Background: Presenilins regulate multiple cellular pathways by controlling transcription.
Results: Presenilins repress neurotrypsin expression by preventing CREB recruitment to its promoter.
Conclusion: A new strategy utilized by presenilins to regulate CREB signaling relies on preventing CREB recruitment to gene promoters.
Significance: Learning how presenilins control CREB signaling will help understanding their physiological/pathological roles.
Keywords: Chromatin Immunoprecipitation (ChIP), CREB, Presenilin, Promoters, Secretases, Drosophila Larval Brains, GSK3, MEFs, Tequila, Neurotrypsin
Abstract
Presenilins, the catalytic components of the γ-secretase complex, are upstream regulators of multiple cellular pathways via regulation of gene transcription. However, the underlying mechanisms and the genes regulated by these pathways are poorly characterized. In this study, we identify Tequila and its mammalian ortholog Prss12 as genes negatively regulated by presenilins in Drosophila larval brains and mouse embryonic fibroblasts, respectively. Prss12 encodes the serine protease neurotrypsin, which cleaves the heparan sulfate proteoglycan agrin. Altered neurotrypsin activity causes serious synaptic and cognitive defects; despite this, the molecular processes regulating neurotrypsin expression and activity are poorly understood. Using γ-secretase drug inhibitors and presenilin mutants in mouse embryonic fibroblasts, we found that a mature γ-secretase complex was required to repress neurotrypsin expression and agrin cleavage. We also determined that PSEN1 endoproteolysis or processing of well known γ-secretase substrates was not essential for this process. At the transcriptional level, PSEN1/2 removal induced cyclic AMP response element-binding protein (CREB)/CREB-binding protein binding, accumulation of activating histone marks at the neurotrypsin promoter, and neurotrypsin transcriptional and functional up-regulation that was dependent on GSK3 activity. Upon PSEN1/2 reintroduction, this active epigenetic state was replaced by a methyl CpG-binding protein 2 (MeCP2)-containing repressive state and reduced neurotrypsin expression. Genome-wide analysis revealed hundreds of other mouse promoters in which CREB binding is similarly modulated by the presence/absence of presenilins. Our study thus identifies Tequila and neurotrypsin as new genes repressed by presenilins and reveals a novel mechanism used by presenilins to modulate CREB signaling based on controlling CREB recruitment.
Introduction
Presenilins are ubiquitously expressed transmembrane aspartyl proteases that play essential roles in development and disease (1, 2). Presenilin functions as the catalytic component of the larger γ-secretase complex, which cleaves hundreds of transmembrane proteins (3). Generally, γ-secretase cleavage of transmembrane substrates results in the release of intracellular domains and small extracellular fragments. Both types of fragments can initiate a cascade of signaling events often culminating in regulation of transcription (3, 4).
Besides presenilin, there are three other γ-secretase components, nicastrin, anterior pharynx-defective 1 (Aph1),4 and presenilin enhancer 2 (Pen2), that are required to form a mature and active γ-secretase complex (1). The prevalent idea for assembly of the complex is that Aph1 functions as a scaffold subunit and initiates a subcomplex together with nicastrin (5). Nicastrin, which might contain the substrate recognition and binding site, promotes maturation and proper trafficking of the other γ-secretase components (5). Subsequently, presenilin binds to the Aph1-Nicastrin subcomplex, and finally, incorporation of Pen2 to the complex appears to trigger autocatalytic endoproteolytic cleavage of the presenilins and therefore formation of a mature and active γ-secretase complex (5). In mammals, the heterogeneity of the γ-secretase complex is achieved by the existence of two presenilins, presenilin-1 (PSEN1) and presenilin-2 (PSEN2), and two Aph1 forms (Aph1A and Aph1B), each being alternatively spliced (5). Additionally and contributing to this heterogeneity, other non-core components (e.g. transmembrane trafficking protein, 21-KD TMP21 and γ-secretase-activating protein gSAP) interact with subsets of γ-secretase complexes and appear to differentially modulate their proteolytic activity over specific γ-secretase substrates (5).
Several signaling pathways regulated by γ-secretase activity involve the transcription factor cAMP response element-binding protein (CREB). Specifically, γ-secretase-generated N-cadherin and E-cadherin intracellular domains (N-cad/CTF2 and E-cad/CTF2, respectively) can bind to and promote proteasomal degradation of the coactivator CREB-binding protein (CBP), which presumably results in poor association with stimulated CREB and therefore reduced CREB-regulated transcription (6, 7). However, this regulatory mechanism must be more complex because inhibition rather than activation of CREB-dependent transcription has been observed in some presenilin-deficient systems (8, 9). Similarly, the γ-secretase-generated amyloid β peptide, which is released from amyloid precursor protein (APP) after sequential processing by β-site APP-cleaving enzyme 1 (BACE1) and γ-secretase, may also impair CREB-dependent transcription via inhibition of protein kinase A (PKA) and other kinases (10, 11). Amyloid β has also been shown to inhibit CREB activity by decreasing intracellular calcium influx, which reduces phosphorylation of the CREB coactivator CRTC1 (12). Whereas a connection between presenilins and CREB signaling has been well established, the list of genes regulated by this connection has not been fully explored and is generally limited to the study of the c-Fos gene, a well known CREB target.
In experiments to identify new presenilin-regulated genes, we discovered that Tequila (Teq) and its mammalian ortholog Prss12 are negatively regulated by presenilins in Drosophila larval brains and mouse embryonic fibroblasts (MEFs), respectively. Prss12 encodes the serine protease neurotrypsin, which cleaves the heparan sulfate proteoglycan agrin. Teq/neurotrypsin is of particular interest as a presenilin target given its likely role in memory and cognition. Specifically, Teq mutants in Drosophila have defects in memory formation, and neurotrypsin itself has been implicated in processing of agrin, which is an important synaptic component (14, 15) Finally, neurotrypsin mutations in humans cause mental retardation (16). Mechanistically, we found that in MEFs the presence of a mature γ-secretase complex, but not the autocatalytic endoproteolysis of PSEN1 or the processing of well known γ-secretase substrates, is required to repress neurotrypsin and agrin cleavage. At the chromatin level, we found that presenilins modulated the transcriptional and epigenetic state of the neurotrypsin promoter. Specifically, presenilins prevented CREB/CBP binding to the neurotrypsin promoter and induced a repressive epigenetic state characterized by binding of MeCP2 in some instances. Furthermore, we found that in Psen1/2 double KO (dKO) cells GSK3 activity was required to maintain high levels of neurotrypsin expression. Intriguingly, we identified hundreds of other promoters whose CREB occupation was also blocked by the presence of presenilins. Altogether, we have uncovered a novel strategy utilized by presenilins to regulate CREB-dependent transcription via suppression of CREB binding to specific promoters in the mouse genome. For Prss12/neurotrypsin, this regulatory process had direct effects on the biological activity of its encoded protein as measured by agrin cleavage.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Antibodies generated against agrin (R132), neurotrypsin (G95), and amyloid precursor protein-like (APPL) as well as purified Wnt3a were described previously (15, 17, 18). The anti-α-tubulin (DM1A), anti-APP (22C11) to detect full-length APP, anti-PSEN1 (MAB5232), anti-GSK3β (07-1413), anti-actin (MAB1501), anti-H3K27me3 (07-449), anti-H3K4me3 (07-473), and anti-H3K9ac (07-352) antibodies were purchased from Millipore. The anti-APP antibody to detect APP C-terminal fragment (CTF) was purchased from Zymed Laboratories Inc./Invitrogen. The anti-nicastrin (PA1758) antibody was purchased from Affinity Bioreagents. The anti-N-cadherin (610920) and β-catenin (610153/4) antibodies were purchased from BD Transduction Laboratories. The anti-PSEN2 (2192) and anti-GSK3β phosphorylated at serine 9 (P-S9-GSK3β) (9336) antibodies were purchased from Cell Signaling Technology. APLP2 (171616) was purchased from Calbiochem. The anti-CREB1 (C-21), anti-CBP (C-20, 451, A-22, and H-300), and anti-RNA polymerase (Pol) II (H-224) antibodies were purchased from Santa Cruz Biotechnology. The anti-MeCP2 (M9317) antibody was purchased from Sigma. The anti-H3K9me3 (ab8898) and anti-H3K27ac (ab4729) antibodies were purchased from Abcam. The mouse cDNA clone containing CMV-neurotrypsin was purchased from Origene. The small interfering RNAs (siRNAs) for mouse Mecp2 and mouse neurotrypsin were purchased from Dharmacon. Compound E (565790) was purchased from Calbiochem and dissolved in DMSO. Lithium chloride (LiCl) (5852) was purchased from Mallinckrodt Baker. Factor XV (Santa Cruz Biotechnology) and IWR (EMD Millipore) were dissolved in DMSO.
Cell Culture and Treatments
Immortalized wild-type (control), Psen1/2 dKO, Psen1/2 dKO (+wild-type human PSEN1), Psen1/2 dKO (+wild-type human PSEN1/2), Psen1/2 dKO (+D257A human PSEN1+wild-type PSEN2), Psen1/2 dKO (+ΔE9 human PSEN1), Psen1/2 dKO (+M146L human PSEN1), Psen1 knock-out (KO), and Psen2 KO MEFs were described previously (19, 20). An independent set of wild-type (control) and Psen1/2 dKO lines also used have been described (13). Nicastrin KO and corresponding wild-type control MEFs were described previously (21). Transfections with plasmids or siRNAs were performed using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Drug concentration and exposure time are indicated in corresponding figure legends.
Drosophila Stocks
Flies were reared in standard Drosophila medium at 25 °C. Drosophila stocks psn143 and controls Oregon R (OrR) were from Bloomington. The psn143 allele contains a short genomic deletion that removes amino acids 136–224 of presenilin (Psn) (22). This stretch of amino acids contains the transmembrane (TM) domains TM2, TM3, and TM4. It was shown previously that this deletion prevents the generation of Notch intracellular domain (22) and similarly induces strong accumulation of APPL CTF (Fig. 1A, right panel). Drosophila UAS-Psn lines and expression of the transgene were induced by ApplGal4 as reported previously (23).
FIGURE 1.
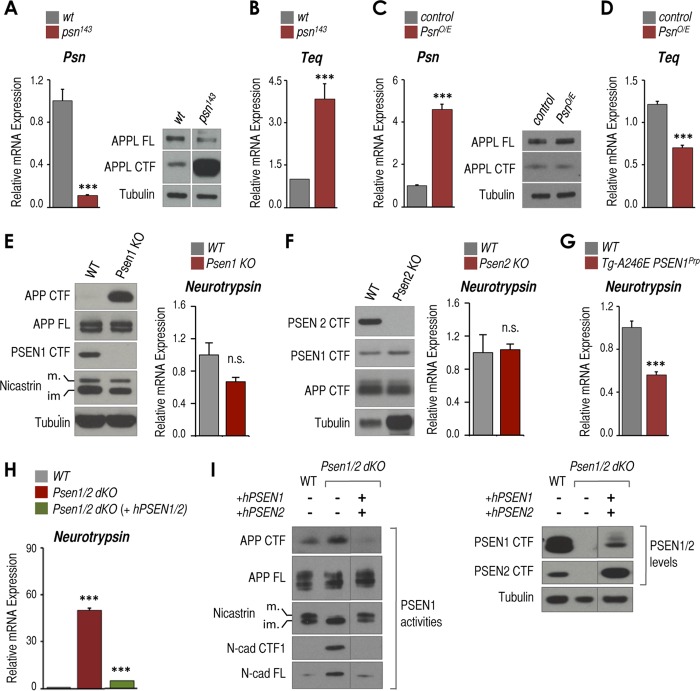
Presenilins negatively regulate expression of neurotrypsin and its Drosophila ortholog Teq. A, relative mRNA expression of Psn measured by RT-qPCR (left panel) and protein levels of full-length APPL (APPL FL), APPL CTFs, and tubulin measured by Western blot (right panel) in OrR (wild-type) and Psn-deficient (psn143) third instar Drosophila larval brains (n = 3 pools of 10 brains each). B, as in A (left panel) but Teq mRNA expression. C, as in A but ApplGAL4 (control) and Psn-overexpressing (PsnO/E) brains. D, as in B but ApplGAL4 (control) and Psn-overexpressing (PsnO/E) brains. E, left panel, Western blot analysis of APP CTFs, full-length APP (APP FL), PSEN1 CTF, mature (m.) and immature (im.) nicastrin, and tubulin as loading controls of whole mouse brains from embryonic day 15 Psen1 KO and wild-type (WT) littermates. Right panel, Tbp-normalized neurotrypsin mRNA expression from the same samples. F, left panel, Western blot analysis of PSEN2 CTF, PSEN1 CTF, APP CTFs, and tubulin as loading controls of whole brains from postnatal day 11 Psen2 KO and aged-matched wild-type (WT) mice. Right panel, Tbp-normalized neurotrypsin mRNA expression from the same samples as in the left panel. G, Tbp-normalized neurotrypsin mRNA expression in hippocampi from postnatal day 14 transgenic mice overexpressing human A246E PSEN1 (Tg-A246E PSEN1Prp), and wild-type (WT) littermates. H, RT-qPCR analysis of Tbp-normalized neurotrypsin mRNA expression in wild-type, Psen1/2 dKO, and Psen1/2 dKO (+hPSEN1/2) cells. I, Western blot analysis of full-length APP (APP FL), APP CTFs, mature (m.) and immature (im.) nicastrin, full-length N-cad (N-cad FL), and N-cad/CTF1 (left panel) and PSEN1 CTF, PSEN2 CTF, and tubulin (right panel) from the same cells as in H. Samples were run in the same gel but were cut and pasted to adjust to layout of panel. All experiments shown in this figure were triplicates (n = 3) except Psen2 KO (n = 2). Representative Western blots are displayed. Mean ± S.E. is shown. p values are shown when significant (Student's t test): ***, p ≤ 0.001. n.s., not significant. Error bars represent S.E.
Mouse Strains
Tg-A246EPSEN1Prp mice expressing the A246E human PSEN1 mutation under the prion protein (PrP) promoter and maintained in a C57/BL/6J background were described previously (24). Transgenic mice were hemizygous with respect to the mutation. Nontransgenic littermates served as controls. Brain samples from neurotrypsin-deficient mice expressing a truncated form of murine neurotrypsin lacking the proteolytic domain and from mice overexpressing human neurotrypsin under the thymus cell surface antigen-1 (Thy-1) promoter (Tg-PRSS12Thy-1) and corresponding controls were generously provided by Dr. Sonderegger (15, 25). Brain samples from embryonic day 15 Psen1, and postnatal day 11 Psen2 KO mice were provided by Dr. Strooper (13) and Jayadev et al. (26), respectively.
Gene Expression Quantification
Total RNA was extracted from a pool of 5–10 third instar Drosophila larval brains, MEFs cells, or mouse dissected hippocampus using the RNeasy kit (Qiagen) or PARIS kit (Ambion). RNA was subjected to DNase I digestion using the TURBO DNA-free kit (Ambion). Reverse transcription was performed with SuperScript First Strand cDNA (Invitrogen). Real time quantitative PCRs (RT-qPCRs) were performed on an Applied Biosystems 7300 real time PCR system using FastStart Universal SYBR Green Master (Roche Applied Science). All primers used in this study were checked for nonspecific genomic cross-reactivity using UCSC Genome Browser PCR tool, and single band amplification was checked by a 1.2% agar, ethidium bromide gel. Primer sequences can be provided upon request.
Chromatin Immunoprecipitation (ChIP) and ChIP Coupled with Next Generation Sequencing (ChIP-seq)
ChIP and ChIP-seq assays were carried out and analyzed as described previously (27, 28). Primer sequences used in these assays can be provided upon request. ChIP-seq data sets for H3K4me3 and RNA Pol II were reported previously (29, 30). The ChIP-seq data have been deposited in the NCBI Gene Expression Omnibus.
Immunoblots
Proteins were extracted using the PARIS kit (Ambion) following the manufacturer's instructions. Equal amounts of cell extracts were loaded on 4–12% gradient NuPAGE Bis-Tris precast gels (Invitrogen), transferred to nitrocellulose (Whatman Protran) or PVDF Immobilon-FL membranes (Millipore), and immunoblotted with corresponding primary antibodies followed by HRP- or infrared (IR) dye-conjugated secondary antibodies. Pierce ECL Western blotting substrate, SuperSignal West Dura extended duration substrate, or SuperSignal West Femto Maximum Sensitive substrate (Thermo Scientific) was used as needed to detect the signal. IRDye-immunoreactive bands were scanned and quantified using Odyssey infrared imaging system (Li-Cor) following the manufacturer's instructions.
Gene Ontology Analysis
Functional annotations were obtained with Genomic Regions Enrichment of Annotations Tool, or GREAT (31). As “test regions,” we used the chromosome location of each identified CREB site. As settings, we used the default “basal plus extension” rules adjusting the distal distance to up to 5 kb and the whole mouse genome as background. GREAT annotations and p values for the three classes were downloaded and visualized with Microsoft Excel.
Statistical Analysis
All results are given as mean ± S.E. and were obtained from at least three biological experiments. Two-tailed unpaired Student's t test was used to determine statistical differences between two groups of data. p ≤ 0.05 was considered to be statistically significant.
RESULTS
Presenilin Inhibits Expression of Teq and Its Mammalian Ortholog Neurotrypsin
In a genome-wide expression analysis (data not shown), we discovered that Teq might be a presenilin-dependent gene in Drosophila larval brains. Drosophila contains a single Psn gene as well as ortholog/homolog genes for each of the other core components of the γ-secretase complex (32–34). To test whether Teq expression is modulated by presenilin, we used larval brains carrying the loss-of-function presenilin mutant psn143 (see “Experimental Procedures”). These brains exhibit low Psn expression (Fig. 1A, left panel) and reduced γ-secretase activity as shown by robust accumulation of the APPL C-terminal fragments (Fig. 1A, right panel). RT-qPCR analysis revealed ∼3.5-fold Teq mRNA up-regulation compared with wild-type brains (Fig. 1B). Conversely, brains of Psn-overexpressing larvae (Fig. 1C, left panel) showed no changes in APPL CTF (Fig. 1C, right panel), and exhibited ∼45% Teq mRNA down-regulation when compared with control (Fig. 1D). Thus, presenilin is a negative regulator of Teq expression in vivo in Drosophila larval brains.
The proposed mammalian ortholog of Teq is Prss12 (also known as neurotrypsin or motopsin (14)), a serine protease recently reported to cleave the heparan sulfate proteoglycan agrin (15). To test whether neurotrypsin expression is modulated by presenilins, we assessed whether neurotrypsin is up-regulated in Psen1 KO mouse brains. Psen1 deficiency resulted in accumulation of APP CTFs in embryonic day 15 brains (Fig. 1E, left panel). However, we did not observe significant changes in neurotrypsin mRNA expression when compared with wild-type littermates (Fig. 1E, right panel). Similarly, Psen2 KO brains (Fig. 1F) exhibited no changes in neurotrypsin expression when compared with age-matched wild-type brains (Fig. 1F, right panel). Similarly, no neurotrypsin up-regulation was observed in MEFs derived from Psen1- or Psen2-deficient mice (data not shown). However, we found reduced neurotrypsin expression in the hippocampi of mice overexpressing the familial Alzheimer disease (FAD) human PSEN1 mutation A246E (Fig. 1G; see also Fig. 2D). These findings suggest that although Psen1 or Psen2 single deficiencies are insufficient to induce neurotrypsin up-regulation in mouse brains, PSEN1 overexpression and/or expression of an FAD PSEN1 mutation represses neurotrypsin expression. We then tested the double Psen1 and Psen2 deficiency in MEFs and observed a >50-fold up-regulation of neurotrypsin expression when compared with wild-type cells (Fig. 1H). Similar results were obtained in a different set of wild-type and Psen1/2 dKO MEFs (data not shown; see “Experimental Procedures”). To unambiguously associate this effect to the combined absence of Psen1/2, we tested neurotrypsin expression in Psen1/2 dKO MEFs in which wild-type human PSEN1 (hPSEN1) and hPSEN2 were simultaneously reintroduced. These conditions restored the well known PSEN1-dependent γ-secretase activities that were abrogated in Psen1/2 dKO cells, including nicastrin maturation and processing of APP CTFs and N-cad/CTF1 (Fig. 1I, PSEN1 activities). The rescue also strongly reduced neurotrypsin expression close to wild-type/basal levels (Fig. 1H). Together, these results suggest that as with its ortholog in flies neurotrypsin expression is negatively regulated by presenilins and that removal of both presenilin forms is required to induce neurotrypsin expression.
FIGURE 2.
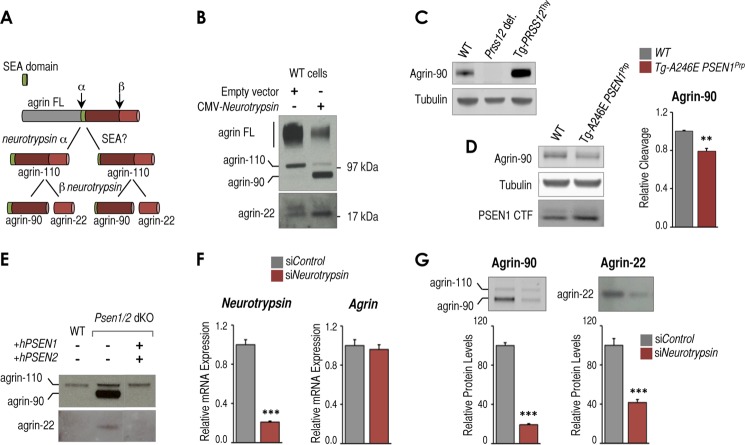
Presenilin-dependent changes in neurotrypsin expression impact neurotrypsin-dependent cleavage of agrin. A, scheme of neurotrypsin-dependent and potential alternative neurotrypsin-independent and SEA-dependent agrin proteolytic fragmentation pathway. B, Western blot analysis of full-length agrin (agrin FL) and proteolytic fragments in conditioned medium from wild-type MEFs transfected with an empty CMV vector or CMV-neurotrypsin-expressing vector. Full-length agrin and agrin proteolytic fragments of ∼110 (agrin-110), ∼90 (agrin-90), and ∼22 kDa (agrin-22) are indicated. C, Western blot analysis showing agrin-90 and tubulin as a loading control in brain samples from wild-type (WT) mice, neurotrypsin-deficient mice (Prss12 def.), and mice overexpressing neurotrypsin (Tg-PRSS12Thy). D, left panel, Western blot analysis showing agrin-90, tubulin as loading control, and PSEN1 CTF in brain samples from postnatal day 14 WT mice, transgenic mice overexpressing the FAD-associated A246E PSEN1 mutation (Tg-A246E PSEN1Prp). Right panel, RT-qPCR analysis of Tbp-normalized neurotrypsin mRNA expression from the same samples. E, Western blot analysis of agrin fragmentation in conditioned medium from wild-type, Psen1/2 dKO, and Psen1/2 dKO (+hPSEN1/2) cells. F, RT-qPCR analysis of Tbp-normalized neurotrypsin and Agrn mRNA expression in control siRNA (siControl)- and neurotrypsin siRNA (siNeurotrypsin)-treated Psen1/2 dKO cells (n = 3, mean ± S.E.). G, top panels, Western blot analysis of conditioned medium from the same cells as in F to detect agrin-90 and agrin-22 proteolytic fragments. Bottom panels, quantification of agrin-90 and agrin-22 levels from triplicate experiments (n = 3, mean ± S.E.). All experiments shown in this figure were at least done in triplicate (n = 3). Mean ± S.E. is shown. p values relative to control/wild type are shown when significant (Student's t test): **, p ≤ 0.01; ***, p ≤ 0.001. Representative Western blots are shown in each panel. Error bars represent S.E.
Unfortunately, we could not complement these results by detecting neurotrypsin protein using multiple available antibodies. Because we were able to detect neurotrypsin in cells exogenously overexpressing neurotrypsin (data not shown), we suspect that although relatively up-regulated in Psen1/2 dKO cells the levels of up-regulation are still below the detection range of our Western blot conditions. Therefore, to assess whether altering levels of neurotrypsin expression via presenilins has a functional impact, we tested presenilin-dependent effects on agrin cleavage, which is the only neurotrypsin-dependent proteolytic activity reported to date (15). Neurotrypsin cleaves agrin at two sites (α and β) to generate two fragments, ∼90 (agrin-90) and ∼22 kDa (agrin-22) (Fig. 2A, left path, and Ref. 15). Agrin is mainly known in a neuronal context; however, we found that MEFs endogenously expressed and secreted full-length agrin, which can be detected in conditioned medium as a smear of bands of ∼250 kDa (Fig. 2B, agrin FL). Exogenous expression of neurotrypsin induced accumulation of agrin-90 and agrin-22 in conditioned medium but reduced levels of full-length agrin (Fig. 2B), which suggests that endogenous agrin is accessible to cleavage by neurotrypsin and that neurotrypsin activity can be monitored in these cells. We also observed an agrin band of ∼110 kDa (Fig. 2B, agrin-110) potentially generated from single neurotrypsin-dependent cleavage at the α-site (Fig. 2A, left path, agrin-110) as reported in other cells (15). However, agrin-110 might also be generated by neurotrypsin-independent mechanisms as for instance an endoproteolytic cleavage at the sea urchin sperm protein, enterokinase, and agrin (SEA) domain (Fig. 2A, right path, and Ref. 35). Thus, to unambiguously monitor neurotrypsin-dependent agrin cleavage, we focused our analysis on agrin-90 and/or agrin-22 (Fig. 2A, right path, agrin-90 and agrin-22). In agreement and confirming neurotrypsin specificity, levels of agrin-90 were undetectable in brain samples from neurotrypsin-deficient mice (Fig. 2C, Prss12 def.), whereas they were increased in neurotrypsin-overexpressing mice compared with wild type (Fig. 2C, Tg-PRSS12Thy). In hippocampi of FAD PSEN1-overexpressing mice, we found that reduced neurotrypsin expression (Fig. 1G) resulted in lower levels of agrin-90 (Fig. 2D). Similarly as for neurotrypsin expression, we found elevated amounts of both fragments in conditioned medium derived from Psen1/2 dKO cells (Fig. 2E). These fragments were drastically reduced in Psen1/2 dKO rescued with hPSEN1/2 (Fig. 2E). To further confirm that agrin fragment accumulation in Psen1/2 dKO cells is mediated by a neurotrypsin-dependent activity, we knocked down neurotrypsin expression by siRNA in these cells (Fig. 2F). Under these conditions, accumulation of agrin fragments was strongly reduced without affecting the levels of Agrn expression (Fig. 2, F and G), which confirms neurotrypsin dependence for formation of agrin fragments in these cells. Taken together, these data suggest that presenilin-dependent changes in neurotrypsin RNA expression in cultured cells and in vivo may have an impact on the production of the active protease as measured by accumulation of agrin fragments.
An Active and Mature γ-Secretase Complex Is Required to Repress Neurotrypsin
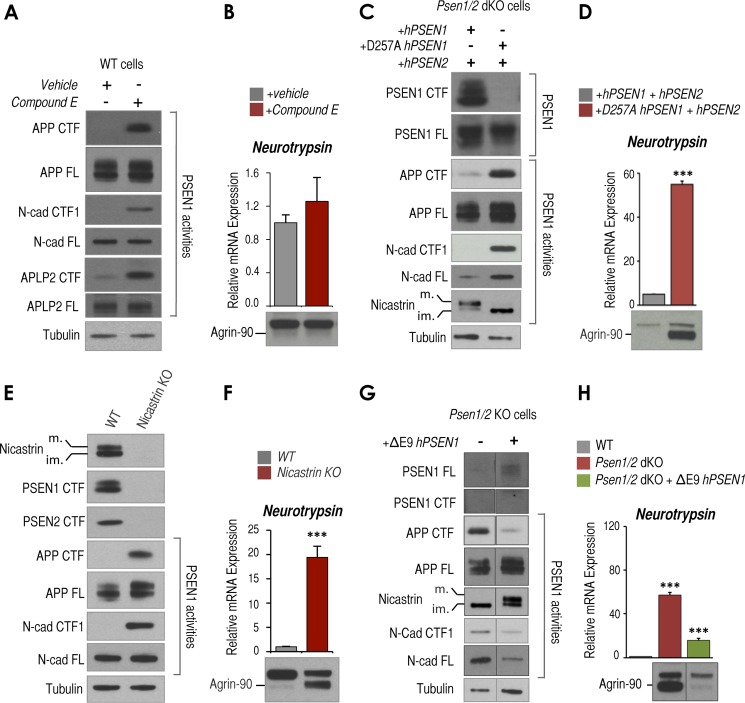
To investigate the underlying mechanisms involved in presenilin-dependent regulation of neurotrypsin expression and function, we focused on MEFs as a model system. To test whether γ-secretase activity is required to regulate neurotrypsin expression, we treated wild-type cells with the commonly used γ-secretase inhibitor, Compound E. After 24 h of treatment with Compound E (0.5 μm), efficient inhibition of γ-secretase activity was supported by robust accumulation of APP, APLP2, and N-cad/CTF1 (Fig. 3A), but treatment had no significant effect on neurotrypsin expression or accumulation of agrin-90 (Fig. 3B). Different concentrations (0.1 and 3 μm) and longer exposure times (48 and 72 h) with Compound E also showed no significant effects on accumulation of agrin-90 (data not shown). Although these results may suggest a γ-secretase-independent mechanism to regulate neurotrypsin expression, previous studies revealed that drug inhibitors of γ-secretase are not necessarily equally efficient in blocking all γ-secretase-dependent activities (36–38). Therefore, to unambiguously test whether γ-secretase is involved in this process, we took advantage of the catalytically dead D257A hPSEN1 mutant, which is unable to undergo autocatalytic endoproteolytic cleavage and unable to form a mature high molecular weight active γ-secretase complex (39, 40). As expected, unlike reintroduction of wild-type hPSEN1/2, reintroduction of D257A hPSEN1 and wild-type hPSEN2 into Psen1/2 dKO cells showed no detectable PSEN1 CTFs or mature nicastrin (Fig. 3C). Also, this mutant, unlike wild type, was unable to restore PSEN1-dependent activities, including cleavage of γ-secretase substrates APP CTFs and N-cad/CTF1 (Fig. 3C) as well as repression of neurotrypsin and agrin cleavage (Fig. 3D). Therefore, these results suggest that an active form of PSEN1-containing γ-secretase complex is required to repress neurotrypsin expression and function. If this is true, we also expect that cells lacking nicastrin, another core component of the γ-secretase complex required for normal γ-secretase complex formation and activity, should show neurotrypsin up-regulation in MEFs. To test this hypothesis, we used nicastrin KO cells, which contain PSEN1 and PSEN2 that cannot be endoproteolytically cleaved or form a mature and active γ-secretase complex (21) (Fig. 3E). As expected, processing of γ-secretase substrates of APP CTFs and N-cad/CTF1 was impaired in these cells as supported by robust accumulation of these fragments (Fig. 3E and Ref. 21). Also in agreement with our prediction, nicastrin KO cells exhibited abnormally elevated neurotrypsin expression and agrin cleavage (Fig. 3F). Together, these data suggest that it is not the lack of PSEN1 or PSEN2 per se but either the impaired formation of a mature active γ-secretase complex or the impaired endoproteolytic cleavage of presenilins that leads to increased neurotrypsin expression and agrin cleavage in Psen1/2 dKO cells. To distinguish between these two possibilities, we took advantage of ΔE9 hPSEN1, an FAD-associated PSEN1 mutation that results in a truncated form of PSEN1 unable to be endoproteolytically cleaved but still able to form a high molecular weight and active γ-secretase complex (41). Reintroduction of ΔE9 hPSEN1 in Psen1/2 dKO cells, even at low levels, efficiently restored APP and N-cadherin processing and nicastrin maturation as reported previously (Fig. 3G and Ref. 42). Furthermore, this mutant was able to robustly reverse neurotrypsin up-regulation induced by the double Psen1/2 deficiency (Fig. 3H). Together, these results suggest that it is the formation of a mature γ-secretase complex, rather than endoproteolytic cleavage of PSEN1 or the proteolytic processing of well known γ-secretase substrates (i.e. APP CTFs, APLP2 CTF, and N-cad/CTF1), that is required to repress neurotrypsin expression and activity.
FIGURE 3.
The γ-secretase complex, but not its proteolytic activity, controls neurotrypsin expression and activity. A, Western blot analysis showing APP, N-cad, and APLP2 full length (FL) and corresponding CTFs in wild-type cells treated with DMSO (vehicle) or 0.5 μm Compound E (CE) for 24 h. Tubulin is shown as a loading control. B, RT-qPCR analysis of Tbp-normalized neurotrypsin mRNA expression in wild-type cells treated as in A (top panel) and Western blot analysis of agrin-90 in conditioned medium analyzed (bottom panel) using the same cells as in A. C, Western blot analysis of Psen1/2 dKO (+hPSEN1/2) or Psen1/2 dKO (+hPSEN1 D257A/hPSEN2) cells to detect PSEN1 CTF, PSEN1 full length (FL), APP full length, APP CTF, N-cad full length (FL), N-cad/CTF1, and mature (m.) and immature (im.) nicastrin. Tubulin is shown as a loading control. D, RT-qPCR analysis of Tbp-normalized neurotrypsin mRNA expression (top panel) and Western blot analysis of agrin-90 (bottom panel) in conditioned medium from the same cells as in C. E, Western blot analysis of wild-type (WT) and nicastrin KO cells of mature (m.) and immature (im.) nicastrin, APP full length (FL), APP CTF, N-cad full length, and N-cad/CTF1. Tubulin is shown as a loading control. F, RT-qPCR analysis of Tbp-normalized neurotrypsin mRNA expression (top panel) and Western blot analysis of agrin-90 in conditioned medium (bottom panel) from the same cells as in E. G, Western blot analysis of Psen1/2 dKO and Psen1/2 dKO cells rescued with ΔE9 mutant hPSEN1 to detect PSEN1 full length (FL), CTF, APP full length, APP CTF, mature (m.) or immature (im.) nicastrin, N-cad full length (FL), N-cad/CTF1, and tubulin as loading control. Both samples were run in the same gel but were cut and pasted to adjust to layout of figure. APP full length (FL) Western blot for Psen1/2 dKO sample shown in this panel is the same as the one displayed in Fig. 1I because samples were run in the same gel. H, RT-qPCR analysis of Tbp-normalized neurotrypsin mRNA expression and Western blot analysis of conditioned medium to detect agrin fragments from cells as in G. Both samples were run in the same gel but were cut and pasted to adjust to layout of figure. All experiments shown in this figure were triplicates (n = 3). Mean ± S.E. is shown. p values are shown when significant (Student's t test): ***, p ≤ 0.001. Error bars represent S.E.
GSK3 Is Involved in the Control of Neurotrypsin Expression
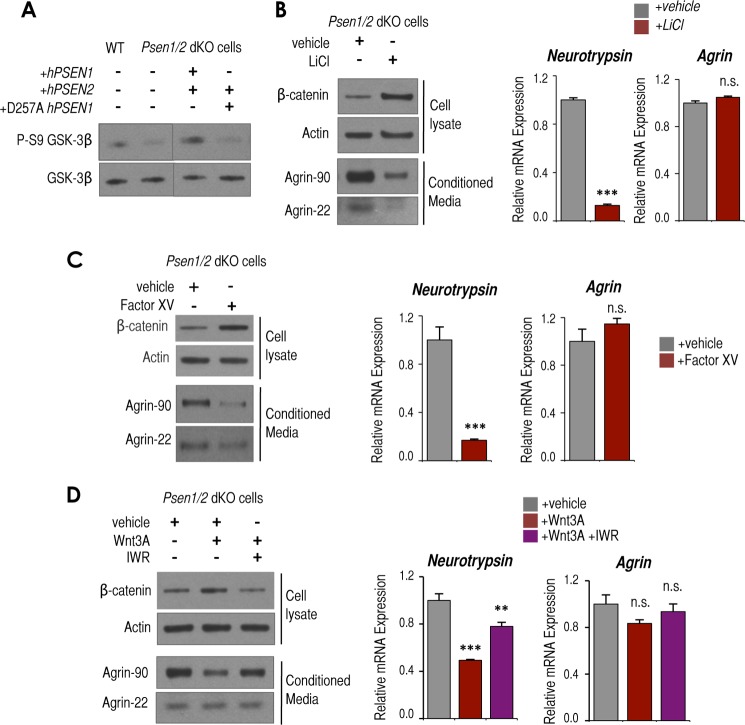
To identify molecules that bridge transmembrane presenilins to downstream effects regulating the expression of neurotrypsin; we focused on the serine/threonine kinase GSK3, whose activity can be modulated by presenilins (19). Psen1/2 dKO cells exhibit “hyperactivation” of GSK3 as measured by lower levels of the inactive P-S9-GSK3 (Fig. 4A) (19). Interestingly, we found that as for neurotrypsin expression and levels of agrin cleavage the low levels of P-S9-GSK3 observed in Psen1/2 dKO cells could be rescued by the reintroduction of hPSEN1/2 but not by reintroduction of D257A hPSEN1 in combination with hPSEN2 (Fig. 4A). Therefore, we next tested whether GSK3 hyperactivation is directly involved in neurotrypsin up-regulation in Psen1/2 dKO cells.
FIGURE 4.
Psen1/2 dKO-dependent hyperactivation of GSK3 contributes to neurotrypsin up-regulation and enhanced activity. A, Western blot analysis of P-S9-GSK3β and total GSK3β in WT, Psen1/2 dKO, and Psen1/2 dKO cells rescued with hPSEN1/2 or catalytically inactive D257A hPSEN1 and wild-type hPSEN2. Samples were run in the same gel but were cut and pasted to adjust the order of lanes displayed in this panel. B, Western blot analysis of β-catenin and actin levels in cellular lysates and agrin-90 and agrin-22 in conditioned medium from Psen1/2 dKO cells treated for 24 h with 50 mm LiCl or vehicle (left panel). RT-qPCR analysis of Tbp-normalized neurotrypsin (middle panel) and Agrn (right panel) mRNA expression from the same samples is shown. C, as in B but treated for 24 h with 0.1 μm factor XV or vehicle. D, as in B but treated for 24 h with purified Wnt3a and DMSO (vehicle) or Wnt3a and 1 μm IWR (IWR). All experiments shown in this figure were triplicates (n = 3). Representative Westerns are shown. Mean ± S.E. is shown. p values relative to control/wild type are shown when significant (Student's t test): **, p ≤ 0.01; ***, p ≤ 0.001. n.s., not significant. Error bars represent S.E.
As expected, inhibition of GSK3 activity in Psen1/2 dKO cells by either the GSK3 inhibitor LiCl or Factor XV led to accumulation of the GSK3 substrate β-catenin (Fig. 4, B and C, left panels) (43–45). Significantly, it also attenuated neurotrypsin up-regulation and agrin cleavage (Fig. 4, B and C, left and middle panels) without altering the levels of Agrn mRNA (Fig. 4, B and C, right panels). To determine whether the GSK3 activity associated with Wnt/β-catenin canonical signaling is also involved in regulation of neurotrypsin expression, we treated Psen1/2 dKO cells with purified Wnt3a, which inhibits this GSK3-dependent pathway (18). As expected, it induced accumulation of β-catenin (Fig. 4D, left panel) as well as attenuation of neurotrypsin up-regulation and attenuation of accumulated agrin-90 with no obvious changes in agrin-22 or Agrn expression (Fig. 4D). Co-incubation of Wnt3 and the Wnt inhibitor IWR, known to stabilize the β-catenin destruction complex (46), partially reversed the Wnt3 effect on β-catenin accumulation and the negative effect on neurotrypsin expression and agrin cleavage (Fig. 4D, left and middle panels). Thus, these results suggest that GSK3 activity specifically associated with the Wnt/β-catenin pathway contributes to some extent to induce neurotrypsin expression and its activity in Psen1/2 dKO cells. In sum, our results establish both Wnt-dependent and -independent GSK3 signaling as intermediate regulators of the induction of neurotrypsin expression when presenilins are absent.
To determine whether GSK3 hyperactivation per se is sufficient to induce neurotrypsin expression and activity in wild-type cells, we inhibited Akt/PKB and PKC activities, which are known to phosphorylate GSK3 at the Ser-9 residue in other cells (47, 48). We found PKC, but not Akt/PKB, to efficiently reduce P-S9-GSK3 levels in wild-type MEFs (data not shown); however, no parallel induction of neurotrypsin expression was observed (data not shown). Therefore, these experiments suggest that although GSK3 is required to maintain induced neurotrypsin expression in the absence of presenilins increasing GSK3 activity per se in wild-type cells (via inhibition of PKC) is not sufficient to induce neurotrypsin expression.
Presenilins Control the Transcriptional and Epigenetic State of the Neurotrypsin Promoter
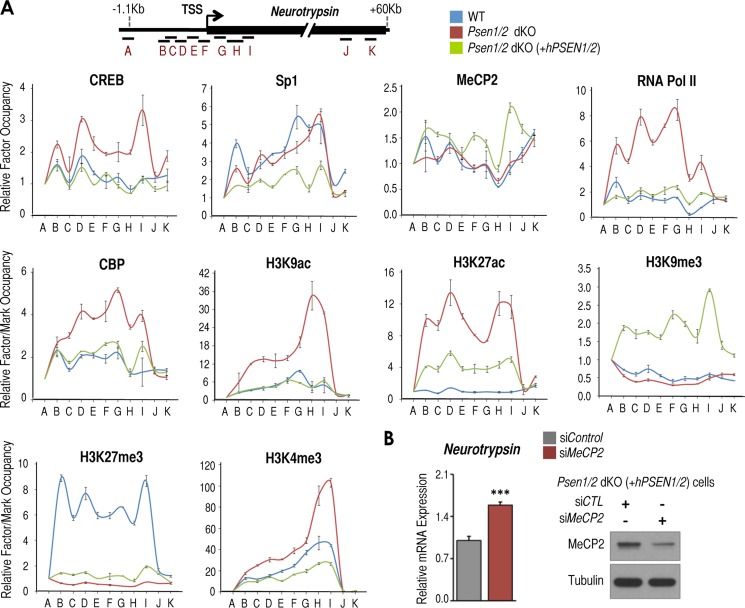
To test whether increased levels of neurotrypsin expression in Psen1/2 dKO cells parallel changes in the transcriptional and epigenetic state of the neurotrypsin promoter under each condition, we first surveyed the genomic sequence surrounding the transcriptional start site (TSS) of this promoter. We identified multiple imperfect binding sites for the transcription factor CREB (49). Intriguingly, we also observed a long GC-rich region containing numerous CpG dinucleotides, which may act as binding sites for the repressive MeCP2 (50, 51), or the transcription factor specificity protein 1 under repressed and active conditions, respectively (e.g. Ref. 52). To test these predictions, we performed ChIP assay in wild-type, Psen1/2 dKO, and Psen1/2 dKO (+hPSEN1/2) cells focusing on genomic regions around the neurotrypsin TSS (Fig. 5A, locus scheme, B–I) and compared them with genomic regions farther away from TSS that most likely would lack binding of proteins tested and would serve as negative controls (Fig. 5A, locus scheme, A, J, and K). Intriguingly, we observed CREB binding around the neurotrypsin TSS exclusively in Psen1/2 dKO cells, correlating with induced neurotrypsin expression in these cells (Fig. 5A, CREB panel). We observed Sp1 recruitment in wild-type and Psen1/2 dKO cells (Fig. 5A, Sp1 panel), and MeCP2 binding was detected primarily in double rescued Psen1/2 dKO cells, correlating with reduced neurotrypsin expression in these cells (Fig. 5A, MeCP2 panel). In correlation with neurotrypsin up-regulation, we observed the strongest recruitment of RNA Pol II and CREB coactivator CBP in Psen1/2 dKO cells (Fig. 5A, RNA Pol II and CBP panels). Likewise, highly elevated levels of CBP-mediated histone post-translational marks (acetylation of histone 3 at lysine 9 (H3K9ac) and lysine 27 (H3K27ac); Ref. 53) were observed at the neurotrypsin promoter in these cells (Fig. 5A, H3K9ac and H3K27ac panels). Also in correlation with reduced neurotrypsin expression, we observed repressive marks H3K9me3 and H3K27me3 in double rescued and wild-type cells, respectively (Fig. 5A, H3K9me3 and H3K27me3 panels). Levels of H3K4me3, which serves as a valuable indicator of transcriptionally active or poised promoters (29), were observed in all three conditions with the strongest levels in Psen1/2 dKO cells in correlation with their up-regulated neurotrypsin expression (Fig. 5A, H3K4me3 panel). These results suggest the existence of two inactive epigenetic states and one active epigenetic state of the neurotrypsin promoter. The two inactive states (H3K27me3-positive in wild-type cells and H3K9me3/MeCP2-positive in PSEN1/2 double rescued cells) coincide with the presence of presenilins, whereas the active state (H3K4me3-H3K9/27ac/CREB/CBP/RNA Pol II-positive) coincided with the absence of presenilins. Together, these results suggest that neurotrypsin is a target for CREB and MeCP2 and that the presence/absence of presenilins determines the transcriptional and epigenetic states of the neurotrypsin promoter in MEFs.
FIGURE 5.
PSEN1/2 regulate the transcriptional and epigenetic states of the neurotrypsin promoter. A, top, scheme of neurotrypsin regions tested by ChIP. A–K indicate genomic regions utilized to detect binding of proteins tested. Bottom panels, ChIP analysis of CREB, Sp1, MeCP2, RNA Pol II, CBP, H3K9ac, H3K27ac, H3K9me3, H3K27me3, and H3K4me3 in wild-type (blue line), Psen1/2 dKO (red line), and Psen1/2 dKO + hPSEN1/2 (green line) cells at the neurotrypsin locus. B, left panel, RT-qPCR analysis of Tbp-normalized neurotrypsin mRNA levels in Psen1/2 dKO (+hPSEN1/2) cells after control (siCTL) or Mecp2-specific (siMeCP2) siRNA-mediated knockdown. Right panel, Western blot analysis of MeCP2 and tubulin in the same cells as in the left panel. This experiment was done in triplicate (n = 3). Mean ± S.E. is shown. p values are shown when significant (Student's t test): ***, p ≤ 0.001. Error bars represent S.E.
If MeCP2 binding, rather than suppression of CREB/CBP recruitment in double rescued cells, is responsible for switching from the active to the inactive state in these cells, we would predict that reducing MeCP2 in PSEN1/2-rescued cells would be sufficient to induce neurotrypsin up-regulation. To test this, we treated double rescued cells with Mecp2 siRNA. We found that efficient MeCP2 reduction induced only modest neurotrypsin up-regulation (Fig. 5B). This finding suggests that CREB/CBP binding to the neurotrypsin promoter might be the main determinant of the active state found in Psen1/2 dKO cells responsible for neurotrypsin up-regulation.
Presenilins Contribute to Defining the Pattern of CREB Binding in the Mouse Genome
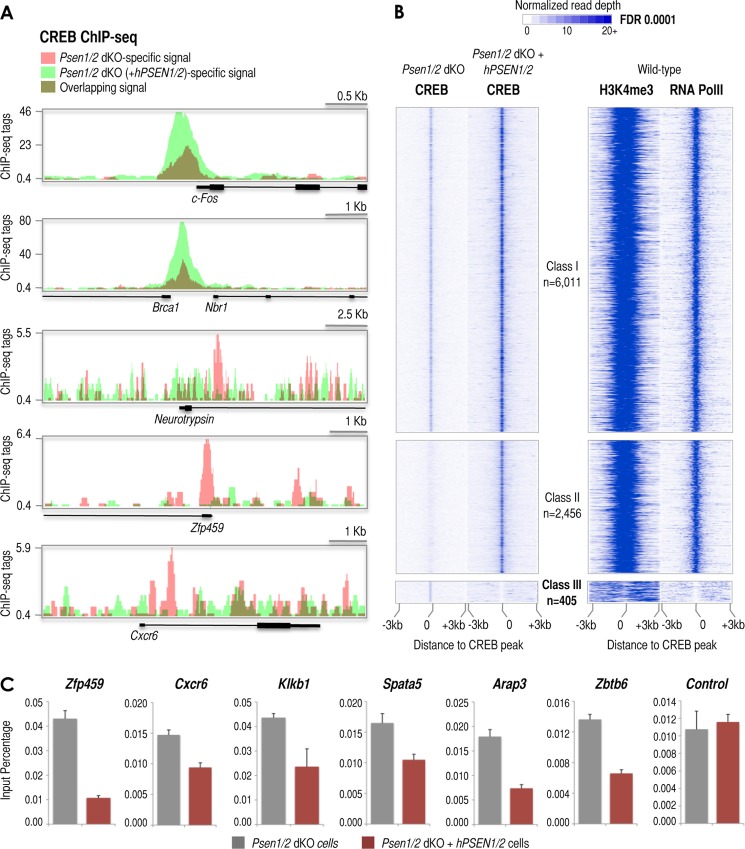
CREB is generally bound constitutively to gene promoters in an inactive form, which upon phosphorylation (stimulation) recruits cofactor CBP and activates transcription of downstream associated genes (54). However, the underlying mechanisms regulating the actual binding of CREB to the genome are largely unknown. Our findings suggest that presenilin might be an upstream regulator controlling CREB recruitment as observed in the neurotrypsin promoter. To investigate whether CREB recruitment to other promoters in the genome is similarly regulated by presenilins, we performed ChIP-seq of CREB in Psen1/2 dKO and Psen1/2 dKO cells rescued with hPSEN1/2. H3K4me3 is a good indicator of promoter regions in the genome (29) as also shown here (Fig. 4A, H3K4me3 panel). Thus, we matched the two CREB ChIP-seq data sets with a published map of H3K4me3 in wild-type MEFs (29) to help identify promoter regions. We detected more than 6,000 CREB peaks associated with H3K4me3-positive regions (n = 6,011), which show less than a 4-fold CREB intensity difference between both genotypes (Fig. 6B). These promoters include known CREB targets such as c-Fos and Brca1, and we defined them as Class I promoters (Fig. 6, A, top two panels, and B, Class I promoters). We also observed a second class of H3K4me3-marked promoters (n = 2,456) that show a >4-fold CREB signal in double rescued when compared with Psen1/2 dKO cells (Fig. 6B, Class II promoters). Finally, we confirmed our findings of CREB binding within a short range of the neurotrypsin TSS exclusively in Psen1/2 dKO cells (Fig. 6A, middle panel). More importantly, we identified an additional set of H3K4me3-marked promoters where CREB peaks were exclusively detected in Psen1/2 dKO cells (n = 405 sites), e.g. Zfp459 and Cxcr6 (Fig. 6, A, bottom two panels, and B, Class III promoters). Some of these peaks were validated by conventional ChIP (Fig. 6C).
FIGURE 6.
ChIP-seq reveals subsets of CREB target promoters defined by the presence or absence of presenilins. A, UCSC Genome Browser-derived images of CREB ChIP-seq signal at the c-Fos, Brca1, neurotrypsin, Zfp459, and Cxcr6 loci in Psen1/2 dKO (pink) and Psen1/2 dKO + hPSEN1/2 (light green) cells (dark green results from overlapping signals derived from both cells). The ChIP-seq signal (tags), annotated genes, and genomic scale for each image are shown. B, distribution of CREB ChIP-seq signal within ±3-kb windows around ChIP-seq-identified CREB sites. ChIP-seq data aligned with respect to the center of the CREB site (left top panels) identified in Psen1/2 dKO cells or identified in Psen1/2 dKO (+hPSEN1/2) cells (left middle and bottom panels). Three classes of CREB binding sites were identified by ChIP-seq: Class I (left top panel), “non”-affected by presenilins (change ≤4-fold; n = 6,011); Class II (left middle panel), promoted/enhanced by the presence of presenilins (change >4; n = 2,456); and Class III (left bottom panel), suppressed by the presence of presenilins (change >4; n = 405). Right panels depict the distribution of H3K4me3 and RNA Pol II ChIP-seq signals within ±3-kb windows around CREB sites in wild-type cells (29, 30). C, relative occupancy of CREB1 determined by ChIP analysis around the indicated loci and compared with genomic control region A (see Fig. 4A, scheme) in Psen1/2 dKO (gray columns) and Psen1/2 dKO + hPSEN1/2 (colored columns) cells. Error bars represent S.E. FDR, false discovery rate.
To further characterize Class III promoters, we matched the three classes of CREB-occupied promoters with available RNA Pol II ChIP-seq data from wild-type MEFs (30). Although Class I and II promoters contain high levels of RNA Pol II and H3K4me3, likely revealing their transcriptionally active state (Fig. 6B, Class I and Class II, RNA PolII and H3K4me3 panels), Class III promoters were essentially negative for RNA Pol II and poorly enriched in H3K4me3 in wild-type cells, thus likely representing transcriptionally inactive promoters (Fig. 6B, Class III, RNA PolII and H3K4me3 panels). These observations agree with our ChIP data in Fig. 5 showing barely detectable RNA Pol II recruitment and lower H3K4me3 intensity at the neurotrypsin promoter in PSEN1/2-rescued and wild-type cells when compared with Psen1/2 dKO cells (Fig. 5A, RNA PolII and H3K4me3 panels). Together, these results suggest that CREB recruitment to most CREB gene targets in MEFs (Class I and II promoters) is unaffected by presenilins. However, for a significant subset of mouse promoters (Class III), presenilins appear to prevent CREB recruitment to their promoters. Finally, functional analysis based on annotated gene ontology and other databases (see “Experimental Procedures”) reveals that the repertoire of genes associated with Class II and III promoters are predicted to be functionally linked to embryonic lethality and developmental defects and appear to be highly expressed in neuronal tissues. Overall, our data suggest that presenilins may induce CREB signaling to a subset of genes (Class II promoters) and suppress the existence of a specific repertoire of CREB binding sites (Class III promoters) in MEFs that includes the neurotrypsin promoter. These findings suggest a potential physiological relevance of this presenilin-dependent regulatory process because these promoters are linked to genes involved in the control of embryogenesis and neuronal functions.
DISCUSSION
Presenilins facilitate multiple normal cellular processes, including transcriptional regulation, often via their associated γ-secretase activity. Some or all of these activities may be altered in Alzheimer and other diseases by mutation or modulation of presenilins. Here, we identify Prss12/neurotrypsin and its Drosophila ortholog Teq as new genes negatively regulated by presenilins in neuronal tissues in vivo and in cultured MEFs. The finding that Teq/neurotrypsin is regulated by presenilins is of particular interest given the likely role of presenilins in Alzheimer disease and thus in memory and cognition. This potential role is strengthened by the finding that Teq mutants in Drosophila have defects in memory formation (14) and by neurotrypsin itself being implicated in processing of the synaptic regulator agrin (15), which may be important in synapse modulation and/or maintenance (55). Finally, the finding that neurotrypsin mutations in humans cause mental retardation (16) further supports a role for the pathway we discovered leading from presenilin to neurotrypsin as potentially playing a role in normal cognition, which may fail in a variety of disease states.
To date, little is known about the mechanisms regulating the expression of neurotrypsin or its orthologs. In situ studies show that levels of neurotrypsin expression are dynamic in non-neuronal and neuronal tissues in developing mice, suggesting roles for neurotrypsin in morphogenesis of non-neuronal tissues as well as in neuronal processes, such as synaptogenesis (56, 57). Our findings reveal that one aspect of neurotrypsin transcription regulation can be traced to its promoter, which in wild-type conditions belongs to a select group of so-called “bivalent” promoters that are defined by the co-occurrence of H3K4me3/H3K27me3 (58). Bivalent promoters frequently control lineage-specific and cell fate genes by being developmentally poised for activation (58). As described in more detail below, our data suggest a model in which presenilins control neurotrypsin expression by preventing CREB/CBP binding to the neurotrypsin promoter and by maintaining an inactive epigenetic state of its promoter (Fig. 7). Consequently, lack of presenilins induces CREB/CBP/RNA Pol II recruitment to the neurotrypsin promoter and induction of an active epigenetic state of the promoter. Under these conditions, an aberrant increase in GSK3 activity is required to maintain high levels of neurotrypsin expression. A functional consequence from changes in neurotrypsin expression likely is the proteolytic processing of agrin. Although neurotrypsin activity and agrin proteolysis have been mainly studied in neuronal systems (59, 60), neurotrypsin and agrin are also expressed in non-neuronal tissues with unknown functional consequences (57).
FIGURE 7.
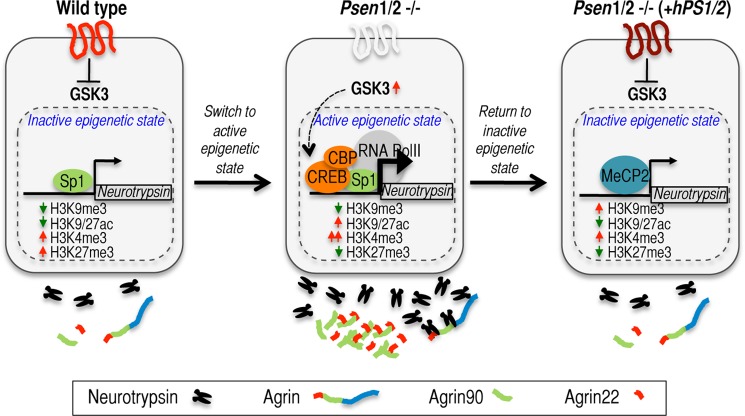
Model for presenilin-dependent regulation of neurotrypsin expression and activity. Wild-type MEFs expressing endogenous PSEN1/2 maintain basal levels of neurotrypsin and neurotrypsin-dependent proteolytic activity (left panel). Under these conditions, Sp1 is recruited to the neurotrypsin promoter (left panel). The simultaneous presence of repressive H3K27me3 and activating H3K4me3 defines neurotrypsin as a bivalent promoter (left panel). Upon presenilin removal, CREB/CBP/RNA Pol II join Sp1 at the neurotrypsin promoter, and repressive H3K27me3 is replaced by the activating H3K9/27ac marks (middle panel). These epigenetic changes promote neurotrypsin expression and activity in these cells as shown by increased extracellular accumulation of the neurotrypsin-dependent generated agrin-90 and agrin-22 fragments (middle panel). GSK3 activity is required for this regulatory shift and to maintain high levels of neurotrypsin expression and activity in Psen1/2 dKO cells (middle panel). Previous reports showing that presenilins inhibit GSK3 activity and that GSK3 can phosphorylate and activate CREB together with our own data suggest that GSK3 might bridge the presenilins and regulation of neurotrypsin expression via CREB modulation in Psen1/2 dKO cells (middle panel). When Psen1/2 dKO cells are rescued with hPSEN1/2, neurotrypsin expression and activity return to basal levels via acquisition of a new repressive epigenetic state at the neurotrypsin promoter such that MeCP2 and the repressive H3K9me3 mark are recruited to the neurotrypsin promoter (right panel).
A Mature γ-Secretase Complex but Not Its Proteolytic Activity Regulates Neurotrypsin Expression and Function
Presenilin-regulated functions often rely on its γ-secretase activity. It is generally expected that treatment with γ-secretase inhibitors will mirror presenilin removal when assaying those presenilin functions involving processing of γ-secretase substrates. Our study, however, shows that although removal of presenilins strongly induced neurotrypsin expression and activity, pharmacological inhibition of γ-secretase activity in wild-type cells did not mimic this effect. Similar results have been reported for other presenilin-dependent functions. For instance, removal of presenilins, but not pharmacological inhibition of γ-secretase activity, delays EGF receptor turnover and signaling and promotes integrin-β1 maturation (20, 61). Our data rule out major contributions of well known γ-secretase substrates, such as APP, APLP2, and N-cadherin, in presenilin-dependent regulation of neurotrypsin expression. However, we cannot completely exclude the possibility that processing of other γ-secretase substrates, unaffected by the inhibitory drug and conditions tested here, could contribute to this regulation. In fact, several reports have suggested that drug inhibitors of γ-secretase activity do not necessarily block all γ-secretase-dependent activities equally efficiently (36–38). Similarly, we cannot exclude the possibility that short term pharmacological treatments might not completely mimic the permanent removal of PSENs in MEFs derived from Psen1/2 dKO mice. For instance, the presence of γ-secretase substrates with turnover kinetics longer than the time of treatment with γ-secretase inhibitors tested here could continue repressing neurotrypsin expression. Alternatively, γ-secretase activity could trigger a delayed rather than an immediate sequence of events culminating in alterations of the transcriptional and epigenetic states of the neurotrypsin promoter at later times than those examined in our pharmacological studies.
Intriguingly, our data suggest an alternative possibility in which formation of a mature γ-secretase complex, but not presenilin endoproteolytic cleavage or proteolytic processing of substrates, is necessary for presenilins to inhibit neurotrypsin expression and activity. The evidence for this view includes the following observations. 1) D257A PSEN1, which could not restore neurotrypsin up-regulation induced by the double deficiency, is incapable of endoproteolytic cleavage and is unable to assemble into a mature γ-secretase complex (40). 2) The uncleavable ΔE9 hPSEN1 mutant form, which is still able to associate and form a mature γ-secretase complex (41), efficiently restored neurotrypsin expression in Psen1/2 dKO cells. 3) Finally, cells containing presenilins but unable to form a mature active γ-secretase complex due to lack of nicastrin also exhibited induced neurotrypsin expression and activity.
In line with this model, most γ-secretase drug inhibitors, including the one used in our study, abrogate cleavage of γ-secretase substrates by interfering with substrate binding or with its transit to the active site in the complex (62) but are unlikely to interfere with presenilin endoproteolysis or formation of a mature γ-secretase complex. Indeed, non-transition state analog inhibitors like Compound E can interact with presenilin fragments when forming a mature γ-secretase complex (63). In the case of presenilin-dependent repression of the neurotrypsin gene, our data may suggest that it is independent of substrate cleavage but that it requires the formation of a mature γ-secretase complex.
Presenilins Regulate CREB Recruitment to Multiple Gene Promoters Including That of Neurotrypsin
Presenilin and CREB signaling were previously linked when the γ-secretase-generated N-cad/CTF2 fragment was shown to inhibit CREB signaling by promoting CBP degradation (6). However, our data suggest that N-cad/CTF2 is unlikely to repress neurotrypsin expression because changes in formation of the N-cad/CTF2 fragment were independent of changes in neurotrypsin expression. Another study found that reduced CREB-dependent signaling in immortalized Psen1/2 dKO MEFs was not rescued by reintroduction of hPSEN1, leading the authors to conclude that CREB-mediated transcriptional changes in these cells were independent of presenilins (64). However, the authors did not test whether simultaneous reintroduction of both PSEN1 and PSEN2 would rescue this effect. In agreement with the aforementioned study, ChIP-seq in Psen1/2 dKO cells compared with PSEN1/2-rescued Psen1/2 dKO cells revealed a general reduction in CREB binding intensity for most CREB sites (Fig. 6, A and B). This reduced CREB binding could result in partial inhibition of CREB signaling in Psen1/2 dKO cells and support the hypothesis that presenilins are positive modulators of CREB signaling for most CREB-regulated promoters in MEFs (Fig. 6B, Class I and II promoters). More importantly, we propose that presenilins might also function as selective suppressors of CREB binding and activity for a previously unreported repertoire of CREB-regulated promoters (Fig. 6B, Class III), including neurotrypsin. In sum, our data provide evidence for a new strategy utilized for presenilins to regulate CREB signaling by preventing CREB recruitment to gene promoters.
Our findings may aid in the understanding of how the CREB binding program (or cistrome) is established and regulated in mammalian cells. The prevalent view is that CREB constitutively sits at its binding sites in the genome and upon phosphorylation triggered by multiple stimuli, including GSK3, CREB recruits CBP and activates transcription (49, 65). However, besides the requirement of CREB-regulatory elements in the genome, how CREB and subsequently CBP recruitment is restricted to a subset of CREB-regulatory element available sites in the genome is not well understood even in well studied targets like c-Fos. Our study adds the new concept that presenilins contribute to define the CREB cistrome in the mouse genome by “masking” a subset of promoters to be occupied. How presenilins accomplish this mechanistically is unknown at this point, but analysis of our ChIP-seq data reveals exclusive enrichment of other cis-regulatory elements in the ∼400 promoters where CREB recruitment is prevented by presenilins (data not shown). We speculate that presenilins may alter the binding of one or more of the transcription factors surrounding CREB sites, which in turn alters accessibility of CREB-regulatory elements specifically in these promoters. Examples supporting similar control of defining cistromes have been recently shown for other transcription factors (66).
Once CREB is bound to the neurotrypsin promoter (i.e. in Psen1/2 dKO cells), we provide evidence implicating GSK3 as a required intermediary player to maintain high levels of neurotrypsin expression and consequently neurotrypsin-dependent agrin cleavage in Psen1/2 dKO cells. GSK3 is known to phosphorylate CREB at Ser-129 to activate CREB signaling (67–70). Indeed, we found reduced levels of P-CREB-S129 in Psen1/2 dKO cells treated with GSK3 inhibitors (data not shown). Also in preliminary experiments, we found P-CREB-S129 bound to the neurotrypsin promoter in Psen1/2 dKO cells (data not shown). Therefore, it is possible that once CREB is bound to the neurotrypsin promoter it can be stimulated by GSK3-dependent phosphorylation. In contrast, in conditions where CREB is not recruited to the neurotrypsin promoter (e.g. wild-type MEFs), we found that increasing GSK3 activity via PKC inhibition was not sufficient to induce neurotrypsin expression (data not shown). Together, our data suggest that the transcriptional and epigenetic state of the neurotrypsin promoter defined by presenilins determines the responsiveness to upstream regulators like GSK3.
Potential Pathological Relevance
Loss of presenilins in mice leads to early embryonic lethality, which is attributed to deficiencies in signaling of the γ-secretase substrate Notch (13, 71, 72). However, loss of presenilins leads to more profound and broad developmental defects that might be caused by additional mechanisms (72). Based on our findings, it is tempting to speculate that besides Notch deficiency a combination of ameliorated CREB-dependent transcription for many genes and de novo CREB-dependent control of other genes caused by presenilin deficiency may contribute to Psen1/2 dKO embryonic lethality and neuronal developmental defects. Perhaps in agreement, the two data sets of genes whose CREB recruitment might be affected by presenilins (Fig. 6B, Class II and III promoters) are linked to embryonic lethality. In this regard, although still controversial, some studies suggest that presenilin deficiency induces premature neuronal differentiation of neuronal precursor cells (73–76). Therefore, an intriguing possibility is that changes in the differentiation state caused by presenilin removal might be induced, at least in part, by presenilin-dependent changes in the CREB cistrome. Cistrome reprogramming events are emerging as important contributors to development and disease (66).
Finally, the FAD PSEN1 mutation (ΔE9 PSEN1) acts as a more potent repressor of neurotrypsin expression and activity than PSEN1 wild type because ΔE9 PSEN1 expression alone was able to rescue the effects induced by the absence of PSEN1/2, whereas wild-type PSEN1 could not unless PSEN2 was also present (data not shown). Recent work using recombinant PSEN1 reconstituted on liposomes suggests that unlike wild type reconstituted ΔE9 PSEN1 exhibits γ-secretase activity on its own (77). Our model suggests that ΔE9 PSEN1 may display unique properties of exacerbated (gain-of-function) PSEN1 not shared by wild-type PSEN1 and therefore could bypass the absence of PSEN2 in the regulation of neurotrypsin expression and agrin cleavage. FAD-associated PSEN1 mutants are generally believed to act as “loss-of-function” forms (78, 79); however, a few “gain-of-function” activities have also been reported (80–82). Similarly, unlike wild-type PSEN1, reintroduction of another FAD-associated PSEN1 mutation, M146L, into Psen1/2 dKO cells also repressed neurotrypsin expression and activity in the absence of PSEN2 (data not shown). Furthermore, we found reduced neurotrypsin expression and agrin cleavage in hippocampi of mice overexpressing another FAD PSEN1 mutation (i.e. A246E PSEN1). These mice overexpress human A246E PSEN1 on top of endogenous levels of Psen1/2 (∼1.5-fold increase in PSEN1 CTF; Fig. 2D), but this small effect only translates to ∼10% reduced APP CTF (data not shown). Therefore, although we cannot rule out the possibility that other γ-secretase substrates might be more affected and lead to γ-secretase-dependent exacerbated repression of neurotrypsin expression, together, our results suggest that some FAD PSEN1 mutations act as γ-secretase-independent gain-of-function forms by being stronger repressors of neurotrypsin expression than wild type. Although it is beyond the scope of this study to establish a direct connection between Alzheimer disease and neurotrypsin dysfunction, these results provide intriguing insights that motivate future research in the context of Alzheimer disease especially because defective CREB signaling has been associated with the disease and neurotrypsin deficiency leads to mental retardation and serious synaptic defects (16, 60, 83).
Acknowledgments
We thank Dr. Edward H. Koo (University of California San Diego (UCSD) for providing a set of wild-type and Psen1/2 dKO cells. We also thank Dr. Bart De Strooper (University of Leuven, Belgium) for providing a different set of wild type and Psen1/2 dKO. We are grateful to Dr. Philip Wong (The Johns Hopkins University School of Medicine) for providing nicastrin KO cells. We also thank Dr. Peter Sonderegger (University of Zurich) for generously providing neurotrypsin and agrin antibodies and for providing us with dissected brains from neurotrypsin-deficient and neurotrypsin-overexpressing mice. We appreciate Dr. Gunter Merdes (Eidgenössische Technische Hochschule (ETH), Zurich, Switzerland) for providing antibodies raised against APPL. We are grateful to Dr. Shermali Gunawardena and Dr. Kristina Henthron for experimental advice and assistance in Drosophila larval brain dissections. We thank Dr. Karl Willert (UCSD) for advice and providing Wnt3a, Factor XV, and IWR. We also thank Jacqueline Luu (UCSD) for experimental help and Dr. Matthew Niederst (UCSD) and Dr. Maya Kunkel (UCSD) for advice and providing Akt and PKC inhibitors, respectively. We also thank Dr. Amir Gamliel (UCSD) for experimental advice on MeCP2 ChIPs. We are also grateful to Dr. Mira Sastri and Dr. Rhiannon Killian for help editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants 1F32AG20039, a National Research Service Award from the NIA (to A. A. Q.); 5R01AG032180 from the NIA (to L. S. B. G.); and 1R01AG033055 (to D. E. K.) and by the Ovarian Cancer Research Fund and the Estate of Agatha Fort (Liz Tilberis Scholar) (to I. G. B.).
- Aph1
- anterior pharynx-defective 1
- PSEN
- presenilin
- CREB
- cyclic AMP response element-binding protein
- CBP
- CREB-binding protein
- Sp1
- specificity protein 1
- GSK3
- glycogen synthase kinase 3
- P-S9-GSK3
- GSK3 phosphorylated at serine 9
- APP
- amyloid precursor protein
- CTF
- C-terminal fragment
- cad
- cadherin
- MeCP2
- methyl CpG-binding protein
- MEF
- mouse embryonic fibroblast
- FAD
- familial Alzheimer disease
- Pol
- polymerase
- RT-qPCR
- real time quantitative PCR
- ChIP-seq
- ChIP coupled with next generation sequencing
- dKO
- double KO
- Teq
- Tequila
- APLP2
- amyloid precursor-like protein 2
- H3
- histone 3
- me3
- trimethylation
- SEA
- sea urchin sperm protein, enterokinase, and agrin
- APPL
- amyloid precursor protein-like
- IWR
- inhibitor of Wnt response
- Psn
- presenilin
- TM
- transmembrane
- PrP
- prion protein
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- hPSEN
- human PSEN
- TSS
- transcriptional start site
- H3K9ac
- acetylation of histone 3 at lysine 9
- H3K27ac
- acetylation of histone 3 at lysine 27
- P-CREB-S129
- CREB phosphorylated at serine 129
- Agrn
- agrin.
REFERENCES
- 1. De Strooper B., Annaert W. (2010) Novel research horizons for presenilins and γ-secretases in cell biology and disease. Annu. Rev. Cell Dev. Biol. 26, 235–260 [DOI] [PubMed] [Google Scholar]
- 2. Ho A., Shen J. (2011) Presenilins in synaptic function and disease. Trends Mol. Med. 17, 617–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kopan R., Ilagan M. X. (2004) γ-Secretase: proteasome of the membrane? Nat. Rev. Mol. Cell Biol. 5, 499–504 [DOI] [PubMed] [Google Scholar]
- 4. Beckett C., Nalivaeva N. N., Belyaev N. D., Turner A. J. (2012) Nuclear signalling by membrane protein intracellular domains: the AICD enigma. Cell. Signal. 24, 402–409 [DOI] [PubMed] [Google Scholar]
- 5. St George-Hyslop P., Fraser P. E. (2012) Assembly of the presenilin γ-/ε-secretase complex. J. Neurochem. 120, Suppl. 1, 84–88 [DOI] [PubMed] [Google Scholar]
- 6. Marambaud P., Wen P. H., Dutt A., Shioi J., Takashima A., Siman R., Robakis N. K. (2003) A CBP binding transcriptional repressor produced by the PS1/ϵ-cleavage of N-cadherin is inhibited by PS1 FAD mutations. Cell 114, 635–645 [DOI] [PubMed] [Google Scholar]
- 7. Raurell I., Codina M., Casagolda D., Del Valle B., Baulida J., de Herreros A. G., Duñach M. (2008) γ-Secretase-dependent and -independent effects of presenilin1 on β-catenin·Tcf-4 transcriptional activity. PLoS One 3, e4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Francis Y. I., Stephanou A., Latchman D. S. (2006) CREB-binding protein activation by presenilin 1 but not by its M146L mutant. Neuroreport 17, 917–921 [DOI] [PubMed] [Google Scholar]
- 9. Saura C. A., Choi S. Y., Beglopoulos V., Malkani S., Zhang D., Shankaranarayana Rao B. S., Chattarji S., Kelleher R. J., 3rd, Kandel E. R., Duff K., Kirkwood A., Shen J. (2004) Loss of presenilin function causes impairments of memory and synaptic plasticity followed by age-dependent neurodegeneration. Neuron 42, 23–36 [DOI] [PubMed] [Google Scholar]
- 10. Vitolo O. V., Sant'Angelo A., Costanzo V., Battaglia F., Arancio O., Shelanski M. (2002) Amyloid β-peptide inhibition of the PKA/CREB pathway and long-term potentiation: reversibility by drugs that enhance cAMP signaling. Proc. Natl. Acad. Sci. U.S.A. 99, 13217–13221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gong B., Cao Z., Zheng P., Vitolo O. V., Liu S., Staniszewski A., Moolman D., Zhang H., Shelanski M., Arancio O. (2006) Ubiquitin hydrolase Uch-L1 rescues β-amyloid-induced decreases in synaptic function and contextual memory. Cell 126, 775–788 [DOI] [PubMed] [Google Scholar]
- 12. España J., Valero J., Miñano-Molina A. J., Masgrau R., Martín E., Guardia-Laguarta C., Lleó A., Giménez-Llort L., Rodríguez-Alvarez J., Saura C. A. (2010) β-Amyloid disrupts activity-dependent gene transcription required for memory through the CREB coactivator CRTC1. J. Neurosci. 30, 9402–9410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Herreman A., Hartmann D., Annaert W., Saftig P., Craessaerts K., Serneels L., Umans L., Schrijvers V., Checler F., Vanderstichele H., Baekelandt V., Dressel R., Cupers P., Huylebroeck D., Zwijsen A., Van Leuven F., De Strooper B. (1999) Presenilin 2 deficiency causes a mild pulmonary phenotype and no changes in amyloid precursor protein processing but enhances the embryonic lethal phenotype of presenilin 1 deficiency. Proc. Natl. Acad. Sci. U.S.A. 96, 11872–11877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Didelot G., Molinari F., Tchénio P., Comas D., Milhiet E., Munnich A., Colleaux L., Preat T. (2006) Tequila, a neurotrypsin ortholog, regulates long-term memory formation in Drosophila. Science 313, 851–853 [DOI] [PubMed] [Google Scholar]
- 15. Reif R., Sales S., Hettwer S., Dreier B., Gisler C., Wölfel J., Lüscher D., Zurlinden A., Stephan A., Ahmed S., Baici A., Ledermann B., Kunz B., Sonderegger P. (2007) Specific cleavage of agrin by neurotrypsin, a synaptic protease linked to mental retardation. FASEB J. 21, 3468–3478 [DOI] [PubMed] [Google Scholar]
- 16. Molinari F., Rio M., Meskenaite V., Encha-Razavi F., Augé J., Bacq D., Briault S., Vekemans M., Munnich A., Attié-Bitach T., Sonderegger P., Colleaux L. (2002) Truncating neurotrypsin mutation in autosomal recessive nonsyndromic mental retardation. Science 298, 1779–1781 [DOI] [PubMed] [Google Scholar]
- 17. Merdes G., Soba P., Loewer A., Bilic M. V., Beyreuther K., Paro R. (2004) Interference of human and Drosophila APP and APP-like proteins with PNS development in Drosophila. EMBO J. 23, 4082–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Willert K., Brown J. D., Danenberg E., Duncan A. W., Weissman I. L., Reya T., Yates J. R., 3rd, Nusse R. (2003) Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423, 448–452 [DOI] [PubMed] [Google Scholar]
- 19. Kang D. E., Yoon I. S., Repetto E., Busse T., Yermian N., Ie L., Koo E. H. (2005) Presenilins mediate phosphatidylinositol 3-kinase/AKT and ERK activation via select signaling receptors. Selectivity of PS2 in platelet-derived growth factor signaling. J. Biol. Chem. 280, 31537–31547 [DOI] [PubMed] [Google Scholar]
- 20. Repetto E., Yoon I. S., Zheng H., Kang D. E. (2007) Presenilin 1 regulates epidermal growth factor receptor turnover and signaling in the endosomal-lysosomal pathway. J. Biol. Chem. 282, 31504–31516 [DOI] [PubMed] [Google Scholar]
- 21. Li T., Ma G., Cai H., Price D. L., Wong P. C. (2003) Nicastrin is required for assembly of presenilin/γ-secretase complexes to mediate Notch signaling and for processing and trafficking of β-amyloid precursor protein in mammals. J. Neurosci. 23, 3272–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mahoney M. B., Parks A. L., Ruddy D. A., Tiong S. Y., Esengil H., Phan A. C., Philandrinos P., Winter C. G., Chatterjee R., Huppert K., Fisher W. W., L'Archeveque L., Mapa F. A., Woo W., Ellis M. C., Curtis D. (2006) Presenilin-based genetic screens in Drosophila melanogaster identify novel notch pathway modifiers. Genetics 172, 2309–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gunawardena S., Goldstein L. S. (2001) Disruption of axonal transport and neuronal viability by amyloid precursor protein mutations in Drosophila. Neuron 32, 389–401 [DOI] [PubMed] [Google Scholar]
- 24. Stokin G. B., Almenar-Queralt A., Gunawardena S., Rodrigues E. M., Falzone T., Kim J., Lillo C., Mount S. L., Roberts E. A., McGowan E., Williams D. S., Goldstein L. S. (2008) Amyloid precursor protein-induced axonopathies are independent of amyloid-β peptides. Hum. Mol. Genet. 17, 3474–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stephan A., Mateos J. M., Kozlov S. V., Cinelli P., Kistler A. D., Hettwer S., Rülicke T., Streit P., Kunz B., Sonderegger P. (2008) Neurotrypsin cleaves agrin locally at the synapse. FASEB J. 22, 1861–1873 [DOI] [PubMed] [Google Scholar]
- 26. Jayadev S., Case A., Eastman A. J., Nguyen H., Pollak J., Wiley J. C., Möller T., Morrison R. S., Garden G. A. (2010) Presenilin 2 is the predominant γ-secretase in microglia and modulates cytokine release. PLoS One 5, e15743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garcia-Bassets I., Kwon Y. S., Telese F., Prefontaine G. G., Hutt K. R., Cheng C. S., Ju B. G., Ohgi K. A., Wang J., Escoubet-Lozach L., Rose D. W., Glass C. K., Fu X. D., Rosenfeld M. G. (2007) Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 128, 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang D., Garcia-Bassets I., Benner C., Li W., Su X., Zhou Y., Qiu J., Liu W., Kaikkonen M. U., Ohgi K. A., Glass C. K., Rosenfeld M. G., Fu X. D. (2011) Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474, 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mikkelsen T. S., Ku M., Jaffe D. B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T. K., Koche R. P., Lee W., Mendenhall E., O'Donovan A., Presser A., Russ C., Xie X., Meissner A., Wernig M., Jaenisch R., Nusbaum C., Lander E. S., Bernstein B. E. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shen Y., Yue F., McCleary D. F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V. V., Ren B. (2012) A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McLean C. Y., Bristor D., Hiller M., Clarke S. L., Schaar B. T., Lowe C. B., Wenger A. M., Bejerano G. (2010) GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boulianne G. L., Livne-Bar I., Humphreys J. M., Liang Y., Lin C., Rogaev E., St George-Hyslop P. (1997) Cloning and characterization of the Drosophila presenilin homologue. Neuroreport 8, 1025–1029 [DOI] [PubMed] [Google Scholar]
- 33. Hu Y., Ye Y., Fortini M. E. (2002) Nicastrin is required for γ-secretase cleavage of the Drosophila Notch receptor. Dev. Cell 2, 69–78 [DOI] [PubMed] [Google Scholar]
- 34. Hong C. S., Koo E. H. (1997) Isolation and characterization of Drosophila presenilin homolog. Neuroreport 8, 665–668 [DOI] [PubMed] [Google Scholar]
- 35. Bogdanik L. P., Burgess R. W. (2011) A valid mouse model of AGRIN-associated congenital myasthenic syndrome. Hum. Mol. Genet. 20, 4617–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang T., Arslanova D., Gu Y., Augelli-Szafran C., Xia W. (2008) Quantification of γ-secretase modulation differentiates inhibitor compound selectivity between two substrates Notch and amyloid precursor protein. Mol. Brain 1, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barthet G., Shioi J., Shao Z., Ren Y., Georgakopoulos A., Robakis N. K. (2011) Inhibitors of γ-secretase stabilize the complex and differentially affect processing of amyloid precursor protein and other substrates. FASEB J. 25, 2937–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Haapasalo A., Kovacs D. M. (2011) The many substrates of presenilin/γ-secretase. J. Alzheimers Dis. 25, 3–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wolfe M. S., Xia W., Ostaszewski B. L., Diehl T. S., Kimberly W. T., Selkoe D. J. (1999) Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and γ-secretase activity. Nature 398, 513–517 [DOI] [PubMed] [Google Scholar]
- 40. Yu G., Chen F., Nishimura M., Steiner H., Tandon A., Kawarai T., Arawaka S., Supala A., Song Y. Q., Rogaeva E., Holmes E., Zhang D. M., Milman P., Fraser P. E., Haass C., George-Hyslop P. S. (2000) Mutation of conserved aspartates affects maturation of both aspartate mutant and endogenous presenilin 1 and presenilin 2 complexes. J. Biol. Chem. 275, 27348–27353 [DOI] [PubMed] [Google Scholar]
- 41. Capell A., Grünberg J., Pesold B., Diehlmann A., Citron M., Nixon R., Beyreuther K., Selkoe D. J., Haass C. (1998) The proteolytic fragments of the Alzheimer's disease-associated presenilin-1 form heterodimers and occur as a 100–150-kDa molecular mass complex. J. Biol. Chem. 273, 3205–3211 [DOI] [PubMed] [Google Scholar]
- 42. Bentahir M., Nyabi O., Verhamme J., Tolia A., Horré K., Wiltfang J., Esselmann H., De Strooper B. (2006) Presenilin clinical mutations can affect gamma-secretase activity by different mechanisms. J. Neurochem. 96, 732–742 [DOI] [PubMed] [Google Scholar]
- 43. O'Brien W. T., Klein P. S. (2009) Validating GSK3 as an in vivo target of lithium action. Biochem. Soc. Trans. 37, 1133–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhong H., Zou H., Semenov M. V., Moshinsky D., He X., Huang H., Li S., Quan J., Yang Z., Lin S. (2009) Characterization and development of novel small-molecules inhibiting GSK3 and activating Wnt signaling. Mol. Biosyst. 5, 1356–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. (2004) WNT and β-catenin signalling: diseases and therapies. Nat. Rev. Genet. 5, 691–701 [DOI] [PubMed] [Google Scholar]
- 46. Chen B., Dodge M. E., Tang W., Lu J., Ma Z., Fan C. W., Wei S., Hao W., Kilgore J., Williams N. S., Roth M. G., Amatruda J. F., Chen C., Lum L. (2009) Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 5, 100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen S., Cao W., Yue P., Hao C., Khuri F. R., Sun S. Y. (2011) Celecoxib promotes c-FLIP degradation through Akt-independent inhibition of GSK3. Cancer Res. 71, 6270–6281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Buttrick G. J., Wakefield J. G. (2008) PI3-K and GSK-3: Akt-ing together with microtubules. Cell Cycle 7, 2621–2625 [DOI] [PubMed] [Google Scholar]
- 49. Hur E. M., Zhou F. Q. (2010) GSK3 signalling in neural development. Nat. Rev. Neurosci. 11, 539–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hong E. J., West A. E., Greenberg M. E. (2005) Transcriptional control of cognitive development. Curr. Opin. Neurobiol. 15, 21–28 [DOI] [PubMed] [Google Scholar]
- 51. Martinowich K., Hattori D., Wu H., Fouse S., He F., Hu Y., Fan G., Sun Y. E. (2003) DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 302, 890–893 [DOI] [PubMed] [Google Scholar]
- 52. Gopisetty G., Xu J., Sampath D., Colman H., Puduvalli V. K. (2013) Epigenetic regulation of CD133/PROM1 expression in glioma stem cells by Sp1/myc and promoter methylation. Oncogene 32, 3119–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tie F., Banerjee R., Stratton C. A., Prasad-Sinha J., Stepanik V., Zlobin A., Diaz M. O., Scacheri P. C., Harte P. J. (2009) CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136, 3131–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Altarejos J. Y., Montminy M. (2011) CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat. Rev. 12, 141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Daniels M. P. (2012) The role of agrin in synaptic development, plasticity and signaling in the central nervous system. Neurochem. Int. 61, 848–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gschwend T. P., Krueger S. R., Kozlov S. V., Wolfer D. P., Sonderegger P. (1997) Neurotrypsin, a novel multidomain serine protease expressed in the nervous system. Mol. Cell. Neurosci. 9, 207–219 [DOI] [PubMed] [Google Scholar]
- 57. Wolfer D. P., Lang R., Cinelli P., Madani R., Sonderegger P. (2001) Multiple roles of neurotrypsin in tissue morphogenesis and nervous system development suggested by the mRNA expression pattern. Mol. Cell. Neurosci. 18, 407–433 [DOI] [PubMed] [Google Scholar]
- 58. Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 59. Matsumoto-Miyai K., Sokolowska E., Zurlinden A., Gee C. E., Lüscher D., Hettwer S., Wölfel J., Ladner A. P., Ster J., Gerber U., Rülicke T., Kunz B., Sonderegger P. (2009) Coincident pre- and postsynaptic activation induces dendritic filopodia via neurotrypsin-dependent agrin cleavage. Cell 136, 1161–1171 [DOI] [PubMed] [Google Scholar]
- 60. Bolliger M. F., Zurlinden A., Lüscher D., Bütikofer L., Shakhova O., Francolini M., Kozlov S. V., Cinelli P., Stephan A., Kistler A. D., Rülicke T., Pelczar P., Ledermann B., Fumagalli G., Gloor S. M., Kunz B., Sonderegger P. (2010) Specific proteolytic cleavage of agrin regulates maturation of the neuromuscular junction. J. Cell Sci. 123, 3944–3955 [DOI] [PubMed] [Google Scholar]
- 61. Zou K., Hosono T., Nakamura T., Shiraishi H., Maeda T., Komano H., Yanagisawa K., Michikawa M. (2008) Novel role of presenilins in maturation and transport of integrin β1. Biochemistry 47, 3370–3378 [DOI] [PubMed] [Google Scholar]
- 62. Wolfe M. S. (2012) γ-Secretase inhibitors and modulators for Alzheimer's disease. J. Neurochem. 120, Suppl. 1, 89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Morohashi Y., Kan T., Tominari Y., Fuwa H., Okamura Y., Watanabe N., Sato C., Natsugari H., Fukuyama T., Iwatsubo T., Tomita T. (2006) C-terminal fragment of presenilin is the molecular target of a dipeptidic γ-secretase-specific inhibitor DAPT (N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester). J. Biol. Chem. 281, 14670–14676 [DOI] [PubMed] [Google Scholar]
- 64. Watanabe H., Smith M. J., Heilig E., Beglopoulos V., Kelleher R. J., 3rd, Shen J. (2009) Indirect regulation of presenilins in CREB-mediated transcription. J. Biol. Chem. 284, 13705–13713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ichiki T. (2006) Role of cAMP response element binding protein in cardiovascular remodeling: good, bad, or both? Arterioscler. Thromb. Vasc. Biol. 26, 449–455 [DOI] [PubMed] [Google Scholar]
- 66. Garcia-Bassets I., Wang D. (2012) Cistrome plasticity and mechanisms of cistrome reprogramming. Cell Cycle 11, 3199–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fiol C. J., Williams J. S., Chou C. H., Wang Q. M., Roach P. J., Andrisani O. M. (1994) A secondary phosphorylation of CREB341 at Ser129 is required for the cAMP-mediated control of gene expression. A role for glycogen synthase kinase-3 in the control of gene expression. J. Biol. Chem. 269, 32187–32193 [PubMed] [Google Scholar]
- 68. Horike N., Sakoda H., Kushiyama A., Ono H., Fujishiro M., Kamata H., Nishiyama K., Uchijima Y., Kurihara Y., Kurihara H., Asano T. (2008) AMP-activated protein kinase activation increases phosphorylation of glycogen synthase kinase 3β and thereby reduces cAMP-responsive element transcriptional activity and phosphoenolpyruvate carboxykinase C gene expression in the liver. J. Biol. Chem. 283, 33902–33910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Böer U., Cierny I., Krause D., Heinrich A., Lin H., Mayr G., Hiemke C., Knepel W. (2008) Chronic lithium salt treatment reduces CRE/CREB-directed gene transcription and reverses its upregulation by chronic psychosocial stress in transgenic reporter gene mice. Neuropsychopharmacology 33, 2407–2415 [DOI] [PubMed] [Google Scholar]
- 70. Tyson D. R., Swarthout J. T., Jefcoat S. C., Partridge N. C. (2002) PTH induction of transcriptional activity of the cAMP response element-binding protein requires the serine 129 site and glycogen synthase kinase-3 activity, but not casein kinase II sites. Endocrinology 143, 674–682 [DOI] [PubMed] [Google Scholar]
- 71. Conlon R. A., Reaume A. G., Rossant J. (1995) Notch1 is required for the coordinate segmentation of somites. Development 121, 1533–1545 [DOI] [PubMed] [Google Scholar]
- 72. Donoviel D. B., Hadjantonakis A. K., Ikeda M., Zheng H., Hyslop P. S., Bernstein A. (1999) Mice lacking both presenilin genes exhibit early embryonic patterning defects. Genes Dev. 13, 2801–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Handler M., Yang X., Shen J. (2000) Presenilin-1 regulates neuronal differentiation during neurogenesis. Development 127, 2593–2606 [DOI] [PubMed] [Google Scholar]
- 74. Wen P. H., De Gasperi R., Gama Sosa M. A., Elder G. A. (2004) Neural progenitor cells do not differentiate prematurely in presenilin-1 null mutant mice. Neurosci. Lett. 371, 249–254 [DOI] [PubMed] [Google Scholar]
- 75. Gadadhar A., Marr R., Lazarov O. (2011) Presenilin-1 regulates neural progenitor cell differentiation in the adult brain. J. Neurosci. 31, 2615–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kim W. Y., Shen J. (2008) Presenilins are required for maintenance of neural stem cells in the developing brain. Mol. Neurodegener. 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ahn K., Shelton C. C., Tian Y., Zhang X., Gilchrist M. L., Sisodia S. S., Li Y. M. (2010) Activation and intrinsic γ-secretase activity of presenilin 1. Proc. Natl. Acad. Sci. U.S.A. 107, 21435–21440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shen J., Kelleher R. J., 3rd (2007) The presenilin hypothesis of Alzheimer's disease: evidence for a loss-of-function pathogenic mechanism. Proc. Natl. Acad. Sci. U.S.A. 104, 403–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. De Strooper B. (2007) Loss-of-function presenilin mutations in Alzheimer disease. Talking point on the role of presenilin mutations in Alzheimer disease. EMBO Rep. 8, 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tu H., Nelson O., Bezprozvanny A., Wang Z., Lee S. F., Hao Y. H., Serneels L., De Strooper B., Yu G., Bezprozvanny I. (2006) Presenilins form ER Ca2+ leak channels, a function disrupted by familial Alzheimer's disease-linked mutations. Cell 126, 981–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nelson O., Tu H., Lei T., Bentahir M., de Strooper B., Bezprozvanny I. (2007) Familial Alzheimer disease-linked mutations specifically disrupt Ca2+ leak function of presenilin 1. J. Clin. Investig. 117, 1230–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kawamura Y., Kikuchi A., Takada R., Takada S., Sudoh S., Shibamoto S., Yanagisawa K., Komano H. (2001) Inhibitory effect of a presenilin 1 mutation on the Wnt signalling pathway by enhancement of β-catenin phosphorylation. Eur. J. Biochem. 268, 3036–3041 [DOI] [PubMed] [Google Scholar]
- 83. Saura C. A., Valero J. (2011) The role of CREB signaling in Alzheimer's disease and other cognitive disorders. Rev. Neurosci. 22, 153–169 [DOI] [PubMed] [Google Scholar]