Abstract
Double strand breaks (DSBs) are the most deleterious form of DNA damage. Natural products that produce them are potent cytotoxic agents. Designing molecules that produce DSBs via a single chemical event is challenging. We determined that formation of a C4′-nucleotide radical in duplex DNA under aerobic conditions gives rise to a DSB. The original radical yields a strand break containing a peroxyl radical, which initiates opposite strand cleavage via C4′-hydrogen atom abstraction. This mechanism provides the impetus to design DNA damaging agents that produce DSBs by abstracting a single hydrogen atom from the biopolymer.
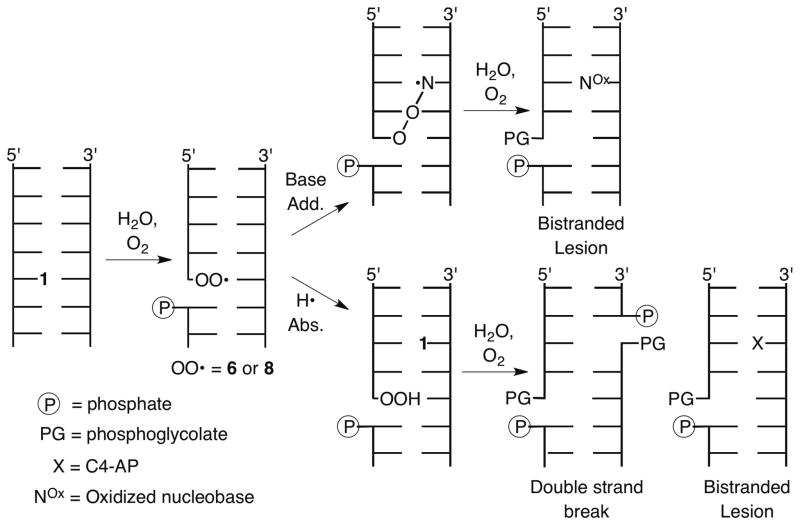
Double strand breaks (DSBs) are difficult to correctly repair and as a result are the most deleterious family of DNA lesions. Consequently, DSB formation is highly desirable when the goal is to induce cell death by damaging DNA. DSBs result from cleaving opposing DNA strands within ~1.5 helical turns of one another. The low probablility that two molecules will react with the biopolymer within the required proximity to generate a DSB makes their formation by molecules that damage a single strand of DNA very inefficient. Potent antitumor natural products, such as the enediynes, generate a biradical that abstracts hydrogen atoms on the opposing strands of DNA in its minor groove to efficiently produce DSBs.1–4 In contrast, a single molecule of bleomycin produces DSBs by a sequence of events in which it oxidatively cleaves one strand of DNA, and is reactivated while remaining bound to its target.5,6 The reactivated molecule then oxidatively cleaves the complementary DNA strand, completing DSB formation. A damaging agent could produce a DSB from a single oxidation event if a DNA reactive intermediate reacted with a nucleotide on the opposing strand. This possibility was considered by radiation chemists based upon the linear dependence of DSBs on hydroxyl radical yield at low radiation doses.7–9 However, such a mechanism was found to be a minor contributor to DSB formation by ionizing radiation.10 Identifying a pathway by which a single oxidation reaction leads to DSBs would be valuable information for designing molecules that produce this family of DNA lesions. We wish to report that a C4′-radical yields a double strand break under aerobic conditions via a multi-step process during which spin is transferred to the opposing strand.
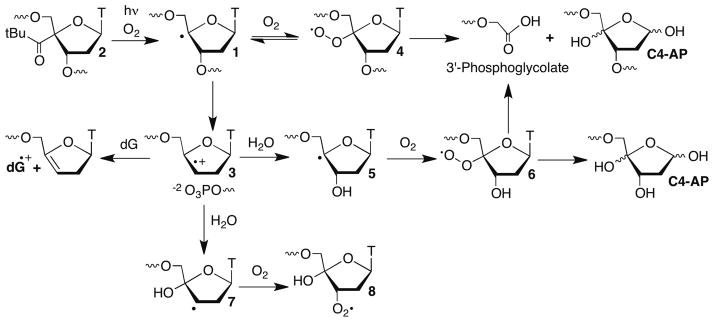
The C4′-hydrogen atom is frequently abstracted by DNA damaging agents, due to its accessibility at the outer edge of the minor groove and the moderate bond dissociation energy of the corresponding C-H bond.11 The C4′-radical (1) was proposed to yield a DNA strand break by β-phosphate elimination (Scheme 1).12 Giese solidified this mechanism by independently generating radical 1 from 2, and utilized the incipient olefin cation radical (3) to study electron transfer in DNA.13–16 Olefin cation radical 3 is trapped sequentially by H2O (5, 7) and O2 to form a mixture of peroxyl radicals 6 and 8 at the 3′-terminus of the cleaved DNA. The former is analogous to 4 (which is formed reversibly).17
Scheme 1.
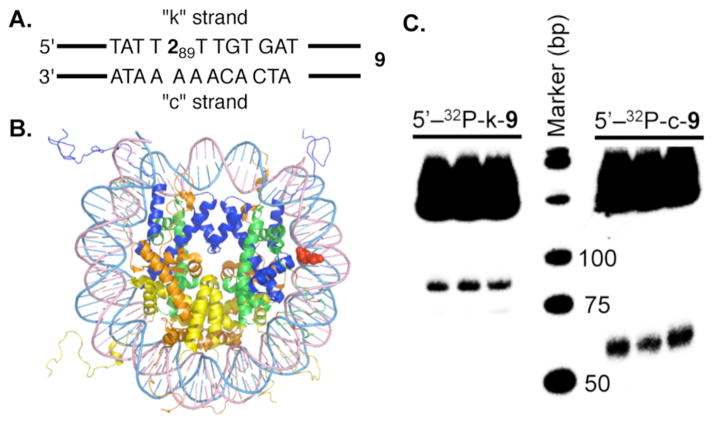
C4′-radical 1 was generated from 2 within a nucleosome core particle (NCP) composed of DNA whose sequence was derived from the strong positioning 601 DNA discovered by Widom (Figure 1A, B).18 Radical 1 was generated within 9 at position 89 of the 145 bp DNA (superhelical location (SHL) 1.5), a known hot spot for molecules that oxidatively damage DNA.19 NCPs containing 2 were produced using previously described methods and one of the DNA strands was 5′-32P-labeled.14,20 (Per Figure 1A, labeling the strand containing 2 is denoted by k, and c for the complementary strand.) Substrate 9 contained 10, which lacked dG in the vicinity of the radical to suppress electron transfer involving 3.21 DSBs were evident by nondenaturing PAGE analysis of photolyzed 5′-32P-c-9 or 5′-32P-k-9 (Figure 1C). The DSB yield was 4.2 ± 1.2 % and the fragment lengths correspond to cleavage in the region where 1 is produced.22 Analysis of free 145 5′-32P-k-9: 6.5 ± 0.9%) and anaerobic conditions (5′-32P-c-bp DNA irradiated under aerobic (5′-32P-c-9: 6.9 ± 1.4%; 9, 5′-32P-k-9: < 1%) revealed that DSB formation does not require generation in a NCP but is O2 dependent.23 In addition, alkaline labile lesions were detected in the complementary strand of free 9 (% Cleavage in 5′-32P-c-9: direct: 7.0 ± 1.1; NaOH: 10.8 ± 0.7; piperidine: 17.8 ± 1.1) by denaturing PAGE.
Figure 1.
Double-strand break formation upon generation of 1 from 2 within a NCP. A. Portion of nucleosomal DNA (9, 145 bp) containing 2 at position 89. B. NCP showing 2 (red) at SHL 1.5. (X-Ray data taken from PDB: 3LZ0.) C. Native PAGE showing DSB formation upon photolysis (3 replicates) of NCP containing 9.
A series of 35 bp duplexes containing 2 were prepared to examine the generality of the transfer of damage from the original strand in which 1 is generated to the complementary strand under aerobic conditions, and to acquire greater detail of this novel reaction. Irradiation of 5′-32P-k-10 produced direct strand breaks and NaOH labile lesions at the position where 1 is produced (Scheme 1). NaOH lability is generally attributed to abasic sites and were formed in 35 ± 9 % relative to direct strand scission.24,25 Specific products from the perox-yl radical (6) derived from photolyzed 2 were identified by LC/MS. Fragments containing 3′-phosphoglycolate (Scheme 1) and 3′-phosphate, as well as a product corresponding to cleaved C4′-oxidized abasic site (C4-AP) were detected upon photolysis of 10.23 These products are consistent with the expected formation of 1.
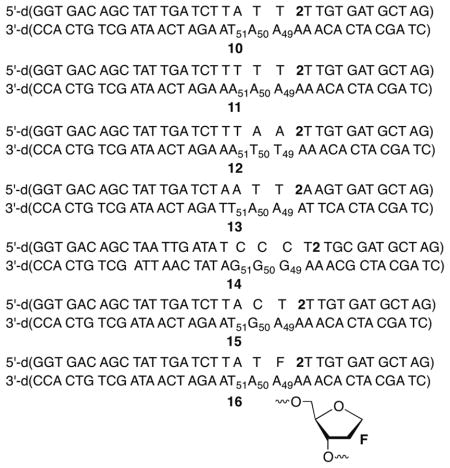
Denaturing PAGE analysis of 5′-32P-c-10 photolyzed under aerobic, but not anaerobic conditions revealed direct strand scission at dA49, dA50, and dT51 (relative yields: 1.0 : 2.4 : 1.0, Table 1). Slightly different ratios were observed following NaOH or piperidine treatment but dA50 remained the major cleavage site.23 In addition, strand scission in the complementary strand was enhanced more than 55% upon mild NaOH (Table 1) or hydrazine treatment. The latter selectively cleaves DNA containing C4-AP.25,26 Formation of cleaved DNA containing 3′-phosphoglycolate (Scheme 1) and 3′-phosphate termini, as well as C4-AP that had undergone β-elimination in the complementary strand was confirmed by LC/MS and denaturing PAGE analysis.23 These products are consistent with C4′-hydrogen atom abstraction from nucleotides in the complementary strand. 3′-Phosphate termini are also produced upon C5′-hydrogen atom abstraction and this may be a minor pathway. Finally, piperidine treatment almost doubled O2 dependent strand scission in the strand opposite 2, indicating that nucleobase damage was incurred as well. Alkali labile lesion on the complementary strand constitute the formation of bis-tranded lesions, which are an important family of DNA damage due to the difficulty of their repair.27,28
Table 1.
Complementary strand cleavage upon photolysis under aerobic conditions of 35 bp duplexes containing 2.
| % Complementary Strand Cleavagea,b, 23 | |||
|---|---|---|---|
| Duplex | Direct | NaOH | Piperidine |
| 10 | 6.8 ± 0.9 | 10.7 ± 1.1 | 19.7 ± 2.6 |
| 11 | 3.9 ± 1.2 | 5.6 ± 0.6 | 11.0 ± 0.3 |
| 12 | 3.7 ± 0.1 | 8.2 ± 0.5 | 16.9 ± 0.8 |
| 13 | 5.5 ± 0.2 | 8.5 ± 0.4 | 14.4 ± 1.4 |
| 14 | < 1 | 1.2 ± 0.5 | 7.4 ± 0.4 |
| 15 | 2.6 ± 0.4 | 4.2 ± 0.6 | 11.0 ± 2.6 |
| 16 | 18.8 ± 2.5 | 37.1 ± 1.9 | 50.2 ± 3.8 |
Yields are averages ± std. dev. of at least 2 experiments, each consisting of 3 independent reactions.
Yield was determined by dividing the percent cleavage at the complementary strand by the percent cleavage in the strand containing 2 following piperidine treatment.
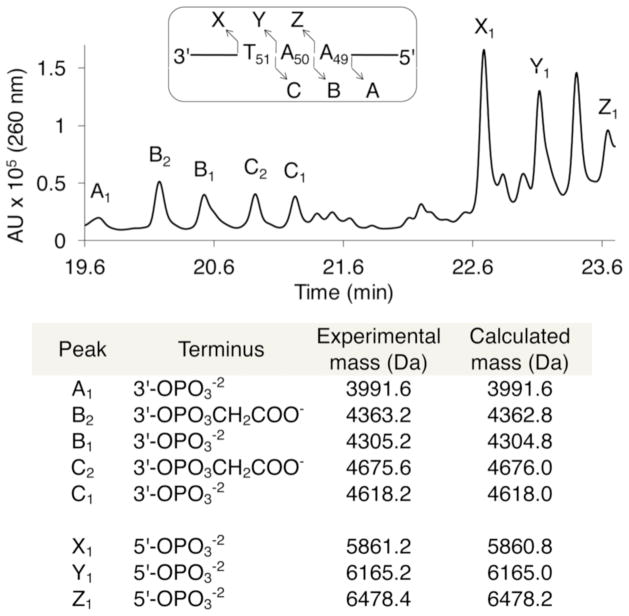
Comparable yields of analogous lesions were observed upon photolysis of duplexes 11–13, indicating that A•T rich sequences in the vicinity of 1 are generally susceptible to this process (Table 1). Replacing the thymidine adjacent to 2 in 10 with a model abasic site (F, 16) significantly increased the yields of all forms of damage (Table 1) in the complementary strand at A49-T51. Enhanced cleavage in 5′-32P-c-16 argues against the involvement of a diffusible species whose reactivity should not be increased by F. Moreover, the increased complementary strand damage in 32P-c-16 is consistent with a reactive species from the strand in which 1 is generated reacting with the opposing strand. The presence of F in 16 weakens the local hydrogen bonding, reducing the barrier(s) that must be overcome to adopt the conformation(s) required for interstrand spin transfer. LC/MS analysis of photolyzed 16 clearly shows the formation of products attributable to C4′-oxidation on the complementary strand (e.g. 3′-phosphoglycolate), as well as phosphate containing fragments that could result from oxidation at C4′ and other positions (Figure 2).
Figure 2.
LC/MS product analysis in the complementary strand of photolyzed 16.
Incorporating a single dG (5′-32P-c-15) at the major damage site (dN50) has the opposite effect on strand damage as F (Table 1). Moreover, substituting a dGGG sequence in the comparable region of the complementary strand (5′-32P-c-14) dramatically reduced the yield of direct strand breaks and NaOH labile products, while reducing the yield of piperidine labile products to a lesser extent (Table 1). dG49 was the major damage site (2 nucleotides removed from 2) in 5′-32P-c-14, but damage was also detected on the complementary strand one nucleotide further removed from the position at which 1 is generated (dG50). The effects of dG on complementary strand damage and the dependency on O2 may explain why DSB formation was not reported in previous studies involving 1 due to the focus on hole migration under anaerobic conditions.16
Based upon the products detected and literature precedent, we suggest that 6 and/or 8 (Scheme 1), which are unrestrained relative to 4 transfer damage to the opposite strand.13,14,29 The peroxyl radical(s) abstracts the C4′- and perhaps C5′-hydrogen atoms from the opposing strand 1–3 nucleotides away and reacts with the corresponding nucleobases (Scheme 2). These radicals are formed from 1 under aerobic conditions via 3, with formation of 6 being favored ~2.2–2.5-fold over 8.29,30 The effect of dG on the product distribution could be attributed to altered partitioning of the peroxyl radical(s) between hydrogen atom abstraction from the sugar and reaction with the nucleobase due to guanine’s more facile oxidation than other native nucleobases and accentuation of this preference by guanine triplets.31–33 Alternatively, dG could intercept olefin cation radical 3 prior to its trapping by water. These mechanisms were differentiated from one another by synthesizing a ternary complex (17) containing 2 at the 3′-terminus of one oligonucleotide.29 This ternary complex can produce 6 but not 3 or 8 due to the absence of a suitable (phosphate) leaving group at the 3′-position. The same yield and distribution of products was observed within experimental error upon photolysis of 5′-32P-c-17 as from 5′-32P-c-14.23 This indicates that radical cation 3 is not responsible for the effect of dG on strand damage products, and that the increase in the proportion of products associated with nucleobase damage in 14 is due to changes in the partitioning of peroxyl radical 6 and/or 8.
Scheme 2.
Finally, the proximity of the peroxyl radical oxygen in 6 (and/or 8) with respect to C4′-hydrogen atoms on the opposing strand was examined using molecular models (Figure 3).23 Significant deformation of the duplex DNA is required for either peroxyl radical to react with the opposing strand. However, the minimum distance between the peroxyl radical oxygens in 6 or 8 and the C4′-hydrogen atoms 1 – 3 base pairs away are significantly shorter than for the respective positions in other nucleotides on the opposing strand. Furthermore, the selectivity for reactivity with nucleotides in the opposite strand correlates with these differences.
Figure 3.
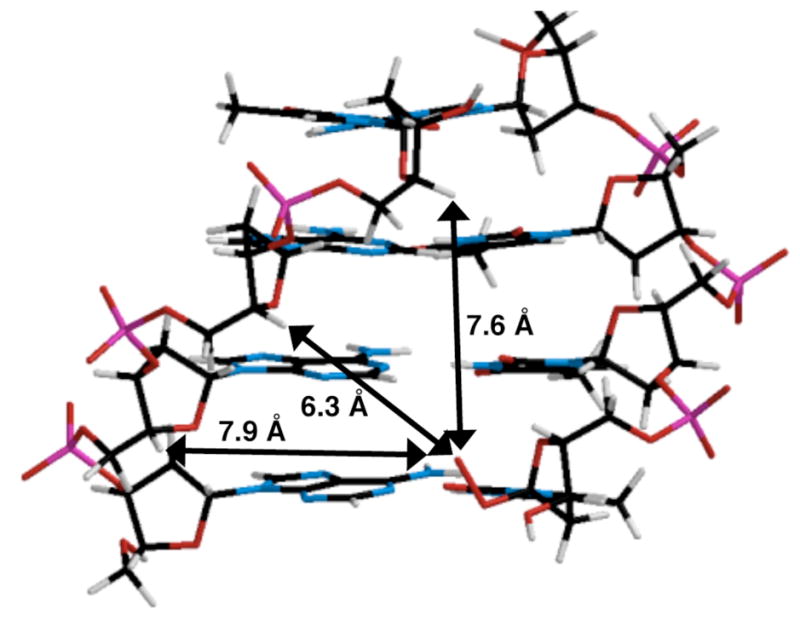
Molecular model of the distance between the peroxyl radical oxygen of 6 and the C4′-hydrogen atoms of nucleotides on the complementary strand in 5′-d(AAAT)/3′-d(6TTA). Models were constructed using Spartan®. (See the Supporting Information for a comparable model containing 8.)
In summary, we have detected an O2 dependent mechanism for DSB formation that is initiated by abstraction of a single hydrogen atom from DNA and is diminished by dG in the opposite strand. To our knowledge, it is the first example of DSB formation from a single initial oxidation reaction. The process occurs in nucleosome core particles, suggesting that it may also be relevant in cells. Based upon these data we suggest that molecules designed to abstract the C4′-hydrogen atom of duplex DNA should produce double strand breaks in dA•T rich sequences and bistranded lesions more generally. Finally, these observations raise the question as to whether the above mechanism is partially responsible for bistranded lesion formation by molecules such as the enediynes.3
Supplementary Material
Acknowledgments
We are grateful for generous financial support from the National Institute of General Medical Science (GM-054996).
Footnotes
Supporting Information. Experimental procedures for all experiments. LC/MS of photolysis of 10, damage in free 9, distribution of damage in 11–17, end group analysis, model of 8 in tetranucleotide duplex. Mass spectra of modified oligonucleotides. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kennedy DR, Lu J, Shen B, Beerman TA. Proc Nat Acad Sci USA. 2007;104:17632–17637. doi: 10.1073/pnas.0708274104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galm U, Hager MH, Van Lanen SG, Ju J, Thorson JS, Shen B. Chem Rev. 2005;105:739–758. doi: 10.1021/cr030117g. [DOI] [PubMed] [Google Scholar]
- 3.Xi Z, Goldberg IH. In: Comprehensive Natural Products Chemistry. Kool ET, editor. Vol. 7. Elsevier; Amsterdam: 1999. pp. 553–592. [Google Scholar]
- 4.Lee MD, Ellestad GA, Borders DB. Acc Chem Res. 1991;24:235–243. [Google Scholar]
- 5.Povirk LF, Han YH, Steighner RJ. Biochemistry. 1989;28:5808–5814. doi: 10.1021/bi00440a016. [DOI] [PubMed] [Google Scholar]
- 6.Absalon MJ, Wu W, Kozarich JW, Stubbe J. Biochemistry. 1995;34:2076–2086. doi: 10.1021/bi00006a030. [DOI] [PubMed] [Google Scholar]
- 7.Siddiqi MA, Bothe E. Radiat Res. 1987;112:449–463. [PubMed] [Google Scholar]
- 8.Frankenberg-Schwager M, Frankenberg D. Int J Radiat Biol. 1990;58:569–575. doi: 10.1080/09553009014551931. [DOI] [PubMed] [Google Scholar]
- 9.Milligan JR, Ng JYY, Wu CCL, Aguilera JA, Fahey RC, Ward JF. Radiat Res. 1995;143:273–280. [PubMed] [Google Scholar]
- 10.Shao C, Saito M, Yu Z. Radiat Environ Biophysics. 1999;38:105–109. doi: 10.1007/s004110050145. [DOI] [PubMed] [Google Scholar]
- 11.Balasubramanian B, Pogozelski WK, Tullius TD. Proc Nat Acad Sci USA. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dizdaroglu M, Von Sonntag C, Schulte-Frohlinde D. J Am Chem Soc. 1975;97:2277–2278. doi: 10.1021/ja00841a051. [DOI] [PubMed] [Google Scholar]
- 13.Giese B, Beyrich-Graf X, Erdmann P, Petretta M, Schwitter U. Chem & Biol. 1995;2:367–375. doi: 10.1016/1074-5521(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 14.Giese B, Beyrich-Graf X, Erdmann P, Giraud L, Imwindelried P, Müller SN, Schwitter U. J Am Chem Soc. 1995;117:6146–6147. [Google Scholar]
- 15.Meggers E, Michel-Beyerle ME, Giese B. J Am Chem Soc. 1998;120:12950–12955. [Google Scholar]
- 16.Giese B. Acc Chem Res. 2000;33:631–636. doi: 10.1021/ar990040b. [DOI] [PubMed] [Google Scholar]
- 17.Dussy A, Meggers E, Giese B. J Am Chem Soc. 1998;120:7399–7403. [Google Scholar]
- 18.Lowary PT, Widom J. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 19.Kuduvalli PN, Townsend CA, Tullius TD. Biochemistry. 1995;34:3899–3906. doi: 10.1021/bi00012a005. [DOI] [PubMed] [Google Scholar]
- 20.Sczepanski JT, Wong RS, McKnight JN, Bowman GD, Greenberg MM. Proc Natl Acad Sci U S A. 2010;107:22475–22480. doi: 10.1073/pnas.1012860108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meggers E, Kusch D, Spichty M, Wille U, Giese B. Angew Chem, Int Ed. 1998;37:460–462. doi: 10.1002/(SICI)1521-3773(19980302)37:4<460::AID-ANIE460>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 22.DSB yields are calculated to account for incomplete conversion of 2. The DSB yield equals the percent double strand cleavage measured by nondenaturing gel divided by the percent cleavage in the strand containing 2 following piperidine treatment measured by denaturing PAGE × 100. The absolute value of the latter ranged from 47 – 55 %.
- 23.See Supporting Information.
- 24.Carter KN, Greenberg MM. J Am Chem Soc. 2003;125:13376–13378. doi: 10.1021/ja036629u. [DOI] [PubMed] [Google Scholar]
- 25.San Pedro JMN, Beerman TA, Greenberg MM. Bioorg Med Chem. 2012;20:4744–4750. doi: 10.1016/j.bmc.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugiyama H, Kawabata H, Fujiwara T, Dannoue Y, Saito I. J Am Chem Soc. 1990;112:5252–5257. [Google Scholar]
- 27.O’Neill P, Wardman P. Int J Radiat Biol. 2009;85:9–25. doi: 10.1080/09553000802640401. [DOI] [PubMed] [Google Scholar]
- 28.Sage E, Harrison L. Mutat Res. 2011;711:123–133. doi: 10.1016/j.mrfmmm.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meggers E, Dussy A, Schäfer T, Giese B. Chem Eur J. 2000;6:485–492. doi: 10.1002/(sici)1521-3765(20000204)6:3<485::aid-chem485>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 30.Giese B, Beyrich-Graf X, Burger J, Kesselheim C, Senn M, Schäfer T. Angew Chem Int Ed. 1993;32:1742–1743. [Google Scholar]
- 31.Steenken S, Jovanovic SV. J Am Chem Soc. 1997;119:617–618. [Google Scholar]
- 32.Saito I, Nakanura T, Nakatani K, Yoshioka Y, Yamaguchi K, Sugiyama H. J Am Chem Soc. 1998;120:12686–12687. [Google Scholar]
- 33.Bergeron F, Auvré F, Radicella JP, Ravanat JL. Proc Nat Acad Sci USA. 2010;107:5528–5533. doi: 10.1073/pnas.1000193107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.