Abstract
Rapidly growing, non-tuberculous mycobacteria (NTM) in the Mycobacterium abscessus (MAB) species are emerging pathogens that cause various diseases including skin and respiratory infections. The species has undergone recent taxonomic nomenclature refinement, and is currently recognized as two subspecies, M. abscessus subsp. abscessus (MAB-A) and M. abscessus subsp. bolletii (MAB-B). The recently reported outbreaks of MAB-B in surgical patients in Brazil from 2004 to 2009 and in cystic fibrosis patients in the United Kingdom (UK) in 2006 to 2012 underscore the need to investigate the genetic diversity of clinical MAB strains. To this end, we sequenced the genomes of two Brazilian MAB-B epidemic isolates (CRM-0019 and CRM-0020) derived from an outbreak of skin infections in Rio de Janeiro, two unrelated MAB strains from patients with pulmonary infections in the United States (US) (NJH8 and NJH11) and one type MAB-B strain (CCUG 48898) and compared them to 25 publically available genomes of globally diverse MAB strains. Genome-wide analyses of 27,598 core genome single nucleotide polymorphisms (SNPs) revealed that the two Brazilian derived CRM strains are nearly indistinguishable from one another and are more closely related to UK outbreak isolates infecting CF patients than to strains from the US, Malaysia or France. Comparative genomic analyses of six closely related outbreak strains revealed geographic-specific large-scale insertion/deletion variation that corresponds to bacteriophage insertions and recombination hotspots. Our study integrates new genome sequence data with existing genomic information to explore the global diversity of infectious M. abscessus isolates and to compare clinically relevant outbreak strains from different continents.
Keywords: Mycobacterium abscessus, comparative genomics, nontuberculous mycobacteria, NTM, bioinformatics
1. Introduction
Rapidly growing, nontuberculous mycobacteria (NTM) species are opportunistic pathogens that cause a range of diseases including lung (Griffith, 2010) and skin (Khan and Khakoo, 2011) infections, and are thought to be primarily acquired from the environment (Primm et al., 2004). NTM often infect immune-compromised patients and are sometimes associated with nosocomial infections due to their occurrence in tap water and ability to form biofilms (Cardoso et al., 2008; Falkinham et al., 2001; Feazel et al., 2009). For example, several outbreaks of post-surgical infections caused by a subtype of Mycobacterium abscessus were reported in various regions of Brazil from 2004 to 2009 (Cardoso et al., 2008; Duarte et al., 2009; Monego et al., 2011; Viana-Niero et al., 2008). These emerging Brazilian outbreak strains were thought to be related to the selection and spread of a particularly virulent epidemic isolate named BRA100, showing a high level of resistance to glutaraldehyde (GTA), the disinfectant used in hospital sites that had reported cases (Duarte et al., 2009; Leao et al., 2009; Shang et al., 2011). The GTA-resistant BRA100 epidemic isolate, CRM-0019, from Rio de Janeiro showed increased virulence in macrophages and mice compared to a GTA-susceptible reference clinical isolate CIP108297 suggesting that BRA100 may represent a particularly invasive and pathogenic strain (Shang et al., 2011). Whether differences in the colonial morphotype of BRA100 isolates relative to CIP108297 (Shang et al., 2011) and, therefore, cell envelope lipid composition (Howard et al., 2006) may account for this difference in pathogenicity is currently under investigation.
The subtypes of M. abscessus (MAB) have undergone recent taxonomical nomenclature changes, previously classified as three separate subspecies including M. abscessus subsp. abscessus (MAB-A), M. abscessus subsp. massiliense (Adekambi et al., 2004) and M. abscessus subsp. bolletii (Adekambi and Drancourt, 2006). The currently accepted nomenclature circumscribes M. abscessus subsp. massiliense and M. abscessus subsp. bolletii into a single subspecies called M. abscessus subsp. bolletii (MAB-B), and preserves the original delineation of the MAB-A subtype (Leao et al., 2011). The ambiguous classification within this clade is due to nearly identical 16s rRNA sequences (Adekambi and Drancourt, 2004; Devulder et al., 2005; Leao et al., 2009), indistinguishable mycolic acid profiles (Leao et al., 2009), and high sequence identities in the housekeeping genes, rpoB (>95%) and hsp65 (>=98%) (Duarte et al., 2009; Leao et al., 2009). Several population studies, however, are supportive of the previous subspecies designation into three distinct subgroups (Bryant et al., 2013; Macheras et al., 2011; Zelazny et al., 2009).
The current era of bacterial genome sequencing by next generation sequencing methods provides the opportunity to evaluate genetic diversity at a much finer resolution than traditional gene sequencing. Recently, whole genome sequencing and large-scale variant analyses resolved the genetic relationships of MAB infections in a cystic fibrosis (CF) clinic in the United Kingdom (UK) from 2006 to 2012 (Bryant et al., 2013). Though both MAB-A and MAB-B were detected, the genomic composition of a subset of MAB-B strains provided the first suggestive evidence of person-to-person transmission and infection of MAB (Bryant et al., 2013). Building on these and other studies, we hypothesized that an investigation of MAB genomic diversity would reveal relationships of strains from geographically different locations and increase our understanding of clinically relevant MAB phylogeny. To this end, we sequenced the genomes of five diverse strains of MAB including two Brazilian epidemic isolates derived from patients with skin infections in Rio de Janeiro (CRM-0019 and CRM-0020), two strains derived from patients with pulmonary infections (NJH8 and NJH11) in the United States (US) and one European type strain originally derived a patient with a pulmonary infection (CCUG 48898) (Adekambi et al., 2004). Together with publically available genomic data, we use an integrative phylogenomics approach to compare our newly sequenced genomes to 25 additional MAB genomes including representative isolates from the recent UK outbreak and unrelated strains from the United States, China, Malaysia and Europe to explore the diversity of infectious MAB isolates in a global context.
2. Materials and Methods
2.1 Bacterial strains sequenced in this study
Five strains of MAB were sequenced in this study, including two Brazilian MAB-B strains, CRM-0019 and CRM-0020 that were associated with an epidemic of post-surgical, cutaneous infections in Rio de Janeiro, Brazil in 2006. These strains were isolated from soft tissue biopsies and were identified as having BRA100 pulse field gel electrophoresis (PFGE) molecular fingerprints (Duarte et al., 2009). Two additional strains were isolated from the sputa of US patients referred to National Jewish Health in Denver, CO, including NJH8 that was derived from a patient with a pulmonary NTM infection in 2011 and was identified as MAB-A by rpoB sequencing, and NJH11 that was derived from a patient with cystic fibrosis and a pulmonary NTM infection in 2009, and was identified as MAB-B (formerly M. abscessus subsp. massiliense) by rpoB sequencing. The type MAB-B strain, CIP108297T/CCUG 48898, that was originally isolated from the sputum of a patient with hemoptoic pneumonia (Adekambi et al., 2004) and which has an existing draft genome (Tettelin et al., 2012) was also re-sequenced in this study.
Mycobacterial cultures were grown in Middlebrook 7H9-OADC broth supplemented with 0.05% Tween 80 or on 7H11-OADC agar, and genomic DNA was extracted as previously described (Pelicic et al., 1997). Genomic sequencing libraries for the MAB strains were prepared using standard protocols and sequenced on Life Technologies SOLiD3 or SOLiD5 platforms (Life Technologies Corporation, Carlsbad, CA) (Table 1). DNA from two strains, CRM-0019 and CCUG 48898, were sequenced as both paired-end and fragment libraries.
Table 1.
Sequencing statistics for five clinical MAB strains sequenced as part of this study.
| MAB Strain | Library Type | Platform | Total reads | Read Length (bp) |
|---|---|---|---|---|
| CRM-0020 | fragment | SOLiD5 | 19,081,264 | 75 |
| CRM-0019 | paired end | SOLiD3 | 4,044,099 | 50,35 |
| CRM-0019 | fragment | SOLiD3 | 20,105,261 | 50 |
| NJH8 | fragment | SOLiD5 | 21,407,961 | 75 |
| NJH11 | fragment | SOLiD5 | 19,151,415 | 75 |
| CCUG 48898 | paired end | SOLiD3 | 7,510,829 | 50,35 |
| CCUG 48898 | fragment | SOLiD3 | 1,764,946 | 50 |
2.2 Taxon sampling, variant analyses and phylogenomics
For genomic comparisons, we selected 25 representative MAB isolates with publically available draft genomes or next generation re-sequencing data including strains from Brazil, China, Malaysia, the United Kingdom, the United States, and type strains originally derived in Europe (Table 2). For MAB strains with publically available draft genomes, we performed multi-genome alignments of each draft genome to the complete reference genome of the MAB-A strain ATCC 19977 (Ripoll et al., 2009). Whole genome alignments and single nucleotide polymorphism (SNP) identification were performed with the progressiveMAUVE algorithm in Mauve 2.3.1 (Darling et al., 2010). For strains with next-generation re-sequencing data, sequence reads were mapped to the complete genome of ATCC 19977 with either Lifescope 2.1 software for SOLiD data (Life Technologies Corporation, Carlsbad, CA) or Bowtie software (Langmead et al., 2009) for Illumina data (Bryant et al., 2013). SNPs were called using the SAMtools pileup program (Li et al., 2009) and were filtered using a custom Perl script using the following parameters: minimum SNP quality score of 20, minimum of 10× read depth, the majority of base calls support the variant base, and less than 25% of variant calls occurring at the beginning or end of the fragment reads.
Table 2.
M. abscessus genome data used for phylogenomic comparisons
| Subspecies | Strain | Location | Isolation Yeara |
Accessionb | Citation |
|---|---|---|---|---|---|
| abscessus | 3A-0119-R | US | 2010 | NZ_AKUY00000000 | unpublished |
| 3A-0122-S | US | 2002 | NZ_AKUZ00000000 | unpublished | |
| 5a | UK | 2006 | ERR115039 | (Bryant et al., 2013) | |
| 6G-0125-R | US | 2010 | NZ_AKUE00000000 | unpublished | |
| 6G-0728-S | US | 2003 | NZ_AKUG00000000 | unpublished | |
| 9808 | China | 1998 | NZ_ANAR00000000 | unpublished | |
| ATCC 19977 | France | 1953 | NC_010397.1 | (Ripoll et al., 2009) | |
| NJH8 | US | 2011 | SRX339602 | this study | |
| M94 | Malaysia | NR | NZ_AJGG00000000 | (Choo et al., 2012) | |
| bolletii | BDT | France | 2004 | NZ_AHAS00000000 | (Choi et al., 2012) |
| M24 | Malaysia | NR | NZ_AJLY00000000 | (Wong et al., 2012) | |
| bolletii | 19a | UK | 2009 | ERR115105 | (Bryant et al., 2013) |
| (formerly massiliense) | 19f | UK | 2009 | ERR115037 | (Bryant et al., 2013) |
| 2f | UK | 2010 | ERR115XXX | (Bryant et al., 2013) | |
| 28f | UK | 2010 | ERR115XXX | (Bryant et al., 2013) | |
| 47J26 | UK | 2009 | NZ_AGQU00000000 | (Chan et al., 2012) | |
| 5S-0304 | US | 1998 | NZ_AKTX00000000 | unpublished | |
| CCUG 48898 | France | 2004 | SRX342940, SRX342942 | this study | |
| CRM-0019 | Brazil | 2006 | SRX339604, SRX339608 | this study | |
| CRM-0020 | Brazil | 2006 | SRX339607 | this study | |
| GO-06 | Brazil | 2006 | NC_018150.1 | (Raiol et al., 2012) | |
| M18 | Malaysia | NR | NZ_AJSC0000000 | (Heydari et al., 2013) | |
| M115 | Malaysia | NR | NZ_AJLZ00000000 | (Heydari et al., 2013) | |
| M139 | Malaysia | NR | NZ_AKVR00000000 | (Heydari et al., 2013) | |
| M148 | Malaysia | NR | NZ_AKVV00000000 | (Heydari et al., 2013) | |
| M154 | Malaysia | 2010 | NZ_AJMA010000000 | (Heydari et al., 2013) | |
| M156 | Malaysia | NR | NZ_AKVU00000000 | (Heydari et al., 2013) | |
| M159 | Malaysia | 2010 | NZ_AJSD00000000 | (Heydari et al., 2013) | |
| M172 | Malaysia | 2010 | NZ_AJSE010000000 | (Heydari et al., 2013) | |
| NJH11 | US | 2009 | SRX339603 | this study |
NR = Not reported
Accession numbers starting with NZ, NC or SRX are housed at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and those starting with ERR are housed at European Nucleotide Archive (http://www.ebi.ac.uk/ena/).
After extracting all variable sites, indels were removed, and a genotype matrix was created for SNP sites in which at least one strain differed from the reference genome, and for which high quality variant and/or reference calls were available for all strains. Base calls were then concatenated and sequences were compared using Neighbor-joining with 100 bootstrap replicates in MEGA 5.1 (Tamura et al., 2011).
2.3 Genome coverage analysis of M. abscessus subsp. bolletii outbreak strains
For six MAB-B epidemic or outbreak strains (CRM-0020, CRM-0019, 19a, 19b, 2f, 28f) and two unrelated strains with next generation sequence data (CCUG 48898 and NJH11), sequence reads were mapped to the complete genome of the MAB-B BRA100 strain, GO-06 (Raiol et al., 2012) as described above. Sequence coverage values were estimated with a custom perl script that counts all mapped reads in sliding, non-overlapping 1Kb windows across the entire GO-06 genome (5068 total windows). A matrix of read counts for all strains was converted to z-scores, and normalized read counts were clustered by hierarchical clustering with a Spearman correlation distance metric and average linkage with the Multiple Experiment Viewer (MeV) Java package (Saeed et al., 2003).
3. Results and Discussion
3.1 Phylogenomics of M. abscessus outbreak and globally diverse strains
After reconstructing the phylogeny using 27,598 concatenated SNPs (Figure 1A), the phylogenomic relationships between MAB strains are congruent with the topology of recently reported phylogenies in the MAB group (Bryant et al., 2013; Heydari et al., 2013; Macheras et al., 2011; Zelazny et al., 2009). Consistent with previously reported relationships, the three formerly described MAB subspecies are monophyletic on the tree and, as expected, NJH8 and NJH11 cluster with the MAB-A and MAB-B (formerly M. abscessus subsp. massiliense) clades, respectively. Although sampling within any locality was not exhaustive, our analyses revealed significant global genomic diversity within MAB. In general, we observed greater genetic diversity within MAB-B compared to MAB-A. A majority of SNPs vary within MAB-B (22,882/27,598=82.9%) compared to the SNP variation observed within MAB-A (3,507/27,598=12.7%). When strictly comparing strains belonging to MAB-B, formerly M. abscessus subsp. massiliense, a majority of SNPs are also variable (17,994/27,598=65.2%). Contrastingly, the SNP variation observed within strains formerly recognized as M. abscessus subsp. bolletii (1,457/27,598=5.3%) is much lower than other MAB subspecies.
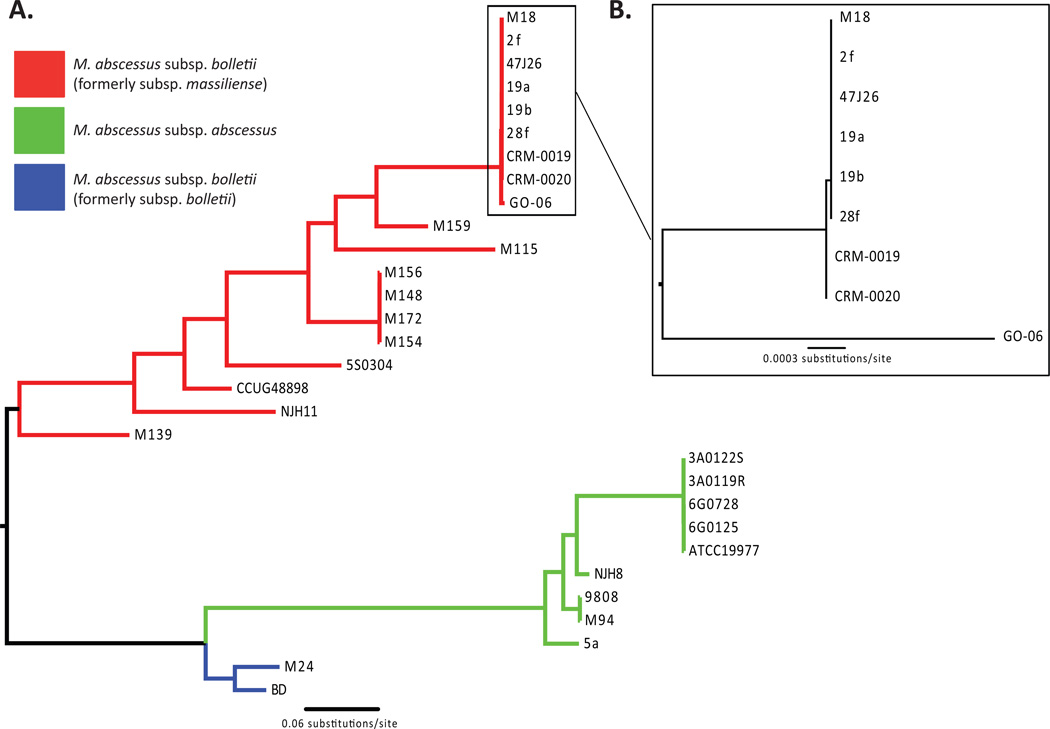
Figure 1.
Phylogenomic relationships of Brazilian epidemic isolates and globally diverse MAB strains A. Neighbor-joining phylogeny generated from 27,598 variable genome-wide SNP sites identified in all Mycobacterium abscessus (MAB) strains compared to the M. abscessus subsp. abscessus (MAB-A) ATCC 19977 complete reference genome. The tree supports the previously recognized subspecies as monophyletic groups. B. A clade composed almost entirely of M. abscessus subsp. bolletii (MAB-B) epidemic or outbreak strains is distinguished by 76 SNPs; among these only a single SNP distinguished the BRA-100 strains CRM-0019 and CRM-0020 from the UK CF-related and Malaysian M18 strains. GO-06 is the basal most member of this clade and is most-closely related to the most recent common ancestor of the MAB-B outbreak strains.
A small number of SNPs (76/27,598=0.27%) are variable within a clade composed almost entirely of previously reported MAB-B epidemic or outbreak strains (Figure 1B). Interestingly, however, the BRA100 epidemic isolates from the Brazilian states of Rio de Janeiro (CRMs) and Goias (GO-06) are not monophyletic. In fact, CRM-0019 and CRM-0020 are more closely related to MAB-B strains isolated in the recently reported UK outbreak in CF patients than they are to the BRA100 outbreak strain GO-06 (Raiol et al., 2012), consistent with previously observed phylogenomic relationships between UK strains and GO-06 (Bryant et al., 2013). A comparison of variable sites within Brazilian strains identified 75 SNPs, all of which differentiate CRM-0019 and CRM-0020 from GO-06 suggesting a low level of variation between epidemic strains from distinct states in Brazil.
Variant analyses between outbreak strains from different geographic locations revealed only one SNP between strains from the Brazilian (CRM-0019 and CRM-0020) and UK (19a, 19b, 2f, and 28f) MAB-B outbreaks. Additionally, our analyses suggest that strains M18 and 47J26 from Malaysia and the UK, respectively, which were not implicated in previous outbreaks are nearly identical to the recent outbreak strains as they have no SNPs compared to the UK strains and only one SNP compared to the Brazilian CRM strains. This lack of variation suggests that a recent circulating strain of MAB-B likely emerged in the Brazilian and UK outbreaks, and further suggests this strain may be emerging in another global location. With CRM-0019, CRM-0020 and GO-06 each isolated in 2006; 19a, 19f and 47J26 isolated in 2009; and 28f isolated in 2010; the isolation dates and phylogenetic pattern collectively suggest the Brazilian MAB-B outbreak as ancestral to the subsequent UK CF clinic MAB-B outbreaks.
The lack of difference between strains from disparate outbreak localities is surprising especially since the CRM strains were derived from cutaneous infections and the UK strains from pulmonary samples. More interesting is the Malaysian strains (M154, M159 and M172) isolated during the same timeframe as the Brazilian and UK outbreaks strains (Table 2) are genetically diverse, and for the most part genetically distinct from the Brazilian and UK outbreak strains. The US-derived MAB-B strains are also genetically different from the Brazilian, UK and Malaysian strains as a comparison of NJH11 and 5S0304 to the MAB-B outbreak strains revealed relatively high SNP variation (12144/27,598=44.0%).
3.2 Large-scale deletions and gene content differences among MAB-B outbreak strains
Given the striking similarities in SNP profiles of MAB-B isolates from geographically distinct outbreaks, we further explored large-scale genomic differences that would not be detected by the SNP analysis. First, we analyzed total genome coverage in six outbreak and two unrelated MAB-B strains compared to the complete genome of the BRA100 strain, GO-06 (Raiol et al., 2012) (Table 3). All samples had substantial sequencing depth (from 57X to 195X), yet the genome mapping analysis showed that only 89.4 to 94.2% of the GO-06 genome is represented in the other MAB-B strains. This suggests that 5.8 to 10.6% of the GO-06 genome is absent in one or more of the other strains, even those isolated from the neighboring region of Rio de Janeiro. This is consistent with genome differences previously described through DNA-DNA hybridization experiments of diverse MAB strains (Leao et al., 2009) and comparable to differences reported between the genomes of pathogenic strains of Helicobacter pylori (Alm et al., 1999) and Streptococcus agalactiae (Tettelin et al., 2005).
Table 3.
Mapping coverage of re-sequenced MAB-B strains versus the complete genome of the MAB-B BRA100 strain, GO-06 (Raiol et al., 2012) and average sequencing depth across the GO-06 genome.
| Strain name | % of GO-06 genome at > 1X coverage | Average sequencing depth (X) |
|---|---|---|
| CRM-0020 | 91.2 | 195 |
| CRM-0019 | 90.0 | 147 |
| 19a | 90.8 | 150 |
| 19b | 90.8 | 143 |
| 28f | 92.2 | 199 |
| 2f | 89.4 | 193 |
| CCUG 48898 | 90.3 | 57 |
| NJH11 | 94.2 | 166 |
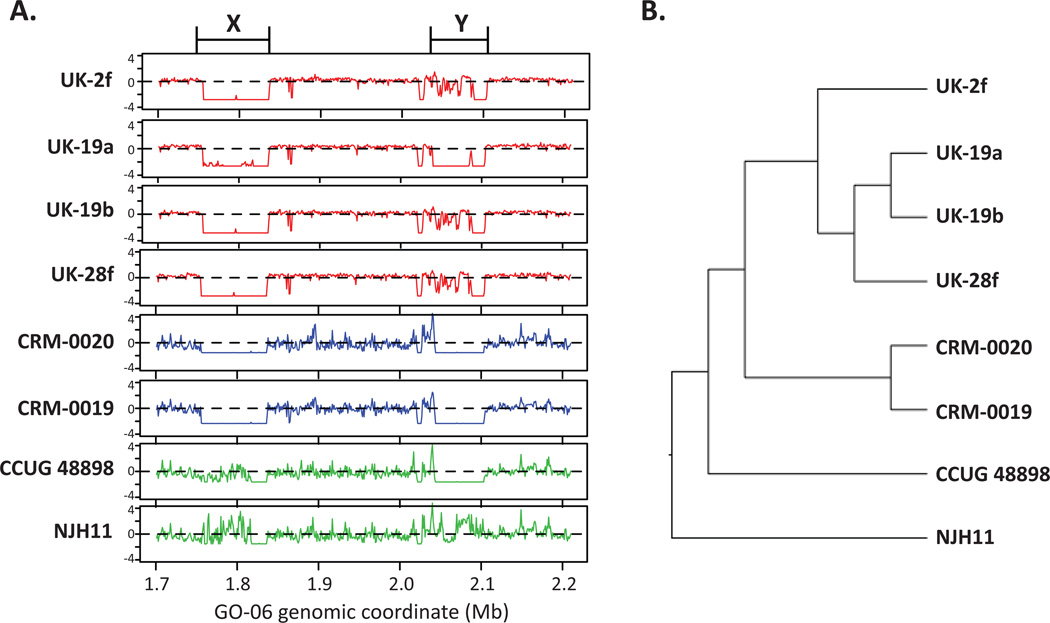
To test the hypothesis that the BRA100 epidemic and UK outbreak strains share similar large-scale deletion patterns, we calculated read coverage values for all strains in sliding 1Kb windows across the entire GO-06 genome (Figure 2). We observed overlapping deletions in some, but not all, regions including a 80Kb deletion that is shared by the CRM and UK strains, but not by NJH11, CCUG 48898 and GO-06 (Figure 2A: X). This region is consistent with a phage insertion as it is flanked by integrase genes and has 111 predicted open reading frames (ORFs) that are primarily annotated as hypothetical or virus-related (Juhas et al., 2009). A different 60Kb deletion that is shared by the CRM strains and CCUG 48898 shows differential mapping patterns in the UK strains and NJH11 (Figure 2A: Y). This region has 64 predicted ORFs including two transposases, a serine-family recombinase and an integrase and is likely a hotspot for homologous recombination (Juhas et al., 2009). A cluster analysis of genome-wide deletion patterns compared to GO-06 revealed that the outbreak strains cluster largely by geographic location (Figure 2B). This suggests that, though highly similar in their core genomes, the CRM epidemic and UK outbreak strains are evolving through local horizontal gene transfer events.
Figure 2.
Large-scale genomic variation among MAB-B outbreak strains. Next-generation sequence reads for six outbreak strains (2f, 19a, 19b, 28f, CRM-0019, CRM-0020) and two epidemiologically unrelated strains (CCUG 48898 and NJH11) were mapped to the complete genome of the BRA100 strain, GO-06. Normalized read coverage counts were estimated for all non-overlapping 1Kb windows and plotted relative to GO-06 genomic coordinates. A. Normalized read mapping coverage (y-axes show z-scores) in a 500Kb region of the GO-06 genome (x-axis). Two contiguous regions with z-scores less than −2.0 represent two large scale deletions including an 80Kb deletion with 111 predicted ORFs (X) and a 60Kb deletion with 64 predicted ORFs (Y) B. Hierarchical cluster analysis of genome-wide deletion patterns for all 5068 windows across the GO-06 genome.
4. Conclusions
Our study explores the phylogenomic relationships of globally diverse MAB isolates including five newly sequenced strains and 25 strains with publically available genomes. Genome wide SNP analysis revealed three distinct subgroups consistent with previous subspecies classifications. We observed more SNP diversity within the formerly recognized subspecies M. abscessus subsp. massiliense than within MAB-A or the formerly recognized subspecies M. abscessus subsp. bolletii. Moreover, our results suggest that the Brazilian BRA100 epidemic isolates from Rio de Janeiro (CRM-0020 and CRM-0019) and the state of Goias (GO-06) belong to a monophyletic clade that also includes representative strains from a recent UK CF clinic outbreak (19a, 19f, 2f and 28f), a single UK CF patient infection (47J26) and one Malaysian strain (M18). In contrast, the CRM epidemic strains show significant variation compared to other MAB-B strains from the US, Malaysia and Europe. Though the CRM and UK strains show considerable similarities in core genome SNPs suggesting a widespread epidemic clone, large-scale variant analysis revealed geographic-specific deletions suggesting independent evolution of outbreak isolates within each outbreak locale. Whether the CRM strains pose a risk of patient-to-patient transmission as suggested for the UK outbreak strains (Bryant et al., 2013) is unclear, as GTA resistance remains the most likely explanation for the rapid transmission of epidemic BRA100 strains across Brazil (Duarte et al., 2009). It is important to underscore the genetic similarities between the sequenced Brazilian BRA100 epidemic and UK outbreak strains. Isolates sampled from multiple outbreak locations on different continents showing no genetic divergence can be caused by: i) a lack of genetic variation in the species; ii) isolates stochastically sharing genetic identity by state; and iii) the possibility of a globally circulating, infectious strain (Achtman, 2008). The infectious nature of the BRA100 epidemic isolates and recent research into the epidemiology of MAB in CF patients warrants these possibilities to be investigated further to better characterize the epidemiological impact.
Highlights.
Phylogenomic relationships of 30 global and diverse M. abscessus strains
Phylogenomics of M. abscessus supports a three subspecies classification
Phylogenomic comparisons reveal similarities among Brazilian and UK outbreak strains
Acknowledgments
RD and MS acknowledge support from the National Jewish Health NTM Center of Excellence funded in part by the Amon G. Carter Foundation, the Colorado Bioscience Discovery Program, the Eppley Foundation, and the Boettcher Foundation Webb-Waring Biomedical Research Program. NH acknowledges support from a NIH Biomedical Informatics training grant 2T15LM009451-06. This work was supported in part by the National Institutes of Health/National Institute of Allergy and Infectious Diseases grant AI089718 (to MJ). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Abbreviations
- NTM
Nontuberculous mycobacteria
- MAB
Mycobacterium abscessus
- MAB-A
Mycobacterium abscessus subsp. abscessus
- MAB-B
Mycobacterium abscessus subsp. bolletii
- PFGE
pulse-field gel electrophoresis
- CF
cystic fibrosis
- SNP
single nucleotide polymorphism
- indel
insertion/deletion
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers: SRX339602 (NJH8), SRX339603 (NJH11), SRX339604, SRX339608 (CRM-0019), SRX339607 (CRM-0020), SRX342940, SRX342942 (CCUG 48898)
REFERENCES
- Achtman M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiol. 2008;62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]
- Adekambi T, Drancourt M. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int J Syst Evol Microbiol. 2004;54:2095–2105. doi: 10.1099/ijs.0.63094-0. [DOI] [PubMed] [Google Scholar]
- Adekambi T, Drancourt M. Isolation of Mycobacterium septicum from the sputum of a patient suffering from hemoptoic pneumonia. Res Microbiol. 2006;157:466–470. doi: 10.1016/j.resmic.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Adekambi T, Reynaud-Gaubert M, Greub G, Gevaudan MJ, La Scola B, Raoult D, Drancourt M. Amoebal coculture of "Mycobacterium massiliense" sp. nov. from the sputum of a patient with hemoptoic pneumonia. J Clin Microbiol. 2004;42:5493–5501. doi: 10.1128/JCM.42.12.5493-5501.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, Carmel G, Tummino PJ, Caruso A, Uria-Nickelsen M, Mills DM, Ives C, Gibson R, Merberg D, Mills SD, Jiang Q, Taylor DE, Vovis GF, Trust TJ. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, Reacher M, Haworth CS, Curran MD, Harris SR, Peacock SJ, Parkhill J, Floto RA. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet. 2013;381:1551–1560. doi: 10.1016/S0140-6736(13)60632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso AM, Martins de Sousa E, Viana-Niero C, Bonfim de Bortoli F, Pereira das Neves ZC, Leao SC, Junqueira-Kipnis AP, Kipnis A. Emergence of nosocomial Mycobacterium massiliense infection in Goias, Brazil. Microbes Infect. 2008;10:1552–1557. doi: 10.1016/j.micinf.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Chan J, Halachev M, Yates E, Smith G, Pallen M. Whole-genome sequence of the emerging pathogen Mycobacterium abscessus strain 47J26. J Bacteriol. 2012;194:549. doi: 10.1128/JB.06440-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi GE, Cho YJ, Koh WJ, Chun J, Cho SN, Shin SJ. Draft genome sequence of Mycobacterium abscessus subsp. bolletii BD(T) J Bacteriol. 2012;194:2756–2757. doi: 10.1128/JB.00354-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo SW, Wong YL, Leong ML, Heydari H, Ong CS, Ng KP, Ngeow YF. Analysis of the genome of Mycobacterium abscessus strain M94 reveals an uncommon cluster of tRNAs. J Bacteriol. 2012;194:5724. doi: 10.1128/JB.01407-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devulder G, Perouse de Montclos M, Flandrois JP. A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int J Syst Evol Microbiol. 2005;55:293–302. doi: 10.1099/ijs.0.63222-0. [DOI] [PubMed] [Google Scholar]
- Duarte RS, Lourenco MC, Fonseca Lde S, Leao SC, Amorim Ede L, Rocha IL, Coelho FS, Viana-Niero C, Gomes KM, da Silva MG, Lorena NS, Pitombo MB, Ferreira RM, Garcia MH, de Oliveira GP, Lupi O, Vilaca BR, Serradas LR, Chebabo A, Marques EA, Teixeira LM, Dalcolmo M, Senna SG, Sampaio JL. Epidemic of postsurgical infections caused by Mycobacterium massiliense. J Clin Microbiol. 2009;47:2149–2155. doi: 10.1128/JCM.00027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkinham JO, 3rd, Norton CD, LeChevallier MW. Factors influencing numbers of Mycobacterium avium Mycobacterium intracellulare and other Mycobacteria in drinking water distribution systems. Appl Environ Microbiol. 2001;67:1225–1231. doi: 10.1128/AEM.67.3.1225-1231.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feazel LM, Baumgartner LK, Peterson KL, Frank DN, Harris JK, Pace NR. Opportunistic pathogens enriched in showerhead biofilms. Proc Natl Acad Sci U S A. 2009;106:16393–16399. doi: 10.1073/pnas.0908446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith DE. Nontuberculous mycobacterial lung disease. Curr Opin Infect Dis. 2010;23:185–190. doi: 10.1097/QCO.0b013e328336ead6. [DOI] [PubMed] [Google Scholar]
- Heydari H, Wee WY, Lokanathan N, Hari R, Mohamed Yusoff A, Beh CY, Yazdi AH, Wong GJ, Ngeow YF, Choo SW. MabsBase: A Mycobacterium abscessus Genome and Annotation Database. PLoS One. 2013;8:e62443. doi: 10.1371/journal.pone.0062443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard ST, Rhoades E, Recht J, Pang X, Alsup A, Kolter R, Lyons CR, Byrd TF. Spontaneous reversion of Mycobacterium abscessus from a smooth to a rough morphotype is associated with reduced expression of glycopeptidolipid and reacquisition of an invasive phenotype. Microbiology. 2006;152:1581–1590. doi: 10.1099/mic.0.28625-0. [DOI] [PubMed] [Google Scholar]
- Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev. 2009;33:376–393. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan FA, Khakoo R. Nontuberculous mycobacterial cutaneous infections: an updated review. Cutis. 2011;88:194–200. [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leao SC, Tortoli E, Euzeby JP, Garcia MJ. Proposal that Mycobacterium massiliense and Mycobacterium bolletii be united and reclassified as Mycobacterium abscessus subsp. bolletii comb. nov., designation of Mycobacterium abscessus subsp. abscessus subsp. nov. and emended description of Mycobacterium abscessus. Int J Syst Evol Microbiol. 2011;61:2311–2313. doi: 10.1099/ijs.0.023770-0. [DOI] [PubMed] [Google Scholar]
- Leao SC, Tortoli E, Viana-Niero C, Ueki SY, Lima KV, Lopes ML, Yubero J, Menendez MC, Garcia MJ. Characterization of mycobacteria from a major Brazilian outbreak suggests that revision of the taxonomic status of members of the Mycobacterium chelonae-M. abscessus group is needed. J Clin Microbiol. 2009;47:2691–2698. doi: 10.1128/JCM.00808-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macheras E, Roux AL, Bastian S, Leao SC, Palaci M, Sivadon-Tardy V, Gutierrez C, Richter E, Rusch-Gerdes S, Pfyffer G, Bodmer T, Cambau E, Gaillard JL, Heym B. Multilocus sequence analysis and rpoB sequencing of Mycobacterium abscessus (sensu lato) strains. J Clin Microbiol. 2011;49:491–499. doi: 10.1128/JCM.01274-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monego F, Duarte RS, Nakatani SM, Araujo WN, Riediger IN, Brockelt S, Souza V, Cataldo JI, Dias RC, Biondo AW. Molecular identification and typing of Mycobacterium massiliense isolated from postsurgical infections in Brazil. Braz J Infect Dis. 2011;15:436–441. doi: 10.1016/s1413-8670(11)70224-0. [DOI] [PubMed] [Google Scholar]
- Pelicic V, Jackson M, Reyrat JM, Jacobs WR, Jr, Gicquel B, Guilhot C. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primm TP, Lucero CA, Falkinham JO., 3rd Health impacts of environmental mycobacteria. Clin Microbiol Rev. 2004;17:98–106. doi: 10.1128/CMR.17.1.98-106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiol T, Ribeiro GM, Maranhao AQ, Bocca AL, Silva-Pereira I, Junqueira-Kipnis AP, Brigido Mde M, Kipnis A. Complete Genome Sequence of Mycobacterium massiliense. J Bacteriol. 2012;194:5455. doi: 10.1128/JB.01219-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll F, Pasek S, Schenowitz C, Dossat C, Barbe V, Rottman M, Macheras E, Heym B, Herrmann JL, Daffe M, Brosch R, Risler JL, Gaillard JL. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One. 2009;4:e5660. doi: 10.1371/journal.pone.0005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Shang S, Gibbs S, Henao-Tamayo M, Shanley CA, McDonnell G, Duarte RS, Ordway DJ, Jackson M. Increased virulence of an epidemic strain of Mycobacterium massiliense in mice. PLoS One. 2011;6:e24726. doi: 10.1371/journal.pone.0024726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O'Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial "pan-genome". Proc Natl Acad Sci U S A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Sampaio EP, Daugherty SC, Hine E, Riley DR, Sadzewicz L, Sengamalay N, Shefchek K, Su Q, Tallon LJ, Conville P, Olivier KN, Holland SM, Fraser CM, Zelazny AM. Genomic insights into the emerging human pathogen Mycobacterium massiliense. J Bacteriol. 2012;194:5450. doi: 10.1128/JB.01200-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana-Niero C, Lima KV, Lopes ML, Rabello MC, Marsola LR, Brilhante VC, Durham AM, Leao SC. Molecular characterization of Mycobacterium massiliense and Mycobacterium bolletii in isolates collected from outbreaks of infections after laparoscopic surgeries and cosmetic procedures. J Clin Microbiol. 2008;46:850–855. doi: 10.1128/JCM.02052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YL, Choo SW, Tan JL, Ong CS, Ng KP, Ngeow YF. Draft genome sequence of Mycobacterium bolletii strain M24, a rapidly growing mycobacterium of contentious taxonomic status. J Bacteriol. 2012;194:4475. doi: 10.1128/JB.00916-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazny AM, Root JM, Shea YR, Colombo RE, Shamputa IC, Stock F, Conlan S, McNulty S, Brown-Elliott BA, Wallace RJ, Jr, Olivier KN, Holland SM, Sampaio EP. Cohort study of molecular identification and typing of Mycobacterium abscessus Mycobacterium massiliense and Mycobacterium bolletii. J Clin Microbiol. 2009;47:1985–1995. doi: 10.1128/JCM.01688-08. [DOI] [PMC free article] [PubMed] [Google Scholar]