Abstract
Cryptosporidiosis, a gastrointestinal disease caused by a protozoan Cryptosporidium hominis is often fatal in immunocompromised individuals. There is little clinical data to show that the existing treatment by nitazoxanide and paromomycin is effective in immunocompromised individuals1, 2. Thymidylate synthase (TS) and dihydrofolate reductase (DHFR) are essential enzymes in the folate biosynthesis pathway and are well established as drug targets in cancer and malaria. A novel series of classical antifolates, 2-amino-4-oxo-5-substituted pyrrolo[2,3-d]pyrimidines have been evaluated as Cryptosporidium hominis thymidylate synthase (ChTS) inhibitors. Crystal structure in complex with the most potent compound, a 2’-chlorophenyl with a sulfur bridge with a Ki of 8.83 ± 0.67 nM is discussed in terms of several Van de Waals, hydrophobic and hydrogen bond interactions with the protein residues and the substrate analog 5-fluorodeoxyuridine monophosphate. Of these interactions, two interactions with the non-conserved residues (A287 and S290) offer an opportunity to develop ChTS specific inhibitors. Compound 6 serves as a lead compound for analog design and its crystal structure provides clues for the design of ChTS specific inhibitors.
Keywords: Pyrrolo[2,3-d]pyrimidines; Thymidylate synthase; Dihydrofolate reductase; Cryptospridium hominis
Cryptosporidiosis is a gastrointestinal disease caused by a protozoan Cryptosporidium hominis (Ch). This infectious disease causes gastrointestinal distress in immunocompetent individuals and is often fatal in immunocompromised individuals such as patients with AIDS, cancer and those who have had organ transplants. There are no effective, approved therapies to treat cryptosporidiosis to date. There is little effort by the pharmaceutical industry in drug development against this disease. The existing treatment includes nitazoxanide and paromomycin. However, there is little clinical data to show that these drugs are effective especially in immunocompromised individuals1, 2. Thus, there is a medical need for discovery of new antiparasitic drugs. Thymidylate synthase (TS) and dihydrofolate reductase (DHFR) are essential enzymes in the folate biosynthesis pathway and are well established as drug targets in cancer and malaria. TS catalyzes the reaction of deoxyuridine monophosphate (dUMP) and methylene tetrahydrofolate (CH2H4F) to yield deoxythymidine monophosphate (dTMP) and dihydrofolate (H2F). DHFR catalyzes the reduction of H2F, in the presence of nicotinamide adenine dinucleotide phosphate (NADPH), to generate tetrahydrofolate (H4F). In C. hominis these enzymes are bifunctional and exist on a single polypeptide chain. The successful development of inhibitors targeting DHFR3–5 from Plasmodium falciparum, Trypanosoma Cruzi, C. hominis3–5 have been well documented in the literature. While there are many TS inhibitors being developed as anticancer agents6, there are fewer developments towards inhibitors targeting TS from parasites. The successful development of potent and selective inhibitors of DHFR for parasites and bacteria encourages the development of species-specific TS inhibitors. A potent TS inhibitor can be administered with a potent DHFR inhibitor as a combination therapy targeting pathogenic parasites. In this study we present a novel series of classical antifolates, 2-amino-4-oxo-5-substituted pyrrolo[2,3-d]pyrimidines with variation in the chain length (1–4) and the side chain phenyl to a thiophene (5) and a 2’-chlorophenyl with a sulfur rather than a carbon bridge (6) as ChTS inhibitors and evaluate these compounds for inhibition of ChTS enzyme activity. We also report the crystal structure of our leading TS inhibitor in complex with ChTS-DHFR. Analysis of the interactions between the inhibitor, cofactor, and TS active site residues can be utilized to develop potent and selective parasite specific TS inhibitor.
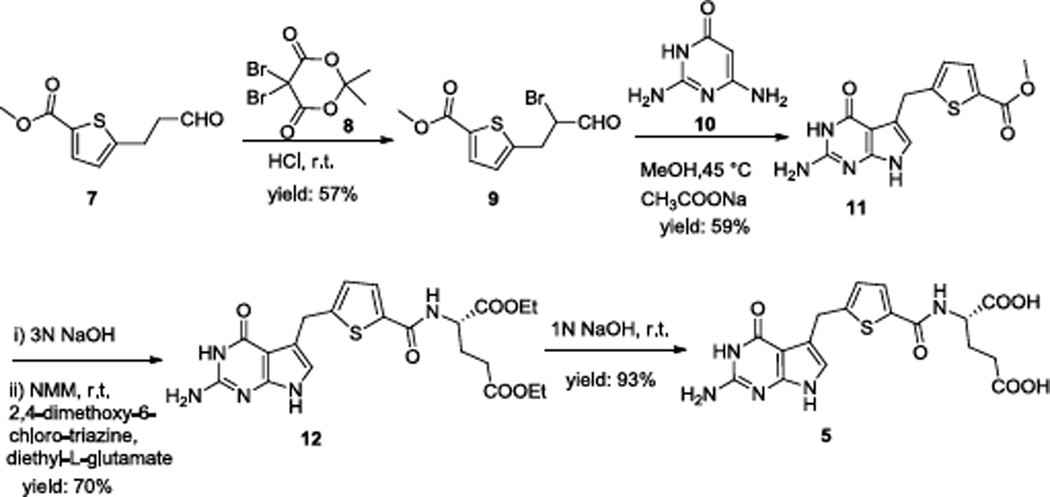
Gangjee et al. reported the synthesis of compounds 1–47 as well as compound 68. Our modification for the synthesis of compound 5 is reported in Scheme 1.
Scheme 1.
α-Bromination of the aldehyde 79 (Scheme 1) with 5,5-dibromo-2,2-dimethyl-4,6-dioxo-1,3-dioxane 8 at room temperature afforded the corresponding α-bromo aldehyde 9. Intermediate 11 was obtained by condensation of 9 with 10 at 45 °C in the presence of sodium acetate. Hydrolysis with 3N NaOH and coupling with diethyl L-glutamate using N-methyl morpholine and 2,4-dimethoxy-6-chlorotriazine as the activating agents afforded the diester 12 (yield: 70% over two steps). Saponification of the diester with 1N NaOH gave 5.
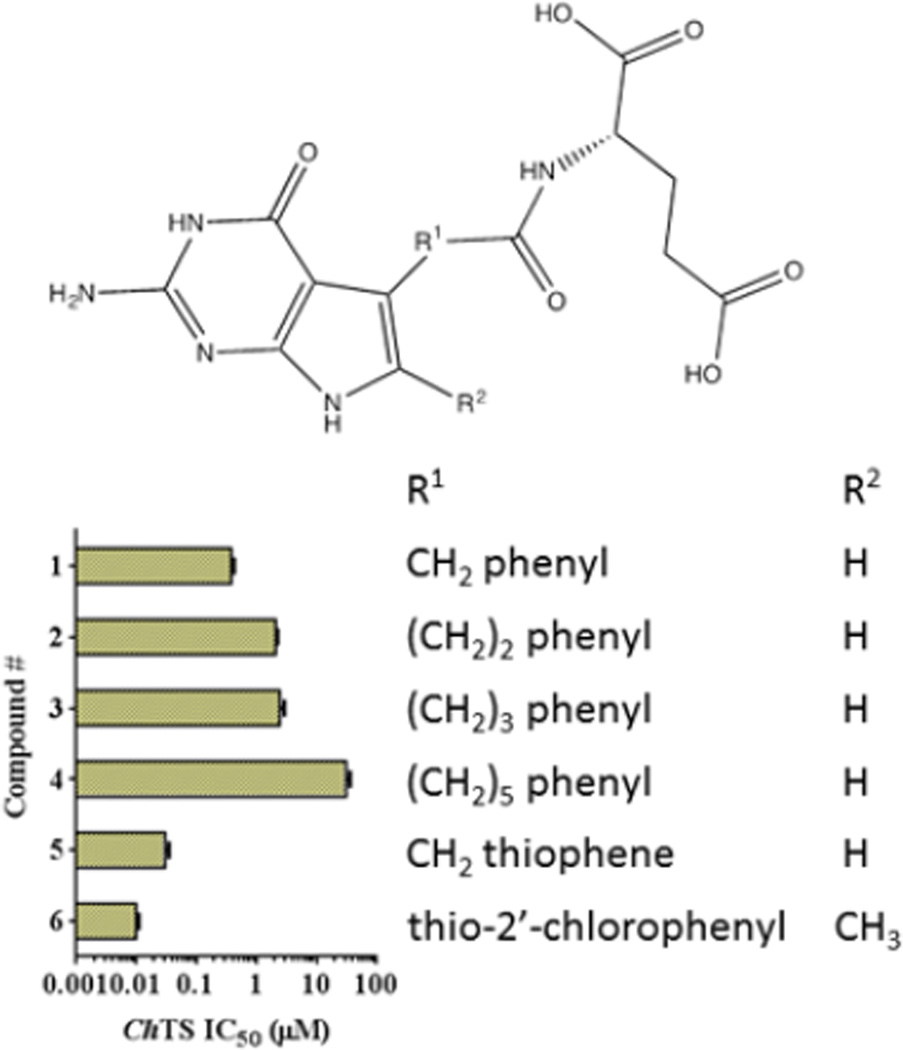
IC50 values for the compounds 1–6 in an in vitro ChTS enzyme assays range between 0.01 µM and 31 µM (Figure 1 and Table S1). Inhibition data suggests that potency of the pyrrolo[2,3-d]pyrimidines is linked to the length and composition of the linker connecting the pyrrolo[2,3-d]pyrimidine scaffold to the glutamate tail. Compound 1 with a one carbon linker has the lowest IC50 (0.38 µM), while IC50 values for Compounds 2 – 4 with increasing length of linkers are higher. Substitution of the side-chain phenyl ring of 1 with an isosteric thiophene in 5 increases potency by 10-fold to IC50 = 0.03 µM. Addition of a 6-methyl and an electron withdrawing 2’-Cl on the phenyl ring of 1 affords increased potency of 38-fold over 1 to the most potent analog of the series, 6. Compounds 5 and 6 with an aromatic ring in the linker show much higher potency.
Figure 1.
Structures of compounds 1 – 6 with their ChTS IC50 values shown as the bar graph.
Compound 6 was further characterized as a tight binding inhibitor with Ki of 8.83 ± 0.67 nM. IC50 values for human TS enzyme are similar to the ones obtained for ChTS and inhibition of ChDHFR shows a similar trend in which the IC50 values increase with linker length, a side chain thiophene and a 6-methyl-2’-Cl phenyl (data not shown).
The most potent ChTS inhibitor compound 6 was cocrystallized along with other three ligands (5-fluorodeoxyuridine monophosphate (FdUMP), NADPH and methotrexate (MTX)) and ChTS-DHFR. Detailed crystallization conditions have been reported in the Supporting Information. The best crystals diffracted to amplitudes extending to a resolution of 3.08 Å (Table S2). Phases were solved via molecular replacement using the program Phaser. The search model used for molecular replacement was the ChTS-DHFR structure in complex with four ligands (CB3717 and deoxyuridine monophosphate at TS site and NADPH and MTX at DHFR site) (PDB code: 1QZF). Root Mean Square deviation (RMSD) between 1QZF and our structure is 0.751Å (for an all atom alignment) suggesting that the overall conformation of the protein is similar (Figure S1). All residues from 3 to 521 except for residues 179 – 192 are clearly defined in the electron density, allowing all of the ligand binding sites of the structure to be visualized (Figure S2).
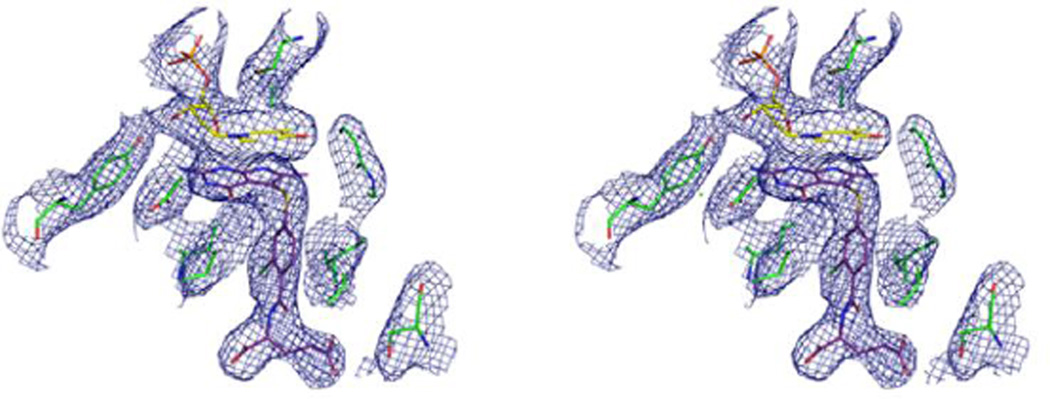
Figure 2 shows a stereo view of a 2Fo-Fc electron density map of the active site region to illustrate the positions of the FdUMP and compound 6 complex.
Figure 2.
Stereo view of a 2Fo-Fc electron density map (contour level at 1.0 σ) for TS active site of ChTS-DHFR: FdUMP: compound 6 complex.
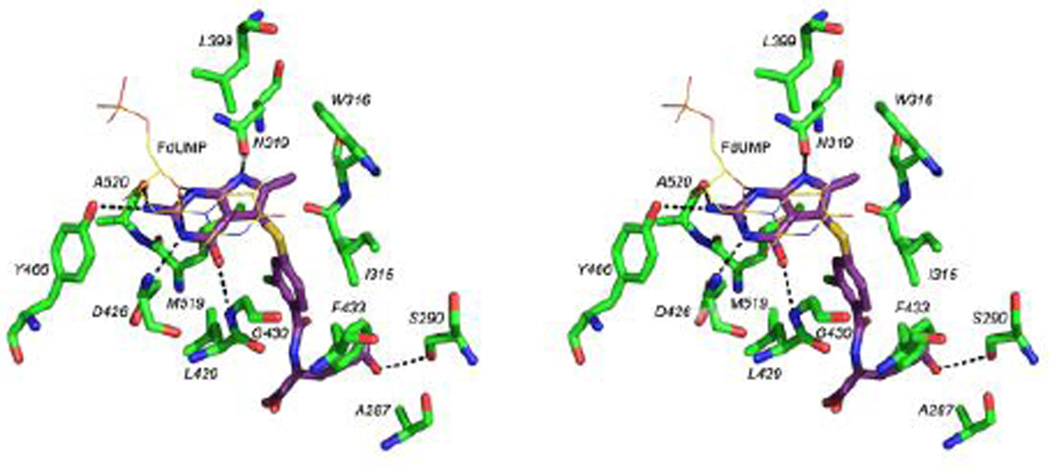
There are several Van der Waals and hydrophobic interactions between compound 6 and ChTS (Figure 3) with residues I315, W316 and F433 on one side of the molecule, L399, Y466, L429, M519 on the other side and A287 at the glutamate tail of compound 6. The sulfur atom of M519 is positioned to interact with N1 as well as 2’-Cl on the phenyl ring of compound 6. The fluorine of FdUMP makes an additional electrostatic contact with hydroxyl of Y342 as well as a non-covalent interaction with sulfur atom of compound 610.
Figure 3.
Stereo view of the active site residues and FdUMP interacting with compound 6. Hydrogen bonds (3.5 Å or less) are shown as dashed lines.
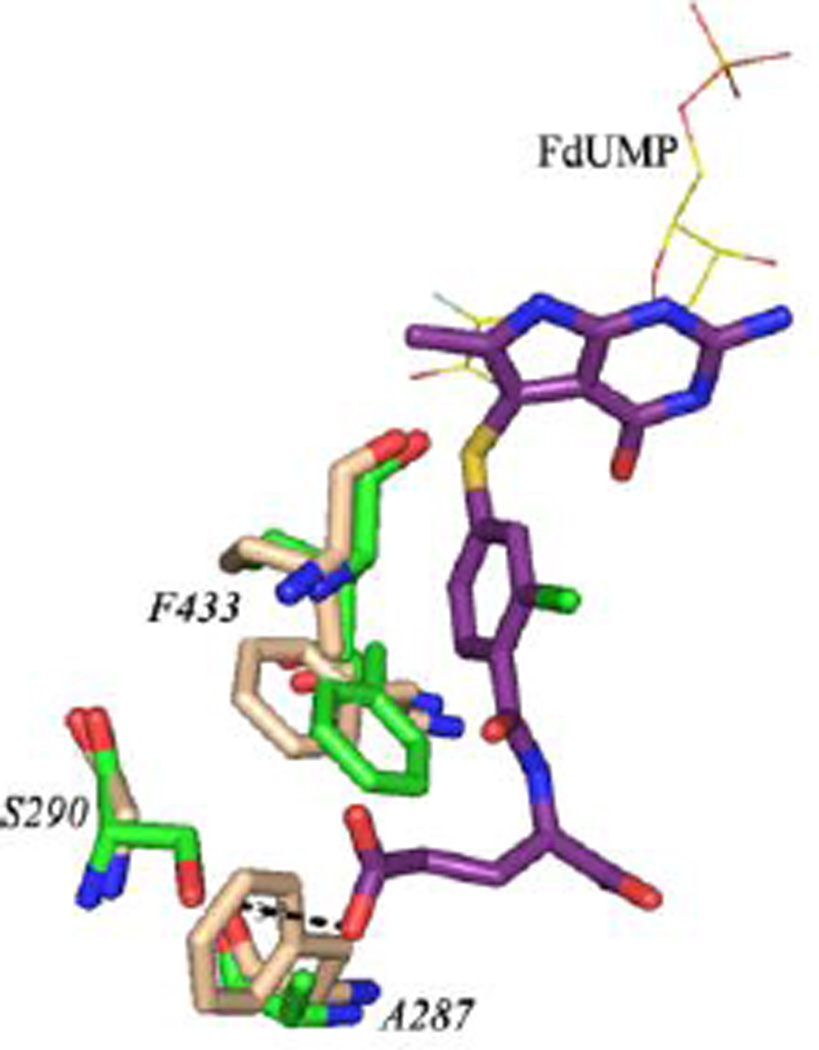
The pyrimidine ring of FdUMP has a stacking interaction with the pyrrolo[2,3-d]pyrimidine scaffold of compound 6. In addition to several Van der Waals and hydrophobic interactions, compound 6 is involved in hydrogen bonding interactions with FdUMP and active site residues. The O2 atom on the ribose ring of FdUMP is hydrogen bonded with N1 and N3 of compound 6. Carbonyl O of D426 hydrogen bonds (3 Å) with N3 and carbonyl O of N319 hydrogen bonds with N7 of compound 6 (2.9 Å). The 4-oxo group of compound 6 interacts with the amino group of G430. Hydrogen bonding interactions between the hydroxyl of Y466 and carbonyl O of A520 with the 2-amino group of compound 6 must add to its binding into the active site. The ChTS active site has two unique residues, A287 and S290. These residues have been shown to be important for optimal positioning of the cofactor CH2H4F and catalysis11. In human TS, the corresponding residues are F80 and G83 (Figure 4). This structural difference can be exploited to design parasite specific potent TS inhibitors by substituting suitable groups to make the binding of inhibitor favorable to ChTS.
Figure 4.
An all atom alignment of ChTS-DHFR:Compound 6 (green:purple) and human TS (tan, PDB code: 1HVY). RMSD = 0.77 Å.
In summary, a series of pyrrolo[2,3-d]pyrimidines are identified as ChTS inhibitors. A direct correlation between inhibition of ChTS enzyme and length of the linker connecting the glutamate tail was established. Shorter alkyl linkers afford better potency. Having aromatic ring structures in the linker provide additional interactions with the active site residues resulting in increased potency of the compound 6 with a Ki of 8.83 ± 0.67 nM. Several hydrophobic, Van der Waals and hydrogen bonding interactions were observed between compound 6 and FdUMP; compound 6 and active site residues. Of these interactions, two are unique as these residues (A287 and S290) are non-conserved residues of ChTS. Interactions with these residues offer an opportunity to develop ChTS specific inhibitors by exploiting the presence of bulky phenylalanine in human TS and an additional hydrogen bond donor with S290 in ChTS as compared with a glycine in human TS. For effective therapy for cryptosporidiosis, especially in immunocompromised individuals, a combination therapy of a potent parasite specific TS inhibitor with a potent DHFR inhibitor can be administered. Compound 6 serves as a lead compound for analog design and its crystal structure provides clues for the design of ChTS specific inhibitors.
Supplementary Material
Acknowledgments
This work is supported, in part, by NIAID grant (AI083146) to KSA, NIAID grant (AI104334) to KMF and NCI grant (CA152316) to AG and the Duquesne University Adrian Van Kaam Chair in Scholarly Excellence (AG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Full details on the enzyme assays, crystallography, experimental section, synthesis and characterization of compound 6. This material is available free of charge via the Internet at http://www.elsevier.com. Data for compound 6 has been deposited in the RCSB Protein Data Bank with codes 4KY8.
References
- 1.Gargala G, Delaunaya A, Lib X, Brasseurb P, Favennecb L, Balleta JJ. J Antimicrob Chemother. 2000;46:57. doi: 10.1093/jac/46.1.57. [DOI] [PubMed] [Google Scholar]
- 2.Abubakar I, Aliyu SH, Arumugam C, Usman NK, Hunter PR. Br J Clin Pharmacol. 2007;63:387. doi: 10.1111/j.1365-2125.2007.02873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolstad DB, Bolstad ESD, Frey KM, Wright DL, Anderson AC. J. Med. Chem. 2008;51:6839. doi: 10.1021/jm8009124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang H, Lu W, Li X, Cong X, Ma H, Liu X, Zhang Y, Che P, Ma R, Li H, Shen X, Jiang H, Huang J, Zhu J. Bioorg Med Chem Lett. 2012;22:958. doi: 10.1016/j.bmcl.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Schormann N, Velu SE, Murugesan S, Senkovich O, Walker K, Chenna BC, Shinkre B, Desai A, Chattopadhyay D. Bioorg Med Chem. 2010;18:4056. doi: 10.1016/j.bmc.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 6.Galvani E, Peters GJ, Giovannetti E. Expert Opin Investig Drugs. 2011;20:1343. doi: 10.1517/13543784.2011.617742. [DOI] [PubMed] [Google Scholar]
- 7.Gangjee A, Wang Y, Desmoulin SK, Cherian C, Matherly LH, Kisliuk RL. 240th American Chemical Society National Meeting; 2010. [Google Scholar]
- 8.Gangjee A, Jain HD, McGuire JJ, Kisliuk RL. J. Med. Chem. 2004;47:6730. doi: 10.1021/jm040144e. [DOI] [PubMed] [Google Scholar]
- 9.Aso K, Imai Y, Yukishige K, Ootsu K, Akimoto H. Chem. & Pharm. Bul. 2001;49:1280. doi: 10.1248/cpb.49.1280. [DOI] [PubMed] [Google Scholar]
- 10.Huston SM, Wang J, Loth MA, Anthony JE, Conrad BR, Dougherty DB. J. Phys. Chem. C. 2012;116:21465. [Google Scholar]
- 11.Doan LT, Martucci WE, Vargo MA, Atreya CE, Anderson KS. Biochemistry. 2007;46:8379. doi: 10.1021/bi700531r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.