Abstract
Posttranscriptional RNA metabolism plays versatile roles in the regulation of gene expression during eukaryotic growth and development. It is mediated by a group of RNA binding proteins with distinct conserved motifs. In this study, an Arabidopsis (Arabidopsis thaliana) gene, designated FLK, was identified and shown to encode a putative RNA binding protein with K homology motifs. A mutant in which FLK was inactivated by T-DNA insertion exhibited a severe late flowering phenotype both in long and short days. The late flowering phenotype was reversed by gibberellin and vernalization treatments. The FLOWERING LOCUS C (FLC) transcription was greatly upregulated, whereas those of FLOWERING LOCUS T and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 decreased in the mutant. These observations demonstrate that FLK regulates the autonomous flowering pathway via FLC. It is now evident that a battery of different RNA binding proteins are involved in the posttranscriptional regulation of flowering time in Arabidopsis.
INTRODUCTION
Molecular genetic studies on the facultative long-day plant Arabidopsis (Arabidopsis thaliana) revealed four major flowering pathways: the photoperiod, autonomous, vernalization, and gibberellin (GA) pathways (Blázquez et al., 2001; Mouradov et al., 2002; Simpson and Dean, 2002). Furthermore, a recently accumulated body of evidence suggests that the flowering pathways are interconnected and converged on a few floral integrators, such as FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) (Chou and Yang, 1999; Lee et al., 2000; Onouchi et al., 2000; Reeves and Coupland, 2001; Rouse et al., 2002; Moon et al., 2003).
Regulation of flowering time in response to seasonal daylength fluctuations is mediated by the interactions between environmental light signals and intrinsic time-keeping mechanisms that are associated with the circadian clock (Doyle et al., 2002; Yanovsky and Kay, 2002; Hayama and Coupland, 2003). Two major photoreceptors, phytochromes and cryptochromes, entrain the circadian clock to oscillate with a period of ∼24 h (Lin, 2000; Suarez-Lopez et al., 2001; Yanovsky and Kay, 2002; Mockler et al., 2003).
The genes in the autonomous pathway, such as FCA, FPA, FVE, FLD, LD, and FY, promote flowering by suppressing FLOWERING LOCUS C (FLC). FCA and FPA encode RNA binding proteins with the RNA recognition motifs (RRMs) (Macknight et al., 1997; Schomburg et al., 2001). The FCA expression is regulated by alternative pre-mRNA processing and polyadenylation. FY, an RNA 3′-end processing factor, is required for this process (Simpson et al., 2003). It is therefore likely that gene regulation at the posttranscriptional level is an essential regulatory component in flowering time control.
The vernalization response is governed by dominant alleles of two genes, FRIGIDA and FLC (Michaels and Amasino, 1999a, 2001). Vernalization induces a developmental state that is mitotically stable, and the mitotic stability is maintained by VERNALIZATION2 (Gendall et al., 2001). In the vernalization2 mutants, the FLC mRNA level is suppressed by vernalization but induced to a high level as in the wild-type plants when they are returned to normal growth temperature. However, vernalization does not directly induce flowering. Seedlings exposed to cold temperature do not flower immediately after vernalization treatment but instead flower weeks later (Gendall et al., 2001).
GA promotes flowering and is absolutely required in noninductive short days. Mutations in GA biosynthesis and signaling result in delayed flowering (Mouradov et al., 2002; Olszewski et al., 2002). Several mutants in GA biosynthesis, such as GA1, GA4, and GA5, have been identified. Genes in GA signaling also control flowering. SPY is a negative regulator of GA responses. The spy mutation causes constitutively active GA signaling and partially suppresses the effects of disrupted GA biosynthesis (Jacobsen et al., 1996). GAI, RGA, and PHOR1 also play key regulatory roles in GA signaling (Mouradov et al., 2002). In addition, experimental evidence has demonstrated genetic interactions between the GA pathway and other flowering pathways (Lee et al., 2000; Moon et al., 2003). SOC1 plays a vital role in the genetic interactions. GA signal is required for the full activation of SOC1, especially in short days, in addition to the FLC repression. This observation suggests that SOC1 is a molecular basis that links the GA pathway with other flowering pathways (Moon et al., 2003). However, the GA pathway is not directly involved in the vernalization response because GA mutant plants (ga1-3) still retain their responsiveness to vernalization (Michaels and Amasino, 1999b).
In this study, we isolated a late flowering Arabidopsis mutant in which a gene encoding a putative RNA binding protein with K homology (KH) motifs is disrupted by T-DNA insertion. The late flowering phenotype is correlated with upregulation of FLC, resulting in downregulation of FT and SOC1. It is now apparent that a range of RNA binding proteins regulate the expression of flowering time genes at the posttranscriptional level.
RESULTS
flk Is Late Flowering
With the aim of further exploring the molecular mechanisms that regulate flowering in Arabidopsis, we generated a mutant pool by randomly integrating the 35S enhancer of Cauliflower mosaic virus (CaMV) into the genome of the Columbia accession (Col-0) (Weigel et al., 2000) and searched for flowering time mutants. Among the isolated mutants, one late flowering mutant (designated flk; see below) was chosen for molecular genetic analysis.
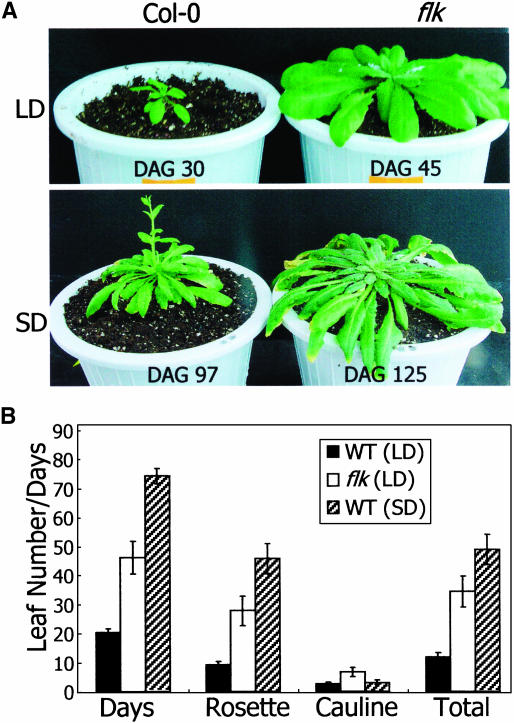
flk showed a severe late flowering phenotype, as measured by the days to bolting and the total leaf numbers at flowering (Figures 1 and 6). However, other growth and developmental aspects, such as leaf and stem morphologies and flower architecture, were essentially normal. This indicates that the flk mutation is primarily related to flowering time control. Genomic DNA gel blot analysis using the CaMV 35S enhancer sequence as probe confirmed that there was a single insertion event into the genome in flk (data not shown).
Figure 1.
Flowering Phenotype of Arabidopsis Plants Described in This Study.
flk1 is SALK 112850 obtained from ABRC. It has a T-DNA insertion in the first intron of the FLK gene. flk x flk1 is a genetic cross between flk and flk1.
Figure 6.
Effects of Vernalization and GA on Flowering Time.
(A) Vernalization effects. Plants were germinated and grown at 4°C for 6 weeks and transferred to normal growth condition.
(B) GA effects. A GA solution of 20 μM was sprayed twice a week on the plants until flowering. Thirty to fifty plants were measured and averaged for each measurement.
FLK Encodes a Putative RNA Binding Protein with Three KH Motifs
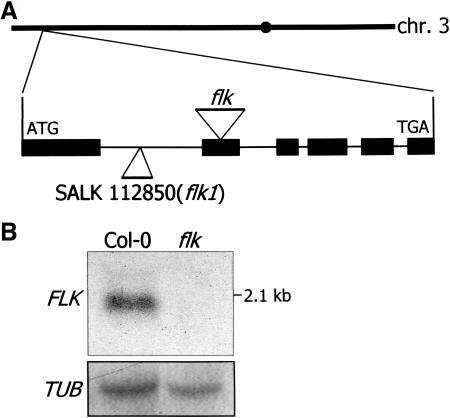
A high-throughput thermal asymmetric interlaced PCR method was employed to map the integration site of the CaMV 35S enhancer, and the amplified DNA was sequenced (Liu et al., 1995). The result revealed that the 35S enhancer was integrated into the second exon of an open reading frame (At3g04610), suggesting that the flk mutation is not a gain-of-function mutation but a loss-of-function mutation (Figure 2A). To confirm this, the expression of the interrupted gene and the adjacent genes on both sides was investigated by reverse transcription (RT)–PCR. The transcript of the interrupted gene was not detected in the mutant as expected (data not shown). The absence of the transcript in flk was further confirmed by RNA gel blot analysis (Figure 2B). By contrast, a transcript with an estimated size of 2.1 kb was detected in the wild-type plant. The transcript levels of the genes on both sides of the interrupted gene were identical in the mutant and the wild-type plants (data not shown). These observations, together with the genomic DNA gel blot analysis, indicate that the late flowering phenotype is specifically caused by the insertion of the 35S enhancer into the locus At3g04610. The mutant was recessive because the flowering time of the heterozygous plants was essentially normal (data not shown). In addition, a segregation ratio of 3:1 (normal flowering:late flowering) was observed from the self-pollinated heterozygous plant.
Figure 2.
FLK Gene Structure and Expression.
(A) FLK gene structure. The FLK gene consists of six exons and is located on chromosome 3. The T-DNA insertion sites in each mutant are indicated by triangles.
(B) FLK expression. The FLK transcript was not detected in flk. A tubulin gene was used as a control for constitutive expression.
We also obtained a loss-of-function mutant in which T-DNA is inserted into the first intron of the same gene (SALK 112850; Figure 2A, flk1). flk1 was also late flowering to a similar degree as flk. An identical result was also observed with the genetic cross between flk and flk1 (flk x flk1; Figure 1), unequivocally confirming that the flk mutations cause the late flowering phenotype. Consistent with this, the FLK transcript was detected neither in flk x flk1 nor in flk1 (data not shown).
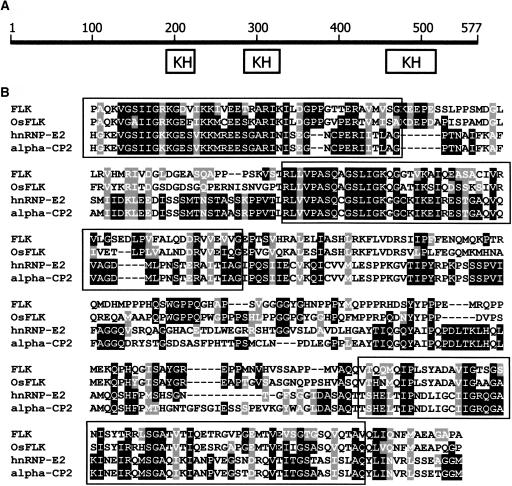
The FLK gene consists of six exons and is located close to the upper end of chromosome 3. It encodes a protein of 577 amino acids with an estimated molecular mass of 63.4 kD. Database searches revealed that it is a putative RNA binding protein with three KH motifs (Figure 3A). The KH motifs of FLK show high sequence homologies to those of the poly(rC/U) binding proteins, such as the αCP1 (hnRNP-E1) and αCP2 (hnRNP-E2) proteins, which have been extensively studied in animals (Chkheidze et al., 1999; reviewed in Makeyev and Liebhaber, 2002; Kong et al., 2003; Figure 3B). The relative location of the KH motifs was also conserved: two motifs in the central region and one in the C-terminal region. However, the N-terminal region of FLK shows no discernible sequence homology to the known eukaryotic proteins in the database. A rice (Oryza sativa) gene was identified in the database to encode a protein (510 amino acids) that has a sequence identity of 43% with FLK throughout the whole sequence (Figure 3B, OsFLK). It may be an FLK homolog in rice, although it remains to be experimentally determined.
Figure 3.
FLK Structure and Multiple Alignment of KH Motif Sequences.
(A) FLK structure. FLK consists of 577 residues and contains three KH motifs.
(B) Multiple alignment of KH motif sequences from FLK and related proteins. OsFLK, a putative FLK homolog in rice (AAL31692); hnRNP-E2, a heterogeneous nuclear ribonucleoprotein E2 in African clawed frog (CAB50743); alpha-CP2, a poly(rC/U) binding protein in human (NP_114366). The sequences were aligned using the CLUSTAL W version 1.7 (Thompson et al., 1994). Three KH motifs are boxed.
FCA and FPA contain multiple RRMs instead of the KH motifs for RNA association (Macknight et al., 1997; Schomburg et al., 2001). The overall amino acid sequence of FLK does not have any homology with them. This is a first example for the involvement of an RNA binding protein with KH motifs in the flowering pathway. The gene was designated FLK in this study, standing for flowering late with KH motifs.
FT and SOC1 Are Regulated by FLK via FLC
The expression of the genes involved in the autonomous and GA pathways was not altered in flk. In addition, the transcription of the photoreceptor genes as well as of the clock genes was also unaffected. However, the FLC transcript level was ∼10 times higher in flk than that in the wild-type plant (Figure 4A), which is in good agreement with the late flowering phenotype. Interestingly, the two floral integrators, FT and SOC1 that are downregulated by FLC, were significantly suppressed by the flk mutation. Altogether, the results imply that FLK promotes flowering through the autonomous flowering pathway. The gene expression patterns are all consistent with the recognized activities of FLC, FT, and SOC1 in flowering time control.
Figure 4.
Expression of Flowering-Related Genes in flk and Genetic Crosses.
(A) Transcript level analyses. The transcript levels were analyzed by RT-PCR–based DNA gel blot analyses. Note that FLC (bold) is upregulated, whereas FT and SOC1 (bold) are downregulated in flk. The other genes are not affected by the flk mutation.
(B) Genetic crosses. flk was genetically crossed with 35S∷CO and 35S∷FT, and homozygotic lines were obtained by segregation ratios and RT-PCR runs. 35S∷CO, a transgenic plant overexpressing CO; 35S∷FT, a transgenic plant overexpressing FT.
(C) FLK transcription in the mutants in the autonomous pathway. Thirty micrograms of total RNA was loaded onto each lane. C, Col-0; L, Landsberg erecta. The blot was probed with digoxigenin-UTP–labeled 18S rDNA as an RNA quality control.
(D) Comparison of FCA transcripts in flk and wild-type plants. Poly(A)+ RNA was isolated from total RNA, and 3 μg was loaded onto each lane.
The FLK transcription was not affected by light wavelengths, and it was transcribed to an equal level in the phytocrome and cryptochrome mutants to that in the wild-type plant (data not shown). In addition, the known mutations in the autonomous pathway did not affect the FLK transcription as verified by RNA gel blot analysis (Figure 4C), suggesting that FLK does not have a direct functional relationship with them. Furthermore, the FLK transcription was also unaltered in the FRIGIDA background (Figure 4C).
The poly(rC/U) binding proteins contain the KH motifs and are represented by the αCP (α-complex proteins), among which αCP1 and αCP2 are best characterized. They form an RNP complex (α-complex) with other RNA binding proteins and RNA helicases (Chkheidze et al., 1999; reviewed in Makeyev and Liebhaber, 2002). Diverse aspects of RNA metabolism, such as mRNA stability, pre-mRNA processing, and translational efficiency, are regulated by the RNP complex. The FCA expression is autoregulated by an alternative splicing and polyadenylation of the primary transcript (Macknight et al., 1997, 2002). FLK functions in the autonomous pathway (Figure 4A). It was therefore assumed that FLK might interact with FCA/FY and affect the splicing and/or polyadenylation of as yet unidentified downstream gene(s). If this is the case, FLK may also be required for the autoregulation of the FCA pre-mRNA processing. To examine this possibility, we analyzed the relative abundance of the FCA transcripts in flk. RNA gel blot analysis was performed using poly(A)+ RNA isolated from total RNA and a 300-bp fragment from the 5′ leader sequence of FCA as probe (Quesada et al., 2003). The relative levels of the FCA transcripts were not altered in flk (Figure 4D), indicating that FLK does not function directly through FCA.
To further explore the FLK-mediated flowering pathway, flk was genetically crossed with a couple of transgenic plants that overexpress CONSTANS (CO) or FT, such as 35S∷CO and 35S∷FT that are early flowering. flk x 35S∷CO was found to be early flowering like 35S∷CO (Figure 4B). This is in harmony with the notion that the photoperiod pathway is more effective than the autonomous pathway in promoting flowering in long days (Reeves and Coupland, 2001). flk x 35S∷FT was also early flowering as expected from the expression patterns of the flowering time genes in flk.
flk Is Late Flowering Both in Long and Short Days
In addition to the upregulation of FLC, the responsiveness to daylength discriminates the mutants in the photoperiod pathway and in the autonomous flowering pathway (Koornneef et al., 1991; Michaels and Amasino, 2001; Mouradov et al., 2002). The mutants in the photoperiod pathway are late flowering in long days but similar to the wild-type plant in short days. By contrast, those in the autonomous pathway are late flowering both in long and short days.
Wild-type and flk mutant plants were grown in long (16 h light and 8 h dark) and short (8 h light and 16 h dark) days, and the flowering times were compared. The flowering of flk was greatly delayed in short days as well as in long days in terms of both the days to flowering and the total leaf numbers at flowering initiation (Figure 5). flk did not flower even at 125 d after germination (DAG) in short days, when some cauline leaves showed senescence. An identical result was also obtained with flk1 (data not shown). This observation further supports the contention that FLK is a genetic component of the autonomous flowering pathway.
Figure 5.
Daylength Effects on Flowering of flk.
(A) Flowering of wild-type and flk plants. Plants were grown either in long days (LD, 16 h light and 8 h dark) or in short days (SD, 8 h light and 16 h dark).
(B) Flowering time measurements. They were measured by days to bolting and by total leaf numbers at flowering. flk did not flower even at 125 DAG in SD, when some cauline leaves exhibited senescence. The experiment was stopped at this point in time.
flk Is Responsive to Vernalization and GA Treatments
The autonomous pathway is represented by a group of mutants that are late flowering under all photoperiods and highly sensitive to vernalization (Martinez-Zapater and Somerville, 1990; Koornneef et al., 1991). The delayed flowering is also reversed by GA (Mouradov et al., 2002; Moon et al., 2003).
To examine the responsiveness of flk to vernalization, the flk plants were germinated and grown at 4°C for 6 weeks and transferred to normal growth temperature (23°C). The vernalization-treated flk plants flowered much earlier than the untreated plants, at a total leaf number of 16 to 17 (Figure 6A), although it is later than the wild-type plant. The untreated plants flowered at a total leaf number of 35 to 40. The days to flowering was also shortened by vernalization. Consistent with this, the transcript level of FLC was high in the untreated flk plants but drastically decreased after vernalization (data not shown). The incomplete recovery of flowering time by vernalization would be because of the dual mechanisms by which vernalization promotes flowering: FLC-dependent and FLC-independent pathways (Michaels and Amasino, 2001).
To examine the GA effects, a GA solution of 20 μM was sprayed twice a week on the growing plants until they flowered. GA also greatly stimulated the flowering of flk (Figure 6B). The GA-treated flk plants initiated flowering at a total leaf number of 11 to 12, which is comparable to that of the wild-type plant. Vernalization promotes flowering by repressing FLC and releasing FT and SOC1 (Michaels and Amasino, 2001). However, only SOC1, not FLC or FT, is regulated by the GA pathway (Moon et al., 2003), explaining the degree of recovery of flowering time by GA in flk. The responsiveness of flk to vernalization and GA is also consistent with the proposed role of FLK in the autonomous flowering pathway.
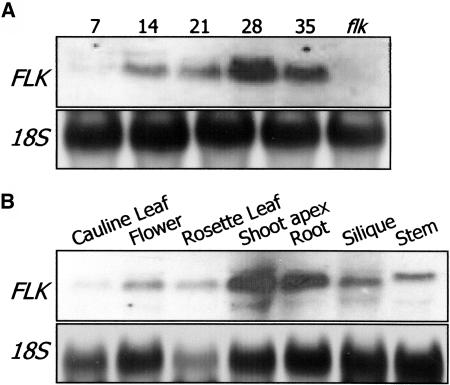
FLK Expression Is Developmentally Regulated
The developmental stage–dependent expression was analyzed using the aerial plant parts that were harvested at different growth stages after germination. The FLK transcript was barely detected until 7 DAG (Figure 7A, DAG 7). However, it was significantly induced at 14 DAG, and the transcript level was maintained through the adult stage with the highest peak at 28 DAG. The highest transcript level at 28 DAG may be as a result of the flowers and the shoot apices in which FLK is actively transcribed to a high level (Figure 7B). This expression pattern is slightly different from that of FPA, which is expressed throughout the whole plant life cycle (Schomburg et al., 2001).
Figure 7.
Growth Stage–Dependent and Tissue-Specific FLK Expression.
(A) Growth stage–dependent expression. Wild-type plants were grown in normal growth condition, and aerial parts were harvested at the indicated growth stages (in DAG) for total RNA isolation. The blot was probed with the FLK gene sequence labeled with digoxigenin-UTP (see Methods). The flk sample was used as a negative control.
(B) Tissue-specific expression. Plants were grown until flowering, and plant parts were separately harvested for total RNA isolation.
The FLK transcript was detected in all plant tissues examined in this study but at the highest levels in the flowers, roots, and shoot apices (Figure 7B). This pattern is similar to those of FCA, FPA, and LD (Schomburg et al., 2001). The high-level transcription in the roots may be linked to some unidentified involvement of FLK in root growth and development, although any related phenotypic changes were not noticeable in flk.
FLK Is Predominantly Localized in the Nucleus
The subcellular localization of an RNA binding protein is closely related to the nature of its biochemical activity (Heintzen et al., 1997; Lorkovic and Barta, 2002). If it is localized in the nucleus, it is more likely to modulate the pre-mRNA processing and/or the nucleocytoplasmic transport of mRNAs. Alternatively, if it is localized in the cytoplasm, it may play a role in the regulation of mRNA stability and/or translation efficiency.
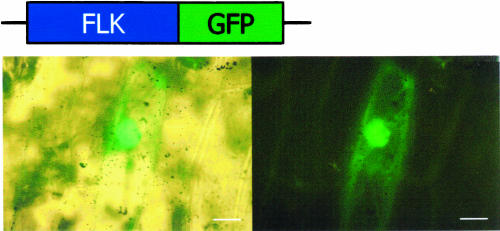
The green fluorescent protein (GFP)–coding sequence was in-frame fused to the 3′ end of the FLK gene, and the distribution of the fusion protein in plant cells was accessed by a transient transfection system using Allium cepa (onion) epidermal cells. The epidermal cells were bombarded with gold particles coated with plasmid DNA containing the gene fusion and subjected to microscopic analysis. The FLK-GFP fusion was predominantly localized in the nucleus as judged by bright-field and fluorescence microscopic images (Figure 8). FLK may modulate the pre-mRNA processing/alternative polyadenylation, as has been proven for FCA (Macknight et al., 1997, 2002; Quesada et al., 2003).
Figure 8.
Subcellular Localization of FLK.
The FLK-GFP gene fusion was transiently expressed in A. cepa epidermal cells, and the subcellular localization was examined by fluorescent microscopy. Note that the FLK-GFP fusion is predominantly localized in the nucleus. Bars = 20 μm.
In conclusion, we demonstrated here that FLK encodes a putative RNA binding protein with KH motifs that serves as a genetic component of the autonomous flowering pathway. It promotes flowering by suppressing a general floral repressor, FLC. FLK may modulate the posttranscriptional gene regulation in flowering time control in Arabidopsis and, moreover, possibly pre-mRNA processing and polyadenylation as has been demonstrated with FCA.
DISCUSSION
FLK Encodes a KH Motif–Containing RNA Binding Protein
It is now generally accepted that posttranscriptional gene regulation is a critical device for the regulation of eukaryotic gene expression during growth and development, especially in flowering time control in plants (Makeyev and Liebhaber, 2002; Bevilacqua et al., 2003; MacDonald and McMahon, 2003). FLK encodes a putative RNA binding protein that contains three copies of the KH motifs. The flk mutation causes late flowering (Figures 1 and 5), which is effectively reversed by vernalization and GA treatments (Figure 6). The FLC expression is upregulated, whereas those of FT and SOC1 are downregulated in flk (Figure 4).
RNA binding proteins are a group of evolutionally conserved proteins that have distinct RNA binding motifs, such as RRMs, KH motifs, zinc fingers, zinc knuckles, RGG boxes, and DEAD boxes (Lorkovic and Barta, 2002; Makeyev and Liebhaber, 2002). Among these, the RRMs and KH motifs are most frequently found in the Arabidopsis proteins. Two RNA binding protein genes, FCA and FPA, have been recently characterized in the flowering pathways (Macknight et al., 1997; Schomburg et al., 2001). Both encode RRM-containing RNA binding proteins and function in the autonomous flowering pathway. The FCA expression is negatively autoregulated through alternative processing and polyadenylation of its own pre-mRNA (Macknight et al., 1997).
The KH motif was originally identified in the human hnRNP K protein (Siomi et al., 1993). It consists of ∼60 amino acids with a highly conserved core sequence of VIGxxGxxI, where x is any residue but with an obvious preference for positive amino acids (Adinolfi et al., 1999; Makeyev and Liebhaber, 2002). It usually exists as multiple copies up to 15 in a protein. The presence of the KH motifs in a diverse range of proteins from bacteria, yeast, plants, and animals suggests that it has a very ancient origin with an important role for cellular function (Lorkovic and Barta, 2002). The KH motif–containing RNA binding proteins have been implicated in a variety of posttranscriptional events, including pre-mRNA processing, nucleocytoplasmic transport of mRNAs, translational silencing, and mRNA stability/turnover. A subgroup of proteins with the KH motifs, such as αCPs and hnRNPs K/J, have a poly(rC/U) binding specificity and bind the C/U-rich stretches in the 3′ untranslated region of target mRNAs (Kong et al., 2003). They have been experimentally associated with mRNA stabilization, translational silencing, and translational enhancement in animal cells.
The Arabidopsis genome has 26 putative RNA binding proteins with KH motifs and 196 proteins with RRMs (Lorkovic and Barta, 2002). Among the proteins with RRMs, a few have been characterized. The functionally studied examples include FCA and FPA. Another example is AtGRP7, which is a nuclear RNA binding protein with RRMs and a component of a circadian-regulated negative feedback loop that functions as a slave oscillator in Arabidopsis (Heintzen et al., 1997; Staiger et al., 2003).
A KH motif–containing protein, HEN4, has recently been demonstrated in AGAMOUS pre-mRNA processing (Cheng et al., 2003). HEN4 physically and functionally interacts with HUA1, another RNA binding protein that contains six tandem CCCH-type zinc fingers and directly binds with AGAMOUS pre-mRNA. Furthermore, it seems that HEN4 and HUA1, together with other proteins, form a complex for the pre-mRNA processing. HEN4 is the only KH motif–containing RNA binding protein functionally characterized in the plant kingdom.
It is currently unclear how FLK regulates the posttranscriptional events in the flowering pathway. All the RNA gel blot analyses using the FLK sequence as probe detected a single band with an appropriate size (Figures 2C and 7). FLK is mainly localized in the nucleus (Figure 8). It is envisioned that FLK may function in a similar way as with HEN4 and αCP2 proteins. The αCP proteins, represented by αCP1 and αCP2, contain three copies of the KH motifs that have a strong sequence homology with those of FLK. They form the α-complex with a group of RNA binding proteins, such as the AU-rich binding factor (AUF1) and an RNA helicase (Chkheidze et al., 1999). FLK may form a functional complex with other RNA binding proteins as well as with other proteins. Although our data indicate that FLK does not function directly via FCA (Figure 4), it is still possible that FLK interacts with other components in the autonomous pathway. Our preliminary experiment by two-dimensional gel electrophoresis showed that the level of a couple of RNA binding proteins, the RRM-containing AtGRP7 (Staiger et al., 2003) and an RNA helicase, is greatly altered in flk but without any changes in the mRNA levels (Y.S. Kim and S.-B. Hur, unpublished data). This observation further supports the notion that FLK may function in a complex with other RNA binding proteins.
FLK Promotes Flowering by Suppressing FLC
All our experimental data indicate that FLK promotes flowering in a similar manner as the genes or loci identified so far in the autonomous pathway. The upregulation of FLC in flk explains the downregulation of the two floral integrators, FT and SOC1 (Figure 4A). A genetic cross of flk with a FT overexpressor (35S∷FT) is early flowering (Figure 4B), supporting the contention that FLK functions by suppressing the FLC transcription.
It is notable that among the seven genes or loci characterized so far in the autonomous pathway, three genes (FCA, FPA, and FLK in this study) encode RNA binding proteins. It is plausible to propose that the regulation of the FLC pre-mRNA processing and/or its mRNA stability by various RNA binding proteins, irrespective of direct or indirect, is a key event in flowering time control. Interestingly, it has been recently reported that the autonomous pathway also mediates the effects of ambient temperature by ultimately regulating FT (Blázquez et al., 2003; Halliday et al., 2003). The posttranscriptional gene regulation may be a molecular event that integrates intrinsic and environmental signals and is required for the fine-tuning of the floral transition.
FLK Functions in the Autonomous Pathway
Two epistatic groups have been described among the autonomous pathway mutants by comparative analysis of a series of double mutants: FPA/FVE and FCA/FY (Koornneef et al., 1998). Each group is represented by one of the two RNA binding protein genes identified so far and functions independently of the other group.
Our demonstration that FLK is an RNA binding protein gene functioning in the autonomous pathway raises several questions concerning the role of posttranscriptional RNA metabolism in flowering time control. Of particular concern is that FLK contains KH motifs, in contrast with the RRM-containing FCA and FPA. FLK seems to function independently of the other genes in the autonomous pathway. The fca and fpa mutations do not affect the FLK expression (Figure 4C). In addition, the flk mutation does not influence the FCA and FPA expressions (Figures 4A and 4D). FLK may be a member of a distinct epistatic group in the autonomous pathway, entailing that the autonomous pathway is not a simple linear pathway but consists of multiple signaling cascades. Flowering time analysis of the fca flk and fpa flk double mutants will clarify this question.
The next question is related to the presence of multiple RNA binding protein genes in the autonomous pathway. They all act to suppress a single floral repressor, FLC, eventually resulting in the FT and SOC1 induction. The autonomous pathway evidently mediates environmental cues, such as ambient growth temperature (Blázquez et al., 2003; Halliday et al., 2003), as well as internal signals. It is therefore assumed that a repertoire of different RNA binding proteins function in tandem to precisely regulate and/or integrate internal and environmental signals. Altogether, our experimental data strongly support the claim that the posttranscriptional gene regulation is a general principle involved in the flowering pathways (Amasino, 2003).
METHODS
Plant Materials and Growth Conditions
All the transgenic and mutant Arabidopsis lines used in this study were in ecotype Col-0 unless indicated otherwise. Plants were grown in a controlled culture room at 23°C with a relative humidity of 60%. The photoperiods were 16 h light and 8 h dark for the long-day condition and 8 h light and 16 h dark for the short-day condition in white light (120 μmol photons m−2 s−1). fkf was obtained from B. Bartel. flk1 (SALK 112850) was isolated from a pool of T-DNA insertion lines (ABRC, Ohio State University).
Screening of Activation Tagging Mutants
Wild-type Arabidopsis ecotype Col-0 was transformed with an activation tagging vector, pSKI015, which contains the CaMV 35S enhancer element. The pSKI015 vector was provided by D. Weigel and used as described previously (Weigel et al., 2000). Transgenic seeds were directly sown on soil, and a Finale solution (AgrEvo, Montvale, NJ), which contains 5.78% Basta, was diluted 1000 times and sprayed twice a week for 3 weeks. The herbicide-resistant plants were then screened for flowering time alterations. The mutants were further examined through two additional generations. Among the T3 plants that showed late flowering, a late flowering mutant (flk) was chosen.
The single insertion event of T-DNA in flk was confirmed by PCR and genomic DNA gel blot analysis using the 35S enhancer sequence as probe. Genomic DNAs were isolated using the DNeasy plant mini kit (Qiagen, Valencia, CA). The flanking sequences of the T-DNA insertion were determined by thermal asymmetric interlaced PCR and DNA sequencing (Liu et al., 1995).
Flowering Time Measurements
Plants were grown on soil in the long-day or short-day condition. Flowering time was measured by counting the total number of rosette and cauline leaves at flowering and the days from sowing to floral bud formation. Thirty to fifty plants were measured and averaged for each measurement and statistical treatment.
Analysis of Transcript Levels
Transcript levels were measured either by RNA gel blots or by RT-PCR–based DNA gel blots. Total RNA was isolated from whole plants or plant materials using the Trizol reagent (Invitrogen, Carlsbad, CA). For RNA gel blot analyses, ∼30 μg of total RNA was separated by 1.2% denaturing formaldehyde-agarose gel electrophoresis. The RNA gel blot probes were labeled with digoxigenin-UTP by in vitro transcription with SP6 or T7 RNA polymerase using the DIG RNA labeling kit (Roche Applied Science, Penzberg, Germany). For RNA gel blot analysis of the FCA transcription, poly(A)+ RNA was isolated from total RNA using the Oligotex mRNA mini kit (Qiagen).
Quantitative RT-PCR was employed to measure the transcript levels of the flowering time genes. Total RNA samples were treated extensively with RNase-free DNase I to remove any contaminating genomic DNAs. The first-strand cDNA was synthesized using Pfu Turbo polymerase (Stratagene, La Jolla, CA) from 2 μg of total RNA in a 20-μL reaction volume, and 2 μL of the reaction mixture was subject to subsequent PCR in a 50-μL reaction volume. The RT-PCR runs were 15 to 30 cycles, depending on the linear range of PCR amplification for each gene, with each cycle at 94°C for 1 min, 58°C for 0.5 min, and 72°C for 4 min, with a final cycle at 72°C for 7 min to allow the completion of the polymerizations.
Vernalization and GA Treatments
For vernalization treatments, plants were germinated and grown at 4°C for 6 weeks in the long-day condition and transferred to the normal growth condition (23°C). To examine GA effects, plants were grown on soil in the long-day condition, and a GA solution of 20 μM was sprayed twice a week until flowering.
Subcellular Localization Analysis
The GFP coding sequence was in-frame fused to the 3′ end of the FLK gene, and the gene fusion was subcloned into the pBI221 vector (Clontech, Palo Alto, CA) for transient expression in A. cepa epidermal cells by DNA-coated gold particle bombardment. After incubation for 24 h at room temperature, the cells were subject to bright-field and fluorescence microscopy. The Olympus BX51 microscope equipped with the BX-URA2 fluorescence illuminator (Tokyo, Japan) was used.
Supplementary Material
Acknowledgments
We thank Bonnie Bartel for the fkf mutant and Detlef Weigel for the pSKI015 vector. We also acknowledge the technical support provided by members of the Molecular Signaling Laboratory, Seoul National University. This work was supported by Korea Research Foundation (KRF) Grant KRF-2000-015-DP0394, the Brain Korea 21 Program (KRF), a grant from the Plant Signaling Network Research Center, Grant CG1231 from the Crop Functional Genomics Center of the 21st Century Frontier Research Program to C.B.H., Korea Science and Engineering Foundation Grant R02-2003-000-10001-0, and Korea Institute of Science and Technology Evaluation and Planning Grant M1-0219-00-0003.
On-line version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Chung-Mo Park (cmpark@snu.ac.kr).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.019331.
References
- Adinolfi, S., Bagni, C., Morelli, M.A.C., Musco, G., and Pastore, A. (1999). Novel RNA-binding motif: The KH module. Biopolymers 51, 153–164. [DOI] [PubMed] [Google Scholar]
- Amasino, R.M. (2003). Flowering time: A pathway that begins at the 3′ end. Curr. Biol. 13, R670–R672. [DOI] [PubMed] [Google Scholar]
- Bevilacqua, A., Ceriani, M.C., Capaccioli, S., and Nicolin, A. (2003). Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J. Cell. Physiol. 195, 356–372. [DOI] [PubMed] [Google Scholar]
- Blázquez, M.A., Ahn, J.H., and Weigel, D. (2003). A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Natl. Genet. 33, 168–171. [DOI] [PubMed] [Google Scholar]
- Blázquez, M.A., Koornneef, M., and Putterill, J. (2001). Flowering on time: Genes that regulate the floral transition. EMBO Rep. 2, 1078–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chkheidze, A.N., Lyakhov, D.L., Makeyev, A.V., Morales, J., Kong, J., and Liebhaber, S.A. (1999). Assembly of the α-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′ untranslated region determinant and poly(C) binding protein αCP. Mol. Cell. Biol. 19, 4572–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y., Kato, N., Wang, W., Li, J., and Chen, X. (2003). Two RNA binding proteins, HEN4 and HUA1, act in the processing of AGAMOUS pre-mRNA in Arabidopsis thaliana. Dev. Cell 4, 53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, M.-L., and Yang, C.-H. (1999). Late-flowering genes interact with early-flowering genes to regulate flowering time in Arabidopsis thaliana. Plant Cell Physiol. 40, 702–708. [DOI] [PubMed] [Google Scholar]
- Doyle, M.R., Davis, S.J., Bastow, R.M., McWatters, H.G., Kozma-Bognar, L., Nagy, F., Millar, A.J., and Amasino, R.M. (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419, 74–77. [DOI] [PubMed] [Google Scholar]
- Gendall, A.R., Levy, Y.Y., Wilson, A., and Dean, C. (2001). The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107, 525–535. [DOI] [PubMed] [Google Scholar]
- Halliday, K.J., Salter, M.G., Thingnaes, E., and Whitelam, G.C. (2003). Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J. 33, 875–885. [DOI] [PubMed] [Google Scholar]
- Hayama, R., and Coupland, G. (2003). Shedding light on the circadian clock and the photoperiodic control of flowering. Curr. Opin. Plant Biol. 6, 13–19. [DOI] [PubMed] [Google Scholar]
- Heintzen, C., Nater, M., Apel, K., and Staiger, D. (1997). ArGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94, 8515–8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S.E., Binkowski, K.A., and Olszewski, N.E. (1996). SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. USA 93, 9292–9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, J., Ji, J., and Liebhaber, S.A. (2003). The KH-domain protein αCP has a direct role in mRNA stabilization independent of its cognate binding site. Mol. Cell. Biol. 23, 1125–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Alonso-Blanco, C., Blankestijn-de Vries, H., Hanhart, C.J., and Peeters, A.J.M. (1998). Genetic interactions among late-flowering mutants of Arabidopsis. Genetics 148, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Hanhart, C.J., and Van Der Veen, J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66. [DOI] [PubMed] [Google Scholar]
- Lee, H., Suh, S.S., Park, E., Cho, E., Ahn, J.H., Kim, S.G., Lee, J.S., Kwon, Y.M., and Lee, I. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C. (2000). Photoreceptors and regulation of flowering time. Plant Physiol. 123, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Lorkovic, Z.L., and Barta, A. (2002). Genomic analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 30, 623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald, C.C., and McMahon, K.W. (2003). The flowers that bloom in the spring: RNA processing and seasonal flowering. Cell 113, 671–672. [DOI] [PubMed] [Google Scholar]
- Macknight, R., Bancroft, I., Page, T., Lister, C., Schmidt, R., Love, K., Westphal, L., Murphy, G., Sherson, S., Cobbett, C., and Dean, C. (1997). FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89, 737–745. [DOI] [PubMed] [Google Scholar]
- Macknight, R., Duroux, M., Laurie, R., Dijkwel, P., Simpson, G., and Dean, C. (2002). Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell 14, 877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev, A., and Liebhaber, S.A. (2002). The poly(C)-binding proteins: A multiplicity of functions and a search for mechanisms. RNA 8, 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Zapater, J.M., and Somerville, C.R. (1990). Effect of light quality and vernalization on late-flowering mutants of Arabidopsis thaliana. Plant Physiol. 92, 770–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (1999. a). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (1999. b). The gibberellic acid biosynthesis mutant ga1–3 of Arabidopsis thaliana is responsive to vernalization. Dev. Genet. 25, 194–198. [DOI] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13, 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler, T., Yang, H., Yu, X., Parikh, D., Cheng, Y.C., Dolan, S., and Lin, C. (2003). Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc. Natl. Acad. Sci. USA 100, 2140–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, J., Suh, S.-S., Lee, H., Choi, K.-R., Hong, J.B., Paek, N.-C., Kim, S.-G., and Lee, I. (2003). The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 35, 613–623. [DOI] [PubMed] [Google Scholar]
- Mouradov, A., Cremer, F., and Coupland, G. (2002). Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell 14, S111–S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski, N., Sun, T.P., and Gublet, F. (2002). Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 14, S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onouchi, H., Igeno, M.I., Perilleux, C., Graves, K., and Coupland, G. (2000). Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12, 885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada, V., Macknight, R., Dean, C., and Simpson, G.G. (2003). Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time. EMBO J. 22, 3142–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves, P.H., and Coupland, G. (2001). Analysis of flowering time control in Arabidopsis by comparison of double and triple mutants. Plant Physiol. 126, 1085–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse, D.T., Sheldon, C.C., Bagnall, D.J., Peacock, W.J., and Dennis, E.S. (2002). FLC, a repressor of flowering, is regulated by genes in different inductive pathways. Plant J. 29, 183–191. [DOI] [PubMed] [Google Scholar]
- Schomburg, F.M., Patton, D.A., Meinke, D.W., and Amasino, R.M. (2001). FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell 13, 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, G.G., and Dean, C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289. [DOI] [PubMed] [Google Scholar]
- Simpson, G.G., Dijkwel, P.P., Quesada, V., Henderson, I., and Dean, C. (2003). FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113, 777–787. [DOI] [PubMed] [Google Scholar]
- Siomi, H., Matunis, M.J., Michael, W.M., and Dreyfuss, G. (1993). The pre-mRNA binding K protein contains a novel evolutionary conserved motif. Nucleic Acids Res. 21, 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger, D., Zecca, L., Kirk, D.A.W., Apel, K., and Eckstein, L. (2003). The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J. 33, 361–371. [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122, 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky, M.J., and Kay, S.A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419, 308–312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.