, and arr8,9 were crossed to generate quadruple mutants arr3,4,5,6, arr5,6,8,9, and arr3,4,8,9. Quadruple mutants arr3,4,5,6 and arr5,6,8,9 were crossed to generate the hextuple mutant arr3,4,5,6,8,9. Double mutants arr4,5 and arr4,6 were generated by crossing the component single mutants. Insertions were confirmed by genomic PCR with gene-specific and T-DNA border primers.
Growth Conditions for Adult Plants and Seedlings
Plants were grown at 23°C in ∼75 μE light under short-day conditions (8-h-light/16-h-dark), long-day conditions (16-h-light/8-h-dark), and constant light as noted.
For seedling assays, seeds were surface-sterilized and cold treated at 4°C for 3 d in the dark and then treated with white light for 3 h. Unless otherwise specified, seedlings were grown on vertical plates containing 1× Murashige and Skoog (MS) salts, 1% sucrose, and 0.6% phytagel (Sigma, St. Louis, MO) at 23°C in ∼100 μE constant light. For growth on horizontal plates, seedlings were grown on 1× MS salts, 1% sucrose, and 0.8% bactoagar at 23°C in ∼75 μE constant light.
Seedling Cytokinin Response Assays
Arabidopsis seeds were grown on vertical plates containing the appropriate concentration of the cytokinin BA or 0.1% dimethyl sulfoxide (DMSO) vehicle control for 10 d. Root lengths at days 4 and 9 were marked on the plates. The plates were photographed at 10 d, and root growth between days 4 and 9 were measured using NIH Image version 1.62 (National Institutes of Health, Bethesda, MD). At 10 d, total lateral roots that emerged from the primary root (stage IV and beyond) were quantified under a dissecting microscope. For chlorophyll assays, seedlings were grown on horizontal plates supplemented with BA. Shoot systems from 2-week-old seedlings were harvested, and chlorophyll was extracted with methanol. Chlorophyll content was determined spectrophotometrically and normalized to fresh weight as described previously (Porra et al., 1989).
Analysis of ARR Expression
For analysis of ARR expression in the T-DNA insertion lines, 5-d-old etiolated seedlings of single mutant lines were treated with 50 μM cycloheximide and 1 μM BA for 40 min, and RNA was extracted and analyzed by RNA gel blot as described previously, using the appropriate type-A cDNAs as hybridization probes (D'Agostino et al., 2000). For RT-PCR, seedlings were grown on horizontal plates layered with Whatman filter paper for 10 d under constant light and harvested for RNA extraction. cDNA was generated using Superscript III RT (Invitrogen, Carlsbad, CA). ARR cDNA was amplified with a 5′ primer at the ATG and a 3′ primer in the third exon for 30 cycles. Primer sequences are listed in the supplemental data online.
Cytokinin Treatment Time Course
Seedlings were grown on horizontal plates layered with Whatman filter paper for 10 d under constant light. Seedlings were treated in liquid MS supplemented with 10 nM BA in 0.1% DMSO for the appropriate duration, and RNA was extracted and analyzed by RNA gel blot as described above. ARR7 and β-tubulin cDNA probes were described previously (D'Agostino et al., 2000); the SST1 probe was generated from full-length cDNA of SST1 (At1g13420).
Complementation Analysis
ARR5 wild-type cDNA was amplified and cloned downstream of the 1.6 kb ARR5 promoter (D'Agostino et al., 2000). The resulting promoter-cDNA construct was inserted into the pCambia1303 binary vector and transformed into arr3,4,5,6 by the floral dip method (Clough and Bent, 1998). Transformants were selected on MS plates supplemented with 30 μg/mL of hygromycin and 50 μg/mL of carbenicillin. Eleven independent T1 hygromycin-resistant lines were selected, and homozygous T3 progeny were examined in seedling cytokinin response assays as described above.
Shoot Initiation Assay
Arabidopsis seedlings were grown on the vertical plates in the dark for 3 d and then in dim light (∼5 μE) for 3 d to produce elongated and firm hypocotyls. Hypocotyls of ∼7 mm were excised from the seedlings. Hypocotyl explants were transferred to MS, 1% sucrose, and 0.4% phytagel plates containing combinations of kinetin and NAA ranging from 0 to 3000 ng/mL for 4 weeks at 23°C in ∼75 μE continuous light. One representative callus at each concentration was selected and arranged to create a composite photograph for each genotype.
Other Assays for Cytokinin Response
For senescence assays, seedlings were grown on horizontal plates for 25 d. Fully expanded leaves (approximately the seventh leaf) were excised from the seedlings. To induce senescence, leaves were floated on water in parafilm-sealed Petri plates supplemented with various concentrations of BA in 0.1% DMSO at 23°C in the dark for 10 d. Chlorophyll was extracted and quantified spectrophotometrically from freshly cut leaves and senesced leaves as in the seedling chlorophyll analysis.
Analysis of ARR Patterns of Expression
Promoter regions 1.6 to 2.0 kb upstream of ATG of ARR3, ARR4, ARR6, ARR8, and ARR9 were amplified by PCR and cloned upstream of the GUS gene in the pCambia3301 binary vector. Primers used are listed in the supplemental data online. The resulting ARR:GUS translational fusion constructs were introduced into wild-type Columbia plants by the floral dip method (Clough and Bent, 1998). Ten plant lines per construct were selected by kanamycin drug resistance and examined for GUS activity. To detect GUS activity, seedlings were grown on horizontal plates supplemented with 10 nM BA or 0.1% DMSO vehicle control. Nine-day-old seedlings were vacuum infiltrated at 130 mbar for 10 min in X-Gluc buffer (100 mM sodium phosphate, pH 7.0, 0.5% Triton X-100, and 100 μM X-Gluc). The color reaction was allowed to proceed at 37°C overnight. Chlorophyll was extracted with three washes of 100% ethanol, and the seedlings were examined under a dissecting microscope. Representative plant lines from each construct were selected. These seedlings, as well as the previously characterized ARR5:GUS line (D'Agostino et al., 2000), were analyzed in parallel.
Analysis of Red Light Response
The response of seedlings to red light was performed as described (Krall and Reed, 2000), with minor modifications. Mutant and wild-type seeds were sown on plates containing 1× MS salts, 0.1% sucrose, and 0.8% Phytagar (Invitrogen/Gibco, Carlsbad, CA). The seeds were cold treated and then pretreated with fluorescent lights for 15 h first or immediately exposed to a red light–emitting diode light source (670 nm) (Quantum Devices, Barneveld, WI) filtered with bronze-tinted Plexiglass filters to obtain a range of light intensities. After 3 d of red light exposure, the seedlings were scanned and the hypocotyls measured using NIH Image (version 1.62).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Science Foundation (J.J.K. and G.E.S.) and the National Institutes of Health (J.J.K.). We thank members of the Kieber Lab for many helpful discussions and critical reading of the manuscript. We also thank C. Fairchild and J. Reed for their invaluable assistance in designing the red light experiments, and S. Metting and S. Bulmer for assistance in conducting these experiments.
On-line version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Joseph J. Kieber (jkieber@unc.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.018978.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Asakura, Y., Hagino, T., Ohta, Y., Aoki, K., Yonekura-Sakakibara, K., Deji, A., Yamaya, T., Sugiyama, T., and Sakakibara, H. (2003). Molecular characterization of His-Asp phosphorelay signaling factors in maize leaves: Implications of the signal divergence by cytokinin-inducible response regulators in the cytosol and the nuclei. Plant Mol. Biol. 52, 331–341. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- D'Agostino, I.B., Deruere, J., and Kieber, J.J. (2000). Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 124, 1706–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlayson, S.A., Lee, I.J., Mullet, J.E., and Morgan, P.W. (1999). The mechanism of rhythmic ethylene production in sorghum. The role of phytochrome B and simulated shading. Plant Physiol. 119, 1083–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan, S., and Amasino, R.M. (1995). Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270, 1986–1988. [DOI] [PubMed] [Google Scholar]
- Hutchison, C.E., and Kieber, J.J. (2002). Cytokinin signaling in Arabidopsis. Plant Cell 14 (suppl.), S47–S59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, I., and Sheen, J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389. [DOI] [PubMed] [Google Scholar]
- Inoue, T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., Tabata, S., Shinozaki, K., and Kakimoto, T. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409, 1060–1063. [DOI] [PubMed] [Google Scholar]
- Kiba, T., Yamada, H., Sato, S., Kato, T., Tabata, S., Yamashino, T., and Mizuno, T. (2003). The type-A response regulator, ARR15, acts as a negative regulator in the cytokinin-mediated signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 44, 868–874. [DOI] [PubMed] [Google Scholar]
- Kieber, J.J. (2002). Cytokinins. In The Arabidopsis Book, E.M. Meyerowitz, ed (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0063, http://www.aspb.org/publications/arabidopsis/.
- Krall, L., and Reed, J.W. (2000). The histidine kinase-related domain participates in phytochrome B function but is dispensable. Proc. Natl. Acad. Sci. USA 97, 8169–8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku, H.M., Vision, T., Liu, J., and Tanksley, S.D. (2000). Comparing sequenced segments of the tomato and Arabidopsis genomes: Large-scale duplication followed by selective gene loss creates a network of synteny. Proc. Natl. Acad. Sci. USA 97, 9121–9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser, O. (2002). Molecular genetics of auxin signaling. Annu. Rev. Plant Biol. 53, 377–398. [DOI] [PubMed] [Google Scholar]
- Li, J., Swanson, R.V., Simon, M.I., and Weis, R.M. (1995). The response regulators CheB and CheY exhibit competitive binding to the kinase CheA. Biochemistry 34, 14626–14636. [DOI] [PubMed] [Google Scholar]
- Li, Y., Hagen, G., and Guilfoyle, T.J. (1992). Altered morphology in transgenic tobacco plants that overproduce cytokinins in specific tissues and organs. Dev. Biol. 153, 386–395. [DOI] [PubMed] [Google Scholar]
- McCormac, A.C., Wagner, D., Boylan, M.T., Quail, P.H., Smith, H., and Whitelam, G.C. (1993). Photoresponses of transgenic Arabidopsis seedlings expressing introduced phytochrome B-encoding cDNAs: Evidence that phytochrome A and phytochrome B have distinct photoregulatory functions. Plant J. 4, 19–27. [Google Scholar]
- Medford, J.I., Horgan, R., El-Sawi, Z., and Klee, H.J. (1989). Alterations of endogenous cytokinins in transgenic plants using a chimeric isopentenyl transferase gene. Plant Cell 1, 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, C.O., Skoog, F., Okomura, F.S., von Saltza, M.H., and Strong, F.M. (1956). Isolation, structure and synthesis of kinetin, a substance promoting cell division. J. Am. Chem. Soc. 78, 1375–1380. [Google Scholar]
- Miller, C.O., Skoog, F., Von Saltza, M.H., and Strong, F. (1955). Kinetin, a cell division factor from deoxyribonucleic acid. J. Am. Chem. Soc. 77, 1392. [Google Scholar]
- Mok, D.W., and Mok, M.C. (2001. a). Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 89–118. [DOI] [PubMed] [Google Scholar]
- Mok, D.W., and Mok, M.C., eds (2001. b). Cytokinins: Chemistry, Activity and Function. (Boca Raton, FL: CRC Press).
- Osakabe, Y., Miyata, S., Urao, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). Overexpression of Arabidopsis response regulators, ARR4/ATRR1/IBC7 and ARR8/ATRR3, alters cytokinin responses differentially in the shoot and in callus formation. Biochem. Biophys. Res. Commun. 293, 806–815. [DOI] [PubMed] [Google Scholar]
- Page, R.D. (1996). TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358. [DOI] [PubMed] [Google Scholar]
- Porra, R.J., Thompson, W.A., and Kriedemann, P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394. [Google Scholar]
- Rabin, R.S., and Stewart, V. (1993). Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J. Bacteriol. 175, 3259–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte, A.M., Carson, S.D., To, J.P., and Kieber, J.J. (2003). Expression profiling of cytokinin action in Arabidopsis. Plant Physiol. 132, 1998–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa, G., Mi, M., He-chun, Y., Ben-ye, L., Guo-feng, L., and Kang, C. (2001). Effects of ipt gene expression on the physiological and chemical characteristics of Artemisia annua L. Plant Sci. 160, 691–698. [DOI] [PubMed] [Google Scholar]
- Sakai, H., Aoyama, T., Bono, H., and Oka, A. (1998). Two-component response regulators from Arabidopsis thaliana contain a putative DNA-binding motif. Plant Cell Physiol. 39, 1232–1239. [DOI] [PubMed] [Google Scholar]
- Sakai, H., Aoyama, T., and Oka, A. (2000). Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 24, 703–711. [DOI] [PubMed] [Google Scholar]
- Sakai, H., Honma, T., Aoyama, T., Sato, S., Kato, T., Tabata, S., and Oka, A. (2001). ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294, 1519–1521. [DOI] [PubMed] [Google Scholar]
- Smigocki, A.C. (1991). Cytokinin content and tissue distribution in plants transformed by a reconstructed isopentenyl transferase gene. Plant Mol. Biol. 16, 105–115. [DOI] [PubMed] [Google Scholar]
- Sourjik, V., and Schmitt, R. (1998). Phosphotransfer between CheA, CheY1, and CheY2 in the chemotaxis signal transduction chain of Rhizobium meliloti. Biochemistry 37, 2327–2335. [DOI] [PubMed] [Google Scholar]
- Stock, A.M., Robinson, V.L., and Goudreau, P.N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215. [DOI] [PubMed] [Google Scholar]
- Su, W., and Howell, S.H. (1995). The effects of cytokinin and light on hypocotyl elongation in Arabidopsis seedlings are independent and additive. Plant Physiol. 108, 1423–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, T., Miwa, K., Ishikawa, K., Yamada, H., Aiba, H., and Mizuno, T. (2001). The Arabidopsis sensor His-kinase, AHK4, can respond to cytokinins. Plant Cell Physiol. 42, 107–113. [DOI] [PubMed] [Google Scholar]
- Sweere, U., Eichenberg, K., Lohrmann, J., Mira-Rodado, V., Baurle, I., Kudla, J., Nagy, F., Schafer, E., and Harter, K. (2001). Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science 294, 1108–1111. [DOI] [PubMed] [Google Scholar]
- Taniguchi, M., Kiba, T., Sakakibara, H., Ueguchi, C., Mizuno, T., and Sugiyama, T. (1998). Expression of Arabidopsis response regulator homologs is induced by cytokinins and nitrate. FEBS Lett. 429, 259–262. [DOI] [PubMed] [Google Scholar]
- Ueguchi, C., Koizumi, H., Suzuki, T., and Mizuno, T. (2001. a). Novel family of sensor histidine kinase genes in Arabidopsis thaliana. Plant Cell Physiol. 42, 231–235. [DOI] [PubMed] [Google Scholar]
- Ueguchi, C., Sato, S., Kato, T., and Tabata, S. (2001. b). The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol. 42, 751–755. [DOI] [PubMed] [Google Scholar]
- Valvekens, D., Van Montagu, M., and Van Lijsebettens, M. (1988). Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85, 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vision, T.J., Brown, D.G., and Tanksley, S.D. (2000). The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117. [DOI] [PubMed] [Google Scholar]
- Werner, T., Motyka, V., Strnad, M., and Schmulling, T. (2001). Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 98, 10487–10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, A.H., and Stock, A.M. (2001). Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26, 369–376. [DOI] [PubMed] [Google Scholar]
- Yamada, H., Suzuki, T., Terada, K., Takei, K., Ishikawa, K., Miwa, K., Yamashino, T., and Mizuno, T. (2001). The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 42, 1017–1023. [DOI] [PubMed] [Google Scholar]
- Zubko, E., Adams, C.J., Machaekova, I., Malbeck, J., Scollan, C., and Meyer, P. (2002). Activation tagging identifies a gene from Petunia hybrida responsible for the production of active cytokinins in plants. Plant J. 29, 797–808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.