Abstract
Recently discovered genome-wide rare copy number variants (CNVs) have unprecedented levels of statistical association with many developmental neuropsychiatric disorders, including schizophrenia, autism spectrum disorders, intellectual disability and attention deficit hyperactivity disorder. However, as CNVs often include multiple genes, causal genes responsible for CNV-associated diagnoses and traits are still poorly understood. Mouse models of CNVs are in use to delve into the precise mechanisms through which CNVs contribute to disorders and associated traits. Based on human and mouse model studies on rare CNVs within human chromosome 22q11.2, we propose that alterations of a distinct set of multiple, noncontiguous genes encoded in this chromosomal region, in concert with modulatory impacts of genetic background and environmental factors, variably shift the probabilities of phenotypes along a predetermined developmental trajectory. This model can be further extended to the study of other CNVs and may serve as a guide to help characterize the impact of genes in developmental neuropsychiatric disorders.
Keywords: autism, copy number variant, intellectual disability, mouse model, schizophrenia, ADHD
The onset of many neuropsychiatric disorders can be traced to childhood. Symptomatic features of autism spectrum disorder (ASD), intellectual disability (ID), attention deficit hyperactivity disorder (ADHD) and anxiety start to appear during infancy and childhood. Although the clinical diagnosis of schizophrenia occurs at an average of 18 years in males and 24 years in females, children and adolescents who later develop schizophrenia often exhibit prodromal symptoms and some exhibit symptoms to the extent that a clinical diagnosis of schizophrenia is appropriate during childhood (i.e., childhood-onset schizophrenia).1,2 We use the term developmental neuropsychiatric disorders to collectively refer to neuropsychiatric disorders whose symptoms are first observed during childhood.
Despite many years of genetics studies of common variants, the etiology of developmental neuropsychiatric disorders is still poorly understood. Although there are many reports of association between common genetic variants and disorders, few have withstood rigorous replication testing. Recent large-scale analyses indicate individual common variants have very small effect on disorders.3–5 It is now clear that rare (<1% population frequency) copy number variants (CNVs) have robust, reproducible impacts; carriers of rare CNVs have shown odd ratios of up to 20 for susceptibility to developmental neuropsychiatric disorders. In contrast to typical syndromes such as Smith–Magenis syndrome, Williams syndrome and Phelan–McDermid syndrome, recently discovered rare CNVs are usually associated with not one but many developmental neuropsychiatric disorders, suggesting the tantalizing possibility that there may be common denominators among symptomatically diverse disorders. Studies of rare chromosomal segment deletions and duplications involving multiple genes present a glimpse of psychiatry’s promised land where clinically defined disorders are reduced to genetically, and possibly pathophysiologically, defined cases.
Although genome-wide analysis of CNVs for ASD and schizophrenia started to appear in the literature in 20076 and 2008,7 respectively, an especially interesting CNV was known long before that time. Shprintzen et al.8 reported that some adult patients with velocardiofacial syndrome developed schizophrenia. In the same year, it was recognized that both velocardiofacial syndrome and DiGeorge syndrome, two clinical syndromes focused on overlapping, yet nonidentical, constellations of heart, thymus and velopharyngeal defects, are associated with human chromosome 22q11.2 deletions,9–11 establishing a previously unknown association between schizophrenia and 22q11.2 deletions.
As extensive analyses have accumulated for 22q11.2 CNV over the past 20 years, we discuss here the current understanding of this rare variant as it relates to mechanisms of developmental neuropsychiatric disorders and describe mechanistic insights we gained from mouse models of this CNV. We first characterize pleiotropic symptoms and traits seen in individuals with this CNV in relation to developmental neuropsychiatric disorders. We then review attempts to identify specific causal genes for behavioral phenotypes seen in 22q11.2 CNV in humans and mice. Finally, we refine and extract general hypothetical principles derived from mouse studies.
CNV-ASSOCIATED DIAGNOSES OF DEVELOPMENTAL NEUROPSYCHIATRIC DISORDERS
In many cases, a CNV is associated with more than one clinical diagnosis.12,13 Schizophrenia, ASD, ID and ADHD are all associated with CNVs at 1q21.1, 15q13.3, 16p11.2 and 22q11.2. Many other CNVs are associated with some, if not all, developmental neuropsychiatric disorders; CNVs at 3q29, 15q11.2, 16p13.11-p13.2 and 17q12 are associated with schizophrenia, ASD and ID. Others are associated with fewer clinical diagnoses, but their clinical characterization is incomplete and additional diagnoses might be added as more clinical evaluations become available, as was the case with 22q11.2 CNV.
Schizophrenia
High rates of schizophrenia in 22q11.2 deletion have been consistently replicated.14–19 A recent estimate indicates that ~25% of adults with 22q11.2 deletions develop schizophrenia, which is considered to represent one of the genetically identifiable causes of schizophrenia.20 This deletion is detected as a rare variant (0.2–0.3%) in the general adult schizophrenic population7,21,22 and might be present at even higher rates among individuals with childhood-onset schizophrenia.23–25 On the other hand, studies report that 22q11.2 duplications are not enriched in the schizophrenic population, 7,21,22,25–28 and there exist few reported cases of schizophrenia among individuals with 22q11.2 duplication.
Autism spectrum disorder
Children with 22q11.2 hemizygosity (with only one copy present in diploid cells) and duplications exhibit many behavioral problems in the social behavior and language development domains.29–36 A series of studies using established ASD scales determined that 14–50% of children with 22q11.2 hemizygosity met diagnostic criteria.33,37–42 Duplications of 22q11.2 are also associated with ASD.43,44 Conversely, studies have identified 22q11.2 duplications and hemizygosity in ASD populations.6,27,45–51
Although, according to some studies, 22q11.2 deletions are not statistically enriched in the general ASD population,52 enrichment of any given CNV in the general ASD population is difficult to establish due to its very rare occurrence. Another technical confounding factor is the sample structure. Although simplex and multiplex do not necessarily equate with de novo and inherited cases, respectively,13 the rate of de novo CNVs tends to be higher in simplex than multiplex samples.51 As deletion and duplication at 22q11.2 are overwhelmingly de novo (~80%) and inherited (>90%), respectively,53 studies including both simplex and multiplex families might underestimate the overall rate of deletion cases. A complementary approach to avoid false negatives involves characterization of clinical phenotypes for any given CNV. In light of consistently replicated, extremely high rates (14–50%) of ASD in 22q11.2 deletion cases,33,37–42 this association is undeniable.
Vorstman et al. reported that many autistic symptoms present during childhood are similar to those seen in 22q11.2 deletion patients with and without schizophrenia. Moreover, childhood ASD features tended to be associated with lower rates of psychosis during adulthood, indicating that ASD and schizophrenia in individuals with 22q11.2 deletion are, at least in some cases, two distinct and unrelated phenotypic manifestations.54
Intellectual disability
ID, defined by a quantitative measure (i.e., intellectual quotient (IQ)), is one of the most prevalent diagnoses among children, adolescents and adults with this CNV. Approximately half of the individuals with 22q11.2 hemizygosity have an IQ ≤70; the majority of the remainder are distributed between 71 and 100, with the overall average IQ near 70.33,35,41,55–60 Children with 22q11.2 deletions also exhibit declines in IQ as they grow.61 Although a majority of 22q11.2 deletion patients have lower performance IQ than verbal IQ, a sizable subpopulation shows the reverse pattern.59 Cognitive impairments, developmental delay and ID have been noted among 22q11.2 duplication cases.43,44,62–75 Screening of ID samples has revealed individuals with 22q11.2 deletions and duplications.53
Attention deficit hyperactivity disorder
ADHD is seen in children and adolescents with 22q11.2 deletion at rates ranging from 30 to 55%,19,33,41,58,76–83 and individuals with this deletion meet clinical diagnostic criteria at much higher rates during childhood than adolescence or adulthood.19 These are significantly elevated rates compared with ~10% in the general population. However, although 22q11.2 CNV has been identified in ADHD cases, its statistical enrichment compared with controls has not been significant in genome-wide searches.84,85
Anxiety
Various forms of anxiety disorders are seen in approximately half of the 22q11.2 hemizygous cases;83 5.6–29% of 22q11.2 hemizygous children, adolescents and adults are diagnosed with generalized anxiety disorder.15,18,19,33,78,81,83 Rates remain stable from childhood to adulthood.19 Rates for obsessive–compulsive disorder in children, adolescents and adults with 22q11.2 hemizygosity range from 9.7 to 32.6%.15,19,83 Various forms of phobias are also seen in this population.15,17,33,78
Depression
Rates of major depressive disorder in 22q11.2 hemizygous individuals of all ages range from 4 to 20%.16,18,19,76–78,80,82 However, the rates of mood disorders are generally higher in individuals with somatic disorders,86 and it remains unclear whether 22q11.2 deletion directly contributes to anxiety and depression.
WHY SO MANY DISORDERS AT THE GROUP BUT NOT AT THE INDIVIDUAL LEVEL?
As the same deletion sizes of 22q11.2 are associated with multiple diagnoses,87 the difference in deletion size does not adequately account for the diagnostic diversity. This begs the question of why the same-sized genetic variant is associated with so many clinically distinct disorders. One possibility is that common mechanisms are shared by many developmental neuropsychiatric disorders, meaning that these developmental neuropsychiatric disorders are not necessarily mechanistically distinct entities.
However, some individuals with the same deletions exhibit not all the disorders but a combination of some diagnoses. For example, in cases of 22q11.2 deletion, subgroups exist for codiagnosis of ASD and ID, ID and ADHD, ASD and ADHD or schizophrenia and ASD;41,54,88 therefore, there must exist factors that make each CNV manifests differently in different individuals.
It has been hypothesized that several sources of genetic background modifiers influence the impact of CNVs. Clinical features of a disorder may depend on the presence of an additional CNV, as shown in the case of 16p12.1 microdeletion.89 The rate of a second CNV hit is higher than that in asymptomatic controls among 16p11.2 distal duplication, 15q11.2 deletion and 17q12 duplication.53 However, rates of a secondary CNV do not differ from asymptomatic control cases for 16p13.11 duplication and deletion, 15q13.3 deletion, 1q21.1 deletion, 16p11.2 deletion, 1q21.1 duplication and 22q11.2 deletion and duplication.53,90 Alternatively or additionally, as a genome-wide set of common single-nucleotide polymorphisms (SNPs) could act as a determinant for disorders,91 such a factor might differentially modify phenotypic expression of a CNV in individuals.
QUANTITATIVE MEASURES OF TRAITS ASSOCIATED WITH DISORDERS
Intelligence, memory, attention and social cognition are not, by themselves, quantitative traits, but they can be treated as such when they are assessed using quantitative experimental tasks. CNV carriers are atypical for these traits in that they deviate from the population average. However, there is not a clear-cut boundary between a case and a control in these traits, and there is no solid basis on which to group individuals as ‘impaired’ or ‘abnormal’.
Many traits deviate from the population average during childhood before diagnosis of adulthood- and childhood-onset schizophrenia, including motor and speech development, academic scores, intelligence and sociability. The average score for social and cognitive tests in preschizophrenic individuals deviates slightly from that of controls,92–95 so that a larger proportion of preschizophrenic children have low-end scores compared with controls.94–97 Low IQ scores during childhood predict the likelihood of developing schizophrenia during adulthood but are also prevalent in unaffected siblings and those who develop affective disorders during adulthood.97 Thus, low scores for these traits do not provide an all-or-none predictor for a disorder but instead provide probabilistic (not categorical) predictors for future schizophrenic onset.
Scores of various cognitive tests and subtests tend to be lower than the population average in children with 22q11.2 hemizygosity. The manner in which 22q11.2 deletions affect IQ is not all or none, and it only lowers the average IQ to approximately 70.33,35,41,55–60 Longitudinal studies show that low verbal IQ during childhood is associated with the emergence of psychosis in adolescence and adulthood,98,99 suggesting that it is a predictor for the emergence of schizophrenia. Moreover, low scores on executive function tests during childhood best predict psychotic symptoms during adolescence.81 Among adults with 22q11.2 deletions, scores of tests designed to evaluate social cognition (i.e., theory of mind), motor skills and verbal learning tend to be lower in 22q11.2 deletion with schizophrenia than without this diagnosis.100
Working memory performance typically shows gradual, but steady, development from childhood (e.g., 6-year old) to adulthood (i.e., 20-year old) in typically developing individuals.101–108 This developmental maturation of working memory capacity is compromised in individuals with ASD.109 Children who later develop idiopathic schizophrenia exhibit stable low scores over time in cognitive tests such as working memory tasks.110–112 Children with idiopathic ADHD score lower in visual–spatial tests than those for auditory–verbal working memory.113 In individuals with idiopathic ID, scores for many forms of working memory capacity are consistently lower than in typically developing individuals throughout development.114 Similarly, children, adolescents and adults with 22q11.2 deletions score lower in various working memory tasks than controls.32,115–120
Attention is another cognitive domain in which test results are atypical in many developmental neuropsychiatric disorders.121 Diminished performance in cognitive flexibility, psychomotor speed and shifting of attention is common in children with ASD.122,123 Scores of subcomponent tests correlate with specific symptomatic elements within a disorder. For example, lower scores on attention tests are correlated with negative, but not positive, symptoms of schizophrenia.124 Children with 22q11.2 deletions score low in mental flexibility and visual attentional focus but not in tasks that test orienting or altering attention.34,36,125–127
Social motivation and skills are psychological processes thought to underlie idiopathic ASD.128 Scores on social cognition tests are lower for both ASD129,130 and schizophrenia131–135 patients than for controls. Compared with non-deletion controls, children with 22q11.2 deletions have more difficulties in understanding facial expression and are developmentally delayed in acquisition of social-cognitive skills.34,136 Average scores on tests designed to evaluate various aspects of social perception skills are lower in children with 22q11.2 deletions than in non-deletion controls.136 Individuals with 22q11.2 hemizygosity exhibit deficits in social motivation as well as social skills.137
Prepulse inhibition (PPI), in which a startle response evoked by a loud sound is attenuated by presentation of a preceding subthreshold sound, is lower in many psychiatric disorders, so it is a nonspecific research tool. For instance, lower-than-average PPI scores were reported in individuals with schizophrenia, obsessive–compulsive disorder, bipolar disorder, ADHD and Huntington’s disease,138 but lower PPI scores are not consistently seen in individuals with ASD.139–141 Children with 22q11.2 deletions score lower than controls on auditory PPI tests.142
IDENTIFYING SPECIFIC GENES WITHIN 22Q11.2 CNV
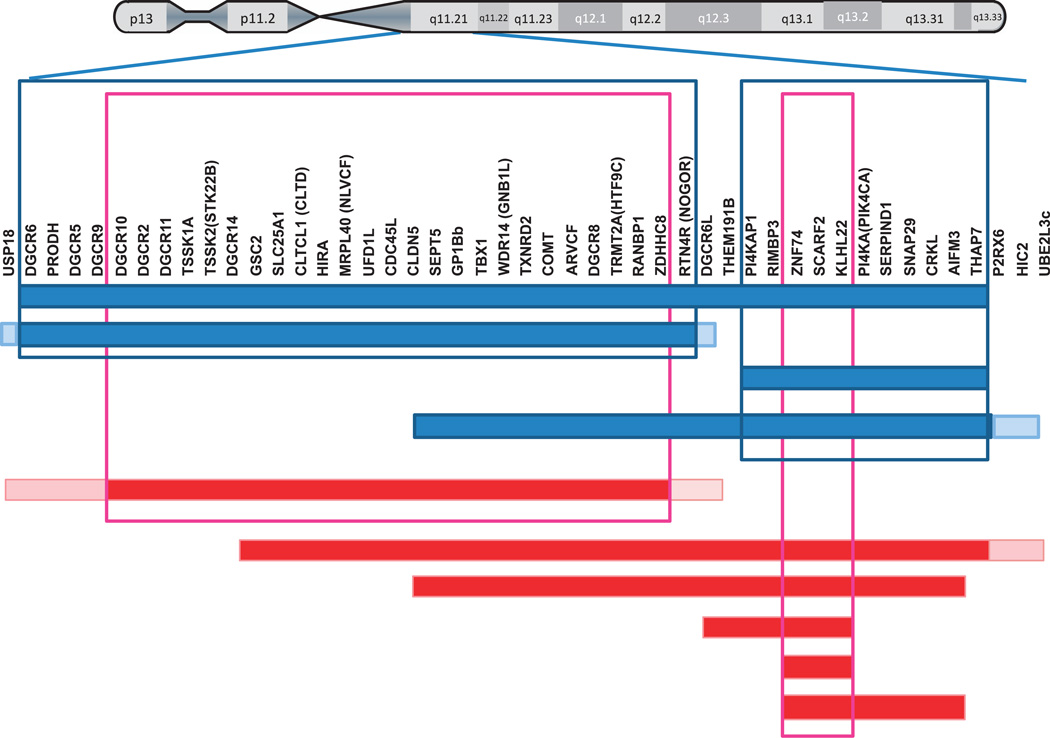
Approximately 90% of 22q11.2 deletions are ~3 Mb, and the remainder includes a nested ~1.5 or ~2Mb deletion or some atypical, non-nested distal deletions. Psychiatric diagnoses and their correlation with various sizes (1.5–3 Mb) of 22q11.2 deletions were previously examined77,87,143 (Figure 1). A potentially informative approach involves correlation of small deletions and various diagnoses. In a study by Michaelovsky et al.,87 the typical 3-Mb deletion was associated with schizophrenia, ADHD, major depressive disorder, anxiety disorders, obsessive–compulsive disorder and ASD. In smaller deletion (~1.5 and ~2.0 Mb) cases, disorders that are present at high frequencies (e.g., ID, ADHD and anxiety) are detected but disorders that occur in less than one-third of in 22q11.2 deletion cases are not (e.g., schizophrenia and ASDs). As pointed out by the authors, this might simply reflect small sample sizes: there were only four cases of ~1.5 Mb deletions and four cases of ~2Mb deletions. Although CNVs smaller than ~1.5Mb at 22q11.2 have been found in individuals with schizophrenia7,144 and in ASD samples,6,48–50 such small CNVs are few in number and are also found in control samples (see http://projects.tcag.ca/variation/?source=hg18).
Figure 1.
Schematic representation of 22q11.2 deletions and duplications in individuals with schizophrenia (blue bars) or ASD (red bars). Cases and boundaries are based on published studies with dense probes.6,7,26,48–51,87,143 The blue and pink frames indicate chromosomal segments that are commonly deleted in schizophrenia and ASD cases, respectively. A boundary with a pale color indicates individually variable ends.
Extremely rare cases of functional mutations at single 22q11.2 genes have been identified with specific clinical features. A single individual with ASD and a frameshift mutation of TBX1 that reduces expression of TBX1 has been identified in a family with multiple carriers.145 A single child with homozygous deletion confined to SEPT5 and GP1BB but not adjacent genes also exhibited social interaction deficits.146 As a mutation of GP1BB alone is characterized by prolonged bleeding and large platelets (i.e., Bernard–Soulier syndrome) but with no psychiatric diagnosis, Sept5 is a possible candidate gene for the behavioral phenotype in this individual. To date, there are no reports of other single 22q11.2 gene mutations associated with psychiatric disorders. Although these findings are suggestive, each mutation was reported in only a single subject. More cases are needed to ascertain the causative role of these mutations in developmental neuropsychiatric disorders. Moreover, as the ENCODE project has identified many more new transcripts at 22q11.2, the role of those additional genes in disorders should also be explored.
Attempts have been made to associate common variants of specific 22q11.2 genes in individuals with developmental neuropsychiatric disorders without 22q11.2 CNV. Results are inconsistent, and both negative and positive associations exist for PRODH,147,148 TBX1,149 catechol-O-methyltransferase (COMT),150–152 ZDHHC8,153 DGCR8154,155 and RTN4R.156–162 Although not many studies are available, reports to date regarding association of GNB1L SNPs with schizophrenia have all been positive.163–165 Comprehensive screenings of SNPs on 22q11.2 in individuals without 22q11.2 hemizygosity have identified association of SNPs on DGCR6, PRODH, ZDHHC8 (KIAA1292) and RTN4R (NOGO-R) with schizophrenia and childhood-onset schizophrenia.162,166 However, none of these common variants was found to be associated with schizophrenia4,167 or ASD3,168,169 in recent large-scale genome-wide association studies, confirming that SNPs on 22q11.2 genes are unlikely to contribute to risk for schizophrenia or ASD.
SNPs on the remaining copy of 22q11.2 hemizygous individuals have also been examined. Many studies consistently report that the Met/Val alleles of COMT are not associated with clinical diagnosis of schizophrenia or schizotypy, depressive disorders or anxiety disorders among adults with 22q11.2 hemizygosity16,170–172 except for one report where the Val and Met alleles were associated with the presence and absence, respectively, of schizophrenia.173 In children and adolescents with 22q11.2 hemizygosity, the Met allele is associated with the incidence of prodromal psychotic symptoms during adolescence in one study98 but not in another.81
Overall, identification of small critical segments or SNPs on single genes within 22q11.2 in humans has not been conclusive. Moreover, it is difficult to know whether weak association of an SNP with a disorder reflects a weak impact of an SNP on protein function or a weak contribution of the protein to specific disorders. As hemizygosity of 22q11.2 is sufficient to cause phenotypes, SNPs on the remaining copy would indicate how such variants modify the phenotype caused by hemizygosity but would not identify single genes whose dominant effect causes the phenotype.
MOUSE MODELS OF DEVELOPMENTAL NEUROPSYCHIATRIC DISORDERS
Mouse models are essential to experimentally evaluate how specific genes causally relate to phenotypes beyond observing correlations in humans. There are three criteria to consider when judging whether a mouse model is or is not valid: construct validity, predictive validity and face validity.
A genetically generated CNV or dose alteration of encoded individual genes satisfies the construct validity of mouse models in that the human genetic alteration is recapitulated in a mouse.
Predictive validity is difficult to satisfy in mouse models, because therapeutic drugs do not provide disorder specificity. Hyman174 pointed out that therapeutic drugs do not respect boundaries defined by the Diagnostic and Statistical Manual of Mental Disorders (DSM). For instance, antidepressants are an effective treatment for depression, anxiety disorders and obsessive–compulsive disorders. Moreover, drugs often do not comprehensively affect all symptomatic elements of a clinically defined disorder. Typical neuroleptics attenuate positive symptoms of schizophrenia (e.g., delusions and hallucinations, disorganized thought and speech, and disorganized behavior) but do not have much effect on negative symptoms (e.g., affective flattening, avolition and social withdrawal) or cognitive impairments (e.g., poor executive functioning, attention and memory problems). Atypical neuroleptics improve the positive and negative symptoms of schizophrenia, but cognitive impairments are resistant to these drugs.
Use of face validity (i.e., phenomenological similarity) in judging a mouse model is equally misguided. Certainly, for some disorders, it is currently not possible to mimic some human symptoms; for example, there is no rodent model that mimics human delusion or hallucination. For other symptomatic elements, humans and mice use different species-specific modes.175 For instance, humans use visual and auditory senses for social interaction, whereas rodents heavily rely on olfactory cues. From an evolutionary perspective, species-specific manifestation of certain functions is still informative to understanding biology across species.175 In cases where some but not all symptomatic elements are exhibited in a single gene deletion mouse model, this is often interpreted as a reason to invalidate face validity. However, as it is questionable to assume that any gene should mechanistically contribute to all symptomatic elements of a given disorder, it is not necessary to attempt achievement of maximal phenotypic similarity in a single mouse model.
GENETIC MOUSE MODELS OF CNVS
Many mice developed to carry large deletions or duplications in murine chromosomes orthologous to human CNVs, including CNVs at 15q11.2-q13.1,176,177 16p11.2,178 17p11.2179,180 and 22q11.2,181–184 do indeed exhibit behavioral phenotypes related to developmental neuropsychiatric disorders. CNV at 22q11.2 has been most extensively and systematically studied in mouse models, and identification of small segments and individual genes is unparalleled compared with other CNVs.
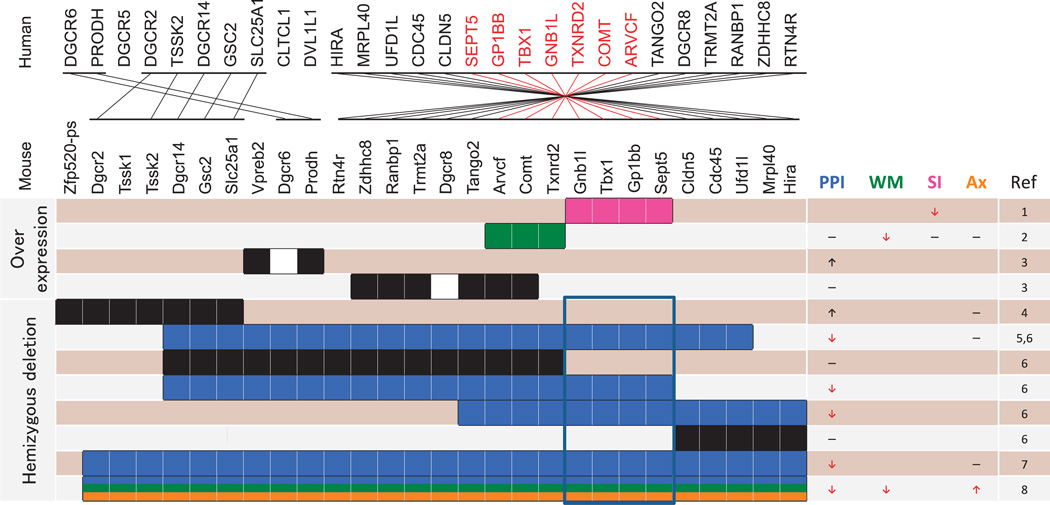
Systematic alteration of a copy number of small segments within 22q11.2 turned out to be a highly effective approach. Both segmental overexpression and hemizygosity at this locus have been modeled in mice (Figure 2). Hiroi et al.183 reported that overexpression of a 200-kb segment of human 22q11.2 that included TBX1, GP1BB, SEPT5 and GNB1L causes social behavioral deficits as well as repetitive hyperactivity and its spontaneous exacerbation, the latter of which was attenuated by chronic treatment with the antipsychotic drug clozapine.
Figure 2.
Organization and location of corresponding genes between human 22q11.2 and murine 16. Mouse models of segmental overexpression and deletions are shown. Purple (social interaction), green (working memory), blue (PPI) and orange (anxiety) bars represent chromosomal segments whose murine copy number variation results in phenotypes consistent (see red arrows) and inconsistent (see black arrows) with 22q11.2 CNVs in humans or no effect (see black horizontal lines). The blue frame represents a segment whose deletion is commonly seen in mouse models with a PPI phenotype consistent with that in humans. PPI, prepulse inhibition; WM, working memory; SI, social interaction; Anx, anxiety. (1) Hiroi et al.;183 (2) Suzuki et al.;184 (3) Stark et al.;185 (4) Kimber et al.;195 (5) Paylor et al.;182 (6) Paylor et al.;145 (7) Long et al.;181 (8) Stark et al.196
Overexpression of an adjacent ~190 kb segment of human 22q11.2 that included TXNRD2, COMT and ARVCF produced a unique set of phenotypes in a BAC transgenic mouse.184 This mouse exhibited lower scores for the rewarded alternation task, an index of working memory. In this task, wild-type (WT) mice showed spontaneous improvement from 1 month to 2 months of age.184 This age-dependent improvement in working memory performance was not due to a carryover effect from previous learning because separate sets of mice were tested at 1 month and 2 months of age. Remarkably, the BAC transgenic mouse did not show such developmental maturation in working memory performance, in which they exhibited lower levels of working memory at 2 months, but not at 1 month, of age than WT mice.184 This phenotype was highly selective; BAC transgenic and WT mice were indistinguishable in results of testing for PPI, social interaction or motor activity.
Stark et al.185 demonstrated that overexpression of a 22q11.2 segment containing Zdhhc8, Ranbp1, Trmt2a (Htf9c), Tango2 (T10), Arvcf and Comt had no effect on PPI; overexpression of another (Vpreb2 and Prodh) instead increased PPI. Relevance of the high PPI level to human phenotypes remains unclear at this point, as PPI in 22q11.2 duplication cases has not been reported.
Taken together, these overexpression studies identified the ~200-kb region and the adjacent ~190-kb region, collectively termed a murine critical region, as causative for distinct sets of behavioral phenotypes relevant to developmental neuropsychiatric disorders.
Paylor et al.145 reported an elegant study that examined the impact of variously sized deletions of partially overlapping segments within murine chromosome 16, the ortholog of human 22q11.2, and found that PPI was reduced only when large deletions encompass a commonly affected segment (Figure 2), which turned out to be the ~200-kb portion of the murine critical region.183
Several genes encoded in the murine critical region were individually examined (Figure 3). Heterozygosity of either Tbx1 or Gnb1l alone lowered auditory PPI levels in non-congenic mice in one study,145 but another study with a different non-congenic mouse line showed that Tbx1 deficiency had no effect on auditory PPI.181
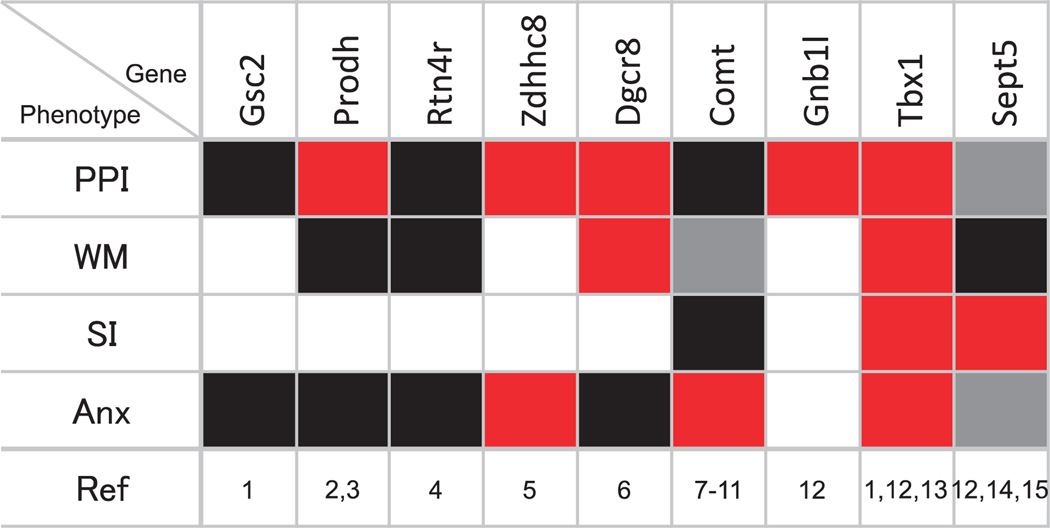
Figure 3.
Mosaic pattern of phenotypes caused by deletion of specific genes in mouse models. Phenotypes consistent (red) and inconsistent (gray) with what is seen in 22q11.2 hemizygous patients are shown; black squares represent cases where a gene deletion caused no effect. Blank squares represent cases where phenotyping was not conducted. PPI, prepulse inhibition; WM, working memory; SI, social interaction; Anx, anxiety. (1) Long et al.;181 (2) Gogos et al.;202 (3) Paterlini et al.;203 (4) Hsu et al.;161 (5) Mukai et al.;197 (6) Stark et al.;196 (7) Gogos et al.;191 (8) Papaleo et al.;190 (9) Babovic et al.;194 (10) O’Tuathaigh et al.;193 (11) O’Tuathaigh et al.;192 (12) Paylor et al.;145 (13) Hiramoto et al.;186 (14) Suzuki et al.;188 (15) Harper et al.187
Hiramoto et al.186 also demonstrated that heterozygosity of Tbx1 lowered reciprocal social interaction and communication and increased anxiety-related behavior (i.e., thigmotaxis) and tendency to repetitive behavior in congenic mice. Moreover, congenic Tbx1 heterozygous (HT) mice exhibited lower levels of working memory performance and heightened levels of interaction with a non-mouse object; there was no change in locomotor activity.186
As an example where not all symptomatic elements are recapitulated in one gene manipulation, Sept5 deficiency lowered reciprocal social interaction but, contrary to 22q11.2 hemizygosity in humans, increased levels of auditory PPI, reduced levels of anxiety-related behavior and had no effect on locomotor activity, working memory or repetitive behavioral tendency in both congenic and non-congenic mice.187,188 Moreover, the phenotypic expression of Sept5 deficiency is attenuated or amplified when genetic background is systematically altered.187–189 As virally guided overexpression of Sept5 alone against a coisogenic genetic background is sufficient to raise social interaction,187 phenotypic differences between WT and Sept5-deficient mice do not simply reflect the collective impact of alleles other than Sept5.
Overexpression of COMT reduced performance on a working memory task184,190 and might be relevant to phenotypes of 22q11.2 duplication. However, deletion of Comt had no effect on PPI190–192 or sociability192–194 and increased working memory performance,190,193,194 whereas humans with 22q11.2 deletion show low levels of PPI,142 social behavior33,37–42 and working memory.120
Although a large hemizygous deletion outside the murine critical region has no effect on PPI145,195 (Figure 2), genes encoded outside the critical region nevertheless might have a weak impact on specific phenotypes. Non-congenic Dgcr8 HT mice exhibited lower levels of working memory and lower PPI levels than WT mice at 74 and 78 dB but not at 82 or 86 dB prepulse levels.196 Non-congenic Zdhhc8-knockout, but not HT, mice exhibited lower PPI levels at 78 dB but not at 82 dB prepulse intensities.197 Given that a large deletion including Dgcr8 and Zdhhc8 has no effect on PPI,145 these partial effects might reflect the impact of unequal alleles (other than the targeted genes) in genetic background of non-congenic mice or other nongenetic factors.198–201 PPI was reduced in non-congenic Prodh-deficient mice in one study;202 however, no PPI deficit was seen in congenic Prodh-deficient mice with 129/SvEv background by the same group.203 Deficiency of Rtn4r161 and Gsc2181 did not affect PPI. Working memory capacity was unaffected by Prodh203 or Rtn4r.161 Anxiety-related behaviors were heightened by deficiency of Zdhhc8197 but not of Gsc2,181 Prodh,202 Rtn4r161 or Dgcr8.196
One important caveat to consider these results is that deletion of two copies (i.e., homozygous deletion) and one copy (i.e., HT deletion) in mice should not be equated with homozygous and hemizygous deletions in humans. Although, in some cases, heterozygosity was not and homozygosity was sufficient to cause a specific phenotype in mice (e.g., Sept5 and Zdhhc8),188,197 the mechanism by which gene-dose alteration causes a phenotype depends on genetic background,187–189 which is not identical in humans and mice.
GENERAL HYPOTHETICAL RULES OF 22Q11.2 CNV–PHENOTYPE RELATION
Mouse behaviors do not necessarily directly address clinically defined disorders. Nonetheless, some general rules governing genotype–phenotype relationships can be extrapolated from mouse studies involving 22q11.2 CNVs (Figure 3). We present these rules as testable hypotheses that can be used to guide analyses of mouse models of this and other CNVs.
Hypothesis 1: noncontiguous gene effect
Dose alteration of a unique set of noncontiguous critical genes causes a phenotype in that dose alteration of other interdigitated genes has no effect on, or even opposes the phenotype.
Mouse studies present a far more complex gene–phenotype relation than expected from a contiguous gene syndrome where a contiguous set of genes contributes to a specific phenotype. This is illustrated for PPI, working memory and anxiety (Figure 3). For any given phenotype, genes whose deficiency results in an expected effect are not continuously distributed; they are intercepted by genes whose deficiency has no apparent or even opposing effect. Moreover, nonidentical sets of genes seem to contribute to different phenotypes. The question remains whether 22q11.2 CNV is still considered a contiguous gene syndrome if all phenotypes are lumped as a syndrome, underscoring the necessity to analyze more than one behavioral phenotype.
It should be noted that the genes listed here have nonidentical anatomical distributions and neuronal functions and, thus, the ways they contribute to behavioral phenotypes are likely diverse. Some genes might have more robust impacts on phenotypes than others, partly because specific neuronal processes, in which each of these genes plays a role, are differentially involved in behavioral phenotypes.
An obvious implication of this hypothesis is that given that there are genes within 22q11.2 whose deficiency has no apparent or even an opposing role in behavioral phenotypes, neuronal, cellular and molecular phenotypes seen in mice with hemizygosity of a large segment or single gene deletions might not necessarily be relevant to behavioral phenotypes, not to mention clinical diagnoses.
Hypothesis 2: gene-dose alteration
The dose of each gene has an optimal range on a linear or nonlinear function for a given phenotype.
Viewed from a simplistic mechanistic perspective, a reduction and an increase in a gene dose would be expected to cause opposite effects. In fact, COMT overexpression and deletion, respectively, lowers and raises levels of working memory-dependent performance.184,190,193,194 Sept5 deficiency and overexpression, respectively, reduces and raises levels of social interaction.187,188 However, given that individuals with 22q11.2 duplications and deletions exhibit similar cognitive atypicality, some genes might induce the same phenotypes with gene-dose deviation in either direction from the optimal level.
Hypothesis 3: pleiotropy
Some, but not all, critical genes have more than one phenotypic target.
Tbx1 heterozygosity, for example, causes phenotypes in social interaction and communication, repetitive behavior tendency and working memory186 (Figure 3). Deletion of Zdhhc8 affects both PPI and anxiety-related behavior,197 and deletion of Dgcr8 lowers scores for PPI and working memory.196 Moreover, various genes within this CNV do not seem to necessarily target the identical set of phenotypes. A corollary of this hypothesis is that the pleiotropic actions of multiple genes within this CNV cause multiple diagnoses.
Hypothesis 4: mass action
The ultimate phenotype of a CNV reflects the impacts of additive and opposing impacts of many genes.
Sept5 deficiency increases PPI and reduces levels of anxiety-related behavior188 (Figure 3). Similarly, Comt deficiency enhances performance levels of working memory (Figure 3). In individuals with 22q11.2 deletions, heightened levels of anxiety15,18,19,33,78,81,83 and lower levels of performance of PPI142 and working memory32,115–120 are seen. Given that Sept5 and Comt are nevertheless deleted in 22q11.2 deletions in humans, their effects are likely overridden by the effects of other genes. These opposing effects and facilitating effects of other genes are likely to sum to the ultimate phenotype.
Hypothesis 5: phenotypic variation
Genetic background and environmental factors modulate phenotypic expression of a CNV.
Genetic background, including common variants throughout the entire genome, might modulate phenotypic expression of a gene deficiency. We demonstrated this by systematically altering genetic background in the setting of Sept5 deficiency.187–189 This modulatory effect of genetic background provides a plausible explanation for the seemingly inconsistent observations that deletion of Prodh or Tbx1 causes PPI deficit in one but not another mouse line with different genetic backgrounds.145,181,202,203 This hypothesis underscores the necessity to analyze mice under several genetic backgrounds.187–189
Environmental factors also modulate phenotypic expression. In mice, a housing condition known to increase stress levels reduced Sept5 protein levels and decreased social interaction.187
This hypothesis is consistent with the observation that CNVs at 22q11.2 are associated with many developmental neuropsychiatric disorders when examined in a large group of patients overall, but individuals with the same CNV exhibit only one or a few clinical diagnoses. If the impact of a CNV or a gene alteration is variably modified by stochastic processes, genetic background and environmental factors, the ultimate summation of various phenotypes would be expected to individually differ, perhaps resulting in variable penetrance and expressivity of many disorders among 22q11.2 CNV carriers.
Hypothesis 6: phenotype beyond average
The impact of 22q11.2 gene alteration is to shift data distribution with or without a change in variance.
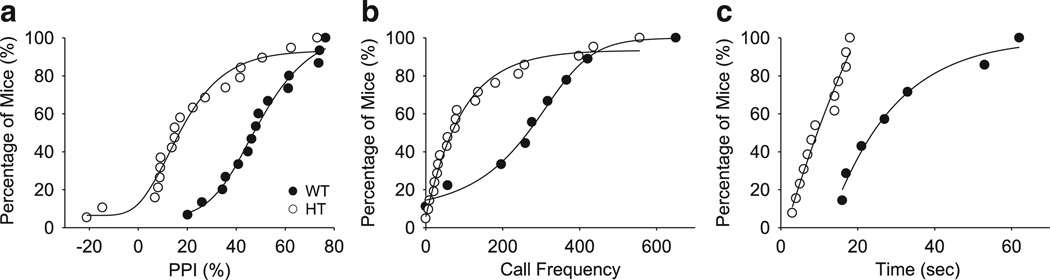
Comparison of averages between a WT and a genetically modified group is the standard practice for evaluating impact of a gene manipulation. However, gene manipulation could alter the data structure in various ways. Even with the same gene deletion, the effect on various phenotypes may not be identical. For example, in our own data, Tbx1 heterozygosity produced a large shift in PPI scores from WT to HT mice so that a sizable portion of HT mice fell below the lowest score of WT mice (Figure 4a). For vocal call frequencies, this shift was not robust, so the results for WT and HT mice are distributed within the same range but the average of HT shifted to the left (Figure 4b). Another type of data restructuring included alteration of data variance and an average shift; the duration of social interaction in HT mice was more tightly clustered below the lowest data point for WT mice (Figure 4c).
Figure 4.
Shifts in data structure of various phenotypes caused by Tbx1 heterozygosity. Figures are based on raw data of our published study.186 Black circles represent WT mice and open circles represent HT mice for the three graphs. (a) For PPI, Tbx1 heterozygosity shifts data lower so that the cumulative data curve shows a parallel shift to the left, with many HT cases below the lowest data point of WT mice. (b) Vocal calls: Tbx1 heterozygosity shifts the average of data slightly to the left, but data largely overlap between WT and HT mice. (c) Social interaction: Tbx1 heterozygosity shifts the data distribution of WT to the left, with a minimal overlap between HT and WT mice. Note that variance is also reduced in HT mice.
Hypothesis 7: developmental trajectory
Gene alteration causes a phenotype at specific developmental stages when the gene function is required.
We demonstrated that overexpression of the ~190 kb region, including COMT, lowers performance of working memory at 2 months of age, but not at 1 month of age. This observation highlights the need to evaluate phenotypes at several developmental time points. Moreover, this hypothesis is consistent with observations in humans that low scores of working memory capacity start to appear at and after adolescence in ASD patients109 and that the high-activity COMT allele is associated with poor visual–spatial working memory after, but not before, 10 years of age compared with the low-activity COMT allele.104
The seven hypotheses outlined above are derived from mouse models of 22q11.2 CNV. Given that some distinct mechanisms are likely to be in play for different CNVs,53 these hypotheses might require revision for other CNVs. Concerning the testability of these hypotheses in humans, unless more single gene deletion cases are discovered, a critical evaluation of hypotheses 1, 2, 3 and 4, where the role of specific genes is concerned, can only be tested in genetically engineered mice. Hypotheses 5, 6 and 7 can be tested in humans insofar as it is assumed that genetic variants are a determinant for symptomatic elements of developmental neuropsychiatric disorders; there is no reason to doubt that these hypothesized gene–phenotype mechanisms exist in humans as well. The ultimate test of mouse-derived hypotheses would be to evaluate the effectiveness of therapeutic options, derived from these hypothetical mechanisms, in humans.
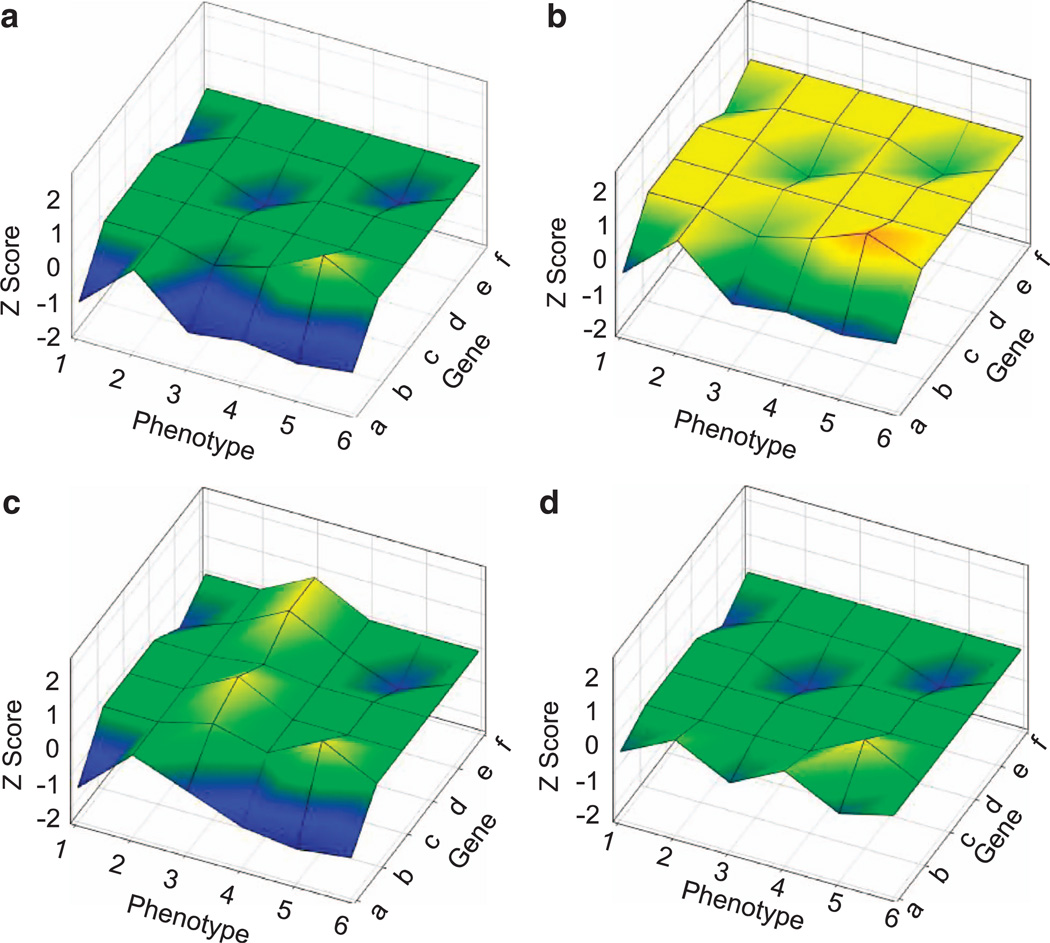
Based on these hypotheses, we propose a quantitative model to explain how deficiency of specific genes encoded in a CNV affects behavioral phenotypes (Figure 5). According to this model, when gene dose is altered, some genes (e.g., gene a) affect more phenotypes more severely than others (genes b, d and e); other genes (c and f) have no impact on phenotypes (green flat plane) or have an effect opposite to that of other genes (yellowish upward protrusion at gene b and phenotype 5, Figure 5a). The ultimate phenotype score is determined by the net effects of all phenotypic deviation of genes or predominantly by genes that have major impacts on a phenotype. Alleles in the genetic background, environmental factors (in some cases, via epigenetic alterations) or both could shift the scores of phenotypes evenly across genes and phenotypes so that phenotypic deviation (blue in the lower plane) from the averages disappears or becomes less severe (Figure 5b). Alternatively, genetic background and/or environmental factors could unevenly affect specific phenotypes (Figure 5c; e.g., phenotype 3) or the phenotypic impacts of specific genes (Figure 5d; e.g., gene a). Any combination of these effects could shift the score plane evenly or tilt it unevenly upward or downward, resulting in unique phenotype sets in individuals with varying genetic backgrounds and environmental influences.
Figure 5.
Hypothetical genotype–phenotype relation. Three dimensions indicate genes, phenotypes and Z-scores of phenotypic expression. The yellow–green zones indicate average scores exhibited in organisms at a normal gene dose. Lower (blue) and higher (red) Z-scores indicate more severe phenotypic deviation. The plane expands as more genes are involved in a CNV and more phenotypes are affected. (a) A hypothetical impact of a CNV on phenotypes. (b) Genetic background and/or environmental factors evenly shift the impact of a CNV on all phenotypes, compared to panel a. (c) Genetic background and/or environmental factors selectively shift the impact of a CNV on a specific phenotype (see phenotype 3), compared to panel a. (d) Genetic background and/or environmental factors selectively shift the impact of a specific gene (see gene a), compared to panel a. Not depicted here is a developmental trajectory along which a gene-dose alteration starts to affect a phenotypic score.
DOES DNA ‘READ’ THE DSM?
The hypothesized mechanisms, based on mouse studies of 22q11.2 CNV, dictate that alterations of a distinct set of multiple, noncontiguous genes encoded in this CNV unequally shift the probabilities of various phenotypes, through modulatory impacts of genetic background and environmental factors, along a predetermined developmental trajectory.
Historically, the nosology of neuropsychiatric disorders, exemplified by those included in the DSM, has evolved largely based on prevailing opinions about how to categorize symptoms and what to consider the primary component of a disorder;204 this trend has continued because of clinical reliability. However, clinical diagnoses based on symptomatic clustering have contributed to many unexplained by-products, such as many comorbid traits and failure to find statistically significant and reproducible genetic associations.174,204
Our overall hypothesis predicts that genes do not necessarily follow the logic by which disorders are clinically categorized. In a sense, the DNA does not ‘read’ the DSM from chapter to chapter; instead, it ‘reads’ the DSM from the index section that ignores the clinical boundaries of developmental neuropsychiatric disorders.
Dimensional measures of neurophysiological, biochemical, endocrinological, neuroanatomical, cognitive or neuropsychological traits have been proposed as a reliable alternative that may better approximate genetic mechanisms in the hope to establish stronger association with gene variants.205 Mouse and human studies of 22q11.2 CNV have reinforced the validity of a quantitative, dimensional approach. However, although a quantitative deviation from the average for each trait is associated with a disorder, such trait atypicality does not predict any clinically defined disorder with sufficient specificity. Is it because we have not devised a quantitative task that adequately taps aspects of perception, attention, memory and social cognition that are so specifically affected in clinically defined disorders? We are left with this question, but instead of initiating a debate, perhaps a more practical approach is to explore mechanisms of causation for quantitative traits and evaluate how therapeutic options devised for such traits affect symptoms of clinically defined developmental neuropsychiatric disorders. Such an approach is likely to provide not only mechanism-based effective therapeutic options but also new insights into the relationship between quantitative traits and symptoms.
Twenty years of 22q11.2 CNV research has provided hypothetical rules and lessons that serve as a guiding torch for a path to psychiatry’s promised land.
ACKNOWLEDGEMENTS
We thank Drs Herb Lachman, Santhosh Girirajan and Edward Brodkin for their invaluable comments on an early draft of this paper. This work was supported by the NIH (R21HD05311 and R01MH099660), NARSAD Independent Investigator Award and the Maltz Foundation to NH; funds from the Ministry of Defense, Japan, to TT; the Uehara fellowship and a Senshin Medical Research Foundation fellowship, Japan, to SB; funds from Kobe University Graduate School of Medicine to AH; and Grants-in-Aid for Scientific Research (24591674) from the Society for Promotion of Science, Japan, to TI.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Jacobsen LK, Rapoport JL. Research update: childhood-onset schizophrenia: implications of clinical and neurobiological research. J Child Psychol Psychiatry. 1998;39:101–113. [PubMed] [Google Scholar]
- 2.McKenna K, Gordon CT, Lenane M, Kaysen D, Fahey K, Rapoport JL. Looking for childhood-onset schizophrenia: the first 71 cases screened. J Am Acad Child Adolesc Psychiatry. 1994;33:636–644. doi: 10.1097/00004583-199406000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Anney R, Klei L, Pinto D, Almeida J, Bacchelli E, Baird G, et al. Individual common variants exert weak effects on the risk for autism spectrum disorders. Hum Mol Genet. 2012;21:4781–4792. doi: 10.1093/hmg/dds301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stergiakouli E, Hamshere M, Holmans P, Langley K, Zaharieva I, Hawi Z, et al. Investigating the contribution of common genetic variants to the risk and pathogenesis of ADHD. Am J Psychiatry. 2012;169:186–194. doi: 10.1176/appi.ajp.2011.11040551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shprintzen RJ, Goldberg R, Golding-Kushner KJ, Marion RW. Late-onset psychosis in the velo-cardio-facial syndrome. Am J Med Genet. 1992;42:141–142. doi: 10.1002/ajmg.1320420131. [DOI] [PubMed] [Google Scholar]
- 9.Driscoll DA, Spinner NB, Budarf ML, Donald-McGinn DM, Zackai EH, Goldberg RB, et al. Deletions and microdeletions of 22q11.2 in velo-cardio-facial syndrome. Am J Med Genet. 1992;44:261–268. doi: 10.1002/ajmg.1320440237. [DOI] [PubMed] [Google Scholar]
- 10.Driscoll DA, Budarf ML, Emanuel BS. A genetic etiology for DiGeorge syndrome: consistent deletions and microdeletions of 22q11. Am J Hum Genet. 1992;50:924–933. [PMC free article] [PubMed] [Google Scholar]
- 11.Scambler PJ, Kelly D, Lindsay E, Williamson R, Goldberg R, Shprintzen R, et al. Velo-cardio-facial syndrome associated with chromosome 22 deletions encompassing the DiGeorge locus. Lancet. 1992;339:1138–1139. doi: 10.1016/0140-6736(92)90734-k. [DOI] [PubMed] [Google Scholar]
- 12.Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Debbane M, Glaser B, David MK, Feinstein C, Eliez S. Psychotic symptoms in children and adolescents with 22q11.2 deletion syndrome: neuropsychological and behavioral implications. Schizophr Res. 2006;84:187–193. doi: 10.1016/j.schres.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 15.Gothelf D, Presburger G, Zohar AH, Burg M, Nahmani A, Frydman M, et al. Obsessive–compulsive disorder in patients with velocardiofacial (22q11 deletion) syndrome. Am J Med Genet B Neuropsychiatr Genet. 2004;126:99–105. doi: 10.1002/ajmg.b.20124. [DOI] [PubMed] [Google Scholar]
- 16.Murphy KC, Jones LA, Owen MJ. High rates of schizophrenia in adults with velocardio- facial syndrome. Arch Gen Psychiatry. 1999;56:940–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- 17.Pulver AE, Nestadt G, Goldberg R, Shprintzen RJ, Lamacz M, Wolyniec PS, et al. Psychotic illness in patients diagnosed with velo-cardio-facial syndrome and their relatives. J Nerv Ment Dis. 1994;182:476–478. doi: 10.1097/00005053-199408000-00010. [DOI] [PubMed] [Google Scholar]
- 18.Fung WL, McEvilly R, Fong J, Silversides C, Chow E, Bassett A. Elevated prevalence of generalized anxiety disorder in adults with 22q11.2 deletion syndrome. Am J Psychiatry. 2010;167:998. doi: 10.1176/appi.ajp.2010.09101463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green T, Gothelf D, Glaser B, Debbane M, Frisch A, Kotler M, et al. Psychiatric disorders and intellectual functioning throughout development in velocardiofacial (22q11.2 deletion) syndrome. J Am Acad Child Adolesc Psychiatry. 2009;48:1060–1068. doi: 10.1097/CHI.0b013e3181b76683. [DOI] [PubMed] [Google Scholar]
- 20.Bassett AS, Chow EW. Schizophrenia and 22q11.2 deletion syndrome. Curr Psychiatry Rep. 2008;10:148–157. doi: 10.1007/s11920-008-0026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LevinsonDF,Duan J, Oh S, Wang K, Sanders AR, Shi J, et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan W, Jacobsen LK, Krasnewich DM, Guan XY, Lenane MC, Paul SP, et al. Chromosome 22q11.2 interstitial deletions among childhood-onset schizophrenics and ‘multidimensionally impaired’. Am J Med Genet. 1998;81:41–43. [PubMed] [Google Scholar]
- 24.Usiskin SI, Nicolson R, Krasnewich DM, Yan W, Lenane M, Wudarsky M, et al. Velocardiofacial syndrome in childhood-onset schizophrenia. J Am Acad Child Adolesc Psychiatry. 1999;38:1536–1543. doi: 10.1097/00004583-199912000-00015. [DOI] [PubMed] [Google Scholar]
- 25.Ivanov D, Kirov G, Norton N, Williams HJ, Williams NM, Nikolov I, et al. Chromosome 22q11 deletions, velo-cardio-facial syndrome and early-onset psychosis. Molecular genetic study. Br J Psychiatry. 2003;183:409–413. doi: 10.1192/bjp.183.5.409. [DOI] [PubMed] [Google Scholar]
- 26.Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guilmatre A, Dubourg C, Mosca AL, Legallic S, Goldenberg A, Drouin-Garraud V, et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66:947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arinami T, Ohtsuki T, Takase K, Shimizu H, Yoshikawa T, Horigome H, et al. Screening for 22q11 deletions in a schizophrenia population. Schizophr Res. 2001;52:167–170. doi: 10.1016/s0920-9964(00)00192-4. [DOI] [PubMed] [Google Scholar]
- 29.Baker KD, Skuse DH. Adolescents and young adults with 22q11 deletion syndrome: psychopathology in an at-risk group. Br J Psychiatry. 2005;186:115–120. doi: 10.1192/bjp.186.2.115. [DOI] [PubMed] [Google Scholar]
- 30.Golding-Kushner KJ, Weller G, Shprintzen RJ. Velo-cardio-facial syndrome: language and psychological profiles. J Craniofac Genet Dev Biol. 1985;5:259–266. [PubMed] [Google Scholar]
- 31.Heineman-de Boer JA, Van Haelst MJ, Cordia-de HM, Beemer FA. Behavior problems and personality aspects of 40 children with velo-cardio-facial syndrome. Genet Couns. 1999;10:89–93. [PubMed] [Google Scholar]
- 32.Kiley-Brabeck K, Sobin C. Social skills and executive function deficits in children with the 22q11 deletion syndrome. Appl Neuropsychol. 2006;13:258–268. doi: 10.1207/s15324826an1304_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niklasson L, Rasmussen P, Oskarsdottir S, Gillberg C. Chromosome 22q11 deletion syndrome (CATCH 22): neuropsychiatric and neuropsychological aspects. Dev Med Child Neurol. 2002;44:44–50. doi: 10.1017/s0012162201001645. [DOI] [PubMed] [Google Scholar]
- 34.Shashi V, Veerapandiyan A, Schoch K, Kwapil T, Keshavan M, Ip E, et al. Social skills and associated psychopathology in children with chromosome 22q11.2 deletion syndrome: implications for interventions. J Intellect Disabil Res. 2012;56:865–878. doi: 10.1111/j.1365-2788.2011.01477.x. [DOI] [PubMed] [Google Scholar]
- 35.Swillen A, Devriendt K, Legius E, Eyskens B, Dumoulin M, Gewillig M, et al. Intelligence and psychosocial adjustment in velocardiofacial syndrome: a study of 37 children and adolescents with VCFS. J Med Genet. 1997;34:453–458. doi: 10.1136/jmg.34.6.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woodin M, Wang PP, Aleman D, Donald-McGinn D, Zackai E, Moss E. Neuropsychological profile of children and adolescents with the 22q11.2 microdeletion. Genet Med. 2001;3:34–39. doi: 10.1097/00125817-200101000-00008. [DOI] [PubMed] [Google Scholar]
- 37.Antshel KM, Aneja A, Strunge L, Peebles J, Fremont WP, Stallone K, et al. Autistic spectrum disorders in velo-cardio facial syndrome (22q11.2 deletion) J Autism Dev Disord. 2007;37:1776–1786. doi: 10.1007/s10803-006-0308-6. [DOI] [PubMed] [Google Scholar]
- 38.Esterberg ML, Ousley OY, Cubells JF, Walker EF. Prodromal and autistic symptoms in schizotypal personality disorder and 22q11.2 deletion syndrome. J Abnorm Psychol. 2013;122:238–249. doi: 10.1037/a0028373. [DOI] [PubMed] [Google Scholar]
- 39.Fine SE, Weissman A, Gerdes M, Pinto-Martin J, Zackai EH, Donald-McGinn DM, et al. Autism spectrum disorders and symptoms in children with molecularly confirmed 22q11.2 deletion syndrome. J Autism Dev Disord. 2005;35:461–473. doi: 10.1007/s10803-005-5036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kates WR, Antshel KM, Fremont WP, Shprintzen RJ, Strunge LA, Burnette CP, et al. Comparing phenotypes in patients with idiopathic autism to patients with velocardiofacial syndrome (22q11 DS) with and without autism. Am J Med Genet A. 2007;143A:2642–2650. doi: 10.1002/ajmg.a.32012. [DOI] [PubMed] [Google Scholar]
- 41.Niklasson L, Rasmussen P, Oskarsdottir S, Gillberg C. Autism, ADHD, mental retardation and behavior problems in 100 individuals with 22q11 deletion syndrome. Res Dev Disabil. 2009;30:763–773. doi: 10.1016/j.ridd.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 42.Vorstman JA, Morcus ME, Duijff SN, Klaassen PW, Heineman-de Boer JA, Beemer FA, et al. The 22q11.2 deletion in children: high rate of autistic disorders and early onset of psychotic symptoms. J Am Acad Child Adolesc Psychiatry. 2006;45:1104–1113. doi: 10.1097/01.chi.0000228131.56956.c1. [DOI] [PubMed] [Google Scholar]
- 43.Mukaddes NM, Herguner S. Autistic disorder and 22q11.2 duplication. World J Biol Psychiatry. 2007;8:127–130. doi: 10.1080/15622970601026701. [DOI] [PubMed] [Google Scholar]
- 44.Ramelli GP, Silacci C, Ferrarini A, Cattaneo C, Visconti P, Pescia G. Microduplication 22q11.2 in a child with autism spectrum disorder: clinical and genetic study. Dev Med Child Neurol. 2008;50:953–955. doi: 10.1111/j.1469-8749.2008.03048.x. [DOI] [PubMed] [Google Scholar]
- 45.Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, Sonnenblick LI, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cai G, Edelmann L, Goldsmith JE, Cohen N, Nakamine A, Reichert JG, et al. Multiplex ligation-dependent probe amplification for genetic screening in autism spectrum disorders: efficient identification of known microduplications and identification of a novel microduplication in ASMT. BMC Med Genomics. 2008;1:50. doi: 10.1186/1755-8794-1-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Christian SL, Brune CW, Sudi J, Kumar RA, Liu S, Karamohamed S, et al. Novel submicroscopic chromosomal abnormalities detected in autism spectrum disorder. Biol Psychiatry. 2008;63:1111–1117. doi: 10.1016/j.biopsych.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moreno-De-Luca D, Sanders SJ, Willsey AJ, Mulle JG, Lowe JK, Geschwind DH, et al. Using large clinical data sets to infer pathogenicity for rare copy number variants in autism cohorts. Mol Psychiatry advance online publication. 2012 Oct 9; doi: 10.1038/mp.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karayiorgou M, Simon TJ, Gogos JA. 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. Nat Rev Neurosci. 2010;11:402–416. doi: 10.1038/nrn2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl J Med. 2012;367:1321–1331. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vorstman JA, Breetvelt EJ, Thode KI, Chow EW, Bassett AS. Expression of autism spectrum and schizophrenia in patients with a 22q11.2 deletion. Schizophr Res. 2013;143:55–59. doi: 10.1016/j.schres.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 55.Bassett AS, Hodgkinson K, Chow EW, Correia S, Scutt LE, Weksberg R. 22q11 deletion syndrome in adults with schizophrenia. Am J Med Genet. 1998;81:328–337. [PMC free article] [PubMed] [Google Scholar]
- 56.Gothelf D, Frisch A, Munitz H, Rockah R, Laufer N, Mozes T, et al. Clinical characteristics of schizophrenia associated with velo-cardio-facial syndrome. Schizophr Res. 1999;35:105–112. doi: 10.1016/s0920-9964(98)00114-5. [DOI] [PubMed] [Google Scholar]
- 57.Moss EM, Batshaw ML, Solot CB, Gerdes M, Donald-McGinn DM, Driscoll DA, et al. Psychoeducational profile of the 22q11.2 microdeletion: a complex pattern. J Pediatr. 1999;134:193–198. doi: 10.1016/s0022-3476(99)70415-4. [DOI] [PubMed] [Google Scholar]
- 58.Niklasson L, Rasmussen P, Oskarsdottir S, Gillberg C. Neuropsychiatric disorders in the 22q11 deletion syndrome. Genet Med. 2001;3:79–84. doi: 10.1097/00125817-200101000-00017. [DOI] [PubMed] [Google Scholar]
- 59.De Smedt B, Devriendt K, Fryns JP, Vogels A, Gewillig M, Swillen A. Intellectual abilities in a large sample of children with velo-cardio-facial syndrome: an update. J Intellect Disabil Res. 2007;51:666–670. doi: 10.1111/j.1365-2788.2007.00955.x. [DOI] [PubMed] [Google Scholar]
- 60.Butcher NJ, Chow EW, Costain G, Karas D, Ho A, Bassett AS. Functional outcomes of adults with 22q11.2 deletion syndrome. Genet Med. 2012;14:836–843. doi: 10.1038/gim.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duijff SN, Klaassen PW, de Veye HF, Beemer FA, Sinnema G, Vorstman JA. Cognitive development in children with 22q11.2 deletion syndrome. Br J Psychiatry. 2012;200:462–468. doi: 10.1192/bjp.bp.111.097139. [DOI] [PubMed] [Google Scholar]
- 62.Alberti A, Romano C, Falco M, Cali F, Schinocca P, Galesi O, et al. 1.5Mb de novo 22q11.21 microduplication in a patient with cognitive deficits and dysmorphic facial features. Clin Genet. 2007;71:177–182. doi: 10.1111/j.1399-0004.2007.00750.x. [DOI] [PubMed] [Google Scholar]
- 63.Brunet A, Gabau E, Perich RM, Valdesoiro L, Brun C, Caballin MR, et al. Microdeletion and microduplication 22q11.2 screening in 295 patients with clinical features of DiGeorge/velocardiofacial syndrome. Am J Med Genet A. 2006;140:2426–2432. doi: 10.1002/ajmg.a.31499. [DOI] [PubMed] [Google Scholar]
- 64.Courtens W, Schramme I, Laridon A. Microduplication 22q11.2: a benign polymorphism or a syndrome with a very large clinical variability and reduced penetrance? — Report of two families. Am J Med Genet A. 2008;146A:758–763. doi: 10.1002/ajmg.a.31910. [DOI] [PubMed] [Google Scholar]
- 65.Descartes M, Franklin J, De Stahl TD, Piotrowski A, Bruder CE, Dumanski JP, et al. Distal 22q11.2 microduplication encompassing the BCR gene. Am J Med Genet A. 2008;146A:3075–3081. doi: 10.1002/ajmg.a.32572. [DOI] [PubMed] [Google Scholar]
- 66.Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, et al. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet. 1999;8:1157–1167. doi: 10.1093/hmg/8.7.1157. [DOI] [PubMed] [Google Scholar]
- 67.Ensenauer RE, Adeyinka A, Flynn HC, Michels VV, Lindor NM, Dawson DB, et al. Microduplication 22q11.2, an emerging syndrome: clinical, cytogenetic, and molecular analysis of thirteen patients. Am J Hum Genet. 2003;73:1027–1040. doi: 10.1086/378818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hassed SJ, Hopcus-Niccum D, Zhang L, Li S, Mulvihill JJ. A new genomic duplication syndrome complementary to the velocardiofacial (22q11 deletion) syndrome. Clin Genet. 2004;65:400–404. doi: 10.1111/j.0009-9163.2004.0212.x. [DOI] [PubMed] [Google Scholar]
- 69.Lo-Castro A, Galasso C, Cerminara C, El-Malhany N, Benedetti S, Nardone AM, et al. Association of syndromic mental retardation and autism with 22q11.2 duplication. Neuropediatrics. 2009;40:137–140. doi: 10.1055/s-0029-1237724. [DOI] [PubMed] [Google Scholar]
- 70.Ou Z, Berg JS, Yonath H, Enciso VB, Miller DT, Picker J, et al. Microduplications of 22q11.2 are frequently inherited and are associated with variable phenotypes. Genet Med. 2008;10:267–277. doi: 10.1097/GIM.0b013e31816b64c2. [DOI] [PubMed] [Google Scholar]
- 71.Portnoi MF, Lebas F, Gruchy N, Ardalan A, Biran-Mucignat V, Malan V, et al. 22q11.2 duplication syndrome: two new familial cases with some overlapping features with DiGeorge/velocardiofacial syndromes. Am J Med Genet A. 2005;137:47–51. doi: 10.1002/ajmg.a.30847. [DOI] [PubMed] [Google Scholar]
- 72.van Campenhout S, Devriendt K, Breckpot J, Frijns J-P, Peters H, van Buggenhout G, et al. Microduplication 22q11.2: a description of the clinical, developmental and behavioral characteristics during childhood. Genet Couns. 2012;23:135–147. [PubMed] [Google Scholar]
- 73.Wentzel C, Fernstrom M, Ohrner Y, Anneren G, Thuresson AC. Clinical variability of the 22q11.2 duplication syndrome. Eur J Med Genet. 2008;51:501–510. doi: 10.1016/j.ejmg.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 74.Yobb TM, Somerville MJ, Willatt L, Firth HV, Harrison K, MacKenzie J, et al. Microduplication and triplication of 22q11.2: a highly variable syndrome. Am J Hum Genet. 2005;76:865–876. doi: 10.1086/429841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu S, Cox K, Friend K, Smith S, Buchheim R, Bain S, et al. Familial 22q11.2 duplication: a three-generation family with a 3-Mb duplication and a familial 1.5-Mb duplication. Clin Genet. 2008;73:160–164. doi: 10.1111/j.1399-0004.2007.00938.x. [DOI] [PubMed] [Google Scholar]
- 76.Arnold PD, Siegel-Bartelt J, Cytrynbaum C, Teshima I, Schachar R. Velo-cardiofacial syndrome: implications of microdeletion 22q11 for schizophrenia and mood disorders. Am J Med Genet. 2001;105:354–362. doi: 10.1002/ajmg.1359. [DOI] [PubMed] [Google Scholar]
- 77.Carlson C, Papolos D, Pandita RK, Faedda GL, Veit S, Goldberg R, et al. Molecular analysis of velo-cardio-facial syndrome patients with psychiatric disorders. Am J Hum Genet. 1997;60:851–859. [PMC free article] [PubMed] [Google Scholar]
- 78.Feinstein C, Eliez S, Blasey C, Reiss AL. Psychiatric disorders and behavioral problems in children with velocardiofacial syndrome: usefulness as phenotypic indicators of schizophrenia risk. Biol Psychiatry. 2002;51:312–318. doi: 10.1016/s0006-3223(01)01231-8. [DOI] [PubMed] [Google Scholar]
- 79.Gothelf D, Presburger G, Levy D, Nahmani A, Burg M, Berant M, et al. Genetic, developmental, and physical factors associated with attention deficit hyperactivity disorder in patients with velocardiofacial syndrome. Am J Med Genet B Neuropsychiatr Genet. 2004;126:116–121. doi: 10.1002/ajmg.b.20144. [DOI] [PubMed] [Google Scholar]
- 80.Papolos DF, Faedda GL, Veit S, Goldberg R, Morrow B, Kucherlapati R, et al. Bipolar spectrum disorders in patients diagnosed with velo-cardio-facial syndrome: does a hemizygous deletion of chromosome 22q11 result in bipolar affective disorder? Am J Psychiatry. 1996;153:1541–1547. doi: 10.1176/ajp.153.12.1541. [DOI] [PubMed] [Google Scholar]
- 81.Antshel KM, Shprintzen R, Fremont W, Higgins AM, Faraone SV, Kates WR. Cognitive and psychiatric predictors to psychosis in velocardiofacial syndrome: a 3-year follow-up study. J Am Acad Child Adolesc Psychiatry. 2010;49:333–344. [PMC free article] [PubMed] [Google Scholar]
- 82.Jolin EM, Weller RA, Jessani NR, Zackai EH, Donald-McGinn DM, Weller EB. Affective disorders and other psychiatric diagnoses in children and adolescents with 22q11.2 deletion syndrome. J Affect Disord. 2009;119:177–180. doi: 10.1016/j.jad.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 83.Young AS, Shashi V, Schoch K, Kwapil T, Hooper SR. Discordance in diagnoses and treatment of psychiatric disorders in children and adolescents with 22q11.2 deletion syndrome. Asian J Psychiatr. 2011;4:119–124. doi: 10.1016/j.ajp.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Elia J, Glessner JT, Wang K, Takahashi N, Shtir CJ, Hadley D, et al. Genome-wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nat Genet. 2012;44:78–84. doi: 10.1038/ng.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Williams NM, Franke B, Mick E, Anney RJ, Freitag CM, Gill M, et al. Genome-wide analysis of copy number variants in attention deficit hyperactivity disorder: the role of rare variants and duplications at 15q13.3. Am J Psychiatry. 2012;169:195–204. doi: 10.1176/appi.ajp.2011.11060822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kroenke K, Rosmalen JG. Symptoms, syndromes, and the value of psychiatric diagnostics in patients who have functional somatic disorders. Med Clin North Am. 2006;90:603–626. doi: 10.1016/j.mcna.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 87.Michaelovsky E, Frisch A, Carmel M, Patya M, Zarchi O, Green T, et al. Genotype–phenotype correlation in 22q11.2 deletion syndrome. BMC Med Genet. 2012;13:122. doi: 10.1186/1471-2350-13-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Niklasson L, Gillberg C. The neuropsychology of 22q11 deletion syndrome. A neuropsychiatric study of 100 individuals. Res Dev Disabil. 2010;31:185–194. doi: 10.1016/j.ridd.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 89.Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, Itsara A, et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bassett AS, Marshall CR, Lionel AC, Chow EW, Scherer SW. Copy number variations and risk for schizophrenia in 22q11.2 deletion syndrome. Hum Mol Genet. 2008;17:4045–4053. doi: 10.1093/hmg/ddn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Klei L, Sanders SJ, Murtha MT, Hus V, Lowe JK, Willsey AJ, et al. Common genetic variants, acting additively, are a major source of risk for autism. Mol Autism. 2012;3:9. doi: 10.1186/2040-2392-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Done DJ, Crow TJ, Johnstone EC, Sacker A. Childhood antecedents of schizophrenia and affective illness: social adjustment at ages 7 and 11. BMJ. 1994;309:699–703. doi: 10.1136/bmj.309.6956.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Neumann CS, Grimes K, Walker EF, Baum K. Developmental pathways to schizophrenia: behavioral subtypes. J Abnorm Psychol. 1995;104:558–566. doi: 10.1037//0021-843x.104.4.558. [DOI] [PubMed] [Google Scholar]
- 94.Jones P, Rodgers B, Murray R, Marmot M. Child development risk factors for adult schizophrenia in the British 1946 birth cohort. Lancet. 1994;344:1398–1402. doi: 10.1016/s0140-6736(94)90569-x. [DOI] [PubMed] [Google Scholar]
- 95.Malmberg A, Lewis G, David A, Allebeck P. Premorbid adjustment and personality in people with schizophrenia. Br J Psychiatry. 1998;172:308–313. doi: 10.1192/bjp.172.4.308. [DOI] [PubMed] [Google Scholar]
- 96.Hollis C. Child and adolescent (juvenile onset) schizophrenia. A case control study of premorbid developmental impairments. Br J Psychiatry. 1995;166:489–495. doi: 10.1192/bjp.166.4.489. [DOI] [PubMed] [Google Scholar]
- 97.Cannon TD, Rosso IM, Bearden CE, Sanchez LE, Hadley T. A prospective cohort study of neurodevelopmental processes in the genesis and epigenesis of schizophrenia. Dev Psychopathol. 1999;11:467–485. doi: 10.1017/s0954579499002163. [DOI] [PubMed] [Google Scholar]
- 98.Gothelf D, Eliez S, Thompson T, Hinard C, Penniman L, Feinstein C, et al. COMT genotype predicts longitudinal cognitive decline and psychosis in 22q11.2 deletion syndrome. Nat Neurosci. 2005;8:1500–1502. doi: 10.1038/nn1572. [DOI] [PubMed] [Google Scholar]
- 99.Gothelf D, Feinstein C, Thompson T, Gu E, Penniman L, Van SE, et al. Risk factors for the emergence of psychotic disorders in adolescents with 22q11.2 deletion syndrome. Am J Psychiatry. 2007;164:663–669. doi: 10.1176/ajp.2007.164.4.663. [DOI] [PubMed] [Google Scholar]
- 100.Chow EW, Watson M, Young DA, Bassett AS. Neurocognitive profile in 22q11 deletion syndrome and schizophrenia. Schizophr Res. 2006;87:270–278. doi: 10.1016/j.schres.2006.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brocki KC, Bohlin G. Executive functions in children aged 6 to 13: a dimensional and developmental study. Dev Neuropsychol. 2004;26:571–593. doi: 10.1207/s15326942dn2602_3. [DOI] [PubMed] [Google Scholar]
- 102.De Luca CR, Wood SJ, Anderson V, Buchanan JA, Proffitt TM, Mahony K, et al. Normative data from the CANTAB. I: development of executive function over the lifespan. J Clin Exp Neuropsychol. 2003;25:242–254. doi: 10.1076/jcen.25.2.242.13639. [DOI] [PubMed] [Google Scholar]
- 103.Demetriou A, Christou C, Spanoudis G, Platsidou M. The development of mental processing: efficiency, working memory, and thinking. Monogr Soc Res Child Dev. 2002;67:1–155. [PubMed] [Google Scholar]
- 104.Dumontheil I, Roggeman C, Ziermans T, Peyrard-Janvid M, Matsson H, Kere J, et al. Influence of the COMT genotype on working memory and brain activity changes during development. Biol Psychiatry. 2011;70:222–229. doi: 10.1016/j.biopsych.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 105.Gathercole SE, Pickering SJ, Ambridge B, Wearing H. The structure of working memory from 4 to 15 years of age. Dev Psychol. 2004;40:177–190. doi: 10.1037/0012-1649.40.2.177. [DOI] [PubMed] [Google Scholar]
- 106.Luna B, Garver KE, Urban TA, Lazar NA, Sweeney JA. Maturation of cognitive processes from late childhood to adulthood. Child Dev. 2004;75:1357–1372. doi: 10.1111/j.1467-8624.2004.00745.x. [DOI] [PubMed] [Google Scholar]
- 107.Luna B. Developmental changes in cognitive control through adolescence. Adv Child Dev Behav. 2009;37:233–278. doi: 10.1016/s0065-2407(09)03706-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Swanson HL. What develops in working memory? A life span perspective. Dev Psychol. 1999;35:986–1000. doi: 10.1037//0012-1649.35.4.986. [DOI] [PubMed] [Google Scholar]
- 109.O’Hearn K, Schroer E, Minshew N, Luna B. Lack of developmental improvement on a face memory task during adolescence in autism. Neuropsychologia. 2010;48:3955–3960. doi: 10.1016/j.neuropsychologia.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cannon M, Caspi A, Moffitt TE, Harrington H, Taylor A, Murray RM, et al. Evidence for early-childhood, pan-developmental impairment specific to schizophreniform disorder: results from a longitudinal birth cohort. Arch Gen Psychiatry. 2002;59:449–456. doi: 10.1001/archpsyc.59.5.449. [DOI] [PubMed] [Google Scholar]
- 111.Erlenmeyer-Kimling L, Rock D, Roberts SA, Janal M, Kestenbaum C, Cornblatt B, et al. Attention, memory, and motor skills as childhood predictors of schizophrenia-related psychoses: the New York High-Risk Project. Am J Psychiatry. 2000;157:1416–1422. doi: 10.1176/appi.ajp.157.9.1416. [DOI] [PubMed] [Google Scholar]
- 112.Niendam TA, Bearden CE, Rosso IM, Sanchez LE, Hadley T, Nuechterlein KH, et al. A prospective study of childhood neurocognitive functioning in schizophrenic patients and their siblings. Am J Psychiatry. 2003;160:2060–2062. doi: 10.1176/appi.ajp.160.11.2060. [DOI] [PubMed] [Google Scholar]
- 113.Martinussen R, Tannock R. Working memory impairments in children with attention-deficit hyperactivity disorder with and without comorbid language learning disorders. J Clin Exp Neuropsychol. 2006;28:1073–1094. doi: 10.1080/13803390500205700. [DOI] [PubMed] [Google Scholar]
- 114.Schuchardt K, Gebhardt M, Maehler C. Working memory functions in children with different degrees of intellectual disability. J Intellect Disabil Res. 2010;54:346–353. doi: 10.1111/j.1365-2788.2010.01265.x. [DOI] [PubMed] [Google Scholar]
- 115.Baker K, Baldeweg T, Sivagnanasundaram S, Scambler P, Skuse D. COMT Val108/158 Met modifies mismatch negativity and cognitive function in 22q11 deletion syndrome. Biol Psychiatry. 2005;58:23–31. doi: 10.1016/j.biopsych.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 116.Campbell LE, Azuma R, Ambery F, Stevens A, Smith A, Morris RG, et al. Executive functions and memory abilities in children with 22q11.2 deletion syndrome. Aust N Z J Psychiatry. 2010;44:364–371. doi: 10.3109/00048670903489882. [DOI] [PubMed] [Google Scholar]
- 117.Goldenberg PC, Calkins ME, Richard J, Donald-McGinn D, Zackai E, Mitra N, et al. Computerized neurocognitive profile in young people with 22q11.2 deletion syndrome compared to youths with schizophrenia and at-risk for psychosis. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:87–93. doi: 10.1002/ajmg.b.32005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lajiness-O’Neill RR, Beaulieu I, Titus JB, Asamoah A, Bigler ED, Bawle EV, et al. Memory and learning in children with 22q11.2 deletion syndrome: evidence for ventral and dorsal stream disruption? Child Neuropsychol. 2005;11:55–71. doi: 10.1080/09297040590911202. [DOI] [PubMed] [Google Scholar]
- 119.Lewandowski KE, Shashi V, Berry PM, Kwapil TR. Schizophrenic-like neurocognitive deficits in children and adolescents with 22q11 deletion syndrome. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:27–36. doi: 10.1002/ajmg.b.30379. [DOI] [PubMed] [Google Scholar]
- 120.Sobin C, Kiley-Brabeck K, Daniels S, Khuri J, Taylor L, Blundell M, et al. Neuropsychological characteristics of children with the 22q11 deletion syndrome: a descriptive analysis. Child Neuropsychol. 2005;11:39–53. doi: 10.1080/09297040590911167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mirsky AF, Duncan CC. A nosology of disorders of attention. Ann N Y Acad Sci. 2001;931:17–32. doi: 10.1111/j.1749-6632.2001.tb05771.x. [DOI] [PubMed] [Google Scholar]
- 122.Pascualvaca DM, Fantie BD, Papageorgiou M, Mirsky AF. Attentional capacities in children with autism: is there a general deficit in shifting focus? J Autism Dev Disord. 1998;28:467–478. doi: 10.1023/a:1026091809650. [DOI] [PubMed] [Google Scholar]
- 123.Goldstein G, Johnson CR, Minshew NJ. Attentional processes in autism. J Autism Dev Disord. 2001;31:433–440. doi: 10.1023/a:1010620820786. [DOI] [PubMed] [Google Scholar]
- 124.Palmer BW, Heaton RK, Paulsen JS, Kuck J, Braff D, Harris MJ, et al. Is it possible to be schizophrenic yet neuropsychologically normal? Neuropsychology. 1997;11:437–446. doi: 10.1037//0894-4105.11.3.437. [DOI] [PubMed] [Google Scholar]
- 125.Bish JP, Ferrante SM, Donald-McGinn D, Zackai E, Simon TJ. Maladaptive conflict monitoring as evidence for executive dysfunction in children with chromosome 22q11.2 deletion syndrome. Dev Sci. 2005;8:36–43. doi: 10.1111/j.1467-7687.2005.00391.x. [DOI] [PubMed] [Google Scholar]
- 126.Sobin C, Kiley-Brabeck K, Daniels S, Blundell M, Anyane-Yeboa K, Karayiorgou M. Networks of attention in children with the 22q11 deletion syndrome. Dev Neuropsychol. 2004;26:611–626. doi: 10.1207/s15326942dn2602_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stoddard J, Beckett L, Simon TJ. Atypical development of the executive attention network in children with chromosome 22q11.2 deletion syndrome. J Neurodev Disord. 2011;3:76–85. doi: 10.1007/s11689-010-9070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chevallier C, Kohls G, Troiani V, Brodkin ES, Schultz RT. The social motivation theory of autism. Trends Cogn Sci. 2012;16:231–239. doi: 10.1016/j.tics.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Baron-Cohen S, Jolliffe T, Mortimore C, Robertson M. Another advanced test of theory of mind: evidence from very high functioning adults with autism or Asperger syndrome. J Child Psychol Psychiatry. 1997;38:813–822. doi: 10.1111/j.1469-7610.1997.tb01599.x. [DOI] [PubMed] [Google Scholar]
- 130.Pellicano E. Individual differences in executive function and central coherence predict developmental changes in theory of mind in autism. Dev Psychol. 2010;46:530–544. doi: 10.1037/a0018287. [DOI] [PubMed] [Google Scholar]
- 131.Sasson NJ, Pinkham AE, Carpenter KL, Belger A. The benefit of directly comparing autism and schizophrenia for revealing mechanisms of social cognitive impairment. J Neurodev Disord. 2011;3:87–100. doi: 10.1007/s11689-010-9068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pinkham AE, Penn DL, Perkins DO, Lieberman J. Implications for the neural basis of social cognition for the study of schizophrenia. Am J Psychiatry. 2003;160:815–824. doi: 10.1176/appi.ajp.160.5.815. [DOI] [PubMed] [Google Scholar]
- 133.Penn DL, Corrigan PW, Bentall RP, Racenstein JM, Newman L. Social cognition in schizophrenia. Psychol Bull. 1997;121:114–132. doi: 10.1037/0033-2909.121.1.114. [DOI] [PubMed] [Google Scholar]
- 134.Frith CD. Schizophrenia and theory of mind. Psychol Med. 2004;34:385–389. doi: 10.1017/s0033291703001326. [DOI] [PubMed] [Google Scholar]
- 135.Craig JS, Hatton C, Craig FB, Bentall RP. Persecutory beliefs, attributions and theory of mind: comparison of patients with paranoid delusions, Asperger’s syndrome and healthy controls. Schizophr Res. 2004;69:29–33. doi: 10.1016/S0920-9964(03)00154-3. [DOI] [PubMed] [Google Scholar]
- 136.Campbell LE, Stevens AF, McCabe K, Cruickshank L, Morris RG, Murphy DG, et al. Is theory of mind related to social dysfunction and emotional problems in 22q11.2 deletion syndrome (velo-cardio-facial syndrome)? J Neurodev Disord. 2011;3:152–161. doi: 10.1007/s11689-011-9082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schneider M, Van der LM, Glaser B, Rizzi E, Dahoun SP, Hinard C, et al. Preliminary structure and predictive value of attenuated negative symptoms in 22q11.2 deletion syndrome. Psychiatry Res. 2012;196:277–284. doi: 10.1016/j.psychres.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 138.Geyer MA. The family of sensorimotor gating disorders: comorbidities or diagnostic overlaps? Neurotox Res. 2006;10:211–220. doi: 10.1007/BF03033358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.McAlonan GM, Daly E, Kumari V, Critchley HD, van AT, Suckling J, et al. Brain anatomy and sensorimotor gating in Asperger’s syndrome. Brain. 2002;125:1594–1606. doi: 10.1093/brain/awf150. [DOI] [PubMed] [Google Scholar]
- 140.Perry W, Minassian A, Lopez B, Maron L, Lincoln A. Sensorimotor gating deficits in adults with autism. Biol Psychiatry. 2007;61:482–486. doi: 10.1016/j.biopsych.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 141.Yuhas J, Cordeiro L, Tassone F, Ballinger E, Schneider A, Long JM, et al. Brief report: sensorimotor gating in idiopathic autism and autism associated with fragile X syndrome. J Autism Dev Disord. 2010;41:248–253. doi: 10.1007/s10803-010-1040-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sobin C, Kiley-Brabeck K, Karayiorgou M. Lower prepulse inhibition in children with the 22q11 deletion syndrome. Am J Psychiatry. 2005;162:1090–1099. doi: 10.1176/appi.ajp.162.6.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Weksberg R, Stachon AC, Squire JA, Moldovan L, Bayani J, Meyn S, et al. Molecular characterization of deletion breakpoints in adults with 22q11 deletion syndrome. Hum Genet. 2007;120:837–845. doi: 10.1007/s00439-006-0242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ye T, Lipska BK, Tao R, Hyde TM, Wang L, Li C, et al. Analysis of copy number variations in brain DNA from patients with schizophrenia and other psychiatric disorders. Biol Psychiatry. 2012;72:651–654. doi: 10.1016/j.biopsych.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Paylor R, Glaser B, Mupo A, Ataliotis P, Spencer C, Sobotka A, et al. Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: implications for 22q11 deletion syndrome. Proc Natl Acad Sci USA. 2006;103:7729–7734. doi: 10.1073/pnas.0600206103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bartsch I, Sandrock K, Lanza F, Nurden P, Hainmann I, Pavlova A, et al. Deletion of human GP1BB and SEPT5 is associated with Bernard–Soulier syndrome, platelet secretion defect, polymicrogyria, and developmental delay. Thromb Haemost. 2011;106:475–483. doi: 10.1160/TH11-05-0305. [DOI] [PubMed] [Google Scholar]
- 147.Kempf L, Nicodemus KK, Kolachana B, Vakkalanka R, Verchinski BA, Egan MF, et al. Functional polymorphisms in PRODH are associated with risk and protection for schizophrenia and fronto-striatal structure and function. PLoS Genet. 2008;4:e1000252. doi: 10.1371/journal.pgen.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li D, He L. Association study of the G-protein signaling 4 (RGS4) and proline dehydrogenase (PRODH) genes with schizophrenia: a meta-analysis. Eur J Hum Genet. 2006;14:1130–1135. doi: 10.1038/sj.ejhg.5201680. [DOI] [PubMed] [Google Scholar]
- 149.Funke BH, Lencz T, Finn CT, DeRosse P, Poznik GD, Plocik AM, et al. Analysis of TBX1 variation in patients with psychotic and affective disorders. Mol Med. 2007;13:407–414. doi: 10.2119/2006-00119.Funke. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD, et al. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry. 2012;17:887–905. doi: 10.1038/mp.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Okochi T, Ikeda M, Kishi T, Kawashima K, Kinoshita Y, Kitajima T, et al. Meta-analysis of association between genetic variants in COMT and schizophrenia: an update. Schizophr Res. 2009;110:140–148. doi: 10.1016/j.schres.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 152.Allen NC, Bagade S, McQueen MB, Ioannidis JP, Kavvoura FK, Khoury MJ, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 153.Xu M, St Clair D, He L. Testing for genetic association between the ZDHHC8 gene locus and susceptibility to schizophrenia: an integrated analysis of multiple datasets. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:1266–1275. doi: 10.1002/ajmg.b.31096. [DOI] [PubMed] [Google Scholar]
- 154.Zhou Y, Wang J, Lu X, Song X, Ye Y, Zhou J, et al. Evaluation of six SNPs of microRNA machinery genes and risk of schizophrenia. J Mol Neurosci. 2013;49:594–599. doi: 10.1007/s12031-012-9887-1. [DOI] [PubMed] [Google Scholar]
- 155.Zhang F, Chen Y, Liu C, Lu T, Yan H, Ruan Y, et al. Systematic association analysis of microRNA machinery genes with schizophrenia informs further study. Neurosci Lett. 2012;520:47–50. doi: 10.1016/j.neulet.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 156.Jitoku D, Hattori E, Iwayama Y, Yamada K, Toyota T, Kikuchi M, et al. Association study of Nogo-related genes with schizophrenia in a Japanese case–control sample. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:581–592. doi: 10.1002/ajmg.b.31199. [DOI] [PubMed] [Google Scholar]
- 157.Pasaje CF, Bae JS, Park BL, Park CS, Kim BJ, Lee CS, et al. Lack of association of the RTN4R genetic variations with risk of schizophrenia and SPEM abnormality in a Korean population. Psychiatry Res. 2011;189:312–314. doi: 10.1016/j.psychres.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 158.Meng J, Shi Y, Zhao X, Guo S, Wang H, Zheng Y, et al. No association between the genetic polymorphisms in the RTN4R gene and schizophrenia in the Chinese population. J Neural Transm. 2007;114:249–254. doi: 10.1007/s00702-006-0538-y. [DOI] [PubMed] [Google Scholar]
- 159.Sinibaldi L, De LA, Bellacchio E, Conti E, Pasini A, Paloscia C, et al. Mutations of the Nogo-66 receptor (RTN4R) gene in schizophrenia. Hum Mutat. 2004;24:534–535. doi: 10.1002/humu.9292. [DOI] [PubMed] [Google Scholar]
- 160.Budel S, Padukkavidana T, Liu BP, Feng Z, Hu F, Johnson S, et al. Genetic variants of Nogo-66 receptor with possible association to schizophrenia block myelin inhibition of axon growth. J Neurosci. 2008;28:13161–13172. doi: 10.1523/JNEUROSCI.3828-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hsu R, Woodroffe A, Lai WS, Cook MN, Mukai J, Dunning JP, et al. Nogo receptor 1 (RTN4R) as a candidate gene for schizophrenia: analysis using human and mouse genetic approaches. PLoS One. 2007;2:e1234. doi: 10.1371/journal.pone.0001234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu H, Abecasis GR, Heath SC, Knowles A, Demars S, Chen YJ, et al. Genetic variation in the 22q11 locus and susceptibility to schizophrenia. Proc Natl Acad Sci USA. 2002;99:16859–16864. doi: 10.1073/pnas.232186099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ishiguro H, Koga M, Horiuchi Y, Noguchi E, Morikawa M, Suzuki Y, et al. Supportive evidence for reduced expression of GNB1L in schizophrenia. Schizophr Bull. 2010;36:756–765. doi: 10.1093/schbul/sbn160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Li Y, Zhao Q, Wang T, Liu J, Li J, Li T, et al. Association study between GNB1L and three major mental disorders in Chinese Han populations. Psychiatry Res. 2011;187:457–459. doi: 10.1016/j.psychres.2010.04.019. [DOI] [PubMed] [Google Scholar]
- 165.Williams NM, Glaser B, Norton N, Williams H, Pierce T, Moskvina V, et al. Strong evidence that GNB1L is associated with schizophrenia. Hum Mol Genet. 2008;17:555–566. doi: 10.1093/hmg/ddm330. [DOI] [PubMed] [Google Scholar]
- 166.Liu H, Heath SC, Sobin C, Roos JL, Galke BL, Blundell ML, et al. Genetic variation at the 22q11 PRODH2/DGCR6 locus presents an unusual pattern and increases susceptibility to schizophrenia. Proc Natl Acad Sci USA. 2002;99:3717–3722. doi: 10.1073/pnas.042700699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Shi Y, Li Z, Xu Q, Wang T, Li T, Shen J, et al. Common variants on 8p12 and 1q24.2 confer risk of schizophrenia. Nat Genet. 2011;43:1224–1227. doi: 10.1038/ng.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Weiss LA, Arking DE, Daly MJ, Chakravarti A. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461:802–808. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bassett AS, Caluseriu O, Weksberg R, Young DA, Chow EW. Catechol-O-methyl transferase and expression of schizophrenia in 73 adults with 22q11 deletion syndrome. Biol Psychiatry. 2007;61:1135–1140. doi: 10.1016/j.biopsych.2006.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.van Amelsvoort T, Zinkstok J, Figee M, Daly E, Morris R, Owen MJ, et al. Effects of a functional COMT polymorphism on brain anatomy and cognitive function in adults with velo-cardio-facial syndrome. Psychol Med. 2008;38:89–100. doi: 10.1017/S0033291707000700. [DOI] [PubMed] [Google Scholar]
- 172.de Koning MB, Boot E, Bloemen OJ, van Duin ED, Abel KM, de Haan L, et al. Startle reactivity and prepulse inhibition of the acoustic startle response are modulated by catechol-O-methyl-transferase Val(158) Met polymorphism in adults with 22q11 deletion syndrome. J Psychopharmacol. 2012;26:1548–1560. doi: 10.1177/0269881112456610. [DOI] [PubMed] [Google Scholar]
- 173.Boot E, Booij J, Abeling N, Meijer J, da Silva AF, Zinkstok J, et al. Dopamine metabolism in adults with 22q11 deletion syndrome, with and without schizophrenia — relationship with COMT Val108/158Met polymorphism, gender and symptomatology. J Psychopharmacol. 2011;25:888–895. doi: 10.1177/0269881111400644. [DOI] [PubMed] [Google Scholar]
- 174.Hyman SE. Can neuroscience be integrated into the DSM-V? Nat Rev Neurosci. 2007;8:725–732. doi: 10.1038/nrn2218. [DOI] [PubMed] [Google Scholar]
- 175.Insel TR. From animal models to model animals. Biol Psychiatry. 2007;62:1337–1339. doi: 10.1016/j.biopsych.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 176.Tamada K, Tomonaga S, Hatanaka F, Nakai N, Takao K, Miyakawa T, et al. Decreased exploratory activity in a mouse model of 15q duplication syndrome; implications for disturbance of serotonin signaling. PLoS One. 2010;5:e15126. doi: 10.1371/journal.pone.0015126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Nakatani J, Tamada K, Hatanaka F, Ise S, Ohta H, Inoue K, et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell. 2009;137:1235–1246. doi: 10.1016/j.cell.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Horev G, Ellegood J, Lerch JP, Son YE, Muthuswamy L, Vogel H, et al. Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. Proc Natl Acad Sci USA. 2011;108:17076–17081. doi: 10.1073/pnas.1114042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Walz K, Paylor R, Yan J, Bi W, Lupski JR. Rai1 duplication causes physical and behavioral phenotypes in a mouse model of dup(17)(p11.2p11.2) J Clin Invest. 2006;116:3035–3041. doi: 10.1172/JCI28953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bi W, Yan J, Shi X, Yuva-Paylor LA, Antalffy BA, Goldman A, et al. Rai1 deficiency in mice causes learning impairment and motor dysfunction, whereas Rai1 heterozygous mice display minimal behavioral phenotypes. Hum Mol Genet. 2007;16:1802–1813. doi: 10.1093/hmg/ddm128. [DOI] [PubMed] [Google Scholar]
- 181.Long JM, Laporte P, Merscher S, Funke B, Saint-Jore B, Puech A, et al. Behavior of mice with mutations in the conserved region deleted in velocardiofacial/ DiGeorge syndrome. Neurogenetics. 2006;7:247–257. doi: 10.1007/s10048-006-0054-0. [DOI] [PubMed] [Google Scholar]
- 182.Paylor R, McIlwain KL, McAninch R, Nellis A, Yuva-Paylor LA, Baldini A, et al. Mice deleted for the DiGeorge/velocardiofacial syndrome region show abnormal sensorimotor gating and learning and memory impairments. Hum Mol Genet. 2001;10:2645–2650. doi: 10.1093/hmg/10.23.2645. [DOI] [PubMed] [Google Scholar]
- 183.Hiroi N, Zhu H, Lee M, Funke B, Arai M, Itokawa M, et al. A 200-kb region of human chromosome 22q11.2 confers antipsychotic-responsive behavioral abnormalities in mice. Proc Natl Acad Sci USA. 2005;102:19132–19137. doi: 10.1073/pnas.0509635102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Suzuki G, Harper KM, Hiramoto T, Funke B, Lee M, Kang G, et al. Over-expression of a human chromosome 22q11.2 segment including TXNRD2, COMT, and ARVCF developmentally affects incentive learning and working memory in mice. Hum Mol Genet. 2009;18:3914–3925. doi: 10.1093/hmg/ddp334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Stark KL, Burt RA, Gogos JA, Karayiorgou M. Analysis of prepulse inhibition in mouse lines overexpressing 22q11.2 orthologues. Int J Neuropsychopharmacol. 2009;12:983–989. doi: 10.1017/S1461145709000492. [DOI] [PubMed] [Google Scholar]
- 186.Hiramoto T, Kang G, Suzuki G, Satoh Y, Kucherlapati R, Watanabe Y, et al. Tbx1: identification of a 22q11.2 gene as a risk factor for autism spectrum disorder in a mouse model. Hum Mol Genet. 2011;20:4775–4785. doi: 10.1093/hmg/ddr404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Harper KM, Hiramoto T, Tanigaki K, Kang G, Suzuki G, Trimble W, et al. Alterations of social interaction through genetic and environmental manipulation of the 22q11.2 gene Sept5 in the mouse brain. Hum Mol Genet. 2012;21:3489–3499. doi: 10.1093/hmg/dds180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Suzuki G, Harper KM, Hiramoto T, Sawamura T, Lee M, Kang G, et al. Sept5 deficiency exerts pleiotropic influence on affective behaviors and cognitive functions in mice. Hum Mol Genet. 2009;18:1652–1660. doi: 10.1093/hmg/ddp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hiroi N, Hiramoto T, Harper KM, Suzuki G, Boku S. Mouse models of 22q11.2- associated autism spectrum disorder. Autism. 2012;S1:1–9. doi: 10.4172/2165-7890.S1-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Papaleo F, Crawley JN, Song J, Lipska BK, Pickel J, Weinberger DR, et al. Genetic dissection of the role of catechol-O-methyltransferase in cognition and stress reactivity in mice. J Neurosci. 2008;28:8709–8723. doi: 10.1523/JNEUROSCI.2077-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Gogos JA, Morgan M, Luine V, Santha M, Ogawa S, Pfaff D, et al. Catechol-O-methyltransferase-deficient mice exhibit sexually dimorphic changes in catecholamine levels and behavior. Proc Natl Acad Sci USA. 1998;95:9991–9996. doi: 10.1073/pnas.95.17.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.O’Tuathaigh CM, Clarke G, Walsh J, Desbonnet L, Petit E, O’Leary C, et al. Genetic vs. pharmacological inactivation of COMT influences cannabinoid-induced expression of schizophrenia-related phenotypes. Int J Neuropsychopharmacol. 2012;15:1331–1342. doi: 10.1017/S1461145711001581. [DOI] [PubMed] [Google Scholar]
- 193.O’Tuathaigh CM, Hryniewiecka M, Behan A, Tighe O, Coughlan C, Desbonnet L, et al. Chronic adolescent exposure to delta-9-tetrahydrocannabinol in COMT mutant mice: impact on psychosis-related and other phenotypes. Neuropsychopharmacology. 2010;35:2262–2273. doi: 10.1038/npp.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Babovic D, O’Tuathaigh CM, O’Connor AM, O’Sullivan GJ, Tighe O, Croke DT, et al. Phenotypic characterization of cognition and social behavior in mice with heterozygous versus homozygous deletion of catechol-O-methyltransferase. Neuroscience. 2008;155:1021–1029. doi: 10.1016/j.neuroscience.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 195.Kimber WL, Hsieh P, Hirotsune S, Yuva-Paylor L, Sutherland HF, Chen A, et al. Deletion of 150 kb in the minimal DiGeorge/velocardiofacial syndrome critical region in mouse. Hum Mol Genet. 1999;8:2229–2237. doi: 10.1093/hmg/8.12.2229. [DOI] [PubMed] [Google Scholar]
- 196.Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 197.Mukai J, Liu H, Burt RA, Swor DE, Lai WS, Karayiorgou M, et al. Evidence that the gene encoding ZDHHC8 contributes to the risk of schizophrenia. Nat Genet. 2004;36:725–731. doi: 10.1038/ng1375. [DOI] [PubMed] [Google Scholar]
- 198.Paylor R, Crawley JN. Inbred strain differences in prepulse inhibition of the mouse startle response. Psychopharmacology (Berl) 1997;132:169–180. doi: 10.1007/s002130050333. [DOI] [PubMed] [Google Scholar]
- 199.Watanabe A, Toyota T, Owada Y, Hayashi T, Iwayama Y, Matsumata M, et al. Fabp7 maps to a quantitative trait locus for a schizophrenia endophenotype. PLoS Biol. 2007;5:e297. doi: 10.1371/journal.pbio.0050297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Willott JF, Tanner L, O’Steen J, Johnson KR, Bogue MA, Gagnon L. Acoustic startle and prepulse inhibition in 40 inbred strains of mice. Behav Neurosci. 2003;117:716–727. doi: 10.1037/0735-7044.117.4.716. [DOI] [PubMed] [Google Scholar]
- 201.Wolfer DP, Crusio WE, Lipp HP. Knockout mice: simple solutions to the problems of genetic background and flanking genes. Trends Neurosci. 2002;25:336–340. doi: 10.1016/s0166-2236(02)02192-6. [DOI] [PubMed] [Google Scholar]
- 202.Gogos JA, Santha M, Takacs Z, Beck KD, Luine V, Lucas LR, et al. The gene encoding proline dehydrogenase modulates sensorimotor gating in mice. Nat Genet. 1999;21:434–439. doi: 10.1038/7777. [DOI] [PubMed] [Google Scholar]
- 203.Paterlini M, Zakharenko SS, Lai WS, Qin J, Zhang H, Mukai J, et al. Transcriptional and behavioral interaction between 22q11.2 orthologs modulates schizophrenia- related phenotypes in mice. Nat Neurosci. 2005;8:1586–1594. doi: 10.1038/nn1562. [DOI] [PubMed] [Google Scholar]
- 204.Keshavan MS, Clementz BA, Pearlson GD, Sweeney JA, Tamminga CA. Reimagining psychoses: an agnostic approach to diagnosis. Schizophr Res. 2013;146:10–16. doi: 10.1016/j.schres.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 205.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]