Abstract
Protein phosphorylation in amyloplasts and chloroplasts of Triticum aestivum (wheat) was investigated after the incubation of intact plastids with γ-32P-ATP. Among the soluble phosphoproteins detected in plastids, three forms of starch branching enzyme (SBE) were phosphorylated in amyloplasts (SBEI, SBEIIa, and SBEIIb), and both forms of SBE in chloroplasts (SBEI and SBEIIa) were shown to be phosphorylated after sequencing of the immunoprecipitated 32P-labeled phosphoproteins using quadrupole-orthogonal acceleration time of flight mass spectrometry. Phosphoamino acid analysis of the phosphorylated SBE forms indicated that the proteins are all phosphorylated on Ser residues. Analysis of starch granule–associated phosphoproteins after incubation of intact amyloplasts with γ-32P-ATP indicated that the granule-associated forms of SBEII and two granule-associated forms of starch synthase (SS) are phosphorylated, including SSIIa. Measurement of SBE activity in amyloplasts and chloroplasts showed that phosphorylation activated SBEIIa (and SBEIIb in amyloplasts), whereas dephosphorylation using alkaline phosphatase reduced the catalytic activity of both enzymes. Phosphorylation and dephosphorylation had no effect on the measurable activity of SBEI in amyloplasts and chloroplasts, and the activities of both granule-bound forms of SBEII in amyloplasts were unaffected by dephosphorylation. Immunoprecipitation experiments using peptide-specific anti-SBE antibodies showed that SBEIIb and starch phosphorylase each coimmunoprecipitated with SBEI in a phosphorylation-dependent manner, suggesting that these enzymes may form protein complexes within the amyloplast in vivo. Conversely, dephosphorylation of immunoprecipitated protein complex led to its disassembly. This article reports direct evidence that enzymes of starch metabolism (amylopectin synthesis) are regulated by protein phosphorylation and indicate a wider role for protein phosphorylation and protein–protein interactions in the control of starch anabolism and catabolism.
INTRODUCTION
The starches produced in plant tissues perform different functions. Transient starches synthesized in chloroplasts during the day are degraded at night to provide carbon for nonphotosynthetic metabolism. Starch produced in tuberous tissues also acts as a carbon store and may need to be accessed as environmental conditions dictate, whereas storage starches in developing seeds are a long-term carbon store for the next generation. Despite the fact that the starches produced in these various tissues share a common biochemical pathway—namely, the production of the immediate soluble precursor ADP-glucose (ADP-Glc) by ADP-Glc pyrophosphorylase (AGPase; EC 2.7.7.27) and its subsequent conversion into insoluble glucans by starch synthases (SS; EC 2.4.1.21) and starch branching enzymes (SBE; EC 2.4.1.18)—it is likely that the regulation of the starch biosynthetic pathway differs in these various tissues.
Recent genetic and biochemical evidence indicates that the synthesis of starch is achieved through the coordinated interactions of several of the enzymes of starch synthesis as well as some enzymes traditionally associated with starch degradation. Evidence for this idea comes from the analysis of genetic mutants that show pleiotropic effects on other enzyme activities. For example, the amylose extender (ae) mutant of maize (Zea mays) lacks SBEIIb (a form of SBE specifically expressed in the endosperm and flowering tissues) but also shows loss of activity of SBEI and altered properties of an isoamylase-type debranching (DBE; Colleoni et al., 2003). Mutations in maize that affect both a pullulanase-type DBE (zpu1-204) and an isoamylase-type DBE (su1-st) both cause a loss in SBEIIa activity, although the amount of SBEIIa protein is apparently unchanged (James et al., 1995; Dinges et al., 2001, 2003). The ae mutation in rice (Oryza sativa) endosperm (lacking SBEIIb) caused a dramatic reduction in the activity of soluble SSI (Nishi et al., 2001). In addition to the pleiotropic loss of SBEIIa activity in the zpu1-204 mutation, a reduction in β-amylase (EC 3.2.1.2) activity and a shift in β-amylase migration have been observed (Colleoni et al., 2003). In both the zpu1-204 and su1-st mutants, the inactive SBEIIa polypeptide accumulated to seemingly normal levels, suggesting the possibility of posttranslational modifications. It has been speculated that the coordination of debranching, branching, and SS activities required for starch synthesis might be accomplished by physical association of the enzymes in a complex or complexes within the amyloplast (Ball et al., 1996; Ball and Morell, 2003). Thus, the various mutations in different components of the putative complex would disrupt or alter the complex and cause a loss or reduction in biosynthetic capacity. Additional evidence exists for functional interactions between starch biosynthetic enzymes. In maize kernel extracts, the activity of SSI was greatly stimulated by the addition of purified SBEI or SBEII (Boyer and Preiss, 1979). Recently, Seo et al. (2002) showed that functional interactions exist between heterologously expressed SBEs from maize and yeast glycogen synthases working in a cyclically interdependent fashion, which is consistent with the idea that starch metabolizing enzymes may function in protein complexes.
Up to now, mechanisms involved in regulating the synthesis of starch from ADP-Glc have been largely confined to consideration of gene expression (Giroux et al., 1994; Gao et al., 1997). Little is known of the potential for posttranslational regulation in the enzymes of starch metabolism. Redox control of AGPase in Solanum tuberosum (potato) tubers has been proposed recently (Fu et al., 1998), and further work demonstrated that this phenomenon is relatively widespread and includes photosynthetic and nonphotosynthetic tissues from other species (Tiessen et al., 2002; Hendriks et al., 2003). There also is evidence for redox regulation of pullulanase-type DBEs in several tissues, including Spinacia oleracea (spinach) leaves and the endosperms of Hordeum vulgare (barley) and maize (Beatty et al., 1999; Cho et al., 1999; Schindler et al., 2001).
Protein kinase cascades play essential roles in diverse intracellular signaling processes in animals and yeast. In plants, there is evidence that protein phosphorylation plays an important role in signaling pathways triggered by abiotic stress, pathogen invasion, and plant hormones (Knetsch et al., 1996; Sheen, 1996; Zhang et al., 1998). Despite the importance and widespread occurrence of protein phosphorylation in plants, to date, there is no direct evidence that any starch metabolizing enzymes are regulated by phosphorylation.
This article describes the analysis of protein phosphorylation in starch synthesizing amyloplasts, with the aim of defining a role for this regulatory mechanism in starch metabolism and therefore bridging the gap in our knowledge between this universal mode of regulation and a fundamentally important storage product and commodity. We have studied phosphorylation in the amyloplasts of developing Triticum aestivum (wheat) endosperm, organelles committed to the synthesis of storage starch. Analysis of some of the phosphoproteins identified in T. aestivum leaf chloroplasts also is described, showing that some of the steps in the starch pathway regulated by phosphorylation are common to enzymes involved in both transitory and storage starch biosynthesis. In particular, we have focused on the discovery that isoforms of SBE, which catalyze the formation of α-(1→6)-branches in amylopectin, are regulated by protein phosphorylation in plastids and that several granule-associated proteins, including one or both forms of SBEII, become phosphorylated when isolated amyloplasts are incubated with γ-32P-ATP. We present evidence that protein phosphorylation regulates the activities of both stromal isoforms of SBEII and the interaction of SBEI with SBEIIb and other phosphoproteins involved in starch metabolism. We also provide evidence of the formation of protein complexes among different enzymes involved in starch synthesis.
RESULTS
ADP-Glc–Dependent Starch Synthesis in Isolated Amyloplasts
Intact amyloplasts isolated from developing T. aestivum endosperm readily synthesize starch from exogenously supplied ADP-[U-14C]-Glc (Tetlow et al., 1994), indicating the operation of an ADP-Glc–specific transport system (Tetlow et al., 2003a). In the current experiments, rates of starch synthesis by isolated plastids (Table 1) were comparable to rates of starch synthesis observed when whole endosperms were incubated with [U-14C]-Glc (Tetlow et al., 1994). When 1 mM ATP was included in incubations containing ADP-[U-14C]-Glc, there was a twofold stimulation in the net synthesis of starch (Table 1). Analysis of the radiolabeled starch suggests that ATP stimulates the synthesis of amylopectin more than amylose. The data for net starch synthesis show that addition of ATP has a small effect on amylose synthesis (P < 0.05) but caused a fourfold increase in amylopectin synthesis (P < 0.001). Under the conditions used in vitro, amylopectin synthesis and amylose synthesis occurred at approximately similar rates in the presence of ATP (Table 1). The concentration of ADP-Glc used (5 mM) may account for the relatively high proportion of amylose, given that it is known that elevated concentrations of ADP-Glc in vivo can increase the proportion of amylose synthesized in storage tissues (Tjaden et al., 1998). Alternatively, the experimental conditions employed in vitro still may be suboptimal for amylopectin synthesis.
Table 1.
ADP-Glc–Dependent Starch Synthesis in Amyloplasts Isolated from T. aestivum Endosperm
| Hexose Incorporation (nmol mg protein−1 h−1)
|
||||
|---|---|---|---|---|
| Incubation | Total Starch | Amylose Fraction | Amylopectin Fraction | Recovery |
| ADP-Glc | ||||
| Intact | 1729 ± 247 | 955 ± 66 | 227 ± 51 | 68% |
| Broken | 679 ± 115 | 382 ± 27 | 91 ± 16 | 70% |
| Net | 1050 ± 87 | 573 ± 35 | 136 ± 27 | — |
| ADP-Glc + ATP | ||||
| Intact | 3705 ± 388 | 1480 ± 413 | 1187 ±120 | 72% |
| Broken | 1551 ± 429 | 669 ± 92 | 551 ± 34 | 78% |
| Net | 2154 ± 318 | 811 ± 194 | 636 ± 48 | — |
Intact amyloplasts were incubated with 5 mM ADP-[U-14C]-Glc with or without 1 mM ATP for 30 min at 25°C. Amyloplasts used were 58 to 67% intact, judged by the latency of the marker enzyme alkaline inorganic pyrophosphatase. Material insoluble in methanol-KCl was fractionated using a thymol precipitation technique, and the amount of 14C recovered in each fraction was determined. Results presented are the mean and standard deviations of three independent experiments after subtracting 14C recovered in incubations stopped at zero time by the addition of 10% (w/v) trichloroacetic acid.
In Vitro Phosphorylation of Amyloplast Stromal Proteins
Protein phosphorylation in amyloplasts was examined using intact organelles prepared from T. aestivum endosperm at a time of maximal starch deposition in the developing grain from 12 to 25 d after pollination (DAP). Amyloplasts incubated with 100 μM γ-32P-ATP showed rapid phosphorylation of a large number of stromal polypeptides (Figure 1A). Preliminary 32P-labeling experiments with amyloplasts were performed between 1 and 30 min at 25°C and showed a rapid rate of phosphorylation for the majority of phosphorylated stromal polypeptides (most proteins were 32P-labeled within 2 min; data not shown). Incubation times with γ-32P-ATP for the experiments described were no longer than 20 min because beyond 30 min the integrity of the amyloplasts begins to diminish (Tetlow et al., 1994). The labeling pattern of stromal proteins was unchanged when 5 mM ADP-Glc was included in incubations (for up to 20 min) and when 1 mM γ-32P-ATP was used in labeling experiments (data not shown).
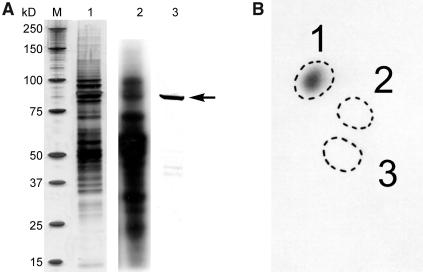
Figure 1.
Phosphorylation of Amyloplast Stromal Proteins from T. aestivum Endosperm.
Intact amyloplasts were isolated from developing T. aestivum endosperm at between 12 and 25 DAP and incubated with 100 μM γ-32P-ATP for 15 min at 25°C. Reactions were terminated, organelles lysed, and stromal phosphoproteins partially purified by affinity chromatography as described in Methods.
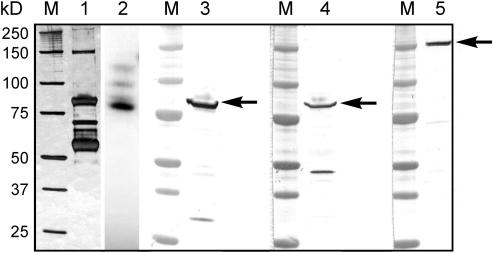
(A) Eluted phosphoproteins were separated by 1D-SDS-PAGE. Lane 1, silver-stained gel of stromal phosphoproteins eluted from affinity column. Lane 2, an autoradiograph of the 32P-labeled stromal phosphoproteins. Lane 3, immunoblot of the protein fraction from lane 1 probed with anti-E. coli branching enzyme antibodies. M, molecular mass markers whose individual sizes are shown at the left in kilodaltons; the arrow at the right indicates the position of the 87- to 88-kD polypeptides (putative SBEs) that were excised for phosphoamino acid analysis.
(B) Autoradiograph of phosphoamino acids separated by 2D thin-layer electrophoresis after acid hydrolysis of the 32P-labeled 87- to 88-kD polypeptide(s) shown in (A). The marked zones are positions of standards: 1, phosphoserine; 2, phosphothreonine; and 3, phosphotyrosine.
Among the more strongly 32P-labeled phosphoproteins in the amyloplast stroma (under the conditions described) were a group of polypeptides of 87 to 88 kD, often seen as a double band when visualized on one-dimensional (1D)-SDS-PAGE. This group of 87- to 88-kD phosphorylated polypeptides also copurified with SBE activity (measured by the phosphorylase a stimulation assay) when phosphoproteins were separated by anion exchange chromatography on HiTrap Q columns (data not shown). The 87- to 88-kD phosphorylated polypeptides also cross-reacted with polyclonal antibodies raised against the Escherichia coli branching enzyme (Figure 1A), leading us to consider that one or more of the forms of SBE could be phosphorylated and that phosphorylation of SBEs in amyloplasts could, at least partly, explain the stimulation in starch synthesis (particularly amylopectin synthesis) observed when plastids were incubated with ADP-Glc in the presence of ATP (Table 1).
Similar in vitro labeling experiments were performed with intact chloroplasts isolated from young T. aestivum leaves, resulting in the phosphorylation of 87- to 88-kD stromal polypeptides that also cross-reacted with the anti-E. coli branching enzyme antibodies (data not shown).
Phosphoamino acid analysis of the phosphorylated 87- to 88-kD polypeptides in amyloplasts and chloroplasts indicated that in all cases, phosphorylation occurred on one or more Ser residues (Figure 1B, data for amyloplasts shown).
Identification of the 87- to 88-kD Phosphorylated Polypeptides in Plastids
To identify the 87- to 88-kD phosphorylated peptides (putative SBEs) from both amyloplast and chloroplast stroma more precisely, peptide-specific antisera raised against the different forms of SBE (Morell et al., 1997; Rahman et al., 2001) were used in immunoprecipitation experiments. Peptide-specific antibodies to SBEI, SBEIIa, and SBEIIb immunoprecipitated phosphorylated 87- to 88-kD polypeptides from the different plastid lysates (Figure 2A), suggesting that all three soluble forms of SBE present in amyloplasts and both SBE forms in chloroplasts (SBEI and SBEIIa) are phosphorylated under the experimental conditions applied. No phosphoproteins were immunoprecipitated when anti-SBEIIb antibodies were used in control immunoprecipitation experiments with chloroplast stroma (data not shown). Further evidence for the anti-SBE antibodies being able to discriminate between the different (native) SBE isoforms is shown in Figure 4. The proteins immunoprecipitated with the different antisera were separated by two-dimensional (2D)-PAGE, electroblotted onto nitrocellulose, and probed with the anti-SBE antisera. The phosphorylated polypeptides that cross-reacted with the antisera were in-gel digested with trypsin (from corresponding gels), and some of the resulting peptides were sequenced using quadrupole-orthogonal acceleration time of flight mass spectrometry (Q-TOF-MS) (Figure 2B). The sequence data in Figure 2B shows that the amyloplast phosphoproteins immunoprecipitated with the different anti-SBE antisera are the corresponding forms of SBE (SBEI, SBEIIa, and SBEIIb). Sequence analysis (by Q-TOF-MS) of the tryptic peptides from the two phosphorylated polypeptides in chloroplasts immunoprecipitated with anti-SBEI and anti-SBEIIa antisera identified the phosphoproteins as SBEI and SBEIIa (data not shown).
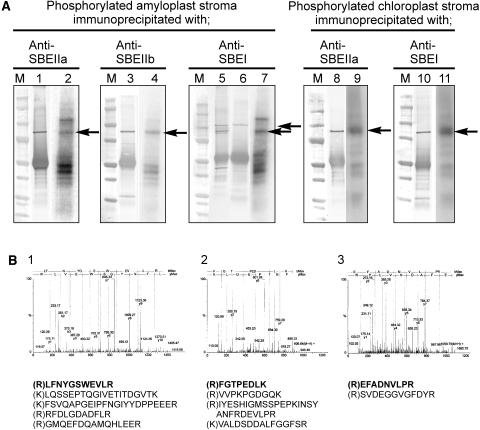
Figure 2.
Identification of the Phosphorylated 87- to 88-kD Polypeptides from Plastids as SBE Isoforms.
Intact amyloplasts (isolated from T. aestivum endosperm 12 to 25 DAP, 1.2 to 1.5 mg protein cm−3) and chloroplasts (isolated from 8- to 10-d-old T. aestivum leaves, 1.4 to 1.8 mg protein cm−3) were incubated with 100 μM γ-32P-ATP for 20 min at 25°C. Reactions were terminated, organelles lysed, and the stromal phosphoproteins partially purified by affinity chromatography as described in Methods.
(A) Stromal phosphoproteins were incubated with different, peptide-specific, anti-SBE antisera, and the 87- to 88-kD phosphoproteins were immunoprecipitated as described in Methods. Lanes 1, 3, 5, 8, and 10, immunoblots of the immunoprecipitated phosphoproteins probed with the respective anti-SBE antisera. Lane 6, an immunoblot of amyloplast stromal proteins immunoprecipitated with anti-SBEI antibodies and probed with anti-H. vulgare endosperm (plastidial) starch phosphorylase antbodies. Lanes 2, 4, 7, 9, and 11, autoradiographs of the respective immunoblots. M, molecular mass markers. Arrows indicate the positions of the phosphorylated 87- to 88-kD polypeptides that cross-reacted with the respective anti-SBE antibodies.
(B) Q-TOF-MS data of peptides from the 87- to 88-kD phosphoproteins immunoprecipitated with different anti-SBE antibodies and separated by 2D-PAGE. Panels 1 to 3, the MS survey acquisition data obtained for the 87- to 88-kD phosphoproteins from amyloplast stroma immunoprecipitated with the following: 1, anti-SBEIIa antibodies; 2, anti-SBEIIb antibodies; and 3, anti-SBEI antibodies. The data presented are for single representative analyses and in each case show the spectra obtained for one of the peptides. Below them are the corresponding sequences from each spectrum (in bold) and the sequences of other peptides acquired from the same sample.
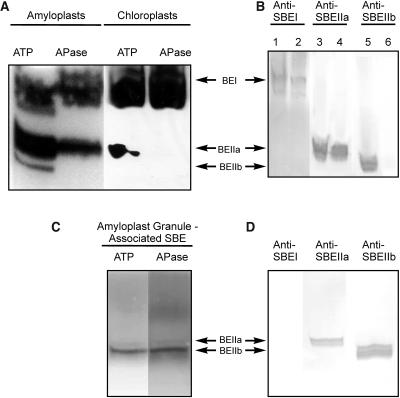
Figure 4.
Activity Gel Analysis of SBE Isoforms Showing Effects of Dephosphorylation on Enzyme Activity.
(A) Zymogram analysis of SBE activity from stromal extracts of amyloplasts (isolated from endosperm 12 to 25 DAP) and chloroplasts (from 8- to 10-d-old leaves) after pretreatment of the samples with 1 mM ATP or 10 units APase for 20 min at 25°C. Approximately 80 μg of amyloplast stromal protein per lane and 120 μg of chloroplast stromal protein per lane were separated on a 7-cm native polyacrylamide gel containing substrates for SBE. SBE activities were visualized by staining with I2/KI.
(B) Immunoblots from SBE zymogram gels of amyloplast and chloroplast stromal proteins (same protein loadings as in [A]) developed with antisera against SBEI (lanes 1 and 2), SBEIIa (lanes 3 and 4), and SBEIIb (lanes 5 and 6), showing the positions of each of the different SBE forms on the zymogram. The antisera used in the immunoblots in Figure 4B were used at the same dilutions as for the immunoprecipitation experiments. Lanes 1, 3, and 5 contain amyloplast proteins, and lanes 2, 4, and 6 contain chloroplast proteins.
(C) Zymogram analysis of granule-associated SBE activity in amyloplasts. Starch granule–associated proteins were extracted after incubation of plastids with ATP and subsequently either untreated or incubated with APase (see above). Samples were separated on native gels (∼80 μg of protein per lane) and SBE activity visualized as above.
(D) Immunoblots from SBE zymogram gels of granule-associated proteins from amyloplasts (ATP treated) developed with various anti-SBE antisera as shown.
When anti-SBEI antibodies were used in immunoprecipitation experiments with amyloplast stroma after labeling of intact plastids with γ-32P-ATP, a weakly labeled phosphoprotein of ∼110 kD was detected in addition to the phosphorylated SBEI (Figure 2A). The identity and significance of this protein will be discussed in the context of later experiments (Figure 5A). The 110-kD phosphoprotein was not detected in the SBEI immunoprecipitates from leaf chloroplast stroma (Figure 2A) or in immunoblots of chloroplast stroma (data not shown). Other 32P-labeled proteins also were present in the different SBE immunoprecipitation complexes from amyloplast stroma (Figure 2A). Notably, phosphoproteins in the region of 45 to 50 kD were immunoprecipitated with all three anti-SBE antibodies, and amyloplast phosphoproteins of ∼120 kD also were present in anti-SBEI and anti-SBEIIa immunocomplexes. None of these phosphoproteins could be identified from 2D gels or using available antibodies because they all comigrated with various components of the immunoprecipitation complexes. Analysis of immunoprecipitation complexes from chloroplasts by autoradiography showed a phosphoprotein of ∼95 kD immunoprecipitated with both phosphorylated SBEI and phosphorylated SBEIIa (Figure 2A, lanes 9 and 11). No further analysis of the 95-kD phosphoprotein(s) was made.
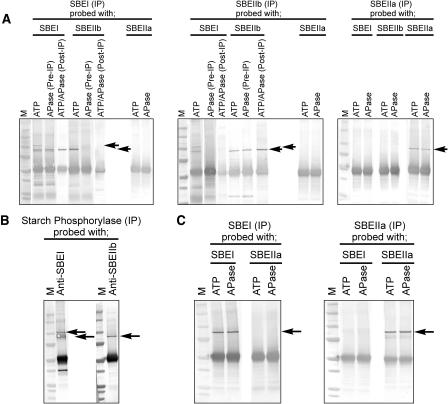
Figure 5.
Coimmunoprecipitation of Plastid Stromal Proteins with Peptide-Specific Anti-SBE Antibodies.
(A) Amyloplast lysates (1.2 to 1.5 mg protein cm−3) were either preincubated with 1 mM ATP or with 10 units of APase (Pre-IP) for 20 min at 25°C. Alternatively, amyloplast lysates were preincubated with 1 mM ATP and immunoprecipitated, and the washed immunoprecipitation products treated with 10 units of alkaline phosphatase (ATP/APase [Post-IP]). After immunoprecipitation (IP) with anti-SBE antibodies, immunoprecipitated proteins were separated by 1D-SDS-PAGE, electroblotted onto nitrocellulose, and developed with various anti-SBE antisera as shown. Bottom arrow indicates positions of 87- to 88-kD SBE proteins cross-reacting with anti-SBE antisera. Top arrow indicates a 110-kD stromal phosphoprotein (identified as starch phosphorylase) that cross-reacted with anti-SBEI antibodies and coimmunoprecipitated with phosphorylated SBEI. M, molecular mass markers.
(B) Amyloplast lysates (1.2 to 1.5 mg protein cm−3) were preincubated with ATP (as above) and immunoprecipitated with anti-starch phosphorylase antibodies, separated by 1D-SDS-PAGE, immunoblotted, and developed with anti-SBEI or anti-SBEIIb antisera.
(C) Chloroplast lysates (1.4 to 1.6 mg protein cm−3) made from plastids harvested at the end of the photoperiod were preincubated with either ATP or APase, immunoprecipitated with anti-SBE antibodies, separated by 1D-SDS-PAGE, immunoblotted, and developed with anti-SBEI and anti-SBEIIa antisera as described for amyloplasts in (A) and (B).
Phosphorylation of Granule-Associated Proteins in Amyloplasts
Granule-associated proteins were extracted from T. aestivum endosperm amyloplast lysates after incubation of plastids with γ-32P-ATP and separated by 1D-SDS-PAGE (Figure 3). At least four phosphorylated granule-associated proteins were visualized by autoradiography after 32P-labeling of intact amyloplasts for 20 min (Figure 3). Immunoblots of the starch granule–associated proteins were probed with the peptide-specific anti-SBE antibodies, and the results confirmed previous reports that SBEI is absent from starch granules in T. aestivum endosperm (Rahman et al., 1995) and that a proportion of each of the SBEIIa and SBEIIb forms in amyloplasts is granule associated (Rahman et al., 2001). Immunoblots of the starch granule–associated proteins also were probed with antibodies raised against other proteins not known to associate with starch granules to check for contamination of the starch granule preparation by stromal proteins. Antibodies raised against the large and small subunits of AGPase were used to probe immunoblots of the granule-associated proteins (both antibodies used at a dilution of 1:1000). No proteins in the starch granule–associated protein preparation cross-reacted with either of the anti-AGPase antisera (data not shown). The phosphorylated 87- to 88-kD polypeptide(s) from starch granules that cross-reacted with both anti-SBEIIa and SBEIIb antibodies were excised from gels and in-gel digested with trypsin. Q-TOF-MS analysis of the tryptic peptides from the phosphorylated 87- to 88-kD polypeptide(s) confirmed the starch granule–associated protein(s) as SBEIIa, SBEIIb, or a mixture of both forms because the peptide sequences obtained were not sufficient to distinguish the two SBEII isoforms (Table 2).
Figure 3.
Analysis and Identification of Granule-Associated Phosphoproteins from T. aestivum Endosperm Amyloplasts.
Intact amyloplasts were isolated from developing endosperm at 12 to 25 DAP and incubated with 100 μM γ-32P-ATP for 20 min at 25°C. Reactions were terminated and organelles lysed and fractionated as described in Methods. Granule-associated proteins were extracted from starch grains after incubation of amyloplasts with γ-32P-ATP and separated by 1D-SDS-PAGE on 4 to 12% gradient gels. Lane 1, silver-stained polyacrylamide gel of polypeptides extracted from starch granules (15 μg of protein per lane); lane 2, autoradiograph of starch granule–associated polypeptides separated by SDS-PAGE (from lane 1), showing phosphorylated polypeptides; lanes 3 to 5, immunoblots of radiolabeled starch granule–associated polypeptides probed with peptide-specific anti-SBE-antibodies: lane 3, anti-SBEIIa; lane 4, anti-SBEIIb; and lane 5, anti-SBEI. M, molecular mass markers. Arrows indicate the positions of the starch granule–associated polypeptides that cross-reacted with the respective anti-SBE antibodies.
Table 2.
Analysis of Starch Granule–Associated Polypeptides from T. aestivum Endosperm Amyloplasts by Q-TOF-MS
| Phosphorylated Starch Granule–Associated Polypeptides | Q-TOF-MS–Derived Sequence | Identity of Protein |
|---|---|---|
| 120 kD | (R)YGDYEEAYDVGVR | SS (fragment) |
| 88-kD phosphoproteins cross-reacting with anti-SBEIIa and anti-SBEIIb antisera | (R)AAIDQHEGGLEAFSR (R)DDYGVWEIFLPNNADGSSAIPHGSR (K)INSYANFRDEVLPR (R)GPQTLPTGK (K)VALDSDDALFGGFSR | SBEIIa and/or SBEIIb |
| 85 kD | (K)A248LSPPAAPAVQQDLWDFKKYIGFEEPVEAKDDGR281 (K)T322GGLGDVAGALPK334 (R)H396RQEDIYGGSR406 (R)G483PVDEFPFTELPEHYLEHFRLYDPVGGEHANYFAAGLK520 (K)L786YEDVLVK793 | SSIIa (wSSII-B) |
| Nonphosphorylated 152-kD polypeptide cross-reacting with anti-SBEI antiserum | (K)F129GINTENDATVYR142 (K)D150AQLIGDFNNWNGSGHR166 (K)D170NFGVWSIRISHVNGKPAIPHNSK193 (K)F223GAPYDGVHWDPPTGER239 (R)S482VDEGGVGFDYR493 | SBEIc |
Silver-stained polypeptides were excised from polyacrylamide gels (Figure 3) and digested with trypsin, and the recovered peptides were sequenced using Q-TOF-MS to identify the phosphoproteins and other polypeptides. The peptide sequences presented for each polypeptide analyzed were acquired from a single representative in-gel digest and, where applicable, show their position within the sequence of the presumptive unprocessed protein. The presence of Gln259 and Val792 in the peptide sequences from SSIIa are shown in bold.
The other granule-associated phosphoproteins, having apparent molecular masses of ∼120, 100, and 85 kD, were in-gel digested with trypsin and analyzed by Q-TOF-MS. Peptide sequences obtained from the digested proteins identified the 120-kD phosphoprotein as a SS, for which only partial amino acid sequence is currently available (accession number O24398), and the 85-kD phosphoprotein as SSIIa (Table 2). The presence of Gln259 and Val792 in the peptide sequences from SSIIa (shown in bold in Table 2) further identified the phosphoprotein as wSSII-B, as described by Li et al. (1999). We were unable to identify the granule-associated phosphoprotein of ∼100 kD using either Q-TOF-MS or mass fingerprinting of the trypsin-derived peptides by matrix-assisted laser-desorption ionization TOF-MS.
The anti-SBEI antibodies also cross-reacted with a 152-kD granule-associated polypeptide (Figure 3). Sequencing of the in-gel trypsin digestion products from the 152-kD polypeptide (Table 2) revealed that this protein is the granule-associated SBEIc recently isolated from T. aestivum endosperm by Båga et al. (2000). SBEIc was not identifiably phosphorylated under the experimental conditions described.
Effects of Protein Phosphorylation on SBEs
Having established that all soluble forms of SBE in amyloplasts and chloroplasts are phosphorylated, as well as one or both granule-associated forms of SBEII in amyloplasts, measurements of total extractable SBE activity were made to determine the possible effects of phosphorylation and dephosphorylation on catalytic activity. SBE activity was measured quantitatively using the phosphorylase a stimulation assay (Table 3). In addition, a semiquantitative zymogram analysis of SBE activity also was employed to visualize the effects of phosphorylation and dephosphorylation on individual SBE isoforms (Figure 4). The results of the phosphorylase a stimulation assays presented in Table 3 show that the extractable SBE activity in stromal preparations of plastid lysates is reduced fourfold in amyloplasts and threefold in chloroplasts after dephosphorylation by alkaline phosphatase-agarose (APase). In addition to direct measurements of APase activity in samples prepared for SBE assays, control experiments were performed that involved mixing equal volumes of ATP-treated and APase-treated samples to determine whether there was any residual APase activity in the latter that would interfere with the phosphorylase a stimulation assay. The results of the direct measurements of APase activity and the mixing experiments (Table 3, legend) indicated that there was no interference from residual APase on the assay of SBE. Zymogram analysis (Figure 4A) shows that the effects of dephosphorylation in stromal lysates are apparently confined to SBEII forms, with no detectable effects on measurable SBEI activity. No APase activity was detected on zymograms, indicating again that the in-gel SBE assay was not subject to any residual APase remaining after dephosphorylation treatments. SBE forms were identified from zymograms using immunoblots of the native gels that had been probed with anti-SBE antibodies (Figure 4B).
Table 3.
Effects of Dephosphorylation on SBE Activity in T. aestivum
| SBE Activity (μmol min−1mg protein−1)
|
||
|---|---|---|
| + ATP | + APase | |
| Amyloplast stroma | 213 ± 20 | 54 ± 7 |
| Chloroplast stroma | 252 ± 14 | 86 ± 6 |
| Amyloplast granule-associated proteins | 179 ± 19 | 184 ± 21 |
Samples were pretreated with 1 mM ATP or 10 units APase for 20 min at 25°C. Agarose-conjugated APase was removed by centrifugation (plastid stroma), and soluble alkaline phosphatase was removed by anion exchange chromatography (granule-associated extracts). SBE activity was measured using the phosphorylase a stimulation assay. Data shown are the mean ± se of six (plastid stroma) and four (granule-associated proteins) individual preparations, respectively. Control experiments with amyloplast and chloroplast stroma were performed whereby equal volumes of ATP-treated and APase-treated stroma were mixed and incubated for 20 min at 25°C before measurement of SBE activity. Values obtained from the mixing experiments were 138 ± 15 μmol min−1mg protein−1 (amyloplast stroma, n = 3) and 176 ± 21 μmol min−1mg protein−1 (chloroplast stroma, n = 3).
The partially purified granule-associated SBE activity extracted from amyloplast starch granules was comparable to the soluble SBE activity measured in the stromal fraction (Table 3). However, dephosphorylation with APase caused no change in measurable SBE activity in the granule-associated protein preparation, and this also was observed in zymograms of granule-associated SBEII activity, which showed no change in activity of either form of SBEII (Figure 4C); SBEIIa activity was barely detectable on these zymograms. When zymograms of granule-associated proteins were overdeveloped with I2/KI solution, a faint band of SBE activity was detected in a similar position to the SBEI activity observed in stromal preparations. This activity also was unchanged after dephosphorylation with APase (data not shown). Based on previous findings that SBEI is not granule-associated (Rahman et al., 1995) and that the anti-SBEI antibody does not recognize SBEII isoforms (Morell et al., 1997), it is concluded that this represents the 152-kD granule-associated SBEIc form. Granule-associated forms of SBE visualized on zymograms also were identified using immunoblots of the native gels with anti-SBE antibodies (Figure 4D). Anti-SBEI antibodies were unable to detect SBEIc on immunoblots of granule-associated proteins from native gels (Figure 4D) despite recognizing SBEIc from denatured granule-associated protein preparations (Figure 3, lane 5).
Soluble alkaline phosphatases (i.e., forms not conjugated to agarose) were incubated with isolated washed starch granules (at 25°C for 1 to 4 h) to test the possibility that starch granule–associated proteins were attached to starch grains directly (or indirectly through protein–protein interactions) in a phosphorylation-dependent manner. Dephosphorylation of starch granule–associated proteins with different soluble alkaline phosphatases caused no change in the amount or complement of starch granule–associated proteins subsequently extracted from the washed starch granules, as visualized by 1D-SDS-PAGE (data not shown). In addition, no proteins from the starch granules could be detected in the alkaline phosphatase extraction buffer after incubations, as visualized by either 1D-SDS-PAGE (silver stained) or immunoblotting using anti-SBE antibodies (data not shown).
Coimmunoprecipitation of Plastid Stromal Proteins
Coimmunoprecipitation experiments were performed with plastid lysates, primarily to examine the possibility of phosphorylation-dependent protein–protein interactions among forms of SBE (Figure 5). Where possible, other interacting proteins were identified by sequencing proteins from immunoprecipitation complexes that had been separated by 2D-PAGE.
The immunoprecipitation experiments shown in Figures 5A and 5C show that each of the peptide-specific anti-SBE antibodies precipitates the respective form of SBE from plastid lysates and that phosphorylation or dephosphorylation (with APase) does not affect the ability of any of the antibodies to bind to the respective forms of SBE. We observed no apparent alterations in the electrophoretic mobility of the different phosphoproteins using the 1D gel systems (Figures 2A and 5) or the native gels used for zymogram analyses (Figures 4A and 4C).
Experiments with amyloplast lysates (Figure 5A) showed that SBEI and SBEIIb coimmunoprecipitate when preparations were preincubated with 1 mM ATP. Notably, the coimmunoprecipitation of SBEI and SBEIIb in amyloplasts also was observed in untreated plastid lysates (i.e., no ATP; data not shown). The coimmunoprecipitation of SBEI and SBEIIb (visualized by the presence of SBEI in the SBEIIb immunoprecipitation complex and the presence of SBEIIb in the SBEI immunoprecipitation complex; Figure 5A) was lost if lysates were first dephosphorylated with APase. SBEIIa showed no interaction with other forms of SBE under the experimental conditions described. A 110-kD phosphoprotein was coimmunoprecipitated with SBEIIb and SBEI in a phosphorylation-dependent manner (Figure 5A). The antibody to SBEI also recognized the 110-kD protein on protein gel blots. This might account for its presence in immunoprecipitates when using the SBEI antibody although, notably, pretreatment with APase led to the disappearance of this protein from immunoprecipitates. However, of greater significance is the observation that the antibody to SBEIIb did not cross-react with the 110-kD protein, yet incubation of ATP-treated samples with this antibody led to the detection of the 110-kD protein in immunoprecipitates. It therefore appears that its coprecipitation is a function of its association with the SBEIIb protein (and, therefore, also SBEI) and is another element of this protein complex. Subsequent Q-TOF-MS analysis of the 110-kD phosphoprotein after separation and in-gel digestion with trypsin identified the protein as α-1,4-glucan phosphorylase (starch phosphorylase; EC 2.4.1.1) after peptide sequencing [(R)AGDSLNWEDFPSK, (R)TVAYTNHTVLPEALE, (K)FADDEDLQSEWR, and (K)AFATYVQAK]. This protein also cross-reacted with anti-H. vulgare endosperm (plastidic) starch phosphorylase antibodies, confirming its identity (Figure 2A). In vitro γ-32P-ATP labeling of starch phosphorylase in amyloplasts followed by phosphoamino acid analysis indicated that starch phosphorylase is phosphorylated at one or more Ser residues (data not shown).
To determine whether the formation of protein complexes could be reversed by dephosphorylation, the coprecipitated products formed after treatment of the stroma with ATP also were treated with soluble alkaline phosphatase (Figure 5A, Post-IP). Treatment of the SBEIIb immunoprecipitation product with alkaline phosphatase caused loss of SBEI and starch phosphorylase. Likewise, in the SBEI immunoprecipitation product, treatment with alkaline phosphatase caused loss of SBEIIb and starch phosphorylase (Figure 5A).
Coimmunoprecipitation experiments with amyloplast lysates similar to those described above were performed with anti-H. vulgare starch phosphorylase antibodies. Gel blots of the starch phosphorylase immunoprecipitation products clearly showed that SBEI (and starch phosphorylase) and SBEIIb proteins were coprecipitated by the phosphorylase antibodies (Figure 5B) despite the fact that starch phosphorylase antibodies do not recognize either SBEI or SBEIIb (Figure 2A, lane 6).
Coimmunoprecipitation experiments with chloroplast lysates (made from chloroplasts harvested at the end of the photoperiod) indicated that the two forms of SBE in chloroplasts did not coprecipitate with each other (Figure 5C). Similar experiments with chloroplasts harvested at the end of the dark period also showed no interaction between the SBE forms (data not shown).
Isolated starch granule–associated proteins from amyloplasts also were used in coimmunoprecipitation experiments with anti-SBE antibodies to test for the presence of protein complexes involving granule-associated SBE forms. No coimmunoprecipitation was observed between SBEII isoforms or between SBEIc and SBEII forms (data not shown) when investigating enzymes released from starch granules.
DISCUSSION
This article reports the involvement of protein phosphorylation in the process of starch biosynthesis and in particular on the effects of posttranslational modification on SBE activity and the association of SBE isoforms with other polypeptides. The observed stimulation of amylopectin synthesis by exogenous ATP in isolated amyloplasts partly may be explained by the activation of SBEII isoforms by protein phosphorylation. Whereas all forms of SBE in isolated plastids were shown to be phosphorylated by exogenous ATP, the effects of phosphorylation on individual forms appear to differ (Figure 4). Indeed, the effects of phosphorylation on SBEII forms appear to depend on their subcellular location within the amyloplast. In addition to the direct measurable effects of protein phosphorylation on the catalytic activity of SBEII forms, we also present evidence that indicates that SBEIIb, SBEI, and starch phosphorylase form a protein complex whose integrity is dependent on protein phosphorylation. Notably, the observed complex also was detected in samples that had not been pretreated with ATP, suggesting that it is present endogenously and can be preserved if gentle extraction techniques, such as organelle purification, are used.
A great deal of recent experimental evidence highlights the potential importance of protein–protein interactions between starch metabolizing enzymes during the synthesis of storage starch in cereal endosperms (James et al., 2003). For example, zymogram analyses of the maize ae mutant, lacking the largely endosperm-specific SBEIIb protein, also show loss in SBEI activity in this material. Such interdependence of enzyme activities has been suggested to be a result of protein–protein interactions, which are disrupted in the mutant plant (Colleoni et al., 2003). The data presented in this report, using T. aestivum endosperm amyloplasts, shows that SBEIIb and SBEI form phosphorylation-dependent protein complexes, which is consistent with the findings and interpretation of the data from the maize ae mutant. Likewise, the 50% reduction in SSI activity observed in rice ae mutants lacking SBEIIb activity suggested that in vivo the SBEIIb and SSI proteins may interact (Nishi et al., 2001). However, after 2D-PAGE analysis of SBE-interacting proteins, we found no detectable evidence for any interactions between SSI and SBEs in T. aestivum amyloplasts. Elegant work by Seo et al. (2002) using heterologously expressed maize SBEs showed that SBEI functions only when expressed with both SBEIIa and SBEIIb, predicting that SBEs act sequentially during glucan polymer synthesis, with the SBEII isoforms producing substrates for SBEI. It is conceivable that the efficiency of polymer construction as described by Seo et al. (2002) would be greatly improved by the physical association of SBEs as well as other components of the biosynthetic machinery (for example, SSs and DBEs). We have begun to describe part of such a biosynthetic complex in amyloplasts, whereby some of the components (SBEI, SBEIIb, and starch phosphorylase) physically interact via a process involving protein phosphorylation. In chloroplasts, which synthesize transient starch, the role of phosphorylation of SBE isoforms is less clear because no interactions were observed between SBEI and SBEIIa or any other identifiable proteins despite the fact that phosphorylation of both enzymes was detected (Figure 2A). If any physical interactions exist between SBEIIa and other SBEs in chloroplasts and amyloplasts, they are relatively weak and must have dissociated during the experimental procedures employed. In chloroplasts, phosphorylation was shown to activate SBEIIa. However, in the absence of any notable change in activity or detection of a protein complex, the function/role of phosphorylation of SBEI in chloroplasts is at present unclear. In general, the role of SBEI in any tissue is not understood. Analysis of a maize transposon-tagged mutant indicated that there was no apparent effect on starch synthesis of deletion of SBEI in either leaves or endosperm (Blauth et al., 2002). Recent analysis of a SBEI-deficient mutant in rice showed a normal phenotype and endosperm starch content; the only measurable effects of the mutation were on the fine structure of amylopectin, which caused some alteration in the physicochemical properties of the mutant starch (Satoh et al., 2003).
The various SBE isoforms differ significantly in their kinetic properties and substrate specificities. For example, in T. aestivum, SBEI (expressed later in development than the SBEII proteins) has a higher affinity for amylose (and longer chained α-glucans) than SBEII forms, suggesting different modes of action and physiological roles for the different SBEs (Morell et al., 1997; McCue et al., 2002). Studies of the regulation of SBE activity in storage tissues mostly have been limited to consideration of gene expression (Burton et al., 1995; Gao et al., 1996, 1997; Jobling et al., 1999) and the previous finding in cereal endosperm that SBE activity (particularly SBEI activity) is stimulated by inorganic orthophosphate (Pi; Morell et al., 1997). We suggest that the phosphorylation-dependent increase in SBEII activity observed in plastid stromal extracts are direct effects of protein kinase(s) on the individual SBEII proteins. The effects of dephosphorylation on the activities of individual SBEII isoforms, which had been separated and visualized on zymograms (Figure 4), points to the effects of phosphorylation acting on the individual proteins rather than being mediated solely through interactions/complex formation with other SBEs. Additional support for this notion comes from analysis of SBEIIa activity on zymograms. SBEIIa did not form protein complexes with other SBEs under the experimental conditions described, but its activity was markedly reduced after dephosphorylation. However, we cannot rule out the possibility that other, as yet unidentified, proteins are able to stimulate phosphorylation-dependent protein–protein interactions with SBEII isoforms, which cause changes in catalytic activity. The finding that total measurable SBE activity in plastids is stimulated by phosphorylation has possible implications for previous measurements of SBE activity in whole cell extracts, where the phosphorylation status of the different SBE isoforms to date has not been considered. Previous measurements of SBE activity in whole cell extracts (plants/bacteria) have not taken into consideration the degree of activation of the SBE isoforms, leading to underestimates of activity if the extracted enzyme was substantially dephosphorylated. Identification of the phosphorylation site motifs recognized by the respective SBE kinases and phosphatases may indicate a role for these regulatory proteins in the posttranslational modification of other members of the α-amylase superfamily of enzymes involved in starch metabolism, of which the SBEs are a part (Jespersen et al., 1993).
In vitro phosphorylation experiments with isolated T. aestivum endosperm amyloplasts showed that several granule-associated proteins were phosphorylated after short incubation periods with γ-32P-ATP, including the granule-associated isoform(s) of SBEII, which comprise up to 45% of total SBEII activity (Mu-Forster et al., 1996). This observation shows that the respective protein kinases (and presumably protein phosphatases) must have some degree of access to the starch granule matrix. However, it is not yet clear whether or not the starch granule–associated proteins being phosphorylated under these conditions are confined to the periphery of the growing starch granule or are distributed throughout the matrix (which is presumably less accessible to soluble proteins). Other (regulatory) proteins not directly involved in starch biosynthesis have been found in starch granules. Sehnke et al. (2001) showed that a form of 14-3-3 protein (from the ɛ subgroup) was present in Arabidopsis (Arabidopsis thaliana) leaf starch and postulated its role in the regulation of transient starch synthesis through protein phosphorylation. The role of 14-3-3 proteins in metabolism is the mediation of protein–protein interactions, whereby the 14-3-3 protein binds to a phosphorylated target protein (Chung et al., 1999). Although 14-3-3 proteins have been identified in Pisum sativum (pea) chloroplasts (Sehnke et al., 2000), their presence in amyloplasts, which make storage starch, has not been demonstrated. Few of the major starch metabolizing enzymes have well-conserved 14-3-3 binding motifs (one notable exception is β-amylase), which suggests that if 14-3-3 proteins are involved in facilitating protein–protein interactions between starch metabolizing enzymes in plastids, then binding to the target phosphoproteins must be at, as yet, uncharacterized binding sites.
The effects of protein phosphorylation on the functions of the granule-associated form(s) of SBEII in vivo are unclear. Zymograms and phosphorylase a stimulation assays of SBE in starch granule–associated protein extracts showed no measurable effects of dephosphorylation with APase, which suggests that phosphorylation of granule-associated SBEII may serve some function other than modulation of catalytic activity. However, we cannot rule out the possibility that the proteins released from the starch granules during the extraction procedure are, or become, insensitive to any possible effects of phosphorylation/dephosphorylation or that the extraction procedures employed do not allow detection of any phosphorylation- or dephosphorylation-dependent changes in activity. The aqueous extraction of starch granule–associated proteins from T. aestivum by this method releases only 10 to 20% of the total (Denyer et al., 1995). The observation that dephosphorylation (using soluble alkaline phosphatases) of the starch granule–associated proteins within the starch matrix failed to release any granule-associated proteins suggests that protein phosphorylation is unlikely to be involved in the attachment of proteins to granules. Coimmunoprecipitation experiments with granule-associated protein extracts showed no evidence of protein–protein interactions between granule-associated SBEs. We are currently pursuing the possibility that phosphorylation of one or both SBEII proteins enables interaction/complex formation with other starch granule–associated proteins within the polymer matrix.
Three other starch granule–associated phosphoproteins were identified from washed starch granules after in vitro phosphorylation experiments with intact amyloplasts. The most strongly phosphorylated phosphoprotein within the starch granules was an 85-kD polypeptide identified as SSIIa (encoded by the ssIIa gene located on the B genome of T. aestivum), the in vivo role/function of which in storage tissues is beginning to be understood by studies of mutants. Loss of SSII (in dicots) or SSIIa (in monocots) activity results in reduced starch content and various alterations in starch structure and morphology (Craig et al., 1998; Umemoto et al., 2002). Interestingly, loss of SSIIa in T. aestivum and H. vulgare endosperms causes a drastic reduction in amylopectin synthesis and abolishes binding of SSI, SBEIIa, and SBEIIb to the starch granules (Yamamori et al., 2000; Morell et al., 2003). Peptide sequence obtained from the 120-kD granule-associated phosphoprotein identified the protein as another SS, although to date this protein has no complete amino acid sequence available in the databases (accession number O24398). The identity of the 100-kD granule-associated phosphoprotein remains unresolved; however, the only known protein of this mass in the starch granules of T. aestivum endosperm was identified as SSIIa (Denyer et al., 1995; Li et al., 1999).
The major findings of this work demonstrate that key stromal- and granule-associated enzymes of the starch (amylopectin) biosynthetic pathway in amyloplasts are phosphorylated. In particular, phosphorylation of SBEII isoforms in both chloroplasts and amyloplasts regulates their catalytic activity. Notably, in amyloplasts, phosphorylation influences the ability of SBEs to form and maintain protein complexes, possibly indicating different mechanisms for polymer construction between storage starch– and transient starch–synthesizing plastids. SBEIIb may play a key role in complex formation because this isoform of SBEII is specific to storage starch–synthesizing tissues (amyloplasts), and the ability to form a protein complex, which was not detected in chloroplasts, may determine to some degree the type of starch synthesized. In addition to phosphorylation-dependent SBE–SBE interactions, we also have identified one other component of a putative complex in amyloplasts: starch phosphorylase. The association of starch phosphorylase with SBEI and SBEIIb suggests that it is involved in starch synthesis. There are two alternative means by which starch phosphorylase can perform such a biosynthetic role. First, SBEs could use the products of starch phosphorylase activity (i.e., branching the glucan chains elongated by starch phosphorylase operating in a glucan-forming direction). A second more likely scenario involves starch phosphorylase operating after SBE activity by modifying the structure of amylopectin via phosphorolysis.
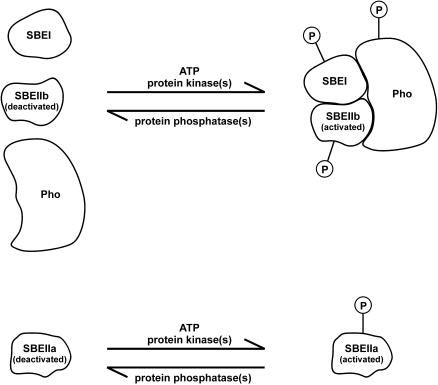
In conclusion, protein phosphorylation appears to have a direct effect on SBEIIa and SBEIIb activity and impacts the rate of measurable starch synthesis in intact organelles (Table 1, Figure 4). Evidence presented in this article clearly shows that protein phosphorylation also is a necessary prerequisite for the formation of a starch-synthesizing protein complex and that reversal by dephosphorylation leads to its disaggregation (Figure 5A). The model in Figure 6 represents a working hypothesis based on the evidence presented in this article. Other interacting proteins are probably present, forming a larger complex than the one identified so far, and these components await identification.
Figure 6.
Model of Phosphorylation-Dependent Protein Complex Formation Involved in Starch Synthesis.
Activation of SBEIIa and activation and complex formation involving SBEI, SBEIIb, and starch phosphorylase (Pho) by protein phosphorylation in the amyloplast stroma stimulate amylopectin biosynthesis. The functional relationships between the different components of the putative protein complex are unclear.
METHODS
Plant Material and Plastid Isolation
Spring T. aestivum cv Axona was grown in glasshouses under conditions described previously (Tetlow et al., 1993). Amyloplasts were isolated from the endosperm obtained from developing grains taken from the mid-ear region of the head (between 12 and 25 DAP) using the methods described by Tetlow et al. (2003b), typically using between 70 and 110 g fresh weight of seed for each preparation. Intact amyloplast preparations were carefully resuspended in amyloplast isolation medium (50 mM Hepes/NaOH, pH 7.5, 1 mM Na2-EDTA, 1 mM KCl, 2 mM MgCl2, and 0.5 M sorbitol) containing 10 μL per cm3 of a protease inhibitor cocktail (catalog number P 9599; Sigma-Aldrich, St. Louis, MO). Amyloplasts prepared in this way were 55 to 70% intact as judged by the latency of the plastid marker enzyme alkaline inorganic pyrophosphatase (EC 3.6.1.1). The maximum cytosolic contamination of the amyloplast preparations was 0.3%, based on the recovery of UDP-Glc pyrophosphorylase (EC 2.7.7.9), and mitochondrial contamination was not >0.2%, based on recoveries of the marker enzymes citrate synthase (EC 4.1.3.7) and cytochrome c oxidase (EC 1.9.3.1) (data not shown). In some experiments, intact amyloplasts were lysed osmotically by the addition of ice-cold rupturing buffer containing 100 mM Tricine/KOH, pH 7.8, 1 mM Na2-EDTA, 1 mM DTT, 5 mM MgCl2, and the protease inhibitor cocktail (above) and starch granules removed by centrifugation at 14,000g for 5 min at 4°C.
Chloroplasts were prepared from the primary leaves of 8- to 10-d-old spring T. aestivum (cv Axona) using the methods described by Bruce et al. (1994). All procedures were performed at 0 to 4°C. Leaves (∼50 g) were harvested at the end of the photoperiod and chopped using a Polytron blender in 50 mL of isolation medium (50 mM Tricine/NaOH, pH 7.9, 330 mM sorbitol, 1 mM Na2-EDTA, 1 mM DTT, 2 mM MgCl2, and 0.1% [w/v] BSA). Isolated chloroplasts were carefully resuspended in isolation medium or lysed osmotically (see above) before use. Chloroplasts were routinely >85% intact, as judged by the latency of alkaline inorganic pyrophosphatase. Measurements of the cytosolic marker enzymes UDP-Glc pyrophosphorylase and alcohol dehydrogenase (EC 1.1.1.1) in chloroplast preparations indicated that cytosolic contamination was <0.2%.
Enzyme Assays
Subcellular marker enzyme assays were performed as described previously (Tetlow et al., 2003b).
SBE activity was assayed semiquantitatively using a phosphorylase a stimulation assay essentially as described by Smith (1988).
Endoamylase (α-amylase; EC 3.2.1.1) was measured using the Ceralpha kit from Megazyme International (Wicklow, Ireland). The specific substrate nonreducing end-blocked ρ-nitrophenol maltoheptaoside (BPNPG7) was used in 20 mM sodium acetate, pH 5.6, in accordance with the manufacturer's instructions.
APase (catalog number P-0762; Sigma-Aldrich), alkaline phosphatase from bovine intestinal mucosa (type VII-L, catalog number P-6772; Sigma-Aldrich), and λ protein phosphatase (λ-Pase from E. coli; New England Biolabs, Beverly, MA) used in dephosphorylation reactions were measured using a quantitative assay following the methods described by Tetlow et al. (1996) for the measurement of acid phosphatase activity. Contaminating alkaline phosphatase on native gels was detected using a 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium liquid substrate system (catalog number B-1911; Sigma-Aldrich). The efficiency of the dephosphorylation of granule-associated proteins by soluble alkaline phosphatases in starch granule preparations was measured by the release of Pi from the starch polymer. Pi was measured colorimetrically using a molybdate-based assay described by Tausky and Shorr (1953). Maximal Pi release occurred after a 2-h incubation of 10 units bovine alkaline phosphatase with 500 mg of washed starch at 25°C (data not shown); these conditions were used for the dephosphorylation of starch granule–associated proteins within intact starch granules.
For zymogram analysis of SBE activities, protein samples were mixed with native gel sample buffer (50% [v/v] glycerol and 0.2% [w/v] bromophenol blue) in a ratio of 20:1 and separated on native 5% (w/v) polyacrylamide gels containing 0.2% (w/v) maltoheptaose, 29 units phosphorylase a (from rabbit muscle, catalog number P-1261; Sigma-Aldrich), and 10 mg of the α-amylase inhibitor acarbose (Bayer, Toronto, Canada) for 2 h at 4°C. Gels were washed, incubated for 2 h at 30°C, and stained as previously described (Nishi et al., 2001). Gels were photographed immediately after staining. The different SBE isoforms from T. aestivum endosperm separated on the native gels were identified by blotting the proteins onto nitrocellulose membranes (see below) and probing with the various peptide-specific anti-SBE antibodies.
Metabolism of [U-14C]-Labeled ADP-Glc
Intact amyloplasts (prepared from endosperm 12 to 20 DAP) were incubated with 5 mM ADP-[U-14C]-Glc (Amersham Biosciences, Quebec, Canada) with or without ATP. Measurement of the incorporation of isotope into starch was as previously described (Tetlow et al., 1994). Material insoluble in methanol-KCl was fractionated using a thymol precipitation technique (Bourne et al., 1948). Amylose and amylopectin fractionated by this technique was verified by running the samples on a 0.7- × 30-cm Sepharose CL-2B column in 10 mM NaOH at room temperature using T. aestivum amylose and amylopectin standards following the methods described by Denyer et al. (1995).
Phosphorylation of Plastid Proteins in Vitro
Intact plastids (containing 1.2 to 1.5 mg protein cm−3) were incubated in the respective isolation medium containing 100 μM ATP with between 600 and 1200 Ci/mmol γ-32P-ATP (Perkin-Elmer, Boston, MA) in a total volume of 0.6 cm3 for 1 to 30 min at 25°C with gentle rocking. Phosphorylation reactions were terminated by the addition of SDS sample buffer (62.5 mM Tris/HCl, pH 6.8, 2% [w/v] SDS, 10% [w/v] glycerol, 5% [v/v] 2-mercaptoethanol, and 0.001% [w/v] bromophenol blue) and boiled for 2 min before electrophoresis. In some cases, the 32P-labeled phosphoproteins were separated into suborganellar fractions and then partially purified as described below.
Plastid Fractionation
After incubation with γ-32P-ATP, intact plastids were lysed by the addition of two volumes of ice-cold rupturing buffer containing protease inhibitors (see above), a phosphatase inhibitor cocktail (Novagen, Madison,WI), and 1 mM each NaF and sodium orthovanadate. All subsequent procedures were performed at 2 to 4°C. Amyloplast lysates were immediately centrifuged at 14,000g for 5 min in a refrigerated microfuge (Sigma 1-15K) to obtain a pellet of starch grains, and the supernatant was immediately desalted on a NAP-10 column (Amersham Biosciences) pre-equilibrated in rupturing buffer in which the Tricine concentration was 10 mM. The desalted plastid lysate then was centrifuged at 120,000g for 15 min in a Beckman Airfuge (Beckman Instruments, Palo Alto, CA) (at 25 p.s.i.) to remove plastid membranes and particulate material. The supernatant from the ultracentrifugation step, termed plastid stroma, was solubilized in either SDS sample buffer for 1D-SDS-PAGE or in rehydration buffer (8 M urea, 2% [w/v] 3-3-cholamidopropyl-dimethylammonio-1-propanesulfonate, 0.5% [v/v] carrier ampholytes [Invitrogen, Carlsbad, CA], pH 3 to 10, and 0.002% [w/v] bromophenol blue) for 2D-PAGE (see below).
Starch granule–associated proteins were isolated from the washed starch grains of amyloplast lysates following the methods described by Denyer et al. (1995). For analysis of solubilized granule-bound SBE activity, the α-amylase used to release the granule-bound proteins was removed by anion exchange chromatography on HiTrap Q columns (Amersham Biosciences) (see below).
Affinity Chromatography
In some experiments, plastid stromal phosphoproteins (prepared as described above) were partially purified by affinity chromatography after labeling with γ-32P-ATP using a phosphoprotein purification kit (Qiagen, Hilden, Germany). Plastid stromal protein (between 2 and 4 mg protein) was applied to affinity columns and eluted following the manufacturer's instructions.
Partial Purification of SBE Activity
SBE activity in plastid stromal samples and solubilized granule-associated SBE activity from amyloplasts was routinely separated from α-amylase activity and, where present, soluble alkaline phosphatases (which both interfere with the SBE assay) when performing measurements of SBE activity. All procedures were performed at 2 to 4°C. Plastid stroma (1 to 1.6 mg protein cm−3) or solubilized granule-associated proteins (0.25 to 0.4 mg protein cm−3) was loaded onto a 1 cm3 HiTrap Q column, which had been pre-equilibrated in column running buffer (100 mM Tricine/NaOH, pH 7.5, 1 mM Na2-EDTA, and 2 mM MgCl2) at a flow rate of 1 cm3 min−1. Protein was eluted using a stepped NaCl gradient (in 0.1 M NaCl increments, made up in column running buffer) at a flow rate of 2 cm3 min−1, collecting 1-cm3 fractions. SBE activity was determined using the phosphorylase a stimulation assay, zymogram analysis, and probing gel blots of column fractions with anti-SBE antibodies. These analyses showed that all forms of stromal- and granule-associated SBE eluted in 0.4 M NaCl, whereas the α-amylase and soluble alkaline phosphatases eluted in 0.2 M NaCl (data not shown).
Preparation of Peptides and Antisera
Polyclonal antibodies were raised in rabbits against the synthetic peptides derived from the N-terminal sequences of T. aestivum SBEI (VSAPRDYTMATAEDGV), T. aestivum SBEIIa (AASPGKVLVPDGESDDLASY) (Rahman et al., 2001), and T. aestivum SBEIIb (AGGPSGEVMIGC). The antigen was prepared by coupling the synthesized peptide to keyhole limpet hemocyanin using the heterobifunctional reagent m-maleimidobenzoyl-N-hydroxysuccinimide ester.
SDS-PAGE and Immunoblotting
Protein samples were separated on 1D-gels using precast 4 to 12% Bis-Tris gradient gels (Invitrogen) with 3-(N-morpholino)-propanesulfonic acid running buffer and following the manufacturer's instructions for sample preparation and electrophoresis. Protein samples also were separated by 2D-PAGE using a Zoom IPGRunner system (Invitrogen) for isoelectric focusing (using pH 3 to 10 strips) and 4 to 12% Bis-Tris gradient gels (as above) for the second dimension following the manufacturer's instructions for sample preparation and electrophoresis. Gels were either stained with a colloidal Coomassie Brilliant Blue G 250 kit (Simply Blue Safestain; Invitrogen) or silver-stained according to methods described by Shevchenko et al. (1996). Stained proteins that were to be analyzed by Q-TOF-MS were excised from gels with a razor blade and stored at −20°C until required.
Samples for immunoblot analysis were transblotted onto nitrocellulose membranes (Pall Life Sciences, Ann Arbor, MI), blocked with 1.5% BSA, and exposed to antibodies using the methods described by Harlow and Lane (1988). The various antisera were used in immunoblot analyses at the following dilutions: anti-E. coli branching enzyme, 1:1000; anti-SBEI and anti-SBEIIa, 1:5000; anti-SBEIIb and anti-H. vulgare (endosperm) starch phosphorylase, 1:2000. Bound antibodies were detected with alkaline phosphatase–conjugated goat anti-rabbit IgG using a 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium liquid substrate system (see above).
Identification of Phosphoproteins by Q-TOF-MS
In-gel digestion with trypsin was performed according to published methods (Jeno et al., 1995; Shevchenko et al., 1996; Wilm et al., 1996) modified for use with a robotic digestion system (Investigator ProGest; Genomic Solutions, Huntington, UK) (Wait et al., 2001). Silver-stained gel pieces were first washed with 30 μL of 15 mM potassium ferricyanide/50 mM sodium thiosulphate (Gharahdaghi et al., 1999) followed by successive rinses with deionized water, 50 mM ammonium hydrogen carbonate buffer, and acetonitrile. This destaining step was omitted in the processing of Coomassie blue–stained bands. Cys residues were reduced with DTT and derivatized by treatment with iodoacetamide. After further washing with ammonium hydrogen carbonate buffer, the gel pieces were again dehydrated with acetonitrile and dried at 60°C before addition of modified trypsin (10 μL at 6.5 ng μL−1 in 25 mM ammonium hydrogen carbonate; Promega, Madison, WI). Digestion proceeded for 8 h at 37°C, and products were recovered by sequential extractions with 25 mM ammonium hydrogen carbonate, 5% formic acid, and acetonitrile. The pooled extracts were lyophilized and dissolved in 0.1% formic acid for MS.
Tandem electrospray mass spectra were recorded using a Q-TOF hybrid Q-TOF spectrometer (Micromass, Manchester, UK) interfaced to a Micromass CapLC capillary chromatograph. Samples were dissolved in 0.1% aqueous formic acid, and 6 μL was injected onto a Pepmap C18 column (300 μm × 0.5 cm; LC Packings, Amsterdam, The Netherlands) and washed for 3 min with 0.1% aqueous formic acid (with the stream select valve diverting the column effluent to waste). The flow rate then was reduced to 1 μL min−1, the stream select valve was switched to the data acquisition position, and the peptides were eluted into the mass spectrometer with an acetonitrile/0.1% formic acid gradient (5 to 70% acetonitrile for 20 min).
The capillary voltage was set to 3500 V, and data-dependent tandem mass spectrometry acquisitions were performed on precursors with charge states of 2, 3, or 4 over a survey mass range of 540 to 1000. Known trypsin autolysis products and keratin-derived precursor ions were automatically excluded. The collision voltage was varied between 18 and 45 V depending on the charge and mass of the precursor. Product ion spectra were charge-state de-encrypted and de-isotoped with a maximum entropy algorithm (MaxEnt 3; Micromass). Proteins were identified by correlation of uninterpreted tandem mass spectra to entries in SwissProt/TrEMBL using the ProteinLynx global server (version 1; Micromass). One missed cleavage per peptide was allowed, and an initial mass tolerance of 50 ppm was used in all searches. Cys was assumed to be carbamidomethylated, but other potential modifications were not considered in the first pass search. When this approach failed, amino acid sequences were deduced manually from the charge-state de-encrypted spectra (Wait et al., 2002) and were used as queries for searches using Basic Local Alignment Search Tool (Altschul et al., 1997) and FASTS (Mackey et al., 2002).
Phosphoamino Acid Analysis
Phosphorylated proteins separated by SDS-PAGE were excised from gels (5 to 10 gel slices per sample), and partial acid hydrolysis was performed on the gel slices in 5.7 N HCl for 1 h at 110°C. The released phosphoamino acids were resolved, together with unlabeled phosphoamino acid standards (Sigma-Aldrich), by 2D thin-layer electrophoresis and autoradiography using methods described by van der Greer et al. (1993). For each phosphorylated protein analyzed, phosphoamino acid analysis was performed twice, and representative results from one experiment (with amyloplasts) are presented.
Immunoprecipitation
Amyloplast and chloroplast lysates were prepared and starch removed (see above). Isoforms of SBE were immunoprecipitated from the plastid stroma using peptide-specific polyclonal antibodies raised against SBEI, SBEIIa, and SBEIIb (see below). Antibodies were added to plastid stroma (3 μL cm−3 for anti-SBEIIb and 2 μL cm−3 for anti-SBEI and anti-SBEIIa) and mixed on a rotating table for 1 h at 4°C. Proteins were immunoprecipitated by adding 40 μL of protein A–Sepharose (Sigma-Aldrich) made up as a 50% (w/v) slurry with PBS (137 mM NaCl, 10 mM Na2HPO4, 2.7 mM KCl, and 1.8 mM KH2PO4) to the samples and incubating for a further 30 min at 4°C. The protein A–Sepharose/protein complex was centrifuged at 2000g for 5 min at 4°C in a refrigerated microfuge, and the supernatant discarded. The protein A–Sepharose/protein complex was washed three times, each with 1.2-cm3 PBS, followed by three similar washes with 10 mM Hepes/KOH, pH 7.5. The washed pellet was mixed with water and SDS-sample buffer, boiled for 2 min, and loaded onto 4 to 12% polyacrylamide gradient gels for 1D-SDS-PAGE (see above). For 2D-PAGE of the protein A–Sepharose/protein complex, the pellet was mixed with 160 μL of rehydration buffer and incubated at room temperature for 1 h and centrifuged at 100,000g for 15 min. The supernatant was used for isoelectric focusing (see above).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number O24398.
Acknowledgments
We thank Mike Burrell, Steve Coates, and Advanced Technologies (Cambridge, UK) for the anti-AGP-L, anti-AGP-S (large and small subunits of AGPase, respectively), and anti-E. coli branching enzyme antisera and K. Gale and J. Higgins (Commonwealth Scientific and Industrial Research Organisation) for the preparation of the anti-SBEIIb and anti-H. vulgare starch phosphorylase antisera, respectively. This work was supported by Biotechnology and Biological Sciences Research Council Grant 34/P10964 awarded to M.J.E, C.G.B, and I.J.T. and Natural Sciences and Engineering Research Council Grant 262209 awarded to M.J.E.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ian J. Tetlow (itetlow@uoguelph.ca).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.017400.
References
- Altschul, S.F., Madden, T.L., Schäffer, A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Båga, M., Nair, R.B., Repellin, A., Scoles, G.J., and Chibbar, R.N. (2000). Isolation of a cDNA encoding a granule-bound 152-kilodalton starch-branching enzyme in wheat. Plant Physiol. 124, 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball, S., Guan, H.-P., James, M., Myers, A., Keeling, P., Mouille, G., Buléon, A., Colonna, P., and Preiss, J. (1996). From glycogen to amylopectin: A model for the biogenesis of the plant starch granule. Cell 86, 349–352. [DOI] [PubMed] [Google Scholar]
- Ball, S.G., and Morell, M.K. (2003). From bacterial glycogen to starch: Understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 54, 207–233. [DOI] [PubMed] [Google Scholar]
- Beatty, M.K., Rahman, A., Cao, H., Woodman, W., Lee, M., Myers, A.M., and James, M.G. (1999). Purification and molecular genetic characterization of ZPU1, a pullulanase-type starch-debranching enzyme from maize. Plant Physiol. 119, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blauth, S.L., Kim, K.N., Klucinec, J., Shannon, J.C., Thompson, D.B., and Guiltinan, M. (2002). Identification of mutator insertional mutants of starch-branching enzyme 1 (sbe1) in Zea mays L. Plant Mol. Biol. 48, 287–297. [DOI] [PubMed] [Google Scholar]
- Bourne, E.J., Donnison, G.H., Haworth, N., and Peat, S. (1948). Thymol and cyclohexanol as fractionation agents for starch. J. Chem. Soc., 1687–1697. [DOI] [PubMed]
- Boyer, C.D., and Preiss, J. (1979). Properties of citrate-stimulated starch synthesis catalyzed by starch synthase I of developing maize kernels. Plant Physiol. 64, 1039–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce, B.D., Perry, S., Froehlich, J., and Keegstra, K. (1994). In vitro import of proteins into chloroplasts. In Plant Molecular Biology Manual, S.B. Gelvin and R.A. Schilperoort, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–15.
- Burton, R.A., Bewley, J.D., Smith, A.M., Bhattacharyya, M.K., Tatge, H., Ring, S., Bull, V., Hamilton, W.D.O., and Martin, C. (1995). Starch branching enzymes belonging to distinct enzyme families are differentially expressed during pea embryo development. Plant J. 7, 3–15. [DOI] [PubMed] [Google Scholar]
- Cho, M.-J., Wong, J.H., Marx, C., Jiang, W., Lemaux, P.G., and Buchanan, B.B. (1999). Overexpression of thioredoxin h leads to enhanced activity of starchy debranching enzyme (pullulanase) in barley grain. Proc. Natl. Acad. Sci. USA 96, 14641–14646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, H.J., Sehnke, P.C., and Ferl, R.J. (1999). The 14-3-3 proteins: Cellular regulators of plant metabolism. Trends Plant Sci. 4, 367–371. [DOI] [PubMed] [Google Scholar]
- Colleoni, C., Myers, A.M., and James, M.G. (2003). One- and two-dimensional native PAGE activity gel analyses of maize endosperm proteins reveal functional interactions between specific starch metabolizing enzymes. J. Appl. Glycosci. 50, 207–212. [Google Scholar]
- Craig, J., Lloyd, J.R., Tomlinson, K., Barber, L., Edwards, A., Wang, T.L., Martin, C., Hedley, C.L., and Smith, A.M. (1998). Mutations in the gene encoding starch synthase II profoundly alter amylopectin structure in pea embryos. Plant Cell 10, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinges, J.R., Colleoni, C., James, M.G., and Myers, A.M. (2003). Mutational analysis of the pullulanase-type debranching enzyme of maize indicates multiple functions in starch metabolism. Plant Cell 15, 666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinges, J.R., Colleoni, C., Myers, A.M., and James, M.G. (2001). Molecular structure of three mutations at the maize sugary1 locus and their allele-specific phenotypic effects. Plant Physiol. 125, 1406–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denyer, K., Hylton, C.M., Jenner, C.F., and Smith, A.M. (1995). Identification of multiple isoforms of soluble and granule-bound starch synthase in developing wheat endosperm. Planta 196, 256–265. [Google Scholar]
- Fu, Y., Ballicora, M.A., Leykam, J.F., and Preiss, J. (1998). Mechanism of reductive activation of potato tuber ADP-glucose pyrophosphorylase. J. Biol. Chem. 273, 25045–25052. [DOI] [PubMed] [Google Scholar]
- Gao, M., Fisher, D.K., Kim, K.-N., Shannon, J.C., and Guiltinan, M.J. (1996). Evolutionary conservation and expression patterns of maize starch branching enzyme I and IIb genes suggests isoform specialization. Plant Mol. Biol. 30, 1223–1232. [DOI] [PubMed] [Google Scholar]
- Gao, M., Fisher, D.K., Kim, K.-N., Shannon, J.C., and Guiltinan, M.J. (1997). Independent genetic control of maize starch-branching enzymes IIa and IIb. Plant Physiol. 114, 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharahdaghi, F., Weinberg, C.R., Meagher, D.A., Imai, B.S., and Mische, S.M. (1999). Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: A method for the removal of silver ions to enhance sensitivity. Electrophoresis 20, 601–605. [DOI] [PubMed] [Google Scholar]
- Giroux, M.J., Boyer, C., Feix, G., and Hannah, L.C. (1994). Coordinated transcriptional regulation of storage product genes in the maize endosperm. Plant Physiol. 106, 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Hendriks, J.H.M., Kolbe, A., Gibon, Y., Stitt, M., and Geigenberger, P. (2003). ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol. 133, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, M.G., Denyer, K., and Myers, A.M. (2003). Starch synthesis in the cereal endosperm. Curr. Opin. Plant Biol. 6, 215–222. [DOI] [PubMed] [Google Scholar]
- James, M.G., Robertson, D.S., and Myers, A.M. (1995). Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell 7, 417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeno, P., Mini, T., Moes, S., Hintermann, E., and Horst, M. (1995). Internal sequences from proteins digested in polyacrylamide gels. Anal. Biochem. 224, 75–82. [DOI] [PubMed] [Google Scholar]
- Jespersen, H.M., MacGregor, E.A., Henrissat, B., Sierks, M.R., and Svensson, B. (1993). Starch- and glycogen-debranching and branching enzymes: Prediction of structural features of the catalytic (β/α)8-barrel domain and evolutionary relationship to other amylolytic enzymes. J. Protein Chem. 12, 791–805. [DOI] [PubMed] [Google Scholar]
- Jobling, S.A., Schwall, G.P., Westcott, R.J., Sidebottom, C.M., Debet, M., Gidley, M.J., Jeffcoat, R., and Safford, R. (1999). A minor form of starch branching enzyme in potato (Solanum tuberosum L.) tubers has a major effect on starch structure: Cloning and characterisation of multiple forms of SBE A. Plant J. 18, 163–171. [DOI] [PubMed] [Google Scholar]
- Knetsch, M.L.W., Wang, M., Snaar-Jagalska, B.E., and Heimovaara-Dijkstra, S. (1996). Abscisic acid induces mitogen-activated protein kinase activation in barley aleurone protoplasts. Plant Cell 8, 1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Chu, X., Mouille, G., Yan, L., Kosar-Hashemi, B., Hey, S., Napier, J., Shewry, P., Clarke, B., Appels, R., Morell, M.K., and Rahman, S. (1999). The localization and expression of the class II starch synthases of wheat. Plant Physiol. 120, 1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, A.J., Haystead, T.A.J., and Pearson, W.R. (2002). Getting more from less: Algorithms for rapid protein identification with multiple short peptide sequences. Mol. Cell Proteomics 1, 139–147. [DOI] [PubMed] [Google Scholar]
- McCue, K.F., Hurkman, W.J., Tanka, C.K., and Anderson, O.D. (2002). Starch-branching enzymes Sbe1 and Sbe2 from wheat (Triticum aestivum cv. Cheyenne): Molecular characterization, developmental expression, and homoeologue assignment by differential PCR. Plant Mol. Biol. Rep. 20, 191a–191m. [Google Scholar]
- Morell, M.K., Blennow, A., Kosar-Hashemi, B., and Samuel, M.S. (1997). Differential expression and properties of starch branching enzyme isoforms in developing wheat endosperm. Plant Physiol. 113, 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell, M.K., Kosar-Hashemi, B., Cmiel, M., Samuel, M.S., Chandler, P., Rahman, S., Buléon, A., Batey, I.L., and Li, Z. (2003). Barley sex6 mutants lack starch synthase IIa activity and contain a starch with novel properties. Plant J. 34, 173–185. [DOI] [PubMed] [Google Scholar]
- Mu-Forster, C., Huang, R., Powers, J.R., Harriman, R.W., Knight, M., Singletary, G.W., Keeling, P.L., and Wasserman, B.P. (1996). Physical association of starch biosynthetic enzymes with starch granules of maize endosperm. Granule-associated forms of starch synthase I and starch branching enzyme II. Plant Physiol. 111, 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi, A., Nakamura, Y., Tanaka, N., and Satoh, H. (2001). Biochemical and genetic effects of amylose-extender mutation in rice endosperm. Plant Physiol. 127, 459–472. [PMC free article] [PubMed] [Google Scholar]
- Rahman, S., Kosar-Hashemi, B., Samuel, M.S., Hill, A., Abbott, D.C., Skerritt, J.H., Preiss, J., Appels, R., and Morell, M.K. (1995). The major proteins of wheat endosperm starch granules. Aust. J. Plant Physiol. 22, 793–803. [Google Scholar]
- Rahman, S., Regina, A., Li, Z., Mukai, Y., Yamamoto, M., Kosar-Hashemi, B., Abrahams, S., and Morell, M.K. (2001). Comparison of starch-branching enzyme genes reveals evolutionary relationships among isoforms. Characterization of a gene for starch-branching enzyme IIa from wheat D genome donor Aegilops tauschii. Plant Physiol. 125, 1314–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, H., Nishi, A., Yamashita, K., Takemoto, Y., Tanaka, Y., Hosaka, Y., Sakurai, A., Fujita, N., and Nakamura, Y. (2003). Starch-branching enzyme I-deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiol. 133, 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler, I., Renz, A., Schmid, F.X., and Beck, E. (2001). Activation of spinach pullulanase by reduction results in a decrease in the number of isomeric forms. Biochim. Biophys. Acta 1548, 175–186. [DOI] [PubMed] [Google Scholar]
- Sehnke, P.C., Chung, H.-J., Wu, K., and Ferl, R.J. (2001). Regulation of starch accumulation by granule-associated plant 14-3-3 proteins. Proc. Natl. Acad. Sci. USA 98, 765–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnke, P.C., Henry, R., Cline, K., and Ferl, R.J. (2000). Interaction of a plant 14-3-3 protein with the signal peptide of a thylakoid-targeted chloroplast precursor protein and the presence of 14-3-3 isoforms in the chloroplast stroma. Plant Physiol. 122, 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, B.S., Kim, S., Scott, M.P., Singletary, G.W., Wong, K.S., James, M.G., and Myers, A.M. (2002). Functional interactions between heterologously expressed starch-branching enzymes of maize and glycogen synthases of brewer's yeast. Plant Physiol. 128, 1189–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen, J. (1996). Ca2+-dependent protein kinases and stress signal transduction in plants. Science 274, 1900–1902. [DOI] [PubMed] [Google Scholar]
- Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Smith, A.M. (1988). Major differences in isoforms of starch-branching enzymes in embryos of round- and wrinkled-seeded peas (Psium sativum L.). Planta 175, 270–279. [DOI] [PubMed] [Google Scholar]
- Tausky, H., and Shorr, H. (1953). A microcolorimetric method for the determination of inorganic phosphate. J. Biol. Chem. 202, 675–685. [PubMed] [Google Scholar]
- Tetlow, I.J., Blissett, K.J., and Emes, M.J. (1993). A rapid method for the isolation of purified amyloplasts from wheat endosperm. Planta 189, 597–600. [Google Scholar]
- Tetlow, I.J., Blissett, K.J., and Emes, M.J. (1994). Starch synthesis and carbohydrate oxidation in amyloplasts from developing wheat endosperm. Planta 194, 454–460. [Google Scholar]
- Tetlow, I.J., Bowsher, C.G., and Emes, M.J. (1996). Reconstitution of the hexosephosphate translocator from the envelope membranes of wheat endosperm amyloplasts. Biochem. J. 319, 717–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow, I.J., Bowsher, C.G., Scrase-Field, E.F.A.L., Davies, E.J., and Emes, M.J. (2003. a). The synthesis and transport of ADPglucose in cereal endosperms. J. Appl. Glycosci. 50, 231–236. [Google Scholar]
- Tetlow, I.J., Davies, E.J., Vardy, K.A., Bowsher, C.G., Burrell, M.M., and Emes, M.J. (2003. b). Subcellular localization of ADPglucose pyrophosphorylase in developing wheat endosperm and analysis of a plastidial isoform. J. Exp. Bot. 54, 715–725. [DOI] [PubMed] [Google Scholar]
- Tiessen, A., Hendriks, J.H.M., Stitt, M., Branscheid, A., Gibon, Y., Farré, E.M., and Geigenberger, P. (2002). Starch synthesis in potato tuber is regulated by post-translational redox modification of ADP-glucose pyrophosphorylase. Plant Cell 14, 2191–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden, J., Möhlmann, T., Kampfenkel, K., Henrichs, G., and Neuhaus, H.E. (1998). Altered plastidic ATP/ADP-transporter activity influences potato (Solanum tuberosum L.) tuber morphology, yield and composition of tuber starch. Plant J. 16, 531–540. [Google Scholar]
- Umemoto, T., Yano, M., Satoh, H., Shomura, A., and Nakamura, Y. (2002). Mapping of a gene responsible for the difference in amylopectin structure between japonica-type and indica-type rice varieties. Theor. Appl. Genet. 104, 1–8. [DOI] [PubMed] [Google Scholar]
- Van der Greer, P., Luo, P., Sefton, B.M., and Hunter, T. (1993). Phosphopeptide mapping and phosphoamino acid analysis on thin layer plates. In Protein Phosphorylation. A Practical Approach, 1st ed., D.G. Hardie, ed (Oxford: Oxford University Press), pp. 31–59.
- Wait, R., Gianazza, E., Eberini, I., Sironi, L., Dunn, M., Gemeiner, M., and Miller, I. (2001). Proteins of rat serum, urine, and cerebrospinal fluid: VI. Further protein identifications and interstrain comparison. Electrophoresis 22, 3043–3052. [DOI] [PubMed] [Google Scholar]
- Wait, R., Miller, I., Eberini, I., Cairoli, F., Veronesi, C., Battocchio, M., Gemeiner, M., and Gianazza, E. (2002). Strategies for proteomics with incompletely characterized genomes: The proteome of Bos taurus serum. Electrophoresis 23, 3418–3427. [DOI] [PubMed] [Google Scholar]
- Wilm, M., Shevchenko, A., Houthaeve, T., Breit, S., Schweigerer, L., Fotsis, T., and Mann, M. (1996). Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379, 466–469. [DOI] [PubMed] [Google Scholar]
- Yamamori, M., Fujita, S., Hayakawa, K., Matsuki, J., and Yasui, T. (2000). Genetic elimination of a starch granule protein, SGP-1, of wheat generates an altered starch with apparent high amylose. Theor. Appl. Genet. 101, 21–29. [Google Scholar]
- Zhang, S., Du, H., and Klessig, D.F. (1998). Activation of the tobacco SIP kinase by both a cell wall-derived carbohydrate elicitor and purified proteinaceous elicitins from Phytophthora spp. Plant Cell 10, 435–449. [DOI] [PMC free article] [PubMed] [Google Scholar]