Background: Chronic PKC activation is the leading pathogenic component of diabetes in the heart.
Results: PKCα/β promotes fetal splicing patterns in adult diabetic hearts via phosphorylation of the RNA-binding proteins CELF1 and Rbfox2.
Conclusion: PKCα/β contributes to diabetes pathogenesis by manipulating developmentally regulated alternative splicing.
Significance: Identifying downstream effectors of PKC can provide novel therapeutics for cardiac pathogenesis of diabetes.
Keywords: Alternative Splicing, Diabetes, Heart Development, Protein Kinase C (PKC), RNA-binding Protein, Signal Transduction
Abstract
Diabetic cardiomyopathy is one of the complications of diabetes that eventually leads to heart failure and death. Aberrant activation of PKC signaling contributes to diabetic cardiomyopathy by mechanisms that are poorly understood. Previous reports indicate that PKC is implicated in alternative splicing regulation. Therefore, we wanted to test whether PKC activation in diabetic hearts induces alternative splicing abnormalities. Here, using RNA sequencing we identified a set of 22 alternative splicing events that undergo a developmental switch in splicing, and we confirmed that splicing reverts to an embryonic pattern in adult diabetic hearts. This network of genes has important functions in RNA metabolism and in developmental processes such as differentiation. Importantly, PKC isozymes α/β control alternative splicing of these genes via phosphorylation and up-regulation of the RNA-binding proteins CELF1 and Rbfox2. Using a mutant of CELF1, we show that phosphorylation of CELF1 by PKC is necessary for regulation of splicing events altered in diabetes. In summary, our studies indicate that activation of PKCα/β in diabetic hearts contributes to the genome-wide splicing changes through phosphorylation and up-regulation of CELF1/Rbfox2 proteins. These findings provide a basis for PKC-mediated cardiac pathogenesis under diabetic conditions.
Introduction
Diabetes is a group of metabolic disorders caused by high blood glucose levels either due to lack of insulin production (type 1) or loss of responsiveness to insulin (type 2) (1). Cardiovascular complications of diabetes, which include coronary heart disease, hypertension, and diabetic cardiomyopathy, are the number one cause of mortality and morbidity (2–4). Diabetic cardiomyopathy occurs in diabetic patients independent of coronary heart disease and hypertension (5). Manifestations include diastolic-systolic dysfunction, cardiomyocyte hypertrophy, and fibrosis, which lead to heart failure (5). Chronic activation of PKC isozymes α, β, and δ promotes diastolic/systolic dysfunction, fibrosis, cardiomyocyte hypertrophy, or apoptosis observed in diabetic cardiomyopathy (6–8). Inhibition of PKC activity improves cardiovascular complications of diabetes in rodent models, indicating a pathogenic role for PKC (9, 10). However, the mechanisms by which PKC induces cardiac pathogenesis in diabetes are not well understood.
PKCα/β has been previously linked to AS4 regulation (11–13) and identified as one of the regulators of a group of AS events in HEK293T cells activated by epidermal growth factor (14). AS regulates eukaryotic gene expression by controlling the inclusion and exclusion of alternative exons within the coding and noncoding RNA transcripts (15, 16). We have previously shown that PKCα/β phosphorylates and increases the steady state levels of an AS regulator called CELF1 (also known as CUG-binding protein 1) in COS M6 cells (11) and contributes to cardiac dysfunction in myotonic dystrophy (11, 17).
CELF1 is one of the RNA-binding proteins involved in AS regulation during heart development together with Rbfox2 and muscleblind-like 1 (MBNL1) (18). Binding sites for these proteins have been identified computationally in adjacent introns of alternative exons that transition during heart development (18). Overexpression of CELF1 in adult mouse hearts causes splicing defects, and they eventually develop heart failure (19). Rbfox2 regulates AS in embryonic stem cells (20). Rbfox1 and Rbfox2 proteins are important for muscle and heart function in zebrafish (21). Tissue-specific Rbfox2-KO in the central nervous system causes developmental defects in the cerebellum (22). MBNL1 is a repressor of embryonic stem cell-specific alternative splicing (23). MBNL1 knock-out mice develop skeletal muscle symptoms of myotonic dystrophy with defects in developmentally regulated splicing (24). It is currently unknown whether these splicing regulators have roles in diabetes pathogenesis.
Because PKC is implicated in AS regulation and diabetic cardiomyopathy pathogenesis, we wanted to determine whether PKC signaling contributes to abnormal gene expression in diabetes via alternative splicing. In this study, transcriptome analysis revealed that conserved AS events revert back to an embryonic pattern in adult diabetic hearts. We demonstrate that PKCα/β signaling regulates embryonic to adult splicing transitions of these genes. PKCα/β induces fetal splicing via phosphorylation of the RNA-binding proteins CELF1 and Rbfox2. Notably, a CELF1 mutant that cannot be phosphorylated by PKC is unable to regulate the splicing event altered in diabetic hearts. Overall, our data indicate that one of the mechanisms for abnormal gene expression in diabetes is due to a shift from an adult to a fetal splicing pattern mediated by PKCα/β.
EXPERIMENTAL PROCEDURES
Cell Culture and Differentiation
H9c2-2 cells (ATCC CRL-1446) originally isolated from E13 (corresponds to ∼E11 in mouse) BDX1 rat heart ventricles were adapted to cell culture via selective serial passaging (25). The cells were cultured and maintained in Dulbecco's modified Eagle's medium (DMEM) (ATCC 30-2002), supplemented with 10% fetal bovine serum (FBS, ATCC 30-2020), 100 units/ml penicillin and streptomycin (Invitrogen 15140) at 37 °C in a 5% CO2 humidified incubator. Media were changed every 2–3 days, and cells were subcultured at 70–75% confluency. The passage number of the cells used in all experiments was 4–6.
Differentiation of H9c2 cells into cardiomyocyte-like cells was induced by adding 1 μm all-trans-retinoic acid (RA) daily to the reduced serum media (1% FBS) for 7 days. H9c2 cells (1 × 106) were plated in a 150-mm dish 1 day before differentiation. Cells were harvested daily or at the end of differentiation for RNA and/or protein extraction. Undifferentiated control cells were mock (DMSO)-treated and harvested after 24 h for RNA and/or protein extraction. In the PKC inhibitor experiments, both undifferentiated and differentiated cells were mock-treated or treated with the 1.0 μm PKC inhibitor BisIX (EMD557529) daily for 7 days. The inhibitor concentration was increased by 0.1 μm each day. Cells were monitored daily by light microscopy to observe morphological changes.
Primary rat cardiomyocytes were isolated from 1-day-old Sprague-Dawley rat heart ventricles (Cell Application Inc., R357-25). Cells were maintained in rat cardiomyocyte growth media (Cell Application Inc., R313-500) at 37 °C in a 5% CO2 humidified incubator and monitored daily by light microscopy. Media were renewed every 2 days. Cells were trypsinized gently (0.05% trypsin/EDTA, Invitrogen 25300-054) prior to plating. For experiments, 2 × 105 cells were plated on gelatin (Sigma G1890)-coated T25 flasks (Nunc 12-565-351). Seven to 10 days after plating, beating cardiomyocytes were either mock-treated or treated with 2.0 μm of the PKC inhibitor BisIX (EMD557529) for 12 h.
Nuclear-Cytoplasmic Fractionation
Cells
The nuclear and cytoplasmic fractionation protocol was modified from Kuyumcu-Martinez et al. (11). Cells were washed twice with PBS and scraped in 500 μl of hypotonic buffer (20 mm HEPES, pH 7.5, 5 mm NaCl, 0.4% Triton X-100) containing protease and phosphatase inhibitors and then passed through a 26-gauge needle. Nuclei were separated by centrifugation at 600 × g at 4 °C for 15 min. The supernatant was kept as the cytoplasmic fraction, and nuclei were washed with the hypotonic buffer to remove the cytoplasmic contaminants, followed by centrifugation at 600 × g at 4 °C for 5 min. The nuclear pellet was resuspended in hypotonic buffer containing 1% SDS, followed by sonication. The protein concentrations of the cytoplasmic and nuclear fractions were determined using BCA protein assay (Sigma B9643).
Heart Tissues
Half of a mouse heart was cut into small pieces followed by homogenization using a glass douncer in 500 μl of cytosolic buffer (10 mm HEPES, pH 7.5, 10 mm MgCl2, 5 mm KCl, 0.1 mm EDTA, 0.2 mm PMSF, 1 mm DTT, and 1× Complete protease inhibitor mixture from Roche Applied Science). The minced heart tissue was dounced six times prior to centrifugation at 3000 × g at 4 °C for 15 min. The supernatant was collected as the cytoplasmic fraction, and the nuclear pellet was washed twice with 1.0 ml of cytosolic buffer and centrifuged at 3000 × g at 4 °C for 5 min to remove cytoplasmic contaminants. Nuclear pellet was resuspended in 200 μl of urea buffer (200 mm Tris, pH 7.4, 4% CHAPS, 7 m urea, and 2 m thiourea) followed by incubation at 90 °C for 5 min (repeated once more). For two-dimensional gel analysis, the nuclear pellet was sonicated gently on ice instead and loaded on two-dimensional gels as described previously (11). Protein concentrations of cytoplasmic and nuclear fractions were estimated using the Bradford protein assay.
Western Blotting
30–50 μg of protein/sample was separated on 10% SDS-PAGE, and proteins were transferred to a PVDF membrane (Immobilon-P, Millipore IPVH00010). Membranes were stained with Ponceau S (Sigma P7170) to assess the quality of the transfer and the loading of the proteins. Membranes were blocked with 5% dry fat-free milk solution in PBST (PBS containing 0.1% Tween 20) followed by overnight incubation with the indicated primary antibodies (Abs) at 4 °C. Membranes were washed three times with PBST 15 min each and incubated with HRP-labeled relevant secondary antibody for 2 h at 4 °C. HRP activity was determined using Immobilon Western chemiluminescent (Millipore P90720) or SuperSignal West Femto Chemiluminescent (Pierce PI34095) HRP substrate followed by exposure to x-ray film (GeneMate F9024) or Kodak Gel Logic 2200. Percent increase or decrease in protein levels was quantified using the Kodak Gel Logic software after normalizing the protein levels to the appropriate loading controls.
Immunohistochemistry
Whole rat (Sprague-Dawley) embryo paraffin sections at E13 and E18 stages and sagittal heart sections of newborn and adult rats (Sprague-Dawley) were purchased from Zyagen. Paraffin sections were incubated at 56 °C for 12–14 h, followed by deparaffinization and dehydration in xylene for 20 min. Slides were washed in decreasing concentrations (100 to 50%) of ethanol for 5 min at each concentration. Antigens were exposed by incubating the sections with sodium citrate buffer (10 mm, pH 6.0) for 20 min in a steam chamber. Blocking was performed in 3% BSA in PBST (0.2% Triton X-100) at RT for 1 h. Sections were then incubated with the appropriate primary Abs as follows: anti-PKCα, anti-CELF1, anti-cTNI, or anti-Nkx2.5 for 14–16 h at RT in a humidifying chamber. Slides were washed with PBS containing 0.1% Triton X-100 and then incubated with fluorescently labeled secondary Abs as follows: anti-mouse Alexa Fluor-488 or anti-rabbit Texas Red for 2 h at 37 °C. Slides were washed four times with PBS + 0.1% Triton X-100, followed by 4′,6-diamino-2-phenylindole dihydrochloride (DAPI) stain for 1 h at room temperature in the dark. Excess stain was washed away with PBS before mounting the coverslips using Mowiol mounting media; the slides were then sealed using nail polish. Fluorescence images were obtained with a confocal laser-scanning microscope (LSM 510META, Carl Zeiss) at the University of Texas Medical Branch imaging core facility. To quantify the number of cardiomyocytes that display nuclear CELF1 or PKCα staining, IHC images obtained from multiple sections for each developmental stage were used to count DAPI-stained nuclei that display both red (cardiomyocytes markers) and green (CELF1 or PKCα) fluorescence using ImageJ software (nuclei>150) (26).
T1D Mice
Nonobese diabetic (NOD) female mice were obtained from The Jackson Laboratory (NOD/ShiLtJ). For STZ-induced T1D mice, C57BL/6J male mice (8 weeks old) were either injected with citrate buffer as a control or with 60 mg/kg STZ daily for 5 days (27). Blood glucose levels of control, NOD, and STZ mice were determined once a week using blood from the tail vein by a one-touch glucometer. Mice with fasting glucose levels >300 mg/dl for 3, 6, and 9 weeks were used as diabetics, and fasting glucose levels <200 were used as controls. Mice were sacrificed, and their hearts were isolated for protein and RNA extraction. All experiments were conducted in accordance with the National Institutes of Health Guidelines for the care and use of laboratory animals and approved by the Institutional Animal Care and Use Committee of University of Texas Medical Branch.
RNA Extraction
RNA was extracted from cells and human and mouse heart tissues using TRIzol (Invitrogen) per the manufacturer's protocol. RNA concentrations were measured using EPOCH Microplate Spectrophotometer (BioTek). Human heart tissues were obtained from the National Disease Research Interchange. Two control human heart tissues were gifts from Dr. Nisha Garg, University of Texas Medical Branch. All human heart tissues used in this study were pre-existing and are exempt from Institute Review Board.
Rat Heart RNA
Sprague-Dawley rat heart RNA at different developmental stages (E13, E18, E20, newborn, and adult) was purchased from Zyagen. At least four pooled hearts were used for RNA extraction at early developmental stages.
Site-directed Mutagenesis
PKC phosphorylation sites were identified using LC-MS/MS on human CELF1 protein expressed in 293T cells. The predicted sites of Ser (28, 52, 178, 179, 241, 300, 302) were mutated to Ala and Thr-173 to Val using site-directed mutagenesis kit (Stratagene 200516) to generate the CELF1M8 construct in p3×FLAG vector.
Quantitative RT-PCR and Statistical Analysis
The qRT-PCR protocol was adapted from Kalsotra et al. (18). 125 ng of oligo(dT) was annealed to the poly(A) tail of total RNA (1 μg) at 65 °C before cDNA synthesis using avian myeloblastosis virus-reverse transcriptase (15 units/μg, Life Biosciences AMV007-1), and 10 μm dNTPs in a 20-μl reaction at 42 °C for an hour. Primer sequences were designed to detect the alternative exons of the following: myotubularin-related protein 3 (Mtmr3); ATPase Ca2+-transporting plasma membrane (Atp2b1); Fragile X mental retardation gene 1 (Fxr1); muscleblind-like 2 (Mbnl2), and heterogeneous nuclear ribonucleoprotein H1 (hnRNPH1) (Table 4). PCR was performed using 5 μl of cDNA and 100 ng of each primer pair with Biolase Taq polymerase (Bioline BIO-21042). PCR amplification of all genes was carried out at the following 95 °C, 45 s; 59 °C, 45 s; 72 °C, 1 min for 25 cycles. PCR products were resolved on 5–8% nondenaturing polyacrylamide gels and stained with ethidium bromide. Bands were quantified for percentage exon inclusion using Kodak Gel Logic 2200 and molecular imaging software as percentage inclusion = ((exon inclusion band)/(exon inclusion band + exon exclusion band)·100) as described previously (18). The quantitation method normalizes the ethidium bromide signal to the size of PCR bands (28). Data from three independent experiments were used for statistical analysis, and significance was calculated using an unpaired t test (Figs. 1–4 and 6).
TABLE 4.
Primer sequences used to amplify alternative exons in qRT-PCR
| Gene name | Gene ID | Forward primer 5′ to 3′ | Reverse primer 5′ to 3′ |
|---|---|---|---|
| Atp2b1 (rat) | ENSRNOG00000004026 | GTGGCCAGATCTTGTGGTTT | CATCGATAAGGGGGATGTGC |
| Atp2b1 (mouse) | ENSMUSG00000019943 | GTGGCCAGATCTTGTGGTTT | CATCAATAAGGGGGATGTGC |
| Mbnl2 (rat) | ENSRNOT00000057557 | CTCCCGGTGCTCTTCACC | CTTGGCATTCCATTCCATTT |
| Mbnl2 (mouse) | ENSMUSG00000022139 | ACTACCAGCAGGCTCTGACC | TGAGGAGGAAACGGGGTAAT |
| Fxr1 (rat) | ENSRNOG00000011938 | GATAATACAGAATCCGATCAG | CTGAAGGACCATGCTCTTCAATCAC |
| Fxr1 (mouse) | ENSMUSG00000027680 | GATAATACAGAATCCGATCAG | CTGAAGGACCATGCTCTTCAATCAC |
| Fxr1 (human) | ENSG00000114416 | GATAATACAGAATCCGATCAG | CTGAAGGACCATGCTCTTCAATCAC |
| Mtmr3 (rat) | ENSRNOG00000007120 | GTTGGCTACCTGACCAC | CTCGACTGGGTTCAAAGAGC |
| Mtmr3 (mouse) | ENSMUSG00000034354 | GTTGGCTACCTGACCACCTG | CTCGACTGGGTTCAAAGAGC |
| Mtmr3 (human) | ENSG00000100330 | GTTGGCTTCCTGACCACCTG | CTCGACTGGGTTCAAAGAGC |
| hnRNPH1 (mouse) | ENSMUST00000069304 | CTCCCTTTTGGATGTAGCAAGG | GGCCATAAGTTTTCGTGGTGG |
| hnRNPH1 (human) | ENSG00000169045 | CTCCCTTTTGGATGTAGCAAGG | GGCCATAAGTTTTCGTGGTGG |
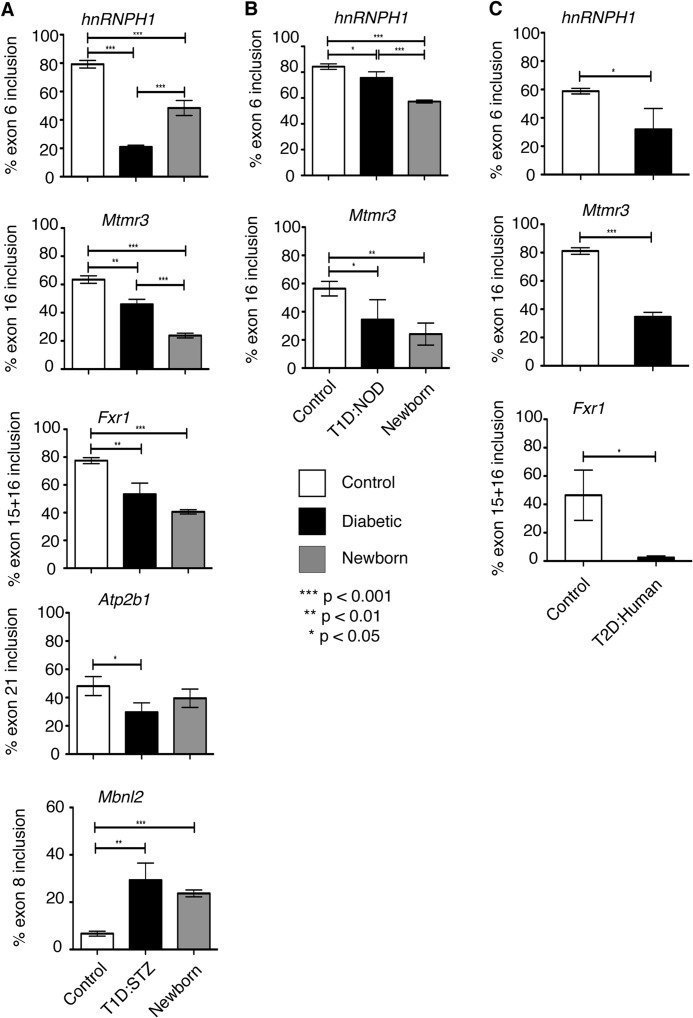
FIGURE 1.
Fetal splicing programs are activated in diabetic hearts. A, qRT-PCR using RNA from mouse heart tissues. Percent inclusion of hnRNPH1 exon6, Mtmr3 exon16, Fxr1 exon15 + 16, Atp2b1 exon21, and Mbnl2 exon8 in hearts from normal C57BL/6 newborn mice (gray bars) and saline (white bars) or STZ-treated C57BL/6 adult mice (black bars) (n ≥ 3). B, percent inclusion of hnRNPH1 exon6 and Mtmr3 exon16 in hearts from wild type newborn FVB mice and in NOD adult mice with low glucose (control) or high glucose (T1D/NOD). Hearts from newborn mice were pooled (n ≥ 3). C, percent inclusion of hnRNPH1 exon6, Mtmr3 exon16, and Fxr1 exon15 + 16 in nondiabetic (control) or type 2 diabetic human (T2D/human) hearts (n = 3). Unpaired t test was used to calculate statistically significant difference between two samples (n ≥ 3).
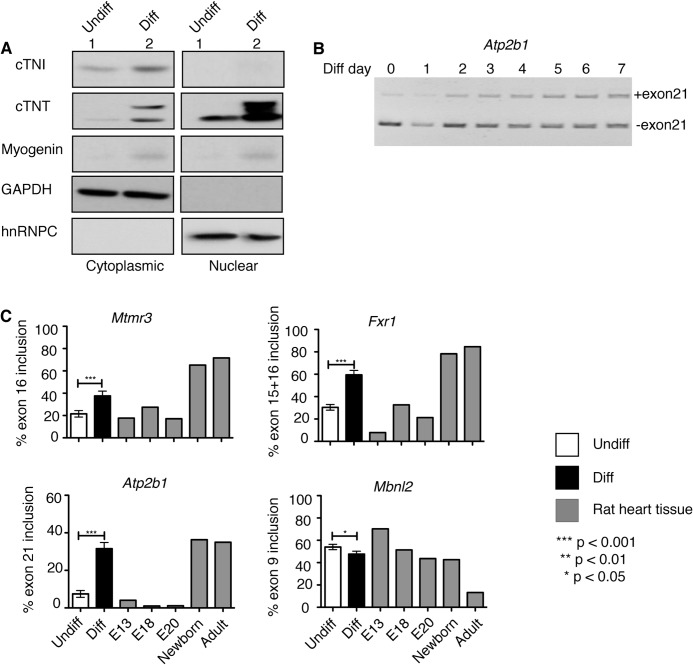
FIGURE 2.
Diabetes-induced alternative splicing events transition during rat heart development and H9c2 differentiation. A, proteins from nuclear and cytoplasmic fractions of H9c2 cells either undifferentiated (undiff) or differentiated (diff) were analyzed by Western blotting using antibodies against cTNI and cTNT (cardiac markers), myogenin (skeletal muscle marker), GAPDH (cytoplasmic marker), hnRNPC (nuclear marker). B and C, qRT-PCR was performed with RNA from H9c2 cells (undiff and diff) and/or from E13*, E18*, E20*, newborn*, and adult rat hearts. B, splicing of Atp2b1 exon21 from day 0 (undiff) to day 7 of H9c2 differentiation. The included band size is 273 bp, and the excluded band size is 186 bp. C, percent inclusion of alternative exons for Mtmr3, Fxr1, Atp2b1, and Mbnl2 during differentiation of H9c2 cells into cardiomyocyte-like cells and at different stages of rat heart development (E13*, E18*, E20*, newborn, and adult rat hearts; * indicates pooled rat hearts ≥4). Significance was calculated using unpaired t test between undiff and diff cells (n ≥ 3).
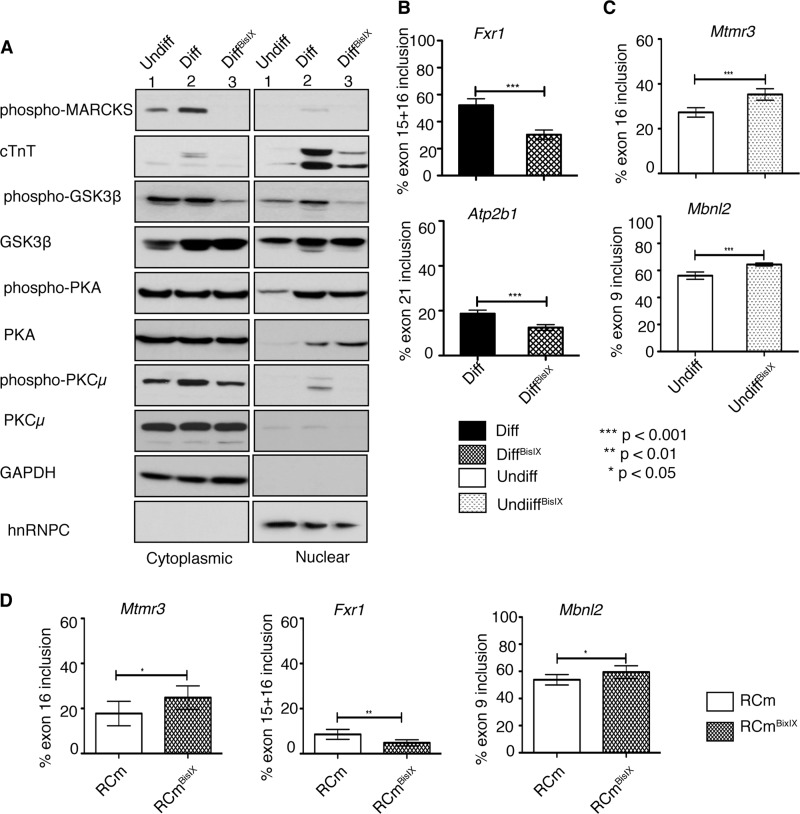
FIGURE 3.
PKC inhibitor BisIX (Ro-31-8220) alters embryonic to adult transitions of splicing events affected in diabetes. Cells were mock-treated (Undiff, Diff) or treated with BisIX (1 μm; UndiffBisIX, DiffBisIX) daily 1 h before RA treatment for 7 days. A, Western blot of protein lysates from cytoplasmic and nuclear fractions using antibodies against phospho-MARCKS, cTNT, phospho-GSK3β (Ser-9), GSK3β, phospho-PKA (PDK1 phosphorylation site Thr-197), PKA, phospho-PKCμ (autophosphorylation site Ser-916), PKCμ, GAPDH and hnRNPC. B, quantification of Fxr1 exon15 + 16 and Atp2b1 exon21 in mock-treated differentiated (Diff, black bars) and BisIX-treated differentiated (DiffBisIX, black-hatched bars) cells. C, percent inclusion of Mtmr3 exon16 and Mbnl2 exon9 in undifferentiated H9c2 cells that are mock-treated (Undiff, white bars) or BisIX-treated (UndiffBisIX, white-hatched bars). D, percent inclusion of Mtmr3 exon16, Fxr1 exon15 + 16, and Mbnl2 exon9 in control (rat cardiomyocytes (RCm), white bars) or PKC inhibitor-treated (RCmBisIX, white-hatched bars) primary neonatal rat cardiomyocytes. Significant changes between two samples were calculated using unpaired t test (n ≥ 3).
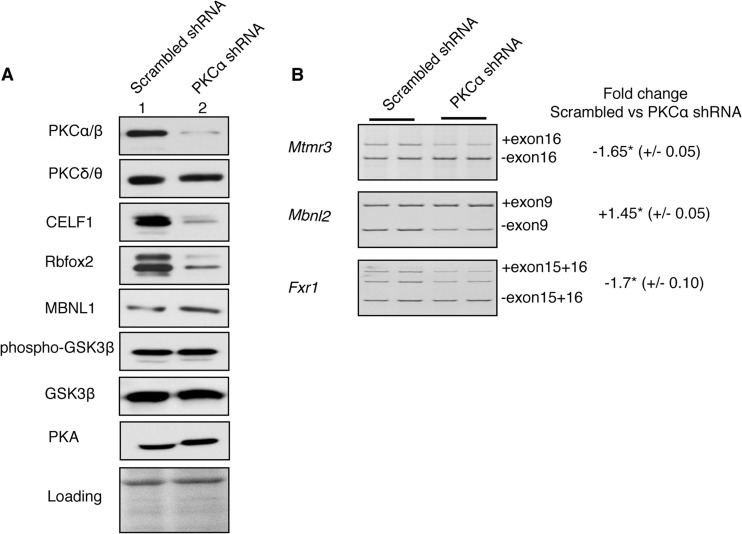
FIGURE 4.
Depletion of PKCα impairs alternative splicing events by regulating CELF1 and Rbfox2 protein levels. H9c2 cells were transfected with scrambled or PKCα-specific shRNA. After PKCα depletion, H9c2 cells were differentiated using RA. A, WB of cell lysates using antibodies against PKCα/β, PKCδ/θ, CELF1, Rbfox2, MBNL1, phospho-GSK3β (Ser-9), GSK3β, and PKA. Ponceau S staining was used to assess protein loading. B, percent inclusion of Mtmr3 exon16, Mbnl2 exon9, and Fxr1 exon15 + 16. qRT-PCR was performed using RNA from H9c2 cells expressing sc-shRNA or PKCα shRNA that were differentiated to cardiomyocyte-like cells. The fold change in alternative exon inclusion was calculated by the following formula, (inclusion in PKCα depleted cells)/(inclusion in sc-shRNA treated cells), and values were represented as negative (−) if inclusion was decreased or as positive (+) if inclusion was increased. (± indicate standard deviation; two independent experiments with duplicate samples were used.) * denotes statistical significance of p < 0.05 determined by unpaired t test (n ≥ 3).
FIGURE 6.
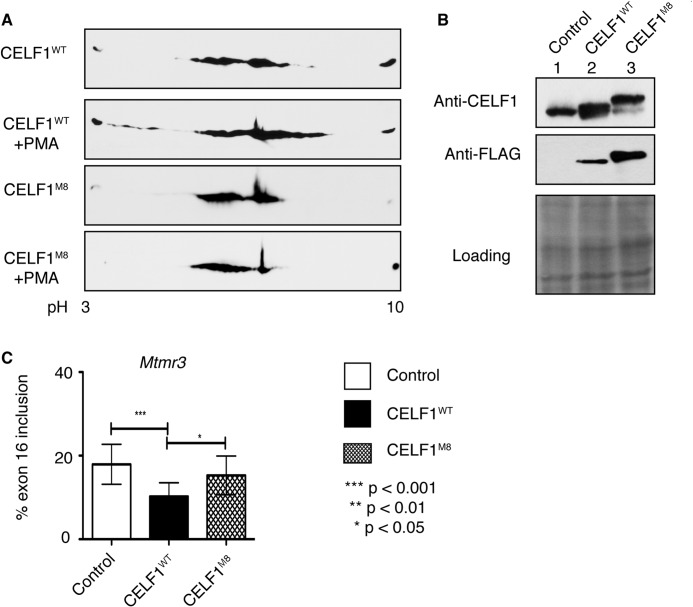
Nonphosphorylatable mutant of CELF1 is unable to promote embryonic splicing of its target pre-mRNA that is reactivated in diabetes. A, two-dimensional/FLAG-specific WB analysis of protein lysates from COS M6 cells expressing either FLAG-tagged wild type (CELF1WT) or nonphosphorylatable mutant of (CELF1M8) CELF1 treated with or without PKC activator phorbol 12-myristate 13-acetate (PMA). B, WB analysis of protein lysates from COS M6 cells transfected with either FLAG-CELF1WT or FLAG-CELF1M8 using anti-CELF1 and anti-FLAG antibodies. Loading was determined by Ponceau S staining. C, percent exon16 inclusion of endogenous Mtmr3 in COS M6 cells transfected with empty vector, CELF1WT, or nonphosphorylatable CELF1M8. Significant changes between two samples were calculated using unpaired t test from three independent experiments, n >12.
RNA Sequencing
RNA extracted from hearts of three diabetic STZ mice (12 weeks old and diabetic for 4 weeks) and two age-matched control mice were used for RNA sequencing (paired end two times for 15 cycles) at the University of Texas Medical Branch Next-Gen core facility using Illumina HiSeq 1000 system. Before library construction, the quality of the RNA was determined using the bioanalyzer. ∼200 m reads per sample were generated. Alternative cassette exon inclusions were determined using the MISO algorithm (29). Splicing event annotations were downloaded from the MISO webserver that was based on the ENSEMBL gene annotation.
Transfections
H9c2 cells (1.25 × 106) were transfected with different plasmids as follows: 1 μg each, encoding an shRNA against PKCα (OriGene TG704205) or encoding a scrambled shRNA (OriGene TR30015) using the Neon Nucleofection System (Invitrogen). Transfected cells were selected in DMEM with 10% FBS containing 1.5 μg/ml puromycin (Sigma P9620) 1 day before differentiation. Cells were maintained in puromycin selection media during 7 days of differentiation. COS M6 cells were transfected using Lipofectamine 2000 according to the manufacturer's protocol. 1 μg of FLAG-tagged wild type human CELF1 (CELF1WT) or mutant CELF1 (CELF1M8) plasmid was transfected. Cells were harvested using urea buffer for protein extraction or TRIzol for RNA extraction 48 h post-transfection.
RESULTS
Reactivation of Embryonic Splicing Events in DiabeticHearts
To determine whether AS is altered in adult diabetic hearts that could contribute to abnormal gene expression, we performed a genome-wide transcriptome analysis to compare alternative splicing profiles between diabetic and control mice. For this analysis, we used a well established type 1 diabetes mouse model that develops diabetes and eventually diabetic cardiomyopathy after injection of STZ that kills pancreatic beta islet cells (5, 30). We chose the STZ model for the ease of diabetes induction and the well characterized diabetic heart phenotype (31). RNA from STZ-treated diabetic and mock-treated control mice was isolated and subjected to RNA-seq library preparation. High throughput RNA-seq data generated from the library were mapped to the mouse genome (mm9) and analyzed for differentially included or excluded cassette exons between control and diabetic mice using the MISO algorithm (29). 967 alternative splicing events that change in diabetic mice hearts were identified and categorized based on their biological function. We found that a group of genes, which are involved in development and RNA metabolism (Table 1), had undergone significant changes in alternative splicing in diabetes (Table 2). Interestingly, these genes have been implicated in the pathogenesis of diabetes and/or heart diseases (Table 3).
TABLE 1.
GO analysis of gene list in Table 2
GO term analysis was performed using DAVID on-line analysis tool (68).
| Category | Term | p value | Count |
|---|---|---|---|
| SP_PIR_Keywords | RNA-binding | 9.3E-05 | 6 |
| Alternative splicing | 2.4E-04 | 13 | |
| Phosphoprotein | 1.6E-03 | 14 | |
| GOTERM_BP_ALL | Cell differentiation | 6.6E-06 | 6 |
| Cellular developmental process | 7.9E-03 | 6 | |
| Post-transcriptional regulation of gene expression | 6.6E-03 | 3 | |
| Regulation of cellular processes | 3.3E-02 | 10 | |
| Regulation of RNA metabolism | 3.0E-02 | 5 |
TABLE 2.
Comparison of alternative splicing events in diabetic mouse hearts to splicing events in developing mouse hearts
+ indicates inclusion, and − indicates exclusion; SH3, Src homology 3 domain.
| Symbol | Gene name | Ensemble ID | Exon no. | Difference (normal vs. diabetic) | Difference (adult vs. embryonic) |
|---|---|---|---|---|---|
| Atp2b1 | Plasma membrane calcium-transporting ATPase 1 | ENSMUSG00000019943 | 21 | −0.18 | −0.28 |
| Fxr1 | FragileX mental retardation gene 1 | ENSMUSG00000027680 | 16 | −0.15 | −0.11 |
| Vegfa | Vascular endothelial growth factor A | ENSMUSG00000023951 | 6 | −0.32 | −0.20 |
| Mbnl1 | Muscleblind-like 1 | ENSMUSG00000027763 | 5 | +0.20 | +0.38 |
| Mbnl2 | Muscleblind-like 2 | ENSMUSG00000022139 | 8 | +0.36 | +0.37 |
| Mef2a | Myocyte enhancer factor 2A | ENSMUSG00000030557 | Intron retention | −0.33 | −0.44 |
| Ablim1 | Actin-binding LIM protein 1 | ENSMUSG00000025085 | 16 | −0.18 | −0.13 |
| Ankrd10 | Ankyrin repeat domain 10 | ENSMUSG00000031508 | 4 | −0.15 | −0.11 |
| Bnip2 | BCL2/adenovirus E1B interacting protein 2 | ENSMUSG00000011958 | 10 | −0.19 | −0.10 |
| Capzb | Capping protein (actin filament) muscle Z-line, β | ENSMUSG00000028745 | 8 | −0.07 | −0.27 |
| G3bp2 | GTPase-activating protein (SH3 domain)-binding protein 2 | ENSMUSG00000029405 | 8 | −0.33 | −0.11 |
| hnRNPH2 | Heterogeneous nuclear ribonucleoprotein H2 | ENSMUSG00000045427 | 3 | − 0.40 | − 0.23 |
| Ldb3 | LIM domain binding 3 | ENSMUSG00000021798 | 8 | + 0.24 | + 0.33 |
| Pbx3 | Pre-B cell leukemia homeobox 3 | ENSMUSG00000038718 | 7 | −0.31 | −0.36 |
| Prrc2b | Proline-rich coiled-coil 2B | ENSMUSG00000039262 | 16 | −0.49 | −0.09 |
| Pum2 | Pumilio 2 | ENSMUSG00000020594 | 13 | +0.12 | +0.11 |
| Rilpl1 | Rab interacting lysosomal protein-like 1 | ENSMUSG00000029392 | 4 | −0.28 | −0.59 |
| Slmap | Sarcolemma-associated protein | ENSMUSG00000021870 | 10 | +0.16 | +0.35 |
| Sorbs1 | Sorbin and SH3 domain containing 1 | ENSMUSG00000025006 | 23 | −0.16 | −0.76 |
| Sorbs1 | Sorbin and SH3 domain containing 1 | ENSMUSG00000025006 | 27 | −0.10 | −0.66 |
TABLE 3.
Genes that undergo developmentally regulated splicing changes in diabetic hearts are implicated in diabetes and/or heart diseases
| ATP2b1 | ATP2b1 is involved in hypertension. Knockout-mice show high systolic blood pressure (69). Several family members of ATP2b1 modulate insulin expression in rats (70). |
| Fxr1 | Reduced Fxr1 levels cause severe embryonic cardiomyopathy resulting in heart failure in zebrafish (71). |
| Vegfa | VEGFA is a regulator of angiogenesis implicated in diabetic retinopathy and microvascular complications of diabetes (72). VEGFA is being tested to treat heart failure (73). |
| Mef2A | Mef2A is a transcription factor and certain mutations cause coronary artery disease and myocardial infarction (74, 75). MEF2A also regulates expression of insulin-responsive glucose transporter GLUT4 (76). |
| Ablim1 | Ablim1 is a candidate gene for congenital heart disease (77) and has also been linked to cardiovascular disease, hypertension, and type 1 and type 2 diabetes by genome association studies (78). |
| Lbd3 | Mutations in Lbd3 gene are associated with familial or idiopathic dilated cardiomyopathy (79). |
| Pbx3 | Pbx3 gene variant (A136V) is a risk allele for congenital heart defects in human (80). Pbx3-KO results in tetralogy of fallot, a congenital heart disease (81). |
| Rilpl1 | Ripl proteins are implicated in obesity (82). |
| Slmap | Slmap is a membrane protein and essential for organization of E-C coupling apparatus (83, 84). Misregulation causes endothelial dysfunction in type 2 diabetic mice (85). |
| Sorbs1 | Sorbs1 SNPs are positively associated with obesity and type 2 diabetes in human studies and play an important role in pathogenesis of insulin resistance (86). |
| Mbnl1 | Unknown |
| Mbnl2 | Unknown |
| Ankrd10 | Unknown |
| Bnip2 | Unknown |
| Cabzb | Unknown |
| G3bp2 | Unknown |
| hnRNPH2 | Unknown |
| Prrc2b | Unknown |
| Pum2 | Unknown |
To determine whether the splicing reverted back to an embryonic pattern in diabetic hearts, we compared the AS events that change in diabetes to 147 developmentally regulated AS events identified in developing mouse hearts (18). 20 AS events displayed an embryonic splicing pattern in diabetic hearts (Table 2). Importantly, these splicing events were not only in the same genes but also were in the exact same exons changing in the same direction (included or excluded) in embryonic versus diabetic hearts (Table 2). We identified two additional AS events that revert to an embryonic pattern using qRT-PCR (Fig. 1A and data not shown). These two events, including hnRNPH1 exon6, were not previously identified as developmentally regulated events in the heart. Overall, we identified a novel set of 22 fetal-specific AS events that are reactivated in diabetic hearts.
To validate RNA-sequencing data, we checked the splicing of several developmentally regulated AS events by qRT-PCR using two well established T1D mouse models and type 2 diabetic human heart tissues. As type 1 diabetes models, both STZ and NOD mice were used. NOD mice develop diabetes spontaneously due to inflammation and death of beta islet cells in the pancreas (32). We found that there was a significant change in the splicing of five AS events as follows: hnRNPH1 exon6, Mtmr3 exon16, Fxr1 exon15 + 16, Atp2b1 exon21, and Mbnl2 exon9 in T1D:STZ mice versus controls (Fig. 1A, white versus black bars). Splicing of hnRNPH1 exon6 and Mtmr3 exon16 changed similarly in T1D:NOD mice (Fig. 1B). In T1D mice, the splicing pattern of all these genes resembled that of newborn hearts (Fig. 1, A and B, black versus gray bars). Importantly, splicing of hnRNPH1, Mtmr3, and Fxr1 also returned to an early developmental splicing pattern in human hearts with confirmed type 2 diabetes (T2D; Fig. 1C). Strikingly, the developmental shift in AS of these genes was observed in diabetic human and mouse, as well as in T1D and T2D, suggesting a conserved phenomenon in diabetes.
Diabetes-induced Alternative Splicing Events Transition during Rat Heart Development and H9c2 Differentiation
To determine whether this network of splicing events induced in diabetes are evolutionarily conserved and regulated during heart development, we tested the splicing of these events during rat heart development and in H9c2 rat embryonic cells.
H9c2 cells can be differentiated into cardiomyocyte-like cells when supplemented with RA in low serum media, which mimic differentiation of embryonic cardiomyocytes into adult-like cardiomyocytes that occur during heart development (33). The cardiac markers cardiac troponin I (cTNI) and T (cTNT) were up-regulated in differentiated H9c2 cells indicating the cardiomyocyte lineage of these cells determined by Western blot (WB) (Fig. 2A). Importantly, cardiac differentiated H9c2 cells were free from myotube contamination as confirmed by myogenin expression when compared with skeletal muscle differentiated cells (Fig. 2A and data not shown).
To test whether H9c2 cells can recapitulate developmentally regulated splicing events that take place in diabetic hearts, we designed rat-specific primers for four genes that undergo AS changes in diabetic mouse hearts (Fig. 1 and Tables 2 and 4). Splicing events were tested by qRT-PCR using RNA from H9c2 cells during differentiation and from rat hearts at E13, E18, E20, newborn, and adult stages (Fig. 2C). Polyacrylamide gel shows the inclusion of alternative exon21 of Atp2b1 in differentiating H9c2 cells (Fig. 2B).
Splicing of four AS events that are altered in diabetes changed in differentiated (diff) cells when compared with (versus) undifferentiated (undiff) cells (Fig. 2C). Three alternative exons (Mtmr3 exon16, Atp2b1 exon21, and Fxr1 exon15 + 16) were differentially included, and one alternative exon (Mbnl2 exon9 in rat is exon8 in mice) was differentially excluded in diff cells versus undiff cells (Fig. 2C, white versus black bars). Splicing changes in Mtmr3, Atp2b1, Fxr1, and Mbnl2 during H9c2 differentiation were similar to the splicing changes between embryonic and postnatal stages of developing rat heart (Fig. 2C, gray versus black bars). We found that most of the alternative exons in these genes correspond to the regulatory protein domains; thus, splicing changes will likely affect protein function (Table 5). These results indicate that diabetes-induced biologically relevant AS events undergo embryonic to postnatal transitions in rat hearts and during H9c2 differentiation.
TABLE 5.
The effect of diabetes-induced splicing changes on gene expression/function
| Symbol | Gene name | Effect of alternative exon inclusion/exclusion |
|---|---|---|
| Atp2b1 | Plasma membrane calcium-transporting ATPase 1 | Exon21 is a part of the C-terminal domain that affects calmodulin binding to the protein. Exclusion of this exon reduces calcium transport affinity of the ATPase (61). |
| Mbnl2 | Muscleblind-like protein 2 | MBNL2 is a splicing regulator involved in pathogenesis of myotonic dystrophy type 1. Exclusion of exon9 (equivalent to exon5 in mouse) is known to cause cytosolic localization of the nuclear protein (87). |
| Mtmr3 | Myotubularin related protein 3 | MTMR3 is a member of the “myotubularin dual specificity protein phosphatase” family. Exon16 is in the RNA-binding FYVE domain of the protein. |
| Fxr1 | Fragile X mental retardation, autosomal homolog 1 | Fxr1 is an RNA-binding protein important for muscle development. Exon (15 + 16) is located in the RGG box. Inclusion of exon (15 + 16) increases the RNA binding repertoire of the protein (88). |
PKC Regulates Fetal to Adult Transitions of AS Events That Are Reactivated in Diabetic Hearts
To test whether PKCα/β regulates fetal to adult-specific AS transitions, we used BisIX, a small molecule inhibitor that preferentially blocks the activity of PKCα/β at low doses (34). H9c2 cells were either mock-treated or treated with BisIX and differentiated for 7 days. Differentiated cells that were mock-treated expressed high levels of the cardiomyocyte marker cTNT and displayed PKCα/β activity as determined by phosphorylation of a known target called MARCKS (Fig. 3A, lane 2 versus 1) (35). In BisIX-treated diff cells, PKCα/β activity was undetectable as shown by the loss of MARCKS phosphorylation (Fig. 3A, lane 2 versus 3). The inhibitor also decreased the phosphorylation of GSK3β at serine 9 (Fig. 3A, lane 2 versus 3), which can be phosphorylated by both PKCα and Akt (36). To ensure that the inhibitor is not affecting other kinases, we checked the ability of PDK-1 kinase to phosphorylate PKA. We found that BisIX did not affect PDK1 activity in H9c2 cells, but PKCμ activity was slightly inhibited as the inhibitor can also block other PKC-related kinases (Fig. 3A). BisIX-treated diff cells expressed low levels of the cardiac marker cTNT when compared with mock-treated diff cells, suggesting that differentiation was not efficient in PKC inhibitor-treated cells (Fig. 3A, lane 2 versus 3). The morphology of the BisIX-treated diff cells was not as elongated as that of mock-treated diff cells, indicating incomplete differentiation (data not shown).
To investigate whether the PKC inhibitor altered AS transitions, we checked four splicing events that are altered in diabetic mouse hearts (Fig. 1 and Tables 2 and 5). Inhibition of PKCα/β activity significantly reduced the exon inclusion of two alternative exons (Fxr1 exon15 + 16 and Atp2b1 exon21) in differentiated cells (Fig. 3B, black versus black hatched bars). Even though PKC inhibitor did not affect splicing of Mtmr3 and Mbnl2 in differentiated cells, it affected their splicing in undifferentiated cells (Fig. 3C and data not shown), indicating that PKC activity is necessary to maintain embryonic splicing pattern of these genes.
Next, we wanted to verify whether PKC controls the same splicing events in beating primary neonatal rat cardiomyocytes. Rat cardiomyocytes were treated with vehicle or the PKC inhibitor BisIX. Splicing of Mtmr3 exon16, Fxr1 exon15 + 16, and Mbnl2 exon9 was tested by qRT-PCR. Treatment with BisIX reduced inclusion splicing of Fxr1 exon15 + 16 and Mtmr3 and increased inclusion of Mbnl2 exon9 similar to the results from H9c2 cells treated with BisIX (Fig. 3, B–D). These results using primary neonatal cardiomyocytes confirm our findings in H9c2 cells. Overall, these results show PKC regulates splicing of AS events that undergo a developmental change in adult diabetic mouse hearts.
PKCα/β Controls Splicing Events Affected in Diabetic Hearts by Regulating RNA-binding Proteins
To verify the results from the PKC inhibitor experiments, we used a more direct assay to target PKC. PKCα was depleted using validated short hairpin RNAs (shRNA). Cells expressing scrambled or PKCα-specific shRNA were selected using puromycin, followed by 7 days of all-trans-retinoic acid treatment. In our experiments, we used an Ab that is raised against the α isoform but can also cross-react with β isoforms of the same molecular weight (βI and βII). Therefore, we cannot definitively distinguish α from β isoforms by WB and immunohistochemistry (IHC) using this antibody. For this reason, we refer to PKCα/β as one kinase, even though the isozymes have distinct functions (6).
Using the most efficient and specific shRNA, we achieved ∼95% depletion of PKCα protein in H9c2 cells compared with scrambled (sc-) shRNA, which was used as a negative control (Fig. 4A). PKCα shRNA was specific to the α isoform and did not affect the levels of PKCδ/θ, GSK3β, or PKA (Fig. 4A).
To determine whether AS transitions are altered in PKCα knocked down cells, we examined the splicing of several events that are misregulated in diabetic hearts (Table 2). Depletion of PKCα affected splicing of Mtmr3, Fxr1, and Mbnl2 (Fig. 4B) similar to the results obtained from the PKC inhibitor experiment (Fig. 3, B–D).
Reviewing the literature for potential splicing regulators that bind to the regulatory regions on the pre-mRNAs mis-spliced in diabetes, we found that PKCα/β -responsive exons have been identified computationally or experimentally as targets of CELF1, MBNL1, and Rbfox2 proteins (18, 20). Splicing of Fxr1 and Mtmr3 reversed to a fetal pattern in CELF1-overexpressing mice (18). Fxr1 and Mtmr3 pre-mRNAs have Rbfox2-binding sites in the adjacent introns of corresponding alternative exons in human embryonic stem cells identified by CLIP -sequencing (20). Mbnl2 exon9 was identified as MBNL1- and CELF1-responsive exon, using MBNL1-knock-out (KO) or CELF1-overexpressing mice (18). Mbnl2 was also identified as a target of Rbfox2 in CLIP-seq experiments with binding sites downstream and upstream intron of exon9 of Mbnl2 (20).
As PKC-regulated AS events affected in diabetes are targets of CELF1, MBNL1, and Rbfox2, we checked the levels of these RNA-binding proteins in PKCα-depleted cells. CELF1 and Rbfox2 levels were reduced by 90 and 80%, respectively, in PKCα knocked down cells (Fig. 4A and data not shown). The significant decrease in Rbfox2 and CELF1 protein levels upon PKCα depletion indicates that PKCα/β isozymes are the major regulators of these proteins. There was no change in MBNL1 levels in PKCα shRNA-treated cells compared with sc-shRNA-treated cells (Fig. 4A). Overall, these results indicate that PKCα/β control CELF1 and Rbfox2 protein levels.
PKCα/β Controls Developmentally Regulated AS via Phosphorylation of RNA-binding Proteins
To determine whether PKC is necessary to maintain Rbfox2 and CELF1 protein levels, we determined the steady state levels and phosphorylation status of these splicing regulators in mock-treated cells or cells treated with the PKC inhibitor BisIX. CELF1 was predominantly nuclear in undiff H9c2 cells, and the protein levels were lower in diff cells (Fig. 5A, nuclear fractions, lane 1 versus 2). Treatment with the PKC inhibitor further down-regulated CELF1 levels in the nuclear fractions of diff cells (Fig. 5A, nuclear fraction, lane 2 versus 3) resembling its low levels in adult heart tissues (Fig. 5B).
FIGURE 5.
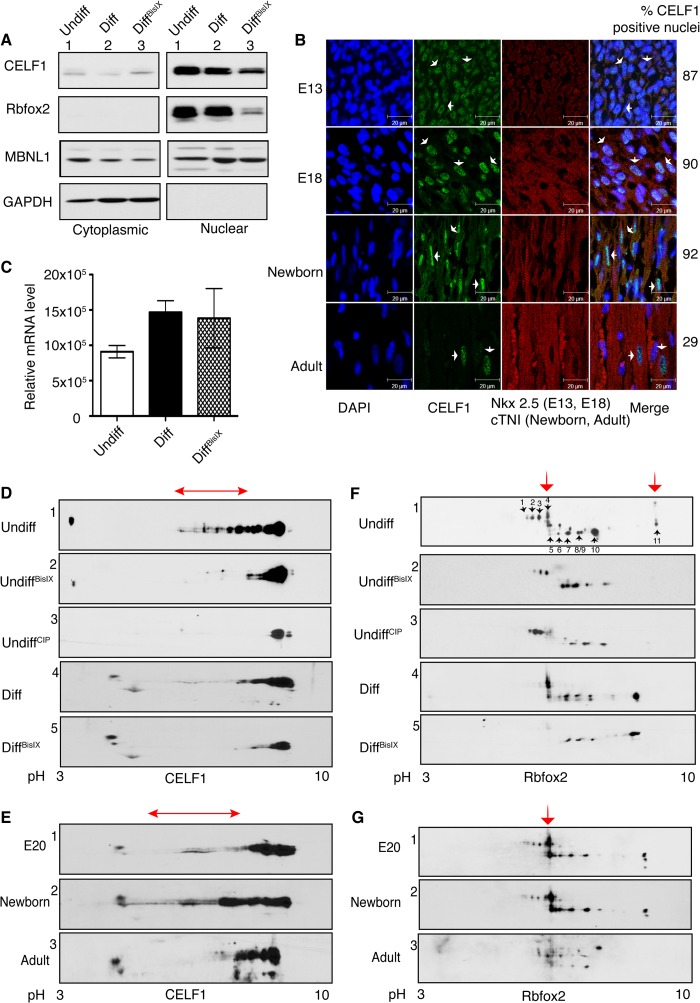
Inhibition of PKC activity reduces the phosphorylation and steady state levels of CELF1 and Rbfox2. A, WB analysis of nuclear and cytoplasmic fractions from undiff, mock-treated (Diff), and PKC inhibitor-treated differentiated (DiffBisIX) H9c2 cells using antibodies against CELF1, Rbfox2, MBNL1, and GAPDH. B, IHC of sagittal rat heart sections using antibodies against CELF1, Nkx2.5 (cardiomyocyte marker for E13 and E18), and cTNI (cardiomyocyte marker for newborn and adult). Nuclei are stained with DAPI. Cardiomyocytes that are CELF1-positive were quantified counting >150 nuclei at each developmental stage. Arrows indicate representative cells. C, relative Rbfox2 mRNA levels was determined by semi-quantitative RT-PCR using RNA from H9c2 cells (Undiff, Diff, and DiffBisIX) by net intensity calculation of the PCR product using Kodak gel Logic 2200. D and F, two-dimensional/Western blot analysis of nuclear fractions from undiff, undiffCIP (calf intestinal phosphatase-treated), diff, and diffBisIX H9c2 cells using anti-CELF1 (D) or anti-Rbfox2 (F) Abs. The red arrows indicate PKC-dependent phospho-isoforms. E and G, protein lysates from E20, newborn, and adult (6-month old) rat hearts were separated on two-dimensional gels followed by WB using anti-CELF1 (E) and anti-Rbfox2 (G) antibodies. The red arrows indicate PKC-induced phosphorylated isoforms. E20 and newborn hearts were pooled, n = 4. Results are representative of at least three independent experiments.
CELF1 expression in developing rat hearts was also tested to ensure changes are not cell culture-specific. Quantification of the IHC data shows that 87% of the cardiomyocyte nuclei were positive for CELF1 at E13 and 90% at E18. In contrast to the considerably low levels of CELF1 in adult hearts (29% CELF1 positive nuclei), CELF1 levels were high in newborn cardiomyocyte nuclei marked with cTNI (92% CELF1-positive nuclei) (Fig. 5B). CELF1 cytoplasmic presence was evident in newborn hearts suggesting that cytoplasmic functions of CELF1 in mRNA stability and translation might be important at neonatal stages (37, 38).
Because BisIX treatment decreased CELF1 levels in H9c2 cells (Fig. 5A), we investigated the phosphorylation status of CELF1 in rat hearts and during H9c2 differentiation. We found that endogenous CELF1 (pI ∼8.5) was hyperphosphorylated in undiff cells as indicated by acidic shifts and the loss of shifted spots after phosphatase (calf intestinal phosphatase, CIP) treatment on two-dimensional gels followed by WB using anti-CELF1 antibody (Fig. 5D, gel 1 versus 3). CELF1 hyperphosphorylation status (Fig. 5D) correlated well with high levels of protein at embryonic and neonatal stages of developing rat hearts (Fig. 5B). In undiff cells, CELF1 was hyperphosphorylated similarly to the pattern observed in embryonic and newborn rat hearts (Fig. 5E) but was less hyperphosphorylated in diff cells as the protein became more basic (Fig. 5D, gel 4). Inhibition of PKCα/β by BisIX reduced CELF1 phosphorylation in both undifferentiated and differentiated cells (Fig. 5D, gels 2 and 5, red arrow) to a hypophosphorylated pattern seen in phosphatase-treated cells and adult rat hearts (Fig. 5, D and E). The hypophosphorylated state of CELF1 in adult rat hearts was consistent with low CELF1 levels (Fig. 5B). CELF1 mRNA levels remain constant throughout heart development (18). These findings indicate that CELF1 hyperphosphorylation and increased steady state levels in embryonic hearts and undifferentiated H9c2 cells are regulated by a PKCα/β-dependent manner.
We also examined the protein levels and localization of Rbfox2, which regulates AS in embryonic stem cells (20) and during brain development (22, 39). Rbfox2 also binds to Mtmr3, Fxr1, and Mbnl2 intronic regions near the alternative exons that are affected in diabetes. Rbfox2 has two isoforms due to alternative promoter and/or cassette exon usage, short and long (40–42). Although there was no obvious change in the protein levels of Rbfox2 isoforms between undiff and diff cells, both isoforms were severely down-regulated in PKC inhibitor-treated cells (Fig. 5A, lanes 1 and 2 versus lane 3) without significant changes in Rbfox2 mRNA levels (Fig. 5C).
To determine whether Rbfox2 down-regulation is due to changes in its phosphorylation by PKC, we used two-dimensional gels followed by WB using Rbfox2-specific antibody. We detected both the long and short isoforms of Rbfox2 with isoelectric points ranging from ∼6.5 to 9 in embryonic rat hearts and undiff cells (Fig. 5, G, gel 1, and F, gel 1). The Rbfox2 pattern in embryonic/newborn heart was similar to that in undiff H9c2 cells, and the pattern in differentiated cells resembled the pattern in adult rat hearts (Fig. 5, F, gel 4, and G, gel 3).
The long isoform of Rbfox2 displayed four spots labeled as 1–4 and pH ranging from 5.5 to 6.5 in undifferentiated H9c2 cells (Fig. 5F, arrows). Treatment with either the phosphatase (calf intestinal phosphatase) or the PKC inhibitor BisIX resulted in loss of spot 4 in undiff cells representing the PKC-dependent phospho-isoform (Fig. 5F, red arrow). The same spot 4 was present in E20 and newborn rat hearts but not in adult rat hearts (Fig. 5G). In differentiated cells, spots 1–3 disappeared in comparison with undiff cells (Fig. 5F, gel 1 versus 4). Spots 1–3 were unaffected in phosphatase and BisIX-treated cells suggesting that they are not phosphorylated species. These spots might be representative of another post-translational modification (i.e. acetylation) that is regulated during differentiation of H9c2 cells.
The short faster migrating isoform displayed six spots labeled from 5 to 11 in undifferentiated H9c2 cells (Fig. 5F, arrows). The levels of spots 10 and 11 were reduced in differentiated cells (Fig. 5F, gel 1 versus 4). Both the PKC inhibitor and the phosphatase treatment led to the loss of spots 5 and 11 in undifferentiated cells indicating that these spots are the PKC-dependent phosphorylated forms (Fig. 5F, gels 2 and 5). In differentiated cells, spot 5 disappeared after BisIX treatment (Fig. 5F, red arrow, gel 4 versus 5). Importantly, spot 5 was present in E20 and newborn rat hearts but not in adult rat hearts, resembling the findings in H9c2 cells (Fig. 5G). These data suggest that spots 4, 5, and 11 are PKC-dependent phosphorylated species of Rbfox2. In sum, these results indicate that Rbfox2 is phosphorylated in a PKC-dependent manner at early developmental stages and also in undifferentiated H9c2 cells.
Next, we checked the steady state levels of MBNL1 during H9c2 differentiation. MBNL1 is identified as a regulator of Mbnl2 exon9 splicing together with Rbfox2 (18, 20). MBNL1 protein levels and localization remained unchanged during the differentiation of H9c2 cells (Fig. 5A, lane 1 versus 2). PKC inhibitor treatment affected neither the protein levels nor the cellular distribution of MBNL1 (Fig. 5A). These results show that PKC promotes phosphorylation of CELF1 and Rbfox2 and regulates their steady state levels at early developmental stages.
To test whether phosphorylation of CELF1 by PKC is necessary for splicing regulation of genes altered in diabetes, we generated a nonphosphorylatable CELF1 by mutating eight phospho-sites identified by mass spectrometry to alanine or valine (CELF1M8) using site-directed mutagenesis. We tested the phosphorylation status of FLAG-tagged wild type (CELF1WT) or nonphosphorylatable mutant of CELF1 (CELF1M8) in cells stimulated by the phorbol ester (phorbol 12-myristate 13-acetate, PMA) that activates PKC. CELF1WT displayed acidic shifts in comparison with its pattern in mock-treated cells representing PKC-induced phospho-isoforms (Fig. 6A). However, CELF1M8 did not shift to an acidic pH in phorbol 12-myristate 13-acetate-treated cells suggesting that mutations blocked the phosphorylation by PKC (Fig. 6A).
Next, we checked whether the nonphosphorylatable mutant CELF1M8 regulates PKCα/β-dependent splicing of Mtmr3 (Fig. 3, C and D, and Fig. 4B) that reverses to a fetal pattern in both T1 and T2 diabetic hearts (Fig. 1B). Overexpression of CELF1WT in COS M6 cells reduced inclusion of exon16 of endogenous Mtmr3 similar (Fig. 6C) to its pattern in neonatal and embryonic rat hearts (Figs. 1A and 2C). Nonphosphorylatable CELF1M8 did not cause exon exclusion in comparison with the wild type protein even though both proteins were expressed at similar levels as determined by both FLAG- and CELF1-specific WBs (Fig. 6B). Importantly, there was no change in endogenous levels of CELF1 (Fig. 6B). As a negative control, we overexpressed the empty vector. Exon inclusion of Mtmtr3 in CELF1M8-expressing cells was similar to that of in empty vector-expressing cells. This result indicates that phosphorylation of CELF1 by PKC is critical for regulation of developmentally regulated splicing.
Under Diabetic Conditions, PKCα/β Localizes to the Nucleus Similar to Its Subcellular Distribution at Early Developmental Stages
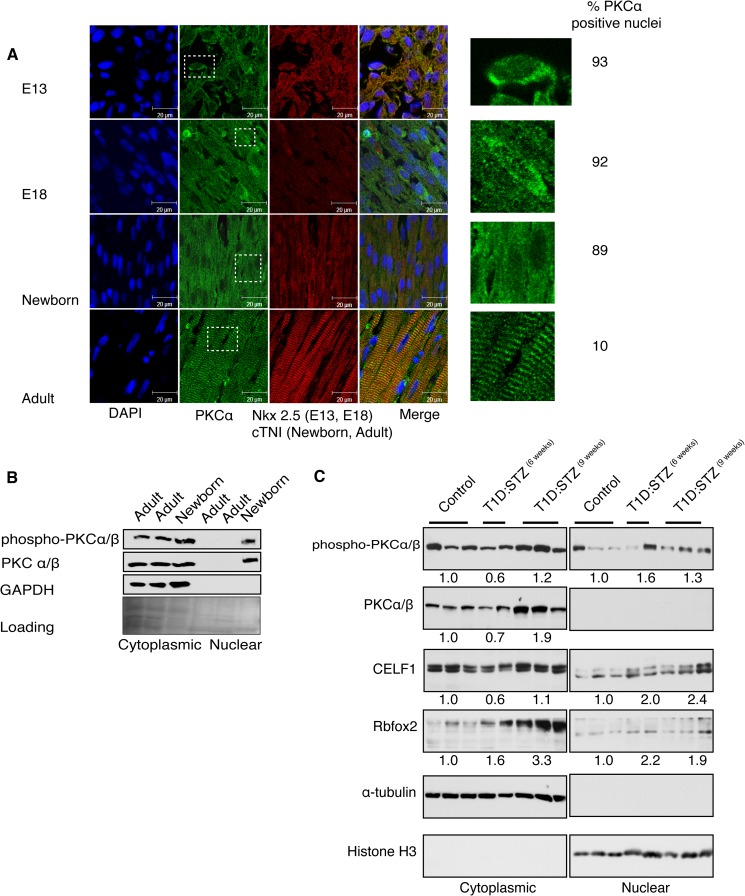
Finally, we tested whether the predominantly cytoplasmic PKCα/β undergoes localization changes during heart development because the splicing regulators CELF1 and Rbfox2 regulate splicing in the nucleus and are hyperphosphorylated by PKCα/β. Using IHC, we labeled the rat cardiomyocytes with Nkx2.5 at embryonic stages and cTNI at newborn and adult stages. Embryonic cardiomyocytes (E13 and E18) became more elongated and branched as they matured into adult cardiomyocytes (Fig. 7A). In embryonic rat cardiomyocytes, PKCα/β could be found in the nucleus and cytoplasm but was slightly more cytoplasmic after birth becoming completely cytoplasmic in adult cardiomyocytes (Fig. 7A, insets). In adult rat left ventricles, PKCα/β colocalized with the contractile apparatus (cTNI) in striations, in agreement with the previous findings that PKCα/β phosphorylates cTNI and cTNT (43, 44) and is important for cardiac contractility (45). At embryonic stages, ∼90% of cardiomyocytes showed nuclear PKCα staining in contrast to adult hearts where only 10% of cardiomyocytes were positive for nuclear PKCα (Fig. 7A).
FIGURE 7.
PKCα/β localizes to the nucleus at embryonic stages and in adult diabetic hearts. A, IHC of PKCα in sagittal sections of rat heart at embryonic (E13 and E18), newborn and adult stages using anti-PKCα Ab. Cardiomyocytes were labeled with Nkx2.5 at embryonic stages and with cTNI at newborn and adult stages. The nucleus was stained with DAPI. Percentage of cardiomyocytes that display nuclear PKCα in the nucleus was calculated using multiple images from each developmental stage (>150 nuclei). Insets indicate representative cells at ×80 magnification. B, WB of nuclear and cytoplasmic fractions in mouse hearts at newborn and adult stages using Abs against PKCα/β, phospho-PKCα/βII, and GAPDH (cytoplasmic marker). Loading was determined by Ponceau S staining. C, nuclear and cytosolic proteins from mice hearts were resolved on 10% acrylamide gel followed by WB using antibodies against phospho-PKCα/β, PKCα/β, CELF1, Rbfox2, α-tubulin, and histone H3 (nuclear marker). To calculate fold change in protein levels, cytoplasmic protein levels were normalized to α-tubulin, and nuclear protein levels were normalized to histone H3. Protein levels in control mice were standardized to 1.0, and the average fold change in protein levels was quantified and summarized for each group of mice (three normal, two T1D/STZ6 weeks, and three T1D/STZ9 weeks).
We also examined the distribution of PKCα/β in mouse heart tissues by WB. Activated PKCα/β was nuclear and cytoplasmic in the newborn but only cytoplasmic in normal adult mouse hearts like in rat hearts (Fig. 7, A and B). The nuclear localization of PKCα/β at embryonic stages (Fig. 7A) correlated well with increased steady state levels and the hyperphosphorylated status of CELF1 and Rbfox2 proteins (Fig. 5, B, E, and G).
These results suggest that the conserved nuclear localization of PKCα/β at early developmental stages may play a role in phosphorylation of splicing regulators in the nucleus at early developmental stages. If this is the case, we expect PKCα/β to be nuclear in diabetic hearts where fetal splicing patterns are induced. To test this, we determined the steady state levels and autophosphorylation status of PKCα/β in STZ-induced diabetic mice and found that phospho-PKCα/β levels were increased ∼1.6-fold in the nuclear fractions of 6-week-old and ∼1.3-fold in 9-week-old diabetic mice (Fig. 7C). We noticed that phospho-PKCα/β levels were reduced in the cytoplasmic fractions of 6-week-old diabetic mice with a corresponding increase in the nuclear fraction of these mice (Fig. 7C), but this was not apparent in 9-week-old diabetic mice.
To examine whether nuclear PKCα/β activity in turn increased the steady state levels of CELF1 and Rbfox2, we measured the protein levels of RNA-binding proteins in the same lysates by WB. Although CELF1 levels were constant in cytoplasmic fractions of control and 9-week-old diabetic mice, protein levels were elevated ≥2-fold in the nuclear fractions of all diabetic mice consistent with increased nuclear activity of PKCα/β (Fig. 7C). CELF1 levels were reduced in the cytoplasmic fractions of 6-week-old diabetic mice consistent with reduced phospho-PKCα/β levels in the cytoplasmic fractions of these mice. The short isoform of Rbfox2 protein levels were also elevated ≥1.9-fold in the nuclear fractions of all diabetic mice as expected (Fig. 7C). Interestingly, the long isoform of Rbfox2, which was predominantly cytoplasmic, was also up-regulated 3.3-fold in 9-week-old diabetic mice (Fig. 7C). These results indicate that increased nuclear PKCα/β levels correlate with up-regulation of splicing regulators in diabetic hearts.
DISCUSSION
Advances in RNA sequencing technology and extensive analysis of the transcriptome indicate that AS is a highly regulated and tissue-specific process affecting >90% of human genes (46–48). AS generates differentially spliced isoforms of primary gene products and also affects gene expression (16, 49, 50). AS of genes that are important for cardiac morphogenesis and maturation undergo significant AS changes during postnatal mouse heart development (18). In addition, AS patterns shift during differentiation of embryonic stem cells into cardiomyocytes in favor of isoforms that are involved in differentiation (51). These studies strongly suggest that AS is vital for embryonic gene expression in the heart.
It has been previously shown that fetal gene expression is up-regulated in diabetic hearts (52). The mechanisms responsible for embryonic gene expression in diabetes are largely unknown. In this study, we demonstrate that an embryonic splicing program is activated in diabetic hearts that could partly explain the expression of fetal genes. Importantly, we identified PKCα/β as regulators of this embryonic network. These findings may have implications in heart conditions induced by ischemia or hypertrophy, which can induce fetal gene expression and PKC activation (53–57).
Prolonged activation of PKCα and/or -β leads to diabetic complications (7, 58, 59). Here, we demonstrate that PKCα/β-regulated alternative exons are mis-spliced in both type 1 and 2 diabetic hearts reverting to an embryonic pattern. It is known that AS of pre-mRNAs transition during heart development such that only embryonic spliced isoforms are expressed in embryonic hearts but not in adult hearts for proper heart function (18). Therefore, these conserved splicing changes likely contribute to cardiac complications in diabetes by promoting the expression of embryonic protein isoforms. For example, Atp2b1 encodes plasma membrane calcium ATPase, an essential enzyme that regulates cytosolic calcium concentrations (Table 5). Exclusion of alternative exon21 makes the ATPase less sensitive to calcium levels (60, 61) that may contribute to abnormal calcium levels observed in diabetic hearts (Table 5). Exon15 + 16 of Fxr1 corresponds to a domain that is responsible for substrate specificity of the RNA-binding protein (Table 5). Exclusion of these exons may shift the substrates of FXR1 in diabetic hearts. Exon16 of Mtmr3 is within the zinc finger FYEV domain of myotubularin-related protein 3, which is important for cell migration by regulating cellular inositol levels and signaling (62, 63). Inclusion of exon16 is predicted to prevent zinc binding, likely affecting substrate binding (Table 5).
In this report, we demonstrate that PKCα/β mediate AS regulation by phosphorylation of Rbfox2 and CELF1. Both protein levels are down-regulated with corresponding changes in the splicing of their target pre-mRNAs in PKCα/β-depleted or -inhibited cells without changes in mRNA levels (Figs. 3, 4, and 5C). Notably, the nonphosphorylatable mutant of CELF1 is unable to regulate the splicing of a PKCα/β target, which displays fetal splicing pattern in both type 1 and 2 diabetic heart tissues (Figs. 1 and 6). CELF mutant M8 has a total of eight residues mutated to alanine/valine to prevent phosphorylation by PKC. Three of these residues are within the divergent domain of the protein and five within the RNA recognition motifs. It is likely that these mutations affect RNA binding ability of CELF1. Further mapping of PKC phospho-sites is necessary to define the exact role of PKC phosphorylation on CELF1 function especially in RNA binding.
Our findings also provide new insights into the novel functions of PKCα/β in the nucleus during heart development and in diabetic hearts. PKCα localization to the nucleus has been reported in cultured cells upon activation (64, 65). For the first time, our findings connect PKCα/β nuclear localization to hyperphosphorylation of RNA-binding proteins and AS regulation in diabetic and embryonic hearts (Figs. 5 and 7).
Further studies are needed to determine the mechanism of PKCα localization to the nucleus and the proteins necessary for this process during development. PKCα does not have a classical nuclear localization/export signal, but its hinge region and C-terminal tail are necessary for its translocation to the nucleus in cells treated with phorbol esters that activate PKCα (66). It is not clear whether the hinge domain or the C-terminal tail is essential for localization to the nucleus at embryonic stages and in diabetic adult hearts. Our data suggest that nuclear localization of PKCα/β might be a trigger for increased phosphorylation and levels of CELF1 and Rbfox2, which in turn activate embryonic AS, as observed in adult diabetic heart tissues (Figs. 3, 4, 5, and 7). Overexpression of CELF1 in heart causes cardiomyopathy (19), and increased steady state levels of CELF1 in diabetic hearts may contribute to diabetic cardiomyopathy. Additional investigations are necessary to determine the role of CELF1 and Rbfox2 in diabetic complications of the heart using cardiac-specific overexpression or knock-out mouse models.
In summary, our results provide insight into the novel role of PKCα/β in fetal gene expression via AS in cardiomyocytes under diabetic conditions. PKC is an attractive target to treat many diseases, including diabetes (7, 67). Identification of RNA-binding proteins or specific splicing events as downstream effectors of PKC may provide us new tools to design safer and novel therapeutics such as the oligo-based therapy to correct these splicing defects.
Acknowledgments
We thank Dr. David Konkel for critically editing the manuscript, Drs. Thai Ho and Rajalaxmi Natarajan for critically reading the manuscript, Dr. Thomas Wood for technical help in RNA-sequencing, and Dr. Adriana Paulucci for help in confocal microscopy. We also thank Dr. Nisha Garg for kindly providing control human heart tissues.
This work was supported, in whole or in part, by National Institutes of Health Grants HG004659 and NS075449 (to G. W. Y.). This work was also supported by American Heart Association Grant SDG-093022N, March of Dimes Basil O'Connor Starter Scholar Award 5 FY12-21, and University of Texas Medical Branch start up funds (to M. N. K.-M.). This work was also supported in part by National Science Council, Taiwan, and Institute of Biomedical Sciences, Academia Sinica (to G.-S. W.).
- AS
- alternative splicing
- NOD
- nonobese diabetic
- STZ
- streptozotocin
- RA
- all-trans-retinoic acid
- qRT
- quantitative RT
- T1D
- type 1 diabetes
- Ab
- monoclonal antibody
- BisIX
- bisindolylmaleimide IX
- IHC
- immunohistochemistry
- WB
- Western blot
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- cTNI
- cardiac troponin I
- cTNT
- cardiac troponin T
- diff
- differentiated
- undiff
- undifferentiated
- MARCKS
- myristoylated alanine-rich C kinase substrate.
REFERENCES
- 1. Salsali A., Nathan M. (2006) A review of types 1 and 2 diabetes mellitus and their treatment with insulin. Am. J. Ther. 13, 349–361 [DOI] [PubMed] [Google Scholar]
- 2. Aneja A., Tang W. H., Bansilal S., Garcia M. J., Farkouh M. E. (2008) Diabetic cardiomyopathy: insights into pathogenesis, diagnostic challenges, and therapeutic options. Am. J. Med. 121, 748–757 [DOI] [PubMed] [Google Scholar]
- 3. Ferrannini E., Cushman W. C. (2012) Diabetes and hypertension: the bad companions. Lancet 380, 601–610 [DOI] [PubMed] [Google Scholar]
- 4. Harcourt B. E., Penfold S. A., Forbes J. M. (2013) Coming full circle in diabetes mellitus: from complications to initiation. Nat. Rev. Endocrinol. 9, 113–123 [DOI] [PubMed] [Google Scholar]
- 5. Boudina S., Abel E. D. (2010) Diabetic cardiomyopathy, causes and effects. Rev. Endocr. Metab. Disord. 11, 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu Q., Chen X., Macdonnell S. M., Kranias E. G., Lorenz J. N., Leitges M., Houser S. R., Molkentin J. D. (2009) Protein kinase Cα, but not PKCβ or PKCγ, regulates contractility and heart failure susceptibility: implications for ruboxistaurin as a novel therapeutic approach. Circ. Res. 105, 194–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Geraldes P., King G. L. (2010) Activation of protein kinase C isoforms and its impact on diabetic complications. Circ. Res. 106, 1319–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Inoguchi T., Battan R., Handler E., Sportsman J. R., Heath W., King G. L. (1992) Preferential elevation of protein kinase C isoform βII and diacylglycerol levels in the aorta and heart of diabetic rats: differential reversibility to glycemic control by islet cell transplantation. Proc. Natl. Acad. Sci. U.S.A. 89, 11059–11063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Connelly K. A., Kelly D. J., Zhang Y., Prior D. L., Advani A., Cox A. J., Thai K., Krum H., Gilbert R. E. (2009) Inhibition of protein kinase C-β by ruboxistaurin preserves cardiac function and reduces extracellular matrix production in diabetic cardiomyopathy. Circ. Heart Fail. 2, 129–137 [DOI] [PubMed] [Google Scholar]
- 10. Liu Y., Lei S., Gao X., Mao X., Wang T., Wong G. T., Vanhoutte P. M., Irwin M. G., Xia Z. (2012) PKCβ inhibition with ruboxistaurin reduces oxidative stress and attenuates left ventricular hypertrophy and dysfunction in rats with streptozotocin-induced diabetes. Clin. Sci. 122, 161–173 [DOI] [PubMed] [Google Scholar]
- 11. Kuyumcu-Martinez N. M., Wang G. S., Cooper T. A. (2007) Increased steady state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell 28, 68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Revil T., Toutant J., Shkreta L., Garneau D., Cloutier P., Chabot B. (2007) Protein kinase C-dependent control of Bcl-x alternative splicing. Mol. Cell. Biol. 27, 8431–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao Y., Koebis M., Suo S., Ohno S., Ishiura S. (2012) Regulation of the alternative splicing of sarcoplasmic reticulum Ca2+-ATPase1 (SERCA1) by phorbol 12-myristate 13-acetate (PMA) via a PKC pathway. Biochem. Biophys. Res. Commun. 423, 212–217 [DOI] [PubMed] [Google Scholar]
- 14. Zhou Z., Qiu J., Liu W., Zhou Y., Plocinik R. M., Li H., Hu Q., Ghosh G., Adams J. A., Rosenfeld M. G., Fu X. D. (2012) The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol. Cell 47, 422–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Black D. L. (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72, 291–336 [DOI] [PubMed] [Google Scholar]
- 16. Kornblihtt A. R., Schor I. E., Alló M., Dujardin G., Petrillo E., Muñoz M. J. (2013) Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 14, 153–165 [DOI] [PubMed] [Google Scholar]
- 17. Wang G. S., Kuyumcu-Martinez M. N., Sarma S., Mathur N., Wehrens X. H., Cooper T. A. (2009) PKC inhibition ameliorates the cardiac phenotype in a mouse model of myotonic dystrophy type 1. J. Clin. Invest. 119, 3797–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kalsotra A., Xiao X., Ward A. J., Castle J. C., Johnson J. M., Burge C. B., Cooper T. A. (2008) A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl. Acad. Sci. U.S.A. 105, 20333–20338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koshelev M., Sarma S., Price R. E., Wehrens X. H., Cooper T. A. (2010) Heart-specific overexpression of CUGBP1 reproduces functional and molecular abnormalities of myotonic dystrophy type 1. Hum. Mol. Genet. 19, 1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yeo G. W., Coufal N. G., Liang T. Y., Peng G. E., Fu X. D., Gage F. H. (2009) An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 16, 130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gallagher T. L., Arribere J. A., Geurts P. A., Exner C. R., McDonald K. L., Dill K. K., Marr H. L., Adkar S. S., Garnett A. T., Amacher S. L., Conboy J. G. (2011) Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev. Biol. 359, 251–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gehman L. T., Meera P., Stoilov P., Shiue L., O'Brien J. E., Meisler M. H., Ares M., Jr., Otis T. S., Black D. L. (2012) The splicing regulator Rbfox2 is required for both cerebellar development and mature motor function. Genes Dev. 26, 445–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han H., Irimia M., Ross P. J., Sung H. K., Alipanahi B., David L., Golipour A., Gabut M., Michael I. P., Nachman E. N., Wang E., Trcka D., Thompson T., O'Hanlon D., Slobodeniuc V., Barbosa-Morais N. L., Burge C. B., Moffat J., Frey B. J., Nagy A., Ellis J., Wrana J. L., Blencowe B. J. (2013) MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature 498, 241–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kanadia R. N., Johnstone K. A., Mankodi A., Lungu C., Thornton C. A., Esson D., Timmers A. M., Hauswirth W. W., Swanson M. S. (2003) A muscleblind knockout model for myotonic dystrophy. Science 302, 1978–1980 [DOI] [PubMed] [Google Scholar]
- 25. Kimes B. W., Brandt B. L. (1976) Properties of a clonal muscle cell line from rat heart. Exp. Cell Res. 98, 367–381 [DOI] [PubMed] [Google Scholar]
- 26. Schneider C. A., Rasband W. S., Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu K. K., Huan Y. (2008) Streptozotocin-induced diabetic models in mice and rats. Curr. Protoc. Pharmacol. Chapter 5, Unit 5.47 [DOI] [PubMed] [Google Scholar]
- 28. Bland C. S., Wang E. T., Vu A., David M. P., Castle J. C., Johnson J. M., Burge C. B., Cooper T. A. (2010) Global regulation of alternative splicing during myogenic differentiation. Nucleic Acids Res. 38, 7651–7664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Katz Y., Wang E. T., Airoldi E. M., Burge C. B. (2010) Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 7, 1009–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van Belle T. L., Taylor P., von Herrath M. G. (2009) Mouse models for type 1 diabetes. Drug Discov. Today Dis. Models 6, 41–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu X., Tesiram Y. A., Towner R. A., Abbott A., Patterson E., Huang S., Garrett M. W., Chandrasekaran S., Matsuzaki S., Szweda L. I., Gordon B. E., Kem D. C. (2007) Early myocardial dysfunction in streptozotocin-induced diabetic mice: a study using in vivo magnetic resonance imaging (MRI). Cardiovasc. Diabetol. 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tochino Y. (1987) The NOD mouse as a model of type I diabetes. Crit. Rev. Immunol. 8, 49–81 [PubMed] [Google Scholar]
- 33. Ménard C., Pupier S., Mornet D., Kitzmann M., Nargeot J., Lory P. (1999) Modulation of L-type calcium channel expression during retinoic acid-induced differentiation of H9C2 cardiac cells. J. Biol. Chem. 274, 29063–29070 [DOI] [PubMed] [Google Scholar]
- 34. Davis P. D., Elliott L. H., Harris W., Hill C. H., Hurst S. A., Keech E., Kumar M. K., Lawton G., Nixon J. S., Wilkinson S. E. (1992) Inhibitors of protein kinase C. 2. Substituted bisindolylmaleimides with improved potency and selectivity. J. Med. Chem. 35, 994–1001 [DOI] [PubMed] [Google Scholar]
- 35. Aderem A. (1992) The MARCKS brothers: a family of protein kinase C substrates. Cell 71, 713–716 [DOI] [PubMed] [Google Scholar]
- 36. Moore S. F., van den Bosch M. T., Hunter R. W., Sakamoto K., Poole A. W., Hers I. (2013) Dual regulation of glycogen synthase kinase 3 (GSK3) α/β by protein kinase C (PKC) α and Akt promotes thrombin-mediated integrin αIIbβ3 activation and granule secretion in platelets. J. Biol. Chem. 288, 3918–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moraes K. C., Wilusz C. J., Wilusz J. (2006) CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA 12, 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Timchenko N. A., Wang G. L., Timchenko L. T. (2005) RNA CUG-binding protein 1 increases translation of 20-kDa isoform of CCAAT/enhancer-binding protein β by interacting with the α and β subunits of eukaryotic initiation translation factor 2. J. Biol. Chem. 280, 20549–20557 [DOI] [PubMed] [Google Scholar]
- 39. Revil T., Gaffney D., Dias C., Majewski J., Jerome-Majewska L. A. (2010) Alternative splicing is frequent during early embryonic development in mouse. BMC Genomics 11, 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kalsotra A., Wang K., Li P. F., Cooper T. A. (2010) MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev. 24, 653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nakahata S., Kawamoto S. (2005) Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 33, 2078–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Underwood J. G., Boutz P. L., Dougherty J. D., Stoilov P., Black D. L. (2005) Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 25, 10005–10016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ramirez-Correa G. A., Cortassa S., Stanley B., Gao W. D., Murphy A. M. (2010) Calcium sensitivity, force frequency relationship and cardiac troponin I: critical role of PKA and PKC phosphorylation sites. J. Mol. Cell. Cardiol. 48, 943–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sumandea M. P., Pyle W. G., Kobayashi T., de Tombe P. P., Solaro R. J. (2003) Identification of a functionally critical protein kinase C phosphorylation residue of cardiac troponin T. J. Biol. Chem. 278, 35135–35144 [DOI] [PubMed] [Google Scholar]
- 45. Braz J. C., Gregory K., Pathak A., Zhao W., Sahin B., Klevitsky R., Kimball T. F., Lorenz J. N., Nairn A. C., Liggett S. B., Bodi I., Wang S., Schwartz A., Lakatta E. G., DePaoli-Roach A. A., Robbins J., Hewett T. E., Bibb J. A., Westfall M. V., Kranias E. G., Molkentin J. D. (2004) PKC-α regulates cardiac contractility and propensity toward heart failure. Nat. Med. 10, 248–254 [DOI] [PubMed] [Google Scholar]
- 46. Keren H., Lev-Maor G., Ast G. (2010) Alternative splicing and evolution: diversification, exon definition and function. Nat. Rev. Genet. 11, 345–355 [DOI] [PubMed] [Google Scholar]
- 47. Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S. F., Schroth G. P., Burge C. B. (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yeo G., Holste D., Kreiman G., Burge C. B. (2004) Variation in alternative splicing across human tissues. Genome Biol. 5, R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lewis B. P., Green R. E., Brenner S. E. (2003) Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Nal. Acad. Sci. U.S.A. 100, 189–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moore M. J., Proudfoot N. J. (2009) Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136, 688–700 [DOI] [PubMed] [Google Scholar]
- 51. Salomonis N., Nelson B., Vranizan K., Pico A. R., Hanspers K., Kuchinsky A., Ta L., Mercola M., Conklin B. R. (2009) Alternative splicing in the differentiation of human embryonic stem cells into cardiac precursors. PLoS Comput. Biol. 5, e1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Depre C., Young M. E., Ying J., Ahuja H. S., Han Q., Garza N., Davies P. J., Taegtmeyer H. (2000) Streptozotocin-induced changes in cardiac gene expression in the absence of severe contractile dysfunction. J. Mol. Cell. Cardiol. 32, 985–996 [DOI] [PubMed] [Google Scholar]
- 53. Taegtmeyer H., Sen S., Vela D. (2010) Return to the fetal gene program: a suggested metabolic link to gene expression in the heart. Ann. N.Y. Acad. Sci. 1188, 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Razeghi P., Young M. E., Alcorn J. L., Moravec C. S., Frazier O. H., Taegtmeyer H. (2001) Metabolic gene expression in fetal and failing human heart. Circulation 104, 2923–2931 [DOI] [PubMed] [Google Scholar]
- 55. Sadoshima J., Izumo S. (1997) The cellular and molecular response of cardiac myocytes to mechanical stress. Annu. Rev. Physiol. 59, 551–571 [DOI] [PubMed] [Google Scholar]
- 56. Ytrehus K., Liu Y., Downey J. M. (1994) Preconditioning protects ischemic rabbit heart by protein kinase C activation. Am. J. Physiol. 266, H1145–H1152 [DOI] [PubMed] [Google Scholar]
- 57. Heineke J., Molkentin J. D. (2006) Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 7, 589–600 [DOI] [PubMed] [Google Scholar]
- 58. Koya D., King G. L. (1998) Protein kinase C activation and the development of diabetic complications. Diabetes 47, 859–866 [DOI] [PubMed] [Google Scholar]
- 59. Brownlee M. (2001) Biochemistry and molecular cell biology of diabetic complications. Nature 414, 813–820 [DOI] [PubMed] [Google Scholar]
- 60. Elwess N. L., Filoteo A. G., Enyedi A., Penniston J. T. (1997) Plasma membrane Ca2+ pump isoforms 2a and 2b are unusually responsive to calmodulin and Ca2+. J. Biol. Chem. 272, 17981–17986 [DOI] [PubMed] [Google Scholar]
- 61. Strehler E. E., Caride A. J., Filoteo A. G., Xiong Y., Penniston J. T., Enyedi A. (2007) Plasma membrane Ca2+ ATPases as dynamic regulators of cellular calcium handling. Ann. N.Y. Acad. Sci. 1099, 226–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Oppelt A., Lobert V. H., Haglund K., Mackey A. M., Rameh L. E., Liestøl K., Schink K. O., Pedersen N. M., Wenzel E. M., Haugsten E. M., Brech A., Rusten T. E., Stenmark H., Wesche J. (2013) Production of phosphatidylinositol 5-phosphate via PIKfyve and MTMR3 regulates cell migration. EMBO Rep. 14, 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Walker D. M., Urbé S., Dove S. K., Tenza D., Raposo G., Clague M. J. (2001) Characterization of MTMR3. An inositol lipid 3-phosphatase with novel substrate specificity. Curr. Biol. 11, 1600–1605 [DOI] [PubMed] [Google Scholar]
- 64. Disatnik M. H., Jones S. N., Mochly-Rosen D. (1995) Stimulus-dependent subcellular localization of activated protein-kinase-C–a study with acidic fibroblast growth-factor and transforming growth-factor-β-1 in cardiac myocytes. J. Mol. Cell. Cardiol. 27, 2473–2481 [DOI] [PubMed] [Google Scholar]
- 65. Martelli A. M., Evangelisti C., Nyakern M., Manzoli F. A. (2006) Nuclear protein kinase C. Biochim. Biophys. Acta 1761, 542–551 [DOI] [PubMed] [Google Scholar]
- 66. James G., Olson E. (1992) Deletion of the regulatory domain of protein kinase Cα exposes regions in the hinge and catalytic domains that mediate nuclear targeting. J. Cell Biol. 116, 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mochly-Rosen D., Das K., Grimes K. V. (2012) Protein kinase C, an elusive therapeutic target? Nat. Rev. Drug Discov. 11, 937–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 69. Kobayashi Y., Hirawa N., Tabara Y., Muraoka H., Fujita M., Miyazaki N., Fujiwara A., Ichikawa Y., Yamamoto Y., Ichihara N., Saka S., Wakui H., Yoshida S., Yatsu K., Toya Y., Yasuda G., Kohara K., Kita Y., Takei K., Goshima Y., Ishikawa Y., Ueshima H., Miki T., Umemura S. (2012) Mice lacking hypertension candidate gene ATP2B1 in vascular smooth muscle cells show significant blood pressure elevation. Hypertension 59, 854–860 [DOI] [PubMed] [Google Scholar]
- 70. Alzugaray M. E., García M. E., Del Zotto H. H., Raschia M. A., Palomeque J., Rossi J. P., Gagliardino J. J., Flores L. E. (2009) Changes in islet plasma membrane calcium-ATPase activity and isoform expression induced by insulin resistance. Arch. Biochem. Biophys. 490, 17–23 [DOI] [PubMed] [Google Scholar]
- 71. Van't Padje S., Chaudhry B., Severijnen L. A., van der Linde H. C., Mientjes E. J., Oostra B. A., Willemsen R. (2009) Reduction in fragile X related 1 protein causes cardiomyopathy and muscular dystrophy in zebrafish. J. Exp. Biol. 212, 2564–2570 [DOI] [PubMed] [Google Scholar]
- 72. Xia X. B., Xiong S. Q., Song W. T., Luo J., Wang Y. K., Zhou R. R. (2008) Inhibition of retinal neovascularization by siRNA targeting VEGF(165). Mol. Vis. 14, 1965–1973 [PMC free article] [PubMed] [Google Scholar]
- 73. Taimeh Z., Loughran J., Birks E. J., Bolli R. (2013) Vascular endothelial growth factor in heart failure. Nat. Rev. Cardiol. 10, 519–530 [DOI] [PubMed] [Google Scholar]
- 74. Bhagavatula M. R., Fan C., Shen G. Q., Cassano J., Plow E. F., Topol E. J., Wang Q. (2004) Transcription factor MEF2A mutations in patients with coronary artery disease. Hum. Mol. Genet. 13, 3181–3188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang L., Fan C., Topol S. E., Topol E. J., Wang Q. (2003) Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science 302, 1578–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mora S., Pessin J. E. (2000) The MEF2A isoform is required for striated muscle-specific expression of the insulin-responsive GLUT4 glucose transporter. J. Biol. Chem. 275, 16323–16328 [DOI] [PubMed] [Google Scholar]
- 77. Ramsdell A. F. (2005) Left-right asymmetry and congenital cardiac defects: getting to the heart of the matter in vertebrate left-right axis determination. Dev. Biol. 288, 1–20 [DOI] [PubMed] [Google Scholar]
- 78. Feng T., Zhu X. (2010) Genome-wide searching of rare genetic variants in WTCCC data. Hum. Genet. 128, 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hershberger R. E., Parks S. B., Kushner J. D., Li D., Ludwigsen S., Jakobs P., Nauman D., Burgess D., Partain J., Litt M. (2008) Coding sequence mutations identified in MYH7, TNNT2, SCN5A, CSRP3, LBD3, and TCAP from 313 patients with familial or idiopathic dilated cardiomyopathy. Clin. Transl. Sci. 1, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Arrington C. B., Dowse B. R., Bleyl S. B., Bowles N. E. (2012) Non-synonymous variants in pre-B cell leukemia homeobox (PBX) genes are associated with congenital heart defects. Eur. J. Med. Genet. 55, 235–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Stankunas K., Shang C., Twu K. Y., Kao S. C., Jenkins N. A., Copeland N. G., Sanyal M., Selleri L., Cleary M. L., Chang C. P. (2008) Pbx/Meis deficiencies demonstrate multigenetic origins of congenital heart disease. Circ. Res. 103, 702–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Novarino G., Akizu N., Gleeson J. G. (2011) Modeling human disease in humans: the ciliopathies. Cell 147, 70–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Guzzo R. M., Salih M., Moore E. D., Tuana B. S. (2005) Molecular properties of cardiac tail-anchored membrane protein SLMAP are consistent with structural role in arrangement of excitation-contraction coupling apparatus. Am. J. Physiol. Heart Circ. Physiol. 288, H1810–H1819 [DOI] [PubMed] [Google Scholar]
- 84. Nader M., Westendorp B., Hawari O., Salih M., Stewart A. F., Leenen F. H., Tuana B. S. (2012) Tail-anchored membrane protein SLMAP is a novel regulator of cardiac function at the sarcoplasmic reticulum. Am. J. Physiol. Heart Circ. Physiol. 302, H1138–H1145 [DOI] [PubMed] [Google Scholar]
- 85. Ding H., Howarth A. G., Pannirselvam M., Anderson T. J., Severson D. L., Wiehler W. B., Triggle C. R., Tuana B. S. (2005) Endothelial dysfunction in type 2 diabetes correlates with deregulated expression of the tail-anchored membrane protein SLMAP. Am. J. Physiol. Heart Circ. Physiol. 289, H206–H211 [DOI] [PubMed] [Google Scholar]
- 86. Lin W. H., Chiu K. C., Chang H. M., Lee K. C., Tai T. Y., Chuang L. M. (2001) Molecular scanning of the human sorbin and SH3-domain-containing-1 (SORBS1) gene: positive association of the T228A polymorphism with obesity and type 2 diabetes. Hum. Mol. Genet. 10, 1753–1760 [DOI] [PubMed] [Google Scholar]
- 87. Terenzi F., Ladd A. N. (2010) Conserved developmental alternative splicing of muscleblind-like (MBNL) transcripts regulates MBNL localization and activity. RNA Biol. 7, 43–55 [DOI] [PubMed] [Google Scholar]
- 88. Blackwell E., Zhang X., Ceman S. (2010) Arginines of the RGG box regulate FMRP association with polyribosomes and mRNA. Hum. Mol. Genet. 19, 1314–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]