Abstract
Breast Cancer Stem (BCS) cells play critical roles in self-renewal, Multi Drug Resistance (MDR), differentiation and generation of secondary tumors. Conventional chemotherapy may efficiently kill the bulk of differentiated drug sensitive breast cancer cells, but not the MDR self-renewable BCS cells, leading to enrichment of the MDR BCS cells. In order to target the MDR BCS cells, we have isolated: 1) BCS cells from either breast cancer cell lines or fresh breast cancer specimens; 2) ATP binding cassette (ABC) transporter group G number 2 (ABCG2)-specific aptamers; and 3) BCS cell-binding aptamers. Interestingly, ABCG2-specific aptamers labeled the membrane surface of the ABCG2-expressing baby hamster kidney (BHK) cells, but stained whole cells of the BCS cells derived from mammospheres, implying that BCS cells might have much higher rate of endocytosis than the ABCG2-expressing BHK cells. In addition, 5D3, a monoclonal antibody that recognizes the extracellular loops of ABCG2 protein, also stained whole BCS cells. Furthermore, BCS cell-binding aptamers stained whole BCS cells, but not the differentiated breast cancer MCF-7 cells. All these results support above conclusion that BCS cells might have high rate of endocytosis. Further experiments performed with aptamers and human transferrin or lactosylceramide showed that BCS cells do have much higher endocytosis rate than the differentiated breast cancer cells. Interestingly, clathrin dependent endocytosis inhibitors, such as monodansylcadaverine or sucrose, or caveolin-dependent endocytosis inhibitors, such as methyl-β-cyclodextrin or genistein, can inhibit the internalization of transferrin or lactosylceramide into the differentiated breast cancer cells, but cannot block the internalization of these compounds into the BCS cells, suggesting that BCS cells undergo clathrin-independent and caveolin-independent endocytosis. Taken together, our data suggest that BCS cells have high rate of endocytosis and open the possibilities for delivering therapeutic agents directly into the MDR BCS cells with aptamer-coated liposomes.
Keywords: Breast cancer, BCS cell, Mammosphere, Aptamer, ABCG2, Endocytosis
Introduction
Breast cancer is the most common malignancy in women [1,2]. Although surgical removal of breast cancer plays a very important role in treating the patients, the treatments with chemotherapy often fail to eradicate the tumor. In fact, many cancers, including breast cancer, initially respond well to chemotherapy. Very often the tumors become resistant to anticancer drugs during or shortly after the chemotherapy [3], presumably because of their inabilities to eradicate the MDR cancer stem cells [4–6].
Cancer stem cells play very important roles in self-renewal, MDR and generation of secondary tumors [7–10]. Their self-renewal, growth, differentiation and epithelial to mesenchymal transition are regulated by Wnt, Notch, Hedgehog and transforming growth factor β signaling pathways [11–17]. For example, Wnt enhances C-myc and cyclin D1 expression [18,19] and induces oncogenic proliferation [20,21]. Wnt and Notch signaling pathways also play a very important role in regulating the expression of ABC transporters [22]. Down-regulating the Wnt signaling significantly decreased the expression of ABCB1 and ABCG2 [23–25]. In contrast, Wnt agonist enhances the expression of β-catenin and increases the protein levels of ABCB1 and ABCG2 [23–25]. ABCG2 [26–28] is universally expressed in the undifferentiated cancer stem cells [29,30] and its expression is shut down in many differentiated cells [29,31]. Thus, conventional chemotherapy may efficiently kill the bulk of differentiated drug sensitive breast cancer cells, but not the MDR self-renewable BCS cells, leading to enrichment of the MDR BCS cells. The enriched MDR BCS cells have abilities to renew themselves and to differentiate to cancer cells. Thus, without eliminating the MDR BCS cells, it is impossible to eradicate breast cancers.
In order to target the ABCG2-expressing MDR BCS cells, we thought about to isolate ABCG2-specific ligands that recognize the extracellular portions of this trans-membrane protein so that we can selectively deliver therapeutic agents into BCS cells by using ABCG2-specific ligand-coated liposomes harboring therapeutic agents. However, since normal stem cells and other tissues, such as hepatic stem cells [32], lung stem cells [33], and placenta cells [34] etc, also express ABCG2, the treatment with ABCG2-specific ligand-coated liposomes harboring therapeutic agents might cause side effects. Thus, we thought about to isolate BCS cell-specific ligands. In this report, we have enriched and cloned ABCG2-specific aptamers and BCS cell-binding aptamers and found that BCS cells have significantly higher rate of clathrin-independent and caveolin-independent endocytosis than the differentiated breast cancer cells.
Materials and Methods
Materials
Aptamer library, primers, Texas Red (TR)-labeled primers and Fluorescein Iso Thio Cyanate (FITC) labeled primers were derived from Mayo Clinic Molecular Biology Core. Fresh human breast cancer specimens were derived from Mayo Clinic Arizona [collected by Dr. Pockaj upon written informed consent, based on her Institutional Review Board (IRB) # 2130-00 00 (Titled “Cancer Tissue Study”)]. Since isolation of BCS cells was performed in Dr. Chang’s laboratory, Dr. Chang was also required to get approval from Mayo Clinic IRB. Dr. Chang’s IRB number for this study is # 10-005974 (Titled “Roles of Wnt Signaling in Breast Cancer Stem Cells Self-renewal and Multidrug Resistance”) and there was a special note to indicate that “Patients must consent under IRB # 2130-00 00 in order to participate in this study. In addition, “the IRB approves waiver of specific informed consent in accordance with 45CFR 46.116 (d) as justified by the investigator and waiver of HIPAA authorization in accordance with applicable HIPAA regulations.” Furthermore, patients’ information was de-identified. Fetal bovine serum (FBS) was from Gemini Bio-Products (Sacramento, California, USA). Collagenase IV, Red-taq DNA polymerase, propidium iodide (PI), endocytosis inhibitors monodansylcadaverine (MDC), sucrose, genistein, methyl-β-cyclodextrin (MCD) and routine chemicals were from Sigma (St. Louis, Missouri, USA). Mounting media with 4′,6-diamidino-2-phenylindole (DAPI) was from Vector Laboratory (Burlingame, California, USA). Aldefluor kit, MammoCult media, supplementary bullet and 100 μm cell strainer were from StemCell Technologies (Vancouver, BC, Canada). HAM’s nutrient mixture F12 was from JRH Biosciences (Lenexa, Kansas, USA). DMEM/F-12, RPMI-1640 and OptiMEM, TOPO TA cloning vector, Alexa fluor 633-conjugated human transferrin (hTF) and BODIPY-labeled lactosylceramide (LacCer) were from Invitrogen (Grand Island, New York, USA). Allophycocyanin (APC)-labeled ABCG2-specific monoclonal antibody (mAb) 5D3 was from Santa Cruz Biotechnology (Santa Cruz, California, USA). APC-labeled CD44 mAb was from BD Biosciences (San Jose, California, USA). Ultralow attachment plates were from Corning (Corning, New York, USA). Millicell chambered cell-culture glass slides were purchased from Millipore (Billerica, Massachusetts, USA).
Cell culture
BHK cells (from ATCC CCL-10) were cultured in DMEM/F-12 media supplemented with 5% FBS at 37°C in 5% CO2. ABCG2 cDNA transfected BHK (BHK/ABCG2) cells were cultured according to the method mentioned previously [35]. MCF-7 (from ATCC HTB-22) and MDA-MB-231 (from ATCC HTB-26) cells were cultured in RPMI-1640 media supplemented with 10% FBS at 37°C in 5% CO2. For staining of these cells, the cells were grown in Millicell chambered cell-culture glass slides.
Aldefluor assay and isolation of BCS cells
The isolation of BCS cells from breast cancer cell lines, including MCF-7 and MDA-MB-231, was performed by using Aldefluor-assay kit [36]. The isolation of BCS cells from fresh human breast cancer specimens was performed according to the method described [36]. Briefly, human primary breast cancer specimens were chopped into small pieces, washed with HAM’s nutrient mixture F12 supplemented with 1% penicillin/streptomycin and 1% fungizone and then digested with collagenase type IV (100 U/ml in RPMI 1640 media supplemented with 5% FBS) and hyaluronidase (100 U/ml in RPMI 1640 media supplemented with 5% FBS) for approximately 2 h at 37°C. The dissociated cells were filtered through a sterile 100 μm cell strainer. The cells were suspended in Aldefluor-assay buffer containing 1.5 μM Aldehyde Dehydrogenase (ALDH) substrate BODIPY-aminoacetaldehyde in the presence or absence of 50 mM ALDH-specific inhibitor diethylaminobenzaldehyde, incubated for 30 min in the dark at 37°C, washed with Aldefluor-assay buffer for three times and added 1 μg/ml PI to check the cell viability. The sorting gates were established based on the corresponding cells treated with ALDH inhibitor. The ALDH positive (ALDH+) and ALDH negative (ALDH−) cells sorted out by BD FACSAria flow cytometer were cultured in MammoCult media with supplementary bullet in ultralow attachment plates at low density (5,000 viable cells/ml).
Enrichment and cloning of ABCG2-specific aptamers or BCS cell-binding aptamers
The systematic evolution of ligands by exponential enriched DNA or RNA aptamers can specifically recognize their corresponding targets [37–42]. Based on the fact that BHK cells do not express human ABCG2 protein, whereas BHK/ABCG2 cells do [35], we have enriched the ABCG2-specific aptamers by using the protocol described [43–45]. Briefly, BHK/ABCG2 cells were washed with wash buffer [(25 mM glucose, 5 mM MgCl2 and 1 mg/ml bovine serum albumin in phosphate buffered saline (PBS)] before incubation with aptamer library 5′-ACGCTCGGATGCCACTACAG-60 randomized nucleotides-CTCATGGACGTGCTGGTGAC-3′. The aptamer library was heated to 95°C for 5 minutes and cooled down in dry-ice to get single stranded DNA (ssDNA). The ssDNA (10 nM) was mixed with aptamer binding buffer (0.1 mg/ml of yeast tRNA in wash buffer) and used to bind to the target BHK/ABCG2 cells on ice for 30 min. Upon washing away of the unbound aptamers with wash buffer (3 times), the bound aptamers were eluted with aptamer binding buffer by incubating the samples at 95°C for 5 minutes. The eluates, after centrifugation at 13,000×g for 5 minutes, were used to bind to the parental BHK cells (counter-selection). The unbound aptamers were amplified by polymerase chain reaction (PCR) with forward primer 5′-ACGCTCGGATGCCACTACAG-3′, reverse primer 5′-GTCACCAGCACGTCCATGAG-3′ and Red-taq DNA polymerase. The PCR products were used to do another cycle of binding and counter-selection. To enrich BCS cell-binding aptamers, the mammosphere cells derived from MCF-7 were used to do the binding (with aptamer library) whereas the differentiated breast cancer MCF-7 cells were used to do the counter-selection. Once the PCR amplified aptamers reached the stage that they cannot bind to the negative control cells, these aptamers were cloned into TOPO TA cloning vector and sequenced.
Aptamer binding and antibody staining
ABCG2-specific aptamers or BCS cell-binding aptamers were amplified by PCR with either FITC- or TR-labeled primers, denatured at 95°C for 5 minutes and cooled down on dry-ice to make ssDNA. The target cells were washed three times with PBS, incubated with aptamer binding buffer containing human Fc blocker on ice for 5 minutes and then mixed with FITC- or TR-labeled aptamers (125 ng/ml) and/or ABCG2 or CD44 specific mAbs on ice for 30 minutes in the dark. The stained cells were washed three times with ice-cold aptamer wash buffer, performed cytospin (for mammosphere cells grown in suspension) to attach the cells to the charged slides, mounted with anti-fade mounting media containing DAPI and then evaluated with Carl-Zeiss confocal microscope. To separate the FITC-ABCG2/A12 aptamer bound and un-bound BHK/ABCG2 cells (Figure 1C and 1D), BHK and BHK/ABCG2 cells grown up in 100 mm plates were detached from the plates by rubber-policeman scratch, stained with FITC-ABCG2/A12 aptamer and sorted out by BD FACSAria flow cytometer. To test whether trypsin digestion (to destroy the extracellular portions of the membrane surface proteins) would abrogate the aptamer or antibody binding, cells were digested with 0.25% trypsin at 37°C for 5 minutes and then stained them with aptamers or antibodies as described above.
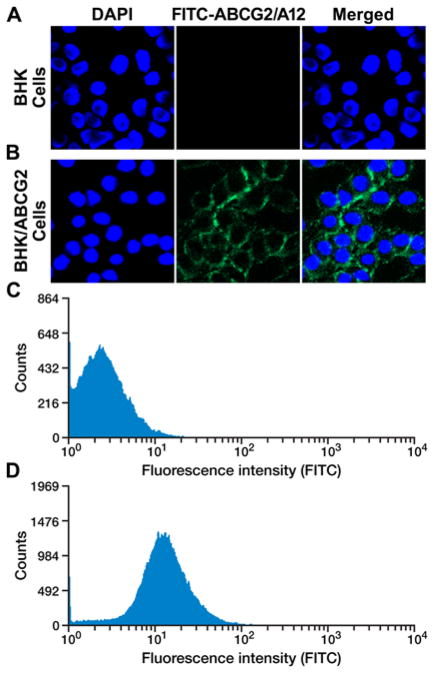
Figure 1.
The cloned ABCG2-specific aptamer ABCG2/A12 clearly stained the surface of ABCG2-expressing BHK/ABCG2 cells. A. ABCG2/A12 cannot stain the parental BHK cells: left section, BHK cells were stained with DAPI; middle section, with FITC-labeled ABCG2/A12; right section, merge of the previous two sections. B. ABCG2/A12 clearly stained the surface of ABCG2-expressing BHK/ABCG2 cells. C. FACS profiles of FITC-labeled ABCG2/A12-stained BHK cells. D. FACS profiles of FITC-labeled ABCG2/A12-stained BHK/ABCG2 cells.
Inhibition of endocytosis
Cells were washed three times with PBS and incubated in optiMEM culture media in the absence or presence of clathrin dependent endocytosis (CDE) inhibitors, such as sucrose (250 mM) or MDC (50 μM), or caveolin-dependent endocytosis (CVDE) inhibitors, such as genistein (25 μM) or MCD (2.5 μM), at 37°C for 30 minutes. The cells were washed with ice-cold aptamer binding buffer once, incubated in the same buffer for 5 min, stained with aptamers on ice for 30 minutes in dark and then incubated with endocytosis markers, such as Alexa fluor 633-conjugated hTF (500 ng/liter) or BODIPY-labeled LacCer (500 nM), at 37°C in dark for 15 minutes. The stained cells were washed three times with ice-cold aptamer wash buffer, performed cytospin to attach the cells to the charged slides, mounted with anti-fade mounting media containing DAPI and then evaluated with Carl-Zeiss confocal microscope.
Mammosphere formation
Mammosphere formation assay was performed according to the method described [46]. Briefly, the cells sorted out by fluorescence-activated cell sorting (FACS) were plated in ultra-low attachment 96-well plates at a density of 1,000 cells per well in MammoCult media supplemented with growth factors. Numbers of mammosphere formation, after 10–14 days culture, were counted under a light microscopy.
Confocal microscopy
Confocal imaging was carried out on a Carl-Zeiss laser scanning microscope equipped with Aprochromat 63×1.40 oil immersion objective lens that is suitable to evaluate fluorescence and differential interference contrast images. Excitation and emission wavelengths (nm) used for the following fluorophores were: DAPI, 360/405; BODIPY and FITC, 488/500–530; Alexa fluor 633, APC and TR, 543/633. Representative confocal images were acquired, analyzed by laser scanning microscope image browser and arranged in Adobe photoshop CS3 software.
Statistical analysis
The results in Figure 3K and Figure S3C were presented as means ± SD. The two-tailed P values were calculated based on the unpaired t test from GraphPad Software Quick Calcs. By conventional criteria, if P value is less than 0.05, the difference between two samples is considered to be statistically significant.
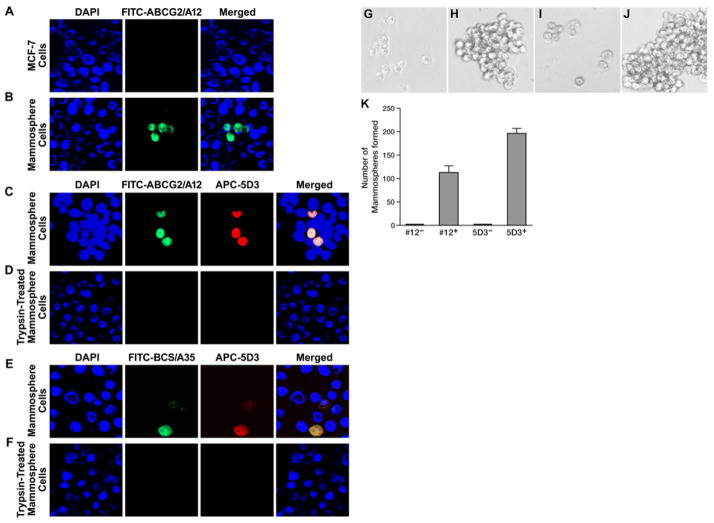
Figure 3.
The cells stained with ABCG2-specific aptamer ABCG2/A12 or ABCG2-specific mAb can form mammospheres. A. ABCG2/A12 did not recognize the differentiated breast cancer MCF-7 cells: left section, cells were stained with DAPI; middle section, with FITC-labeled ABCG2/A12; right section, merge of the previous two sections. B. ABCG2/A12 recognizes the ABCG2-expressing BCS cells in mammospheres derived from MCF-7. C. ABCG2/A12 and ABCG2-specific mAb 5D3 recognized the same BCS cells in mammospheres: left section, cells were stained with DAPI; second section, with FITC-labeled ABCG2/A12; third section, with APC-conjugated ABCG2-specific mAb 5D3; right section, merge of the previous three sections. D. Short time trypsin digestion completely abolished the binding of ABCG2/A12 and ABCG2-specific mAb 5D3 to BCS cells in mammospheres. E. BCS cells in mammospheres derived from MCF-7 were co-stained with BCS/A35 and ABCG2-specific mAb 5D3: left section, cells were stained with DAPI; second section, with FITC-labeled BCS/A35; third section, with APC-conjugated human ABCG2-specific mAb 5D3; right section, merge of the previous three sections. F. Short time trypsin digestion completely abolished the binding of BCS/A35 and ABCG2-specific mAb 5D3 to BCS cells in mammospheres. G. FITC-ABCG2/A12 negative (#12−) cells (4,000), sorted out by FACS from mammospheres derived from MCF-7, can barely form mammospheres. H. FITC-ABCG2/A12 positive (#12+) cells (4,000), sorted out by FACS from mammospheres derived from MCF-7, can form mammospheres. I. APC-conjugated ABCG2-specific mAb 5D3 negative (5D3−) cells (4,000) can barely form mammospheres. J. APC-conjugated ABCG2-specific mAb 5D3 positive (5D3+) cells (4,000) can form mammospheres. K. Numbers of mammospheres formed from above four cell populations.
Results
Enrichment and cloning of ABCG2-specific aptamers
We had found that the parental BHK cells did not have ability to transport methotrexate across the biological membranes whereas the human ABCG2 cDNA transfected BHK cells did [35], suggesting that: 1) ABCG2 protein is located on the membrane surface; 2) the parental BHK cells do not express endogenous counterpart transporter. Indeed, it had been proved that the majority of human ABCG2 protein expressed heterogeneously was located on the cell surface [47]. Therefore, the pair of BHK cells, i.e., the parental BHK cells and the BHK/ABCG2 cells, provided good source of materials for us to enrich the ABCG2-specific aptamers. In order to do so, the ABCG2-expressing BHK/ABCG2 cells (used as binding materials) and the parental BHK cells (used as counter-selection materials) were used to enrich the human ABCG2-specific aptamers by employing the aptamer library and the protocol mentioned in Materials and Methods. After 5, 10 or 15 cycles of binding and counter-selection, the enriched aptamers were labeled with FITC and used to stain the parental BHK cells and the ABCG2- expressing BHK/ABCG2 cells. The results in Figure S1 indicated that, after 15 cycles of binding and counter-selection, the enriched aptamers bound to the ABCG2-expressing BHK/ABCG2 cells (Figure S1B), but not to the parental BHK cells (Figure S1A). In addition, upon short time trypsin digestion, the enriched ABCG2-specific aptamers cannot stain the ABCG2-expressing BHK/ABCG2 cells any more (Figure S1C), suggesting that trypsin digestion destroyed the aptamers’ binding sites.
The enriched ABCG2-specific aptamers were cloned into TOPO TA cloning vector and used to stain the parental BHK cells and the ABCG2-expressing BHK/ABCG2 cells. The results in Figure 1 clearly indicated that ABCG2-specific aptamer clone #12 (ABCG2/A12, its nucleotide sequence is shown in Figure S2) bound to the surface of the ABCG2-expressing BHK/ABCG2 cells (Figure 1B), but not to the parental BHK cells (Figure 1A). Flow cytometry analysis of the ABCG2/A12-labeled cells indicated that the majority of the ABCG2-expressing BHK/ABCG2 cells were labeled with this aptamer (Figure 1D), whereas the parental BHK cells were not (Figure 1C).
Enrichment and cloning of BCS cell-binding aptamers
In order to enrich BCS cell-binding aptamers, we should have BCS cells. BCS cells had been enriched from breast cancer cell lines, such as MCF-7 or MDA-MB-231, or fresh breast cancer specimens by using Aldefluor assay which is based on the intracellular ALDH activity. The results shown in Figure S3 indicated that the ALDH negative (ALDH−) cells cannot form mammospheres in vitro (Figure S3A) whereas the ALDH positive (ALDH+) cells can (Figure S3B), indicating that ALDH+ cells contain BCS cells.
Since we planned to use the cells derived from mammospheres as binding materials for enrichment of BCS cell-binding aptamers, we’d like to know what percentage of the cells in mammospheres were ALDH+ cells. Interestingly, we found that only approximately 18% of the cells derived from mammospheres were ALDH+ cells (Figure S3C), implying that the majority of the cells in mammospheres were differentiated breast cancer cells. This result also suggested that we should separate the ALDH+ cells from the ALDH− cells and then use them to do the enrichment of BCS cell-binding aptamers. However, since we had found that MCF-7 cells have only approximately 0.7% of ALDH+ cells (Figure S3C), we could use them as counter-selection materials to bypass the FACS step. Indeed, by using the same protocol as the one used in the enrichment of ABCG2-specific aptamers, except that the cells derived from mammospheres were used as binding materials whereas the cells derived from MCF-7 were used as counter-selection materials, we had enriched (after 15 cycles of binding and counter-selection) the BCS cell-binding aptamers that bind to some of the cells derived from mammospheres (Figure S4B), but not to the cells derived from MCF-7 (Figure S4A). In addition, some of the cells recognized by BCS cell-binding aptamers also detected by mAb against ABCG2 (Figure S4B), suggesting that they might be the ABCG2-expressing BCS cells. Furthermore, the enriched BCS cell-binding aptamers detected more cells (indicated by the arrowheads in Figure S4B) than the human ABCG2-specific mAb 5D3, implying that some of the mammosphere cell-enriched aptamers might detect the early progenitor cells that did not over-express ABCG2, but were distinguishable from the differentiated breast cancer cells. Of note, the aptamers bound to the cells or the mAb 5D3 bound to the cells were efficiently internalized (Figure S4B), suggesting that BCS cells might have high rate of endocytosis.
Regardless of whether they were BCS cells or early progenitor cells, the treatment with trypsin for short time completely abolished the aptamer binding and the ABCG2-specific mAb binding (Figure S4C), suggesting that trypsin digestion completely destroyed the mAb 5D3 binding site and the aptamers binding sites located on the BCS cell surface.
It has been demonstrated that human breast cancers contain a cell population with stem cell properties bearing the surface markers CD44+/CD24−/lin− [48]. In order to test whether these aptamers would bind to the CD44+ BCS cells or not, the FITC-labeled BCS cell-enriched aptamers and the APC-conjugated CD44-specific mAb were used to co-stain the cells derived from mammospheres. As shown in Figure S5, the cells stained with BCS cell-enriched aptamers were also detected by CD44-specific mAb, suggesting that the BCS cell-enriched aptamers might bind to human BCS cells.
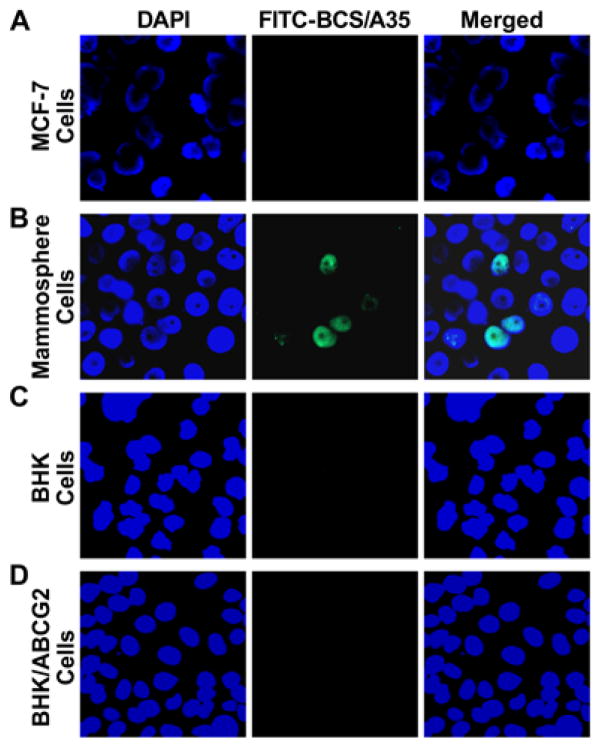
The enriched BCS cell-binding aptamers were cloned into TOPO TA cloning vector and used to stain the control MCF-7 cells and the cells derived from mammospheres. The results in Figure 2A clearly indicated that BCS cell-binding aptamer # 35 (BCS/A35, its nucleotide sequence is shown in Figure S6) stained some cells (internalized) derived from mammospheres (Figure 2B), but not the cells derived from MCF-7 (Figure 2A). In addition, the results shown in Figure 2 indicate that BCS/A35 can neither stain the parental BHK cells (Figure 2C) nor the ABCG2-expressing BHK/ABCG2 cells (Figure 2D), suggesting that BCS/A35 recognized a BCS cell surface protein that is distinguishable from ABCG2.
Figure 2.
The cloned BCS cell-binding aptamer BCS/A35 recognized a BCS cell membrane surface protein that was distinguishable from ABCG2. A. BCS/A35 cannot stain the differentiated breast cancer MCF-7 cells: left section, cells were stained with DAPI; middle section, with FITC-labeled BCS/A35; right section, merge of the previous two sections. B. BCS/A35 stained some of the mammosphere cells derived from MCF-7. C. BCS/A35 cannot stain the parental BHK cells. D. BCS/A35 cannot stain the ABCG2-expressing BHK/ABCG2 cells.
Aptamer-stained cells can form mammospheres
Since ABCG2 is universally expressed in the undifferentiated cancer stem cells [29,30], we speculated that our ABCG2-specific aptamers should recognize the undifferentiated BCS cells. In order to test this hypothesis, FITC-labeled ABCG2/A12 was used to stain the control MCF-7 cells and the cells derived from mammospheres. The results in Figure 3A showed that ABCG2/A12 cannot stain the differentiated breast cancer MCF-7 cells, but clearly detected low percentage of the BCS cells derived from mammospheres (Figure 3B). In addition, those cells stained with ABCG2/A12 were also recognized by ABCG2-specific mAb 5D3 (Figure 3C). Of note, in contrast to the surface labeling of BHK/ABCG2 cells with ABCG2/A12 (Figure 1B), the bound ABCG2/A12 aptamers were efficiently internalized into the BCS cells (Figure 3B and 3C), suggesting that human BCS cells might have much higher endocytosis rate than the ABCG2-expressing BHK/ABCG2 cells.
In order to test whether this internalization is via the binding of ABCG2/A12 or ABCG2-specific mAb 5D3 to the extra-cellular loops of ABCG2 protein, the cells derived from mammospheres were digested with trypsin for a short time and then did the binding analysis. As shown in Figure 3D, short time trypsin digestion completely abrogated the co-staining of ABCG2/A12 and ABCG2-specific mAb 5D3, suggesting that trypsin digestion completely destroyed the ABCG2/A12 and the ABCG2-specific mAb 5D3 binding sites.
If the cells recognized by aptamers and/or ABCG2-specific mAb 5D3 are BCS cells, they should form mammospheres in vitro. Indeed, ABCG2/A12 negative cells (Figure 3G and 3K) or 5D3 negative cells (Figure 3I and 3K) sorted out by FACS barely formed mammospheres, whereas the ABCG2/A12 positive cells (Figure 3H and 3K) or the ABCG2-specific mAb 5D3 positive cells (Figure 3J and 3K) clearly formed multiple mammospheres, suggesting that the ABCG2/A12 bound cells or the ABCG2-specific mAb 5D3 bound cells are the undifferentiated BCS cells.
CDE inhibitors can inhibit the endocytosis in differentiated breast cancer cells, but not in BCS cells
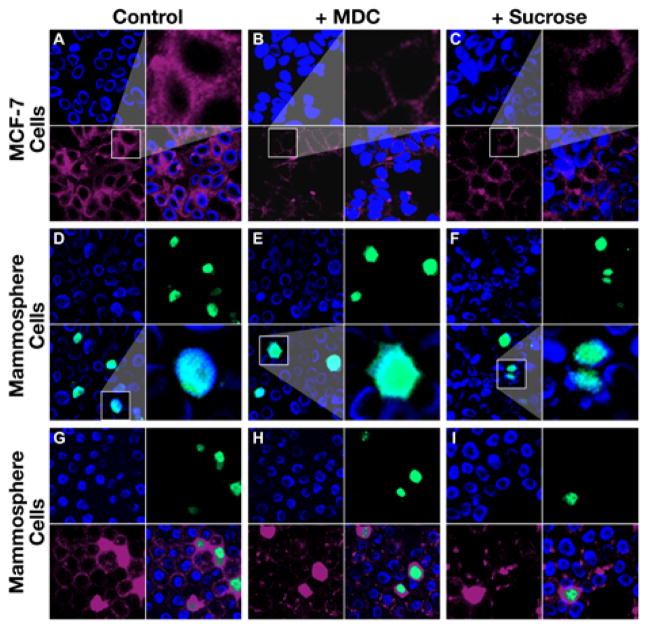
The results in Figures 2, 3, S4B and S5 suggested that BCS cells might have high rate of endocytosis. In order to test whether this high endocytosis rate undergoes CDE or not, Alexa fluor 633-conjugated hTF and FITC-labeled BCS/A35 or FITC-labeled ABCG2/A12 were used to stain the cells derived from either MCF-7 or mammospheres in the presence or absence of CDE inhibitors. In the absence of CDE inhibitors, hTF clearly labeled the surface of MCF-7 cells, with certain amount of hTF located in cytoplasmic portion (Figure 4A). However, the labeling intensity in the cytoplasmic portion of MCF-7 cells in the presence of CDE inhibitors, such as MDC (amplified images in Figure 4B and S7B) or sucrose (amplified images in Figure 4C and S7C), was significantly lower than in the absence of CDE inhibitors (amplified images in Figure 4A and S7A). In contrast, the labeling of the cells derived from mammospheres with FITC-labeled BCS/A35 or ABCG2/A12 in the presence of CDE inhibitors, such as MDC (amplified images in Figure 4E or S7E) or sucrose (amplified images in Figure 4F or S7F), was similar to that the labeling in the absence of CDE inhibitor (amplified images in Figure 4D or S7D), suggesting that the internalization of the FITC-labeled aptamers in BCS cells may not be mediated by CDE. This conclusion was further confirmed by the co-staining of the cells derived from mammospheres with Alexa fluor 633-conjugated hTF and FITC-labeled BCS/A35 or ABCG2/A12 in the presence (Figure 4H and 4I or S7H and S7I) or absence of CDE inhibitors (Figure 4G or S7G) in which the cells heavily stained with aptamer BCS/A35 or ABCG2/A12 were, regardless of whether the CDE inhibitor is present or not, also heavily stained with hTF. In addition, the cells not stained with FITC-labeled aptamer BCS/A35 or ABCG2/A12 were lightly labeled with hTF (Figure 4G, 4H and 4I or S7G, S7H and S7I), suggesting that BCS cells have significantly higher rate of clathrin-independent endocytosis than the corresponding differentiated breast cancer cells.
Figure 4.
BCS cells have significantly higher rate of clathrin-independent endocytosis than the differentiated breast cancer cells. A. Staining of the differentiated breast cancer MCF-7 cells with hTF in the absence of CDE inhibitor: top left, cells were stained with DAPI; bottom left, with Alexa fluor 633-conjugated hTF; top right, amplified image of the cells from bottom left section; bottom right, merge of the top left section and bottom left section. B. Staining of the differentiated breast cancer MCF-7 cells with hTF in the presence of CDE inhibitor MDC. C. Staining of the differentiated breast cancer MCF-7 cells with hTF in the presence of CDE inhibitor sucrose. D. Staining of the mammosphere cells derived from MCF-7 with BCS/A35 in the absence of CDE inhibitor: top left, cells were stained with DAPI; top right, with FITC-labeled BCS/A35; bottom left, merge of the top left section and top right section; bottom right, amplified image of the cells from bottom left section. E. Staining of the cells derived from mammospheres with BCS/A35 in the presence of CDE inhibitor MDC. F. Staining of the cells derived from mammospheres with BCS/A35 in the presence of CDE inhibitor sucrose. G. Co-staining of the cells derived from mammospheres with BCS/A35 and hTF in the absence of CDE inhibitor: top left, cells were stained with DAPI; top right, with FITC-labeled BCS/A35; bottom left, with Alexa fluor 633-conjugated hTF; bottom right, merge of the previous three sections. H. Co-staining of the cells derived from mammospheres with BCS/A35 and hTF in the presence of CDE inhibitor MDC. I. Co-staining of the cells derived from mammospheres with BCS/A35 and hTF in the presence of CDE inhibitor sucrose.
CVDE inhibitors can inhibit the endocytosis in differentiated breast cancer cells, but not in BCS cells
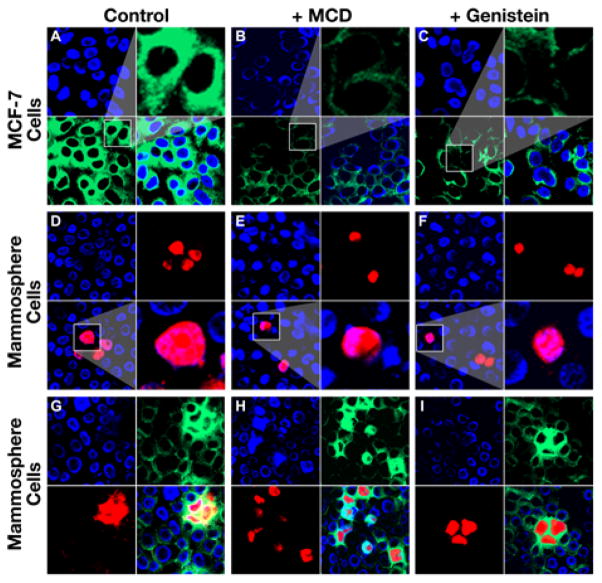
In order to test whether the high rate of endocytosis in BCS cells undergoes CVDE or not, BODIPY-labeled LacCer and TR-conjugated BCS/A35 or ABCG2/A12 were used to stain the cells derived from either MCF-7 or mammospheres in the presence or absence of CVDE inhibitors. In the absence of CVDE inhibitors, LacCer clearly labeled the surface and intracellular portion of MCF-7 cells (amplified images in Figure 5A or S8A). However, in the presence of CVDE inhibitors, such as MCD (amplified images in Figure 5B or S8B) or genistein (amplified images in Figure 5C or S8C), the labeling intensity in the cytoplasmic portion of MCF-7 cells was significantly lower than in the absence of CVDE inhibitors (amplified images in Figure 5A or S8A). Interestingly, the labeling of the cells derived from mammospheres with TR-conjugated aptamer BCS/A35 or ABCG2/A12 in the presence of CVDE inhibitors, such as MCD (amplified images in Figure 5E or S8E) or genistein (amplified images in Figure 5F or S8F), was similar to that the labeling in the absence of CVDE inhibitor (amplified images in Figure 5D or S8D), suggesting that the internalization of the TR-conjugated aptamers in BCS cells was not mediated by CVDE. This conclusion was further confirmed by the labeling of the cells derived from mammospheres with BODIPY-labeled LacCer and TR-conjugated BCS/A35 or ABCG2/A12 in the presence (Figure 5H and 5I or S8H and S8I) or absence of CVDE inhibitors (Figure 5G or S8G) in which the cells heavily stained with aptamer BCS/A35 or ABCG2/A12 were, regardless of whether the CVDE inhibitor is present or not, also heavily stained with LacCer. In addition, the cells not stained with TR-conjugated aptamer BCS/A35 or ABCG2/A12 were lightly labeled with LacCer (Figure 5G, 5H and 5I or S8G, S8H and S8I), suggesting that BCS cells may have significantly higher rate of caveolin-independent endocytosis than the corresponding differentiated breast cancer cells.
Figure 5.
BCS cells have a significantly higher rate of caveolin-independent endocytosis than the differentiated breast cancer cells. A. Staining of the differentiated breast cancer MCF-7 cells with LacCer in the absence of CVDE inhibitor: top left, cells were stained with DAPI; bottom left, with BODIPY-labeled LacCer; top right, amplified image of the cells from bottom left section; bottom right, merge of the top left section and bottom left section. B. Staining of the differentiated breast cancer MCF-7 cells with LacCer in the presence of CVDE inhibitor MCD. C. Staining of the differentiated breast cancer MCF-7 cells with LacCer in the presence of CVDE inhibitor genistein. D. Staining of the mammosphere cells derived from MCF-7 with BCS/A35 in the absence of CVDE inhibitor: top left, cells were stained with DAPI; top right, with TR-labeled BCS/A35; bottom left, merge of the top left section and top right section; bottom right, amplified image of the cells from bottom left section. E. Staining of the cells derived from mammospheres with BCS/A35 in the presence of CVDE inhibitor MCD. F. Staining of the cells derived from mammospheres with BCS/A35 in the presence of CVDE inhibitor genistein. G. Co-staining of the cells derived from mammospheres with BCS/A35 and LacCer in the absence of CVDE inhibitor: top left, cells were stained with DAPI; top right, with BODIPY-labeled LacCer; bottom left, with TR-labeled BCS/A35; bottom right, merge of the previous three sections. H. Co-staining of the cells derived from mammospheres with BCS/A35 and LacCer in the presence of CVDE inhibitor MCD. I. Co-staining of the cells derived from mammospheres with BCS/A35 and LacCer in the presence of CVDE inhibitor genistein.
Discussion
As mentioned in introduction, cancer stem cells over-express ABC transporters, such as ABCB1 and ABCG2, and play very important roles in self-renewal, growth and generation of secondary tumors. Due to over-expression of ABC transporters, they become resistant to multiple anticancer drugs. Thus, conventional chemotherapy may kill the bulk of drug sensitive differentiated breast cancer cells, but not the MDR BCS cells. Their self-renewal, growth, differentiation and epithelial to mesenchymal transition are regulated by Wnt, Notch, Hedgehog and transforming growth factor β signaling pathways. Thus, knocking down, such as using small interfering siRNA, the crucial factor(s) in these signaling pathways might prevent their self-renewal and growth. However, the question is how to deliver these therapeutic agents selectively into cancer stem cells. The report described here is to develop BCS cell-specific ligands so that we can selectively deliver therapeutic agents into the MDR BCS cells.
The following results clearly demonstrated that both BCS cell-binding aptamers and ABCG2-specific aptamers can be considered to be BCS cell-binding ligands: 1) BCS cell-enriched aptamers co-stained the potential BCS cells derived from mammospheres with either ABCG2-specific mAb 5D3 (Figure S4B) or CD44-specific mAb (Figure S5); 2) BCS/A35 (Figure 2B) or ABCG2/A12 (Figure 3B) can bind to some of the cells derived from mammospheres, but not to the differentiated MCF-7 breast cancer cells (Figure 2A or 3A); 3) ABCG2/A12 or BCS/A35 and ABCG2-specific mAb 5D3 recognized the same cells derived from mammospheres (Figure 3C or 3E); 4) ABCG2/A12 positive cells or ABCG2-specific mAb 5D3 positive cells, sorted by FACS, can form mammospheres in vitro (Figure 3H, 3J and 3K), whereas ABCG2/A12 negative cells or 5D3 negative cells cannot (Figure 3G, 3I and 3K). Thus, these aptamers could be used as ligands to make the aptamer-coated liposomes to selectively deliver therapeutic agents into the MDR BCS cells.
Interestingly, the bound ABCG2-specific mAb 5D3 or the bound aptamers, regardless of whether ABCG2/A12 or BCS/A35 aptamer was used, were efficiently internalized into the BCS cells derived from mammospheres (Figure 2B, 3B, 3C, 3E, S4B and S5). In addition, hTF or LacCer mainly bind to the surface of the differentiated breast cancer cells (Figure 4A and S7A or 5A and S8A), whereas the bound hTF or LacCer were efficiently internalized into the BCS cells derived from mammospheres (Figure 4G and S7G or 5G and S8G). These results strongly suggest that the un-differentiated BCS cells may have a significantly higher endocytosis rate than the differentiated breast cancer cells. This might be a common feature in cancer stem cells regardless of whether the cancer stem cells were derived from cancer cell line (Figure 2–5), fresh breast cancer specimen (Figure S5) or BT-12 neurospheres (data not shown). Unlike the endocytosis occurred in murine L cells [49] or in retinal pigment epithelial D407 cells [50], the treatments of the BCS cells with either CDE inhibitors, such as MDC (Figure 4E, 4H, S7E and S7H) or sucrose (Figure 4F, 4I, S7F and S7I), or CVDE inhibitors, such as MCD (Figure 5E, 5H, S8E and S8H) or genistein (Figure 5F, 5I, S8F and S8I), did not inhibit the efficient internalization of the bound aptamers, hTF or LacCer, suggesting that BCS cells may have a high rate of clathrin-independent and caveolin-independent endocytosis [51].
ABCG2-specific aptamers, such as ABCG2/A12, clearly recognized the ABCG2-expressing BCS cells (Figure 3, S7 and S8). Thus, they could be used to make ABCG2-specific aptamer-coated liposomes to selectively deliver therapeutic agents (harbored in the liposomes) into the ABCG2-expressing cells, such as BCS cells. However, since normal stem cells and other tissues, such as hepatic stem cells [32], lung stem cells [33], cardiac side population cells [52], mammary epithelia side population cells [53], skeletal muscle side population cells [54], neural stem cells [55], corneal side population cells [55], and placenta [34], also express ABCG2, the treatment with ABCG2-specific aptamer-coated liposomes harboring therapeutic agents might cause side effects. In contrast, if the aptamers, such as BCS/A35, enriched from human BCS cells (Figure S4B) mainly bind to human BCS cells, but not other cells (this point needs to be further confirmed), the treatment with BCS cell-specific aptamer-coated liposomes harboring therapeutic agents might have minimal side effect. Taken together, we have cloned BCS cell-binding aptamers and proved that BCS cells efficiently internalized the bound ABCG2-specific mAb, aptamers, hTF protein and LacCer. Thus, our findings opened the door for us to develop a novel therapeutic approach to target the pluripotent and MDR BCS cells.
Supplementary Material
Acknowledgments
We thank Irene Beauvais for processing of the manuscript and Marv Ruona for preparation of the graphics. This study was supported by Dr. Xiu-Bao Chang, NIH Grant number: 5R01CA089078-05 for Mechanism of ATP Dependent Transport by MRP1 Protein.
Abbreviations
- ABC
ATP Binding Cassette
- ABCG2
ABC Transporter group G number 2
- ABCG2/A12
the cloned ABCG2-specific aptamer #12
- ALDH
Aldehyde Dehydrogenase
- APC
Allo Phyco Cyanin
- BCS
Breast Cancer Stem
- BCS/A35
the cloned BCS cell-binding aptamer # 35
- BHK
Baby Hamster kidney
- BHK/ABCG2
human ABCG2 cDNA transfected BHK cells
- CDE
Clathrin-Dependent Endocytosis
- CVDE
Caveolin-Dependent Endocytosis
- DAPI
4′,6-diamidino-2-phenylindole
- FACS
Fluorescence-Activated Cell Sorting
- FBS
Fetal Bovine Serum
- FITC
Fluorescein Isothiocyanate
- hTF
Human Transferrin
- IRB
Institutional Review Board
- LacCer
Lactosylceramide
- mAb
Monoclonal Antibody
- MCD
Methyl-β-Cyclodextrin
- MDC
Monodansylcadaverine
- MDR
Multi Drug Resistance
- PBS
Phosphate Buffered Saline
- PCR
Polymerase Chain Reaction
- PI
Propidium Iodide
- ssDNA
Single Stranded DNA
- TR
Texas Red
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Nooter K, Stoter G. Molecular mechanisms of multidrug resistance in cancer chemotherapy. Pathol Res Pract. 1996;192:768–780. doi: 10.1016/S0344-0338(96)80099-9. [DOI] [PubMed] [Google Scholar]
- 4.Stockler M, Wilcken NR, Ghersi D, Simes RJ. Systematic reviews of chemotherapy and endocrine therapy in metastatic breast cancer. Cancer Treat Rev. 2000;26:151–68. doi: 10.1053/ctrv.1999.0161. [DOI] [PubMed] [Google Scholar]
- 5.Song LL, Miele L. Cancer stem cells--an old idea that’s new again: implications for the diagnosis and treatment of breast cancer. Expert Opin Biol Ther. 2007;7:431–438. doi: 10.1517/14712598.7.4.431. [DOI] [PubMed] [Google Scholar]
- 6.Morrison BJ, Schmidt CW, Lakhani SR, Reynolds BA, Lopez JA. Breast cancer stem cells: implications for therapy of breast cancer. Breast Cancer Res. 2008;10:210. doi: 10.1186/bcr2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clayton H, Titley I, Vivanco M. Growth and differentiation of progenitor/stem cells derived from the human mammary gland. Exp Cell Res. 2004;297:444–460. doi: 10.1016/j.yexcr.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 8.Patrawala L, Calhoun T, Schneider-Broussard R, Zhou J, Claypool K, et al. Side population is enriched in tumorigenic, stem-like cancer cells, whereas Abcg2+ and Abcg2- cancer cells are similarly tumorigenic. Cancer Res. 2005;65:6207–6219. doi: 10.1158/0008-5472.CAN-05-0592. [DOI] [PubMed] [Google Scholar]
- 9.Pommier SJ, Quan GG, Christante D, Muller P, Newell AE, et al. Characterizing the Her2/Neu status and metastatic potential of breast cancer stem/progenitor cells. Ann Surg Oncol. 2009;17:613–623. doi: 10.1245/s10434-009-0730-z. [DOI] [PubMed] [Google Scholar]
- 10.Lawson JC, Blatch GL, Edkins AL. Cancer stem cells in breast cancer and metastasis. Breast Cancer Res Treat. 2009;118:241–254. doi: 10.1007/s10549-009-0524-9. [DOI] [PubMed] [Google Scholar]
- 11.Huang YH, Zugates GT, Peng W, Holtz D, Dunton C, et al. Nanoparticledelivered suicide gene therapy effectively reduces ovarian tumor burden in mice. Cancer Res. 2009;69:6184–6191. doi: 10.1158/0008-5472.CAN-09-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, et al. A Role for Wnt Signalling in Self-Renewal of Haematopoietic Stem Cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 13.Saxena M, Stephens MA, Pathak H, Rangarajan A. Transcription factors that mediate epithelial-mesenchymal transition lead to multidrug resistance by upregulating abc transporters. Cell Death Dis. 2011;2:e179. doi: 10.1038/cddis.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spink KE, Polakis P, Weis WI. Structural Basis of the Axin-Adenomatous Polyposis Coli Interaction. EMBO J. 2000;19:2270–2279. doi: 10.1093/emboj/19.10.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taipale J, Beachy PA. The hedgehog and wnt signalling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 17.Srivastava S, Ramdass B, Nagarajan S, Rehman M, Mukherjee G, et al. Notch1 regulates the functional contribution of rhoc to cervical carcinoma progression. Br J cancer. 2010;102:196–205. doi: 10.1038/sj.bjc.6605451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katoh M. Expression and regulation of Wnt1 in human cancer: upregulation of Wnt1 by beta-estradiol in Mcf-7 cells. Int J Oncol. 2003;22:209–212. [PubMed] [Google Scholar]
- 19.Katoh Y, Katoh M. Comparative genomics on Dkk2 and Dkk4 orthologs. Int J Mol Med. 2005;16:477–481. [PubMed] [Google Scholar]
- 20.Prathapam T, Tegen S, Oskarsson T, Trump A, Martin GS. Activated Src abrogates the Myc requirement for the G0/G1 transition but not for the G1/S transition. Proc Natl Acad Sci U S A. 2006;103:2695–2700. doi: 10.1073/pnas.0511186103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Q, Ciemerych MA, Sicinski P. Ras and Myc can drive oncogenic cell proliferation through individual D-Cyclins. Oncogene. 2005;24:7114–7119. doi: 10.1038/sj.onc.1208853. [DOI] [PubMed] [Google Scholar]
- 22.Cho S, Lu M, He X, Ee PL, Bhat U, et al. Notch1 regulates the expression of the multidrug resistance gene Abcc1/Mrp1 in cultured cancer cells. Proc Natl Acad Sci U S A. 2011;108:20778–20783. doi: 10.1073/pnas.1019452108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim JC, Kania KD, Wijesuriya H, Chawla S, Sethi JK, et al. Activation of Beta-Catenin Signalling by Gsk-3 Inhibition Increases P-Glycoprotein Expression in Brain Endothelial Cells. J Neurochem. 2008;106:1855–1865. doi: 10.1111/j.1471-4159.2008.05537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flahaut M, Meier R, Coulon A, Nardou KA, Niggli FK, et al. The Wnt receptor Fzd1 mediates chemoresistance in neuroblastoma through activation of the Wnt/beta-catenin pathway. Oncogene. 2009;28:2245–2456. doi: 10.1038/onc.2009.80. [DOI] [PubMed] [Google Scholar]
- 25.Bisson I, Prowse DM. Wnt signaling regulates self-renewal and differentiation of prostate cancer cells with stem cell characteristics. Cell Res. 2009;19:683–697. doi: 10.1038/cr.2009.43. [DOI] [PubMed] [Google Scholar]
- 26.Allikmets R, Schriml LM, Hutchinson A, Romano-Spica V, Dean M. A human placenta-specific ATP-binding cassette gene (ABCP) on chromosome 4q22 that is involved in multidrug resistance. Cancer Res. 1998;58:5337–5339. [PubMed] [Google Scholar]
- 27.Miyake K, Mickley L, Litman T, Zhan Z, Robey R, et al. Molecular cloning of cDNAs which are highly overexpressed in mitoxantrone-resistant cells: demonstration of homology to ABC transport genes. Cancer Res. 1999;59:8–13. [PubMed] [Google Scholar]
- 28.Doyle LA, Yang W, Abruzzo LV, Krogmann T, Gao Y, et al. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci U S A. 1998;95:15665–15670. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou S, Schuetz JD, Bunting KD, Colapietro AM, Sampath J, et al. The ABC transporter Bcrp1/Abcg2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat Med. 2001;7:1028–1034. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
- 30.Bunting KD. ABC transporters as phenotypic markers and functional regulators of stem cells. Stem Cells. 2002;20:11–20. doi: 10.1002/stem.200011. [DOI] [PubMed] [Google Scholar]
- 31.Apati A, Orban TI, Varga N, Nemeth A, Schamberger A, et al. High level functional expression of the ABCG2 multidrug transporter in undifferentiated human embryonic stem cells. Biochim Biophys Acta. 2008;1778:2700–2709. doi: 10.1016/j.bbamem.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 32.Nakauchi H. Isolation and clonal characterization of hematopoietic and liver stem cells. Cornea. 2004;23:S2–S7. doi: 10.1097/01.ico.0000136664.18129.4f. [DOI] [PubMed] [Google Scholar]
- 33.Summer R, Kotton DN, Sun X, Ma B, Fitzsimmons K, et al. Side Population Cells and Bcrp1 Expression in Lung. Am J Physiol Lung Cell Mol Physiol. 2003;285:L97–L104. doi: 10.1152/ajplung.00009.2003. [DOI] [PubMed] [Google Scholar]
- 34.Ni Z, Mao Q. ATP-binding cassette efflux transporters in human placenta. Curr Pharm Biotechnol. 2011;12:674–685. doi: 10.2174/138920111795164057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hou YX, Li CZ, Palaniyandi K, Magtibay PM, Homolya L, et al. Effects of putative catalytic base mutation E211Q on ABCG2-mediated methotrexate transport. Biochemistry. 2009;48:9122–9131. doi: 10.1021/bi900675v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellington AD, Szostak JW. In vitro selection of rna molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 38.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 39.Hesselberth J, Robertson MP, Jhaveri S, Ellington AD. In vitro selection of nucleic acids for diagnostic applications. J Biotechnol. 2000;74:15–25. doi: 10.1016/s1389-0352(99)00005-7. [DOI] [PubMed] [Google Scholar]
- 40.Lee JF, Stovall GM, Ellington AD. Aptamer Therapeutics Advance. Curr Opin Chem Biol. 2006;10:282–289. doi: 10.1016/j.cbpa.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 41.Famulok M, Hartig JS, Mayer G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem Rev. 2007;107:3715–3743. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 42.Mayer G. The chemical biology of aptamers. Angew Chem Int Ed Engl. 2009;48:2672–2689. doi: 10.1002/anie.200804643. [DOI] [PubMed] [Google Scholar]
- 43.Chen HW, Medley CD, Sefah K, Shangguan D, Tang Z, et al. Molecular recognition of small-cell lung cancer cells using aptamers. Chem Med Chem. 2008;3:991–1001. doi: 10.1002/cmdc.200800030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shangguan D, Meng L, Cao ZC, Xiao Z, Fang X, et al. Identification of liver cancer-specific aptamers using whole live cells. Anal Chem. 2008;80:721–728. doi: 10.1021/ac701962v. [DOI] [PubMed] [Google Scholar]
- 45.Herr JK, Smith JE, Medley CD, Shangguan D, Tan W. Aptamer-conjugated nanoparticles for selective collection and detection of cancer cells. Anal Chem. 2006;78:2918–2924. doi: 10.1021/ac052015r. [DOI] [PubMed] [Google Scholar]
- 46.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orban TI, Seres L, Ozvegy-Laczka C, Elkind NB, Sarkadi B, et al. Combined localization and real-time functional studies using a Gfp-tagged Abcg2 multidrug transporter. Biochem Biophys Res Commun. 2008;367:667–673. doi: 10.1016/j.bbrc.2007.12.172. [DOI] [PubMed] [Google Scholar]
- 48.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blitzer JT, Nusse R. A critical role for endocytosis in wnt signaling. BMC Cell Biol. 2006;7:28. doi: 10.1186/1471-2121-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vercauteren D, Vandenbroucke RE, Jones AT, Rejman J, Demeester J, et al. The use of inhibitors to study endocytic pathways of gene carriers: optimization and pitfalls. Mol Ther. 2010;18:561–569. doi: 10.1038/mt.2009.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 52.Martin CM, Meeson AP, Robertson SM, Hawke TJ, Richardson JA, et al. Persistent expression of the ATP-binding cassette transporter, Abcg2, identifies cardiac SP cells in the developing and adult heart. Dev Biol. 2004;265:262–275. doi: 10.1016/j.ydbio.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 53.Alvi AJ, Clayton H, Joshi C, Enver T, Ashworth A, et al. Functional and molecular characterisation of mammary side population cells. Breast Cancer Res. 2003;5:R1–R8. doi: 10.1186/bcr547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meeson AP, Hawke TJ, Graham S, Jiang N, Elterman J, et al. Cellular and molecular regulation of skeletal muscle side population cells. Stem Cells. 2004;22:1305–1320. doi: 10.1634/stemcells.2004-0077. [DOI] [PubMed] [Google Scholar]
- 55.Mouthon MA, Fouchet P, Mathieu C, Sii-Felice K, Etienne O, et al. Neural stem cells from mouse forebrain are contained in a population distinct from the ‘side population’. J Neurochem. 2006;99:807–817. doi: 10.1111/j.1471-4159.2006.04118.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.