Background: Chlamydomonas contains six chloroplast ferredoxins (FDXs) whose function is still unclear.
Results: A global FDX interactome was obtained where FDX1 has a predominant role and is the most relevant electron donor to FNR1 and HYDA1.
Conclusion: FDXs have distinct but also overlapping function.
Significance: We discovered new FDX interaction partners and specific roles for each FDX isoform.
Keywords: Bioenergetics/Electron Transfer Complex, Chlamydomonas, Energy Metabolism, Protein-Protein Interactions, Redox, Ferredoxin, Network
Abstract
Ferredoxins (FDXs) can distribute electrons originating from photosynthetic water oxidation, fermentation, and other reductant-generating pathways to specific redox enzymes in different organisms. The six FDXs identified in Chlamydomonas reinhardtii are not fully characterized in terms of their biological function. In this report, we present data from the following: (a) yeast two-hybrid screens, identifying interaction partners for each Chlamydomonas FDX; (b) pairwise yeast two-hybrid assays measuring FDX interactions with proteins from selected biochemical pathways; (c) affinity pulldown assays that, in some cases, confirm and even expand the interaction network for FDX1 and FDX2; and (d) in vitro NADP+ reduction and H2 photo-production assays mediated by each FDX that verify their role in these two pathways. Our results demonstrate new potential roles for FDX1 in redox metabolism and carbohydrate and fatty acid biosynthesis, for FDX2 in anaerobic metabolism, and possibly in state transition. Our data also suggest that FDX3 is involved in nitrogen assimilation, FDX4 in glycolysis and response to reactive oxygen species, and FDX5 in hydrogenase maturation. Finally, we provide experimental evidence that FDX1 serves as the primary electron donor to two important biological pathways, NADPH and H2 photo-production, whereas FDX2 is capable of driving these reactions at less than half the rate observed for FDX1.
Introduction
Ferredoxins (FDXs)3 are [FeS]-containing proteins that are widely distributed in micro-organisms, plants, and animals. They serve as electron shuttles and take part in diverse redox-driven metabolic pathways. In plants and photosynthetic micro-organisms, FDXs usually harbor [2Fe-2S] clusters and act as central molecules distributing electrons originating from photosynthetic water oxidation and fermentation to specific FDX-dependent enzymes in the chloroplast. FDXs can also be involved in redox reactions linked to hydroxylases, oxygenases, desaturases and oxidoreductases (1). In higher plants, FDXs are phylogenetically grouped into leaf and root isoforms, which differ in their redox potentials; the former typically have more negative redox potentials (2). Functionally, the differences in redox potentials between the different isoforms correlate with higher rates of NADP+ photo-reduction by leaf-type FDXs and efficient oxidation of NADPH by root-type FDXs (3, 4). Plant-type FDXs present in higher plants, algae, and cyanobacteria are proteins with a molecular mass ranging from about 10 to 35 kDa. They serve as low potential, single electron carriers with Em values ranging from −310 to −455 mV (5–9). In general, these FDXs are acidic proteins with three solvent-exposed flexible regions responsible for mediating protein-protein interactions with binding partners (10). The amino acid motif required for [2Fe-2S] cluster ligation (CX4CX2CXnC) forms a highly conserved and rigid structure that facilitates cluster insertion to the apoprotein and electron transfer to/from the mature protein (10, 11). The production of NADPH by photosynthetically reduced FDX provides a reductant for important pathways such as nitrogen fixation, sulfate assimilation, and glutamate synthesis via either nitrite reductase, sulfite reductase, or glutamate synthase (12).
In Chlamydomonas reinhardtii (hereafter referred to as Chlamydomonas), the FDX genes are encoded in the nucleus, translated in the cytosol, and translocated into the chloroplast stromal space (13). The actual maturation process (insertion of the [2Fe2S] cluster) is not known yet. In plants, FDX apoproteins are translocated (probably via an ATP- or GTP-dependent process) into the chloroplast, where they undergo signal peptide cleavage, which generates the mature and active protein (14, 15). Analysis of the Chlamydomonas genome revealed at least six chloroplast FDX genes (PETF, referred to as FDX1 in this work, and FDX2 through FDX6). All six genes encode proteins with typical [2Fe-2S]-binding domains, containing the conserved cysteine residues required for [FeS] cluster ligation. Phylogenetic analysis indicates that FDX1 and FDX5 are most closely related to photosynthetic leaf-type FDXs, whereas FDX2 appears to be a nonphotosynthetic root-type FDX (16). FDX3, FDX4, and FDX6 phylogenetically cluster with FDXs of unknown function, as well as with cyanobacterial FDXs. It is thought that this alternative lineage may indicate evolution into specialized functions independent of those of typical leaf- or root-type FDXs (16).
All six FDXs have putative chloroplast transit peptides. The transit peptide cleavage sites for FDX1 and FDX5 have been determined by protein sequencing (17, 18). In addition, Western blot analysis of chloroplast fractions provided evidence that five FDXs (with the exception of FDX4) are localized to the chloroplast (16, 17). Transcript abundance analysis indicates that FDX1 is the predominant isoform transcript found in mid-log phase Chlamydomonas cells grown in standard Tris acetate/phosphate (TAP) medium (16). Interestingly, both FDX2 and FDX5 transcripts are induced during anaerobic growth and in the presence of added nickel (17, 19). Moreover, FDX2 is also up-regulated when cells are grown on nitrate rather than ammonium or when they are exposed to H2O2 (16). Finally, FDX5 has been shown to be induced during copper- and sulfur-deficient growth conditions (16, 17, 20).
Thus far, there is little direct evidence to indicate the range and specificity of FDX interactions with specific metabolic pathways. Of particular interest is the linkage of FDXs to three distinct hydrogen evolution pathways, including two photo-production pathways (PSII-dependent and PSII-independent, also known as photo-fermentation), and one dark fermentative pathway in Chlamydomonas (21). FDXs represent the physiological electron donor for hydrogenases in all known hydrogen production pathways (22), and FDX1 has been proposed to be the sole electron donor during Chlamydomonas hydrogen production (17, 23). The Chlamydomonas genome encodes two [FeFe] hydrogenase enzymes, HYDA1 and HYDA2, that catalyze the reduction of two protons to hydrogen in all three pathways. In silico docking analysis and site-directed mutagenesis combined with in vitro hydrogen production assays have identified two amino acid residues (HYDA1 Lys-396 and FDX1 Glu-122) thought to mediate the electrostatic interaction between the two proteins (23, 24). However, these site-directed mutagenesis and in silico docking studies were only informed by homology models of the Chlamydomonas hydrogenases and ferredoxins. Nevertheless, these studies represent the best investigation of FDX and hydrogenase interaction thus far, and they suggest that HYDA1 Lys-396 and FDX1 Glu-122 (amino acid numbers represent the position in the apoprotein) make major contributions to the formation of an electron transfer complex (23). The two Chlamydomonas hydrogenases share conservation of the required lysine, and five FDXs (all except FDX3) have the conserved glutamic acid. However, a rigorous structural examination of FDX-hydrogenase complexes has not been performed due to the lack of crystal structures for the respective Chlamydomonas proteins.
Here, we present data from yeast two-hybrid assays, using each FDX as bait to screen against a Chlamydomonas cDNA library. Although extensively used to screen in vivo protein binding partners, the yeast two-hybrid assay has limitations (25) and can yield false positives and false negatives. Furthermore, some transcripts present at low levels in the library might not be detected, and interactions that require folding/assembly of Chlamydomonas proteins in yeast might not be efficient. Despite its limitations, the results from the assay allowed us to start highlighting components of the FDX interactome.
Each of the FDXs was also examined in pairwise yeast two-hybrid interaction assays with proteins from relevant biochemical pathways to further validate interactions known to occur, from the literature or detected through the library screening. In addition, we developed a pulldown assay to further characterize the in vivo binding partners of FDX1, given the above-mentioned limitations of the yeast two-hybrid assay.
Finally, we examined the in vitro production of two important FDX-dependent products (NADPH and hydrogen), as mediated by each of the FDX isoforms. Taken together, these data provide a preliminary map of the global interaction networks for each of the Chlamydomonas FDXs.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
C. reinhardtii CC-124 cells were grown in TAP medium, pH 7.2. Algal cultures were maintained at 25 °C, vigorously bubbled with air enriched with 3% CO2, stirred using a magnetic stir bar, and illuminated with continuous light of 80 μmol photon m−2 s−1 photosynthetically active radiation at the surface of 1-liter Roux culture bottles (255 × 55 × 120 mm). Cell densities ranged from 1 × 105 to 3 × 106 cells ml−1 of culture. Aerobic cell cultures were harvested at 16–24 μg ml−1 total chlorophyll by centrifugation and were immediately frozen in liquid nitrogen. For anaerobic induction, Chlamydomonas cultures were grown and harvested as above and resuspended in one-tenth the volume (50 ml) of anaerobic induction buffer containing 50 mm potassium phosphate, pH 7.2, and 3 mm MgCl2 (26). The cells were then transferred to a sealed anaerobic vial in the dark and sparged with argon for 30 min. To measure fermentative hydrogen production, 400 μl of head-space gas was withdrawn from the sealed anaerobic vials and analyzed by gas chromatography (Hewlett Packard 5890 series II) using a Supelco column (60/80 mol sieve 5A 6 feet × 1/8 inch) coupled to a TCD detector. After 2 h of anaerobic induction, the cells were harvested by centrifugation and immediately frozen in liquid nitrogen.
RNA Extraction
Cells were grown as above, and RNA was extracted from dark anaerobically adapted cells using the method described in Ref. 19.
Chlorophyll Measurements
Chlorophyll a and b contents were determined spectrophotometrically in 95% ethanol (27).
Yeast Two-hybrid cDNA Library Screening
Yeast two-hybrid screening was performed by Hybrigenics (Paris, France). The DNA coding sequences for each FDX were PCR-amplified and cloned into pB27 as C-terminal fusions to LexA (N-LexA-FDX1-C). Each construct was checked by sequencing the entire insert and used as a bait to screen a random-primed Chlamydomonas cDNA library. For the construction of the cDNA library, 1.2 mg of total RNA isolated from Chlamydomonas cells grown under dark anaerobiosis (to include hydrogenase-related transcripts) were used for the extraction of messenger RNA. In total, 7.6 μg of poly(A)+ RNA were obtained using the IllustraTM mRNA purification kit (GE Healthcare). The poly(A)+ RNA (5 μg) was used as template for the cDNA synthesis (just cDNA double-stranded cDNA synthesis kit, Stratagene). A linker containing SfiI restriction sites was ligated to the products of the cDNA synthesis. The ligation reaction was then purified by a size fractionation column to remove free linkers and fragments smaller than 200 nucleotides. A large scale ligation of cDNA fragments amplified from Chlamydomonas poly(A)+ RNA was performed in the yeast two-hybrid prey vector pP6, downstream of the Gal4 activation domain (28). The plasmid DNA of 50 million independent clones was harvested and transformed into yeast. Around 11 million independent clones were obtained, corresponding to over 10 million clones containing Chlamydomonas mRNA sequences. The clones were then harvested, and aliquots of the library were stored at −80 °C, allowing 1 aliquot of the same library to be used for each screen. A total of 62 million clones (6-fold the complexity of the library) were screened, using a mating approach with Y187 and L40ΔGal4 yeast strains, as described previously (29). His+ colonies were selected on a medium lacking tryptophan, leucine, and histidine. The prey fragments present in the positive clones were amplified by PCR and sequenced at their 5′ and 3′ junctions. The resulting sequences were used to identify the corresponding interacting proteins in GenBankTM. Computationally defined confidence scores (Predicted Biological Score) were assigned to all of the identified clones.
Interaction Scoring
The method for calculating the Predicted Biological Score (PBS), previously described for genomic libraries (30), has been adapted for randomly primed cDNA libraries (31). Briefly, the PBS takes into account the redundancy and independency of prey fragments (number of times an interaction was observed with one given bait fragment), the distributions of reading frames and stop codons in overlapping fragments, and the local topology of the interaction network, including the presence of (a) highly connected prey regions and (b) interactions confirmed by two independent screens in the bait/prey and prey/bait orientations. The PBS E value ranges from 0 to 1 and has been classified in five distinct categories, A–E, with A representing very high confidence and E representing not assigned scores. Intercategory thresholds were chosen manually with respect to a training data set containing known true-positive and false-positive interactions as follows: A < 1e−10 < B < 1e−5 < C < 1e−2.5 < D < 1. The E category corresponds to prey domains nonspecifically selected by baits for which the corresponding PBS has been set to 1, and most probably it represents false positives due to cloning artifacts (out of frame or antisense sequences). The latter should be confirmed independently to substantiate the potential interaction. Categories A, B, and C represent probable true-positive interactions at different levels of confidence. In between, the D category gathers protein interactions detected by only one prey fragment for each given bait; it may represent false positives (prey fragment selected nonspecifically by the bait) or interesting rare events (interaction with a protein encoded by a rare mRNA or interaction difficult to detect in classical two-hybrid assays). Those categories have been shown to be positively correlated to the biological significance of interactions (30, 32).
Yeast Two-hybrid Pairwise Interactions
Gene sequences were codon-optimized (Mr. Gene, Germany) for expression in yeast. All inserts were amplified with Advantage HD polymerase (Clontech) and cloned into desired yeast two-hybrid vectors (DualSystems Biotech, Switzerland) using the In-Fusion Advantage PCR cloning kit (Clontech), and the inserts were fully sequenced (Davis Sequencing). The PCR primers (IDT) used to amplify each gene are compiled in supplemental Table S1. Yeast strain NMY51 was transformed using the DSY yeast transformation kit (DualSystems, Biotech, Switzerland). Transgenic yeast strains were maintained on Synthetic Dropout (SD) media lacking appropriate nutrients (Leu, leucine; Trp, tryptophan; His, histidine, Ura, uracil). In some cases, the inhibitor of the HIS3 enzyme, 3-amino-1,2,4-triazole (3AT), was also included. Growth assays were performed by resuspending cells in sterile double distilled H2O at an A600 of 0.5. Four 1:5 serial dilutions were created and replica-plated onto appropriate SD agar plates using a 32-spoke inoculating manifold (Dan-Kar, Reading, MA). Cells were maintained at 30 °C until growth was observed.
Protein Overexpression and Purification
The DNA sequence corresponding to the mature FDX1 protein of Chlamydomonas was cloned into a modified pRSET-A expression vector (33). The PCR fragment was amplified from the pMAPetF plasmid that contained a codon-optimized version (Mr. Gene) of the FDX1 cDNA using Phusion DNA polymerase (New England Biolabs) and primers FDX1E-S-Fw GGATCCTATAAAGTCACCTTGAAAACCCCATCCGG and FDX1E-Rv GAATTCTCAATACAAGGCTTCTTCTTGATGGGTTTGAATT. After double digestion with EcoRI and BamHI, the fragment was ligated (Quick Ligation kit, New England Biolabs) into the EcoRI/BamHI-linearized pRSET-A vector. The resulting plasmid was transformed into KRX cells (Promega). For expression in Escherichia coli, a 5-ml Terrific Broth (VWR) starter culture was grown overnight at 37 °C and diluted 1:100 in 100 ml of fresh Terrific Broth the following morning. After the sub-culture had reached an A600 of ∼0.4, 10 ml of it were used to inoculate a 1-liter Terrific Broth culture (supplemented with 0.4% (w/v) glycerol). Once the 1-liter culture had reached an A600 of ∼0.6, l-rhamnose and ferric ammonium citrate were added to final concentrations of 0.05% (w/v) and 2.5 mm, respectively. The induced culture was then incubated overnight at RT on a shaker. The next day, cells were harvested and resuspended in 50 ml of lysis buffer (50 mm NaPO4 pH 7.0, 300 mm NaCl, 5% (w/v) glycerol) to which 1 tablet of complete protease inhibitor (Roche Applied Science), 100 μl of 50 mg ml−1 lysozyme (Sigma), and 250 μg of DNase I (Thermo Scientific) were added. The cells were then broken by two passages through a French press cell (1200 p.s.i.; American Instrument Co.), and the cell lysate was incubated in a water bath for 15 min at 55 °C. Subsequently, cell debris was pelleted out in an ultracentrifuge (Optima XL-100k ultracentrifuge, Beckman Coulter) at 100,000 × g and 4 °C for 1 h. The supernatant was incubated for 1 h at 4 °C with 5 ml of Talon metal affinity resin (Clontech). After the incubation period, the resin was washed with 20 column volumes (CV) of lysis buffer, 10 CV of lysis buffer containing 10 mm imidazole, and 10 CV of lysis buffer containing 20 mm imidazole. Protein elution was performed with 2 CV of lysis buffer containing 300 mm imidazole. Eluted FDX was concentrated to a final volume of 2 ml using 5-kDa molecular mass cutoff concentrators (Satorius Stedim Biotech). The 2-ml sample was then loaded on a HiLoadTM 26/60 SuperdexTM 75 prep grade (GE Healthcare) size exclusion column coupled to an Äkta FPLC (GE Healthcare) and using 25 mm NaPO4, pH 7.0, 100 mm NaCl, and 5% (w/v) glycerol as the running buffer at a flow rate of 2 ml min−1. The run was monitored at A280 nm, and the main peak was eluted after ∼195 ml. The pooled fractions (185–205 ml) were concentrated again, and the final concentration was determined using the DC protein assay (Bio-Rad) according to the manufacturer's instructions.
Chlamydomonas FDX2 and FDX3 were purified using the same procedure as for FDX1, although the above protocols were modified slightly for the purifications of FDX4, FDX5, and FNR1. Our attempts to purify FDX6 were unsuccessful. FDX4 was bound, washed, and eluted from Talon metal affinity resin using an imidazole gradient elution and an Äkta FPLC (GE Healthcare) rather than by the batch method described above. For FDX5, all steps after cell harvesting were performed anaerobically in a Coy chamber. For FNR1, the protein was released from the Talon metal affinity resin by overnight incubation at 4 °C with 500 units of thrombin. HYDA1 (from C. reinhardtii) was heterologously expressed in E. coli and purified as described previously (34, 35). Purification was carried out under strict anaerobic conditions in either a Coy Chamber or a MBRAUN glove box. Hydrogenase activity of pure HYDA1 was verified using the reduced methyl viologen assay as earlier reported (34).
Pulldown Experiment
FDX coupled to cyanogen bromide-activated Sepharose beads (GE Healthcare; 5 mg of protein per ml of resin) were prepared according to the manufacturer's instructions. Cell extract generation and pulldown experiments were performed under anaerobic conditions in a Coy Chamber. Typically, cells harvested from 150 ml of culture were resuspended in 10 ml of ACA buffer (50 mm ϵ-aminocaproic acid, 50 mm BisTris/HCl, pH 7.0, 0.5 mm EDTA, 1 complete protease inhibitor tablet per 50 ml of ACA) and passed twice through a cell disruptor (Aero Magics) at 40,000 p.s.i. After cell breakage, debris and unbroken cells were pelleted out (3000 × g, 5 min, 4 °C), and the supernatant was adjusted to 15 ml. Prior to incubation with cell extracts, 200 μl of FDX beads for each sample were washed once with 1 ml of ACA buffer plus 1 mm sodium dithionite (to pre-reduce FDX) and then equilibrated twice with 1 ml of ACA buffer. Washed FDX were incubated with 1.5 ml of cell extracts for 1 h on ice, and the beads were then washed with washing buffer (25 mm Tris, pH 7.5, 50 mm NaCl, 2 mm EDTA) until the flow-through was clear. Elution of FDX was typically performed with 200 μl of elution buffer (50 mm Tris, pH 6.8, 50 mm DTT, 1% (w/v) SDS). Protein concentration was determined using an RC/DC protein assay kit (Bio-Rad) according to the manufacturer's instructions.
Mass Spectrometry Analysis
Protein samples (25 μg) were acetone-precipitated, dried, and solubilized with 8 m urea and 0.1% ProteaseMAX surfactant (Promega) in 25 mm ammonium bicarbonate. Cysteines were reduced with DTT and alkylated with iodoacetamide. Proteins were then digested with 1.8 μg of trypsin at 37 °C for 3 h, followed by precipitation of surfactant with 0.5% TFA. The resulting peptide mixtures were desalted with PepClean C18 spin columns (Pierce). The resulting peptide mixtures were analyzed by reversed-phase chromatography using a Waters nanoAcquity UPLC pump coupled on line to an LTQ/Orbitrap mass spectrometer. Peptides were loaded onto a Symmetry 180-μm × 2-cm C18 5.0-μm trap column, which was washed and placed in line with a Waters BEH130 C18 analytical column (75-μm × 25-cm, 1.7-μm beads). As the peptides elute off the column, they are ionized into the gas phase of the mass spectrometer. The peptide ion masses are measured with resolution of 60,000 at m/z 400 by the Orbitrap, followed by MS/MS fragmentation of the six most intense precursors with charge state ≥2 above an intensity threshold of 10,000 in the LTQ ion trap. Dynamic exclusion was used to avoid repeat sequencing, with a repeat count of one and exclusion duration of 30 s for precursors within 10 ppm. The MS/MS spectra were extracted and searched against the C. reinhardtii protein database version 4 from the Department of Energy Joint Genome Institute (36) using the MASCOT search engine, version 2.2 (Matrix Science), with a mass tolerance on precursor ions of 25 ppm and 0.5 Da for fragment ions. The following variable modifications were considered: carbamidomethyl-Cys, methionine oxidation, and pyro-glutamic acid for N-terminal Gln. Peptide identifications were accepted at a 1% false discovery rate threshold based on a reversed protein database search. Protein assembly and label-free quantification by spectral counting were performed using Isoform Resolver (37), and protein abundance differences between FDX pulldowns and controls were evaluated using a G-test (38).
NADPH Photo-production
Buffer A (0.9 ml, 50 mm Tris-HCl, pH 7.4, 3.35 mg ml−1 bovine serum albumin, 10 mm MgCl2, 200 mg ml−1 sucrose) was added to a 1-ml cuvette with 2 μl of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (0.3 mm in DMSO), 5 μl of dichlorophenolindophenol (0.01 mm), and 10 μl of sodium ascorbate (1 m). After mixing, 10 μm FDX, 1 μm FNR1 (from Chlamydomonas), and purified thylakoids (25 μg) were added to the solution in the dark. After mixing, 50 μl of NADP+ (0.04 mm) was added, and the cuvette was sealed with parafilm. The reaction was performed a using halogen lamp as a light source. The concentration of NADPH was determined by the change of absorbance at 340 nm measured by the Cary 4000 UV-visible spectrometer (Agilent Technologies) using an extinction coefficient of 6.22 mm−1.
Hydrogen Photo-production
Buffer A (0.9 ml, 50 mm Tris-HCl, pH 7.4, 3.35 mg ml−1 bovine serum albumin, 10 mm MgCl2, 200 mg ml−1 sucrose) was added to a 15-ml serum vial with 2 μl of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (0.3 mm in DMSO), 5 μl of dichlorophenolindophenol (0.01 mm), 10 μl of sodium ascorbate (1 m), 10 μl of glucose oxidase (30 mg/ml), 50 μl of glucose (1 m), 10 μl of catalase (10 mg ml−1), and 50 μl of 96% ethanol. After mixing, purified FDX was added to a final concentration of 10 μm. If appropriate, 50 μl of NADP+ (0.04 mm) was added, followed by purified FNR1 to a final concentration of 1 μm. Only 25 μg of purified thylakoids were added to the solution in the dark. Serum vials were sealed with rubber septa, and the vial was wrapped in foil, followed by argon sparging for 15 min. HYDA1 was added to a final concentration of 100 nm via a gas-tight syringe, and the 0 time point was read using a gas by a gas chromatograph. The reaction was exposed to light from a halogen lamp for 20 h, and headspace samples were drawn at different time points.
RESULTS
Yeast Two-hybrid Library Screen
To obtain an interaction network for Chlamydomonas chloroplast FDXs, we utilized yeast two-hybrid screens, with each FDX isoform as bait and a Chlamydomonas cDNA library as prey. The cDNA library was constructed from dark anaerobic adapted samples (as described under “Experimental Procedures”). We expect Chlamydomonas FDX proteins to mature correctly in yeast, because yeast harbors [2Fe-2S] FDXs and the maturation factors required for their activity (39).
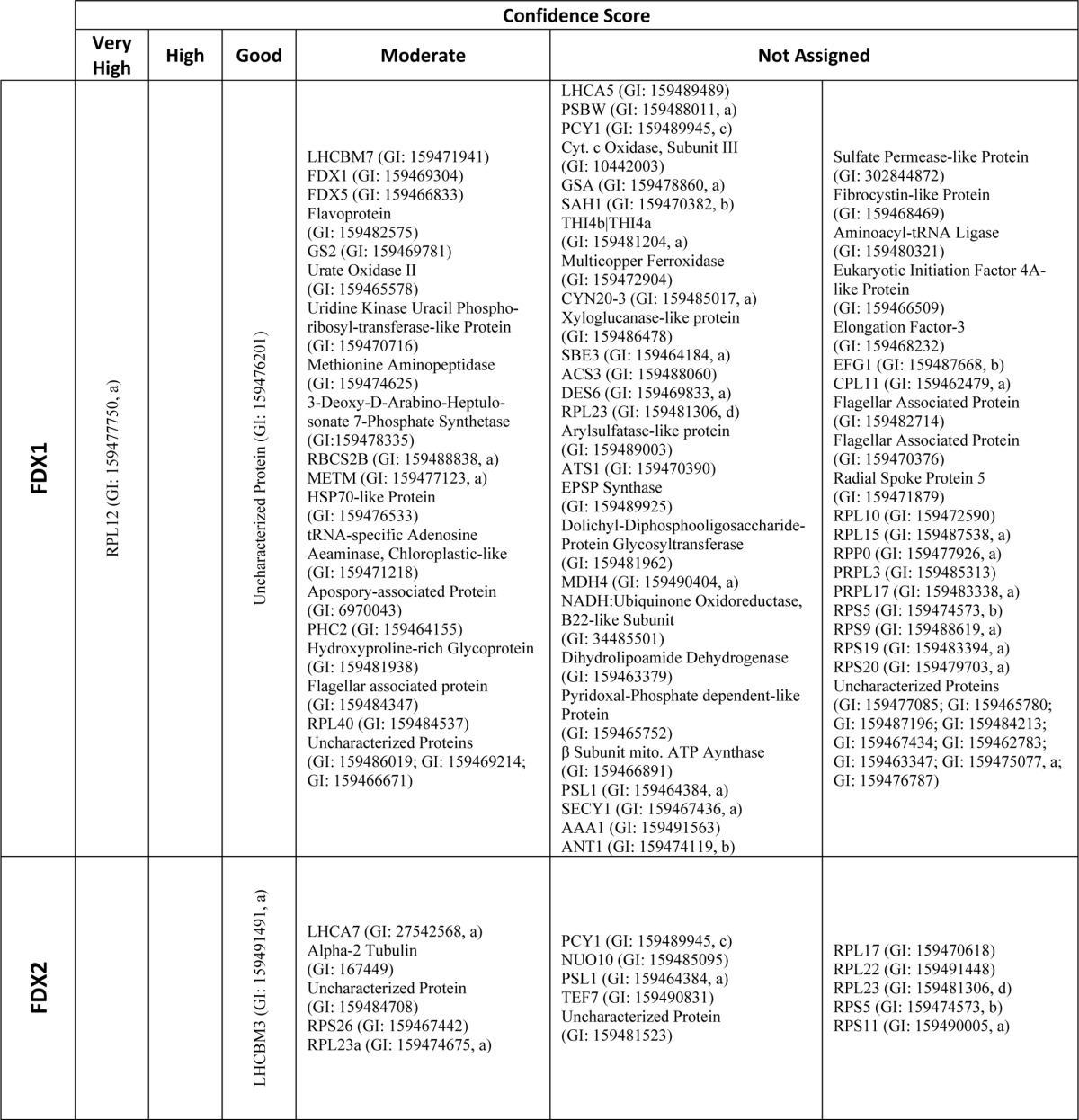
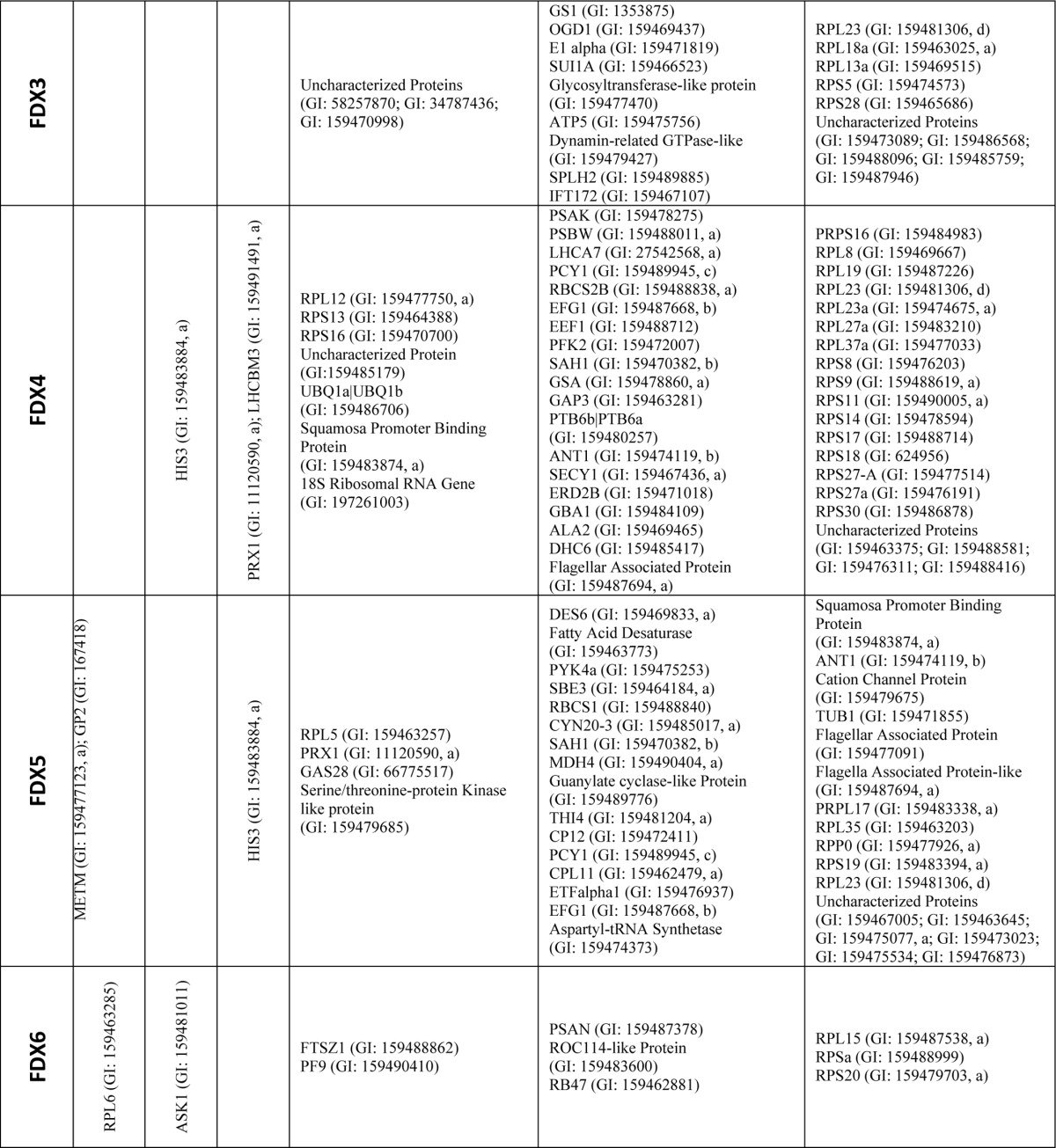
In total, our yeast two-hybrid library screens resulted in the identification of 1204 prey clones capable of supporting yeast growth when co-expressed with at least one of the FDX isoforms (supplemental Tables S2–S7; FDX1, 267; FDX2, 326; FDX3, 150; FDX4, 230; FDX5, 163; and FDX6, 68). After sequencing, it was determined that these clones represent 216 unique putative FDX-interacting proteins (Table 1, FDX1, 78; FDX2, 16; FDX3, 22; FDX4, 49; FDX5, 40; and FDX6, 11). Of the putative interaction partners, 79 proteins were identified as potential interaction partners with more than one FDX isoform (Table 1). The identified proteins are involved in a number of important biochemical pathways, including photosynthesis, pyruvate metabolism, carbon, and nitrogen fixation. Table 1 lists the interacting proteins identified for each FDX and their assigned confidence score.
TABLE 1.
Yeast two-hybrid library screen hits and scores
All hits identified by yeast two-hybrid screening of a C. reinhardtii cDNA library using each FDX isoform as bait are shown. Computationally assigned confidence scores provided by Hybrigenics (Paris, France) are provided for each hit (see “Experimental Procedures”). Hits identified for more than one of the FDX baits are indicated with superscript letters according to the number of FDXs they were identified with as interaction partners for (a-2, b-3, c-4, and d-5 bait-prey interactions). 29 hits were identified in two bait-prey interaction tests.
Pairwise Yeast Two-hybrid Assays
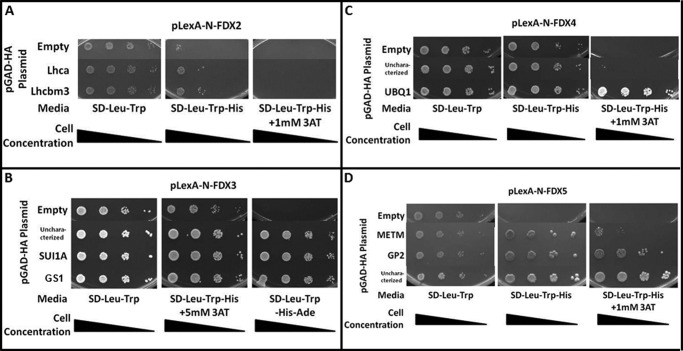
To validate some of the results obtained from the yeast two-hybrid library screening, we performed pairwise two-hybrid assays for a limited number of the identified prey proteins. In these assays, we fused each FDX isoform to the N terminus of the LexA DNA-binding domain (LexA-DB) and the selected protein to the GAL4-activation domain. The selected preys included LHCA7 (light harvesting protein 7 of the photosystem I) and LHCBM3 (light harvesting protein M3 of the photosystem II) for FDX2 (Fig. 1A); GS1 (glutamate synthase), SUI1A (putative translation initiation factor 1) and an uncharacterized protein (GI: 159470998) for FDX3; UBQ1a|b (B-ubiquitine), and another uncharacterized protein (GI: 159485179) for FDX4; and METM (S-adenosylmethionine synthetase), GP2 (hydroxyproline-rich glycoprotein), and a third uncharacterized protein (GI: 159475534) for FDX5. Similar assays could not be performed with FDX6 due to auto-activation of the HIS3 reporter expression (data not shown). Compared with the yeast two-hybrid screen presented above, our pairwise assays used yeast codon-optimized LexA::FDX fusion proteins cloned into independent expression vectors. Furthermore, the assay was performed in an independent yeast strain and screened on additional selective media, supporting the observed interactions (see “Experimental Procedures”). Each of the pairwise interaction results corroborated the findings of the library screen. It is important to highlight that some proteins (previously mentioned) without assigned confidence scores (GS1, SUI1A, and the uncharacterized protein GI: 159475534) were tested with FDX3, FDX4, and FDX5 in these pairwise yeast two-hybrid assays and were shown to interact with the bait FDX, suggesting that this category of interaction partners may harbor valuable interaction data (Fig. 1). Furthermore all the pairwise interactions tested to validate the library screen were positive.
FIGURE 1.
Yeast two-hybrid pairwise confirmation of hits from the cDNA library screen. Yeast transformants harboring pLexA plasmid (baits, indicated above the image) and the pGAD-HA plasmid (preys, indicated in the left column) were grown on SD-Leu-Trp (growth indicates the presence of each indicated plasmid) as well as SD-Leu-Trp-His (growth indicates synthesis of the HIS3 enzyme, suggesting an interaction between the expressed proteins), SD-Leu-Trp-His+3AT (3AT is a competitive inhibitor of the HIS3 gene product and is used to titrate the lowest amount of HIS3 expression required for growth on media lacking histidine) or SD-Leu-Trp-His-Ade (the most stringent selection media) plates. A, both of the tested proteins interact with FDX2. B, all of the tested proteins interact equally with FDX3. C, all of the tested proteins (uncharacterized protein GI: 159470998) interact with FDX4. UBQ1 interacts the most strongly resulting in robust growth, although the uncharacterized protein (GI: 159485179) results in weak but detectible growth. D, all of the tested proteins interact with FDX5 (uncharacterized protein GI: 159475534).
Finally, it must be noted that none of the hydrogenase structural (HYDA1/2) or maturation proteins (HYDEF and HYDG) were identified in our two-hybrid library screens. This may be due to low abundance, lack of these cDNAs in the library, or instability of these proteins in yeast. In addition, during dark anaerobiosis, the transcripts for the hydrogenase proteins are relatively low in abundance (19), because hydrogen production is a minor fermentative pathway, and the transcripts are probably under-represented in our library once transformed into yeast. Thus, to determine the interaction profile(s) between each FDX isoform and the biosynthetic and enzymatic proteins associated with hydrogen evolution, we undertook pairwise two-hybrid assays using the two-hydrogenase enzymes, HYDA1 and HYDA2, and their maturation factors, HYDEF and HYDG, as either bait or prey.
FDX Interactions with the Hydrogen Production Metabolic Network
HYDA1 and HYDA2
We used HYDA1 and HYDA2 as baits, and each of the six FDXs (FDX1 to FDX6) as preys. We found that yeast strains HYDA1 interacted with FDX1, FDX2, FDX3, FDX4, and FDX5 (Fig. 2), although HYDA2 interacted with FDX1 and FDX3 only in the presence of 1 mm 3AT, a competitive inhibitor of the product of the HIS3 transcriptor inhibitor (Fig. 3). In summary, HYDA1 can interact strongly with FDX1, moderately with FDX2, and weakly with FDX3 or FDX4. HYDA2 also interacts strongly with FDX1 and less strongly with FDX3
FIGURE 2.
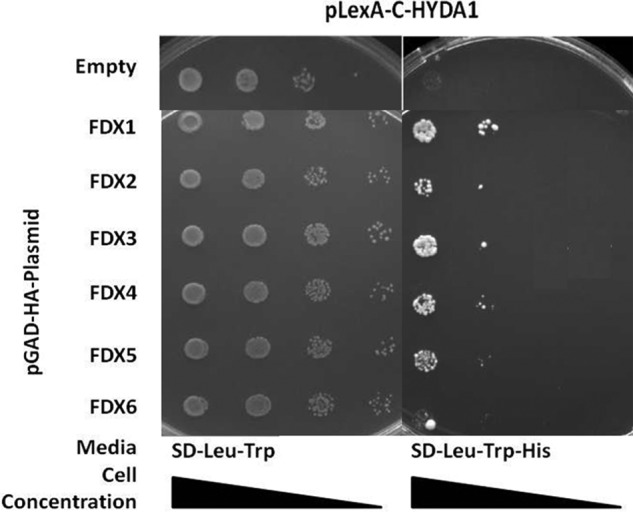
Pairwise yeast two-hybrid interactions using HYDA1 as bait. Yeast transformants harboring pLexA-C-HYDA1 and the pGAD-HA plasmid indicated in the left column were grown on SD-Leu-Trp and SD-Leu-Trp-His (see Fig. 1 legend). Increased growth is observed for yeast harboring HYDA1 and either FDX1, FDX2, FDX3, FDX4, or FDX5 when compared with the negative control yeast harboring the empty pGAD-HA plasmid. No increase in growth is observed for yeast strains harboring HYDA1 and FDX5 or FDX6.
FIGURE 3.
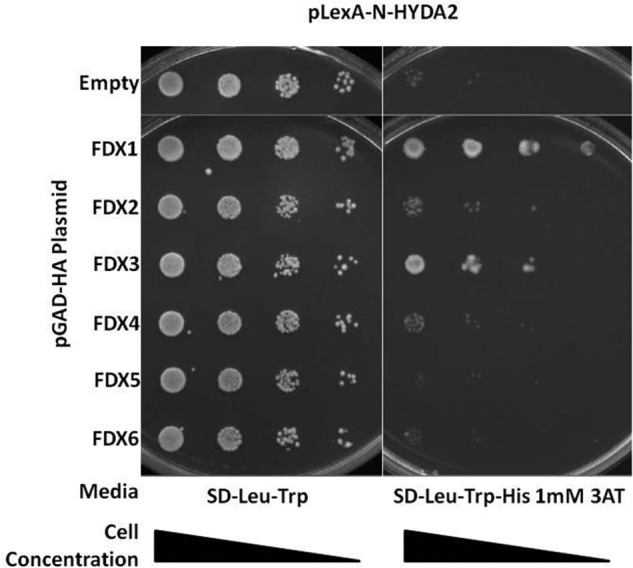
Pairwise yeast two-hybrid interactions using HYDA2 as bait. Yeast transformants harboring pLexA-N-HYDA2 and the pGAD-HA plasmid indicated in the left column were grown on SD-Leu-Trp and on SD-Leu-Trp-His with 1 mm 3AT. Increased growth is observed for yeast harboring HYDA2 and either FDX1 or FDX3.
HYDEF and HYDG
Each FDX isoform was used as bait and tested for interactions against the hydrogenase maturation factors HYDEF and HYDG. Because of auto-activation of the HIS3 promoter by LexA fusions with FDX1, FDX3, and FDX6, we could not determine their interaction profiles with the maturation factors (data not shown). However, we determined that FDX2, FDX4, or FDX5, interacted with HYDEF and with FDX5 showing the strongest interaction (Fig. 4).
FIGURE 4.
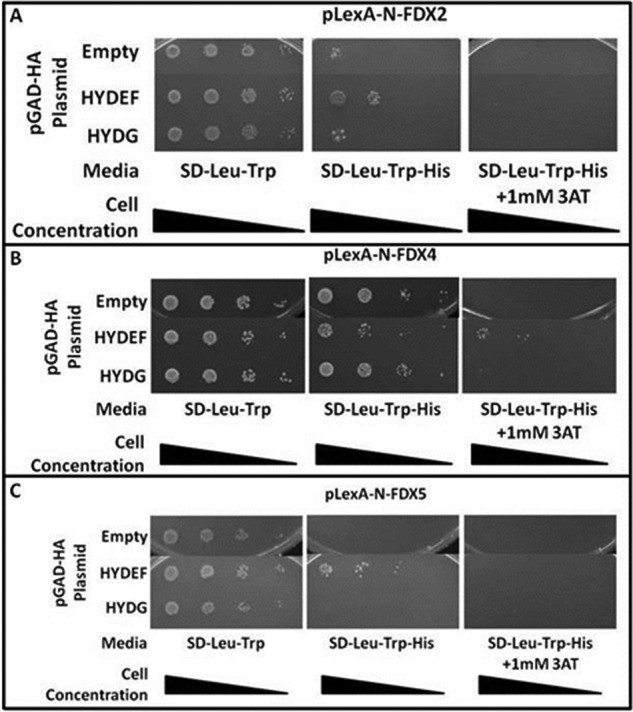
Yeast two-hybrid interactions between FDXs (bait) and the hydrogenase maturation factors HYDEF and HYDG (prey). Yeast transformants harboring pLexA plasmid indicated above the image and the pGAD-HA plasmid indicated in the left column were grown on SD-Leu-Trp and SD-Leu-Trp-His. A, interaction is observed between HYDEF and FDX2. B, increased growth in 1 mm AT medium indicates an interaction between HYDEF and FDX4. C, HYDEF and FDX5 interaction is suggested by the increased growth. All other FDX interaction tests could not be interpreted due to autoactivation by the LexA-FDX fusion protein (data not shown).
Hydrogenase Maturation Factor Interaction Profiles
To further understand the hydrogen production network, we examined the direct interactions between the hydrogenases and their maturation factors. We could not detect any direct pairwise interactions between HYDA1 and either HYDEF or HYDG (data not shown). We hypothesized that interactions between the hydrogenase and a single maturation factor (either HYDEF or HYDG) might be too transient to be detected alone and that a tripartite protein complex might be more stable. To test this assumption, we performed a yeast two-hybrid bridging experiments in which a third expression plasmid (pY3H) was introduced to express the additional maturation factor. We found that in these experiments HYDA1 interacted with HYDEF in the presence of HYDG (and vice versa) (Fig. 5). Likewise, we showed that HYDA2 interacted with HYDG in the presence of 5 mm 3AT (Fig. 6), but we were not able to detect interactions between HYDA2 and HYDEF. This might be due to the fact that LexA::HYDA2 exhibits auto-activation of the HIS3 transcription (data not shown). We also attempted yeast two-hybrid bridging experiments with HYDA2, but again we were not able to detect growth at a level greater than that of the negative control (data not shown).
FIGURE 5.
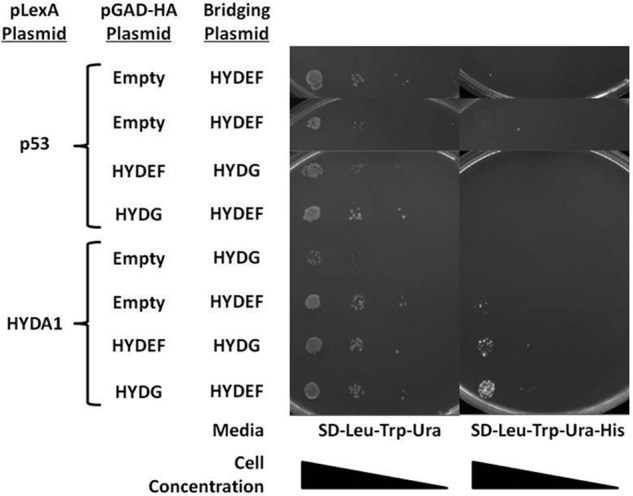
Yeast two-hybrid bridging assay for HYDA1 (bait) and the maturation factors HYDEF and HYDG (preys). Yeast transformants harboring the indicated pLexA plasmid (pLexA-p53 or pLexA-C-HYDA1), pGAD-HA plasmids (empty, pGAD-HA-HYDEF, or pGAD-HA-HYDG), and bridging plasmid (pY3H-HYDEF or pY3H-HYDG) were grown on SD-Leu-Trp-Ura and SD-Leu-Trp-Ura-His. Increased growth is observed for yeast harboring HYDA1, HYDEF and HYDG.
FIGURE 6.
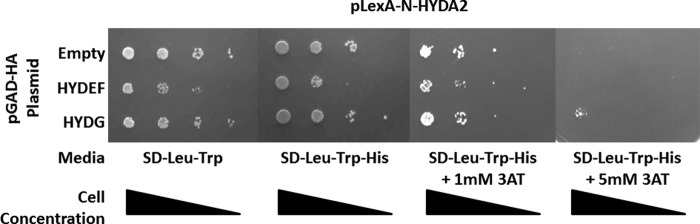
Pairwise yeast two-hybrid interactions for HYDA2 (bait) and the maturation factors HYDEF and HYDG (preys). Yeast transformants harboring pLexA-N-HYDA2 and the pGAD-HA plasmid indicated in the left column were grown on SD-Leu-Trp, SD-Leu-Trp-His, and SD-Leu-Trp-His with 1 mm 3AT and SD-Leu-Trp-His with 5 mm 3AT. All yeast transformants, including the negative control strain harboring the empty pGAD-HA plasmid, grow on SD-Leu-Trp-His and SD-Leu-Trp-His with 1 mm 3AT indicating autoactivation of HIS3 synthesis by HYDA2. When 5 mm 3AT is included, no growth is observed for the negative control strain, although a modest increase in growth is observed for yeast harboring HYDA2 and HYDG.
These findings suggest that maturation of Chlamydomonas HYDA1, and possibly HYDA2, requires each of the maturation factors to act in concert for proper function. These results corroborate recent data showing that Clostridium acetobutylicum [FeFe] hydrogenase can interact with HydF only when co-expressed with the other two maturation factors (40).
FDX Interaction Profile with Metabolic Pathways Competing with Hydrogen Photo-production
Ferredoxin-NADP reductase (FNR) is the primary recipient of reductant from FDXs under normal photosynthetic growth conditions (41). The Chlamydomonas chloroplast FNR, FNR1, was not identified in our yeast two-hybrid screen as a putative interaction partner with any of the FDX isoforms, perhaps due to the low abundance of FNR1 transcript in the library constructed from samples isolated in dark anaerobiosis (Table 1). To determine an interaction profile between the FDX isoforms and FNR1, we conducted pairwise interaction assays using FNR1 as bait and all FDXs as preys. We found FNR1 primarily interacts with FDX1 and FDX2. Furthermore, our data show that FDX1 is the strongest interacting protein with HYDA1, HYDA2, and FNR1 (Fig. 2, 3, and 7).
FIGURE 7.
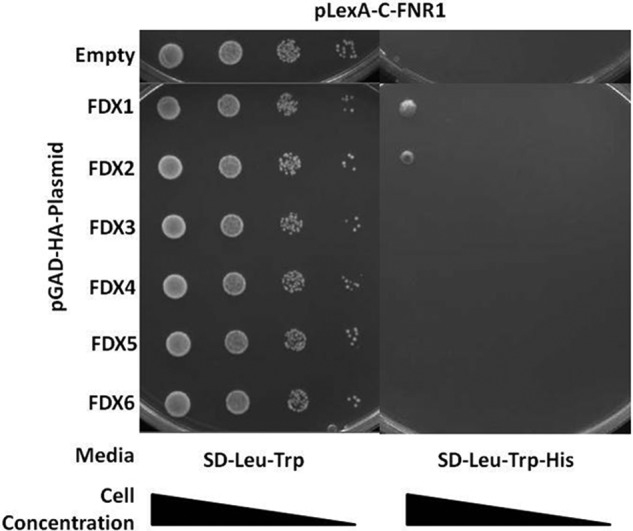
Pairwise yeast two-hybrid interactions between FNR1 (bait) and each FDX (preys). Yeast transformants harboring pLexA-C-FNR1 and the pGAD-HA plasmid indicated in the left column were grown on SD-Leu-Trp and SD-Leu-Trp-His. Growth is observed for yeast transformants harboring FNR1 and FDX1 or.
FDX/FDX Interactions
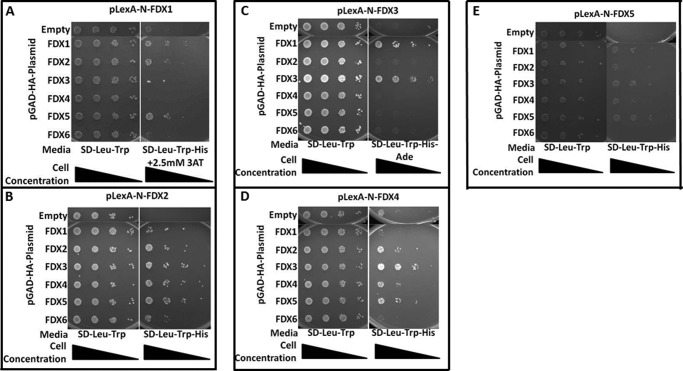
Because FDX1 and FDX5 were identified as putative interaction partners for FDX1 in our yeast two-hybrid screen, we tested for the ability of each FDX to interact with themselves and/or with the other FDXs. Using the two-hybrid system, we used each of the six FDXs as baits and tested their interactions with each other as preys (Fig. 8, A–E). In summary, our data supports homodimerization for FDX1, FDX2, FDX3, and FDX4, although heterodimerization is observed for the following pairs of FDX isoforms as follows: FDX1:FDX2, FDX1:FDX3, FDX1:FDX5, FDX2:FDX3, FDX2:FDX4, FDX2:FDX5, FDX2:FDX6, FDX3:FDX4, FDX3:FDX5, and FDX4:FDX5. Most interestingly, FDX1:FDX5 heterodimers were detected in the yeast two-hybrid screen, as well as in our pulldown experiment (Table S9), and these results are supported by pairwise yeast two-hybrid testing in all possible orientations.
FIGURE 8.
Pairwise yeast two-hybrid assays for FDX/FDX interactions. Yeast transformants harboring pLexA plasmid (bait) indicated above each image and the pGAD-HA plasmid (prey) indicated in the left column were grown on SD-Leu-Trp and selective media, either SD-Leu-Trp-His, SD-Leu-Trp-His + 2.5 mm 3AT, or SD-Leu-Trp-His-Ade depending on the required level of selection to overcome autoactivation resulting from the transformed plasmids. A, FDX1 interacts with FDX1, FDX2, and FDX5. B, FDX2 interacts with every FDX isoform. C, FDX3 interacts with FDX1 and FDX3. D, FDX4 interacts with FDX2, FDX3, FDX5, and itself. E, FDX5 interacts with FDX1 and FDX3. FDX6 dimerization tests could not be interpreted due to un-suppressible autoactivation by the LexA-FDX fusion protein (data not shown).
Pulldown Analysis
In an attempt to independently verify the potential protein/protein interactions and possibly extend the list of targets for FDX1 and FDX2, we performed affinity pulldown experiments using overexpressed and purified His6-tagged FDX1 and FDX2, and cell lysates generated from aerobically or anaerobically grown Chlamydomonas cultures. The eluted proteins that bound to the beads were analyzed by liquid chromatography and high resolution mass spectrometry and quantified by the label-free method of spectral counting (38). Initially, for the least stringent sorting, we compared the spectral counts for each of the identified proteins to those found in the negative control (beads without protein coupled to them, but otherwise treated the same), and we used a ratio of 1.5 (sample/negative control) as the cutoff limit, which includes proteins that were not identified in the negative control. Subsequently, based on the respective spectral counts, p values were calculated for each of the identified proteins as a measure of confidence (see “Experimental Procedures”).
Specific FDX1 Interactions with Cell Extract from Aerobic Growth Condition
After application of the 1.5 cutoff ratio, a total of 174 putative FDX1-interacting proteins were pulled down and identified from cell extracts generated from aerobically grown Chlamydomonas cells (supplemental Table S8). Table 2 lists the six identified proteins with a computed p value of less than 0.01 (see “Experimental Procedures”). A receptor of activated protein kinase C (RCK1) had the highest scores (56 peptides identified/0 peptides in the negative control/∼0 p value), whereas an uncharacterized transcription factor was at the other end of the spectrum (16/9/0.968). This list of enzymes includes a flavoprotein and FBP1, supporting the important role that FDX1 is predicted to play in the allocation of photosynthetic reductant in light-grown aerobic cultures. We also identified a protein involved in protein stability, CEP1, the tricarboxylic acid cycle enzyme FUM1, and an additional photosynthetic protein, PSAF. Some of the hits here were also detected by the yeast two-hybrid assay (i.e. METM and a flavoprotein). However, the pulldown technique is a more sensitive and useful method to detect interaction partners that are expressed at low levels.
TABLE 2.
Aerobic Pull-Down Results Using Immobilized FDX1
| # | Accession no. | Protein | p value | Peptide count |
Biological pathway | |
|---|---|---|---|---|---|---|
| Sample | -ctrl | |||||
| 1 | GI:159479982 | RCK1-Receptor of Activated Protein Kinase C | 1.10179E−12 | 56 | 0 | Regulatory Proteins |
| 2 | GI:159478523 | FNR1-Ferredoxin-NADP Reductasea | 1.92780E−05 | 21 | 0 | Photosynthesis |
| 3 | GI:159478266 | ATO1-Acetyl-CoA Acyltransferase | 8.02177E−05 | 18 | 0 | Citric Acid Cycle |
| 4 | GI:159469919 | GCST-Glycine Cleavage System-T Proteina | 3.35985E−05 | 15 | 0 | Photorespiration |
| 5 | GI:159489124 | DEAD-Box ATP-dependent RNA Helicase | 5.42789E−05 | 14 | 0 | RNA Metabolism/Protein Synthesis/RNA Binding Protein |
| 6 | GI:159485508 | ZDS1-Zeta-Carotene Desaturase | 2.31201E−04 | 11 | 0 | Carotenoid Biosynthesis |
a Indicates hits in the pull-down experiment that were also detected in Synechocystis (43).
Specific FDX1 Interactions with Cell Extract from Anaerobic Growth Condition
Using the 1.5 peptide ratio cutoff, a total of 181 putative FDX-interacting proteins were identified from cell extracts generated from anaerobically grown Chlamydomonas cells (supplemental Table S9). Table 3 lists the eight identified proteins with a computed p value of less than 0.01 (see “Experimental Procedures”). Again, RCK1 was identified as the most strongly interacting protein (61 peptides identified/0 peptides in the negative control/∼0 p value). Included in this subset was FDX5, which has been demonstrated to be up-regulated under anaerobic growth conditions (17). We also found interaction of FDX1 with the phosphorylation protein ATP2 and the photosynthetic protein LHCA9.
TABLE 3.
Anaerobic Pull-Down Results Using Immobilized FDX1
| # | Accession no. | Protein | p value | Peptide count |
Biological pathway | |
|---|---|---|---|---|---|---|
| Sample | -ctrl | |||||
| 1 | GI:159479982 | RCK1-Receptor of Activated Protein Kinase C | 1.13243E−14 | 61 | 0 | Regulatory Proteins |
| 2 | GI:159478523 | FNR1-Ferredoxin-NADP Reductasea | 6.95742E−07 | 26 | 0 | Photosynthesis |
| 3 | GI:159478266 | ATO1-Acetyl-CoA Acyltransferase | 6.85993E−05 | 17 | 0 | Citric Acid Cycle |
| 4 | GI:159469919 | GCST-Glycine Cleavage System-T Proteina | 3.20858E−05 | 14 | 0 | Photorespiration |
| 5 | GI:159469684 | GCSP-Glycine Cleavage System-P protein | 5.38077E−05 | 13 | 0 | Photorespiration |
| 6 | GI:159485508 | ZDS1-Zeta-Carotene Desaturase | 2.57016E−04 | 10 | 0 | Carotenoid Biosynthesis |
| 7 | GI:159468063 | Cytoplasmic DExD/H-box RNA Helicase | 7.40611E−04 | 8 | 0 | Fatty acid metabolism |
| 8 | GI:158278388 | Uncharacterized Protein | 7.40611E−04 | 8 | 0 | RNA Metabolism/protein synthesis/RNA Binding Proteins |
a Indicates hits in the pull-down experiment that were also detected in Synechocystis (43).
Common FDX1 Interactions with Cell Extracts from Both Aerobic and Anaerobic Growth Conditions
We identified 104 common proteins between the aerobic and anaerobic samples. This subset of proteins includes several known FDX-dependent enzymes such as FTRA (aerobic, 1/0/0.452; anaerobic, 2/0/0.220), GSF1 (aerobic, 3/0/0.135; anaerobic, 2/0/0.220), and FNR1 (aerobic, 21/0/0.00002; anaerobic, 26/0/0.0000007), supporting the effectiveness of the assay and highlighting the strongest interactions of all those proteins with FDX1. Photosynthetic proteins, including PETO (aerobic, 1/0/0.45; anaerobic, 2/0/0.22) and PSII stability/assembly factor HCF136 (aerobic, 3/0/0.14; anaerobic, 2/0/0.22) were found to be common to both conditions, although the nature of their interaction with FDX remains unexplained. When the proteins identified by each of the pulldown experiments were compared with the proteins identified by yeast two-hybrid screening, a small number of overlapping interaction candidates emerged. In total, 20 proteins were co-identified in at least one of the two pulldown assays and in the yeast two-hybrid screening. These proteins include FDX5, several ribosomal proteins (RPS4/9, RPL12, RPL14, RPL18, RPL3, and RPP0), ATP2 (mitochondrial F1F0 ATP synthase β subunit) and MDH4 (malic dehydrogenase).
Common FDX1 and FDX2 Interactions with Cell Extracts from Both Aerobic and Anaerobic Growth Conditions
Pulldown assays were also performed with FDX2 to identify specific and common targets with FDX1 under aerobiosis and anaerobiosis. The results show (supplemental Tables S10 and S11) that under both conditions FDX1 and FDX2 share common binding partners such as RCK1 (reception of activated protein kinase C), ATO1 (acetyl-CoA acyltransferase), and GCST (glycine cleavage system T-protein) (the strongest interacting proteins), and also some unique targets such as FDX2, citrate synthase (CIS3), and the glyceraldehyde-3-phosphate dehydrogenase (GAP1) under anoxic conditions. Altogether, the pulldown technique allowed us to identify new potential interactions, suggesting that the yeast two-hybrid and pulldown techniques can be used to validate (in some cases) and complement each other's results.
Physiological Assays Using Purified Recombinant FDXs, FDX-driven NADP+ Photo-reduction
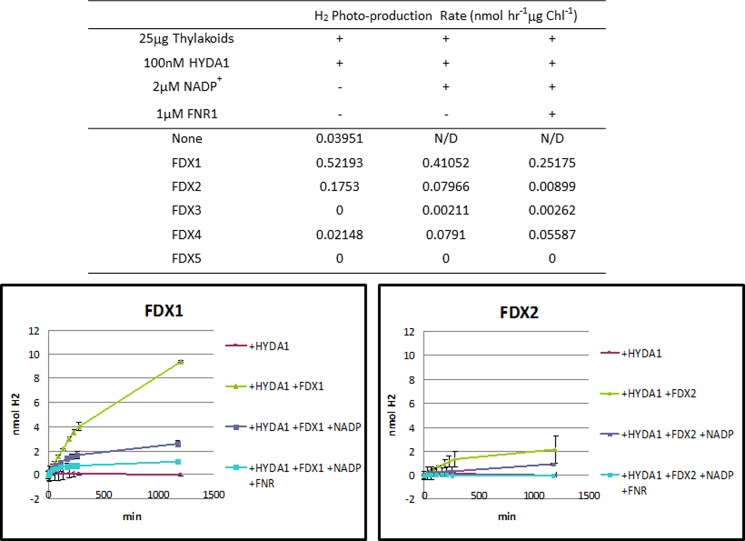
To confirm the exclusive interaction of FNR with FDX1 and FDX2, we tested the in vitro specificity of each FDX as electron donors to FNR1 by measuring NADP+ photo-reduction activity mediated by purified recombinant FDXs (Table 4 and “Experimental Procedures”). We prepared in vitro reactions using Chlamydomonas thylakoid membranes, which harbor membrane-associated FNR activity (42). FDX1 mediated the highest rates of NADPH photo-production (7.896 μmol of NADPH h−1 mg of Chl−1), although the rate observed for FDX2 was 2-fold lower (3.569 μmol of NADPH h−1 mg of Chl−1). The other FDXs tested (FDX3, FDX4, and FDX5) were not able to support this enzymatic reaction at detectable rates. It must be noted that FDX6 could not be successfully purified to the same quantity or level of purity as the other FDXs and thus was not included in the in vitro assays (see “Experimental Procedures”). Addition of exogenous Chlamydomonas FNR1 to the thylakoid assay dramatically boosted NADP+ photo-reduction rates by both FDX1 (41.54 μmol of NADPH h−1 mg of Chl−1) and FDX2 (27.7 μmol of NADPH h−1 mg of Chl−1), indicating that the residual thylakoid-bound FNR1 was insufficient to saturate NADP+ photo-reduction kinetics. These data confirm that FDX1 is the primary electron donor to FNR1 and that FDX2 can also perform this function.
TABLE 4.
NADPH photo-production mediated by purified Chlamydomonas FDXs. Individual FDX isoforms were added to a reaction mixture (see Experimental Procedures) and production of NADPH from NADP+ was monitored FDX1 drives NADPH production at a rate of 7.896 μmol NADPH h−1 mg Chl−1 while FDX2 drives NADPH production at a rate of 3.569 μmol NADPH h−1 mg Chl−1. None of the other FDX isoforms tested (FDX3, FDX4 and FDX5) generated NADPH at detectable levels
| 1 μm FNR1 | NADPH Photo-production Rate (μmol NADPH h−1 μg Chl−1) |
|
|---|---|---|
| − | + | |
| FDX1 | 7.896 | 41.54 |
| FDX2 | 3.569 | 27.7 |
| FDX3 | 0 | 0 |
| FDX4 | 0 | 0 |
| FDX5 | 0 | 0 |
FDX Driven Hydrogen Photo-production
We assayed hydrogen photo-production in vitro in the presence of each individual FDX. Reactions using isolated Chlamydomonas thylakoid membranes (42) were prepared by adding purified recombinant HYDA1 (34) and purified recombinant FDX (see “Experimental Procedures”). We found that FDX1 drives the highest rates of hydrogen photo-production (0.52 nmol of H2 h−1), although FDX2-mediated hydrogen photo-production was 3-fold lower (0.18 nmol of H2 h−1) (Fig. 9). The other FDX isoforms (FDX3, FDX4, and FDX5) were not able to mediate this reaction (Fig. 9). Addition of either NADP+ alone or NADP+ plus exogenous Chlamydomonas FNR1 to this assay drastically reduced hydrogen photo-production rates (Fig. 9) with either FDX1 or FDX2. The data are consistent with competition between FNR and HYDA1 for FDX1. Moreover, in this assay FDX1 is a more efficient electron donor to HYDA1 and FNR than FDX2.
FIGURE 9.
Hydrogen photo-production mediated by Chlamydomonas FDXs. To evaluate hydrogen production mediated by each FDX isoform, we performed assays utilizing purified Chlamydomonas thylakoid membranes and purified HYDA1. Individual FDX isoforms (10 μm final concentration) were added to the reaction mixture (see “Experimental Procedures”), and production of hydrogen was monitored with a gas chromatograph by measuring headspace hydrogen accumulation over time. FDX1 drives hydrogen production at a rate of 0.52 nmol of H2 h−1 although FDX2 drives hydrogen production at a rate of 0.17 nmol of H2 h−1. None of the other FDX isoforms tested (FDX3, FDX4, and FDX5) facilitated hydrogen production at rates higher than the negative control. N/D, not determined.
DISCUSSION
In this study, we generated a first generation protein/protein interaction map for each of the six Chlamydomonas FDXs to establish their functional role in metabolism. Previously, most of the FDX interaction predictions in Chlamydomonas were inferred from higher plant data.
We initially performed yeast two-hybrid library screens to gain insight into the full interactome of ferredoxins in Chlamydomonas. Despite the high sequence similarity displayed by Chlamydomonas FDXs, the screens identified highly variable numbers of interaction partners for each FDX (Table 1). These results revealed a large number of false positives and false negatives; however, they suggest that each FDX displays a unique interaction profile. Some FDX isoforms may mediate many redox reactions (e.g. FDX1 has 78 putative interaction partners), although they may be limited in the number of redox reactions in which they participate. For example, FDX6 shows the least interaction partners, with 10 putative hits, whereas FDX4 interacts 49 targets; FDX5 with 40; FDX3 with 22; and FDX2 with 17. Our findings include a large number of ribosomal proteins detected in both the yeast two-hybrid screens and in pulldown experiments (Table 1 and Tables S8–S11). Pulldown assays in Synechocystis similarly identified suites of ribosomal proteins (43). It remains to be seen if these putative interactions represent biologically relevant functions.
Previous publications indicate that chloroplast FDXs from Chlamydomonas are phylogenetically related to either plant leaf-type, found primarily in photosynthetic tissues (FDX1 and FDX5), plant root-type, related to assimilatory metabolism (FDX2), and FDXs with unknown function that are most closely related to other microbial [2Fe-2S] FDXs (FDX3, FDX4, and FDX6) (16). To understand FDX function within the cell, we analyzed our data with these groupings in mind.
Microbial Type FDX Interaction Networks
Three Chlamydomonas FDXs (FDX3, FDX4, and FDX6) are phylogenetically unrelated to the typical leaf-type and root-type FDXs of higher plants. FDX3 and FDX6 have been determined to function in plastid-based metabolism but are not well characterized (16). The transcript levels of FDX3 and FDX6 were shown by others to be elevated under iron-limited growth conditions, although FDX3 and FDX6 protein abundance decreases under the same growth conditions. These contradictory data have led to some uncertainty about their role in Chlamydomonas metabolism (16).
In our library screens, FDX3 was found to interact primarily with ribosomal proteins and translation factors (7 of 22 hits), as well as with eight uncharacterized proteins (Table 1). Interestingly, an oxo-glutarate dehydrogenase subunit (OGD1) and a glutamine synthase (GS1) were identified as binding partners for FDX3, and the GS1 interaction was confirmed by pairwise interaction (Fig. 1). Because GS1 contains a putative chloroplast transit peptide and is predicted to be chloroplast-localized (data not shown), we suggest that so is FDX3. Nitrogen assimilation is a multistep process involving the following: (a) NADPH-dependent reduction of nitrate into nitrite by the nitrate reductase; (b) reduction of nitrite into ammonia by the FDX-dependent enzyme nitrite reductase (NiR or NII1 in Chlamydomonas); (c) incorporation of ammonia and glutamate to form glutamine, catalyzed by glutamine synthase; and (d) conversion of glutamine and 2-oxoglutarate into two glutamates by FDX-dependent glutamate synthase (GSF1) (44). In plants, steps b, c, and d occur in the chloroplast (45); in algae, NiR has been localized to the chloroplast as well; however, the location of the remainder of the nitrogen assimilation process remains to be studied in this organism. The specific role of different FDXs in nitrogen assimilation is unclear. Previously, FDX2 was proposed to be involved in the nitrogen assimilation pathway, and in vitro data showed that FDX2 exhibits substrate specificity for NiR (16, 23), which our data confirm. Moreover, our FDX1 library screen identified GS2, a homolog of GS1 as a possible interacting partner for FDX1, expanding its role in nitrogen assimilation beyond previous observations of its interaction with NiR or GSF1 in crude Chlamydomonas extracts by cross-linking experiments (46). Thus, based on our current results, GS1 and GS2 represent novel binding partners for FDXs, and three FDXs (FDX1, FDX2, and FDX3) may play a role in nitrogen assimilation.
In this work, FDX4 was found to interact with two key enzymes involved in glycolysis, both of which have putative chloroplast transit peptides, phosphofructokinase (PFK2) and glyceraldehyde-3-phosphate dehydrogenase (GAP3), as well as with two proteins sharing an interaction with FDX5 (PRX1, a peroxiredoxin family protein; SBP, a squamosa promoter-binding protein) (Table 1). PRX1 has been found to protect chloroplasts from reactive oxygen species, which are induced by many environmental factors, including light, and result in the damage of the D1 protein (47). Our findings suggest a role for FDX4 in redox sensing and in protecting from reactive oxygen species, although the particular mechanism involved is currently unknown (48). SBP proteins are characterized by a zinc finger domain responsible for interaction with the cis-acting DNA sequence GTAC. Recently, it was reported that the SBP domain of a transcriptional activator, CRR1, was able to bind the typical GTAC cores and that FDX5 was a CRR1 target (20). Our data suggest that both FDX4 and FDX5 can interact with the SBP transcription factor, which might have implications for their own transcriptional regulation (in the nucleus) or for the transcriptional regulation of other genes.
Finally, the FDX6 library screen yielded only 10 hits, with ribosomal proteins representing a majority of the interacting partners (5 out of 10) (Table 1). These limited results are due to the strong auto-activation of the FDX6 fusion protein used as bait in the yeast two-hybrid screen and in the pairwise interaction assays. FDX6 is the most divergent FDX, characterized by long N- and C-terminal extensions that result in a much larger protein when compared with the other FDX isoforms. More work is needed to elucidate the specific role of this FDX and to identify the conditions under which FDX6 is functional.
These combined data suggest that the microbial type FDXs bind to targets in common with the leaf-type FDXs, but they also have additional interaction partners, therefore implying unique functions for these nontypical FDXs in Chlamydomonas.
Root-type FDX Interaction Network
The library screen using FDX2 as bait identified 16 proteins, including seven ribosomal proteins, two uncharacterized proteins, and two light harvesting complex (LHC) proteins (LHCBM3 and LHCA7). The LHC proteins represent two of the highest confidence hits from the screen (Table 1). LHCs are components of the photosynthetic machinery that bind chlorophyll molecules responsible for harvesting light energy. Some of the LHC proteins become phosphorylated under certain growth conditions (such as exposure to high light intensity), which causes over-reduction of the plastoquinone pool and triggers state transitions (48), a process that leads to a redistribution of excitation energy between PSII and PSI by reorganization of the antennae, thereby regulating energy flow between the photosystems. Anaerobiosis also leads to the influx of reducing equivalents into the plastoquinone pool and to the activation of the LHCII kinase (49). LHCBM3 has recently been identified as a target for phosphorylation by this kinase, and it must thus be directly involved in state transitions (48). Both LHCBM3 and LHCBM7 were identified as binding partners for FDX4 and FDX1, respectively, strengthening our suggestion of the role for FDXs in state transitions (50).
One last interaction partner of interest for FDX2 is TEF7, which has recently been identified in a quantitative proteomic study of the Chlamydomonas chloroplasts as induced under both normal light anaerobic (argon bubbling) conditions and sulfur-deficient anaerobiosis (51). Because FDX2 transcripts also accumulate during anaerobiosis (19), our results suggest a functional link between the two proteins during acclimation to anaerobic stress. Interestingly, TEF7 seems to be required for efficient cyclic photosynthetic electron transfer in Chlamydomonas, a function that needs to be demonstrated for FDX2 (52).
In summary, the pulldown assays performed with FDX2 confirmed some of the hits obtained with the library screen (mainly LHC and ribosomal proteins) but also raised a number of new potential interacting targets, both under oxic and anoxic conditions, such as CIS3 and GAP1, highlighting a possible link with the citric acid cycle.
Leaf-type FDX Interaction Network
By phylogenetic analysis, FDX1 and FDX5 are classified as photosynthetic or leaf-type FDXs. However, FDX5 is much more divergent from plant leaf-type FDXs, casting uncertainty on its physiological role (16). Based on previous and current results, FDX5 seems to be involved in anaerobic adaptation and fermentation processes. The FDX5 transcript is more abundant during anaerobiosis and in sulfur- or copper-deficient growth conditions (16). In our study, FDX5 interacts with enzymes involved in pyruvate synthesis (PYK4; pyruvate kinase) and fermentation (MDH4; malate dehydrogenase, which contains a putative chloroplast transit peptide), further suggesting that FDX5 has a role in metabolic pathways other than typical photosynthetic pathways (Table 1).
Our work also proposes a role for FDX5 in the hydrogenase maturation process. Indeed, our library screen identified METM (with a very high confidence score) and THI4 (with a putative chloroplast transit peptide), which are both SAM-radical proteins, as interacting partners with FDX5 (Table 1 and Fig. 1). Because the hydrogenase maturation proteins HYDEF and HYDG are also SAM-radical enzymes, we tested the interaction of FDX5 and other FDXs with HYDEF/HYDG in pairwise yeast two-hybrid assays (Fig. 4) and showed that HYDEF does indeed interact with FDX5, FDX4, and FDX2, further supporting a potential role for these FDXs in hydrogenase metallocluster biosynthesis. In bacteria, FDX is known to be involved in [FeS] cluster synthesis (along with other genes from the isc operon) and could be required for the first step in [FeFe] hydrogenase maturation (53). Moreover, FDX1 also interacts with METM and THI4 (confirmed by the yeast two-hybrid and pulldown experiments), suggesting that it also could be directly involved in metallocluster biosynthesis. Unfortunately, auto-activation of the reporter gene triggered by FDX1 in the pairwise yeast two-hybrid assay did not allow us to test its direct interaction with HYDEF and HYDG. At present, it is not known where hydrogenase assembly takes place, although the hydrogenase structural and assembly genes carry a putative chloroplast transit peptide.
Until now FDX1, was proposed to be the primary FDX involved in NADP+ reduction and the sole electron donor to hydrogenases. Several pieces of direct evidence from our research support the predominant role of FDX1 in FNR1-mediated NADPH production and in hydrogen production by HYDA1 (Fig. 9): (a) FDX1 and FDX2 are the only chloroplast FDXs able to interact with FNR1 in pairwise yeast two-hybrid assays (Fig. 7); (b) the FDX1 pulldown assay (supplemental Table S8) confirms a strong interaction with FNR (p value = 1.92 × 10−5 or p value = 6.95 × 10−7 representing the second strongest hit detected in both aerobic and anaerobic assay respectively); (c) FDX1 supports the highest rate of in vitro NADPH photo-production with FNR1 (Table 4); and (d) the highest rate of in vitro hydrogen photo-production with HYDA1 (Fig. 8). Previous studies demonstrated that FDX2 mediates 6-fold higher catalytic rates than FDX1 when coupled to Chlamydomonas FNR (16). Those assays were based on the transfer of electrons from oxidation of NADPH through FNR to FDX leading to the reduction of cytochrome c, and they are consistent with the proposal that FDX2 is a nonphotosynthetic root-type FDX that is reduced (rather than oxidized) by FNR.
Analysis of in vitro hydrogen photo-production mediated by each of the FDX isoforms showed that FDX1 mediates efficient hydrogen production, although FDX2 can mediate hydrogen production but at a 3-fold lower rate (Fig. 9). Furthermore, we confirmed that the previously observed competition (42) between HYDA1 (hydrogen production) and FNR1 (NADP+ reduction) for photo-reduced FDX1 also occurred with FDX2. However, for the first time, under our condition, we demonstrate that among all the FDXs, FDX1 is the kinetically preferred electron donor to HYDA1 and FNR1, although FDX2 is capable of acting as an alternative mediator under specific conditions (Fig. 9 and Table 4).
Although FDX2 seems to share similar functions with FDX1, the FDX2 pulldown experiment under aerobic and anaerobic condition data did not reveal any major common interacting partners (supplemental Tables S10 and S11). The best common hits were proteins involved in photorespiration (GSTC) and regulatory processes (RCK1). FNR1, for instance, was not detected in this assay, suggesting that due to the FDX1 predominance under these conditions and probably the higher affinity for FNR, all the binding sites are already taken by FDX1 preventing FDX2 from associating with this protein. It must be noted that the FDX2 transcript is only up-regulated when cells are grown on nitrate rather than ammonium, suggesting a stronger role of FDX2 under this condition (13).
As mentioned above, the aerobic FDX1 library screen identified the largest number of interacting partners (78 hits) (Table 1). Furthermore, the aerobic FDX1 affinity pulldown experiment identified 174 putative interaction partners, including many well known FDX-dependent enzymes and interaction partners identified by similar approaches in Synechocystis (supplemental Tables S8) (43). Altogether, our data show that FDX1 has a major role in mediating electron transfer reactions in aerobically grown Chlamydomonas compared with the other FDX isoforms. This is in agreement with published transcript abundance analyses indicating that FDX1 includes up to 98% of all FDX transcripts in aerobically grown cells on TAP media (16).
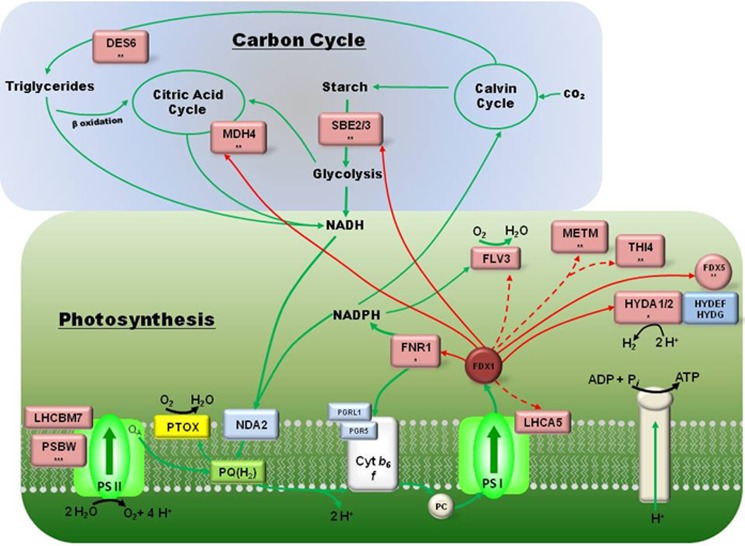
Fig. 10 shows the network of FDX1-interacting partners and highlights the most important pathways directly or indirectly linked to, or in competition with, photo-hydrogen production. Our yeast two-hybrid screen identified a Chlamydomonas homolog to the cyanobacterial flavoprotein Flv3 as a putative interaction partner (Table 1). In addition, an uncharacterized flavoprotein (GI: 158277718) with a putative chloroplast transit peptide, which is most closely related to the cyanobacterial gene sll1521, was also identified in our assay (supplemental Table S8). The Flv3 from Synechocystis sp. PCC 6803 was shown to function as an NAD(P)H:oxygen oxidoreductase (54) and has a role in the Mehler reaction (Fig. 10) (55); thus, they are proposed to be involved in detoxification of oxidative compounds such as oxygen and may act as a safety valve allowing excessive reducing power to be dissipated under aerobic or microaerobic environments (54). Our interaction results suggest that FDX1 could also have a role in these functions.
FIGURE 10.
FDX1 putative interaction partners identified in Chlamydomonas. FDX1 binding partners are indicated with red boxes and categorized into two primary physiological functions (carbon cycle and photosynthesis). Green lines indicate reductant flow through the relevant pathways. Solid red lines represent interactions supported by two assays (interaction experiments, i.e. yeast two-hybrid or pulldown, and in vitro experiments, i.e. biochemical assay) and dashed red lines, or the absence of lines (for convenience), indicate interactions supported by a single interaction experiment (i.e. ribulose-1,5-bisphosphate carboxylase or pulldown). The * identifies interaction partners shared with FDX2; ** highlights interaction partners shared with FDX5, and *** indicates interaction partners shared with FDX4. HYDEF/HYDG interacts with HYDA1 and -2 but are colored in blue because they were detected to interact with FDX5 but not FDX1 due to autoactivation. LHCBM7 is shown as associated with PSII but could be associated with PSI during state transition.
FDX1 was also shown to interact with the light harvesting protein LHCA5, which in plants is involved in the supercomplex formation with PSI-LHCI, cytochrome b6f, FNR, and PGRL1 and plays a major role in cyclic electron flux (Table 1 and Fig. 10) (56). However, the Chlamydomonas LHCA5 protein is not a real ortholog of the Arabidopsis LHCA5, and its function is not known.
TRXs are central to regulation of both photosynthetic and nonphotosynthetic cellular metabolisms. FDXs reduce TRXs by interaction with a specific iron-sulfur-containing enzyme, FDX thioredoxin reductase. Under our experimental conditions, both FDX1 and FDX5 showed interaction with chloroplastic FDX thioredoxin reductase (GI:158284302) and with malic dehydrogenase 4 (MDH4). FDX thioredoxin reductases and FDXs form complexes with fructose-1,6-biphosphatase, which is linked to the Calvin/Bassham cycle (57) and is a well known carbon fixation regulatory mechanism, but also with the NADP+-dependent malate dehydrogenase that is used as a redox valve in cyanobacteria (58) (Fig. 10). The latter interaction was recently confirmed by Terauchi et al. (16), who reconstituted a complete light activation system and evaluated the electron transfer from FDX1 or FDX2 to NADP-malate dehydrogenase by following malate dehydrogenase activation; they showed that FDX2 was a better mediator than FDX1; FDX5 was not tested in their assay. Our results (Table 1), combined with theirs, suggest an additional role for these FDXs in maintaining redox balance in Chlamydomonas (59, 60).
Besides its role in the regulation of carbohydrate synthesis through the activation of the fructose-1,6-biphosphatase system, FDX1 is also involved in the TRX-mediated regulation of pyrophosphorylase and ribulose-1,5-bisphosphate carboxylase in response to phosphoglycerate concentrations. Here, we show that FDX1 and FDX4 interact with GAP3, FBA3, and PFK2 (see “Results” and Table 1), confirming the proposed function of these FDXs in phosphoglycerate sensing and subsequent regulatory events. Interestingly, it has been observed that phosphoglycerate activates yet another regulatory enzyme, adenosine diphosphoglucose pyrophosphorylase, which is important in the synthesis of starch (61). In our study, FDX1 and FDX5 interact with SBE3, a starch branching enzyme with a putative chloroplast transit peptide (Table 1 and Fig. 10), the nonreducing end of the cleaved glucan to an α-1,6-position.
It is also known that reduced FDX is the electron donor in the reductive synthesis of pyruvate from carbon dioxide and acetyl coenzyme A (62), and here we show that FDX1 interacts with acetate CoA ligase (ACS3) and the dihydrolipoyl dehydrogenase subunit of the pyruvate dehydrogenase involved in pyruvate synthesis. It is also important to note that pyruvate decarboxylation is a source of acetyl-CoA that can be use as a substrate by the FDX-dependent enzyme, pyruvate-ferredoxin oxidoreductase, which in turn transfers electrons to the hydrogenase for hydrogen production under dark anaerobiosis. It has been shown that FDX1 is the best mediator (compared with the other five FDXs) for providing electrons to the pyruvate-ferredoxin oxidoreductase enzyme and drives the best hydrogen production rate (63, 64).
Finally, FDXs have been shown to interact with desaturase enzymes involved in fatty acid biosynthesis (65). Desaturase enzymes play critical roles in adjusting the physiological properties of membrane lipids and in several essential cellular processes (66). The two reducing equivalents for catalysis (67) are delivered by FDX in the case of acyl-ACP desaturase and of the integral membrane acyl lipid desaturases (68). We found that both FDX1 and FDX5 interact with DES6, the chloroplast isoform of a desaturase, suggesting a potential role of these FDXs in fatty acid biosynthesis as well (Table 1 and Fig. 10).
This work describes a large scale screening and characterization effort to comprehensively describe the FDX interaction network in Chlamydomonas. The research identified the FDXs that are primarily responsible for supporting hydrogen photo-production and NADP reduction. We found that only FDX1 and FDX2 are capable of donating reductant to hydrogenases, with FDX1 driving a rate of hydrogen and NADPH production at greater than 2-fold higher rates than FDX2. Furthermore, we discovered other FDX-linked pathways that are related to central metabolism and discussed their possible implication in Chlamydomonas. We also presented novel evidence for the formation of FDX dimers as observed in Pseudomonas (69). Finally, we suggest that the FDXs have overlapping activity in multiple metabolic pathways. Our study opens the door for future experiments aimed at understanding in detail the novel interactions revealed by our research.
Acknowledgments
We thank Dr. Kathleen Ratcliff and Dr. Iftach Yacoby for thoughtful discussions regarding our in vitro NADPH and hydrogen photo-production assays. We thank Dr. Venkat Subramanian for the constant help in the laboratory. In addition, we thank the Proteomics and Metabolomics Facility at Colorado State University for performing in-gel digest of FDX pulldowns.
This work was supported by the United States Department of Energy under Contract DE-AC36-08GO28308 from the National Renewable Energy Laboratory, a grant from the Biological and Environmental Research program, Division of Energy Biosciences, Office of Science, the United States Department of Energy (to M. L. G.), and the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the United States Department of Energy (to P. W. K.).

This article contains supplemental Table S1–S11.
- FDX
- ferredoxin
- FNR
- ferredoxin-NADP reductase
- 3AT
- 3-amino-1,2,4-triazole
- PSI and PSII
- photosystem I or II
- SBP
- squamosa promoter-binding protein
- LHC
- light harvesting complex
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PBS
- Predicted Biological Score
- Ura
- uracil
- CV
- column volume
- NiR
- nitrite reductase
- Chl
- chlorophyll.
REFERENCES
- 1. Hanke G., Mulo P. (2013) Plant type ferredoxins and ferredoxin-dependent metabolism. Plant Cell Environ. 36, 1071–1084 [DOI] [PubMed] [Google Scholar]
- 2. Gou P., Hanke G. T., Kimata-Ariga Y., Standley D. M., Kubo A., Taniguchi I., Nakamura H., Hase T. (2006) Higher order structure contributes to specific differences in redox potential and electron transfer efficiency of root and leaf ferredoxins. Biochemistry 45, 14389–14396 [DOI] [PubMed] [Google Scholar]
- 3. Hase T., Mizutani S., Mukohata Y. (1991) Expression of maize ferredoxin cDNA in Escherichia coli: Comparison of photosynthetic and nonphotosynthetic ferredoxin isoproteins and their chimeric molecule. Plant Physiol. 97, 1395–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Onda Y., Matsumura T., Kimata-Ariga Y., Sakakibara H., Sugiyama T., Hase T. (2000) Differential interaction of maize root ferredoxin:NADP+ oxidoreductase with photosynthetic and nonphotosynthetic ferredoxin isoproteins. Plant Physiol. 123, 1037–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hase T., Wada K., Matsubara H. (1976) Amino-acid sequence of major component of Aphanothece-sacrum ferredoxin. J. Biochem. 79, 329–343 [DOI] [PubMed] [Google Scholar]
- 6. Hutson K. G., Rogers L. J., Haslett B. G., Boulter D., Cammack R. (1978) Comparative studies on two ferredoxins from the cyanobacterium Nostoc strain MAC. Biochem. J. 172, 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Matsubara H., Sasaki R. M. (1968) Spinach ferredoxin. II. Tryptic, chymotryptic, and thermolytic peptides, and complete amino acid sequence. J. Biol. Chem. 243, 1732–1757 [PubMed] [Google Scholar]
- 8. Orme-Johnson W. H. (1973) Iron-sulfur proteins: structure and function. Annu. Rev. Biochem. 42, 159–204 [DOI] [PubMed] [Google Scholar]
- 9. Wada K., Kagamiyama H., Shin M., Matsubara H. (1974) Ferredoxin from a blue-green alga, Aphanothece sacrum (Suringar) Okada. J. Biochem. 76, 1217–1225 [DOI] [PubMed] [Google Scholar]
- 10. Kameda H., Hirabayashi K., Wada K., Fukuyama K. (2011) Mapping of protein-protein interaction sites in the plant-type [2Fe-2S] ferredoxin. PLoS One 6, e21947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bertini I., Luchinat C., Provenzani A., Rosato A., Vasos P. R. (2002) Browsing gene banks for Fe2S2 ferredoxins and structural modeling of 88 plant-type sequences: an analysis of fold and function. Proteins 46, 110–127 [DOI] [PubMed] [Google Scholar]
- 12. Yonekura-Sakakibara K., Onda Y., Ashikari T., Tanaka Y., Kusumi T., Hase T. (2000) Analysis of reductant supply systems for ferredoxin-dependent sulfite reductase in photosynthetic and nonphotosynthetic organs of maize. Plant Physiol. 122, 887–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huisman J. G., Moorman A. F., Verkley F. N. (1978) In vitro synthesis of chloroplast ferredoxin as a high molecular weight precursor in a cell-free protein synthesizing system from wheat germs. Biochem. Biophys. Res. Commun. 82, 1121–1131 [DOI] [PubMed] [Google Scholar]
- 14. Smeekens S., Weisbeek P., Robinson C. (1990) Protein-transport into and within chloroplasts. Trends Biochem. Sci. 15, 73–76 [DOI] [PubMed] [Google Scholar]
- 15. Takahashi Y., Mitsui A., Hase T., Matsubara H. (1986) Formation of the iron-sulfur cluster of ferredoxin in isolated chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 83, 2434–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Terauchi A. M., Lu S. F., Zaffagnini M., Tappa S., Hirasawa M., Tripathy J. N., Knaff D. B., Farmer P. J., Lemaire S. D., Hase T., Merchant S. S. (2009) Pattern of expression and substrate specificity of chloroplast ferredoxins from Chlamydomonas reinhardtii. J. Biol. Chem. 284, 25867–25878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jacobs J., Pudollek S., Hemschemeier A., Happe T. (2009) A novel, anaerobically induced ferredoxin in Chlamydomonas reinhardtii. FEBS Lett. 583, 325–329 [DOI] [PubMed] [Google Scholar]
- 18. Schmitter J. M., Jacquot J. P., de Lamotte-Guéry F., Beauvallet C., Dutka S., Gadal P., Decottignies P. (1988) Purification, properties and complete amino acid sequence of the ferredoxin from a green alga, Chlamydomonas reinhardtii. Eur. J. Biochem. 172, 405–412 [DOI] [PubMed] [Google Scholar]
- 19. Mus F., Dubini A., Seibert M., Posewitz M. C., Grossman A. R. (2007) Anaerobic acclimation in Chlamydomonas reinhardtii: anoxic gene expression, hydrogenase induction, and metabolic pathways. J. Biol. Chem. 282, 25475–25486 [DOI] [PubMed] [Google Scholar]
- 20. Lambertz C., Hemschemeier A., Happe T. (2010) Anaerobic expression of the ferredoxin-encoding FDX5 gene of Chlamydomonas reinhardtii is regulated by the Crr1 transcription factor. Eukaryot. Cell 9, 1747–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ghirardi M. L., Dubini A., Yu J., Maness P. C. (2009) Photobiological hydrogen-producing systems. Chem. Soc. Rev. 38, 52–61 [DOI] [PubMed] [Google Scholar]
- 22. Meuser J. E., D'Adamo S., Jinkerson R. E., Mus F., Yang W., Ghirardi M. L., Seibert M., Grossman A. R., Posewitz M. C. (2012) Genetic disruption of both Chlamydomonas reinhardtii [FeFe]-hydrogenases: Insight into the role of HYDA2 in H-2 production. Biochem. Biophys. Res. Commun. 417, 704–709 [DOI] [PubMed] [Google Scholar]
- 23. Winkler M., Hemschemeier A., Jacobs J., Stripp S., Happe T. (2010) Multiple ferredoxin isoforms in Chlamydomonas reinhardtii–their role under stress conditions and biotechnological implications. Eur. J. Cell Biol. 89, 998–1004 [DOI] [PubMed] [Google Scholar]
- 24. Long H., Chang C. H., King P. W., Ghirardi M. L., Kim K. (2008) Brownian dynamics and molecular dynamics study of the association between hydrogenase and ferredoxin from Chlamydomonas reinhardtii. Biophys. J. 95, 3753–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brückner A., Polge C., Lentze N., Auerbach D., Schlattner U. (2009) Yeast two-hybrid, a powerful tool for systems biology. Int. J. Mol. Sci. 10, 2763–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ghirardi M. L., Togasaki R. K., Seibert M. (1997) Oxygen sensitivity of algal H2- production. Appl. Biochem. Biotechnol. 63, 141–151 [DOI] [PubMed] [Google Scholar]
- 27. Harris E. H. (2009) Introduction to Chlamydomonas and Its Laboratory Use, 2nd Ed., Academic Press, Oxford [Google Scholar]
- 28. Bartel P., Chien C. T., Sternglanz R., Fields S. (1993) Elimination of false positives that arise in using the two-hybrid system. BioTechniques 14, 920–924 [PubMed] [Google Scholar]
- 29. Fromont-Racine M., Rain J. C., Legrain P. (1997) Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16, 277–282 [DOI] [PubMed] [Google Scholar]
- 30. Rain J. C., Selig L., De Reuse H., Battaglia V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., Schächter V., Chemama Y., Labigne A., Legrain P. (2001) The protein-protein interaction map of Helicobacter pylori. Nature 409, 211–215 [DOI] [PubMed] [Google Scholar]
- 31. Formstecher E., Aresta S., Collura V., Hamburger A., Meil A., Trehin A., Reverdy C., Betin V., Maire S., Brun C., Jacq B., Arpin M., Bellaiche Y., Bellusci S., Benaroch P., Bornens M., Chanet R., Chavrier P., Delattre O., Doye V., Fehon R., Faye G., Galli T., Girault J. A., Goud B., de Gunzburg J., Johannes L., Junier M. P., Mirouse V., Mukherjee A., Papadopoulo D., Perez F., Plessis A., Rossé C., Saule S., Stoppa-Lyonnet D., Vincent A., White M., Legrain P., Wojcik J., Camonis J., Daviet L. (2005) Protein interaction mapping: a Drosophila case study. Genome Res. 15, 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wojcik J., Boneca I. G., Legrain P. (2002) Prediction, assessment and validation of protein interaction maps in bacteria. J. Mol. Biol. 323, 763–770 [DOI] [PubMed] [Google Scholar]
- 33. Michoux F., Takasaka K., Boehm M., Nixon P. J., Murray J. W. (2010) Structure of CyanoP at 2.8 A: implications for the evolution and function of the PsbP subunit of photosystem II. Biochemistry 49, 7411–7413 [DOI] [PubMed] [Google Scholar]
- 34. Yacoby I., Tegler L. T., Pochekailov S., Zhang S., King P. W. (2012) Optimized expression and purification for high-activity preparations of algal [FeFe]-hydrogenase. PLoS One 7, e35886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. King P. W., Posewitz M. C., Ghirardi M. L., Seibert M. (2006) Functional studies of [FeFe] hydrogenase maturation in an Escherichia coli biosynthetic system. J. Bacteriol. 188, 2163–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., Witman G. B., Terry A., Salamov A., Fritz-Laylin L. K., Maréchal-Drouard L., Marshall W. F., Qu L. H., Nelson D. R., Sanderfoot A. A., Spalding M. H., Kapitonov V. V., Ren Q., Ferris P., Lindquist E., Shapiro H., Lucas S. M., Grimwood J., Schmutz J., Cardol P., Cerutti H., Chanfreau G., Chen C. L., Cognat V., Croft M. T., Dent R., Dutcher S., Fernández E., Fukuzawa H., González-Ballester D., González-Halphen D., Hallmann A., Hanikenne M., Hippler M., Inwood W., Jabbari K., Kalanon M., Kuras R., Lefebvre P. A., Lemaire S. D., Lobanov A. V., Lohr M., Manuell A., Meier I., Mets L., Mittag M., Mittelmeier T., Moroney J. V., Moseley J., Napoli C., Nedelcu A. M., Niyogi K., Novoselov S. V., Paulsen I. T., Pazour G., Purton S., Ral J. P., Riano-Pachon D. M., Riekhof W., Rymarquis L., Schroda M., Stern D., Umen J., Willows R., Wilson N., Zimmer S. L., Allmer J., Balk J., Bisova K., Chen C. J., Elias M., Gendler K., Hauser C., Lamb M. R., Ledford H., Long J. C., Minagawa J., Page M. D., Pan J., Pootakham W., Roje S., Rose A., Stahlberg E., Terauchi A. M., Yang P., Ball S., Bowler C., Dieckmann C. L., Gladyshev V. N., Green P., Jorgensen R., Mayfield S., Mueller-Roeber B., Rajamani S., Sayre R. T., Brokstein P., Dubchak I., Goodstein D., Hornick L., Huang Y. W., Jhaveri J., Luo Y., Martinez D., Ngau W. C., Otillar B., Poliakov A., Porter A., Szajkowski L., Werner G., Zhou K., Grigoriev I. V., Rokhsar D. S., Grossman A. R. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meyer-Arendt K., Old W. M., Houel S., Renganathan K., Eichelberger B., Resing K. A., Ahn N. G. (2011) IsoformResolver: A peptide-centric algorithm for protein inference. J. Proteome Res. 10, 3060–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Old W. M., Meyer-Arendt K., Aveline-Wolf L., Pierce K. G., Mendoza A., Sevinsky J. R., Resing K. A., Ahn N. G. (2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- 39. Lill R., Mühlenhoff U. (2006) Iron-sulfur protein biogenesis in eukaryotes: components and mechanisms. Annu. Rev. Cell Dev. Biol. 22, 457–486 [DOI] [PubMed] [Google Scholar]
- 40. Vallese F., Berto P., Ruzzene M., Cendron L., Sarno S., De Rosa E., Giacometti G. M., Costantini P. (2012) Biochemical analysis of the interactions between the proteins involved in the [FeFe]-hydrogenase maturation process. J. Biol. Chem. 287, 36544–36555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bukhov N., Carpentier R. (2004) Alternative photosystem I-driven electron transport routes: mechanisms and functions. Photosynth. Res. 82, 17–33 [DOI] [PubMed] [Google Scholar]
- 42. Yacoby I., Pochekailov S., Toporik H., Ghirardi M. L., King P. W., Zhang S. (2011) Photosynthetic electron partitioning between [FeFe]-hydrogenase and ferredoxin:NADP+-oxidoreductase (FNR) enzymes in vitro. Proc. Natl. Acad. Sci. U.S.A. 108, 9396–9401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hanke G. T., Satomi Y., Shinmura K., Takao T., Hase T. (2011) A screen for potential ferredoxin electron transfer partners uncovers new, redox-dependent interactions. Biochim. Biophys. Acta 1814, 366–374 [DOI] [PubMed] [Google Scholar]
- 44. Guerrero M. G., Vega J. M., Losada M. (1981) The assimilatory nitrate-reducing system and its regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 32, 169–204 [Google Scholar]
- 45. Hewitt E. J. (1975) Assimilatory nitrate-nitrite reduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 26, 73–100 [Google Scholar]
- 46. García-Sánchez M. I., Gotor C., Jacquot J. P., Stein M., Suzuki A., Vega J. M. (1997) Critical residues of Chlamydomonas reinhardtii ferredoxin for interaction with nitrite reductase and glutamate synthase revealed by site-directed mutagenesis. Eur. J. Biochem. 250, 364–368 [DOI] [PubMed] [Google Scholar]
- 47. Baier M., Dietz K. J. (1999) Protective function of chloroplast 2-cysteine peroxiredoxin in photosynthesis. Evidence from transgenic Arabidopsis. Plant Physiol. 119, 1407–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lemeille S., Turkina M. V., Vener A. V., Rochaix J. D. (2010) Stt7-dependent phosphorylation during state transitions in the green alga Chlamydomonas reinhardtii. Mol. Cell. Proteomics 9, 1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bulte L., Gans P., Rebeille F., Wollman F. A. (1990) Atp control on state transitions in vivo in Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1020, 72–80 [Google Scholar]
- 50. Elrad D., Niyogi K. K., Grossman A. R. (2002) A major light-harvesting polypeptide of photosystem II functions in thermal dissipation. Plant Cell 14, 1801–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Terashima M., Specht M., Naumann B., Hippler M. (2010) Characterizing the anaerobic response of Chlamydomonas reinhardtii by quantitative proteomics. Mol. Cell. Proteomics 9, 1514–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Terashima M., Petroutsos D., Hüdig M., Tolstygina I., Trompelt K., Gäbelein P., Fufezan C., Kudla J., Weinl S., Finazzi G., Hippler M. (2012) Calcium-dependent regulation of cyclic photosynthetic electron transfer by a CAS, ANR1, and PGRL1 complex. Proc. Natl. Acad. Sci. U.S.A. 109, 17717–17722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Peters J. W., Broderick J. B. (2012) Emerging paradigms for complex iron-sulfur cofactor assembly and insertion. Annu. Rev. Biochem. 81, 429–450 [DOI] [PubMed] [Google Scholar]
- 54. Allahverdiyeva Y., Ermakova M., Eisenhut M., Zhang P., Richaud P., Hagemann M., Cournac L., Aro E. M. (2011) Interplay between flavodiiron proteins and photorespiration in Synechocystis sp. PCC 6803. J. Biol. Chem. 286, 24007–24014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Helman Y., Tchernov D., Reinhold L., Shibata M., Ogawa T., Schwarz R., Ohad I., Kaplan A. (2003) Genes encoding A-type flavoproteins are essential for photoreduction of O2 in cyanobacteria. Curr. Biol. 13, 230–235 [DOI] [PubMed] [Google Scholar]
- 56. Peng L., Fukao Y., Fujiwara M., Takami T., Shikanai T. (2009) Efficient operation of NAD(P)H dehydrogenase requires supercomplex formation with photosystem I via minor LHCI in Arabidopsis. Plant Cell 21, 3623–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jacquot J. P., Lopez-Jaramillo J., Miginiac-Maslow M., Lemaire S., Cherfils J., Chueca A., Lopez-Gorge J. (1997) Cysteine 153 is required for redox regulation of pea chloroplast fructose-1,6-bisphosphatase. FEBS Lett. 401, 143–147 [DOI] [PubMed] [Google Scholar]
- 58. Lemaire S. D., Quesada A., Merchan F., Corral J. M., Igeno M. I., Keryer E., Issakidis-Bourguet E., Hirasawa M., Knaff D. B., Miginiac-Maslow M. (2005) NADP-malate dehydrogenase from unicellular green alga Chlamydomonas reinhardtii. A first step toward redox regulation? Plant Physiol. 137, 514–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Krömer S., Scheibe R. (1996) Function of the chloroplastic malate valve for respiration during photosynthesis. Biochem. Soc. Trans. 24, 761–766 [DOI] [PubMed] [Google Scholar]
- 60. Scheibe R., Dietz K. J. (2012) Reduction-oxidation network for flexible adjustment of cellular metabolism in photoautotrophic cells. Plant Cell Environ. 35, 202–216 [DOI] [PubMed] [Google Scholar]
- 61. Van den Koornhuyse N., Libessart N., Delrue B., Zabawinski C., Decq A., Iglesias A., Carton A., Preiss J., Ball S. (1996) Control of starch composition and structure through substrate supply in the monocellular alga Chlamydomonas reinhardtii. J. Biol. Chem. 271, 16281–16287 [DOI] [PubMed] [Google Scholar]
- 62. Buchanan B. B., Bachofen R., Arnon D. I. (1964) Role of ferredoxin in the reductive assimilation of Co2 and acetate by extracts of the photosynthetic bacterium, chromatium. Proc. Natl. Acad. Sci. U.S.A. 52, 839–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Noth J., Krawietz D., Hemschemeier A., Happe T. (2013) Pyruvate: Ferredoxin oxidoreductase is coupled to light-independent hydrogen production in Chlamydomonas reinhardtii. J. Biol. Chem. 288, 4368–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. van Lis R., Baffert C., Couté Y., Nitschke W., Atteia A. (2013) Chlamydomonas reinhardtii chloroplasts contain a homodimeric pyruvate: ferredoxin oxidoreductase that functions with FDX1. Plant Physiol. 161, 57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chazarreta-Cifre L., Martiarena L., de Mendoza D., Altabe S. G. (2011) Role of ferredoxin and flavodoxins in Bacillus subtilis fatty acid desaturation. J. Bacteriol. 193, 4043–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Aguilar P. S., de Mendoza D. (2006) Control of fatty acid desaturation: a mechanism conserved from bacteria to humans. Mol. Microbiol. 62, 1507–1514 [DOI] [PubMed] [Google Scholar]
- 67. Shanklin J., Cahoon E. B. (1998) Desaturation and related modifications of fatty acids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 611–641 [DOI] [PubMed] [Google Scholar]
- 68. Wada H., Schmidt H., Heinz E., Murata N. (1993) In vitro ferredoxin-dependent desaturation of fatty acids in cyanobacterial thylakoid membranes. J. Bacteriol. 175, 544–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Iwasaki T., Kappl R., Bracic G., Shimizu N., Ohmori D., Kumasaka T. (2011) ISC-like [2Fe-2S] ferredoxin (FdxB) dimer from Pseudomonas putida JCM 20004: structural and electron-nuclear double resonance characterization. J. Biol. Inorg. Chem. 16, 923–935 [DOI] [PubMed] [Google Scholar]