Background: Cytosolic glutaredoxin 2 (Grx2) is essential for neuronal development in zebrafish; collapsin response mediator protein 2 (CRMP2) was identified as Grx2 substrate.
Results: Oxidation of CRMP2 by hydrogen peroxide induces an intermolecular disulfide, changes in α-helical content, and hydrophobicity.
Conclusion: The dithiol-disulfide redox switch defines two conformations of CRMP2.
Significance: This switch may be functional during axonal outgrowth.
Keywords: Disulfide, Neurodifferentiation, Redox Regulation, Redox Signaling, Thiol, CRMP2, Glutaredoxin, Neuronal Differentiation, Redox Switch
Abstract
Vertebrate-specific glutaredoxin 2 (Grx2) is expressed in at least two isoforms, mitochondrial Grx2a and cytosolic Grx2c. We have previously shown that cytosolic Grx2 is essential for embryonic development of the brain. In particular, we identified collapsin response mediator protein 2 (CRMP2/DPYSL2), a mediator of the semaphorin-plexin signaling pathway, as redox-regulated target of Grx2c and demonstrated that this regulation is required for normal axonal outgrowth. In this study, we demonstrate the molecular mechanism of this regulation, a specific and reversible intermolecular Cys-504-Cys-504 dithiol-disulfide switch in homotetrameric CRMP2. This switch determines two conformations of the quaternary CRMP2 complex that controls axonal outgrowth and thus neuronal development.
Introduction
Glutaredoxins are glutathione-dependent oxidoreductases that catalyze the reduction of protein disulfides first, for instance in ribonucleotide reductase (1) and, second, the reversible formation and reduction of protein-glutathione mixed disulfides (2). The first reaction requires both cysteinyl residues in the CXXC active site, the latter only the more N-terminal (see Refs. 3–5 for detailed discussions). Vertebrate-specific glutaredoxin 2 (Grx2)2 was originally identified as mitochondrial protein (6, 7); however, alternative splicing and transcription initiation give rise to additional isoforms of the protein as demonstrated in human and mouse (8–10). The human GLRX2 gene gives rise to three transcript variants encoding the mitochondrial Grx2a and the cytosolic Grx2b and Grx2c. The mouse glrx2 gene gives rise to five transcript variants encoding three protein isoforms. Grx2a and Grx2c are conserved, and Grx2d is unique to mice (10).
The zebrafish genome contains a single gene (glrx2) that encodes the equivalent to human and mouse Grx2c. Recently, we demonstrated that embryonic brain development depends on the enzymatic activity of this cytosolic Grx2 (11). Zebrafish with morpholino-silenced expression of the cytosolic Grx2 lost virtually all types of neurons by apoptotic cell death and the ability to develop an axonal scaffold. As early as 18 h post fertilization, the axon scaffold was severely reduced. At 24 h post fertilization, essentially all embryos depleted of cytosolic Grx2 lost HuC-positive (an early neuronal marker) cells in the central nervous system. These severe defects could be rescued by co-injection of morpholino-resistant glrx2 mRNA but not by co-injection of mRNA encoding redox-inactive mutants. Notably, even a mutant that was still capable of catalyzing the Grx-specific monothiol reaction mechanism, which is sufficient for the catalysis of protein de/glutathionylation, did not rescue the phenotype to any degree. In a cellular model of neuronal differentiation, SH-SY5Y neuroblastoma cells overexpressing Grx2c during differentiation displayed 1.5- to 2-fold longer axons with almost twice as many branching points. Thus, Grx2c is essential for neuronal differentiation through the control of axonal outgrowth in a process that requires the dithiol reaction mechanism, implying the reduction of at least one specific protein disulfide (11).
To uncover the mechanism through which Grx2c acts in axon guidance, we deployed a proteomic approach: enzymatic intermediate trapping. This strategy allowed us to identify proteins that can undergo dithiol-disulfide exchange reactions catalyzed by Grx2 in a dithiol reaction mechanism (12). Among the potential targets, the association of Grx2c with the collapsin response mediator protein 2 (CRMP2/DPYSL2/ULIP2), could be confirmed by co-immunoprecipitation. CRMP2 is a part of the semaphorin-3A-plexin signaling cascade whose critical functions in axon guidance are well documented (13, 14). The protein was reported to be regulated post-translationally by phosphorylation at multiple sites and calpain-catalyzed proteolysis (see, for instance, Refs. 15–17). Phosphorylation was suggested to be facilitated by interaction of CRMP2 with thioredoxin 1 (Trx1), in a redox-dependent mechanism (18).
Using differential labeling of oxidized and reduced thiol groups with N-ethylmaleimide (NEM) and 5-maleimido isophthalic acid, the latter compound shifts the isoelectric point of proteins, followed by two-dimensional gel electrophoresis and specific Western blotting, revealed the reduction of cysteinyl residues in CRMP2 in SH-SY5Y and HeLa cells overexpressing Grx2c (11, 12). Corroboratively, CRMP2 in zebrafish embryos depleted of Grx2 was significantly more oxidized (11). Altogether, these results strongly suggest the regulation of CRMP2 through a specific protein dithiol-disulfide switch. Here, we aimed at the identification of this dithiol-disulfide switch, how it controls the function of CRMP2, and whether it could play a role during neuronal differentiation.
EXPERIMENTAL PROCEDURES
Materials
Chemicals and enzymes were purchased from Sigma-Aldrich unless otherwise stated and were of analytical grade or better. The antibodies used in this work were as follows: CRMP2 (Sigma-Aldrich, C2993), CRMP2 P-T514 (Cell Signaling Technologies, catalog no. 9397), neurofilament m (Cell Signaling Technologies, catalog no. 2838), glial fibrillary acidic protein (Thermo Scientific, Waltham, MA; PA3-16727), and GAPDH (Sigma-Aldrich, G9545).
Electrophoresis and Western Blotting
Protein concentrations were determined using Bradford reagent from Bio-Rad. SDS-PAGE and Western blots were run using precasted Precise gels (4–20% (w/v), Thermo Scientific) or mini-Protean TGX Stain-free gels (4–20% (w/v), Bio-Rad), and PVDF membranes (Macherey & Nagel, Düren, Germany) according to the manufacturers' instructions. Horseradish peroxidase-conjugated anti-rabbit and anti-mouse IgGs were obtained from Bio-Rad. Blue native electrophoresis was performed using precasted NativeBlueNovex gels and buffers as suggested by the manufacturer (Life Technologies, Paisley, UK). Western blots were developed by enhanced chemiluminescence staining and documented using a Chemidoc XRS+ documentation system (Bio-Rad).
Cloning of Expression Constructs
CRMP2 was amplified by PCR from human cDNA (Life Technologies) using the oligonucleotides 5′-CACACACATGCTAGCATGTCTTATCAGGGGAAGAAAAATATTCC and 5′-GTGTGTTCTAGACTGCCCAGGCTGGTGATG or 5′-CACACAAGATCTGCCCAGGCTGGTGATGTTG, respectively, ligated into the vector pGEM-T (Promega, Madison, BI), excised with the restriction endonucleases NdeI and BglII, and ligated into the NdeI and BamHI sites of the vectors pET15b and pET24a (Millipore, Darmstadt, Germany), respectively. Correct sequences were confirmed by sequencing at Seqlab (Göttingen, Germany). The plasmids for the recombinant expression and purification of human Grx2c, Grx2c C40S, and thioredoxin 1 have been described previously (19–21).
Quantitative PCR
The cells were treated with trypsin, harvested, and washed once with PBS. RNA isolation was performed according to the manual provided for the NucleoSpin® RNA II kit (Macherey-Nagel, Düren, Germany). RNA concentration was measured photometrically using the NanoDrop 2000c spectrophotometer (Thermo Scientific). First strand cDNA was prepared using the RevertAid First Strand cDNA synthesis kit according to the protocol provided using 1 μg of RNA as template and oligo dT18 primer (Thermo Scientific). 1 μl of the cDNA was used as template for quantitative PCR. The SensiMix SYBR HI-ROX contained SYBR Green I dye, dNTPs, stabilizers, and a hot start DNA polymerase was provided as 2× Master Mix (Bioline, London, UK). The primer concentration used for the quantitative PCR was 0.5 μm each (5′-GGAGAGAGATGTCTTATCAGG and 3′-CTGCATAGAACGACTGGTCATC), and the volume was adjusted to 20 μl with double distilled H2O. All quantitative PCRs were performed using the CFX96 Real Time System from Bio-Rad. For optimization, a gradient quantitative PCR from 55 to 65 °C was performed, and samples were analyzed by agarose gel electrophoresis. At 60 °C annealing temperature, the reaction resulted in a specific product at the expected size. GAPDH was used as a reference (primer pair: 5′-CAAGGTCATCCATGACAACTTTG and 3′-GTCCACCACCCTGTTGCTGTAG) in the ΔΔCq mode.
Recombinant Expression and Purification of CRMP2
For recombinant protein expression, E. coli cells were propagated in a 5 liters of Bioreactor (R'ALF; Bioengineering, Wald, Switzerland). The LB medium (5 liters), containing the selective antibiotics, was inoculated at 37 °C with 1% (v/v) of an overnight culture of E. coli BL21 (DE3) codon-plus (Life technologies) harboring the pET15b-CRMP2 or pET24a-CRMP2. At OD 0.6, expression was induced by addition of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside and by lowering the temperature to 14 °C. The cells were harvested after 16–20 h by centrifugation. Bacteria were lysed by incubation with 1 mg/ml lysozyme and 0.05 mg/ml Dnase I for 20 min at room temperature and subsequent ultrasonic treatment for 3 min, 80% intensity, 0.5s cycle time (50% cycling) using a UP200S ultrasonic processor (Hielscher; Teltow, Germany). Recombinant His-tagged CRMP2 proteins were purified by the immobilized metal affinity chromatography technique using an Äkta FPLC System as suggested by the manufacturer (GE Healthcare) at 4 °C.
Gel Filtration Chromatography
Gel filtration chromatography was performed using a HiLoad 16/60 Superdex 200 prep grade column and an Äkta System (both GE healthcare) equilibrated with PBS. 85 μm purified CRMP2 were reduced with 5 mm DTT, 5 mm pH-neutralized Tris(2-chlorethyl)phosphate (TCEP), and 0.8 μm human Grx2c and incubated for 30 min at room temperature. To remove all reductants, the protein solution was re-buffered using either NAP-5 or PD10 Sephadex G25 prepacked columns (GE Healthcare). After desalting, 16 nmol of the reduced protein in 200–500 μl were loaded on the column and separated at a flow rate of 1 ml/minute. The remaining protein was oxidized by 200 μm H2O2 for 30 min at room temperature. H2O2 was removed by rebuffering as described previously, and 16 nmol of oxidized protein were loaded on the column and separated. The remaining protein was rereduced as before and also separated by gel filtration. The Superdex 200 column was equilibrated with PBS between the runs. Fractions were analyzed by Western blotting. A calibration curve for apparent molecular masses was produced using the manufacturer's calibration standards.
Mass Spectrometry
Recombinant CRMP2 (85 μm) was reduced with 5 mm DTT and 5 mm pH-neutralized TCEP or oxidized with 200 μm H2O2 for 30 min at room temperature each. Subsequently, the proteins were denatured, and all thiols were blocked by specific alkylation with 10 mm NEM. A standard SDS-PAGE was performed with the samples considered for mass spectrometry, and the gel was stained with Coomassie staining solution (Thermo Scientific). Excised gel fragments were destained in 100 mm NH4HCO3 in 50% (v/v) acetonitrile. Following drying, proteins were digested in 20 μl trypsin (10 ng/μl in 10 mm NH4HCO3 solution) at 37 °C overnight. Peptides were collected in 50% (v/v) acetonitrile containing 0.1% (v/v) acetic acid. Next, samples were mixed with 0.7 μl of α-cyano-4-hydroxycinnamic acid matrix in 50% acetonitrile/0.5% TFA on the target and measured by MS using a 4800 MALDI TOF/TOF analyzer (Thermo Scientific). Spectra were aligned manually with data of an in silico analysis of the theoretical amino acid sequence, with and without modifications, to confirm the presence of the peptides of interest. Peptide mass fingerprint peak lists were compared with the SwissProt database rel. 57, restricted to human taxonomy, using the Mascot search engine (version 1.9, Matrix Science Ltd, London, UK).
CD Spectroscopy
Circular dichroism spectra were recorded in a Jasco instrument. Freshly purified CRMP2 (80 μm) was reduced with 5 mm DTT, 5 mm pH-neutralized TCEP, and 0.8 μm human Grx2c for 30 min at room temperature. To remove all reductants, the protein solution was rebuffered using NAP-5 columns (GE Healthcare). Next, the protein was oxidized by 200 μm H2O2 for 30 min at room temperature. H2O2 was removed by rebuffering in 10 mm Hepes, pH 7.3, containing 20 mm MgCl2 as described previously. The oxidized sample (6 μm) was analyzed in a 1-mm cuvette, scanning 0.2-nm steps, averaging 10 iterations. Subsequently, the protein was reduced by addition of 1 mm DTT, 1 mm TCEP, and 0.06 μm Grx2c, incubated for 30 min, and analyzed again. All spectra were corrected by subtraction of the associated baselines.
Differential Scanning Fluorimetry (Thermofluor Assay)
The thermal stability of reduced and oxidized CRMP2 was assayed using the thermofluor assay as outlined in Ref. 22. In brief, 10 μm protein, reduced and oxidized as outlined above and rebuffered in PBS containing 20 mm MgCl2, were mixed with SYPRO Orange (1:100 diluted, Sigma-Aldrich) and heated in a CFX96 Real Time System from Bio-Rad in 0.5 K increments from 20 to 95 °C in 1 h. The increase in fluorescence due to binding of the dye to hydrophobic regions exposed during denaturation was recorded using the FRET settings of the instrument. The curves presented represent the average of five experiments.
Cell Culturing
All media, FCS, antibiotics (penicillin, streptomycin), trypsin, and PBS were purchased from PAA (Cölbe, Germany), disposable plastics from Sarstedt (Nümbrecht, Germany). SH-SY5Y cells were propagated in MEM, low glucose, supplemented with penicillin (100 units/ml)/streptomycin (0.1 mg/ml), 10% (v/v) FCS, and 2 mm l-glutamine. For differentiation into a neuron-like phenotype, cells were treated with 10 μm retinoic acid (solved in dimethyl sulfoxide) up to 7 days (23). SH-SY5Y cells treated with dimethyl sulfoxide served as control. At 6, 24, 48, 72, 120, and 168 h, cells were detached by trypsin treatment, washed with PBS, and lysed by adding 40 mm Hepes, 50 mm NaCl, 1 mm EDTA, 1 mm EGTA, 2% (w/v) CHAPS, 1-fold protease inhibitor mixture (Roche Applied Science), and 1-fold phosphatase inhibitor mixture 3 (Sigma) at pH 7.4 and frozen in liquid nitrogen. Extracts were clarified by centrifugation at 13,000 rpm (21,000 × g) for 10 min at 4 °C in a microcentrifuge.
Immunocytochemistry and Confocal Microscopy
Cells were seeded on fibronectin-covered glass slides, cultivated as indicated, washed with PBS, fixated with 4% (w/v) paraformaldehyde at 4 °C for 30 min, washed with PBS, permeabilized for 30 min at room temperature in 0.4% (v/v) Triton X-100 in PBS, and blocked with 3% (w/v) BSA in PBS for another 30 min. The primary antibody (CRMP2, 1:1000) was incubated overnight at 4 °C, the secondary antibody (anti-rabbit Alexa Fluor 488; Life Technologies, 1:400) for 1 h at room temperature. Nuclei were counterstained with 1 μg/ml DAPI. Slides were mounted with Mowiol (Calbiochem, Darmstadt, Germany) and analyzed by confocal microscopy using a Leica TCS SP5 instrument with a 63-fold/1.4 oil lens.
RESULTS
Two isoforms of human CRMP2 have been described, the canonical isoform CRMP2B and CRMP2A with an alternative N-terminal section. Here, we always refer to CRMP2B when addressing CRMP2. Human CRMP2 consists of 572 amino acid residues, it contains eight cysteinyl residues, from which six are conserved from zebrafish to man (see supplemental Fig. 1). The cDNA of human CRMP2 was cloned by PCR from a commercial cDNA library and ligated into the expression vectors pET15b and pET24a for expression as N- and C-terminal His6 fusion protein, respectively. Expression was optimized in a 5-liter bioreactor; propagation was performed at 14 °C for ∼18 h following the induction of expression at an OD of 0.6 typically yielded 5 (pET24a) to 15 mg (pET15b) soluble protein per liter of culture without the appearance of insoluble portions. We could not observe any significant differences between the behavior of the N- and C-terminally tagged proteins in initial gel filtration chromatography and blue native electrophoresis (see below). We have thus chosen the N-terminally tagged protein for in-depth analysis because of the higher yield in expression.
Quaternary State of Reduced and Oxidized CRMP2
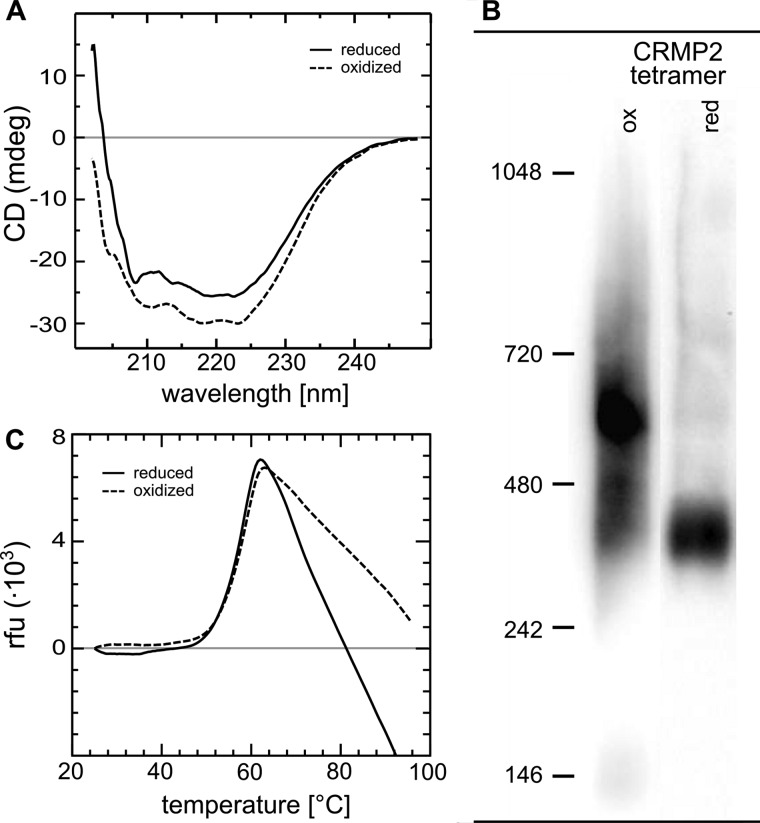
CRMP2 was reported to form homotetramers (24). We first hypothesized that redox regulation, i.e. oxidation and reduction of CRMP2, could alter this quaternary structure. Freshly purified CRMP2 was reduced by DTT, TCEP, and a catalytic amount of purified human Grx2c (1/100th molar ratio). CRMP2 was efficiently reduced by DTT and TCEP alone; however, the addition of Grx2c significantly increased the velocity of this reaction when followed by CD spectroscopy (see below). Reducing agents were quickly removed by size exclusion chromatography (Sephadex G25), and the quaternary structures of the reduced and oxidized proteins were analyzed by gel filtration chromatography (Superdex 200, Fig. 1A). In parallel, the reduced and rebuffered protein was oxidized by H2O2 (5-fold molecular excess) and subsequently analyzed by gel filtration chromatography (Fig. 1A). To confirm the reversibility, oxidized CRMP2 was also rebuffered, rereduced as done previously and analyzed by gel filtration chromatography (Fig. 1A). As depicted in Fig. 1 and Table 1, reduced, oxidized, and re-reduced CRMP2 fractions with the proteins in their native, folded states were primarily detected in the homotetrameric form, despite a minor fraction of dodecameric protein and the transient appearance of a minor fraction of monomeric CRMP2 in the oxidized form. In conclusion, oxidation and reduction did not change the tetrameric structure of CRMP2 when present in the native state. When analyzed by non-reducing SDS-PAGE (Fig. 1B), denatured oxidized CRMP2 migrated in two bands corresponding to dimers, i.e. two disulfide-linked monomers and monomeric proteins at a ratio of 1:2. Denatured reduced CRMP2 migrated in a single band at the expected size of monomeric CRMP2. In conclusion, oxidation of CRMP2 did not change the tetrameric quaternary structure of the folded protein, but it led to the formation of up to one intermolecular disulfide in the tetrameric complex.
FIGURE 1.
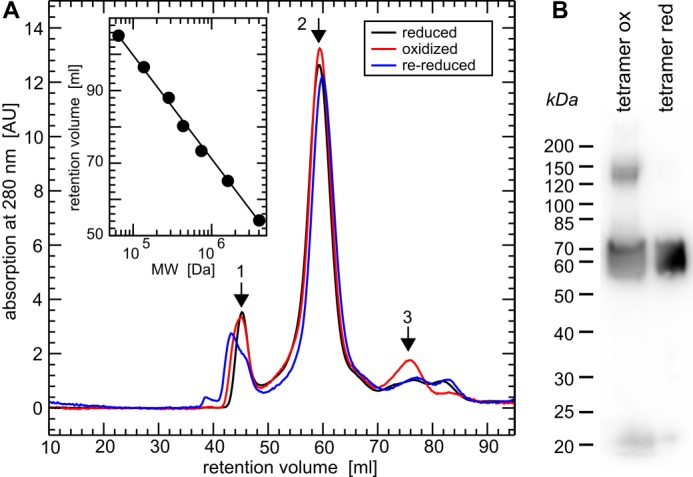
Quaternary structure of reduced, oxidized, and re-reduced CRMP2. A, gel filtration chromatography (HiLoad 16/60 Superdex 200 column, GE Healthcare) of (Grx2c+DTT+TCEP) reduced, H2O2-oxidized, and re-reduced recombinant CRMP2 in their native, folded states. Calibration (inset) was done using the manufacturer's instructions: 1, ∼800 kDa, i.e. dodecamer (theoretical mass, 770.4 kDa); 2, 256 kDa, i.e. (homo)tetramer (theoretical mass, 256.8 kDa); 3, 65 kDa, i.e. monomeric CRMP2 (theoretical mass, 64.2 kDa). B, non-reducing SDS-PAGE and Western blot of the tetrameric fractions of oxidized and reduced CRMP2 following the gel-filtration chromatography depicted in A and denaturation of the protein complexes.
TABLE 1.
Apparent molecular weight and quaternary structure of reduced, oxidized, and re-reduced CRMP2 determined by gel filtration chromatography (see Fig. 1) (n = 3)
| Fraction | MW | ± S.D. | Subunits | MW/subunit |
|---|---|---|---|---|
| kDa | kDa | |||
| 1 | 812.9 | 4.8 | 12 | 67.7 |
| 2 | 256.3 | 3.3 | 4 | 64.1 |
| 3 | 66.0 | 3.5 | 1 | 66.0 |
| 4 | 39.9 | 2.1 | ||
| CRMP2 | 64.2 |
Grx2c Reduces CRMP2 in a Dithiol Reaction Mechanism
To confirm that the reduction of the CRMP2 disulfide by Grx2c is catalyzed in a dithiol reaction mechanism, i.e. that it requires both thiols in the CXXC active site of Grx2c, oxidized CRMP2 was incubated with reduced wild-type and the CXXS active site mutant (C40S) Grx2c and analyzed by SDS-PAGE. As predicted for the dithiol reaction mechanism, CRMP2 did not form a complex with wild-type Grx2c, but with the mutant lacking the resolving cysteinyl residue (Fig. 2, see arrowhead). Thus, Grx2c reduces CRMP2 by attack of the N-terminal cysteinyl active site thiol on the CRMP2 disulfide; this intermediate mixed disulfide is immediately reduced by the second active site thiol. Noteworthy, also wild-type Trx1 was able to reduce the CRMP2 disulfide in vitro and did not form a stable complex with CRMP2.
FIGURE 2.
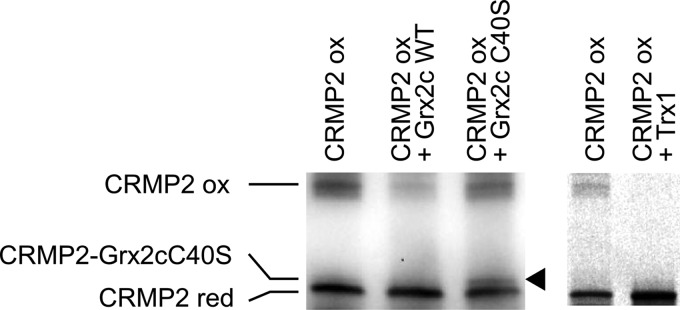
Grx2c reduces CRMP2 in a dithiol reaction mechanism. Incubation of reduced (red) Grx2c with oxidized (ox) CRMP2 did not lead to a complex between the proteins when analyzed by non-reducing SDS-PAGE. Only incubation with a Grx2c mutant lacking the second (resolving) cysteinyl residue in the CXXC (C40S) active site led to the formation of a mixed disulfide (see arrowhead). In addition, wild-type Trx1 was able to reduce the protein in vitro and did not form a stable complex with CRMP2.
Oxidation and Reduction of CRMP2 Induce a Significant Conformational Change in the Protein
First information for a switch between two distinct conformations of the CRMP2 tetramer in consequence of oxidation and reduction came from the analysis of zebrafish CRMP2 by CD spectroscopy (11). For human CRMP2, we recorded a reversible decrease in ellipticity upon reduction of CRMP2 at 210–220 nm (Fig. 3A), indicative for a reversible decrease in α-helical content of the reduced protein. To confirm and further characterize this switch in conformations, we analyzed the changes induced by reduction and oxidation of tetrameric CRMP2 (in parallel to gel filtration chromatography, see above) in blue native gel electrophoresis (Fig. 3B). The mobility of proteins in blue native electrophoresis is determined by the binding of the Coomassie dye to hydrophobic regions of the protein. As shown in Fig. 3, reduced CRMP2 migrates significantly faster in blue native electrophoresis, indicating conformational changes in the reduced tetramer that lead to the exposition of hydrophobic regions. We also tested the influence of the redox switch on the thermal stability of the protein applying differential scanning fluorimetry (Fig. 3C) but did not record significant differences between the two species (Tm, 56 versus 57 °C).
FIGURE 3.
Conformational changes of CRMP2 upon reduction. A, CD spectroscopy of oxidized and subsequently reduced CRMP2. The decrease in ellipticity between 210–220 nm indicates a decrease in α-helical content of reduced CRMP2. B, blue native gel electrophoresis of reduced and oxidized tetrameric CRMP2. 1 μg of the protein was reduced by a catalytic amount of Grx2c (1:100, +DTT+TCEP) or oxidized by H2O2 treatment. Both fractions were analyzed by blue native gel electrophoresis. The higher migration velocity of reduced CRMP2 indicated binding of more Coomassie dye molecules and thus the exposure of more hydrophobic surfaces compared with the oxidized CRMP2 tetramer. The marks depicted were introduced only for orientation purposes and do not reflect the actual molecular weight of the complexes (see Fig. 1). C, differential scanning fluorimetry of reduced and oxidized CRMP2. No significant differences were observed in the thermal stability of reduced and oxidized CRMP2.
Identification of Thiol Modifications in CRMP2 upon Oxidation
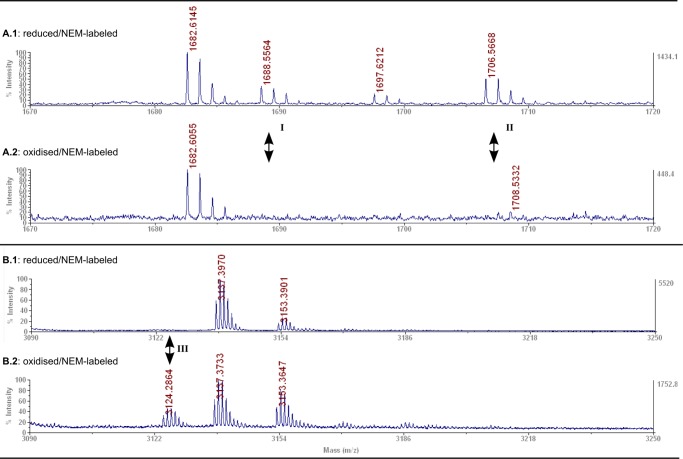
To identify the cysteinyl residues forming the dithiol-disulfide switch, we labeled the thiol groups of oxidized and reduced CRMP2 with NEM to prevent further redox modifications. Tryptic fragments were analyzed by MALDI-TOF mass spectrometry. Among the peptides containing some of the eight cysteinyl residues, we identified only one difference between the reduced and oxidized protein (see also supplemental Tables 1 and 2). Reduced and NEM-labeled CRMP2 contained two peptides corresponding to the masses of a fragment in which Cys-504 was labeled with NEM or NEM/H2O (Fig. 4, A.1, I and II), which are absent from oxidized CRMP2 (Fig. 4, A.2). Oxidized CRMP2 contains one peptide (Fig. 4, B.2, III) corresponding to the mass of two disulfide-linked Cys-504-containing peptides that was not present in the reduced protein (Fig. 4, B.1). Interestingly, we were not able to induce glutathionylation of the protein by treatment with excess GSSG (data not shown). Thus, oxidation specifically induced intermolecular disulfides between the Cys-504 residues of two CRMP2 molecules in the homotetrameric complex.
FIGURE 4.
Identification of a specific dithiol-disulfide switch in CRMP2 by MALDI-TOF mass spectrometry. Mass spectra details of reduced (A.1/B.1) or oxidized (A.2/B.2) NEM-labeled CRMP2. A.1/A.2 peptide I, GLYDGPVCEVSVTPK+NEM; peptide II, GLYDGPVCEVSVTPK + NEM/H2O at Cys-504 (A.1, I and II, theoretical masses, 1688.89/1706.81; observed, 1688.56/1706.57); B.1/B.2 peptide III, 2× GLYDGPVCEVSVTPK in form of disulfide-linked peptides (B.2, III, theoretical mass, 3124.52; observed, 3124.28) not present in the reduced protein (B.1).
The Conformational Change Can Be Observed in a Model of Neuronal Differentiation
To confirm the relevance of the redox-induced conformational switch in vivo, we have analyzed CRMP2 in a cellular model of neuronal differentiation. The neuroblastoma cell line SH-SY5Y can be transformed into a neuron-like cell culture by treatment with retinoic acid. During this process, the cells display axonal outgrowth, phase out cell division, and express several neuron-specific markers (23). Predominantly, these cells develop a mature dopaminergic-like neurotransmitter phenotype (25). Relative to controls, we did not observe changes in the levels of CRMP2 when analyzed by SDS-PAGE and Western blotting (Fig. 5, B and C). The phosphorylation of the residue Thr-514 of CRMP2 by cyclin-dependent kinase 5 has been described as a prerequisite for subsequent phosphorylation steps by glycogen synthase kinase 3 that were implied in regulating the function of CRMP2 (26). Using an antibody specific for this modification, we observed a tendency for a decrease in Thr-514 phosphorylation (∼50%) at t = 120 and 168 h (Fig. 5B); however, the analysis of three independent experiments did not indicate statistical significance. The analysis of CRMP2 by blue native electrophoresis demonstrated an increase of the oxidized conformation of CRMP2 between days three and five (Fig. 5A). This is the time frame when the developing protrusions of the cells found their way to other cells and began to establish connections. The success of the differentiation protocol was controlled by analysis of the typical morphological changes, i.e. neurite outgrowth and a decrease in the volume of the cell somata (Fig. 5, D and E) (see Refs. 11 and 23), the presence of neurofilament m, and the loss of the glia marker glial fibrillary acidic protein (Fig. 5, B and C). Unlike the levels of CRMP2 protein, the levels of the CRMP2 mRNA, analyzed by quantitative PCR, increased continuously during the 7 days (Fig. 5F). In case this increased amount of mRNA would lead to an increase in translation, this could indicate a higher turnover of the protein during the process of differentiation.
FIGURE 5.
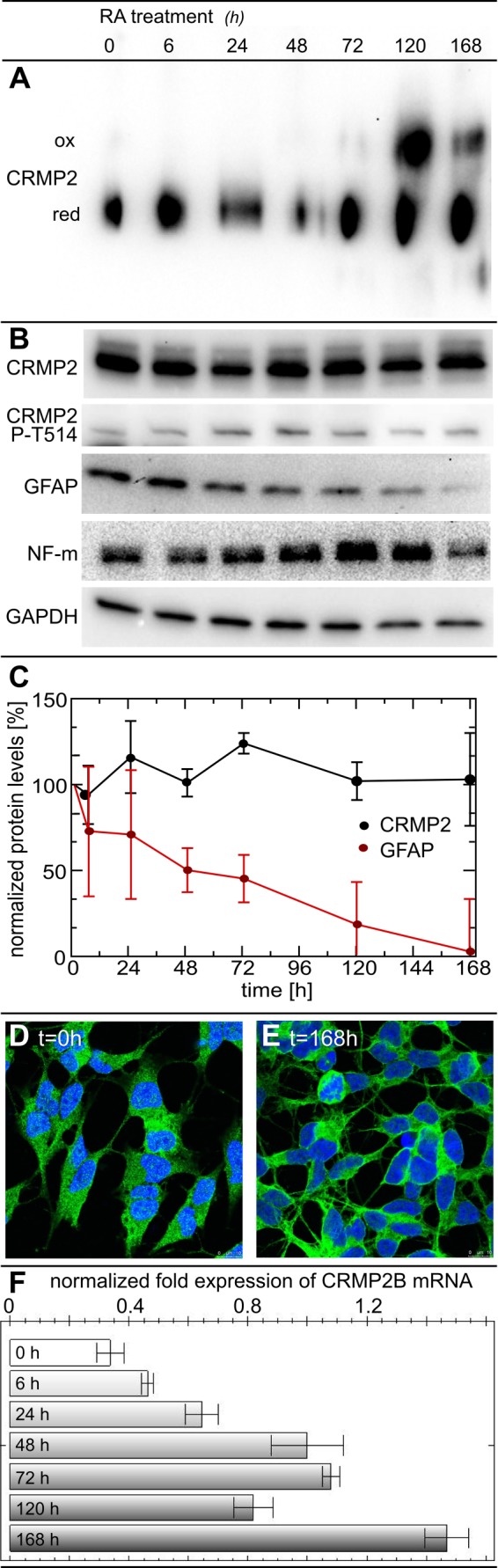
The CRMP2 conformational switch in a cellular model of neuronal differentiation. For differentiation into a neuron-like phenotype, SH-SY5Y cells were treated with 10 μm retinoic acid (RA). At 6, 24, 48, 72, 120, and 168 h, cells were detached by trypsin treatment, washed with PBS, lysed, separated by blue native gel electrophoresis, and analyzed by anti-CRMP2 Western blotting (A). Western blots of CRMP2, CRMP2 phospho-Thr-514, glial fibrillary acidic protein (GFAP), neurofilament m (NF-m), and the GAPDH loading control following SDS-PAGE (B). The presence of the neuronal marker neurofilament m confirmed the neuronal character of the cells. Quantification of CRMP2 and glial fibrillary acidic protein levels by densitometry of Western blots from three independent experiments, normalized to GAPDH levels and the untreated controls (C). The phenotype and localization of CRMP2 were analyzed by immunofluorescence and confocal microscopy (D and E). CRMP2B mRNA was quantified by quantitative PCR and normalized to GAPDH mRNA levels (F). ox, oxidized; red, reduced.
Together, our results demonstrate the presence of a specific Cys-504-Cys-504 thiol-disulfide switch in recombinant CRMP2 that controls and induces two distinct conformations of the CRMP2 tetramer that can also be observed in vivo in a model of neuronal differentiation.
DISCUSSION
Both human and zebrafish CRMP2 contain eight cysteinyl residues, and six of these are conserved between all vertebrate species: Cys-132, -133, -248, -323, -334, and -504 (see supplemental Fig. 1). Here, we demonstrated the oxidation of two adjacent Cys-504 thiols in the homo-tetrameric quaternary complex to a disulfide by treatment with hydrogen peroxide at low levels, i.e. 3–5-fold molar excess over CRMP2. Despite rigorous data analysis, we did not find evidence for the oxidation of any of the other seven cysteinyl thiols by treatment, to either disulfides, mixed disulfides with glutathione, sulfenic, sulfinic, or sulfonic acids. Although we cannot exclude that some of the respective peptides could not be ionized and detected by the MALDI-TOF/TOF analysis. The principal product of the reaction between a protein cysteinyl thiol and hydrogen peroxide is a protein sulfenic acid. In some cases, this sulfenic acid is rather stable at physiological conditions; often, however, this sulfenic acid reacts with a second thiol of a protein or with GSH, yielding disulfides. The intermolecular Cys-504-Cys-504 disulfide that resulted from hydrogen peroxide oxidation here most likely formed by oxidation of one Cys-504 residue to a sulfenic acid and the subsequent formation of the intermolecular disulfide. The formation of hydrogen peroxide has been implied in semaphorin signaling before (26), supporting the evidence that such a mechanism may also be relevant in vivo.
In this study, we provide clear evidence for a conformational change of tetrameric CRMP2 controlled by the dithiol-disulfide switch. Such conformational switches have been proposed before for CRMP2 and other CRMPs, however, dependent on the phosphorylation state of the proteins (27). At least six phosphorylation sites for CRMP2 have been characterized in the literature. Following an initial priming phosphorylation of Thr-514 by cyclin-dependent kinase 5 in response to semaphorin signaling, glycogen synthase kinase 3 β was reported to phosphorylate Thr-509, Thr-518, and Ser-522 (28). The Src-type Tyr kinase FYN phosphorylates Tyr-38 (29), Rho kinase and calmodulin-dependent kinase II residue Thr-555 (30, 31). Cys-504 is located in the C terminus of CRMP2, apparently the hotspot for the regulation of the function of the protein through post-translational modifications. Five of the previously characterized phosphorylation sites as well as the putative calpain recognition and cleavage site (17) are located immediately downstream of Cys-504 (supplemental Fig. 1). Note that Cys-504 is conserved in CRMP2s from all available vertebrate genomes but not present in any of the other four CRMPs (1, 3, 4, and 5). Unfortunately, the high resolution structures of CRMP2 and all closely related proteins lack the C-terminal regulatory hot spot (32). Here, we did not record changes in the phosphorylation of Thr-514 during differentiation, however, mechanistically the redox switch and phosphorylation of C-terminal amino acid residues could act in a similar way. The formation of a disulfide in CRMP2 must bring the C-terminal regions of two CRMP2 subunits in close proximity; reduction will lead to a more open conformation of the complex, partial unfolding, and the exposure of hydrophobic regions as demonstrated by CD spectroscopy and blue native electrophoresis (Fig. 3). These newly exposed surface areas could, for instance, function in the interaction with subsequent effector molecules of cytoskeletal dynamics. The addition of negative charge to the C termini in form of phosphate moieties might also lead to a more open conformation through electrostatic repulsion. However, these charged areas might favor interactions with other effector molecules.
It was proposed that the phosphorylation state of CRMP2 may be redox regulated by formation of a mixed disulfide of CRMP2 with the N-terminal active site cysteinyl residue of Trx1, which was supposed to facilitate neuronal growth cone collapse (18). The evidence presented for this hypothesis was primarily deduced from an active site mutant of Trx1 and some faint Western blot bands. Because of the different pKa values of the active site cysteine residues and the resulting reaction mechanism and kinetics of Trx1 and related oxidoreductases, a stable mixed disulfide complex between wild-type Trx1 and CRMP2 is highly unlikely, if not excluded (33, 34). We did not detect a complex between wild-type Grx2c (or Trx1, data not shown) and CRMP2 in vivo or in vitro, independent of the experimental conditions and the methods applied (electrophoresis, chromatography, mass spectrometry, or reduced or oxidized species; see also Fig. 2). We did observe, however, a tendency for an inverse correlation between the appearance of the oxidized conformation and the phosphorylation state of the Thr-514 residue of CRMP2 (Fig. 5, A and B). We can thus not exclude that redox and phosphorylation signaling acting on CRMP2 are functionally connected.
We have previously demonstrated that cytosolic Grx2 is essential for the formation of neuronal networks and the survival of differentiating neurons in zebrafish. Animals with silenced expression of Grx2 lacked virtually all types of neurons investigated, i.e. secondary motor neurons, dopaminergic neurons, and glutamatergic excitatory interneurons. Axonal length and number of branching points were significantly reduced in embryos lacking Grx2, neuronal networks did not mature. These functions of cytosolic Grx2 were dependent on the protein disulfide reductase activity of redoxin (11). We thus propose that the redox switch identified here controls CRMP2 function during neuronal development. CRMP2 itself is essential for brain development (16); it is an effector protein of the semaphorin/plexin pathway that controls axon/neurite branching and guidance (35–37). CRMP2 was implied in several neurological disorders. Alzheimer disease is characterized by the presence of neurofibrillary tangles, senile plaques, and loss of axonal networks, and these pathologies have all been attributed to oxidative stress. In a study focused on the identification of oxidized proteins in Alzheimer disease, CRMP2 was identified as significantly more oxidized protein (38). Moreover, hyperphosphorylation of CRMP2 is an early event in Alzheimer disease progression that can be induced by amyloid β (39), and CRMP2 accumulates in the brains of mice that developed neurofibrillary tangles and amyloid β plaques (40). It has been proposed that the manipulation of the biological functions of CRMP2, e.g. by the epilepsy drug lacosamide (41), may offer new strategies for therapy against neurological disorders (16, 42).
Axonal outgrowth and thus neuronal connectivity is facilitated by orchestrated dynamics of the cytoskeleton, directed by proteins such as CRMP2 in response to external signals. Redox regulation mechanisms of cytoskeletal dynamics and neuronal development have been suggested previously (e.g. Refs. 43–45). For instance, a transcript variant (TXNRD1_v3) of the selenoprotein thioredoxin reductase 1 encodes an atypical N-terminal Grx domain. Expression of complete TXNRD1_v3 protein or this Grx domain alone induced cell membrane protrusions (46). The Grx domain of TXNRD1_v3 localized first in the emerging protrusion and was then accompanied into the protrusions by actin and tubulin. In a subsequent study, TXNRD1_v3 was shown to induce dynamic cytoplasmic filaments itself. Actin polymerization was required for filopodia formation triggered by TXNRD1_v3 but not for generation of cytoplasmic filaments (47). The authors concluded that the Grx domain of TXNRD1_v3 is an atypical regulator of the cytoskeleton (46). Note that targeted deletion of the TXNRD1 gene leads to severe defects in the development of the brain (48). Grx2 and the Grx domain of TXNRD1_v3 share a higher homology to each other than to any other Grx of the mammalian cell. It is thus not unlikely that they act on the same or connected signaling pathways. Importantly, in vitro, Trx1 was able to reduce the CRMP2 disulfide (Fig. 2); however, in contrast to Grx2c (11, 12), neither the (over)expression of Trx1 nor Grx1 led to changes in cell morphology or the redox state of CRMP2 in cell cultures.3
In this work, we identified a dithiol-disulfide switch in CRMP2, a critical regulator of cytoskeletal dynamics, connecting two Cys-504 residues of neighboring subunits in the tetrameric quaternary complex. This switch controls two distinct conformations of the tetramer, its reduction is controlled specifically by Grx2c in a dithiol reaction mechanism, it is triggered during neuronal differentiation, and it controls axonal outgrowth. The major open questions that remain to be solved in the future are as follows: first, the mechanism of CRMP2 oxidation and, second, how the conformational switch affects subsequent effector proteins of cytoskeletal dynamics.
Acknowledgments
We express our gratitude to Kai Masur and Kristian Wende (INP Greifswald) for providing access to confocal microscopy and Winfried Hinrichs and Gottfried Palm for access to CD spectroscopy.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG SFB593-N01) and start-up funding by the Science Networks (Forschungsverbünde) Neuroscience and Molecular Medicine, University Medicine Greifswald.

This article contains supplemental Tables 1 and 2 and Fig. 1.
M. Gellert, S. Venz, J. Mitlöhner, C. Cott, E.-M. Hanschmann, and C. H. Lillig, unpublished results.
- Grx2
- glutaredoxin 2
- CRMP
- collapsin response mediator protein
- Grx
- glutaredoxin
- NEM
- N-ethylmaleimide
- TCEP
- Tris(2-chlorethyl)phosphate
- Trx
- thioredoxin.
REFERENCES
- 1. Holmgren A. (1979) Glutathione-dependent synthesis of deoxyribonucleotides. Purification and characterization of glutaredoxin from Escherichia coli. J. Biol. Chem. 254, 3664–3671 [PubMed] [Google Scholar]
- 2. Gravina S. A., Mieyal J. J. (1993) Thioltransferase is a specific glutathionyl mixed disulfide oxidoreductase. Biochemistry 32, 3368–3376 [DOI] [PubMed] [Google Scholar]
- 3. Mieyal J. J., Gallogly M. M., Qanungo S., Sabens E. A., Shelton M. D. (2008) Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid. Redox Signal. 10, 1941–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lillig C. H., Berndt C., Holmgren A. (2008) Glutaredoxin systems. Biochim. Biophys. Acta 1780, 1304–1317 [DOI] [PubMed] [Google Scholar]
- 5. Lillig C. H., Berndt C. (2013) Glutaredoxins in thiol/disulfide exchange. Antioxid. Redox Signal. 18, 1654–1665 [DOI] [PubMed] [Google Scholar]
- 6. Lundberg M., Johansson C., Chandra J., Enoksson M., Jacobsson G., Ljung J., Johansson M., Holmgren A. (2001) Cloning and expression of a novel human glutaredoxin (Grx2) with mitochondrial and nuclear isoforms. J. Biol. Chem. 276, 26269–26275 [DOI] [PubMed] [Google Scholar]
- 7. Gladyshev V. N., Liu A., Novoselov S. V., Krysan K., Sun Q. A., Kryukov V. M., Kryukov G. V., Lou M. F. (2001) Identification and characterization of a new mammalian glutaredoxin (thioltransferase), Grx2. J. Biol. Chem. 276, 30374–30380 [DOI] [PubMed] [Google Scholar]
- 8. Jurado J., Prieto-Alamo M. J., Madrid-Rísquez J., Pueyo C. (2003) Absolute gene expression patterns of thioredoxin and glutaredoxin redox systems in mouse. J. Biol. Chem. 278, 45546–45554 [DOI] [PubMed] [Google Scholar]
- 9. Lönn M. E., Hudemann C., Berndt C., Cherkasov V., Capani F., Holmgren A., Lillig C. H. (2008) Expression Pattern of Human Glutaredoxin 2 Isoforms: Identification and Characterization of Two Testis/Cancer Cell-Specific Isoforms. Antioxid. Redox Signal. 10, 547–557 [DOI] [PubMed] [Google Scholar]
- 10. Hudemann C., Lönn M. E., Godoy J. R., Zahedi Avval F., Capani F., Holmgren A., Lillig C. H. (2009) Identification, Expression Pattern, and Characterization of Mouse Glutaredoxin 2 Isoforms. Antioxid. Redox Signal. 11, 1–14 [DOI] [PubMed] [Google Scholar]
- 11. Bräutigam L., Schütte L. D., Godoy J. R., Prozorovski T., Gellert M., Hauptmann G., Holmgren A., Lillig C. H., Berndt C. (2011) Vertebrate-specific glutaredoxin is essential for brain development. Proc. Natl. Acad. Sci. U.S.A. 108, 20532–20537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schütte L. D., Baumeister S., Weis B., Hudemann C., Hanschmann E. M., Lillig C. H. (2013) Identification of potential protein dithiol-disulfide substrates of mammalian Grx2. Biochim. Biophys. Acta 1830, 4999–5005 [DOI] [PubMed] [Google Scholar]
- 13. Arimura N., Menager C., Fukata Y., Kaibuchi K. (2004) Role of CRMP-2 in neuronal polarity. J. Neurobiol. 58, 34–47 [DOI] [PubMed] [Google Scholar]
- 14. Vincent P., Collette Y., Marignier R., Vuaillat C., Rogemond V., Davoust N., Malcus C., Cavagna S., Gessain A., Machuca-Gayet I., Belin M. F., Quach T., Giraudon P. (2005) A role for the neuronal protein collapsin response mediator protein 2 in T lymphocyte polarization and migration. J. Immunol. 175, 7650–7660 [DOI] [PubMed] [Google Scholar]
- 15. Arimura N., Inagaki N., Chihara K., Ménager C., Nakamura N., Amano M., Iwamatsu A., Goshima Y., Kaibuchi K. (2000) Phosphorylation of collapsin response mediator protein-2 by Rho-kinase. Evidence for two separate signaling pathways for growth cone collapse. J. Biol. Chem. 275, 23973–23980 [DOI] [PubMed] [Google Scholar]
- 16. Charrier E., Reibel S., Rogemond V., Aguera M., Thomasset N., Honnorat J. (2003) Collapsin response mediator proteins (CRMPs): involvement in nervous system development and adult neurodegenerative disorders. Mol. Neurobiol. 28, 51–64 [DOI] [PubMed] [Google Scholar]
- 17. Bretin S., Rogemond V., Marin P., Maus M., Torrens Y., Honnorat J., Glowinski J., Prémont J., Gauchy C. (2006) Calpain product of WT-CRMP2 reduces the amount of surface NR2B NMDA receptor subunit. J. Neurochem. 98, 1252–1265 [DOI] [PubMed] [Google Scholar]
- 18. Morinaka A., Yamada M., Itofusa R., Funato Y., Yoshimura Y., Nakamura F., Yoshimura T., Kaibuchi K., Goshima Y., Hoshino M., Kamiguchi H., Miki H. (2011) Thioredoxin mediates oxidation-dependent phosphorylation of CRMP2 and growth cone collapse. Sci. Signal. 4, ra26. [DOI] [PubMed] [Google Scholar]
- 19. Lillig C. H., Berndt C., Vergnolle O., Lönn M. E., Hudemann C., Bill E., Holmgren A. (2005) Characterization of human glutaredoxin 2 as iron-sulfur protein: A possible role as redox sensor. Proc. Natl. Acad. Sci. U.S.A. 102, 8168–8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hanschmann E. M., Lönn M. E., Schütte L. D., Funke M., Godoy J. R., Eitner S., Hudemann C., Lillig C. H. (2010) (2010) Both thioredoxin 2 and glutaredoxin 2 contribute to the reduction of the mitochondrial 2-CYS peroxiredoxin PRX3. J. Biol. Chem. 285, 40699–40705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johansson C., Lillig C. H., Holmgren A. (2004) Human mitochondrial glutaredoxin reduces S-glutathionylated proteins with high affinity accepting electrons from either glutathione or thioredoxin reductase. J. Biol. Chem. 279, 7537–7543 [DOI] [PubMed] [Google Scholar]
- 22. Ericsson U. B., Hallberg B. M., Detitta G. T., Dekker N., Nordlund P. (2006) Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal. Biochem. 357, 289–298 [DOI] [PubMed] [Google Scholar]
- 23. Påhlman S., Ruusala A. I., Abrahamsson L., Mattsson M. E., Esscher T. (1984) Retinoic acid-induced differentiation of cultured human neuroblastoma cells: a comparison with phorbolester-induced differentiation. Cell Differ. 14, 135–144 [DOI] [PubMed] [Google Scholar]
- 24. Wang L. H., Strittmatter S. M. (1997) Brain CRMP forms heterotetramers similar to liver dihydropyrimidinase. J. Neurochem. 69, 2261–2269 [DOI] [PubMed] [Google Scholar]
- 25. Korecka J. A., van Kesteren R. E., Blaas E., Spitzer S. O., Kamstra J. H., Smit A. B., Swaab D. F., Verhaagen J., Bossers K. (2013) Phenotypic characterization of retinoic acid differentiated SH-SY5Y cells by transcriptional profiling. PLoS One 8, e63862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nadella M., Bianchet M. A., Gabelli S. B., Barrila J., Amzel L. M. (2005) Structure and activity of the axon guidance protein MICAL. Proc. Natl. Acad. Sci. U.S.A. 102, 16830–16835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schmidt E. F., Strittmatter S. M. (2007) The CRMP family of proteins and their role in Sema3A signaling. Adv. Exp. Med. Biol. 600, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Uchida Y., Ohshima T., Sasaki Y., Suzuki H., Yanai S., Yamashita N., Nakamura F., Takei K., Ihara Y., Mikoshiba K., Kolattukudy P., Honnorat J., Goshima Y. (2005) Semaphorin3A signalling is mediated via sequential Cdk5 and GSK3β phosphorylation of CRMP2: implication of common phosphorylating mechanism underlying axon guidance and Alzheimer's disease. Genes Cells 10, 165–179 [DOI] [PubMed] [Google Scholar]
- 29. Uchida Y., Ohshima T., Yamashita N., Ogawara M., Sasaki Y., Nakamura F., Goshima Y. (2009) Semaphorin3A signaling mediated by Fyn-dependent tyrosine phosphorylation of collapsin response mediator protein 2 at tyrosine 32. J. Biol. Chem. 284, 27393–27401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Patrakitkomjorn S., Kobayashi D., Morikawa T., Wilson M. M., Tsubota N., Irie A., Ozawa T., Aoki M., Arimura N., Kaibuchi K., Saya H., Araki N. (2008) Neurofibromatosis type 1 (NF1) tumor suppressor, neurofibromin, regulates the neuronal differentiation of PC12 cells via its associating protein, CRMP-2. J. Biol. Chem. 283, 9399–9413 [DOI] [PubMed] [Google Scholar]
- 31. Hou S. T., Jiang S. X., Aylsworth A., Ferguson G., Slinn J., Hu H., Leung T., Kappler J., Kaibuchi K. (2009) CaMKII phosphorylates collapsin response mediator protein 2 and modulates axonal damage during glutamate excitotoxicity. J. Neurochem. 111, 870–881 [DOI] [PubMed] [Google Scholar]
- 32. Deo R. C., Schmidt E. F., Elhabazi A., Togashi H., Burley S. K., Strittmatter S. M. (2004) Structural bases for CRMP function in plexin-dependent semaphorin3A signaling. EMBO J. 23, 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kallis G. B., Holmgren A. (1980) Differential reactivity of the functional sulfhydryl groups of cysteine-32 and cysteine-35 present in the reduced form of thioredoxin from Escherichia coli. J. Biol. Chem. 255, 10261–10265 [PubMed] [Google Scholar]
- 34. Berndt C., Lillig C. H., Holmgren A. (2007) Thiol-based mechanisms of the thioredoxin and glutaredoxin systems: implications for diseases in the cardiovascular system. Am. J. Physiol. Heart. Circ. Physiol. 292, H1227–H1236 [DOI] [PubMed] [Google Scholar]
- 35. Goshima Y., Nakamura F., Strittmatter P., Strittmatter S. M. (1995) Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature 376, 509–514 [DOI] [PubMed] [Google Scholar]
- 36. Inagaki N., Chihara K., Arimura N., Ménager C., Kawano Y., Matsuo N., Nishimura T., Amano M., Kaibuchi K. (2001) CRMP-2 induces axons in cultured hippocampal neurons. Nat. Neurosci. 4, 781–782 [DOI] [PubMed] [Google Scholar]
- 37. Nishimura T., Fukata Y., Kato K., Yamaguchi T., Matsuura Y., Kamiguchi H., Kaibuchi K. (2003) CRMP-2 regulates polarized Numb-mediated endocytosis for axon growth. Nat. Cell Biol. 5, 819–826 [DOI] [PubMed] [Google Scholar]
- 38. Sultana R., Boyd-Kimball D., Poon H. F., Cai J., Pierce W. M., Klein J. B., Merchant M., Markesbery W. R., Butterfield D. A. (2006) Redox proteomics identification of oxidized proteins in Alzheimer's disease hippocampus and cerebellum: an approach to understand pathological and biochemical alterations in AD. Neurobiol. Aging 27, 1564–1576 [DOI] [PubMed] [Google Scholar]
- 39. Cole A. R., Noble W., van Aalten L., Plattner F., Meimaridou R., Hogan D., Taylor M., LaFrancois J., Gunn-Moore F., Verkhratsky A., Oddo S., LaFerla F., Giese K. P., Dineley K. T., Duff K., Richardson J. C., Yan S. D., Hanger D. P., Allan S. M., Sutherland C. (2007) Collapsin response mediator protein-2 hyperphosphorylation is an early event in Alzheimer's disease progression. J. Neurochem. 103, 1132–1144 [DOI] [PubMed] [Google Scholar]
- 40. Takata K., Kitamura Y., Nakata Y., Matsuoka Y., Tomimoto H., Taniguchi T., Shimohama S. (2009) Involvement of WAVE accumulation in Aβ/APP pathology-dependent tangle modification in Alzheimer's disease. Am. J. Pathol. 175, 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Beydoun A., D'Souza J., Hebert D., Doty P. (2009) Lacosamide: pharmacology, mechanisms of action and pooled efficacy and safety data in partial-onset seizures. Expert Rev. Neurother. 9, 33–42 [DOI] [PubMed] [Google Scholar]
- 42. Hensley K., Venkova K., Christov A., Gunning W., Park J. (2011) Collapsin response mediator protein-2: an emerging pathologic feature and therapeutic target for neurodisease indications. Mol. Neurobiol. 43, 180–191 [DOI] [PubMed] [Google Scholar]
- 43. Giles G. I. (2006) The redox regulation of thiol dependent signaling pathways in cancer. Curr. Pharm. Des. 12, 4427–4443 [DOI] [PubMed] [Google Scholar]
- 44. Giridharan S. S., Rohn J. L., Naslavsky N., Caplan S. (2012) Differential regulation of actin microfilaments by human MICAL proteins. J. Cell Sci. 125, 614–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang J., Boja E. S., Tan W., Tekle E., Fales H. M., English S., Mieyal J. J., Chock P. B. (2001) Reversible glutathionylation regulates actin polymerization in A431 cells. J. Biol. Chem. 276, 47763–47766 [DOI] [PubMed] [Google Scholar]
- 46. Dammeyer P., Damdimopoulos A. E., Nordman T., Jiménez A., Miranda-Vizuete A., Arnér E. S. (2008) Induction of cell membrane protrusions by the N-terminal glutaredoxin domain of a rare splice variant of human thioredoxin reductase 1. J. Biol. Chem. 283, 2814–2821 [DOI] [PubMed] [Google Scholar]
- 47. Damdimopoulou P. E., Miranda-Vizuete A., Arnér E. S., Gustafsson J. A., Damdimopoulos A. E. (2009) The human thioredoxin reductase-1 splice variant TXNRD1_v3 is an atypical inducer of cytoplasmic filaments and cell membrane filopodia. Biochim. Biophys. Acta 1793, 1588–1596 [DOI] [PubMed] [Google Scholar]
- 48. Soerensen J., Jakupoglu C., Beck H., Förster H., Schmidt J., Schmahl W., Schweizer U., Conrad M., Brielmeier M. (2008) The role of thioredoxin reductases in brain development. PLoS One 3, e1813. [DOI] [PMC free article] [PubMed] [Google Scholar]