Abstract
The mammalian brain is a complex multicellular system involving enormous numbers of neurons. The neuron is the basic functional unit of the brain, and neurons are organized by specialized intercellular connections into circuits with many other neurons. Physiological studies have revealed that individual neurons have remarkably selective response properties, and this individuality is a fundamental requirement for building complex and functionally diverse neural networks. Recent molecular biological studies have revealed genetic bases for neuronal individuality in the mammalian brain. For example, in the rodent olfactory epithelium, individual olfactory neurons express only one type of odorant receptor (OR) out of the over 1000 ORs encoded in the genome. The expressed OR determines the neuron's selective chemosensory response and specifies its axonal targeting to a particular olfactory glomerulus in the olfactory bulb. Neuronal diversity can also be generated in individual cells by the independent and stochastic expression of autosomal alleles, which leads to functional heterozygosity among neurons. Among the many genes that show autosomal stochastic monoallelic expression, approximately 50 members of the clustered protocadherins (Pcdhs) are stochastically expressed in individual neurons in distinct combinations. The clustered Pcdhs belong to a large subfamily of the cadherin superfamily of homophilic cell-adhesion proteins. Loss-of-function analyses show that the clustered Pcdhs have critical functions in the accuracy of axonal projections, synaptic formation, dendritic arborization, and neuronal survival. In addition, cis-tetramers, composed of heteromultimeric clustered Pcdh members, represent selective binding units for cell-cell interactions, and provide exponential numbers of possible cell-surface relationships between individual neurons. The extensive molecular diversity of neuronal cell-surface proteins affects neurons’ individual properties and connectivities. The molecular features of the diverse clustered Pcdh molecules suggest that they provide a genetic basis for neuronal individuality and appropriate neuronal wiring in the brain.
Keywords: epigenetic, neural network, protocadherin, stochastic expression
INTRODUCTION
The mammalian brain contains an enormous number of neurons and complex topological neural networks, in which differences at the level of individual neurons play important roles. The neuron is the basic functional unit of the complex brain system. Neuronal individuality was demonstrated physiologically by the remarkably selective response properties of single neurons in specific functional situations and is related to the specific patterns of synaptic connections made by the huge numbers of neurons that form functional circuits. That is, the organization of neuronal networks, which is directed in part by the properties of individual neurons, determines the processing flow of information in neural circuits and has important implications for regulating brain functions. Recent physiological studies demonstrated that the bidirectional and highly clustered connections that typify local neuronal networks exhibit specific features more frequently than would be expected if the circuits formed randomly (Markram, 1997; Kalisman et al., 2005; Song et al., 2005; Yoshimura et al., 2005; Koulakov et al., 2009; Yassin et al., 2010; Perin et al., 2011). Furthermore, individual neurons with similar response properties are highly selective in their synaptic connections and show preferential connectivity (Yoshimura & Callaway, 2005). These networks are characterized not only by their complex topology, but also by their non-Gaussian (long-tail) distributions of synaptic weight in the cortex and hippocampus (Song et al., 2005; Lefort et al., 2009; Ikegaya et al., 2013). In theoretical model, the long-tail distributions of connection weights can generate noise useful for associative memory (Hiratani et al., 2013). Recent research shows that cell lineage plays a key role in specifying neuronal connections and functional properties in the cortex (Yu et al., 2009, 2012; Li et al., 2012; Ohtsuki et al., 2012). Therefore, the establishment of neuronal individuality, which helps determine neuronal connectivity and physiological function during development, is a fundamental mechanism underlying the building of complex neural networks in the brain.
At the molecular level, the mechanism of stochastic monoallelic expression generates cellular individuality and provides potential functional variation among the individual cells of a complex system. For example, the immune system is a complex system in which enormous numbers of diverse cells are developmentally generated by the random DNA rearrangement of immune receptors (Tonegawa, 1983; Lieber, 1992). This system can newly acquire specific immune responses to a nearly limitless number of antigens that attack an animal, and “memorize” the antigen. Thus, immune cell individuality is generated by the expression of a specific immune receptor, random somatic DNA rearrangements, the combination of heteromeric receptors, and selection by the specific response to an attacking antigen. This molecular mechanism that regulates the expression and function of immune receptors constitutes the predetermined immune memory system.
As another example, diverse olfactory receptors are expressed in the olfactory sensory system. In 1991, Buck and Axel discovered over 1000 odorant receptors (ORs) in olfactory sensory neurons in the olfactory epithelium in mice (Buck & Axel, 1991). This discovery led to further investigation of the complex olfactory system, and the individuality of olfactory sensory neurons. These investigations showed that an olfactory sensory neuron can express only one functional OR gene, and the specific OR protein expressed suppresses the expression of other OR genes and induces the neuron's axonal targeting of the appropriate glomerulus in the olfactory bulb by topological mapping. Thus, the individuality of olfactory sensory neurons is determined genetically by the OR expression and function (Sakano, 2010).
Thus, in both the immune and olfactory systems, cellular individuality is generated by the expression of diverse receptor-type molecules, and involves random allelic expression and specific functional properties. The elucidation of these mechanisms has been invaluable for increasing our understanding of these complex systems.
In humans, at the single-cell level, large numbers of genes with diverse functions are regulated by stochastic monoallelic expression (Gimelbrant et al., 2007), and random autosomal expression is a common mechanism in rodents and primate orthologs of these genes (Zwemer et al., 2012). Their genes with random monoallelic expression give a possible mechanism for generating cellular diversity. When there is their functional heterozygosity, the random allelic expression apparently leads to cellular functional diversity at the individual cell level. Studies of random allelic expression patterns should shed light on the roles of cellular individuality. Interestingly, stochastic allelic expression is also observed in immortalized neural clones from human fetal brain, indicating that neurons exhibit clonal diversity in the developing brain (Jeffries et al., 2012).
In the brain, the complexity and diversity of neurons led to the suggestion that a somatic mutational mechanism was responsible for the evolution of diversified neuronal functions (Muotri & Gage, 2006; Rehen et al., 2005), and the endogenous retrotransposition of LINE-1 (L1) elements was proposed as a potential mechanism for generating neuronal genome diversity (Singer et al., 2010). Although it was estimated that in the amplification of the genome of a single neuron in the human brain, < 0.6 unique somatic insertions would occur, most neurons lack detectable somatic insertions, suggesting that L1 is not a major generator of neuronal diversity (Evrony et al., 2012).
Like the immune receptors and odorant receptors, the clustered protocadherins (Pcdhs), which are organized as gene clusters, undergo random allelic expression and elicit specific functional properties in individual neurons (Esumi et al., 2005; Kaneko et al., 2006; Hirano et al., 2012). The clustered Pcdhs are predominantly expressed in vertebrate brains and have functions in mice in neuronal survival, axonal targeting, dendrite arborization, synaptogenesis, neuronal migration, cortical activity, and learning and memory (Yagi, 2008; Zipursky & Sanes, 2010; Yamashita et al., 2012; Hirayama & Yagi, 2013; Weiner & Jontes, 2013; Ledderose et al., 2013). Similarly, clustered Pcdh genes are susceptibility genes for autism, bipolar disorder, and musical aptitude (Anitha et al., 2012; Redies et al., 2012; Ukkola-Vuoti et al., 2013). This review describes the molecular characteristics of the clustered Pcdh families and how they generate neuronal individuality, and it discusses how these findings increase our comprehension of neural circuit formation and function.
THE CLUSTERED PROTOCADHERINS
Cadherins are type I transmembrane proteins that were originally characterized as calcium-dependent cell-adhesion molecules (Takeichi, 2007). Cadherin family members are identified by the cadherin sequence repeats in their extracellular (EC) domain. Several subgroups of cadherins are classified by their sequence similarity: the classical (type I) and closely related type II cadherins, desmosomal cadherins, and protocadherins. The sequencing of vertebrate genomes has revealed more than 100 different cadherin-related genes (Morishita & Yagi, 2007). Protocadherins (Pcdhs) were first discovered by their sequence homology with the extracellular repeats of classical cadherins. Of the cadherin superfamily, which has over 80 members, the Pcdhs constitute a major family, and are further divided into clustered Pcdhs and nonclustered Pcdhs. The clustered Pcdhs are divided into three smaller gene clusters: the Pcdh-α, -β, and -γ families (Wu & Maniatis, 1999). The Pcdh-α family was originally discovered in mouse brain and designated “cadherin- related neuronal receptors” (CNRs) (Kohmura et al., 1998). With over 50 members, the clustered Pcdhs are the largest subgroup within the cadherin superfamily. The nonclustered Pcdhs have 13 members: 4 in the Pcdh-δ1, 5 in the Pcdh-δ2, and 4 in other Pcdh subgroups. Interestingly all of the Pcdh-δ2 proteins have a similar structure to the clustered protocadherin members: five EC repeats and a Cys-(X)5-Cys sequence in the EC1 domain (Morishita & Yagi, 2007). The structural characterization of the EC1 domain revealed that the protocadherin-specific disulfide-bounded Cys-(X)5-Cys sequence is exposed to solvent as a loop (Morishita et al., 2006). Similarly, some Pcdh-δ1 proteins have Cys-(X)8 or 10-Cys sequence in the EC1 domain.
GENOMIC STRUCTURES OF CLUSTERED PROTOCADHERINS
Clustered Pcdh genes have been identified in vertebrates. They have an unusual genomic organization, similar to that of the immunoglobulin and T-cell receptor gene clusters. The N-terminal extracellular domains are different, whereas the cytoplasmic tail of each protein is identical among the Pcdh-α or -γ members (Kohmura et al., 1998; Wu & Maniatis, 1999). The Pcdh-α and -γ cytoplasmic tails can bind directly to non-receptor-type tyrosine kinases, FAK and PYK2, which play crucial roles in synaptic and dendrite maturation (Chen et al., 2009; Garret et al., 2012; Suo et al., 2012). Therefore, distinct extracellular interactions can be transferred to a common signaling pathway in the cytoplasm through the cytoplasmic tails of these molecules. The Pcdh-β members do not share a common constant cytoplasmic tail, but this region is highly conserved. In mouse chromosome 18, a total of 58 genes are arranged in the Pcdh-α, -β, and -γ clusters, which have 14, 22, and 22 members, respectively. In humans, a total of 53 Pcdh-α, -β, and -γ members are located at 5q31. There are many polymorphisms in the Pcdh gene cluster, including amino acid exchanges, extensive linkage disequilibrium, and deletions, in the human population (Noonan et al., 2003; Miki et al., 2005).
ROLES OF CLUSTERED PROTOCADHERINS IN NEURAL CIRCUIT FORMATION
The extracellular domains of the Pcdhs contain cadherin motifs. Many of the cadherin superfamily members have important roles in developmental processes, including synapse formation (Yagi & Takeichi, 2000). Mice lacking Pcdh-α are viable but exhibit abnormal learning and memory (Fukuda et al., 2008), and abnormal cortical activity (Yamashita et al., 2012). In addition, the precise targeting of olfactory and serotonergic axonal terminals is impaired in Pcdh-α knockout (KO) mice (Hasegawa et al., 2008, 2012; Katori et al., 2009). The loss of Pcdh-γ leads to neonatal death with neurological impairments, including the cell death of spinal cord interneurons and decreased numbers of synapses (Wang et al., 2002b). Conditional KO experiments for Pcdh-γ revealed that this subfamily has roles in the self-avoidance function in the dendrite formation of retinal starburst amacrine cells and cerebellar Purkinje neurons (Lefebvre et al., 2012). The dendritic phenotype (reduced arborization) was also observed in cortical neurons and granule cells in which Pcdh-γ was conditionally deleted (Garrett et al., 2012; Ledderose et al., 2013). The loss of Pcdh-γ in astrocytes and in neurons in the spinal cord and olfactory granule cells leads to a reduction in synapses (Garrett & Weiner, 2009; Ledderose et al., 2013). These results suggest that the clustered Pcdhs have important function for the proper building of neural networks in the brain. However, it does not have completely revealed their molecular mechanism for building specific connection of neural networks (including synapse formation) at an individual neuron level.
STOCHASTIC AND COMBINATORIAL GENE REGULATION IN INDIVIDUAL NEURONS
One of the most fascinating characteristics of the clustered Pcdh families is their differential and combinatorial expression in neurons in the brain. Over 50 clustered Pcdhs (i.e., all of the clustered Pcdhs except the 5 “C” members, Pcdh-αC1, -αC2, -γC3, -γC4, and -γC5) are expressed in a scattered pattern throughout the brain (Esumi et al., 2005; Kaneko et al., 2006; Noguchi et al., 2009; Yokota et al., 2011; Hirano et al., 2012). In addition, at the single Purkinje cell level, each Pcdh-α, -β, or -γ member is expressed monoallelically, although both the paternal and maternal cluster alleles are transcriptionally active in individual neurons (Esumi et al., 2005; Kaneko et al., 2006; Hirano et al., 2012). The promoter of each variable exon is independently chosen from both alleles. The exceptions to this rule are the constitutively expressed Pcdh-αC1, -αC2, -γC3, -γC4, and -γC5 variable exons, which are expressed from both alleles in individual neurons (Esumi et al., 2005; Kaneko et al., 2006).
Single-cell reverse transcriptase–polymerase chain reaction (RT-PCR) analysis of cerebellar Purkinje neurons indicated that individual neurons stochastically and monoallelically express ∼2 of the 12 Pcdh-α, ˜4 of the 22 Pcdh-β, and ˜4 of the 19 Pcdh-γ genes (Figure 1). Therefore, 78 possible combinations from the 12 Pcdh-α, 26,796 from the 22 Pcdh-β, and 14,706 from the 19 Pcdh-γ genes are generated, and a total of 78 × 26,796 × 14,706 = 30,736,834,128 (approximately 3 × 1010) variations are possible in each individual neuron (Yagi, 2012). Notably, the cerebral cortex of the human brain contains approximately 1010 neurons. In addition, the 5 “C” members are constitutively and biallelically expressed in neurons, and increase the total number of clustered Pcdh members expressed in each neuron, but do not contribute to neuronal variation.
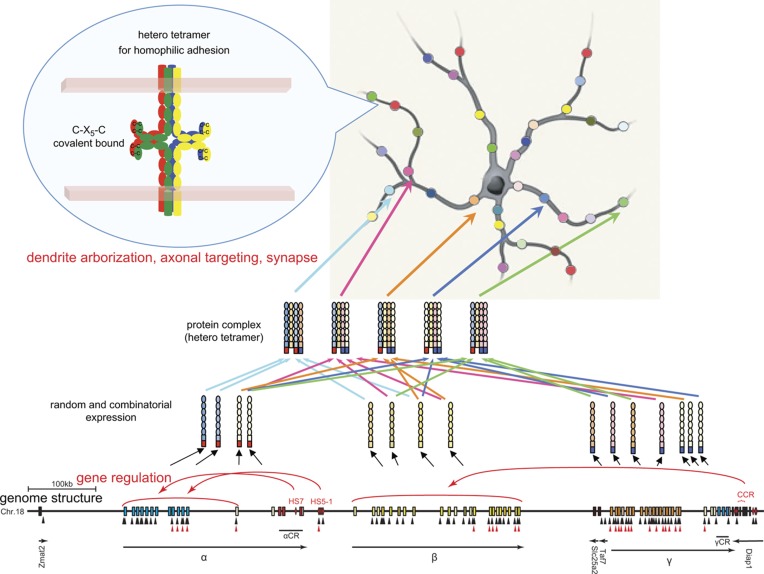
Figure 1.
Neuronal individuality determined by the random and combinatorial expression of clustered Pcdh members and the random production of heteromultimeric cis-tetramers. From each clustered Pcdh allele, about 10 α, β, and γ members are randomly expressed, and 5 “C” members are constitutively expressed. The random expressions are regulated by the cis-elements HS5-1 for the Pcdh-α cluster and CCR for the Pcdh-β cluster; the CCR is located downstream from the Pcdh-γ cluster. The random expressions are also regulated by the trans-factors CTCF and cohesin-SA. Black and red triangles show CTCF-binding and cohesin-SA-binding sites, respectively. The expressed Pcdh member proteins form random heteromultimeric cis-tetramers. The C-X5-C motif, which is important for forming the cis-tetramers, is conserved among all the clustered Pcdh and nonclustered Pcdh-δ2 members. The heteromultimeric cis-tetramers can bind homophilically between cells. Genetic analyses in mice suggested that these interactions influence dendritic arborization, axonal targeting, and synaptogenesis. This figure is modified from Yagi (2012).
COMBINATORIAL COMPLEXITY OF THE CLUSTERED PROTOCADHERINS AT THE PROTEIN LEVEL
The clustered Pcdh proteins show a punctate localization in neurons in vitro and in vivo (Phillips et al., 2003; Murata et al., 2004; Femandez-Monreal et al., 2009). Pcdh-α, -β, and -γ protein members form a protein complex in the cytoplasmic membrane of cultured cells and in the brain (Murata et al., 2004; Han et al., 2010). These clustered Pcdh protein complexes have cell-adhesion activities, as reported by Schreiner and Weiner (2010), who showed that seven Pcdh-γ members exhibit isoform-specific homophilic binding. These authors suggested that heteromultimeric cis-tetramers function as a homophilic binding unit (Schreiner & Weiner, 2010).
In fact, the clustered Pcdhs form cis-tetramers before their translocation to the cell membrane. Therefore, the binding properties of the cis-tetramers of clustered Pcdhs are very different from those of classical cadherins. If two different clustered Pcdh isoforms are expressed in a cell, six different cis-tetramers are expressed on the cell membrane. If one isoform is common between two cells, only one of six tetramers will be common between the two cell surfaces, resulting in weak cell-cell aggregation (Schreiner & Weiner, 2010; Yagi, 2012). Thus, by forming functional units of hetero cis-tetramers, the Pcdhs exponential increase the number of variations in the protein complexes at the cell surface.
Individual neurons are estimated to express 15 clustered Pcdh members (i.e., 10 randomly expressed ones: 2 Pcdh-α, 4 Pcdh-β, and 4 Pcdh-γ; and 5 constitutively expressed ones: αC1, αC2, γC3, γC4, and γC5) (Figure 1). Random combinations could produce 12,720 15-member sets of Pcdhs and 154 (50,625) possible tetramers (Yagi, 2012). Notably, the probability of matching tetramers occurring between a pair of neurons decreases exponentially as the number of different isoforms increases. If only 1 of the 15 clustered Pcdh members is different (i.e., 14 of the 15 are the same) between a pair of neurons, only 76% (144/154) of the tetramers would be matched. Even though the 5 “C” members are always expressed in common, between a pair of neurons only 1.2% (54/154) of their tetramers will match. Therefore, the tetramers of the clustered Pcdh members can account for an exponential diversity in Pcdh tetramers between any given pair of neurons (Yagi, 2012). Interestingly, recent physiological studies revealed that the features of the neuronal connections in vivo include a non-Gaussian (long-tail) distribution of synaptic weight (Song et al., 2005; Lefort et al., 2009; Ikegaya et al., 2013).
REGULATION OF CLUSTERED PROTOCADHERIN EXPRESSION
The clustered Pcdh genes, except for the 5 “C” members, are randomly expressed from individual alleles in individual neurons. The sequence upstream of each variable exon contains its own promoter, which includes a conserved sequence element (CSE) (Wu & Maniatis, 1999). The CSE is required for the expression of the variable exons (Tasic et al., 2002; Wang et al., 2002a). The cis-regulatory elements are identified by their sequence conservation and hypersensitivity to DNase I (DNase I hypersensitive site; HS). Interestingly, the cis-regulatory elements independently control each gene cluster (Ribich et al., 2006; Yokota et al., 2011). Deletion of the HS5-1 site in mice leads to significantly reduced expressions of Pcdh-α6 to -α12 and -αC1 (Kehayova et al., 2011; Yokota et al., 2011). Deletion of the HS16–20 site (called the cluster control region; CCR) leads to a nearly complete loss of random allelic expression across the Pcdh-β cluster. Notably, deletion of the CCR has a much smaller effect on the closer Pcdh-γ cluster, and no effect on the Pcdh-α cluster in mice (Yokota et al., 2011), demonstrating that the CCR is an important cis-element for long-range independent regulation of the Pcdh-β cluster (Figure 1).
The random expressions of the Pcdh-α isoforms depend on the number of variable exons in the Pcdh-α cluster. In cells bearing an allele in which variable exons α2 to α11 (from the total α1 to α12) are deleted, the expressional frequencies of the remaining variable exons α1 and α12 are increased, compared with those of the wild-type allele. That is, each neuron expresses α1, α12, or both from the deletion allele, whereas α1 and α12 are expressed only rarely (i.e., stochastically) from the wild-type allele. Interestingly, the total Pcdh-α expression level is maintained even in the deletion allele (Noguchi et al., 2009). Thus, the expressions of the variable exons are stochastic, like the results of throwing dice.
Results from several mouse lines with deletions or duplications within the Pcdh-α variable exon cluster support the hypothesis that the variable isoforms are stochastically expressed, that the frequency of their expression depends on the number of variable exons in the cluster, and that the total expression level is maintained by long-range regulation by the outer cis-regulatory elements. Interestingly, in these deletion constructs, the 3’-most variable exon in each construct assumed a ubiquitous and biallelic expression pattern similar to that of αC2 in the Pcdh-α cluster in the wild-type allele.
The CCCTC-binding factor (CTCF), a zinc finger transcription factor, completely regulates the random allelic expression of the clustered Pcdhs at the individual neuron level, but does not down-regulate the constitutively expressed αC2, γC3, γC4, and γC5 (Hirayama et al., 2012). CTCF-binding sites are found in the randomly expressed Pcdh promoters and in HS5-1 and HS16–20 (Figure 1) (Kehayova et al., 2011; Golan-Mashiach et al., 2012; Hirayama et al., 2012). CTCF is known to mediate enhancer and promoter interactions by DNA looping (Merkenschlager & Odom, 2013). In long-range regulation, the DNA looping between a promoter and enhancer mediated by CTCF account for the “random choice” of clustered Pcdh genes in individual neurons. Notably, neurons lacking CTCF exhibit abnormal dendritic arborization and decreased spine density (and therefore decreased numbers of synapses), but show no effects on synapse maturation or the amplitude of excitatory synaptic currents. In addition, mice with a conditional CTCF KO in differentiated excitatory neurons exhibit a somatosensory map defect (a loss of barrel structure) in the brain. Thus, CTCF affects the final refinement of functional neural circuits during postnatal brain development (Hirayama et al., 2012).
The Pcdh expression in individual neurons is epigenetically controlled, independently and monoallelically (Tasic et al., 2002; Kawaguchi et al., 2008). Using two mouse cell lines that express different Pcdh-α isoform combinations, the DNA methylation of each Pcdh-α promoter was found to be negatively correlated with its expression level (Kawaguchi et al., 2008). In addition, the induction of DNA demethylation with 5-azacytidin increases the Pcdh-α transcription. Consistent with their constitutive expression in neurons, the promoters of Pcdh-αC1 and -αC2 are hypomethylated in the brain (Kawaguchi et al., 2008).
The hypermethylation of multiple clustered Pcdh genes is observed in mouse offspring receiving a low frequency of maternal care (licking and grooming behaviors), whereas many of the clustered Pcdh genes show significantly higher expression in offspring receiving a high frequency of maternal care (McGowan et al., 2011). Similar results were found in humans; hippocampal samples from suicide victims with a history of severe child abuse exhibit hypermethylation across the clustered Pcdh genes (Suderman et al., 2012). It is possible that the clustered Pcdh genes play an important role in the response to environmental stress. Interestingly, Tet1 protein, a DNA hydroxylase that converts 5-methyl cytosine to 5-hydroxymethylcytosine in DNA demethylation, binds across the clustered Pcdh genes (Xu et al., 2011). These epigenetic studies and the molecular functions of the clustered Pcdhs in axonal targeting and dendritic arborization suggest that they are important mediators of neural circuit formation that respond to environmental stress during brain development. Furthermore, since epigenetic modifications in individual cells are maintained during cell proliferation, the neuronal individuality based on the random and combinatorial expression of the clustered Pcdh genes could be responsible for the clonal diversity of differentiated neurons related to cell lineage.
DETERMINANTS OF NEURONAL INDIVIDUALITY
Combinatorial explosion can generate a nearly limitless variety of information through the genetic code simply through variations in ATGC sequence combinations. In the immune system, combinations of random DNA rearrangements of variable regions, somatic mutations, and receptor heteromultimers generate the predetermined immune response to an unpredictable antigen. In the brain, neurons connect to form specific neural networks. Donald Hebb hypothesized the “cell assembly” hypothesis for the encoding of neural information by functional neuronal groups (Hebb, 1949). In Hebb's cell assembly hypothesis, a nearly limitless number of combinatorial neuronal groups is produced from a limited number of neurons by combinatorial explosion. Recent physiological studies involving large-scale recordings from individual neurons have experimentally defined putative cell assemblies (reviewed by Buzsaki, 2010). Neuronal individuality is likely to be generated by both a neuron's Pcdh expression pattern and its participation in circuits.
CONCLUSION
Genetic studies of the clustered Pcdhs have gradually unveiled their functions in axonal targeting, dendritic arborization, self-avoidance of dendrites, and synapse formation. In addition, the clustered Pcdhs contribute to neuronal individuality (approximate 1010 neuronal diversity) by random allelic expression. This individuality is generated by epigenetic regulation in the genome during development. Furthermore, heterotetramers of the clustered Pcdhs provide specific cell-adhesion activities and exponential neuronal diversity at the cell surface.
Recent physiological studies have revealed that local networks form complex topological neuronal ensembles in which there are differences at the individual neuron level (Markram et al., 1997; Kalisman et al., 2005; Song et al., 2005; Yoshimura et al., 2005; Koulakov et al., 2009; Yassin et al., 2010; Perin et al., 2011). In addition, the synaptic weight of the local connectivity has a non-Gaussian (long-tail) distribution (Song et al., 2005; Lefort et al., 2009; Ikegaya et al., 2013). Specific local connectivities develop preferentially among clonally related excitatory cortical neurons (Yu et al., 2009, 2012; Li et al., 2012; Ohtsuki et al., 2012). Theoretical analyses of neural networks have suggested that complex topological networks exist in the brain (Sporns, 2011). Interestingly, Watts and Strogatz showed that “small-world” networks with high clustering coefficients and short characteristic path lengths, but without strong habbs, emerge between random and regular networks (Watts & Strogatz, 1998). It is estimated that the neural wiring in the brain is a Watts and Strogatz–type “small-world” network. The molecular features of the clustered Pcdhs, which permit random and selective connectivity, provide a fascinating mechanism underlying the building of complex networks in the brain.
ACKNOWLEGMENTS
I thank members of our laboratory and our CREST team for discussions.
Declaration of interest: The author reports no conflicts of interest. The author alone is responsible for the content and writing of the paper.
This work was supported in part by a Grant- in-Aid for Scientific Research (S), a Grant-in-Aid for Scientific Research on an Innovative Area “Mesoscopic Neurocircuitry” from the Ministry of Education, Science, Sports, and Culture of Japan (MEXT), and CREST from Japan Science and Technology Agency (JST) (T.Y.).
REFERENCES
- Anitha A., Thanseem I., Nakamura K., Yamada K., Iwayama Y., Toyota T., Iwata Y., Suzuki K., Sugiyama T., Tsujii M., Yoshikawa T., Mori N. Protocadherin alpha (PCDHA) as a novel susceptibility gene for autism. J Psychiatry Neurosci. (2012);37:12. doi: 10.1503/jpn.120058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L., Axel R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell. (1991);65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Buzsaki G. Neural syntax: Cell assemblies, synapsembles, and readers. Neuron. (2010);68:362–385. doi: 10.1016/j.neuron.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Lu Y., Meng S., Han M. H., Lin C., Wang X. alpha- and gamma-Protocadherins negatively regulate PYK2. J Biol Chem. (2009);284:2880–2890. doi: 10.1074/jbc.M807417200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esumi S., Kakazu N., Taguchi Y., Hirayama T., Sasaki A., Hirabayashi T., Koide T., Kitsukawa T., Hamada S., Yagi T. Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat Genet. (2005);37:171–176. doi: 10.1038/ng1500. [DOI] [PubMed] [Google Scholar]
- Evrony G. D., Cai X., Lee E., Hills L. B., Elhosary P. C., Lehmann H. S., Parker J. J., Atabay K. D., Gilmore E. C., Poduri A., Park P. J., Walsh C. A. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. (2012);151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Monreal M., Kang S., Phillips G. R. Gamma-protocadherin homophilic interaction and intracellular trafficking is controlled by the cytoplasmic domain in neurons. Mol Cell Neurosci. (2009);40:344–353. doi: 10.1016/j.mcn.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda E., Hamada S., Hasegawa S., Katori S., Sanbo M., Miyakawa T., Yamamoto T., Yamamoto H., Hirabayashi T., Yagi T. Down-regulation of protocadherin-alpha A isoforms in mice changes contextual fear conditioning and spatial working memory. Eur J Neurosci. (2008);28:1362–1376. doi: 10.1111/j.1460-9568.2008.06428.x. [DOI] [PubMed] [Google Scholar]
- Garrett A. M., Schreiner D., Lobas M. A., Weiner J.A. gamma-protocadherins control cortical dendrite arborization by regulating the activity of a FAK/PKC/MARCKS signaling pathway. Neuron. (2012);74:269–276. doi: 10.1016/j.neuron.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett A. M., Weiner J. A. Control of CNS synapse development by {gamma}-protocadherin-mediated astrocyte-neuron contact. J Neurosci. (2009);29:11723–11731. doi: 10.1523/JNEUROSCI.2818-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelbrant A., Hutchinson J. N., Thompson B. R., Chess A. Widespread monoallelic expression on human autosomes. Science. (2007);318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- Golan-Mashiach M., Grunspan M., Emmanuel R., Gibbs-Bar L., Dikstein R., Shapiro E. Identification of CTCF as a master regulator of the clustered protocadherin genes. Nucleic Acids Res. (2012);40:3378–3391. doi: 10.1093/nar/gkr1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M. H., Lin C., Meng S., Wang X. Proteomics analysis reveals overlapping functions of clustered protocadherins. Mol Cell Proteomics. (2010);9:71–83. doi: 10.1074/mcp.M900343-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa S., Hamada S., Kumode Y., Esumi S., Katori S., Fukuda E., Uchiyama Y., Hirabayashi T., Mombaerts P., Yagi T. The protocadherin-alpha family is involved in axonal coalescence of olfactory sensory neurons into glomeruli of the olfactory bulb in mouse. Mol Cell Neurosci. (2008);38:66–79. doi: 10.1016/j.mcn.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Hasegawa S., Hirabayashi T., Kondo T., Inoue K., Esumi S., Okayama A., Hamada S., Yagi T. Constitutively expressed Protocadherin-alpha regulates the coalescence and elimination of homotypic olfactory axons through its cytoplasmic region. Front Mol Neurosci. (2012);5:97. doi: 10.3389/fnmol.2012.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebb D.O. The organization of behavior. New York: John Wiley & Sons; (1949). [Google Scholar]
- Hirano K., Kaneko R., Izawa T., Kawaguchi M., Kitsukawa T., Yagi T. Single-neuron diversity generated by Protocadherin-beta cluster in mouse central and peripheral nervous systems. Front Mol Neurosci. (2012);5:90. doi: 10.3389/fnmol.2012.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani N., Teramae J. N., Fukai T. Associative memory model with long-tail-distributed Hebbian synaptic connections. Front Comput Neurosci. (2013);6:102. doi: 10.3389/fncom.2012.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T., Tarusawa E., Yoshimura Y., Galjart N., Yagi T. CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep. (2012);2:345–357. doi: 10.1016/j.celrep.2012.06.014. [DOI] [PubMed] [Google Scholar]
- Hirayama T., Yagi T. Clustered protocadherins and neuronal diversity. Prog Mol Biol Transl Sci. (2013);116:145–167. doi: 10.1016/B978-0-12-394311-8.00007-8. [DOI] [PubMed] [Google Scholar]
- Ikegaya Y., Sasaki T., Ishikawa D., Honma N., Tao K., Takahashi N., Minamisawa G., Ujita S., Matsuki N. Interpyramid Spike transmission stabilizes the sparseness of recurrent network activity. Cereb Cortex. (2013);23:293–304. doi: 10.1093/cercor/bhs006. [DOI] [PubMed] [Google Scholar]
- Jeffries A. R., Perfect L. W., Ledderose J., Schalkwyk L. C., Bray N. J., Mill J., Price J. Stochastic choice of allelic expression in human neural stem cells. Stem Cells. (2012);30:1938–1947. doi: 10.1002/stem.1155. [DOI] [PubMed] [Google Scholar]
- Kalisman N., Silberberg G., Markram H. The neocortical microcircuit as a tabula rasa. Proc Natl Acad Sci U S A. (2005);102:880–885. doi: 10.1073/pnas.0407088102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko R., Kato H., Kawamura Y., Esumi S., Hirayama T., Hirabayashi T., Yagi T. Allelic gene regulation of Pcdh-alpha and Pcdh-gamma clusters involving both monoallelic and biallelic expression in single Purkinje cells. J Biol Chem. (2006);281:30551–30560. doi: 10.1074/jbc.M605677200. [DOI] [PubMed] [Google Scholar]
- Katori S., Hamada S., Noguchi Y., Fukuda E., Yamamoto T., Yamamoto H., Hasegawa S., Yagi T. Protocadherin-alpha family is required for serotonergic projections to appropriately innervate target brain areas. J Neurosci. (2009);29:9137–9147. doi: 10.1523/JNEUROSCI.5478-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi M., Toyama T., Kaneko R., Hirayama T., Kawamura Y., Yagi T. Relationship between DNA methylation states and transcription of individual isoforms encoded by the protocadherin-alpha gene cluster. J Biol Chem. (2008);283:12064–12075. doi: 10.1074/jbc.M709648200. [DOI] [PubMed] [Google Scholar]
- Kehayova P., Monahan K., Chen W., Maniatis T. Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster. Proc Natl Acad Sci U S A. (2011);108:17195–17200. doi: 10.1073/pnas.1114357108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohmura N., Senzaki K., Hamada S., Kai N., Yasuda R., Watanabe M., Ishii H., Yasuda M., Mishina M., Yagi T. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron. (1998);20:1137–1151. doi: 10.1016/s0896-6273(00)80495-x. [DOI] [PubMed] [Google Scholar]
- Koulakov A. A., Hromadka T., Zador A. M. Correlated connectivity and the distribution of firing rates in the neocortex. J Neurosci. (2009);29:3685–3694. doi: 10.1523/JNEUROSCI.4500-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledderose J., Dieter S., Schwarz M. K. Maturation of postnatally generated olfactory bulb granule cells depends on functional gamma-protocadherin expression. Sci Rep. (2013);3:1514. doi: 10.1038/srep01514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre J. L., Kostadinov D., Chen W. V., Maniatis T., Sanes J. R. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature. (2012);488:517–521. doi: 10.1038/nature11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort S., Tomm C., Floyd Sarria J. C., Petersen C. C. The excitatory neuronal network of the C2 barrel column in mouse primary somatosensory cortex. Neuron. (2009);61:301–316. doi: 10.1016/j.neuron.2008.12.020. [DOI] [PubMed] [Google Scholar]
- Li Y., Lu H., Cheng P.L., Ge S., Xu H., Shi S.H., Dan Y. Clonally related visual cortical neurons show similar stimulus feature selectivity. Nature. (2012);486:118–121. doi: 10.1038/nature11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M. R. The mechanism of V(D)J recombination: A balance of diversity, specificity, and stability. Cell. (1992);70:873–876. doi: 10.1016/0092-8674(92)90237-7. [DOI] [PubMed] [Google Scholar]
- Markram H. A network of tufted layer 5 pyramidal neurons. Cereb Cortex. (1997);7:523–533. doi: 10.1093/cercor/7.6.523. [DOI] [PubMed] [Google Scholar]
- McGowan P. O., Suderman M., Sasaki A., Huang T. C., Hallett M., Meaney M. J., Szyf M. Broad epigenetic signature of maternal care in the brain of adult rats. PLoS ONE. (2011);6:e14739. doi: 10.1371/journal.pone.0014739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M., Odom D. T. CTCF and cohesin: Linking gene regulatory elements with their targets. Cell. (2013);152:1285–1297. doi: 10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- Miki R., Hattori K., Taguchi Y., Tada M. N., Isosaka T., Hidaka Y., Hirabayashi T., Hashimoto R., Fukuzako H., Yagi T. Identification and characterization of coding single-nucleotide polymorphisms within human protocadherin-alpha and -beta gene clusters. Gene. (2005);349:1–14. doi: 10.1016/j.gene.2004.11.044. [DOI] [PubMed] [Google Scholar]
- Morishita H., Umitsu M., Murata Y., Shibata N., Udaka K., Higuchi Y., Akutsu H., Yamaguchi T., Yagi T., Ikegami T. Structure of the cadherin-related neuronal receptor/protocadherin-alpha first extracellular cadherin domain reveals diversity across cadherin families. J Biol Chem. (2006);281:33650–33663. doi: 10.1074/jbc.M603298200. [DOI] [PubMed] [Google Scholar]
- Morishita H., Yagi T. Protocadherin family: Diversity, structure, and function. Curr Opin Cell Biol. (2007);19:584–592. doi: 10.1016/j.ceb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Muotri A. R., Gage F. H. Generation of neuronal variability and complexity. Nature. (2006);441:1087–1093. doi: 10.1038/nature04959. [DOI] [PubMed] [Google Scholar]
- Murata Y., Hamada S., Morishita H., Mutoh T., Yagi T. Interaction with protocadherin-gamma regulates the cell surface expression of protocadherin-alpha. J Biol Chem. (2004);279:49508–49516. doi: 10.1074/jbc.M408771200. [DOI] [PubMed] [Google Scholar]
- Noguchi Y., Hirabayashi T., Katori S., Kawamura Y., Sanbo M., Hirabayashi M., Kiyonari H., Nakao K., Uchimura A., Yagi T. Total expression and dual gene-regulatory mechanisms maintained in deletions and duplications of the Pcdha cluster. J Biol Chem. (2009);284:32002–32014. doi: 10.1074/jbc.M109.046938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan J. P., Li J., Nguyen L., Caoile C., Dickson M., Grimwood J., Schmutz J., Feldman M. W., Myers R. M. Extensive linkage disequilibrium, a common 16.7-kilobase deletion, and evidence of balancing selection in the human protocadherin alpha cluster. Am J Hum Genet. (2003);72:621–635. doi: 10.1086/368060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki G., Nishiyama M., Yoshida T., Murakami T., Histed M., Lois C., Ohki K. Similarity of visual selectivity among clonally related neurons in visual cortex. Neuron. (2012);75:65–72. doi: 10.1016/j.neuron.2012.05.023. [DOI] [PubMed] [Google Scholar]
- Perin R., Berger T. K., Markram H. A synaptic organizing principle for cortical neuronal groups. Proc Natl Acad Sci U S A. (2011);108:5419–5424. doi: 10.1073/pnas.1016051108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips G. R., Tanaka H., Frank M., Elste A., Fidler L., Benson D. L., Colman D. R. Gamma-protocadherins are targeted to subsets of synapses and intracellular organelles in neurons. J Neurosci. (2003);23:5096–5104. doi: 10.1523/JNEUROSCI.23-12-05096.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redies C., Hertel N., Hubner C. A. Cadherins and neuropsychiatric disorders. Brain Res. (2012);1470:130–144. doi: 10.1016/j.brainres.2012.06.020. [DOI] [PubMed] [Google Scholar]
- Rehen S. K., Yung Y. C., McCreight M. P., Kaushal D., Yang A. H., Almeida B. S., Kingsbury M. A., Cabral K. M., McConnell M.J., Anliker B., et al. Constitutional aneuploidy in the normal human brain. J Neurosci. (2005);25:2176–2180. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribich S., Tasic B., Maniatis T. Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proc Natl Acad Sci U S A. (2006);103:19719–19724. doi: 10.1073/pnas.0609445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H. Neural map formation in the mouse olfactory system. Neuron. (2010);67:530–542. doi: 10.1016/j.neuron.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Schreiner D., Weiner J. A. Combinatorial homophilic interaction between gamma-protocadherin multimers greatly expands the molecular diversity of cell adhesion. Proc Natl Acad Sci U S A. (2010);107:14893–14898. doi: 10.1073/pnas.1004526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T., McConnell M. J., Marchetto M. C., Coufal N. G., Gage F.H. LINE-1 retrotransposons: Mediators of somatic variation in neuronal genomes? Trends Neurosci. (2010);33:345–354. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Sjostrom P. J., Reigl M., Nelson S., Chklovskii D. B. Highly nonrandom features of synaptic connectivity in local cortical circuits. PLoS Biol. (2005);3:e68. doi: 10.1371/journal.pbio.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O. Networks of the brain. Cambridge: The MIT Press; (2011). [Google Scholar]
- Suderman M., McGowan P. O., Sasaki A., Huang T. C., Hallett M. T., Meaney M. J., Turecki G., Szyf M. Conserved epigenetic sensitivity to early life experience in the rat and human hippocampus. Proc Natl Acad Sci U S A. (2012);109((Suppl 2)):17266–17272. doi: 10.1073/pnas.1121260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo L., Lu H., Ying G., Capecchi M. R., Wu Q. Protocadherin clusters and cell adhesion kinase regulate dendrite complexity through Rho GTPase. J Mol Cell Biol. (2012);4:362–376. doi: 10.1093/jmcb/mjs034. [DOI] [PubMed] [Google Scholar]
- Takeichi M. The cadherin superfamily in neuronal connections and interactions. Nat Rev Neurosci. (2007);8:11–20. doi: 10.1038/nrn2043. [DOI] [PubMed] [Google Scholar]
- Tasic B., Nabholz C. E., Baldwin K. K., Kim Y., Rueckert E. H., Ribich S. A., Cramer P., Wu Q., Axel R., Maniatis T. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol Cell. (2002);10:21–33. doi: 10.1016/s1097-2765(02)00578-6. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. (1983);302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Ukkola-Vuoti L., Kanduri C., Oikkonen J., Buck G., Blancher C., Raijas P., Karma K., Lahdesmaki H., Jarvela I. Genome-wide copy number variation analysis in extended families and unrelated individuals characterized for musical aptitude and creativity in music. PLoS One. (2013);8:e56356. doi: 10.1371/journal.pone.0056356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Su H., Bradley A. Molecular mechanisms governing Pcdh-gamma gene expression: Evidence for a multiple promoter and cis-alternative splicing model. Genes Dev. (2002a);16:1890–1905. doi: 10.1101/gad.1004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Weiner J. A., Levi S., Craig A. M., Bradley A., Sanes J. R. Gamma protocadherins are required for survival of spinal interneurons. Neuron. (2002b);36:843–854. doi: 10.1016/s0896-6273(02)01090-5. [DOI] [PubMed] [Google Scholar]
- Watts D. J., Strogatz S. H. Collective dynamics of ‘small-world’ networks. Nature. (1998);393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Weiner J. A., Jontes J. D. Protocadherins, not prototypical: A complex tale of their interactions, expression, and functions. Front Mol Neurosci. (2013);6:4. doi: 10.3389/fnmol.2013.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. (1999);97:779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- Xu Y., Wu F., Tan L., Kong L., Xiong L., Deng J., Barbera A. J., Zheng L., Zhang H., Huang S., et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. (2011);42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T. Clustered protocadherin family. Dev Growth Differ. (2008);50:S131–S140. doi: 10.1111/j.1440-169X.2008.00991.x. [DOI] [PubMed] [Google Scholar]
- Yagi T. Molecular codes for neuronal individuality and cell assembly in the brain. Front Mol Neurosci. (2012);5:45. doi: 10.3389/fnmol.2012.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T., Takeichi M. Cadherin superfamily genes: Functions, genomic organization, and neurologic diversity. Genes Dev. (2000);14:1169–1180. [PubMed] [Google Scholar]
- Yamashita H., Chen S., Komagata S., Hishida R., Iwasato T., Itohara S., Yagi T., Endo N., Shibata M., Shibuki K. Restoration of contralateral representation in the mouse somatosensory cortex after crossing nerve transfer. PLoS ONE. (2012);7:e35676. doi: 10.1371/journal.pone.0035676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassin L., Benedetti B. L., Jouhanneau J. S., Wen J. A., Poulet J. F., Barth A. L. An embedded subnetwork of highly active neurons in the neocortex. Neuron. (2010);68:1043–1050. doi: 10.1016/j.neuron.2010.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota S., Hirayama T., Hirano K., Kaneko R., Toyoda S., Kawamura Y., Hirabayashi M., Hirabayashi T., Yagi T. Identification of the cluster control region for the protocadherin-beta genes located beyond the protocadherin-gamma cluster. J Biol Chem. (2011);286:31885–31895. doi: 10.1074/jbc.M111.245605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura Y., Callaway E. M. Fine-scale specificity of cortical networks depends on inhibitory cell type and connectivity. Nat Neurosci. (2005);8:1552–1559. doi: 10.1038/nn1565. [DOI] [PubMed] [Google Scholar]
- Yoshimura Y., Dantzker J. L., Callaway E. M. Excitatory cortical neurons form fine-scale functional networks. Nature. (2005);433:868–873. doi: 10.1038/nature03252. [DOI] [PubMed] [Google Scholar]
- Yu Y. C., Bultje R. S., Wang X., Shi S. H. Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature. (2009);458:501–504. doi: 10.1038/nature07722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y. C., He S., Chen S., Fu Y., Brown K. N., Yao X. H., Ma J., Gao K. P., Sosinsky G. E., Huang K., Shi S. H. Preferential electrical coupling regulates neocortical lineage-dependent microcircuit assembly. Nature. (2012);486:113–117. doi: 10.1038/nature10958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipursky S. L., Sanes J. R. Chemoaffinity revisited: Dscams, protocadherins, and neural circuit assembly. Cell. (2010);143:343–353. doi: 10.1016/j.cell.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Zwemer L. M., Zak A., Thompson B. R., Kirby A., Daly M. J., Chess A., Gimelbrant A. A. Autosomal monoallelic expression in the mouse. Genome Biol. (2012);13:R10. doi: 10.1186/gb-2012-13-2-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]