Background: IFITM3 is a general antiviral host restriction factor against RNA viruses.
Results: SET7-mediated monomethylation of IFITM3 at Lys-88 negatively affected its antiviral activity toward vesicular stomatitis virus (VSV) and influenza A virus (IAV) infection.
Conclusion: The monomethylation of antiviral host restriction factors may perturb their function.
Significance: Targeting the SET7 pathway could provide new antiviral therapeutic strategies.
Keywords: Antiviral Agents, Host Defense, Host-pathogen Interactions, Post-translational Modification, Protein Methylation, Antiviral Host Restriction Factors, IFITM3, Lysine Methylation, SET7
Abstract
Although lysine methylation is classically known to regulate histone function, its role in modulating antiviral restriction factor activity remains uncharacterized. Interferon-induced transmembrane protein 3 (IFITM3) was found monomethylated on its lysine 88 residue (IFITM3-K88me1) to reduce its antiviral activity, mediated by the lysine methyltransferase SET7. Vesicular stomatitis virus and influenza A virus infection increased IFITM3-K88me1 levels by promoting the interaction between IFITM3 and SET7, suggesting that this pathway could be hijacked to support infection; conversely, IFN-α reduced IFITM3-K88me1 levels. These findings may have important implications in the design of therapeutics targeting protein methylation against infectious diseases.
Introduction
Antiviral host restriction factors defend against infection by preventing viral entry and replication (1). Interferon-induced transmembrane protein 3 (IFITM3)3 is one such factor that belongs to the CD225 gene family, inclusive of IFITM1, IFITM2, and IFITM3, which are highly inducible by both type I (IFN-α/IFN-β) and II (IFN-γ) interferons (2, 3); these are also known as interferon-stimulated genes (ISGs). IFITM3 plays a fundamental role in protecting the host against morbidity and mortality due to infection (4, 5). Recent studies have shown how IFITM3 inhibits the early replication of influenza A, West Nile, and dengue viruses at both basal and induced levels (6–8). The current literature of the mode of action of IFITM3 includes the inhibition of viral cytosolic entry at the late endocytic stage and through blockade of viral membrane hemifusion to enodosomal membranes (9–11).
The posttranslational modification (PTM) of proteins by processes such as phosphorylation, ubiquitination, and acetylation is widely known to dynamically alter protein function through modulation of enzymatic function, localization, protein interactions, or determination of protein degradative fate. IFITM3 can be subjected to S-palmitoylation and ubiquitination to fine tune its function by placing IFITM3 into the pathway of viral invasion (12, 13). The N-terminal region of IFITM3 has also been shown to be essential for its trafficking, where the Tyr-20 residue is key in mediating the correct subcellular localization of IFITM3 (14, 15). Although some IFITM3 binding partners have been identified (16, 17), the enzymes responsible for the current known PTMs of IFITM3 remain elusive; once discovered, they could potentially serve as therapeutic targets against viral infection.
During our efforts to uncover novel binding partners and enzymatic modulators of IFITM3, a monomethylation signal at its Lys-88 residue was detected. Until recently, lysine methylation was predominantly seen as a histone modification to regulate gene expression (18). Although it is now appreciated that lysine methylation may affect non-histone proteins, including viral effectors (19–22), there have yet to be any reports that indicate a role for lysine methylation in controlling the activity of antiviral host restriction factors, and a PTM enzyme has not been identified that targets IFITM3. As such, we focused on uncovering the role of monomethylation on IFITM3 function and finding the PTM enzyme responsible for this process.
EXPERIMENTAL PROCEDURES
Plasmids, Antibodies, and Reagents
Coding sequences of IFITM1, IFITM2, IFITM3, and SET7 were amplified by PCR from cDNA generated from purified RNA of HeLa cells using TRIzol reagent (Invitrogen) and the cDNA reverse transcription kit (TaKaRa) as per the manufacturer's instructions. These were subsequently cloned into the tandem affinity purification (TAP) tag (pCDNA4/to-N-TAP), HA or FLAG-tagged (pIPHA2 or pIPFLAG2; modified from pIRES2-puro) or non-tagged (pcDNA3.1) vectors by standard procedures. SET7-H297A (23) and IFITM3-K88R and K88A mutants were generated using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's standard procedures and confirmed by DNA sequencing. The shRNA expressing vector pLKO.1 was purchased from Open Biosystems and modified for insertion of custom-made shRNAs. His-tagged (pET-28a) and GST-tagged (pGEX-4T1) plasmids were used for the production of recombinant proteins in Escherichia coli. The plasmids del8.9 and VSV-G were gifts from Ke Lan (Institut Pasteur of Shanghai, Chinese Academy of Sciences). The antibodies used in this study were as follows: anti-HA (F-7, Santa Cruz Biotechnology), anti-FLAG (M2, Sigma-Aldrich), anti-calnexin (H-70, Santa Cruz Biotechnology), anti-SET7 (Cell Signaling), anti-IFITM3 (Protein Tech Group, Inc.), anti-histone 3 (Cell Signaling), anti-H3K4me1 (D1A9, Cell Signaling), and anti-α-tubulin (DM1A, Sigma). Protein A/G-agarose beads (A10001) were purchased from Santa Cruz Biotechnology. The αK88me1 rabbit polyclonal antibody was generated by Abmart, raised toward the polypeptide SRDR(Kme1)MVGD. IFN-α (recombinant human interferon α-2b, Shanghai Hua Xin High Biotechnology Inc.). Anti-influenza A nucleoprotein (9G8, Sc-101352, Santa Cruz Biotechnology).
Tandem Affinity Purification
1 × 109 U2OS cells stably expressing TAP (2× protein A and a calmodulin binding protein linked by a tobacco etch virus protease cleavage site) or TAP-tagged IFITM3 were harvested and washed with PBS, and then lysed in lysis buffer (10 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.5% (v/v) Nonidet P-40, protease inhibitors (Roche Applied Science), 1 mm PMSF). For first affinity purification, cell lysates were clarified by centrifugation. The supernatants were incubated with rabbit IgG-agarose (A2909, Sigma). The IgG-agarose was then resuspended in tobacco etch virus cleavage buffer (10 ml Tris-HCl, pH 8.0, 150 mm NaCl, 0.5 mm EDTA, 0.1% (v/v) Nonidet P-40, and 1 mm DTT) and cleaved by TEV protease (expressed in E. coli and purified by nickel-nitrilotriacetic acid) at 4 °C overnight. The supernatants were harvested for second affinity purification. Here, calmodulin binding buffer (10 mm Tri-HCl, 0.1% (v/v) Nonidet P-40, 1 mm magnesium acetate, 1 mm imidazole, 2 mm CaCl2, and 1 mm 2-mercaptoethanol) and calmodulin beads (214303, Stratagene) were incubated with supernatants for 1 h at 4 °C. Finally, IFITM3 complexes were eluted by calmodulin elution buffer (50 mm (NH4)2CO3, 25 mm EGTA). Elution fractions were precipitated by TCA and were subjected to SDS-PAGE and silver staining. Visible protein bands were excised and subjected to trypsin digestion followed by mass spectrometry analysis.
Cell Culture
HEK-293T, U2OS, MRC5, A549, MEF, and Vero cells were cultured in DMEM containing 10% (v/v) FBS, 50 units/ml of penicillin, and 50 μg/ml of streptomycin at 37 °C, 5% CO2 in a humidified incubator. HEK-293T cells expressing HA-IFITM3 were selected using puromycin.
Transfection
HEK-293T cells were transfected using polyethylenimine (Polysciences) according to the manufacturer's instructions.
Quantitative PCR
Total RNA was extracted using TRIzol Reagent (Invitrogen) following the manufacturer's instructions. cDNA was synthesized using a reverse transcriptase kit (TaKaRa) following the manufacturer's instructions. cDNA samples were then used as templates (25 ng/well) in a 384-well plate and run in triplicate. PCR reactions were set up in 20-μl total volumes using SYBR green (TaKaRa). Real-time PCR was carried out on the Prism 7500 Fast System Sequence Detection System (Applied Biosystems). Cycling reaction conditions consisted of the following: 95 °C for 30 s, followed by 95 °C for 15 s, 60 °C for 30 s, with a total of 40 cycles. GAPDH was used as the reference control for the target genes. Real-time primers are as follows (for WSN33 target primers, see Ref. 24): TCCCACGTACTCCAACTTCCA (IFITM3-F); AGCACCAGAAACACGTGCACT (IFITM3-R); TCTTCTTGAACTGGTGCTGTC (IFITM1-F); GTCGCGAACCATCTTCCTGT (IFITM1-R); CCTTGACCTGTATTCCACT (IFITM2-F); GCCATTGTAGAAAAGCGT (IFITM2-R); AGTTCTCCAGGGCACGTATG (SET7-F); TCTCCAGTCATCTCCCCATC (SET7-R); TGATACAGTACAATTATTTTGGGAC (VSV-L-F); GAGACTTTCTGTTACGGGATCTGG (VSV-L-R); GTGACGGACGAATGTCTCATAA (VSV-P-F); TTTGACTCTCGCCTGATTGTAC (VSV-P-R); GAGTCAACGGATTTGGTCGT (GAPDH-F); GACAAGCTTCCCGTTCTCAG (GAPDH-R).
In Vitro Methylation Assay
GST, GST-IFITM3, His-SET7, or His-SET7-H297A were expressed in BL21/pLysS cells and purified using glutathione-Sepharose 4B (GE Healthcare) or nickel-nitrilotriacetic acid. After dialysis, proteins were quantified by SDS-PAGE using BSA as a standard. In vitro methylation assays were performed in a 30-μl reaction volume, with 2 μg of purified recombinant SET7 or SET7-H297A together with 2 μg of recombinant IFITM proteins in the presence of 0.1 mm S-adenosylmethionine (Sigma, A2408) in reaction buffer (50 mm Tris-Cl, pH 8.5, 5 mm MgCl2, 4 mm DTT) at 30 °C for 1 h. The reaction products were analyzed by Western blotting.
Western Blotting and Immunoprecipitation
Cells were harvested and washed with ice-cold PBS and lysed on ice for 30 min in 1× radioimmune precipitation assay buffer containing 50 mm Tris/HCl, pH 7.4, 150 mm NaCl, 1% (v/v) Triton X-100, 0.5% (v/v) sodium deoxycholate, 0.1% (v/v) SDS, 1 mm EDTA, 10% (v/v) glycerol with 1 mm PMSF, 1 mm Na3 VO4, 1 mm NaF, and protease inhibitor (1:100, P8340, Sigma-Aldrich). Cell lysates were cleared by centrifugation, and the supernatants were immunoprecipitated with the appropriate antibodies on protein A/G-agarose beads for 1 h at 4 °C. The immunocomplexes were then washed with radioimmune precipitation assay buffer four times and examined with Western blotting by standard procedures.
GST Pulldown Assay
2 μg of purified recombinant GST, GST-IFITM3, and 2 μg of His-SET7 were incubated in reaction buffer (20 mm HEPES, pH 8.0; 150 mm KCl; 1 mm EDTA; 0.1% (v/v) Nonidet P40; 10% (v/v) glycerol) for 2 h at 4 °C and then incubated with glutathione-Sepharose 4B for another 2 h. The reaction products were then washed 4× with 1 ml of reaction buffer. Bound proteins were analyzed by SDS-PAGE and stained using Coomassie Blue.
VSV Virus Production and Infection
VSV (Indiana serotype, ATCC). Recombinant VSV expressing green fluorescent protein (VSV-GFP) (25) was propagated in Vero cells after infection at a MOI of 0.01. 48 h later, supernatant was collected after centrifugation and then filtered through a 0.45-μm filter (Millipore) and stored at −80 °C. Virus infections were performed in the absence of serum for 1h, and then replaced with fully supplemented growth medium. End point dilution assay was used to quantify VSV titers with log dilutions on 100% confluent Vero cells plated in 96-well plates.
IAV Production and Infection
Recombinant viruses were generated by plasmid-based reverse genetics followed by two-round plaque purification and propagation in MDCK cells as described previously (26). Briefly, eight viral cDNAs from A/WSN/33 (H1N1) were cotransfected into HEK-293T cells. Supernatant was then transferred onto MDCK cells 72 h post transfection. Virus multiplied from MDCK cells was further purified by a two-round plaque assay followed by multiplication. To examine the methylation of IFITM3 in infected cells, virus titers for infection were determined by plaque assay titration in A549 cells. Cells were transfected with shRNAs followed by virus infection at a MOI of 2 for 30 min in 0.2% BSA in PBS, 3× washes with PBS and then cultured in complete DMEM culture media supplemented with 2% FBS. Cell extracts were collected 24 h post infection and analyzed by Western blotting and qPCR.
WSN33 Plaque Assays by Immunostaining
Plaque assays by immunostaining were performed as described previously (27, 28). Monolayers of MDCK cells in 12-well plates were infected with 20–40 pfu per well. After a 1-h incubation at 37 °C in 5% CO2, the medium was removed, and 2 ml of Avicel medium (DMEM with 0.2% BSA) was added. Cells were incubated for a further 3 days and then fixed in 4% paraformaldehyde. Immunostaining was performed using an anti-nucleoprotein polyclonal primary antibody (Antibody Research Centre, Shanghai Institute of Biological Science) and an HRP-conjugated anti-rabbit IgG secondary antibody. True Blue substrate (KPL Industry Co., Ltd.) was added to visualize the plaques.
shRNA Lentivirus Packaging and Transduction
Oligonucleotides were generated at Shanghai Sunny Biotechnology Co., Ltd. of the shRNA sequences (below), annealed, and then inserted between the EcoRI and NheI sites of the pLKO.1 plasmid. 12.5 μg of plasmids pLKO.1-shCK, pLKO.1-shSET7, or pLKO.1-shIFITM were co-transfected with 11.125 μg of del8.9 and 1.375 μg of VSV-G into 1.2 × 107 HEK-293T cells using polyethylenimine (Polysciences) according to the manufacturer's instructions. 48 h post-transfection, viral supernatants (8 ml total for each dish) were harvested after centrifugation and filtration for downstream use. shRNA sequences were as follows: CAACAAGATGAAGAGCACCAA (shCK); CCTGTTCAACACCCTCTTCAT (shIFITM); GCAAACTGGCTACCCTTATGT (shSET7–1); and GGGAGTTTACACTTACGAAGA (shSET7–2).
GFP-VSV MEF Cell Sorting
2 × 106 MEF cells infected with GFP-VSV at a MOI of 5 for 8 h were resuspended in FACS buffer (1 mm EDTA, 25 mm HEPES (pH 7.0) and 1% FBS in PBS). GFP-VSV(+/−) MEF cells were sorted on a FACS ARIA II system (BD Biosciences).
Endogenous Immunoprecipitation
MEF cells were seeded onto six-well plates. 20 h later, cells were pretreated with IFN-α for 24 h and then treated with VSV or IFN-α for another 6 h. Cells were lysed using radioimmune precipitation assay buffer for 1 h on ice and then precleared with protein A/G-Sepharose for 30 min at 4 °C. Supernatants were then transferred to a new tube and incubated with 2 μg of anti-IFITM3 antibody overnight on a rotor at 4 °C. Protein A/G-Sepharose was then added and rotated at 4 °C for 2 h. Beads were washed five times with radioimmune precipitation assay buffer and then examined by Western blotting using standard procedures.
Statistical Analysis
Statistical significance was calculated by the paired Student's t test (*, p < 0.05; **, p < 0.005) on GraphPad Prism, unless otherwise indicated. Data are representative of at least three independent experiments.
RESULTS
SET7 Regulates Monomethylation of IFITM3 at Lys-88
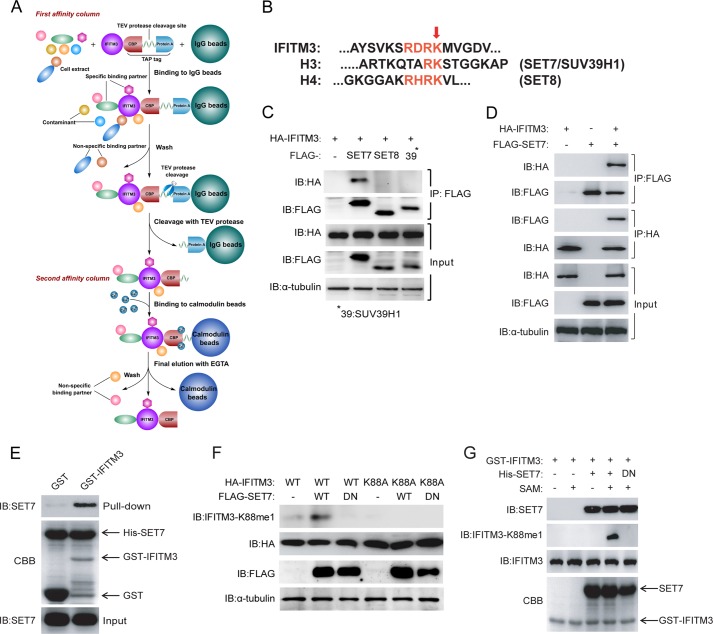
Our initial approach was to purify IFITM3 using the TAP approach (29) to identify novel binding partners and putative regulators of IFITM3 (Fig. 1A). Mass spectrometric analysis of the purified products revealed high association of IFITM3 with candidate proteins, such as those from the CCT (chaperonin containing T-complex polypeptide (TCP1), also called TRiC (TCP1 ring complex)).4 However, most surprisingly, we detected that Lys-88 of IFITM3 (IFITM3-K88) was subject to monomethylation (IFITM3-K88me1) (data not shown). As monomethylation has yet to be described to affect the function of antiviral host restriction factors, we decided to characterize the role of monomethylation in regulating IFITM3. Upon alignment of the monomethylated region of IFITM3 to those from histone proteins H3 and H4, the lysine methyltransferases SET7, SET8, and SUV39H1 were cloned for interaction studies as they were identified as highly likely candidates for mediating the monomethylation of IFITM3 at Lys-88 (Fig. 1B).
FIGURE 1.
Monomethylation of IFITM3 at Lys-88 is regulated by SET7. A, the strategy of tandem affinity purification. B, alignment of amino acid sequences of SET protein substrates: IFITM3 (amino acids 79–93) against histones H3 and H4. The red arrow indicates similarly conserved SET protein methylation sites. C, identification of the interaction between IFITM3 and SET7. 1 μg of FLAG-tagged SET7, SET8 or SUV39H1 were co-transfected into HEK-293T cells grown in six-well plates with 0.5 μg of HA-tagged IFITM3. Co-immunoprecipitation was performed with 1 μg of anti-FLAG antibody plus protein A/G beads. Protein blots were probed with the antibodies as indicated. D, reciprocal immunoprecipitation of IFITM3 and SET7. 1 μg of FLAG-tagged SET7 and 0.5 μg of HA-tagged IFITM3 were co-transfected into HEK-293T cells grown in six-well plates. Immunoprecipitation was performed with 1 μg of anti-HA or anti-FLAG antibodies plus protein A/G beads. Protein blots were probed with the antibodies as indicated. E, IFITM3 interacts with SET7 in vitro. A pulldown assay was performed with recombinant His-SET7, GST, and GST-IFITM3 as indicated. Immunoblotting (IB) and Coomassie Brilliant Blue staining (CBB) were used to show protein levels. F, IFITM3 is monomethylated by SET7 at Lys-88. Different combinations of 0.5 μg of HA-IFITM3, 0.5 μg of HA-IFITM3-K88A, 1 μg of FLAG-SET7 and 1 μg of FLAG-SET7-DN (H297A catalytically inactive mutant) were co-transfected into HEK-293T cells grown in six-well plates. Protein blots were probed with the antibodies as indicated. G, SET7 methylates IFITM3 in a cell-free system. In vitro methylation was performed with S-adenosylmethionine (SAM), recombinant His-SET7, His-SET7-DN, and GST-IFITM3 as indicated. Immunoblotting and Coomassie Brilliant Blue staining (CBB) were used to show methylation and protein levels, respectively. All figures are representative of >3 independent experiments. CBP, calmodulin binding protein; DN, dominant negative.
To test the interaction between IFITM3 and the candidate lysine methyltransferases, HA-tagged IFITM3 and FLAG-tagged SET7, SET8, or SUV39H1 were overexpressed in HEK-293T cells followed by co-immunoprecipitation using anti-FLAG antibody and then immunoblotting. An increase in signal was detected relative to the FLAG-only control that represented the successful pulldown and interaction between HA-IFITM3 and FLAG-SET7 (Fig. 1C). The interaction between IFITM3 and SET7 was confirmed by reciprocal co-immunoprecipitation using HA-IFITM3 or FLAG-SET7 independently as bait (Fig. 1D) and by in vitro GST pulldown with recombinant GST-IFITM3 and His-SET7 proteins that were purified from E. coli (Fig. 1E).
Next, an antibody was generated against IFITM3-K88me1 (designated αK88me1) as a tool to detect whether SET7 could specifically monomethylate IFITM3 at Lys-88. The overexpression of WT HA-tagged IFITM3 with FLAG-tagged SET7 in HEK-293T cells, followed by cell lysis and immunoblotting, showed an increase in signal detected by αK88me1, but not when the dominant negative form of SET7 (dominant negative, SET7-H297A) or IFITM3 containing a point mutation at Lys-88 to alanine (IFITM3-K88A), were used (Fig. 1F). We further showed the specificity of αK88me1 toward IFITM3 via an in vitro cell-free methylation reaction with purified recombinant GST-IFITM3 and His-SET7 in the presence of S-adenosylmethionine (Fig. 1G). These results showed the specificity of αK88me1 in detecting SET7-mediated monomethylation of IFITM3 at Lys-88.
VSV/IAV and Type I Interferon Control Monomethylation Levels of IFITM3
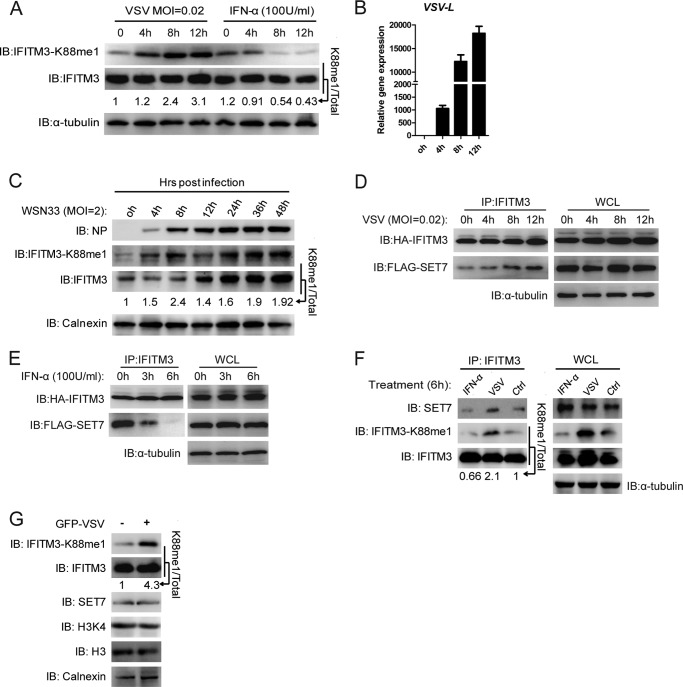
A VSV infection model in HEK-293T cells, which express low basal levels of endogenous IFITM3 at steady state, was employed to test the effects of SET7-mediated IFITM3-K88me1 modification on antiviral activity. Interestingly, in HEK-293T cells overexpressing IFITM3, IFITM3-K88me1 levels increased over time after VSV infection, which coincided with an increase in VSV RNA levels (VSV-L) but no change in total IFITM3 protein expression (Fig. 2, A and B). In contrast, IFITM3-K88me1 levels were effectively reduced upon short term IFN-α treatment, which also did not increase levels of IFITM3 (Fig. 2A and qPCR (data not shown)). These results indicated that both viral and host signals may influence the monomethylation of IFITM3 at the posttranslational level.
FIGURE 2.
Monomethylation of IFITM3 at Lys-88 is promoted by viral infection but reduced by IFN-α treatment. A and B, monomethylation of IFITM3 at Lys-88 is induced by VSV infection but reduced by IFN-α treatment. HEK-293T cells overexpressing IFITM3 grown in 12-well plates were infected with VSV at a MOI of 0.02 and treated with IFN-α (100 units/ml). Cells were collected at the indicated time points post-infection or treatment for immunoblotting with the indicated antibodies (A) or qPCR for detection of VSV-L (B). C, WSN33 infection increases monomethylation of IFITM3 at Lys-88. A549 cell lines grown in 12-well plates are infected with WSN33 at a MOI of 2 and collected at the indicated time points post-infection for immunoblotting (IB) analysis with the indicated antibodies. Numbers below the IFITM3-K88me1 and IFITM3 bands (in A and C) represent their relative density normalized to 0 h. D, virus infection increases the interaction between SET7 and IFITM3. HEK-293T cells grown in six-well plates were transfected with 0.5 μg of HA-IFITM3 and 1 μg of FLAG-SET7. 48 h later, cells were infected with VSV at a MOI of 0.02. Cells were then collected at the indicated time-points post-infection and were immunoprecipitated with 1 μg of anti-IFITM3 antibody plus protein A/G beads. Protein blots were probed with the antibodies as indicated. E, IFN-α reduces the interaction between SET7 and IFITM3. The experiment was performed as described in D but treated with IFN-α (100 units/ml) without virus for the indicated time periods. F, endogenous interaction between IFITM3 and SET7 is up-regulated by VSV infection but down-regulated by IFN-α treatment. MEF cells grown in six-well plates were infected with VSV (tested 6 h post-infection) or treated with IFN-α (6 h) as indicated and then collected for endogenous co-immunoprecipitation (co-IP). WCL, whole cell lysate; NP, nucleoprotein. G, VSV infection increases IFITM3-K88me1 directly. 2 × 106 MEF cells were pretreated with IFN-α (100 units/ml) for 24 h and then infected with GFP-VSV at a MOI of 1 followed by downstream experiments 8 h post-infection. GFP-VSV negative and positive cells were sorted (labeled “−” and “+”, respectively) and analyzed for IFITM3-K88me1 expression by Western blotting. Numbers below the IFITM3-K88me1 and IFITM3 bands represent their relative density normalized to the controls (Ctrl). Error bars represent the S.D. from the mean (n = 3). All figures are representative of >3 independent experiments. Note that the HA tag epitope is derived from an H3 influenza virus strain and is not present in the WSN33 strain of H1N1 influenza virus.
In an IAV infection assay using the strain WSN33 in A549 cells, which express higher basal levels of endogenous IFITM3 at steady state conditions compared with 293T cells, an increase in IFITM3-K88me1 levels was also found during early stages of infection (<8 h), during which endogenous IFITM3 levels did not significantly change (Fig. 2C). At 12 h post infection, the level of IFITM3-K88me1 slightly reduced, and endogenous IFITM3 levels started to increase. This was most likely caused by IFN-α signals as the up-regulation of IFNa transcription was observed at 8 h (data not shown). However, IFITM3-K88me1 levels subsequently increased after this period alongside IFITM3 expression.
In line with the observations above, co-immunoprecipitation of overexpressed HA-IFITM3 and FLAG-SET7 in HEK-293T cells followed by treatment with VSV or IFN-α revealed how VSV infection increased the interaction between IFITM3 and SET7 (Fig. 2D) but reduced upon IFN-α treatment (Fig. 2E). This phenomenon was confirmed by endogenous pulldown of SET7 by endogenous IFITM3 in MEF cells infected or treated either with VSV or IFN-α, respectively, for 6 h, in which the interaction between SET7 and IFITM3 was up-regulated upon VSV infection but reduced with IFN-α treatment, with IFITM-K88me1 levels up- and down-regulated, respectively (Fig. 2F). To show that infected cells up-regulated levels of IFITM3-K88me1, we infected MEF cells with VSV that include the coding sequence for GFP and tested the levels of IFITM3 and IFITM3-K88me1 by Western blotting in the GFP− and GFP+ populations. Here, we found that IFITM3 was more highly methylated when cells were infected with VSV (GFP+) (Fig. 2G). Because these data suggested that viral infection increased, and host responses decreased IFITM3 monomethylation, we speculated that this could be a method adopted by the virus to compromise the antiviral activity of IFITM3 for its own benefit.
Monomethylated IFITM3 Has Reduced Antiviral Activity
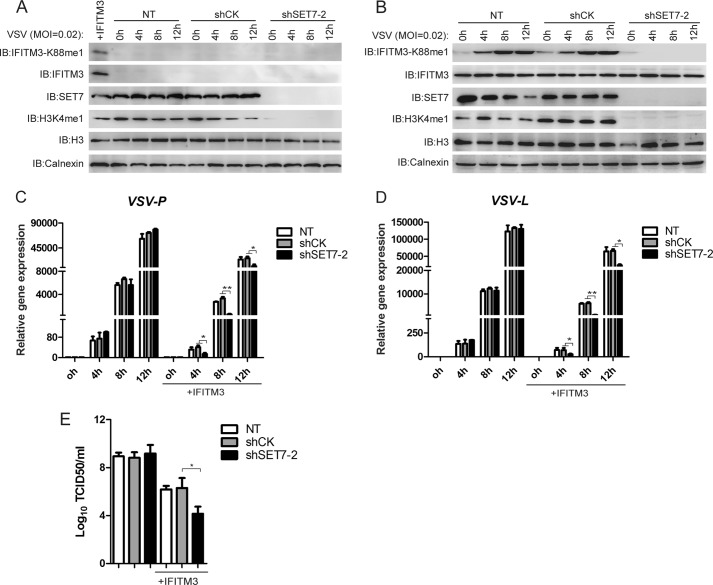
To test the effect of SET7 on IFITM3 activity toward infection, HEK-293T or HEK-293T cells overexpressing HA-IFITM3 (selected using puromycin) were transduced with lentivirus delivering either control shRNA (CK) or shRNA targeting SET7 (shSET7); after 3 days, cells were infected with VSV, and after 12 h, the expression levels of VSV-L and VSV-P RNA were detected. In WT HEK-293T cells with low basal levels of endogenous IFITM3 (Fig. 3A), the depletion of SET7 had no effect on the levels of viral RNA detected by qPCR after infection (Fig. 3, C and D), indicating no changes in the susceptibility of SET7-depleted cells toward viral infection. In HEK-293T cells overexpressing HA-IFITM3, as expected, VSV infection increased IFITM3-K88me1 levels over time; however, upon the depletion of SET7, both the IFITM3-K88me1 signal (Fig. 3B) and VSV RNA levels (Fig. 3, C and D) were effectively reduced, which indicated the possibility that the demethylated form of IFITM3 harbored superior antiviral activity as compared with its K88me1 form, and that the effect of SET7 on antiviral responses was dependent on the expression of IFITM3. In agreement with the above, VSV titers were reduced upon depletion of SET7 only in the presence of IFITM3 (Fig. 3E).
FIGURE 3.
Knockdown of SET7 promotes the antiviral activity of IFITM3. A–D, knockdown of SET7 affects VSV infection only in the presence of IFITM3 expression. Lentivirus packaged with shRNA was transduced into HEK-293T (A) or HEK-293T cells overexpressing IFITM3 (B) grown in 12-well plates. 72 h later, cells were infected with VSV at a MOI of 0.02. Cells were collected at the indicated time points post-infection for Western blot analysis with the indicated antibodies (A and B) or qPCR (C and D). E, VSV titers in the supernatants of the infected cells were determined by a plaque assay with Vero cells. Error bars represent the S.D. from the mean (n = 3). All figures are representative of >3 independent experiments. IB, immunoblot; NT, no treatment.
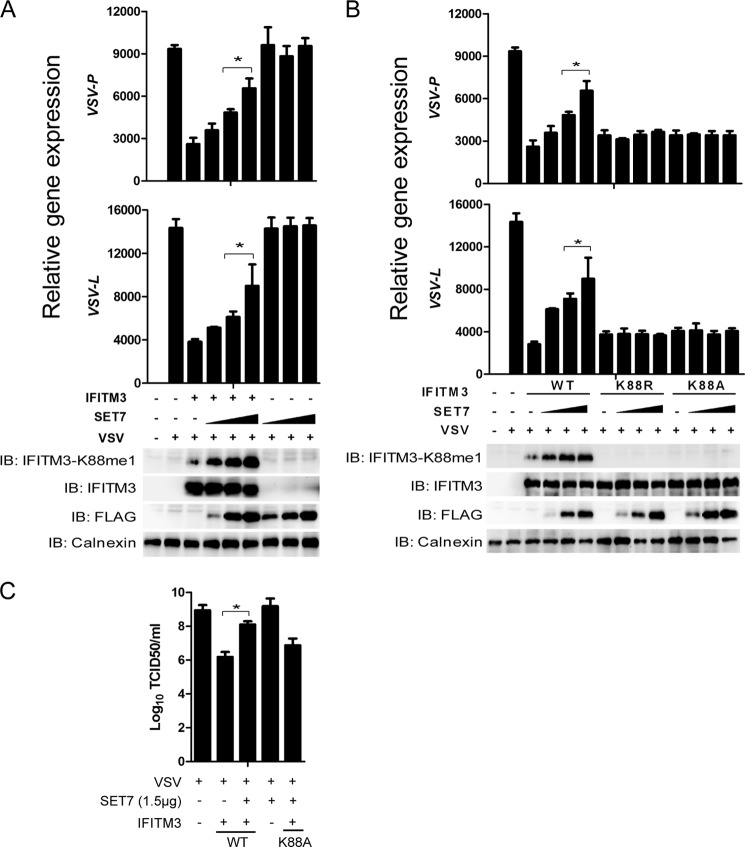
Next, we carried out studies to analyze the overexpression of SET7 and its impact on virus infection. The overexpression of SET7 in HEK-293T cells up-regulated levels of IFITM3-K88me1 and reduced antiviral activity in a dose/IFITM3-dependent manner (Fig. 4A), but in the absence of ectopically expressed IFITM3, the levels of VSV infection did not change (Fig. 4A). Moreover, the K88A and K88R mutants of IFITM3 rendered IFITM3 insensitive toward SET7-mediated monomethylation and modulation of its antiviral function (Fig. 4B), and SET7 overexpression in the presence of IFITM3 increased virus titers (Fig. 4C), thus supporting the notion that Lys-88 monomethylation of IFITM3 negatively affects its activity.
FIGURE 4.
Overexpression of SET7 reduces the antiviral activity of IFITM3. A, overexpression of SET7 reduces IFITM3 antiviral function. HEK-293T cells grown in 12-well plates were transfected with 1 μg of pcDNA3.1-IFITM3 and 1 μg of FLAG-SET7 plasmids as indicated. 48 h later, cells were infected with VSV at a MOI of 0.02 and analyzed 12 h post-infection. Cells were then collected and tested by Western blotting with the indicated antibodies (bottom) or subjected to qPCR analysis for VSV-P and VSV-L expression (top). B, mutation of Lys-88 of IFITM3 removes SET7-mediated modulation of its antiviral function. HEK-293T cells grown in 12-well plates were transfected with 0.5 μg of pcDNA3.1-IFITM3 or 0.5 μg of pcDNA3.1-IFITM3-K88R/K88A mutants and increasing amounts of FLAG-SET7 plasmids (0.3 μg, 0.9 μg, or 1.5 μg). 48 h later, cells were infected with VSV at a MOI of 0.02 and collected for analyses 12 h post-infection. Cells were collected and tested by Western blotting with the indicated antibodies (bottom) or subjected to qPCR analysis for VSV-P and VSV-L expression (top). C, VSV titers in the supernatants of the indicated infected cells were determined by a plaque assay with Vero cells using 1.5 μg of FLAG-SET7 and 0.5 μg of pcDNA3.1-IFITM3 WT or K88A plasmids. Error bars represent the S.D. from the mean (n = 3). All figures are representative of >3 independent experiments.
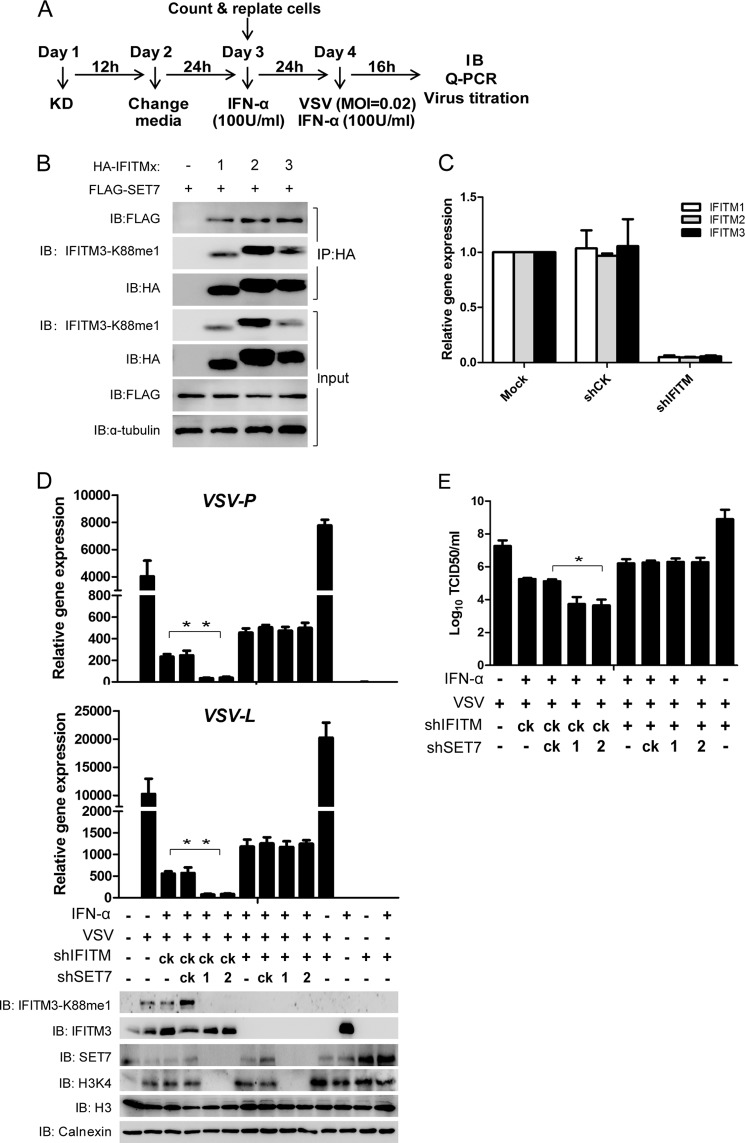
SET7 Modulates ISG-mediated Anti-VSV/IAV Responses in an IFITM-dependent Manner
To test the effect of SET7 on VSV infection using a more physiological model, we induced ISGs with IFN-α in the secondary cell line MRC5 and depleted SET7 to examine its effect on viral infection. SET7 was depleted (day 1) followed by the induction of ISGs with IFN-α (day 3) and then infected with VSV and tested after 16h (day 4) (Fig. 5A). IFN-α may up-regulate a host of ISGs, including IFITM1, 2, and 3 (IFITM1–3), which all share high homology. We found that SET7 could also interact with IFITM1 and IFITM2, and signals were detected using the αK88me1 antibody that indicated the presence of monomethylation on IFITM1 and IFITM2 (Fig. 5B). Thus, to determine whether the effect of SET7 on the antiviral activity of these ISGs is IFITM-dependent, IFITM1–3 were all simultaneously depleted (Fig. 5C). The depletion of SET7 alone (Fig. 5D) coincided with the significant reduction of IFITM3-K88me1 levels (Fig. 5D, Western blot) and also caused a dramatic reduction in VSV RNA levels as compared with no significant changes in the shCK control (Fig. 5D, qPCR). This indicated that SET7 had a negative effect on ISG-mediated antiviral activity against VSV infection. The effect of SET7 on ISG-mediated antiviral function was effectively abolished when IFITM1–3 were depleted (Fig. 5D), and the effects of depleting SET7 and/or the IFITMs were also seen via a virus titration assay (Fig. 5E). SET7, however, seemed to not affect antiviral responses when ISGs were not induced, suggesting that at least in this system SET7 does not regulate antiviral responses of low/basal levels of IFITM3 (data not shown).
FIGURE 5.
SET7 inhibits anti-VSV potency of ISGs in an IFITM-dependent manner. A, experimental procedure. B, IFITM1 and IFITM2 can interact with SET7 and are sensitive to SET7-mediated monomethylation. KD, knockdown. 1 μg of FLAG-tagged SET7 was co-transfected into HEK-293T cells grown in six-well plates with 0.5 μg of HA-tagged IFITM1, -2, or -3. Co-immunoprecipitation was performed with 1 μg of anti-HA antibody plus protein A/G beads. Protein blots were probed with the antibodies as indicated. C, IFITM shRNA targets IFITM1, -2, and -3. MRC5 cells were tested for knockdown of IFITM1, -2, and -3 by lentivirus containing shIFITM followed by analysis via qPCR. D, knockdown of SET7 promotes antiviral activity of ISGs in an IFITM-dependent manner. Lentivirus-packaged shRNAs were transduced into MRC5 cells grown in six-well plates on day 1. After lentivirus infection for 12 h, the medium was changed to fresh DMEM (day 2). 24 h later (day 3), 5 × 105 cells were counted and transferred to 12-well plates and then treated with IFN-α (100 units (U)/ml) to induce the expression of ISGs. After IFN-α treatment for 24 h, cells were infected with VSV at a MOI of 0.02 and then collected at 16 h post-infection for quantitative PCR (top) or Western blot analysis with the indicated antibodies (bottom). E, VSV titers in the supernatants of the indicated infected cells were determined by a plaque assay with Vero cells. Error bars represent the S.D. from the mean (n = 3). All figures are representative of >3 independent experiments. IB, immunoblot.
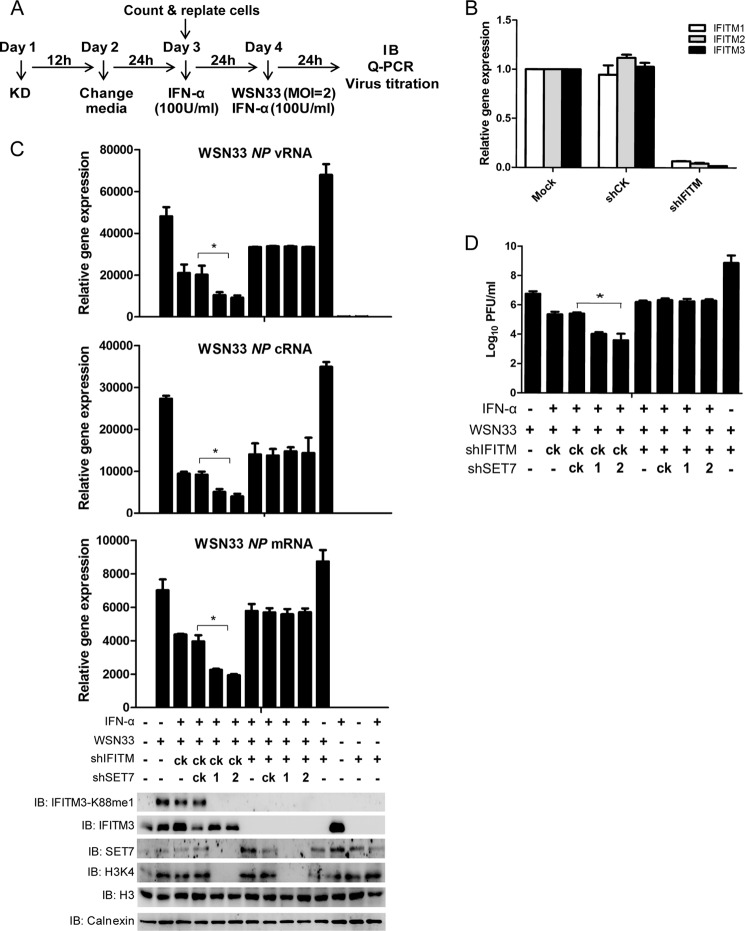
These observations were also reproducible in A549 cells infected by WSN33 and tested after 24 h (WSN33 strain) (Fig. 6A), in which RNA levels of nucleoprotein (vRNA, cRNA, and mRNA) were reduced upon SET7 depletion (Fig. 6C). Once IFITM1–3 levels were depleted (Fig. 6B), the knockdown of SET7 no longer had an effect on the levels of WSN33 RNAs detected in the A549 cells (Fig. 6C) corroborated via a plaque assay in MDCK cells (Fig. 6D). These results indicate how the IFITM antiviral factors may be modulated in a SET7-depdendent manner to determine host susceptibility toward viral infection, showing a once unappreciated role of lysine methylation in controlling the antiviral activity of host restriction factors at the posttranslational level.
FIGURE 6.
SET7 inhibits anti-IAV potency of ISGs in an IFITM-dependent manner. A, experimental procedure. B, knockdown (KD) efficiency of IFITM1–3 examined by qPCR. A549 cells were tested for knockdown of IFITM1, -2, and -3 by lentivirus containing shIFITM followed by analysis via qPCR. C, knockdown of SET7 promotes antiviral activity of ISGs in an IFITM-dependent manner. Lentivirus-packaged shRNAs were transduced into A549 cells grown in six-well plates on day 1. After lentivirus infection for 12 h, the medium was changed to fresh DMEM (day 2). 24 h later (day 3), 5 × 105 cells were counted and transferred to 12-well plates and then treated with IFN-α (100 units (U)/ml) to induce the expression of ISGs. After IFN-α treatment for 24 h, infected cells with WSN33 at a MOI of 2 and collected 24 h post-infection for quantitative PCR (top) or Western blot analysis with the indicated antibodies (bottom). D, WSN33 titers in the supernatants of the indicated infected cells were determined by a plaque assay with MDCK cells. Error bars represent the S.D. from the mean (n = 3). All figures are representative of >3 independent experiments. IB, immunoblot; NP, nucleoprotein gene.
DISCUSSION
This work reveals how lysine monomethylation of IFITM3 at Lys-88, which is positively regulated by viral infection/SET7 and negatively regulated by type I IFN, could play an important role in tipping the balance of infection toward benefiting the virus. This is the first study to identify an enzyme that directly modifies IFITM3 or targets an antiviral restriction factor using monomethylation to ultimately affect its function. Although other lysine methyltransferases were not screened for their interaction with IFITM3, we believe that the methyltransferase SET7 directly mediates the Lys-88 modification of IFITM3 because the depletion of SET7 significantly reduces the K88me1 signal as detected by the αK88me1 antibody generated toward this PTM.
It still remains unclear as to how the monomethylation of IFITM3 is regulated. In previous studies, SET7 was shown to shuttle between the cytoplasm and nucleus upon infection or treatment with inflammatory cytokines (30). However, we did not detect any significant changes in SET7 localization (found in the nucleus and cytoplasm) or expression upon viral infection or IFN-α treatment.4 Another factor that remains unclear is how the monomethylation of IFITM3 modulates its antiviral activity. Although some studies have shown that SET7 mediates the stability of its non-histone targets (31), our study found no significant effect of monomethylation on IFITM3 expression; we did not detect any differences in localization between WT IFITM3 and its K88R or K88A mutants, even in the presence of ectopically expressed SET7 to drive increased monomethylation of IFITM3 (data not shown). Both of the K88R and K88A mutants of IFITM3 have a mild or at least a slightly negative effect on its antiviral function. These observations would agree with a previous study in which the K88A mutant of IFITM3 caused a mild reduction in its antiviral activity toward dengue virus infection (15). However, in the same study, the activity of the IFITM3-K88A mutant had a slight improvement on anti-IAV restriction. We initially hypothesized that the K88R or K88A mutations would improve the function of IFITM3 as they would render IFITM3 insensitive to monomethylation. However, as Yount et al. (12) showed that Lys-88 of IFITM3 could be ubiquitinated, the mutation of Lys-88 may affect both ubiquitination and monomethylation, so it may not be appropriate to directly relate these mutations to the disruption of monomethylation. The Lys-88 residue of IFITM3 is found in the conserved intracellular loop region of IFITM3 (12, 15). The Lys-88 residue is close to Arg-87 and Arg-85, which were found important for IAV restriction (15). The direct functional consequences of mutating these arginine residues remain unclear, but we speculate that the monomethylation of IFITM3 at Lys-88 may confer similar mechanisms leading to the loss of function of IFITM3 to disrupt its antiviral activity.
Because monomethylation of IFITM3 does not seem to affect its localization or expression, we hypothesize that the monomethylation of IFITM3 may affect its direct binding to other cofactors. To this end, IFITM3 has recently been shown to interact with the cholesterol homeostatic controller VAPA (VAMP (vesicle-associated membrane protein)-associated protein A) (17) or other IFITM paralogs (15). Our preliminary data has indicated that the interaction between IFITM3 and VAPA does not change upon monomethylation of IFITM3 (data not shown). However, the changes in interaction between IFITM3 and other potentially unknown molecules in the presence or absence of SET7-mediated monomethylation are currently being explored. Also, the individual role of monomethylation on IFITM1 and IFITM2 require examining.
In summary, the uncovering of signaling pathways that increase the monomethylation levels of IFITM3 could serve as potential targets for treatment against viral infection. This study may also have implications toward others in which protein methylation pathways are targeted for anti-cancer therapies (32), such that the targeting of methyltransferases or demethylases for the treatment of cancer may affect host susceptibility toward viral infection due to the inadvertent perturbation of antiviral restriction factors.
This work was supported by National Natural Science Foundation of China Grants 30972702, 31170825, 81270083, 31150110337, 31200646, 31200647, and 81271835; National Institutes of Health NSFC Collaborative Grant 81161120417; Shanghai “Rising Star” Program Grant 10QA1407900; the Novo Nordisk-Chinese Academy of Sciences Foundation; and National Science and Technology Major Project Grants 2012ZX10002007-003 and 2013ZX10003009-002. This work was also supported by the Sanofi-Aventis-Shanghai Institutes for Biological Sciences scholarship program and the Knowledge Innovation Program of Shanghai Institutes for Biological Sciences and Chinese Academy of Sciences Grants 2010KIP205 and 2012KIP204.
Z. Shan, Q. Han, J. Nie, X. Cao, Z. Chen, S. Yin, Y. Gao, F. Lin, X. Zhou, K. Xu, H. Fan, Z. Qian, B. Sun, J. Zhong, B. Li, and A. Tsun, unpublished data.
- IFITM3
- interferon-induced transmembrane protein 3
- ISG
- interferon-stimulated gene
- TAP
- tandem affinity purification
- qPCR
- quantitative PCR
- PTM
- posttranslational modification
- IAV
- influenza A virus
- F
- forward
- R
- reverse
- MOI
- multiplicity of infection
- MEF
- mouse embryonic fibroblast.
REFERENCES
- 1. Yan N., Chen Z. J. (2012) Intrinsic antiviral immunity. Nat. Immunol. 13, 214–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lewin A. R., Reid L. E., McMahon M., Stark G. R., Kerr I. M. (1991) Molecular analysis of a human interferon-inducible gene family. Eur. J. Biochem. 199, 417–423 [DOI] [PubMed] [Google Scholar]
- 3. Diamond M. S., Farzan M. (2013) The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 13, 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Everitt A. R., Clare S., Pertel T., John S. P., Wash R. S., Smith S. E., Chin C. R., Feeley E. M., Sims J. S., Adams D. J., Wise H. M., Kane L., Goulding D., Digard P., Anttila V., Baillie J. K., Walsh T. S., Hume D. A., Palotie A., Xue Y., Colonna V., Tyler-Smith C., Dunning J., Gordon S. B., GenISIS Investigators, MOSAIC Investigators, Smyth R. L., Openshaw P. J., Dougan G., Brass A. L., Kellam P. (2012) IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484, 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bailey C. C., Huang I. C., Kam C., Farzan M. (2012) Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog 8, e1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brass A. L., Huang I. C., Benita Y., John S. P., Krishnan M. N., Feeley E. M., Ryan B. J., Weyer J. L., van der Weyden L., Fikrig E., Adams D. J., Xavier R. J., Farzan M., Elledge S. J. (2009) The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiang D., Weidner J. M., Qing M., Pan X. B., Guo H., Xu C., Zhang X., Birk A., Chang J., Shi P. Y., Block T. M., Guo J. T. (2010) Identification of five interferon-induced cellular proteins that inhibit west nile virus and dengue virus infections. J. Virol. 84, 8332–8341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weidner J. M., Jiang D., Pan X. B., Chang J., Block T. M., Guo J. T. (2010) Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J. Virol. 84, 12646–12657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li K., Markosyan R. M., Zheng Y. M., Golfetto O., Bungart B., Li M., Ding S., He Y., Liang C., Lee J. C., Gratton E., Cohen F. S., Liu S. L. (2013) IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 9, e1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feeley E. M., Sims J. S., John S. P., Chin C. R., Pertel T., Chen L. M., Gaiha G. D., Ryan B. J., Donis R. O., Elledge S. J., Brass A. L. (2011) IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 7, e1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang I. C., Bailey C. C., Weyer J. L., Radoshitzky S. R., Becker M. M., Chiang J. J., Brass A. L., Ahmed A. A., Chi X., Dong L., Longobardi L. E., Boltz D., Kuhn J. H., Elledge S. J., Bavari S., Denison M. R., Choe H., Farzan M. (2011) Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 7, e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yount J. S., Karssemeijer R. A., Hang H. C. (2012) S-palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus. J. Biol. Chem. 287, 19631–19641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yount J. S., Moltedo B., Yang Y. Y., Charron G., Moran T. M., López C. B., Hang H. C. (2010) Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat. Chem. Biol. 6, 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jia R., Pan Q., Ding S., Rong L., Liu S. L., Geng Y., Qiao W., Liang C. (2012) The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J. Virol. 86, 13697–13707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. John S. P., Chin C. R., Perreira J. M., Feeley E. M., Aker A. M., Savidis G., Smith S. E., Elia A. E., Everitt A. R., Vora M., Pertel T., Elledge S. J., Kellam P., Brass A. L. (2013) The CD225 domain of IFITM3 is required for both IFITM protein association and inhibition of influenza A virus and dengue virus replication. J. Virol. 87, 7837–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wee Y. S., Roundy K. M., Weis J. J., Weis J. H. (2012) Interferon-inducible transmembrane proteins of the innate immune response act as membrane organizers by influencing clathrin and v-ATPase localization and function. Innate Immun. 18, 834–845 [DOI] [PubMed] [Google Scholar]
- 17. Amini-Bavil-Olyaee S., Choi Y. J., Lee J. H., Shi M., Huang I. C., Farzan M., Jung J. U. (2013) The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe 13, 452–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Black J. C., Van Rechem C., Whetstine J. R. (2012) Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell 48, 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang X., Wen H., Shi X. (2012) Lysine methylation: beyond histones. Acta Biochim. Biophys. Sin 44, 14–27 [DOI] [PubMed] [Google Scholar]
- 20. Huang J., Berger S. L. (2008) The emerging field of dynamic lysine methylation of non-histone proteins. Curr. Opin. Genet. Dev. 18, 152–158 [DOI] [PubMed] [Google Scholar]
- 21. Pagans S., Kauder S. E., Kaehlcke K., Sakane N., Schroeder S., Dormeyer W., Trievel R. C., Verdin E., Schnolzer M., Ott M. (2010) The Cellular lysine methyltransferase Set7/9-KMT7 binds HIV-1 TAR RNA, monomethylates the viral transactivator Tat, and enhances HIV transcription. Cell Host Microbe 7, 234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sakane N., Kwon H. S., Pagans S., Kaehlcke K., Mizusawa Y., Kamada M., Lassen K. G., Chan J., Greene W. C., Schnoelzer M., Ott M. (2011) Activation of HIV transcription by the viral Tat protein requires a demethylation step mediated by lysine-specific demethylase 1 (LSD1/KDM1). PLoS Pathog. 7, e1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nishioka K., Chuikov S., Sarma K., Erdjument-Bromage H., Allis C. D., Tempst P., Reinberg D. (2002) Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 16, 479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kawakami E., Watanabe T., Fujii K., Goto H., Watanabe S., Noda T., Kawaoka Y. (2011) Strand-specific real-time RT-PCR for distinguishing influenza vRNA, cRNA, and mRNA. J. Virol. Methods 173, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Boritz E., Gerlach J., Johnson J. E., Rose J. K. (1999) Replication-competent rhabdoviruses with human immunodeficiency virus type 1 coats and green fluorescent protein: entry by a pH-independent pathway. J. Virol. 73, 6937–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoffmann E., Neumann G., Kawaoka Y., Hobom G., Webster R. G. (2000) A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U.S.A. 97, 6108–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matrosovich M., Matrosovich T., Garten W., Klenk H. D. (2006) New low-viscosity overlay medium for viral plaque assays. Virol J. 3, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu C., Hu W. B., Xu K., He Y. X., Wang T. Y., Chen Z., Li T. X., Liu J. H., Buchy P., Sun B. (2012) Amino acids 473V and 598P of PB1 from an avian-origin influenza A virus contribute to polymerase activity, especially in mammalian cells. J. Gen. Virol. 93, 531–540 [DOI] [PubMed] [Google Scholar]
- 29. Kitao H., Takata M. (2006) Purification of TAP-tagged proteins by two-step pull down from DT40 cells. Subcell Biochem. 40, 409–413 [DOI] [PubMed] [Google Scholar]
- 30. Li Y., Reddy M. A., Miao F., Shanmugam N., Yee J. K., Hawkins D., Ren B., Natarajan R. (2008) Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-κB-dependent inflammatory genes. Relevance to diabetes and inflammation. J. Biol. Chem. 283, 26771–26781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Estève P. O., Chin H. G., Benner J., Feehery G. R., Samaranayake M., Horwitz G. A., Jacobsen S. E., Pradhan S. (2009) Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 106, 5076–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. He Y., Korboukh I., Jin J., Huang J. (2012) Targeting protein lysine methylation and demethylation in cancers. Acta Biochim. Biophys. Sin. 44, 70–79 [DOI] [PMC free article] [PubMed] [Google Scholar]