Background: Although the ubiquitin-proteasome pathway is thought to regulate the level of SOX9, the ubiquitin ligase involved has not yet been determined.
Results: E6-AP/UBE3A was found to bind and ubiquitinate SOX9.
Conclusion: E6-AP/UBE3A functions as a ubiquitin ligase toward SOX9 both in vitro and in vivo.
Significance: These data provide the regulatory mechanism of the SOX9 level in chondrocytes by ubiquitin ligase.
Keywords: Chondrocytes, E3 Ubiquitin Ligase, Protein Degradation, Proteomics, Ubiquitin, Ubiquitination, E6-AP, SOX9, UBE3A
Abstract
SOX9 is a transcription factor that acts as a key regulator at various stages of cartilage differentiation. There is ample evidence that intracellular SOX9 protein levels are tightly regulated both by sumoylation and by degradation through the ubiquitin-proteasome pathway. Using a proteomics approach, here we report the identification of a SOX9-binding protein, E6-AP/UBE3A, that may act as a ubiquitin ligase toward Sox9. E6-AP bound SOX9 through the region consisting mostly of its high mobility group domain in vitro. In nuclear lysates, FLAG-tagged E6-AP coprecipitated with Sox9 and its high mobility group domain. This finding was estimated using nuclear lysates from a chondrocytic cell line that endogenously expresses E6-AP and SOX9. Accordingly, ectopically expressed E6-AP and SOX9 colocalized in the nucleus. We show that E6-AP ubiquitinates SOX9 in vitro and in vivo and that SOX9 levels are enhanced after addition of the proteasome inhibitor bortezomib. Similar, siRNA knockdown of E6-AP and the E2 ligase Ubc9 increased cellular SOX9 amounts, supporting the notion that SOX9 may be ubiquitinated in hypertrophic chondrocytes by E6-AP and degraded by proteasomes. This is in accordance with the distribution of SOX9 levels, which are high in proliferating and prehypertrophic chondrocytes but low in hypertrophic chondrocytes, whereas E6-AP levels are high in hypertrophic chondrocytes and low in prehypertrophic chondrocytes. Furthermore, E6-AP-deficient mice showed SOX9 accumulation in chondrocytes and the brain. These findings support the concept that E6-AP regulates SOX9 levels in developing cartilage by acting as a ubiquitin ligase.
Introduction
SOX9 is a transcription factor of the sex-determining region of the Y chromosome (SRY) family and has been grouped with SOX8 and SOX10 into the subgroup E on the basis of the amino acid sequences of the high mobility group (HMG)3 domain, a transactivation and dimerization domain (1). The role of SOX9 in chondrocyte differentiation has been well documented. Specifically, SOX9 is expressed in chondroprogenitor cells and fully differentiated chondrocytes (2, 3) and regulates the transcription of cartilage-specific matrix molecules such as collagen types II, IX, XI, XXVII, and aggrecan (4–9). Mutations in the SOX9 gene in humans cause campomelic dysplasia (10, 11), a severe skeletal malformation syndrome. Results from SOX9 inactivation studies in mice (12, 13) indicate that SOX9 plays a major role in chondrogenic differentiation, starting with mesenchymal condensation and continuing until the early stages of endochondral bone formation. Overexpression of SOX9 in mouse chondrocytes produced by knock-in into a Col2a1 allele showed a distinct skeletal phenotype characterized by a marked delay in endochondral bone formation (14). In addition, SOX9 overexpression in hypertrophic chondrocytes using the Bac-Col10a1 promoter showed delayed endochondral bone formation that was associated with reduced bone growth (15). These results strongly suggest that proper SOX9 levels are essential for chondrocyte differentiation. In addition to its role in chondrocyte differentiation, SOX9 has been shown to have distinct roles in the differentiation of other tissues, including neural crest cells (16), hair follicles (17), testicular Sertoli cells (18), pancreatic cells (19), prostate epithelia (20), and Paneth cells in the small intestine (21). In mouse embryos, SOX9 is highly expressed in chondrocytes of the proliferating and prehypertrophic zone but is barely detected in the hypertrophic zone (9, 12). However, the level of SOX9 in chondrocytes is regulated not only by mRNA down-regulation but also by its degradation through the ubiquitin-proteasome pathway (22, 23). In this pathway, at first the enzyme E1 activates a ubiquitin moiety and forms a thioester bond between the cysteine residue of E1 and the C terminus of ubiquitin in an ATP-dependent manner. The activated ubiquitin is then transferred to the ubiquitin-conjugating enzyme E2 via the thioester bond. Finally, the ubiquitin ligase E3 recognizes the target protein to be ubiquitinated and transfers the ubiquitin moieties from itself or E2 to the target. The resulting polyubiquitin chain serves as a recognition signal for proteasomal degradation (24, 25). There are two groups of ubiquitin ligase E3: HECT (homologous to the E6-AP carboxyl terminus) ligases and the RING (group of the really interesting new gene) ligases. HECT ligases bind ubiquitin to themselves and transfer the ubiquitin molecule to their substrates. RING/Ubox ligases do not bind ubiquitin directly and activate the transfer of the ubiquitin molecule of E2 to the substrates (26). All ubiquitin ligases recognize their own substrates and are specific for these substrates. The ubiquitin ligase for SOX9 has not yet been identified, and its identification is essential to dissolving the mechanism of the degradation of SOX9 in this pathway.
Using a proteomics approach, we aimed to find SOX9-binding proteins and detected several kinds of PIAS (protein inhibitor of activated STAT family) protein (23). Here, in addition to PIAS, we characterized E6-AP/UBE3A as a SOX9-binding protein and proved that it functions as a ubiquitin ligase in relation to SOX9.
EXPERIMENTAL PROCEDURES
Detection of SOX9-binding Proteins in Chondrocytes
Bovine chondrocytes were prepared from epiphyseal cartilage of calf fetuses (vertex breech length, 50–70 cm) by digestion with 0.04% clostridial collagenase P (Roche Applied Science) in the presence of 5% fetal calf serum as described (27). Within 6 h after plating, the freshly isolated chondrocytes were infected with adenoviruses at a multiplicity of infection of 1 for 48 h. The adenovirus used has a Tet-Off-inducible system and a SOX9 open reading frame that has a FLAG tag at its N terminus and a His6 tag at its C terminus. For inducible expression under the Tet-Off system, chondrocytes were coinfected with the Tet-Off virus. Nuclear extracts were prepared from 1.8 × 109 cells and loaded onto nickel-agarose. The imidazole eluates were further purified on anti-FLAG-agarose (Sigma-Aldrich). The proteins bound to anti-FLAG-agarose were precipitated with 10% TCA, dissolved in 40 ml of SDS-PAGE sample buffer, and resolved by 10% SDS-PAGE (Invitrogen). Protein bands were detected by silver staining. In the control experiment, the same amount of nuclear extract was prepared from cells not expressing FLAG-Sox-His6 protein in the absence of tetracycline. Additional bands in SDS-PAGE coeluting with SOX9 were excised from the gel, and “subproteome” components were degraded by trypsin. The peptides were separated by HPLC and eluted into an electrospray ion source for mass analysis. Selected peptides were further analyzed by MS/MS experiments and identified by computer analysis.
GST Pull-down and Immunoprecipitation Assays
HA-tagged, full-length SOX9 and truncated SOX9 cDNAs were transcribed and translated in vitro in 50 μl of reaction mixture using a T7 quick-coupled transcription/translation kit (Promega, Madison, WI). Ten percent of the reaction mixtures was directly analyzed by SDS-PAGE. The rest was used for a pull-down assay with GST-E6-AP- or GST-expressing cell lysates (1 μg of protein in each reaction) and glutathione-Sepharose beads (GE Health Science). Reactions were performed in 10 volumes of immunoprecipitation buffer containing 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1% Triton X-100, 0.1% SDS, and 1 mm PMSF for 4 h at 4 °C. The gel was washed five times with the same buffer. HA-SOX9 proteins were released by boiling with 2× SDS sample buffer, fractionated by SDS-PAGE, and immunoblotted with anti-HA antibody. For coimmunoprecipitation experiments, cells were harvested in coimmunoprecipitation buffer (10 mm Tris-HCl (pH8.0), 1 mm EDTA, 20 mm NaCl, 10% glycerol, 1 mm DTT, 1 mm PMSF, and 0.5% Nonidet P-40) and centrifuged. The supernatants were collected as cytosolic fractions, and the pellets (nuclear fraction) were resuspended with nuclear extract buffer (50 mm Hepes (pH 7.9), 3 mm MgCl2, 20 mm NaCl, 1 mm DTT, and 1 mm PMSF) containing 2.5 units of benzonase nuclease (Novagen, Darmstadt, Germany) and incubated for 2 h at 4 °C. After centrifugation, the resulting supernatants were corrected as nuclear extracts. The presence of target proteins in nuclear extracts was monitored by Western blot analysis using specific antibodies.
Ubiquitination of SOX9 in Cultured Cells
HEK293 cells were cotransfected during the logarithmic growth phase with 1 μg of pcDNA3.1/HA-ubiquitin, 0.5 μg of pcDNA4/HisMax-Sox9 that expressed His-Xpress-Sox9, and 2 μg of pFLAG-E6-AP using Lipofectamine reagent (Invitrogen). Twenty-four hours after transfection, the cells were harvested using 10 mm Tris-HCl (pH7.4) containing 0.1 m NaCl, 1 mm PMSF, 0.1% Nonidet P-40, and protease inhibitor mixture (Roche Applied Science) and disrupted by sonication. Anti-Xpress antibody (Invitrogen) was added to the cleared supernatant obtained by centrifugation from the disrupted cells. The SOX9 complexes were recovered by addition of protein G-Sepharose 4B (GE Health Science), loaded onto SDS 4–20% polyacrylamide gel, transferred to a PVDF membrane, and detected by Western blotting using anti-SOX9 antibody, HRP-labeled anti-mouse goat secondary antibody, and the ECL method (GE Health Science).
Inhibition of Proteasomal Degradation by Proteasome Inhibitor
Human chondrocytic HCS-2/8 cells (29) were treated with bortezomib (Cell Signaling Technology, Danvers, MA) for 3 h before harvesting with SDS buffer. Sox9 concentration was analyzed by Western blot analysis using anti-Sox9 antibody (Millipore, Darmstadt, Germany) and anti-actin antibody (Sigma) for standardization.
In Vitro Ubiquitination
GST-E1, GST-E6-AP, or GST-Sox9 was expressed in a baculovirus expression system and purified using glutathione-Sepharose 4B as described previously (28). Briefly, Sf-9 cells expressing GST-E1 were suspended in 10 mm Tris-HCl (pH7.4) containing 3 mm MgCl2, 0.1% Nonidet P-40, 1 mm PMSF, and protease inhibitor mixture (Roche Applied Science) and disrupted by sonication. Then, the sonicated cells were centrifuged at 12,000 × g for 10 min. Glutathione-Sepharose 4B resin was added to the resultant supernatant, which was then incubated for 30 min at 4 °C. The resin was recovered by centrifugation and washed three times with 10 mm Tris-HCl (pH7.4) containing PMSF and protease inhibitor mixture. The GST-E1 was recovered from the resin by incubation with 5 mm reduced glutathione and dialyzed against 10 mm Tris-HCl (pH 7.4) as described above. In this in vitro assay, these proteins binding to the resin were used without elution from the resin. Ubiquitin (Sigma-Aldrich) was labeled with biotin using sulfo-LC-biotin (Pierce). UbcH5c was expressed in a pET expression system and purified as described previously. We incubated, at 30 °C for 30 min, 50 ml of assay mixture containing 50 mm Tris-HCl (pH 7.4), 5 mm MgCl2, 5 mm ATP, 1 μg of GST-E1, 0.1 μg of UbcH5, and 1 μg of biotinylated ubiquitin; the 1 μg of GST-E6-AP (bound to the resin); and 1 μg of GST-SOX9 (bound to the resin). After incubation, the resin was washed once by centrifugation with 1 ml of 10 mm Tris-HCl (pH 7.4) and resuspended in SDS-PAGE sample buffer. The proteins were separated on SDS-PAGE (4–20%, Dai-ichi Chemical) and transferred to a PVDF membrane (Bio-Rad). Biotinylated ubiquitin was detected by Western blotting using HRP streptavidin (Extravidin, Sigma-Aldrich) and the ECL method (GE Health Science).
Indirect Immunofluorescence and Laser Microscopic Analysis
COS7 monkey kidney cells and human chondrocytic HCS-2/8 cells were cultured in DMEM supplemented with 10% fetal bovine serum. For transient expression of GFP-SOX9 and FLAG-E6-AP or Halo-E6AP, cells were seeded on coverglass and transfected with the corresponding expression vectors using X-treameGENE 9 reagent (Roche Applied Science) according to the instructions of the manufacturer. Twenty-four hours later, the cells that transfected GFP-Sox9- and/or Halo-E6-AP-expressing vectors were incubated with HaloTag TMR fluorescent ligands (Promega) according to the protocol of the manufacturer, fixed and mounted on slide glasses, and observed under a laser microscope. The cells that harbored GFP-Sox9 and FLAG-E6-AP were first fixed with 4% formaldehyde in PBS for 15 min at room temperature. The cells were then permeabilized with 20 mm Tris-HCl (pH7.6), 137 mm NaCl, and 0.1% Tween 20 (TBST) containing 5% skim milk for 30 min at room temperature before being incubated with 1:100 diluted anti-FLAG M2 monoclonal antibody (Sigma-Aldrich) for FLAG-tagged E6-AP for 90 min at 4 °C. After being washed three times, the cells were incubated with 1:5000-diluted Alexa Fluor 555 anti-mouse IgG. Images of FLAG-tagged E6-AP and GFP-tagged SOX9 were obtained with a laser confocal microscope (Bio-Rad and Zeiss).
Immunohistochemical Analysis
Mouse embryo forelimbs (E15.5) were fixed in 4% paraformaldehyde in PBS for 16 h. After dehydration, tissues were embedded in paraffin and sectioned. Immunohistochemical staining was done with a PicTure kit (Zymed Laboratories Inc.) for anti-SOX9 rabbit IgG (30) or an MM biotinylation kit (Biocare Medical) for E6-AP mouse monoclonal antibody (Sigma).
E6-AP Knockdown with siRNA
E6-AP and control siRNA were obtained from Fasmac Co. The E6-AP siRNA was derived from the human sequence but also from recognized mouse E6-AP. Control siRNA does not recognize any human and mouse mRNA. The sequence of the top strand of each siRNA was as follows: E6-AP, 5′-UAUCUCAGAGCAGGAGUUGTT-3′; and control, 5′-CAUGUCAUGUGUCACAUCUTT-3′. Chondrocytes from newborn mice were prepared from rib cages. After the removal of soft tissues with 0.1% trypsin at 37 °C for 10 min, cartilage parts were digested with 0.01% collagenase A (Roche Applied Science) at 37 °C for 16 h. For siRNA analysis, mouse primary chondrocytes (7.5 × 105 cells) and HCS-2/8 cells (1 × 106 cells) were transfected with 2.5 μl of 100 pmol/μl double-stranded siRNA (final 100 nm) using 25 μl of Lipofectamine 2000 according to the instructions of the manufacturer. COS7 cells were cotransfected with 1 μl of siRNA (final 40 nm) and 1 μg of pEGFP-SOX9, 0.75 μg of pFLAG-E6-AP, and 1 μg of HA-Ub. Forty-eight hours after transfection, the cells were lysed in 1× SDS-PAGE sample buffer and subjected to Western blot analysis. For detection of endogenous SOX9 and E6-AP, anti-SOX9 rabbit antibody (Abcam) and anti-E6-AP monoclonal antibody (Sigma-Aldrich) were used. For the detection of the expressed GFP-SOX9, FLAG-E6-AP, and HA-ubiquitin in COS7 cells, anti-GFP antibody (MBL), anti-FLAG antibody (Sigma-Aldrich), and anti-HA antibody were used, respectively. For monitoring e6-ap and sox9 mRNA, total RNA was isolated by RNeasy kit (Qiagen, Hilden, Germany), cDNA was generated with reverse transcriptase (AMV, Takara, Shiga, Japan), and the mRNA levels of e6-ap and sox9 were monitored using Cyber Green (Toyobo, Tokyo, Japan) and StepOne Plus (Applied Biosystems). Primers used were as follows: for human e6-ap, tgcttgaggttgagcctttt and tggtctacaaatggtggcaa; for human sox9, aggcaagcaaaggagatgaa and tggtgttctgagaggcacag; and for human gapdh, caatgaccccttcattgacc and gacaagcttcccgttctcag.
Detection of SOX9 in E6-AP-deficient Mouse Embryonic Heads and Newborn Cartilage
Tissue lysates from E14 heads of E6-AP-deficient littermates (31) were prepared by homogenization in 10 mm sodium phosphate buffer (pH7.2), 150 mm NaCl, 1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS, 1 mm DTT, 0.1 mm PMSF, 1 mm NaF, and 0.4 mm sodium orthovanadate. Fifty micrograms of tissue lysate was analyzed by Western blotting using anti-SOX9 antibody. Primary chondrocytes from newborn E6-AP-deficient mice were cultured until confluent, and cell lysates were prepared as above.
RESULTS
Identification of E6-AP/UBE3A as a SOX9-binding Protein
To characterize SOX9-binding proteins in fetal bovine chondrocytes, the nuclear extracts infected with AdeCMV-FLAG-SOX9-His6 were loaded onto nickel resin. After elution of FLAG-Sox9-His6 protein complexes from nickel resin with imidazole, the eluate was directly loaded onto anti-FLAG resin, and the binding proteins to the resin were eluted by FLAG peptides. When the proteins coimmunoprecipitated with SOX9 were analyzed by SDS-PAGE, as shown under “Experimental Procedures,” about 10 protein bands, including FLAG-SOX9-His6, were visible in silver-stained gel (Fig. 1). Each protein band was excised from the gel and subjected to proteolysis with trypsin. The tryptic digest was subjected to a combination of peptide mass analysis using electrospray ion spectrometry and mass spectrometry. As a result, from one of the bands shown in Fig. 1, we identified three peptides corresponding to the ubiquitin ligase E6-AP when the sequences obtained were searched against a mammalian protein database. All three peptides were completely aligned to bovine, human, and mouse E6-AP.
FIGURE 1.
Proteomics analysis of FLAG-SOX9-His-binding nuclear proteins. A, after nuclear extracts from fetal bovine rib chondrocytes expressing FLAG- and His-tagged SOX9 were loaded onto nickel-agarose (Ni-agarose), the eluates were further purified on anti-FLAG-agarose. Bound proteins were precipitated with 10% TCA, resolved by SDS gel electrophoresis on 10% BisTris gel, and stained with silver. For the control, the same amount of nuclear extract was prepared from non-transfected chondrocytes, but no significant bands were detected (not shown). All SOX9-binding bands were excised from the gel individually, and eluted proteins were digested with trypsin and subjected to MS/MS analysis. MW, molecular weight. B, three peptides that were sequenced corresponded to the ubiquitin ligase E6-AP. The locations of these peptides are shown as gray shadows in the entire amino acid sequence of E6-AP. One peptide is located in the N terminus, and the other two are in the HECT domain.
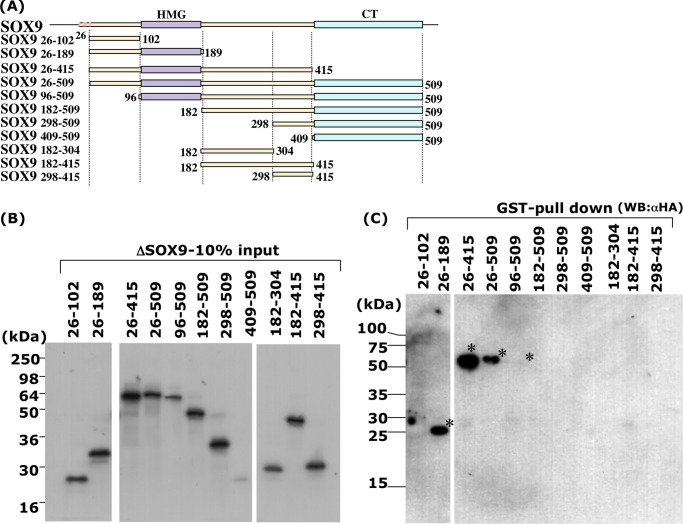
To identify the region of SOX9 necessary for interaction with E6-AP, we performed a pull-down assay using HA-SOX9 of various lengths and GST-E6-AP (Fig. 2). Each full-length and truncated HA-SOX9 fragment synthesized by in vitro transcription/translation was mixed with recombinant GST-E6-AP expressed in Sf-9 cells, and GST-E6-AP was pulled down by glutathione-Sepharose beads. Although the expression levels of some truncated fragments of HA-SOX9, including HA-SOX9 (96–509) and HA-SOX9 (409–509), were weak (Fig. 2B), each of the forms of HA-SOX9 (HA-SOX9 (26–189), HA-SOX9 (26–415), HA-SOX9 (26–509), and HA-SOX9 (96–509)) was pulled down with GST-E6-AP (Fig. 2C). The precipitated band of HA-SOX9 (96–509) was very weak because of its low expression but is clearly visible in Fig. 2C. Judging from the results, the minimum E6-AP interaction region in HA-SOX9 seemed to be SOX9 (96–189), which corresponded to the HMG DNA-binding domain of SOX9 (Fig. 2A). Only the control GST fragment did not show any binding to HA-SOX9.
FIGURE 2.
In vitro association of SOX9 and E6-AP. Several truncated SOX9 protein fragments were generated by in vitro translation as described previously (23), applied onto SDS-PAGE, and detected by Western blotting using an HA antibody. A, schematic representation of N-terminal- or C-terminal-truncated SOX9 fragments. SOX9 consists of a high mobility group DNA-binding domain (HMG box), a C-terminal transactivation (CT) domain, and four ATG codons at the N terminus portion in-frame. Numbers indicate the location of the fragments in amino acid number from the N terminus. B, 10% of the input SOX9 proteins was applied to the gel. C, pull-down assay of SOX9 fragments with GST-E6-AP. Significant pull-down of SOX9 fragments (asterisk) was seen with SOX9 fragments 26–189, 26–415, 26–509, and 96–509, which contain amino acids 96–189, corresponding to the HMG domain.
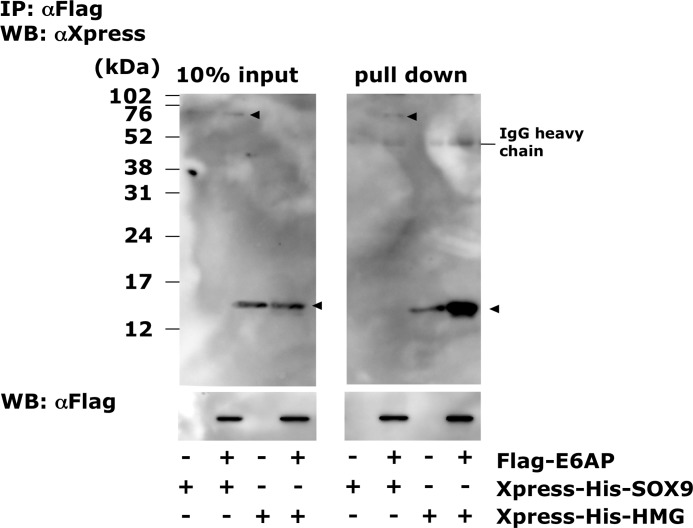
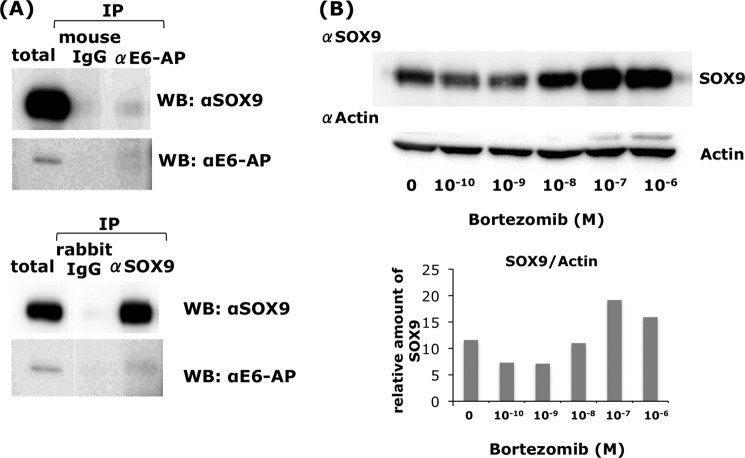
To confirm the binding between the Sox9-HMG domain and E6-AP, we performed a coimmunoprecipitation assay using ectopically expressed FLAG-E6-AP and Xpress-His-Sox9 or Xpress-His-HMG in COS7 cells (Fig. 3). The Xpress-His-Sox9- or Xpress-His-HMG-expressing vectors were cotransfected with FLAG-E6AP- or FLAG (mock)-expressing vectors, and the nuclear extracts were mixed with FLAG-agarose. Only in the nuclear lysates expressing FLAG-E6-AP, Xpress-His-SOX9, and Xpress-His-HMG were precipitated effectively (Fig. 3). Cellular binding of endogenous SOX9 and E6-AP was also confirmed using HCS-2/8 chondrocytic cells (Fig. 4A). Nuclear Sox9 and E6-AP were coprecipitated by anti E6AP and SOX9 antibody, respectively (Fig. 4A).
FIGURE 3.
In vivo association of E6-AP with SOX9 and the HMG domain of SOX9. Expression vectors that harbor Xpress-tagged full-length SOX9 or the HMG domain of SOX9 (HMG) cDNA were cotransduced with the FLAG-E6-AP expression vector to COS7 cells, and E6-AP binding proteins were pulled down with anti-FLAG antibody. Ten percent of the input and pull-down fraction were immunoblotted with anti-Xpress and anti-FLAG antibodies. In the presence of FLAG-E6AP, Xpress-SOX9 and Xpress-HMG were pulled down effectively. IP, immunoprecipitation; WB, Western blot.
FIGURE 4.
In vivo association of endogenous SOX9 and E6-AP in chondrocytes and accumulation of SOX9 by proteasomal inhibitor. A, nuclear proteins from the chondrocytic cell line HCS-2/8 were extracted by benzonase digestion of chromatin DNA and E6-AP binding proteins, and Sox9 binding proteins were pulled down by anti-E6-AP and Sox9 antibodies, respectively. E6-AP-bound Sox9 and Sox9-bound E6-AP were detected by immunoblotting using the respective antibodies. IP, immunoprecipitation; WB, Western blot. B, HCS-2/8 cells were treated with bortezomib, a proteasome inhibitor, for 3 h, and the cells were lysed with SDS buffer. Bortezomib significantly increased the SOX9 concentration. Lower panel, relative amounts of Sox9 to actin. M, mol concentration.
Ubiquitination of SOX9 by E6-AP
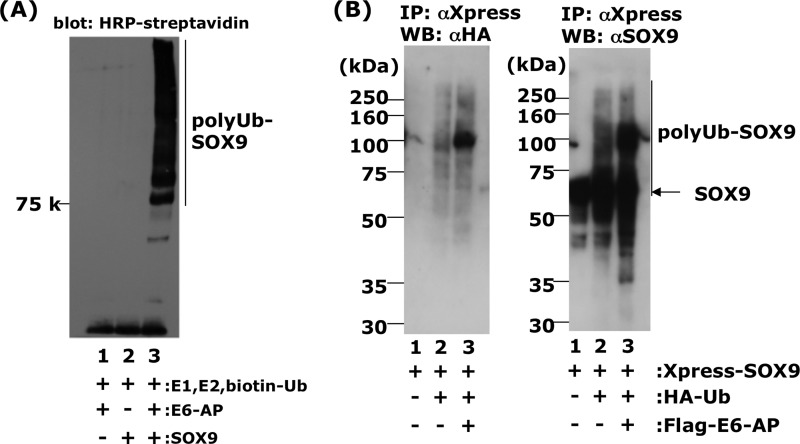
We showed that E6-AP could physically interact with SOX9, which suggests that this might ubiquitinate SOX9. However, the Wwp2, Nedd4-like HECT domain containing ubiquitin ligase was also shown to be bound to SOX9, although it could not ubiquitinate SOX9 in in vitro assays (32). Therefore, it was very important to show whether E6-AP/UBE3A ubiquitinates SOX9 using an in vitro assay. The substrates of WWp2 have been shown to be RNA polymerase II (33), tumor suppressor PTEN (phosphatase and tensin homologue deleted on chromosome 10) (34), and Goosecoid (35). First, we inhibited the proteasome degradation by various concentration of bortezomib to see whether SOX9 was involved in the ubiquitin-proteasomal pathway. As shown in Fig. 4B, we observed a strong accumulation of SOX9 in the presence of 10−7 or 10−8 m of bortezomib in HCS-2/8 cells. In contrast, a lower concentration of bortezomib (10−10 and 10−9 m) rather decreased Sox9 concentration (Fig. 4B). Next, we confirmed E6-AP ubiquitination activity toward to SOX9 in vitro using E6-AP protein purified from insect cells, SOX9, biotinylated ubiquitin, E1, UbcH5C (E2), and ATP. Only the reaction mixture containing the complete set of these factors showed a high molecular weight ladder of ubiquitin (Fig. 5A), whereas, in the absence of E6-AP or SOX9, no high molecular bands were seen. These observations strongly suggest that E6-AP functions as a ubiquitin ligase toward SOX9. Further, to confirm this in vitro result, we performed ubiquitination assays using cultured cells. HEK293T cells were cotransfected with FLAG-E6-AP with Xpress-His-SOX9 and HA-ubiquitin. The cell lysates were prepared 48 h after the incubation. When Xpress-His-SOX9 was immunoprecipitated using an anti-Xpress tag antibody, HA-ubiquitin was coprecipitated in the presence of FLAG-E6-AP, showing a weak ladder flanking the strongest band at 110 kDa (Fig. 5B, lane 3). However, a small amount of HA-ubiquitin was also coprecipitated in the absence of FLAG-E6-AP (Fig. 5B, lane 2). The molecular weight of the HA-ubiquitin conjugate was ∼110 kDa. Because the molecular weights of HA-ubiquitin and Xpress-SOX9 are ∼8.5 and 73 kDa, respectively, the strongest band of HA-ubiquitin conjugate (Fig. 5B, lane 3) seemed to be Xpress-SOX9 conjugated with four ubiquitin moieties. When the precipitates obtained by Xpress antibody were further Western blotted with anti-SOX9 antibody, the high molecular weight form of SOX9 was observed. These observations strongly support the notion that E6-AP acts as a ubiquitin ligase toward SOX9.
FIGURE 5.
E6-AP enhances SOX9 ubiquitination in vivo and in vitro. A, ubiquitination of recombinant SOX9 in vitro in the presence of E1, E2 (UbcH5c), biotinylated ubiquitin, ATP, and E6-AP (lane 3). In lane 2, E6-AP was omitted from the reaction mixture. In lane 1, SOX9 was omitted from the reaction mixture. B, ubiquitination of SOX9 in vivo. HEK293 cells were cotransfected with 0.5 μg of pcDNA4/HisMax-SOX9 (lane 1), 0.5 μg of pcDNA4/HisMax-SOX9, plus 1 μg of pcDNA3.1-HA-ubiquitin (HA-Ub) (lane 2), 0.5 μg of pcDNA4/HisMax-SOX9, 1 μg of pcDNA3.1-HA-ubiquitin, and 4 μg of pFLAG-E6-AP (lane 3). Twenty-four hours after transfection, the cells were treated with 2 μm MG132, a proteasome inhibitor, and further incubated for 15 h. After 15 h, the cells were disrupted by sonication, immunoprecipitated (IP) with anti-Xpress antibody to precipitate Xpress-SOX9 complexes, and Western blotted (WB) with anti-HA antibody for detection of ubiquitinated protein (B, left panel) and anti-SOX9 antibody (B, right panel) as described under “Experimental Procedures.” In lane 3, some anti-SOX9-positive bands (polyUb-Sox9) were migrating more slowly than the major SOX9 band.
Colocalization of SOX9 and E6-AP in Cells
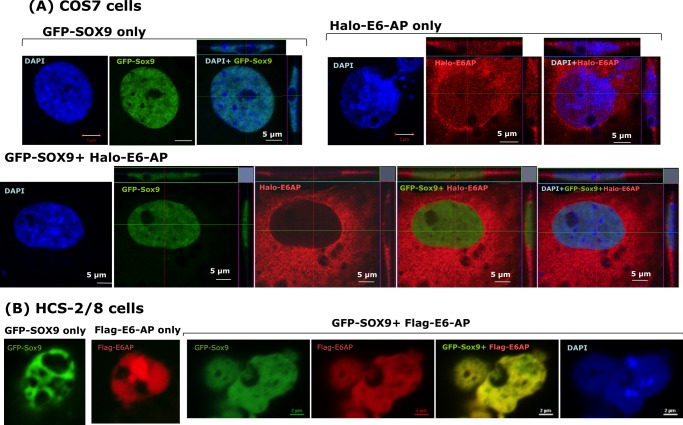
If binding of SOX9 to E6-AP occurs in cells in vivo, SOX9 and E6-AP should colocalize in intact cells. Therefore, we used COS7 cells and HCS-2/8 cells, a human chondrosarcoma cell line that maintains many features of chondrocytes, including a high expression of matrix proteins. COS7 cells or HCS-2/8 cells were transfected with expression plasmids encoding GFP-SOX9 and/or Halo-E6-AP. Under a confocal fluorescence microscope, GFP-SOX9 was observed directly, and Halo-E6-AP was visualized through the TMR ligand. When the cells were transfected with each protein alone or cotransfected with both proteins, subcellular localization was observed (Fig. 6A). Colocalized proteins appeared in yellow when we merged GFP (green) and TMR (red) under a confocal microscope. In COS7 cells, ectopically expressed SOX9 alone displayed a speckled pattern of nuclear distribution, whereas E6-AP displayed a nuclear, perinuclear, and cytosolic distribution. However, coexpression of SOX9 with E6-AP resulted in fused nuclear localization of SOX9, indicating that E6-AP was bound to SOX9 in intact cells. In HCS-2/8 cells, both SOX9 and E6-AP were expressed in nuclei, and most of them were proven to be colocalized, showing a yellow color image (Fig. 6B).
FIGURE 6.
Colocalization of ectopic overexpressed Sox9 and E6-AP in COS7 cells and chondrocytic cell line HCS-2/8 cells. GFP-fused SOX9 and Halo-tagged or FLAG-tagged E6-AP were transiently overexpressed in COS7 cells (A) or HCS-2/8 cells (B) either individually or together. Halo-tagged E6-AP was detected with TMR ligand, and FLAG-tagged E6-AP was detected with anti-FLAG antibody. A, in COS7 cells, SOX9 shows a slightly punctuated subnuclear localization, whereas E6-AP is seen in the nucleus, perinucleus, and cytoplasm when it is expressed alone. In some photos, z axis images of the horizontal and vertical line in the photos are shown. B, in HCS-2/8 cells, SOX9 and E6-AP localize in nucleus and mostly colocalized together.
Knockdown of E6-AP Inhibits SOX9 Degradation
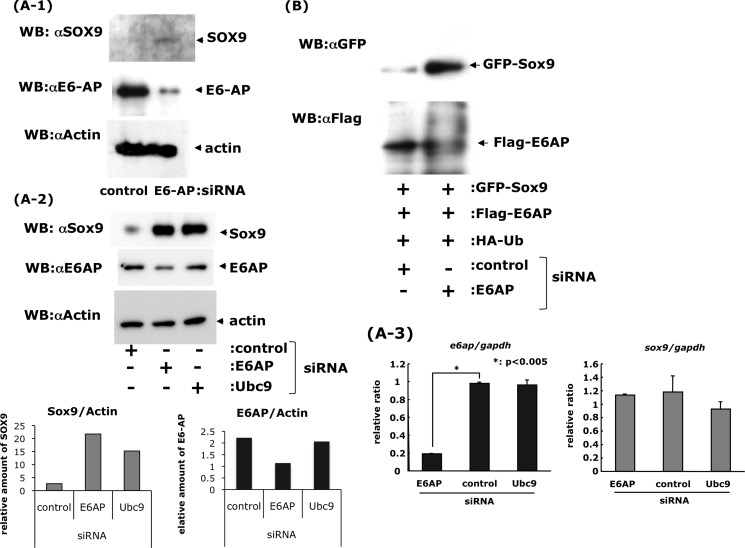
To verify the possibility of endogenous SOX9 degradation by E6-AP in chondrocytes, synthetic, double-stranded siRNAs were designed to target mRNAs encoding E6-AP. When E6-AP siRNA or non-targeting siRNA (as control) was transfected into primary mouse chondrocytes, the levels of E6-AP were decreased, and the levels of SOX9 were increased relative to control siRNA (Fig. 7A-1). These experiments were confirmed using HCS-2/8 cells (Fig. 7A-2). E6-AP, Ubc9, a ubiquitin E2 ligase-encoding gene, and/or control siRNA was transfected into HCS-2/8 cells, and the level of E6-AP was only decreased by E6-AP siRNA, whereas the SOX9 protein level increased by E6-AP and Ubc9 siRNA (Fig. 7A-2) without changing the sox9 mRNA level (A-3). This confirmed that SOX9 degradation occurred at least partially through an E6-AP-mediated proteasomal pathway. Similar experiments were performed using COS7 cells that overexpressed GFP-SOX9, FLAG-E6-AP, and HA-ubiquitin. The cells were treated with E6-AP siRNA or control siRNA, and changes in SOX9 and E6-AP protein levels were detected with antibodies against the GFP and FLAG tag, respectively. Similarly, in primary chondrocytes and HCS-2/8 cells, E6-AP siRNA treatment resulted in lower levels of E6-AP and higher levels of SOX9 than produced by control siRNA treatment (Fig. 7B). These results suggest that SOX9, in chondrocytes or COS7 cells, is subject to degradation by proteasomes after ubiquitination by E6-AP ubiquitin ligase.
FIGURE 7.
Inhibition of E6-AP degradation of Sox9 in vivo by E6-AP siRNA. A-1, primary chondrocytes prepared from the rib cages of newborn mice were treated with control siRNA or E6-AP siRNA for 24 h. Proteins were extracted and analyzed by Western blotting (WB) with specific antibodies against SOX9 and E6-AP. The SOX9 protein level increased after down-regulation of E6-AP with specific siRNA. A-2, HCS-2/8 cells were also treated with control, E6-AP, and Ubc9 siRNA and treated for 48 h. The cells were treated with 1× 10−8 m bortezomib for last 3 h. Both E6-AP and Ubc9 siRNA inhibited SOX9 degradation. Band intensities were measured and standardized to actin. A-3, mRNA was collected after 24-h treatment of siRNA from HCS-2/8 cells, and the mRNA levels of e6ap and sox9 mRNA were measured by real-time PCR and standardized to their GAPDH. The e6ap mRNA was decreased by treatment of E6-AP siRNA compared with the control and Ubc9 siRNA. Sox9 mRNA, however, did not show any significant changes. B, COS7 cells overexpressing GFP-tagged SOX9, FLAG-tagged E6-AP, and HA-tagged ubiquitin were incubated with control siRNA or E6-AP siRNA. The amount of SOX-GFP increased after the addition of E6-AP siRNA, which inhibited ubiquitination.
E6-AP Is Expressed in Hypertrophic Chondrocytes
Previous studies on the localization of SOX9 in cartilage and long bone development in mouse embryos revealed that SOX9 is expressed in cartilage blastema and in early chondrocytes. Furthermore, SOX9 expression is high in proliferating and prehypertrophic cartilage but disappears from hypertrophic chondrocytes and bone (9, 15, 36). To compare the expression pattern of E6-AP with SOX9 during cartilage development in mouse embryos, limb sections from E15.5 were analyzed by immunohistochemistry with specific antibodies against E6-AP and SOX9. In the limb of 15.5-day-old embryos, SOX9 was expressed in resting, proliferating, and prehypertrophic chondrocytes and in the prechondrogenic front but was absent from hypertrophic chondrocytes (Fig. 8). In contrast, E6-AP is expressed in hypertrophic as well as resting and proliferating chondrocytes but absent from SOX9-expressing prehypertrophic chondrocytes and the prechondrogenic front (Fig. 8). This implies that high levels of SOX9 protein in prehypertrophic chondrocytes are consistent with low-level expression of E6-AP in that zone. In hypertrophic chondrocytes, however, E6-AP may contribute to SOX9 degradation by proteasomes following its ubiquitination by E6-AP.
FIGURE 8.
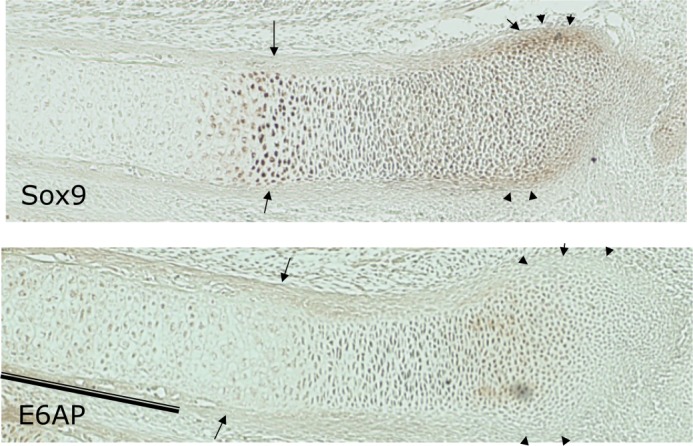
SOX9 and E6-AP expression in developing cartilage. At E15.5 of embryonic development, SOX9 is expressed in the resting, proliferating, and prehypertrophic cartilage zones of a radius, with the strongest expression in prehypertrophic chondrocytes (arrow) and the prechondrogenic front (arrowhead) but not in hypertrophic cartilage. In contrast, E6-AP is also expressed in hypertrophic cartilage (bar) in addition to the resting and proliferating zone but it is absent from the SOX9-expressing prehypertrophic zone (arrows) and the prechondrogenic front (arrowheads).
Deficiency of E6-AP in Neural Tissues of E6-AP-null Mice Is Associated with Elevated Levels of SOX9
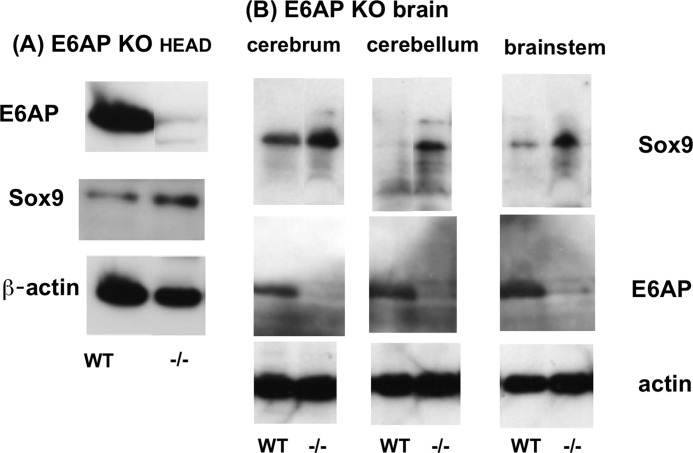
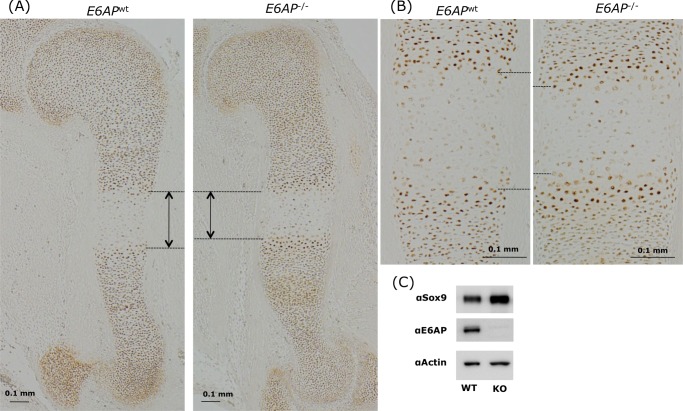
The above findings suggests that SOX9 levels are reduced in tissues of E6-AP-deficient mice. To test this possibility, head tissues, calvariae, brain, and soft tissues from E6-AP-deficient embryos and wild-type littermates were extracted and analyzed for E6-AP and SOX9 using Western blotting. Fig. 9 shows that E6-AP expression was eliminated in E6-AP-deficient tissues, whereas SOX9 was increased relative to wild-type tissues (Fig. 9A). Furthermore, in the cerebrum, brain stem, and cerebellum of E6-AP-deficient adult brains, higher levels of SOX9 had accumulated than in adult wild-type brains (Fig. 9B). These results support our in vitro findings and show that E6-AP reduces the amount of SOX9 in head tissues. In E15.5 humeri, the SOX9-negative hypertrophic zone was slightly, but significantly, shorter in E6-AP-deficient homozygotes (−/−) compared with the wild type (Fig. 10, A and B). The Sox9 amount in primary chondrocytes from E6-AP−/− newborn mice was increased compared with the wild type (Fig. 10C).
FIGURE 9.
Increased levels of SOX9 in the tissues of E6-AP-deficient mice. A, SOX9 protein levels in brain tissues from homozygous E6-AP null mice and WT littermates were analyzed by Western blotting with SOX9 antibody. The amount of SOX9 was increased in brain tissues of E6-AP null mice. B, SOX9 in the brains of adult E6-AP-deficient mice. The cerebrum, cerebellum, and brain stem from a 7-month-old WT and 6-month-old homozygote E6-AP-deficient head were collected and lysed in lysis buffer, and 100 μg of protein was analyzed by Western blotting using specific antibody.
FIGURE 10.
The SOX9-absent hypertrophic zone is shorter in humeri from E6-AP-deficient mice. Embryonic 15-day-old humeri from E6-AP-deficient littermates were stained with anti-SOX9 antibody. Slight but significant differences in the SOX9-absence hypertrophic zone were observed in embryos with WT and homozygote E6-AP deficiencies (A and B). C, protein amount of SOX9 in primary chondrocytes from WT and E6-AP-deficient newborn cartilage. The SOX9 amount was increased in E6-AP chondrocytes as compared with the wild-type.
DISCUSSION
SOX9 is a transcription factor that acts as a key regulator in various stages of cartilage differentiation (2, 12, 13, 37). SOX9 promotes mesenchymal condensation and accelerates chondrocyte growth but inhibits the final maturation of chondrocytes and disappears in bone. In addition to regulating chondrocyte differentiation, SOX9 has been shown to regulate differentiation in various tissues, including the neural crest, hair follicles, testes, pancreas, prostate, and Paneth cells in the small intestine.
Given that SOX9 haploinsufficiencies or overexpression show specific phenotypes, it would seem that levels of SOX9 are tightly regulated in vivo (12, 37). At least part of the regulation of SOX9 levels could be achieved through the ubiquitin-proteasome pathway. Here, we identified E6-AP as a SOX9-binding protein using proteomic analysis. The region of SOX9 necessary for interaction with E6-AP was shown to be SOX9 (96–186), which includes the entire HMG DNA binding domain, SOX9 (104–174) (Fig. 2). With coimmunoprecipitation experiments and immunofluorescence studies we also confirmed the interaction of E6-AP and SOX9 in cultured cells. Furthermore, we demonstrated that E6-AP acts as a specific ubiquitin ligase for SOX9. In contrast, the ubiquitin ligase Wwp2 protein, which also binds to SOX9, cannot ubiquitinate SOX9 (32). This indicates that binding of ubiquitin ligase is necessary, but not sufficient, to ubiquitinate the protein.
These findings suggest that E6-AP may play a major role in the control of SOX9 levels in vivo. We confirmed this hypothesis by analyzing SOX9 expression in developing cartilage and neural tissues of E6-AP/UBE3A null mice. During normal fetal and postnatal bone development, SOX9 mRNA disappears almost completely in the hypertrophic zone (15). Also, SOX9 protein levels decreased rapidly in hypertrophic chondrocytes, where E6-AP/UBE3A is present (Fig. 8). In the growth plate of E6-AP null long bones, the SOX9-negative hypertrophic zone was shorter than in WT littermates. In other words, SOX9 continued to be expressed in the transition zone between prehypertrophic and hypertrophic cartilage longer than in WT bones. This may be the result of delayed proteasomal degradation of SOX9 in the absence of E6AP.
Similarly, the comparison of Sox9 expression in brain tissues from WT and E6-AP/UBE3A knockout mice clearly showed that the absence of E6-AP increased the amount of SOX9 (Fig. 9), indicating again that E6-AP supports proteasomal degradation of SOX9 (Fig. 10). However, it is possible that E6-AP is not the only ligase that ubiquitinates SOX9 and that an unknown ligase might compensate the activity of E6-AP/UBE3A, at least partially, in the knockout mice. In Fig. 4B, when bortezomib was added to HCS-2/8 cells in a low concentration, the SOX9 amount was decreased slightly. Some homeostasis mechanism to keep the Sox9 amount may exist.
SOX9 has been shown to be a key transcription factor that regulates neural and skeletal development. Here, we have shown that it is a specific target for the E6-AP ubiquitin ligase. E6-AP/UBE3A functions as a HECT-type ubiquitin ligase E3. It has been shown to ubiquitinate tumor suppressor protein p53 in the presence of the E6 protein of human papillomavirus types 16 and 18 (38). Furthermore, several substrates have been identified, including HHR23A (39), MCM7 (40), annexin A1 (41), and the promyelocytic leukemia (PML) tumor supressor (42).
Further, E6-AP has been shown to ubiquitinate itself (auto-ubiquitination) and to be degraded by proteasomes (43). The degradation by self-ubiquitination could be a mechanism to regulate E6-AP levels. Posttranslational substrate modification(s), such as phosphorylation, sumoylation, and acetylation, could be important regulators of ubiquitination of E6-AP. Also, SOX9 is subject to these modifications. Therefore, it is important to clarify whether these modifications affect the ubiquitinations of Sox9 to further understand the entire mechanism of SOX9 degradation through ubiquitination.
Acknowledgments
We thank Diane Hackett and Amelia Scholtz (Scientific Publications, University of Texas M. D. Anderson Cancer Center) for editing the manuscript and Drs. Ryuji Kobayashi and David Hawke for technical help in performing peptide sequencing by mass spectrometry.
This work was supported, in whole or in part, by National Institutes of Health grant RO1AR309854 (to B. d. C.). This work was also supported by Scientific Research Grants 21592359 and 25462888 from the Ministry of Education, Culture, and Sports, Japan (to T. H., M. T., and H. Y.).
- HMG
- high mobility group
- E15.5
- embryonic day 15.5
- Ub
- ubiquitin.
REFERENCES
- 1. Schepers G. E., Teasdale R. D., Koopman P. (2002) Twenty pairs of Sox. Extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev. Cell 3, 167–170 [DOI] [PubMed] [Google Scholar]
- 2. Bi W., Deng J. M., Zhang Z., Behringer R. R., de Crombrugghe B. (1999) Sox9 is required for cartilage formation. Nat. Genet. 22, 85–89 [DOI] [PubMed] [Google Scholar]
- 3. Ng L. J., Wheatley S., Muscat G. E., Conway-Campbell J., Bowles J., Wright E., Bell D. M., Tam P. P., Cheah K. S., Koopman P. (1997) SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev. Biol. 183, 108–121 [DOI] [PubMed] [Google Scholar]
- 4. Bridgewater L. C., Lefebvre V., de Crombrugghe B. (1998) Chondrocyte-specific enhancer elements in the Col11a2 gene resemble the Col2a1 tissue-specific enhancer. J. Biol. Chem. 273, 14998–15006 [DOI] [PubMed] [Google Scholar]
- 5. Genzer M. A., Bridgewater L. C. (2007) A Col9a1 enhancer element activated by two interdependent SOX9 dimers. Nucleic Acids Res. 35, 1178–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Han Y., Lefebvre V. (2008) L-Sox5 and Sox6 drive expression of the aggrecan gene in cartilage by securing binding of Sox9 to a far-upstream enhancer. Mol. Cell Biol. 28, 4999–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jenkins E., Moss J. B., Pace J. M., Bridgewater L. C. (2005) The new collagen gene COL27A1 contains SOX9-responsive enhancer elements. Matrix Biol. 24, 177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oh C. D., Maity S. N., Lu J. F., Zhang J., Liang S., Coustry F., de Crombrugghe B., Yasuda H. (2010) Identification of SOX9 interaction sites in the genome of chondrocytes. PloS ONE 5, e10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao Q., Eberspaecher H., Lefebvre V., De Crombrugghe B. (1997) Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev. Dyn. 209, 377–386 [DOI] [PubMed] [Google Scholar]
- 10. Foster J. W., Dominguez-Steglich M. A., Guioli S., Kwok C., Weller P. A., Stevanović M., Weissenbach J., Mansour S., Young I. D., Goodfellow P. N. (1994) Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 372, 525–530 [DOI] [PubMed] [Google Scholar]
- 11. Wagner T., Wirth J., Meyer J., Zabel B., Held M., Zimmer J., Pasantes J., Bricarelli F. D., Keutel J., Hustert E., Wolf U., Tommerup N., Schempp W., Scherer G. (1994) Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79, 1111–1120 [DOI] [PubMed] [Google Scholar]
- 12. Akiyama H., Chaboissier M. C., Martin J. F., Schedl A., de Crombrugghe B. (2002) The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16, 2813–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bi W., Huang W., Whitworth D. J., Deng J. M., Zhang Z., Behringer R. R., de Crombrugghe B. (2001) Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc. Natl. Acad. Sci. U.S.A. 98, 6698–6703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Akiyama H., Lyons J. P., Mori-Akiyama Y., Yang X., Zhang R., Zhang Z., Deng J. M., Taketo M. M., Nakamura T., Behringer R. R., McCrea P. D., de Crombrugghe B. (2004) Interactions between Sox9 and β-catenin control chondrocyte differentiation. Genes Dev. 18, 1072–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hattori T., Müller C., Gebhard S., Bauer E., Pausch F., Schlund B., Bösl M. R., Hess A., Surmann-Schmitt C., von der Mark H., de Crombrugghe B., von der Mark K. (2010) SOX9 is a major negative regulator of cartilage vascularization, bone marrow formation and endochondral ossification. Development 137, 901–911 [DOI] [PubMed] [Google Scholar]
- 16. Stolt C. C., Lommes P., Sock E., Chaboissier M. C., Schedl A., Wegner M. (2003) The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 17, 1677–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vidal V. P., Chaboissier M. C., Lützkendorf S., Cotsarelis G., Mill P., Hui C. C., Ortonne N., Ortonne J. P., Schedl A. (2005) Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr. Biol. 15, 1340–1351 [DOI] [PubMed] [Google Scholar]
- 18. Chaboissier M. C., Kobayashi A., Vidal V. I., Lützkendorf S., van de Kant H. J., Wegner M., de Rooij D. G., Behringer R. R., Schedl A. (2004) Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development 131, 1891–1901 [DOI] [PubMed] [Google Scholar]
- 19. Seymour P. A., Freude K. K., Tran M. N., Mayes E. E., Jensen J., Kist R., Scherer G., Sander M. (2007) SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc. Natl. Acad. Sci. U.S.A. 104, 1865–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thomsen M. K., Butler C. M., Shen M. M., Swain A. (2008) Sox9 is required for prostate development. Dev. Biol. 316, 302–311 [DOI] [PubMed] [Google Scholar]
- 21. Mori-Akiyama Y., van den Born M., van Es J. H., Hamilton S. R., Adams H. P., Zhang J., Clevers H., de Crombrugghe B. (2007) SOX9 is required for the differentiation of Paneth cells in the intestinal epithelium. Gastroenterology 133, 539–546 [DOI] [PubMed] [Google Scholar]
- 22. Akiyama H., Kamitani T., Yang X., Kandyil R., Bridgewater L. C., Fellous M., Mori-Akiyama Y., de Crombrugghe B. (2005) The transcription factor Sox9 is degraded by the ubiquitin-proteasome system and stabilized by a mutation in a ubiquitin-target site. Matrix Biol. 23, 499–505 [DOI] [PubMed] [Google Scholar]
- 23. Hattori T., Eberspaecher H., Lu J., Zhang R., Nishida T., Kahyo T., Yasuda H., de Crombrugghe B. (2006) Interactions between PIAS proteins and SOX9 result in an increase in the cellular concentrations of SOX9. J. Biol. Chem. 281, 14417–14428 [DOI] [PubMed] [Google Scholar]
- 24. Bedford L., Paine S., Sheppard P. W., Mayer R. J., Roelofs J. (2010) Assembly, structure, and function of the 26S proteasome. Trends Cell Biol. 20, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ciechanover A., Gonen H. (1990) The ubiquitin-mediated proteolytic pathway: enzymology and mechanisms of recognition of the proteolytic substrates. Semin. Cell Biol. 1, 415–422 [PubMed] [Google Scholar]
- 26. Dikic I., Robertson M. (2012) Ubiquitin ligases and beyond. BMC Biol. 10, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Riemer S., Gebhard S., Beier F., Pöschl E., von der Mark K. (2002) Role of c-fos in the regulation of type X collagen gene expression by PTH and PTHrP. Localization of a PTH/PTHrP-responsive region in the human COL10A1 enhancer. J. Cell. Biochem. 86, 688–699 [DOI] [PubMed] [Google Scholar]
- 28. Honda R., Tanaka H., Yasuda H. (1997) Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27 [DOI] [PubMed] [Google Scholar]
- 29. Takigawa M., Tajima K., Pan H. O., Enomoto M., Kinoshita A., Suzuki F., Takano Y., Mori Y. (1989) Establishment of a clonal human chondrosarcoma cell line with cartilage phenotypes. Cancer Res. 49, 3996–4002 [PubMed] [Google Scholar]
- 30. Lefebvre V., Huang W., Harley V. R., Goodfellow P. N., de Crombrugghe B. (1997) SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro α1(II) collagen gene. Mol. Cell Biol. 17, 2336–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miura K., Kishino T., Li E., Webber H., Dikkes P., Holmes G. L., Wagstaff J. (2002) Neurobehavioral and electroencephalographic abnormalities in Ube3a maternal-deficient mice. Neurobiol. Dis. 9, 149–159 [DOI] [PubMed] [Google Scholar]
- 32. Nakamura Y., Yamamoto K., He X., Otsuki B., Kim Y., Murao H., Soeda T., Tsumaki N., Deng J. M., Zhang Z., Behringer R. R., Crombrugghe B., Postlethwait J. H., Warman M. L., Nakamura T., Akiyama H. (2011) Wwp2 is essential for palatogenesis mediated by the interaction between Sox9 and mediator subunit 25. Nat. Commun. 2, 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H., Zhang Z., Wang B., Zhang J., Zhao Y., Jin Y. (2007) Wwp2-mediated ubiquitination of the RNA polymerase II large subunit in mouse embryonic pluripotent stem cells. Mol. Cell Biol. 27, 5296–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maddika S., Kavela S., Rani N., Palicharla V. R., Pokorny J. L., Sarkaria J. N., Chen J. (2011) WWP2 is an E3 ubiquitin ligase for PTEN. Nat. Cell Biol. 13, 728–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zou W., Chen X., Shim J. H., Huang Z., Brady N., Hu D., Drapp R., Sigrist K., Glimcher L. H., Jones D. (2011) The E3 ubiquitin ligase Wwp2 regulates craniofacial development through mono-ubiquitylation of Goosecoid. Nat. Cell Biol. 13, 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Murakami S., Lefebvre V., de Crombrugghe B. (2000) Potent inhibition of the master chondrogenic factor Sox9 gene by interleukin-1 and tumor necrosis factor-α. J. Biol. Chem. 275, 3687–3692 [DOI] [PubMed] [Google Scholar]
- 37. de Crombrugghe B., Lefebvre V., Nakashima K. (2001) Regulatory mechanisms in the pathways of cartilage and bone formation. Curr. Opin. Cell Biol. 13, 721–727 [DOI] [PubMed] [Google Scholar]
- 38. Huibregtse J. M., Scheffner M., Howley P. M. (1993) Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol. Cell Biol. 13, 4918–4927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kumar S., Talis A. L., Howley P. M. (1999) Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J. Biol. Chem. 274, 18785–18792 [DOI] [PubMed] [Google Scholar]
- 40. Kühne C., Banks L. (1998) E3-ubiquitin ligase/E6-AP links multicopy maintenance protein 7 to the ubiquitination pathway by a novel motif, the L2G box. J. Biol. Chem. 273, 34302–34309 [DOI] [PubMed] [Google Scholar]
- 41. Shimoji T., Murakami K., Sugiyama Y., Matsuda M., Inubushi S., Nasu J., Shirakura M., Suzuki T., Wakita T., Kishino T., Hotta H., Miyamura T., Shoji I. (2009) Identification of annexin A1 as a novel substrate for E6AP-mediated ubiquitylation. J. Cell. Biochem. 106, 1123–1135 [DOI] [PubMed] [Google Scholar]
- 42. Louria-Hayon I., Alsheich-Bartok O., Levav-Cohen Y., Silberman I., Berger M., Grossman T., Matentzoglu K., Jiang Y. H., Muller S., Scheffner M., Haupt S., Haupt Y. (2009) E6AP promotes the degradation of the PML tumor suppressor. Cell Death Differ. 16, 1156–1166 [DOI] [PubMed] [Google Scholar]
- 43. Nuber U., Schwarz S. E., Scheffner M. (1998) The ubiquitin-protein ligase E6-associated protein (E6-AP) serves as its own substrate. Eur. J. Biochem. 254, 643–649 [DOI] [PubMed] [Google Scholar]