Background: Ezrin is a conformationally regulated microfilament-membrane linker restricted to the apical aspect of epithelial cells.
Results: Quantitative mass spectrometry identified proteins that associate with different ezrin conformations; the interactors include novel apical microvilli-associated proteins, with classes perceiving ezrin's conformation differentially.
Conclusion: Different proteins selectively associate with three distinct conformational states of ezrin.
Significance: This study extends the conformational activation model of ezrin and identifies new interacting partners.
Keywords: Cytoskeleton, Ezrin, Phosphorylation, Plasma Membrane, Proteomics
Abstract
Ezrin, a member of the ezrin-radixin-moesin family (ERM), is an essential regulator of the structure of microvilli on the apical aspect of epithelial cells. Ezrin provides a linkage between membrane-associated proteins and F-actin, oscillating between active/open and inactive/closed states, and is regulated in part by phosphorylation of a C-terminal threonine. In the open state, ezrin can bind a number of ligands, but in the closed state the ligand-binding sites are inaccessible. In vitro analysis has proposed that there may be a third hyperactivated form of ezrin. To gain a better understanding of ezrin, we conducted an unbiased proteomic analysis of ezrin-binding proteins in an epithelial cell line, Jeg-3. We refined our list of interactors by comparing the interactomes using quantitative mass spectrometry between wild-type ezrin, closed ezrin, open ezrin, and hyperactivated ezrin. The analysis reveals several novel interactors confirmed by their localization to microvilli, as well as a significant class of proteins that bind closed ezrin. Taken together, the data indicate that ezrin can exist in three different conformational states, and different ligands “perceive” ezrin conformational states differently.
Introduction
The establishment and maintenance of cell polarity are carried out through the asymmetrical distribution of macromolecules. An important contributor to the structure of microvilli on the apical aspect of epithelial cells is the F-actin-binding protein ezrin. Ezrin, the founding member of the conserved ezrin-radixin-moesin (ERM) protein family, was originally isolated from the intestinal brush border (1) and later shown to play essential roles in cell polarity throughout animal development (2). Ezrin adopts a highly polarized distribution in epithelial cells where it is found both in the cytoplasm and specifically associated with the microvillar plasma membrane. Consistently, it is essential for the normal morphology of microvilli in vivo (3–6) and in cultured cells (6, 7).
In their inactive state, ERMs undergo an intramolecular head-to-tail association, masking binding sites for both plasma membrane-associated proteins on their N-terminal four-point-one ezrin-radixin-moesin (FERM) domain and the F-actin-binding site in the C-terminal tail. The appearance of ezrin in its active state on the microvillar plasma membrane requires direct interaction with the membrane phospholipid PI(4,5)P2 through its N-terminal FERM domain (8–12) followed by phosphorylation on a conserved C-terminal threonine (Thr-567 in ezrin (7, 8, 13)). In epithelial cells, kinase and phosphatase activity drives constant, dynamic interconversion between membrane-bound phosphorylated ezrin and cytoplasmic dormant, unphosphorylated ezrin, with each state having a half-life of 1–2 min (7, 14, 15).
Although ezrin is generally considered to simply oscillate between open/active or closed/inactive states, there are likely to be varying degrees of ezrin openness, reflecting the existence of multiple conformational states. Notably, in vitro analysis has suggested that phosphorylation of the C-terminal threonine in ezrin creates a partial but not fully open state (16). Thus, we explored this possibility by examining two forms of open ezrin in our analysis.
Upon reaching the plasma membrane, ERM proteins engage a number of membrane-associated factors through the N-terminal FERM domain. Numerous binding partners of mammalian ERMs have been identified (Table 1). In most of these interactions, the interacting protein has been proposed to be the effector as opposed to being the regulator of ERMs. Conversely, one of these, the scaffolding ERM-binding phosphoprotein 50 (EBP50, also known as NHERF1 or SLC9A3R1), has been shown to regulate the ERM-dependent formation of microvilli (17–19). However, transmembrane ERM-binding proteins have also been proposed to play a role in ERM recruitment or clustering in the apical domain leading to the formation of microvilli (20), although no such protein has yet been identified in epithelial cells. Moreover, in vitro analyses show that although the surfaces on the FERM domain for EBP50, PI(4,5)P2, and transmembrane proteins are distinct (10, 21–25), there is likely to be a complex interplay among all of these ligands (26). Thus, multiple regulatory ERM binding partners might be identified by an unbiased proteomic screen for ERM-binding proteins in epithelial cells.
TABLE 1.
Reported ERM-interacting proteins
| Interaction partner | Selected refs. | Proposed binding mode | Proposed function |
|---|---|---|---|
| Aquaporin-0 | 50 | Unknown | Unknown |
| ARHGAP-18/conundrum | 33 | FERM domain | Active RhoA level |
| Bitesize | 51 | FERM domain (lobe F3) | Adherens junction morphology |
| CD146 | 52 | Unknown | Melanoma cell line migration through control of ERM-RhoGDI interaction |
| CD43 | 20, 53–56 | FERM domain | Lymphocyte migration via PKC-mediated phosphorylation of CD43 tail; tethering of ERMs to plasma membrane for microvillus formation |
| CD44 | 9, 20, 53, 57–62 | FERM domain | Migration speed and directionality; lymphocyte polarization; microvilli length and number |
| CLIC3/4/5 | 32, 42 | C-terminal region of ezrin | Microvillus formation in RPE cells (CLIC4) |
| Dbl | 48, 63–67 | FERM domain with Dbl PH | Active Cdc42 level |
| DCC | 68, 69 | FERM domain | Netrin-induced axon guidance |
| Dlg1 | 70, 71 | FERM domain | Immune synapse formation |
| SLC9A3R1/2 (EBP50/E3KARP) | 17, 18, 21, 28, 30, 49, 72 | FERM domain | Microvilli formation; clustering of various receptors |
| Eps8/−L1a | 73 | Coiled-coil domain NPXY motif with Eps8 SH3 | Microvillus length and spacing |
| Fes | 74 | Coiled-coil domain NPXY motif with Fes SH3 | Cell spreading and migration in response to HGF |
| ICAM-1/-2/-3 | 24, 25, 75–79 | FERM domain | Polarization of ICAMs to the uropod in migrating lymphocytes; microvilli length and number |
| L1CAM | 80, 81 | FERM domain | Neurite outgrowth |
| Myo18aα | 82 | Unknown | Unknown |
| NHE-1 | 83, 84 | FERM domain | Migration speed and directionality |
| NHE-3 | 85 | FERM domain | NHE-3 surface distribution in OK cells |
| SCYL3/PACE-1 | 34 | C-terminal region of ezrin | Unknown |
| Palladin | 86, 87 | Ezrin (278–585) with palladin Ig2/3 | Unknown |
| PDZD8 | 88 | Unknown | Glutamylated microtubule abundance and HSV-1 viral entry |
| PLEKHG6 | 89 | FERM domain with PLEKHG6 C-terminal region | Macropinocytosis |
| Podocalyxin | 90 | Unknown | Unknown |
| PSGL-1 | 78, 91–95 | FERM domain | Lymphocyte rolling on P-selectin |
| Rab11 | 96 | Unknown | Collective cell migration |
| Ras | 97 | FERM domain | Active Ras level |
| RhoGDI | 9, 66, 98 | FERM domain | Active Rho GTPase level |
| S100P | 99, 100 | FERM domain | Cellular calcium response |
| SAP97 | 101 | Unknown | Unknown |
| SOS | 97 | FERM domain with SOS DH/PH | Active Ras level |
| SYND2/Syndecan-2 | 35, 36 | FERM domain | Unknown |
| Vps11/HOPS | 102 | Coiled-coil domain | Endocytosis |
| WWOX | 103 | Coiled-coil domain NPXY motif with WWOX SH3 | Secretion in gastric parietal cells |
| WWP-1 | 104 | Coiled-coil domain NPXY motif with WWP-1 SH3 | Cell migration in response to HGF |
| ZAP-70 | 55 | Unknown | Immune synapse formation |
| ITGB4/β4 integrin | 105 | FERM domain | β4 integrin protein level |
| DAG1/β-Dystroglycan | 41, 48 | FERM domain | Filopodia formation in C2C12 cells co-transfected with active Cdc42 |
Here, we report the first global analysis of ezrin binding partners in an epithelial cell line. We first established a reversible cross-linking strategy that preserves the transient interaction between ezrin and its strongest known interactor, EBP50. We then used mass spectrometry to determine the ezrin interactome under these optimized conditions. Next, we examined how the ezrin interactome changes depending on its conformational state, and we documented the changes in response to ezrin conformation. The analysis reveals many novel components of microvilli that discriminate between different open forms, as well as an unexpected category of proteins binding to the closed form of ezrin.
EXPERIMENTAL PROCEDURES
Plasmids
Plasmids for stable or transient transfection of ezrin-iFLAG and variants (in pQCXIP, Clontech) have been previously described (7). The “K4N” mutation (22) was generated by inverse PCR (using primers 5′-CCC ATC GAC AAC AAC GCA CCT GAC TTT GTG TTT TAT GCC CCA C-3′ and 5′-TTT AAT GAC AAA GTT ATT GTC ATT GAA AGA GAT GTT CCT GAT TTC ACT CC-3′). To clone TACTSTD2, BASP1, SLC1A5, FAM129B, and EPCAM, Jeg-3 RNA was first extracted using the RNeasy kit (Qiagen) and then reverse-transcribed using the SuperScript III reverse transcriptase poly(dT) primer (Invitrogen). The open reading frames were cloned from the cDNA using PCR (with the following primers: TACSTD2, 5′-GTT CGA GGA TCC ATG GCT CGG GGC CCC GGC-3′ and 5′-TCG AAC GAA TTC CAA GCT CGG TTC CTT TCT CAA CTC CC-3′; BASP1, 5′-GTT CGA GGA TCC ACC ATG GGA GGC AAG CTC AGC AAG AAG AAG-3′ and 5′-TCG AAC GAA TTC CTC TTT CAC GGT TAC GGT TTG GTC GG-3′; SLC1A5, 5′-GTT CGA GAA TTC ACC ATG GTG GCC GAT CCT CCT CGA GAC-3′ and 5′-TCG AAC GCG GCC GCt tCA TGA CTG ATT CCT TCT CAG AGG CGA CC-3′) and then inserted into pEGFP-N2 (Clontech) and pcDNA3.1/MycHisA (Invitrogen). GFP-FHOD1 was generated by PCR from an ATCC cDNA clone and insertion into pEGFP-C2. GFP-CLIC4 expression plasmid was a gift of Dr. M. Berryman, Ohio University, Athens, OH. SCYL3-myc expression plasmid was a gift of Dr. R. Thorne, University of Newcastle, Australia. The following constructs were examined and reported in Fig. 5H without images: FAM129B, cloned from Jeg-3 cDNA by PCR (using primers 5′-GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT TAT GGG GGA CGT GCT GTC CAC GC-3′ and 5′-GGG GAC CAC TTT GTA CAA GAA AGC TGG GTA GAA CTC AGT CTG CAC CCC TGC ACT G-3′), inserted into pDONR221 (Invitrogen), and then recombined into pcDNA-DEST47 (Invitrogen); EPCAM, cloned from Jeg-3 cDNA by PCR (using primers 5′-GTT CGA CTC GAG ATG GCG CCC CCG CAG GTC C-3′ and 5′-TCG AAC GAA TTC TGC ATT GAG TTC CCT ATG CAT CTC ACC C-3′) and inserted into pcDNA3.1/V5HisA; ATP11C-HA, a gift of Dr. H. W. Shin, University of Kyoto, Japan, which was co-expressed with Cdc50A as described (27); LOK and SLK, reported previously (7); F11R (also known as junction adhesion molecule A) with an internal HA tag, a gift of Dr. U. Naik, University of Delaware; ARHGAP18, ID HsCD00379004, from the Dana-Farber/Harvard Cancer Center recombined into pcDNA-DEST47.
FIGURE 5.
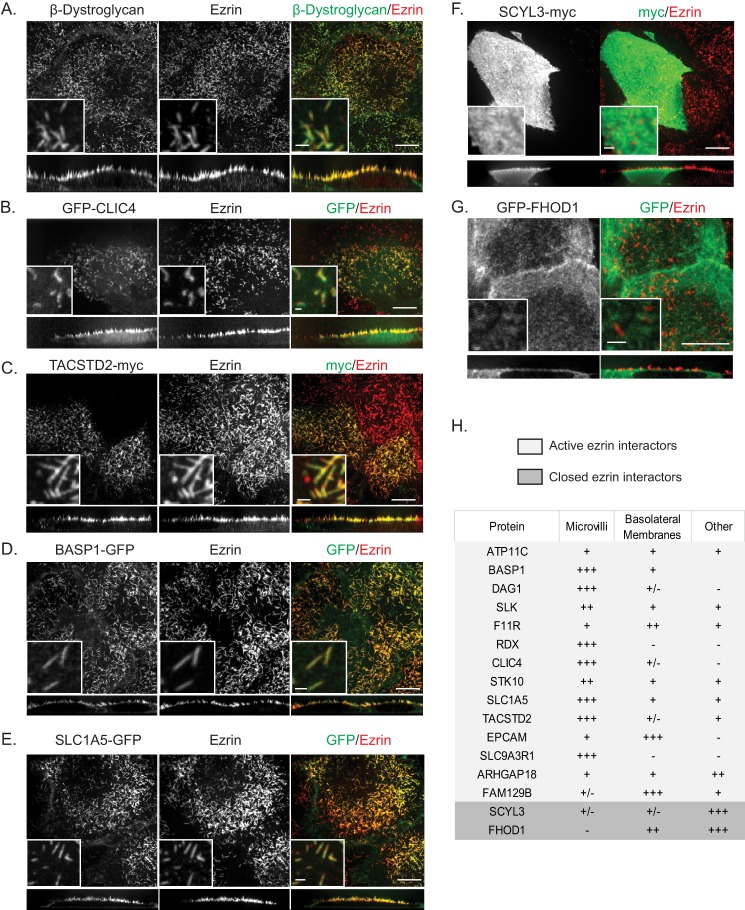
Active ezrin interactors localize in microvilli, whereas closed ezrin interactors localize in the cytoplasm. A–G, maximum intensity (XY) projection, 10-fold magnified image, and side view (XZ) of cells stained for endogenous β-dystroglycan (A) or transfected to express the indicated tagged interactor and counterstained for ezrin. Selected active ezrin interactors (A–E) localize in the microvilli. F and G, two interactors of closed ezrin do not localize to microvilli but are found in the cytoplasm. Scale bars are 10 μm for full maximum intensity projections and side views and 1 μm for insets. Side views have been stretched vertically 3-fold for clarity. H, summary of localization of all interactors examined.
Antibodies
FLAG antibody and resin were M2 from Sigma. Antibody against ezrin was CPCT-Ezrin-1; moesin was CPCT-Moesin-1, and β-dystroglycan (DAG1) was MANDAG2, all from the Developmental Studies Hybridoma Bank. Antibody against an epitope present in all ERMs was from Cell Signaling Technologies. Antibodies against radixin and EBP50 have been described (7, 28). Antibody against FHOD-1 was from Abcam. Antibody against Myc was 9E10 from Roche Applied Science. Antibody against GFP was from Santa Cruz Biotechnology. Antibody against HA was HA.11 from Covance. Antibody against TACSTD2 was GA733.1 (immunofluorescence) from Abcam or H-85 from Santa Cruz Biotechnology (Western blot).
Cell Culture, Transfection, and Stable Cell Line Creation
Jeg-3 cells were obtained from the American Type Culture Collection and cultured in minimal Eagle's medium supplemented with GlutaMAX-1 (Invitrogen), penicillin/streptomycin (Invitrogen), and 10% FBS (Invitrogen) and maintained at 37 °C and 5% CO2.
All transient transfections were by polyethyleneimine (PolyPlus) as described previously (17). Following transfection, cells were grown for ∼18 h prior to use.
Stable cell lines expressing ezrin-iFLAG variants were transfected as described previously (7) and then batch-selected using puromycin (2 μg/ml). Stable cell lines expressing BASP-1-GFP and SLC1A5-GFP were generated by transfection with the appropriate plasmid, grown in G418 (400 μg/ml) for 2 weeks, and ring-cloned and pooled at least three GFP-expressing colonies.
SILAC3 Labeling
Jeg-3 cells stably transfected with pQCXIP and derivatives and then selected for at least 2 weeks were grown for at least 2 additional weeks in SILAC minimal Eagle's medium (Thermo) containing 0.1 mg/ml normal lysine and arginine (“light”) or [13C6,15N2]lysine and [13C6,15N4]arginine (“heavy,” Sigma) containing 10% dialyzed FBS (Invitrogen), penicillin/streptomycin (Invitrogen), and puromycin (2 μg/ml). Incorporation was verified by mixing lysate preparations from heavy and light cells (1:1) followed by mass spectrometry. Expected morphology and ezrin-iFLAG localization after labeling was verified by immunofluorescence (data not shown).
Dithiobis(succinimidyl Propionate) (DSP) Cross-linking
Cells grown to ∼80% confluence were washed three times with PBS and treated with 1.25 mm DSP (Thermo) at 37 °C for 2 min. The cells were then removed to room temperature and washed three times with TBS. They were incubated in the last TBS wash for 15–20 min prior to use. Expected morphology and ezrin-iFLAG localization after labeling were verified by immunofluorescence (data not shown).
Immunoprecipitation and Mass Spectrometry
For SILAC mass spectrometry, ∼4 × 107 DSP-treated heavy and the same number of light cells were independently scraped into cold IP buffer (25 mm Tris, pH 7.4, 5% glycerol, 150 mm NaCl, 50 mm NaF, 0.1 mm sodium orthovanadate, 10 mm β-glycerophosphate, 8.7 mg/ml para-nitrophenyl phosphate, 0.5% Triton X-100, 0.1 μm calyculin A, protease inhibitor table from Roche Applied Science), incubated for 2–3 h in the cold using 20 μl of FLAG M2 affinity gel (Sigma), and then washed four times in IP wash buffer (25 mm Tris, pH 7.4, 5% glycerol, 150 mm NaCl, 50 mm NaF, 0.2% Triton X-100). Gel was then eluted by boiling in 50 mm Tris, pH 8.0, containing 1% SDS, reduced, and alkylated. Proteins were precipitated with 50:49.9:0.1, acetone/ethanol/acetic acid, reconstituted in urea solution, and trypsin-digested overnight. Peptides were then purified on a C-18 column (Waters), dried, and reconstituted in 80% acetonitrile and 1% formic acid and fractionated by hydrophilic interaction chromatography. Fractions were dried, reconstituted in 0.1% trifluoroacetic acid, and analyzed by LC-MS/MS using an Orbitrap XL mass spectrometer (Thermo). Database search and peptide quantification of heavy/light isotope ratios were performed as described previously (29). Criteria for selection of true interactors are presented under “Results.”
For all other immunoprecipitations, ∼8 × 106 cells transfected with 7 μg of appropriate plasmid(s) were lysed in IP buffer, immunoprecipitated with 4 μl of FLAG M2 affinity gel for 2–3 h, washed four times with IP wash buffer, eluted by addition of 3×FLAG peptide to 200 μg/ml, and incubated for 10 min at room temperature. Soluble material was separated in a spin column (Sigma), denatured by boiling in Laemmli buffer, and resolved by SDS-PAGE.
Immunofluorescence
Cells grown on glass coverslips were fixed in 3.7% formaldehyde/PBS for 15 min at room temperature. Cells were then washed with PBS and blocked with IF buffer (PBS + 0.5% BSA + 0.5% goat serum + 0.1% Triton X-100) for 10 min. Primary and secondary antibodies were then applied in IF buffer with 1% FBS. After antibody addition, the coverslips were washed three times in PBS, mounted in Vectashield (Vector Laboratories), imaged using a CSU-X spinning disk microscope (Intelligent Imaging Innovations) with a spherical aberration correction device, 63 × 1.4 NA objective on an inverted microscope (Leica), and acquired with a QuantEM EMCCD camera using Slidebook software (Intelligent Imaging Innovations). Maximum intensity projections were assembled in Slidebook and exported to Adobe Illustrator software. For clarity, side projections were vertically expanded 3-fold using Illustrator.
RNA Interference
Jeg-3 cells were transfected with 30 nm of siRNA for 72 h prior to use by using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. siRNAs were from Ambion, Dharmacon, or Integrated DNA Technologies as follows: GL2 luciferase (17), ezrin (7), radixin (5′-UGAAGAUGUUUCUGAGGAAdUdU-3′), TACSTD2 (−1, 5′-GCUUAAAUGAGUUUAGAUGGGAAAT-3′ and −2, 5′-GCUUAAAUGAGUUUAGAUGGGAAAT-3′), and DAG1 (5′-CCCUAGAGCCUGACUUUAAdTdT-3′).
RESULTS
Cross-linking Prior to Immunoprecipitation Is Required to Recover the Ezrin-EBP50 Interaction
Since its discovery in 1983 (1), numerous proteins have been shown to interact with ezrin (Table 1). Among the most tenacious in vitro interaction partner is EBP50 (SLC9A3R1), a scaffolding protein that binds with very high affinity through its C-terminal region to the FERM domain of ezrin (28). The binding site for EBP50 on the FERM domain is masked in the closed, inactive form of ezrin (21, 28, 30). Our goal was to perform an unbiased screen for proteins present in epithelial cells that bind to active ezrin in vivo, and EBP50 was used as a positive control to establish optimal conditions.
Our strategy centers around the generation of cell lines expressing various mutants of ezrin representing various conformational states and then analyzing the bound proteins that are co-recovered with ezrin. As reported recently, we have placed a FLAG epitope into an internal region of ezrin that is poorly conserved in ERM proteins (7). Using this tagged version designated ezrin-iFLAG, we have generated stable Jeg-3 choriocarcinoma cell lines that express wild-type ezrin (ezrin-iFLAG), a variant with the activating T567E phosphomimetic mutation (ezrin-T567E-iFLAG), an inactive variant with the nonphosphorylatable T567A mutation (ezrin-T567A-iFLAG), and a variant containing a truncation of the C terminus that compromises the normal FERM/C-terminal tail interaction to expose the fully unmasked FERM domain of ezrin (ezrin(1–583)-iFLAG). The level of the ezrin-iFLAG variant expression in these cells is about the same as endogenous ezrin (7). As expected, ezrin-iFLAG localization was strictly apical, co-localizing with another ERM member radixin, whereas ezrin-T567A-iFLAG is cytoplasmic, and as reported recently, ezrin-T567E-iFLAG hyperlocalizes to the plasma membrane and localizes to the apical and basolateral domains of epithelial cells (Fig. 1A) (7).
FIGURE 1.
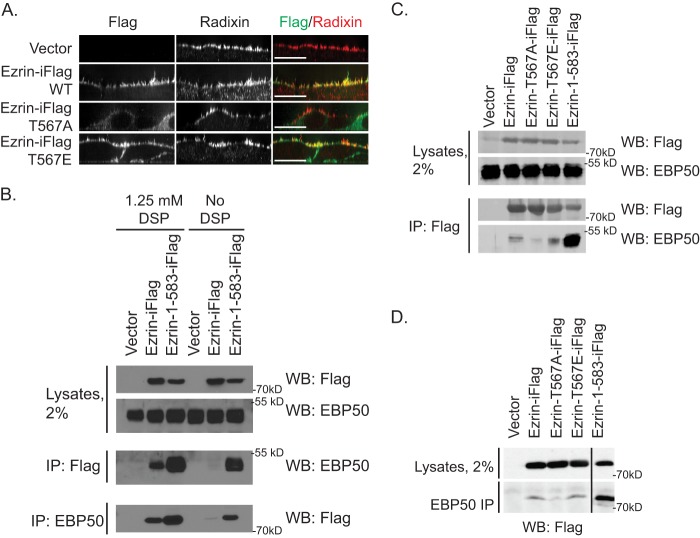
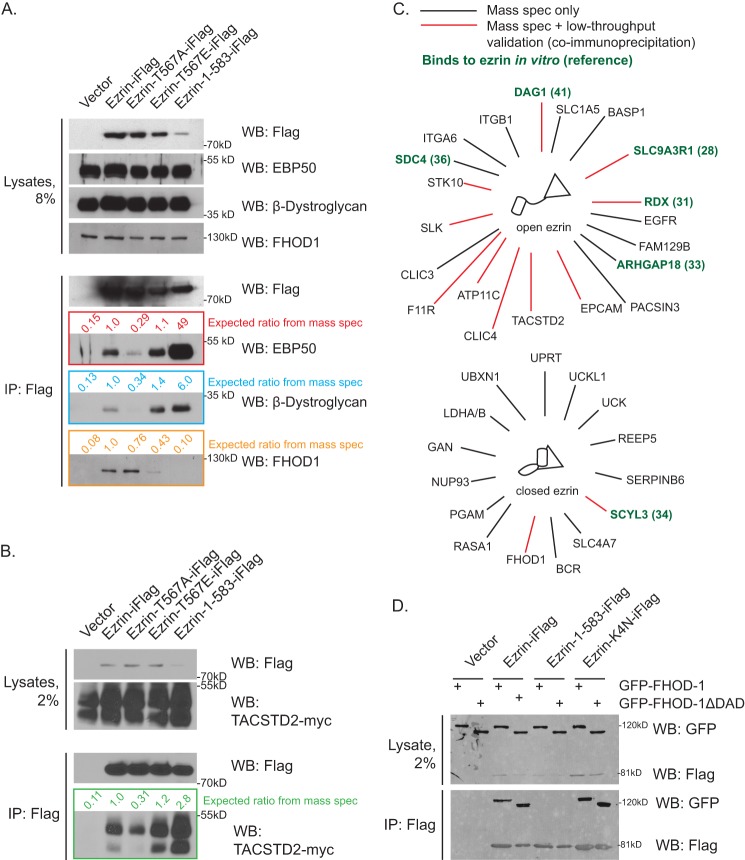
Ezrin/EBP50 interaction can be preserved by enhancing the open state of ezrin or by DSP cross-linking. A, Jeg-3 cells stably expressing the indicated ezrin-iFLAG variant were subjected to immunofluorescence to detect transfected FLAG-tagged ezrin and endogenous radixin, the other ERM protein present in this cell line. Scale bar, 10 μm. B, stable cell lines expressing empty vector control, ezrin-iFLAG, or hyperactive ezrin-iFLAG truncation 1–583 as indicated were subjected to immunoprecipitation (IP) with the indicated antibody with or without DSP pretreatment, and the immunoprecipitates were Western blotted (WB) for EBP50 or FLAG. C, stable cell lines expressing the indicated ezrin-iFLAG variant were subjected to DSP cross-linking followed by FLAG immunoprecipitation, and the immunoprecipitates were Western-blotted for EBP50. D, stable cell lines expressing the indicated ezrin-iFLAG variant were subjected to DSP cross-linking followed by EBP50 immunoprecipitation, and the immunoprecipitates were Western-blotted for FLAG.
As about 50% of the ezrin-iFLAG is phosphorylated in vivo (7), we were initially surprised to find that immunoprecipitation of ezrin-iFLAG failed to recover detectable EBP50. By contrast, the construct with an unmasked FERM domain due to the truncation of the final two residues, ezrin(1–583)-iFLAG, recovered EBP50 efficiently (Fig. 1B). To see if the interaction of EBP50 with ezrin-iFLAG can be stabilized, we explored the use of the reversible cross-linking reagent DSP. Cells were subjected to different concentrations of cross-linker for 2 min, and the recovery of EBP50 with ezrin-iFLAG was monitored. These results showed that brief cross-linking with 1.25 mm DSP allowed for modest recovery of EBP50 with ezrin-iFLAG and vice versa (Fig. 1B). To explore how the ezrin/EBP50 interaction was affected by ezrin Thr-567 phosphosite mutations, we detected EBP50 in unphosphorylatable ezrin-T567A-iFLAG or phosphomimetic ezrin-T567E-iFLAG immunoprecipitations (Fig. 1C). The modest recovery of EBP50 was significantly reduced by mutation to T567A but was unaffected by mutation to T567E (Fig. 1C). We obtained similar results by immunoprecipitating EBP50 and detecting ezrin-iFLAG phosphomutants (Fig. 1D). Thus, the DSP-preserved interaction between EBP50 and ezrin is dependent on the availability of Thr-567 for phosphorylation and can be greatly increased by fully unmasking the FERM domain of ezrin as in ezrin-(1–583)-iFLAG (Fig. 1C).
Use of SILAC-Mass Spectrometry to Identify the Ezrin Interactome
To use mass spectrometry to identify proteins other than EBP50 that bind to wild-type ezrin, the ezrin-iFLAG cells were subject to SILAC relative to a reference culture expressing just the empty vector control. The control cells were grown in light medium, and the cells expressing ezrin-iFLAG were grown for sufficient time in heavy medium containing [13C/15N]arginine and [13C/15N]lysine to uniformly label all proteins. Both samples were subject to DSP cross-linking followed by FLAG immunoprecipitation. The immunoprecipitates were combined and trypsin-digested, and the digest was subjected to quantitative mass spectrometry. By comparing the abundance of specific peptides between the reference and ezrin-iFLAG samples, background material can be minimized and proteins enriched in the ezrin-iFLAG sample identified (Fig. 2A).
FIGURE 2.
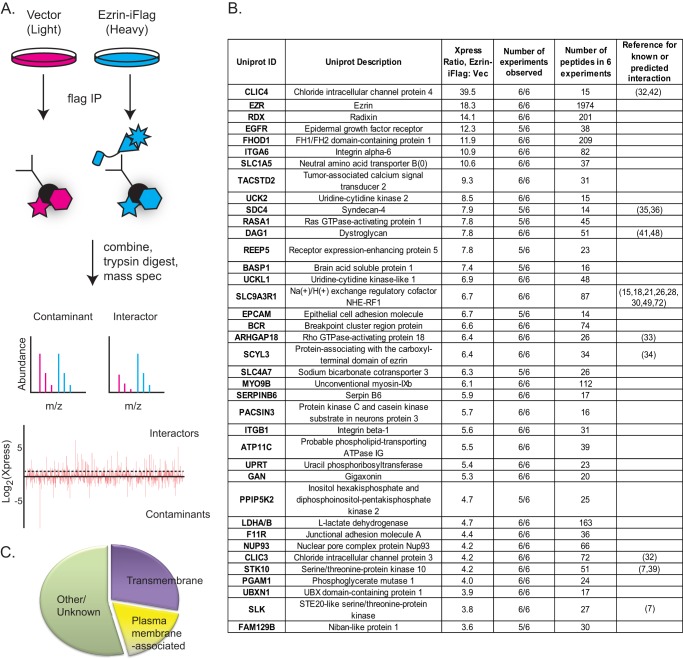
Identification of 38 high confidence interactors of ezrin. A, schematic of SILAC experiment. Jeg-3 cells stably transfected with vector control or expressing ezrin-iFLAG were differentially labeled in SILAC medium, independently subjected to cross-linking, and subjected to FLAG immunoprecipitation (IP). The immunoprecipitates were combined, trypsin-digested, and subjected to mass spectrometry, where heavy-to-light ratio was determined for each peptide. Diagram of mass spectrometry results for peptides from a typical background protein and an interactor is shown. Plot of the Xpress SILAC ratio for all recovered proteins (plotted along the horizontal axis) is shown. Proteins above the dashed line were considered interactors. (Most proteins enriched in the light sample are antibody fragments.) B, proteins with a heavy-to-light Xpress ratio greater than 3.6 for which 14 or more peptides were identified reproducibly in at least 5/6 experiments are listed. C, analysis of fraction of interactors containing transmembrane domains or shown to be membrane-associated (supplemental Table 1, columns U and V).
In each mass spectrometry run, 7,000–13,000 high confidence peptides (representing 2,000–3,500 unique proteins) were detected with most binding the FLAG antibody beads as background. To identify ezrin-iFLAG-bound proteins, we computationally selected those present at a heavy/light or “Xpress” ratio of greater than 3.6:1, and then to ensure reproducibility, we further selected only those that were detected in wild-type ezrin-iFLAG immunoprecipitations in at least 5/6 mass spectrometry experiments (biological replicates). These were considered as true interactors (Fig. 2B; supplemental Table 1). Of the 38 reproducibly interacting proteins, 9 were expected as either these proteins or their orthologs had been previously implicated in binding ERMs. These included the other ERM protein in these cells, radixin (“RDX,” see Ref. 31), EBP50 (28), the chloride ion channel-like proteins CLIC3 and CLIC4 (32), the Drosophila ortholog of RhoA GTPase-activating protein Conundrum, ARHGAP18 (33), the pseudokinase SCYL3 (34), and syndecan-4 (“SDC4”), an adhesion molecule that contains the minimal syndecan ERM-binding motif first identified in syndecan-2 (35, 36). Additionally, the kinases LOK and SLK were identified, which were shown to be the major C-terminal threonine kinases for ERMs in Drosophila and mammalian epithelial and immune cells (7, 37–39). Consistent with ezrin localization at the plasma membrane and the nature of established interaction partners, 18/38 (47%) of the interaction partners were known or predicted to be transmembrane or membrane-associated proteins (supplemental Table 1).
Interactome of Phosphorylated Versus Unphosphorylated Ezrin
To identify which of these interactors bind preferentially to phosphorylated ezrin, we made use of SILAC comparing Jeg-3 cells expressing the nonphosphorylatable ezrin-T567A-iFLAG (labeled light) versus wild-type ezrin-iFLAG (labeled heavy; Fig. 3A). Because ezrin-iFLAG can be phosphorylated in vivo, this approach allowed us to identify proteins that selectively associate with active, phosphorylated ezrin and, conversely, ones that preferentially associate with inactive ezrin-T567A-iFLAG. The degree of enrichment for each candidate was normalized to the degree of enrichment of ezrin peptides. Biological replicates were performed, and correlation analysis confirmed that the enrichment statistics were reproducible (Fig. 3B). As another approach to identify proteins that distinguish the activity state of ezrin, we also undertook the SILAC procedure with cells expressing ezrin-iFLAG (labeled light), of which about 50% should be in the active state, and cells expressing ezrin-T567E-iFLAG (labeled heavy), a mimetic for the fully phosphorylated form. Again, correlation analysis between biological duplicates confirmed that enrichment statistics were reproducible (Fig. 3C). Moreover, most of the proteins identified in the two approaches were the same and were quite well correlated (Fig. 3D). The results of this analysis are presented in Fig. 3E (left), where a consensus of proteins (from these and subsequent experiments, listed by Uniprot identifier) that bind more active ezrin is shown on the left. These data reveal that binding of many interactors to ezrin was enhanced for wild-type versus ezrin-T567A, and approximately two-thirds of these were further enhanced by the phosphomimetic ezrin-T567E mutation. Among these proteins, as expected, is radixin (RDX), the other ERM protein present in these cells, EBP50 (“SLC9A3R1”), β-dystroglycan (“DAG1”), CLIC4, and the kinases LOK (“STK10”) and SLK. In addition, about a dozen new proteins were identified.
FIGURE 3.
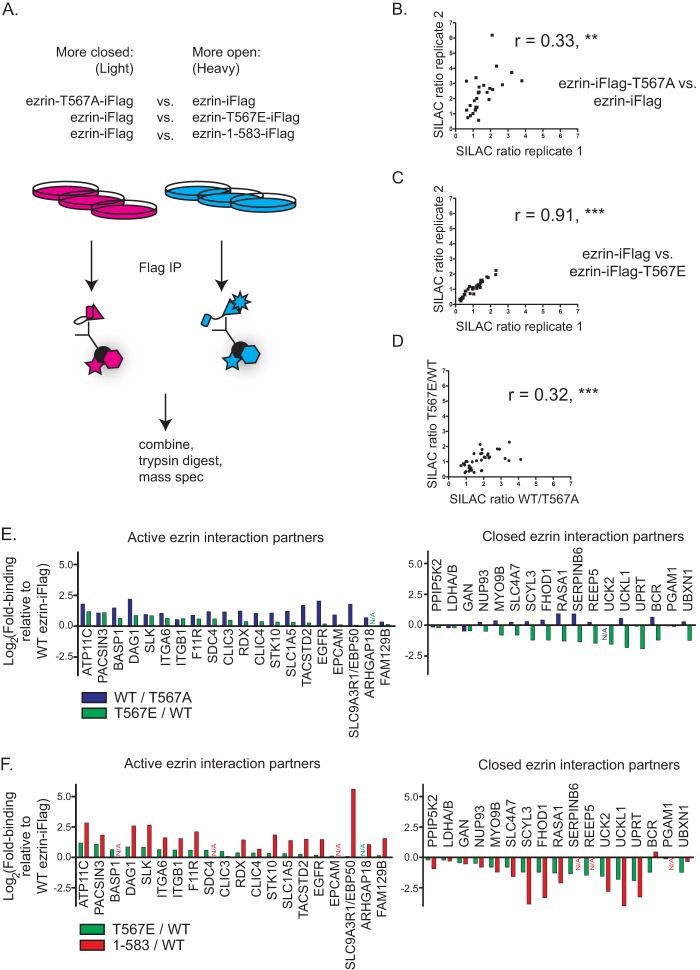
Comparison of phosphorylated versus unphosphorylated ezrin interactome. A, schematic of SILAC experiment. Jeg-3 cells transfected to express the indicated ezrin-iFLAG variant were differentially labeled in SILAC medium and independently subjected to cross-linking immunoprecipitation (IP). The immunoprecipitates were combined, trypsin-digested, and subjected to mass spectrometry (spec), where heavy-to-light ratio was determined for each peptide. B and C, correlation analysis between two biological replicates of the indicated SILAC experiment showing reproducible SILAC quantification. **, p < 0.01; ***, p < 0.001. D, correlation analysis between the average Xpress ratios from indicated SILAC comparisons showing that interactors preferring wild-type ezrin to unphosphorylatable ezrin tend to also prefer constitutively phosphorylated ezrin over wild type. ***, p < 0.001. E, graph of average Xpress ratio in indicated SILAC experiment by ezrin interactor. Based on their preference for ezrin-T567E-iFLAG over wild-type ezrin-iFLAG, proteins were classed as either interactors of active ezrin or closed ezrin. F, graph of average Xpress ratio in T567E versus wild-type as compared with 1–583 versus wild type for each ezrin interactor. For the 1–583 truncation, the preference of active ezrin interactor is further enhanced, although the preference of closed ezrin interactors is further decreased.
We also identified a subclass of proteins that were recovered at approximately similar levels with inactive ezrin-T567A versus wild-type ezrin, yet they were more strongly identified as binding wild-type ezrin versus ezrin-T567E (Fig. 3E, right). We designated these proteins as interactors with closed ezrin. These include the F-actin nucleating formin FHOD-1, the Ras GTPase activating protein RASA1, and SCYL3 (also known as PACE-1 for protein-associating with the carboxyl-terminal domain of ezrin, see Ref. 34). We believe this is the first report of such proteins, which constitute a significant fraction of all ezrin-binding proteins.
Interactome Shows Enhanced Binding to Fully Unmasked Ezrin FERM Domain
The analysis above identified proteins that preferentially associate with phosphorylated versus unphosphorylated ezrin. In the current understanding of ezrin regulation, the phosphorylated form is regarded as fully active. However, it is clear from the results presented in Fig. 1 that EBP50 is much more efficiently recovered with an ezrin construct when the FERM domain is completely unmasked ezrin(1–583)-iFLAG, than with either wild-type ezrin or ezrin-T567E. We therefore wanted to examine in an unbiased manner how general this type of interaction is.
We again performed SILAC to compare proteins recovered from cells expressing ezrin-iFLAG (light) with those recovered from cells expressing ezrin(1–583)-iFLAG (heavy). We found essentially the same set of proteins we did with ezrin-T567E-iFLAG but with a stronger preference for ezrin(1–583)-iFLAG (Fig. 3F). A notable exception was EBP50, being vastly more efficiently recovered with ezrin(1–583)-iFLAG, which is discussed further below.
Biochemical and Localization Validation of the Mass Spectrometry Results
Our proteomics approach identified three classes of ezrin interactors as follows: those that had a preference for phosphorylated ezrin (and ezrin-T567E), EBP50 (SLC9A3R1), which is recovered much more efficiently on unmasked ezrin than with phosphorylated ezrin, and an unexpected group of proteins that bind preferentially to inactive ezrin-T567A. To see if these classes of interactors could be confirmed by analysis of selected candidates, we immunoprecipitated ezrin from cells expressing the different variants and then Western-blotted for specific binding partners.
A close correspondence with the mass spectrometry data was found for EBP50 (SLC9A3R1), DAG1, and FHOD1 (FH1/FH2 domain-containing protein 1, a formin protein, see Fig. 4A). As we do not have reagents to test most of the other identified interactors, we expressed tagged versions in Jeg-3 cells and examined their recovery with co-expressed ezrin-iFLAG variants. For example, TACSTD2-myc shows a similar pattern of recovery as was found with the proteomics analysis (Fig. 4B). We summarize the results of this analysis in Fig. 4C.
FIGURE 4.
Biochemical validation of SILAC experiments. A, stable cell lines expressing empty vector control or indicated ezrin-iFLAG variants were subjected to cross-linking FLAG immunoprecipitation (IP) followed by Western blot (WB) for the indicated interactor. Predicted yield relative to wild-type ezrin-iFLAG is computed from mass spectrometry (spec) data in supplemental Table 1. B, parental Jeg-3 cell line was co-transfected to express empty vector or indicated ezrin-iFLAG variant along with TACSTD2-myc and then subjected to cross-linking FLAG immunoprecipitation and Myc Western blot. Predicted yield relative to wild-type ezrin-iFLAG is computed from mass spectrometry data in supplemental Table 1. C, summary of mass spectrometry interactors confirmed in low throughput methods. Black lines indicate proteins recovered in mass spectrometry only; red lines indicate proteins that were recovered and confirmed through low throughput co-immunoprecipitation followed by Western blotting experiments. Green text highlights proteins expected to bind directly with ERM proteins in vitro by others. D, cells transiently transfected to express empty vector or indicated ezrin-iFLAG variant and co-transfected to express full-length or hyperactivated (ΔDAD) GFP-FHOD-1 were subjected to FLAG immunoprecipitation, and the immunoprecipitates were resolved and Western-blotted for FLAG and GFP. FHOD-1 binds to wild-type or closed ezrin but not truncated and hyperactivated ezrin. The K4N mutant (K253N,K254N,K262N,K263N) of ezrin is completely closed in cells due a mutation in basic residues that bind to membrane PI(4,5)P2 (10, 22).
We were surprised to recover a formin with ezrin, so we examined whether the interaction was affected by its interaction state. Many formins are negatively regulated by a head-to-tail intramolecular interaction involving a region known as the diaphanous association domain (DAD) (40). Equivalent amounts of GFP-FHOD1 or GFP-FHOD1ΔDAD (lacking the DAD domain) were selectively recovered with inactive forms of ezrin, indicating no preference for the activity state of the formin. No interaction was found with hyperactive ezrin(1–583)-iFLAG (Fig. 4D).
Another avenue to test the validity of the proteomic analysis is based on subcellular localization. Because active ezrin is found specifically in microvilli, candidates that show a preference for active ezrin might be expected to be enriched in microvilli. Conversely, those proteins that show a preference for inactive ezrin might be expected to be found predominantly in the cytoplasm, like ezrin-T567A-iFLAG (Fig. 1A).
The proteins DAG1 and CLIC4 have been shown previously to localize in microvilli (32, 41–43), so these serve as positive controls. Indeed, endogenous β-dystroglycan and transiently expressed GFP-CLIC4 both co-localize precisely with ezrin in the apical microvilli of Jeg-3 cells in maximum intensity projections through the entire cell or projections through a cross-section (Fig. 5, A and B). We then tested some of the other candidates, and as predicted, TACSTD2-myc, BASP1-GFP, and SLC1A5-GFP were all preferentially localized in microvilli (Fig. 5, C–E). Expression of proteins showing a preference for inactive ezrin, SCYL3-myc, and GFP-FHOD1 was not enriched in microvilli but was found diffusely in the cytoplasm or enriched on basolateral membranes (Fig. 5, F and G). A summary of all the candidates examined is shown in Fig. 5H. Thus, both the co-immunoprecipitation and localization results strongly support the proteomic data.
Function of Novel Ezrin Interaction Partners
Ezrin is known to interact with many proteins (Table 1), and in this study we have documented yet more. Ezrin is important for the presence of microvilli on Jeg-3 cells (7), so we asked if altering the expression of ezrin-interacting proteins had any effect on microvilli.
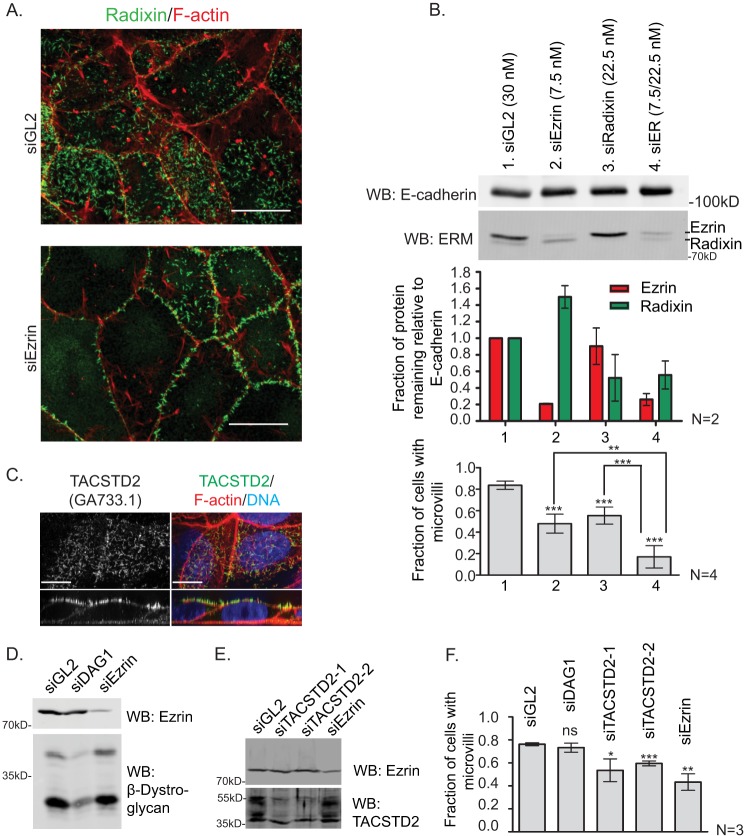
Jeg-3 epithelial cells express both ezrin and radixin. When ezrin levels are reduced 80% by siRNA treatment, about half the cells lose microvilli. The number of cells with microvilli can be further reduced to 20% by simultaneously knocking down radixin, supporting a critical role for ERM proteins in maintaining microvilli in Jeg-3 cells (Fig, 6, A and B). We next tested the effect of knocking down two of our candidates, DAG1 and TACSTD2. Both endogenous proteins were found to localize in microvilli (Figs. 5A and 6C). Although reduction of the levels of β-dystroglycan has no discernible effect, reduction of TACSTD2 reduced the number of cells with microvilli modestly (Fig. 6, D–F).
FIGURE 6.
TACSTD2 suppression by RNA interference results in a moderate loss-of-microvilli phenotype. A, Jeg-3 cells were transfected with nontargeting control siRNA (siGL2) or ezrin siRNA (siEzrin) for 72 h and then stained for F-actin and radixin to reveal a loss of microvilli after to ezrin knockdown. Scale bars, 10 μm. B, Jeg-3 cells were transfected with indicated siRNAs for 72 h, and cell lysates were prepared and Western-blotted (WB) for ezrin and radixin. The remaining protein was quantified. The cells were also fixed and stained for F-actin, β-dystroglycan, and EBP50 (data not shown). The presence of microvilli by any of these markers was scored using a previously described scheme (7, 15, 17, 18, 49). Error bars are standard deviation. p values were computed using a two-tailed t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant. C, endogenous TACSTD2 was detected by immunofluorescence using the GA733.1 antibody. Scale bars, 10 μm. D and E, cells were transfected with indicated siRNA for 72 h, then lysates were prepared and Western-blotted (WB) as indicated. F, Jeg-3 cells transfected as described in B were stained for radixin, and the presence of microvilli was scored as in B. Error bars are standard deviation. p values were computed using a two-tailed t test. *, p < 0.05; **, p < 0.01, ***; p < 0.001; ns, not significant.
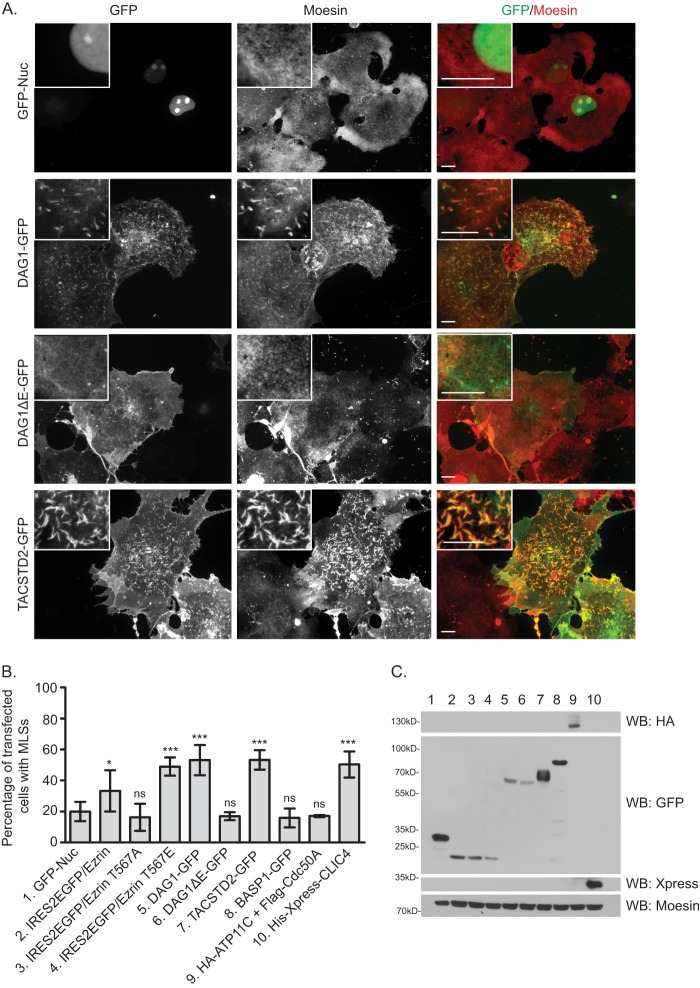
The overexpression of activated ERM proteins or ERM-binding proteins in fibroblastic cells has been shown to cause an increase in the number of microvilli-like structures (MLSs) (20, 44). We therefore examined the effect of overexpression of our candidates, DAG1, TACSTD2, BASP1, and ATP11C (which must be co-overexpressed with its chaperone Cdc50A for plasma membrane enrichment, see Ref. 27), and CLIC4 in COS7 fibroblastic cells. Overexpression of wild-type or activated ezrin but not the more closed ezrin mutant, T567A, caused an elevation in the percentage of cells containing MLSs as indicated by F-actin and moesin staining (data not shown) as expected (quantified in Fig. 7B). Like moesin, ezrin was also enriched in these structures (data not shown). In the case of our candidates, DAG1, TACSTD2, and CLIC4 each caused an elevation in the percentage of cells containing MLSs (Fig. 7, A and B). By contrast, the overexpression of DAG1ΔE, a mutant previously shown to be incapable of binding to ezrin (41), BASP1, or ATP11C (expressed with its chaperone as above) had no effect (Fig. 7B). Western blotting showed that each transfected protein was present at the expected size (Fig. 7C). The combined analysis suggests that some of our interactors are capable of promoting the formation of dorsal microvilli-like structures in fibroblastic cells. Taken together with our knockdown experiments, we propose that ezrin anchorage to the plasma membrane is likely due to the concerted activity of many binding partners.
FIGURE 7.
DAG1, TACSTD2, or CLIC4 overexpression in fibroblastic cells promotes dorsal ERM-containing surface structures. A, COS7 fibroblastic cells were transfected with indicated protein and stained for endogenous moesin (an ERM protein present in these cells). Panels, maximum intensity projections through a field of cells; insets, maximum intensity projections through a small dorsal region of one transfected cell. Bar, 10 μm. B, cells transfected as in A were scored for the presence of MLSs. GFP-Nuc is nuclear-localized GFP, intended as a negative-control. IRES2EGFP allows expression of free GFP as a reporter of untagged ezrin construct transfection. Error bars are standard deviation. p values were computed using a two-tailed t test. *, p < 0.05; ***; p < 0.001; ns, not significant. C, cells expressing the transfected proteins as listed in B were subjected to Western blotting (WB) with indicated antibodies.
DISCUSSION
ERMs function as regulated linking proteins, so to learn more about them, we took a global proteomics view, not only to uncover new ligands and regulators but also to understand the role of conformational change in ERM function.
Our use of cross-linking immunoprecipitation rather than a direct interaction assay, as reported previously (28), biased the analysis toward proteins bound to ezrin in vivo, including transient interactors, rather than those with tenacious in vitro affinity. This revealed many new interactors. We provide three validations of our approach as follows: 1) greater than one-quarter of the interactors were expected based on previous studies, and a large fraction (almost half) of the interactors contained transmembrane helices or membrane-binding domains; 2) where tested, we were able to biochemically validate the mass spectrometry results; 3) many of the interactors, including three novel interactors, accumulate not just on the plasma membrane, but also strongly in microvilli.
Our analysis defined three conformational states of ezrin (closed, active, and fully open) as perceived by the 38 high confidence interactors. One group, designated as active ezrin interactors, bound more to wild-type than unphosphorylatable T567A ezrin, more to phosphomimetic T567E than wild-type ezrin, and more to truncated 1–583 than T567E ezrin. A second group, closed ezrin interactors, bound roughly equally to wild-type and T567A ezrin, less to T567E ezrin, and still less to 1–583 ezrin. Most ERM-binding proteins had a modest (1.2–4.2-fold) preference for truncated 1–583 ezrin over phosphomimetic T567E ezrin. However, EBP50, which binds to the FERM domain more intimately than other known FERM interaction partners (21), was unique in having an extreme (45-fold) preference for 1–583 ezrin over T567E ezrin. This result supports in vitro data that, even when phosphorylated, the tail of ezrin still serves as a potent competitive inhibitor of FERM domain accessibility (16). Moreover, it argues that the influence of the tail on FERM domain binding differs in degree for different interaction partners and highlights the uniqueness of EBP50's interaction with ezrin.
Surprisingly, we identified multiple partners of the inactive state of ezrin. As expected, these are not enriched in microvilli but are found in the cytoplasm. To date, the only known interactor of inactive ezrin is the lipid PI(4,5)P2, which plays a role in the transition to an activated state. Thus, we are looking into the function of this large class of ezrin interactors in ezrin regulation.
The conformational activation model (45) stipulates that interactors binding ezrin on the plasma membrane should bind preferentially to more open forms of ezrin. Consistently, most interactors containing transmembrane helices were active ezrin interactors. But is this merely an effect of the localization of open versus closed ezrin? We can determine this by comparing the binding partners of the two constitutively open forms of ezrin, truncated ezrin(1–583)-iFLAG versus phosphomimetic ezrin-T567E-iFLAG, because both localize identically in a depolarized manner on the plasma membrane (7). Strikingly, all but one transmembrane interactor (8/9) bound truncated ezrin(1–583)-iFLAG to a greater extent than phosphomimetic ezrin-T567E-iFLAG (for instance, see TACSTD2, Fig. 5B). Therefore, we argue that the extent to which an interactor binds to ezrin is not only a function of the localization of the proteins but also ezrin conformation.
This work adds to several recent reports showing that protein complexes involving ERMs are unusually dynamic. Ezrin turns over from microvilli faster than microvilli themselves turnover (14, 15, 46), and EBP50 turns over even faster than ezrin (15). The turnover rate of EBP50 is regulated by PDZ interactions, and impairing these interactions decreases EBP50's turnover rate from microvilli and increases the recovery of co-immunoprecipitated ezrin (15). Moreover, cross-linking was also used to preserve the interactions between the Drosophila orthologs of ERMs, EBP50, and the LOK/SLK kinase (47). Thus, rapid turnover seems to be a general feature of ERM interactions.
Some known interactors were notably absent from our analysis, possibly reflecting cell type-specific expression differences. None of the peptides in our analysis corresponded to CD44, ICAM-2, ICAM-3, syndecan-2, CLIC5, and S100P (Table 1; data not shown). Thus, these are not major ezrin-associated proteins in Jeg-3 cells and may not be expressed. ICAM-1 is present in Jeg-3 cells but appeared in our immunoprecipitates in amounts too low to accurately quantify.
RNA interference knockdown of two of the top candidate ERM-binding transmembrane proteins in Jeg-3 epithelial cells failed to yield a striking phenotype, although TACSTD2 suppression partially reduced the number of microvilli-bearing cells in Jeg-3 cells (Fig. 6F), and dominant-negative β-dystroglycan (DAG1ΔE) inhibited the formation of active-Cdc42-induced filopodia in a myoblast cell line (41). Conversely, overexpression of TACSTD2, DAG1, or CLIC4 caused the enumeration of MLSs in overexpressing fibroblastic cells, which normally produce dorsal ERM-containing protrusions only rarely (Fig. 7). The lack of overt phenotype in the epithelial cell line can be interpreted in two ways as follows: 1) transmembrane ERM-binding proteins are not required as ERM tethering or clustering agents as has been proposed (20); 2) ERM binding to numerous transmembrane proteins coordinately regulates their tethering or clustering. In support of the second possibility, ERM-binding motifs are minimal and present in many transmembrane proteins, and the proteins containing them have evolved entirely separate functions (e.g. β-dystroglycan functions as a receptor for the extracellular matrix component laminin), making it possible that ERMs evolved promiscuous affinity to a wide range of available ligands. Also in support of this model, our calculations suggest that ezrin is stoichiometrically present in a similar abundance to actin (data not shown), suggesting that no one membrane protein is in sufficient abundance to bind all the ezrin molecules, such that many transmembrane proteins would be needed to serve such a function. Thus, our current efforts are to ask whether simultaneous perturbation of multiple transmembrane ezrin interactors described here affects ezrin function.
Acknowledgments
We are grateful to Dr. A. Hanono for the cross-linking immunoprecipitation protocol. We also thank Drs. D. Garbett, D. Lalonde, and C. Sauvanet for critically reading this manuscript. Dr. M. Berryman (Ohio University), Dr. R. Thorne (University of Newcastle), Dr. H. W. Shin (University of Kyoto), and Dr. U. Naik (University of Delaware) generously provided plasmids for this study. The CPCT-Ezrin-1 and CPCT-Moesin-1 antibodies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD, National Institutes of Health, and maintained by the Department of Biology, University of Iowa, Iowa City.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-036652 (to A. B.).

This article contains supplemental Table S1.
- SILAC
- stable isotope labeling in cell culture
- MLS
- microvilli-like structure
- DSP
- dithiobis(succinimidyl propionate)
- PI(4,5)P2
- phosphatidylinositol 4,5-biphosphate.
REFERENCES
- 1. Bretscher A. (1983) Purification of an 80,000-dalton protein that is a component of the isolated microvillus cytoskeleton, and its localization in nonmuscle cells. J. Cell Biol. 97, 425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fehon R. G., McClatchey A. I., Bretscher A. (2010) Organizing the cell cortex: the role of ERM proteins. Nat. Rev. Mol. Cell Biol. 11, 276–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saotome I., Curto M., McClatchey A. I. (2004) Ezrin is essential for epithelial organization and villus morphogenesis in the developing intestine. Dev. Cell 6, 855–864 [DOI] [PubMed] [Google Scholar]
- 4. Bonilha V. L., Rayborn M. E., Saotome I., McClatchey A. I., Hollyfield J. G. (2006) Microvilli defects in retinas of ezrin knockout mice. Exp. Eye Res. 82, 720–729 [DOI] [PubMed] [Google Scholar]
- 5. Karagiosis S. A., Ready D. F. (2004) Moesin contributes an essential structural role in Drosophila photoreceptor morphogenesis. Development. 131, 725–732 [DOI] [PubMed] [Google Scholar]
- 6. Bonilha V. L., Finnemann S. C., Rodriguez-Boulan E. (1999) Ezrin promotes morphogenesis of apical microvilli and basal infoldings in retinal pigment epithelium. J. Cell Biol. 147, 1533–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Viswanatha R., Ohouo P. Y., Smolka M. B., Bretscher A. (2012) Local phosphocycling mediated by LOK/SLK restricts ezrin function to the apical aspect of epithelial cells. J. Cell Biol. 199, 969–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fievet B. T., Gautreau A., Roy C., Del Maestro L., Mangeat P., Louvard D., Arpin M. (2004) Phosphoinositide binding and phosphorylation act sequentially in the activation mechanism of ezrin. J. Cell Biol. 164, 653–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hirao M., Sato N., Kondo T., Yonemura S., Monden M., Sasaki T., Takai Y., Tsukita S., Tsukita S. (1996) Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane association: possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J. Cell Biol. 135, 37–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Niggli V., Andréoli C., Roy C., Mangeat P. (1995) Identification of a phosphatidylinositol-4,5-bisphosphate-binding domain in the N-terminal region of ezrin. FEBS Lett. 376, 172–176 [DOI] [PubMed] [Google Scholar]
- 11. Matsui T., Yonemura S., Tsukita S., Tsukita S. (1999) Activation of ERM proteins in vivo by Rho involves phosphatidylinositol 4-phosphate 5-kinase and not ROCK kinases. Curr. Biol. 9, 1259–1262 [DOI] [PubMed] [Google Scholar]
- 12. Yonemura S., Matsui T., Tsukita S., Tsukita S. (2002) Rho-dependent and -independent activation mechanisms of ezrin/radixin/moesin proteins: an essential role for polyphosphoinositides in vivo. J. Cell Sci. 115, 2569–2580 [DOI] [PubMed] [Google Scholar]
- 13. Gautreau A., Louvard D., Arpin M. (2000) Morphogenic effects of ezrin require a phosphorylation-induced transition from oligomers to monomers at the plasma membrane. J. Cell Biol. 150, 193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coscoy S., Waharte F., Gautreau A., Martin M., Louvard D., Mangeat P., Arpin M., Amblard F. (2002) Molecular analysis of microscopic ezrin dynamics by two-photon FRAP. Proc. Natl. Acad. Sci. U.S.A. 99, 12813–12818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garbett D., Bretscher A. (2012) PDZ interactions regulate rapid turnover of the scaffolding protein EBP50 in microvilli. J. Cell Biol. 198, 195–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chambers D. N., Bretscher A. (2005) Ezrin mutants affecting dimerization and activation. Biochemistry 44, 3926–3932 [DOI] [PubMed] [Google Scholar]
- 17. Hanono A., Garbett D., Reczek D., Chambers D. N., Bretscher A. (2006) EPI64 regulates microvillar subdomains and structure. J. Cell Biol. 175, 803–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garbett D., LaLonde D. P., Bretscher A. (2010) The scaffolding protein EBP50 regulates microvillar assembly in a phosphorylation-dependent manner. J. Cell Biol. 191, 397–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morales F. C., Takahashi Y., Kreimann E. L., Georgescu M. M. (2004) Ezrin-radixin-moesin (ERM)-binding phosphoprotein 50 organizes ERM proteins at the apical membrane of polarized epithelia. Proc. Natl. Acad. Sci. U.S.A. 101, 17705–17710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yonemura S., Tsukita S., Tsukita S. (1999) Direct involvement of ezrin/radixin/moesin (ERM)-binding membrane proteins in the organization of microvilli in collaboration with activated ERM proteins. J. Cell Biol. 145, 1497–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Finnerty C. M., Chambers D., Ingraffea J., Faber H. R., Karplus P. A., Bretscher A. (2004) The EBP50-moesin interaction involves a binding site regulated by direct masking on the FERM domain. J. Cell Sci. 117, 1547–1552 [DOI] [PubMed] [Google Scholar]
- 22. Barret C., Roy C., Montcourrier P., Mangeat P., Niggli V. (2000) Mutagenesis of the phosphatidylinositol 4,5-bisphosphate (PIP2) binding site in the NH2-terminal domain of ezrin correlates with its altered cellular distribution. J. Cell Biol. 151, 1067–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ben-Aissa K., Patino-Lopez G., Belkina N. V., Maniti O., Rosales T., Hao J. J., Kruhlak M. J., Knutson J. R., Picart C., Shaw S. (2012) Activation of moesin, a protein that links actin cytoskeleton to the plasma membrane, occurs by phosphatidylinositol 4,5-bisphosphate (PIP2) binding sequentially to two sites and releasing an autoinhibitory linker. J. Biol. Chem. 287, 16311–16323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hamada K., Shimizu T., Matsui T., Tsukita S., Hakoshima T. (2000) Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain. EMBO J. 19, 4449–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hamada K., Shimizu T., Yonemura S., Tsukita S., Tsukita S., Hakoshima T. (2003) Structural basis of adhesion-molecule recognition by ERM proteins revealed by the crystal structure of the radixin-ICAM-2 complex. EMBO J. 22, 502–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Terawaki S., Maesaki R., Hakoshima T. (2006) Structural basis for NHERF recognition by ERM proteins. Structure 14, 777–789 [DOI] [PubMed] [Google Scholar]
- 27. Takatsu H., Baba K., Shima T., Umino H., Kato U., Umeda M., Nakayama K., Shin H. W. (2011) ATP9B, a P4-ATPase (a putative aminophospholipid translocase), localizes to the trans-Golgi network in a CDC50 protein-independent manner. J. Biol. Chem. 286, 38159–38167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reczek D., Berryman M., Bretscher A. (1997) Identification of EBP50: A PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. J. Cell Biol. 139, 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smolka M. B., Albuquerque C. P., Chen S. H., Zhou H. (2007) Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. U.S.A. 104, 10364–10369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reczek D., Bretscher A. (1998) The carboxyl-terminal region of EBP50 binds to a site in the amino-terminal domain of ezrin that is masked in the dormant molecule. J. Biol. Chem. 273, 18452–18458 [DOI] [PubMed] [Google Scholar]
- 31. Gary R., Bretscher A. (1993) Heterotypic and homotypic associations between ezrin and moesin, two putative membrane-cytoskeletal linking proteins. Proc. Natl. Acad. Sci. U.S.A. 90, 10846–10850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Berryman M., Bretscher A. (2000) Identification of a novel member of the chloride intracellular channel gene family (CLIC5) that associates with the actin cytoskeleton of placental microvilli. Mol. Biol. Cell 11, 1509–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Neisch A. L., Formstecher E., Fehon R. G. (2013) Conundrum, an ARHGAP18 orthologue, regulates RhoA and proliferation through interactions with moesin. Mol. Biol. Cell 24, 1420–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sullivan A., Uff C. R., Isacke C. M., Thorne R. F. (2003) PACE-1, a novel protein that interacts with the C-terminal domain of ezrin. Exp. Cell Res. 284, 224–238 [DOI] [PubMed] [Google Scholar]
- 35. Granés F., Urena J. M., Rocamora N., Vilaró S. (2000) Ezrin links syndecan-2 to the cytoskeleton. J. Cell Sci. 113, 1267–1276 [DOI] [PubMed] [Google Scholar]
- 36. Granés F., Berndt C., Roy C., Mangeat P., Reina M., Vilaró S. (2003) Identification of a novel ezrin-binding site in syndecan-2 cytoplasmic domain. FEBS Lett. 547, 212–216 [DOI] [PubMed] [Google Scholar]
- 37. Hipfner D. R., Keller N., Cohen S. M. (2004) Slik Sterile-20 kinase regulates moesin activity to promote epithelial integrity during tissue growth. Genes Dev. 18, 2243–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hughes S. C., Fehon R. G. (2006) Phosphorylation and activity of the tumor suppressor merlin and the ERM protein moesin are coordinately regulated by the Slik kinase. J. Cell Biol. 175, 305–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Belkina N. V., Liu Y., Hao J. J., Karasuyama H., Shaw S. (2009) LOK is a major ERM kinase in resting lymphocytes and regulates cytoskeletal rearrangement through ERM phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 106, 4707–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Alberts A. S. (2001) Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J. Biol. Chem. 276, 2824–2830 [DOI] [PubMed] [Google Scholar]
- 41. Spence H. J., Chen Y. J., Batchelor C. L., Higginson J. R., Suila H., Carpen O., Winder S. J. (2004) Ezrin-dependent regulation of the actin cytoskeleton by β-dystroglycan. Hum. Mol. Genet. 13, 1657–1668 [DOI] [PubMed] [Google Scholar]
- 42. Chuang J. Z., Chou S. Y., Sung C. H. (2010) Chloride intracellular channel 4 is critical for the epithelial morphogenesis of RPE cells and retinal attachment. Mol. Biol. Cell 21, 3017–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Berryman M. A., Goldenring J. R. (2003) CLIC4 is enriched at cell-cell junctions and colocalizes with AKAP350 at the centrosome and midbody of cultured mammalian cells. Cell Motil. Cytoskeleton 56, 159–172 [DOI] [PubMed] [Google Scholar]
- 44. Oshiro N., Fukata Y., Kaibuchi K. (1998) Phosphorylation of moesin by rho-associated kinase (Rho-kinase) plays a crucial role in the formation of microvilli-like structures. J. Biol. Chem. 273, 34663–34666 [DOI] [PubMed] [Google Scholar]
- 45. Bretscher A., Reczek D., Berryman M. (1997) Ezrin: a protein requiring conformational activation to link microfilaments to the plasma membrane in the assembly of cell surface structures. J. Cell Sci. 110, 3011–3018 [DOI] [PubMed] [Google Scholar]
- 46. Gorelik J., Shevchuk A. I., Frolenkov G. I., Diakonov I. A., Lab M. J., Kros C. J., Richardson G. P., Vodyanoy I., Edwards C. R., Klenerman D., Korchev Y. E. (2003) Dynamic assembly of surface structures in living cells. Proc. Natl. Acad. Sci. U.S.A. 100, 5819–5822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hughes S. C., Formstecher E., Fehon R. G. (2010) Sip1, the Drosophila orthologue of EBP50/NHERF1, functions with the sterile 20 family kinase Slik to regulate moesin activity. J. Cell Sci. 123, 1099–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Batchelor C. L., Higginson J. R., Chen Y. J., Vanni C., Eva A., Winder S. J. (2007) Recruitment of Dbl by ezrin and dystroglycan drives membrane proximal Cdc42 activation and filopodia formation. Cell Cycle 6, 353–363 [DOI] [PubMed] [Google Scholar]
- 49. LaLonde D. P., Garbett D., Bretscher A. (2010) A regulated complex of the scaffolding proteins PDZK1 and EBP50 with ezrin contribute to microvillar organization. Mol. Biol. Cell 21, 1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang Z., Schey K. L. (2011) Aquaporin-0 interacts with the FERM domain of ezrin/radixin/moesin proteins in the ocular lens. Invest. Ophthalmol. Vis. Sci. 52, 5079–5087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pilot F., Philippe J. M., Lemmers C., Lecuit T. (2006) Spatial control of actin organization at adherens junctions by a synaptotagmin-like protein Btsz. Nature 442, 580–584 [DOI] [PubMed] [Google Scholar]
- 52. Luo Y., Zheng C., Zhang J., Lu D., Zhuang J., Xing S., Feng J., Yang D., Yan X. (2012) Recognition of CD146 as an ERM-binding protein offers novel mechanisms for melanoma cell migration. Oncogene 31, 306–321 [DOI] [PubMed] [Google Scholar]
- 53. Yonemura S., Hirao M., Doi Y., Takahashi N., Kondo T., Tsukita S., Tsukita S. (1998) Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J. Cell Biol. 140, 885–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cannon J. L., Mody P. D., Blaine K. M., Chen E. J., Nelson A. D., Sayles L. J., Moore T. V., Clay B. S., Dulin N. O., Shilling R. A., Burkhardt J. K., Sperling A. I. (2011) CD43 interaction with ezrin-radixin-moesin (ERM) proteins regulates T-cell trafficking and CD43 phosphorylation. Mol. Biol. Cell 22, 954–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ilani T., Khanna C., Zhou M., Veenstra T. D., Bretscher A. (2007) Immune synapse formation requires ZAP-70 recruitment by ezrin and CD43 removal by moesin. J. Cell Biol. 179, 733–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Serrador J. M., Nieto M., Alonso-Lebrero J. L., del Pozo M. A., Calvo J., Furthmayr H., Schwartz-Albiez R., Lozano F., González-Amaro R., Sánchez-Mateos P., Sánchez-Madrid F. (1998) CD43 interacts with moesin and ezrin and regulates its redistribution to the uropods of T lymphocytes at the cell-cell contacts. Blood 91, 4632–4644 [PubMed] [Google Scholar]
- 57. Legg J. W., Isacke C. M. (1998) Identification and functional analysis of the ezrin-binding site in the hyaluronan receptor, CD44. Curr. Biol. 8, 705–708 [DOI] [PubMed] [Google Scholar]
- 58. Legg J. W., Lewis C. A., Parsons M., Ng T., Isacke C. M. (2002) A novel PKC-regulated mechanism controls CD44 ezrin association and directional cell motility. Nat. Cell Biol. 4, 399–407 [DOI] [PubMed] [Google Scholar]
- 59. Li Y., Harada T., Juang Y. T., Kyttaris V. C., Wang Y., Zidanic M., Tung K., Tsokos G. C. (2007) Phosphorylated ERM is responsible for increased T cell polarization, adhesion, and migration in patients with systemic lupus erythematosus. J. Immunol. 178, 1938–1947 [DOI] [PubMed] [Google Scholar]
- 60. Mori T., Kitano K., Terawaki S., Maesaki R., Fukami Y., Hakoshima T. (2008) Structural basis for CD44 recognition by ERM proteins. J. Biol. Chem. 283, 29602–29612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hao J. J., Liu Y., Kruhlak M., Debell K. E., Rellahan B. L., Shaw S. (2009) Phospholipase C-mediated hydrolysis of PIP2 releases ERM proteins from lymphocyte membrane. J. Cell Biol. 184, 451–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tsukita S., Oishi K., Sato N., Sagara J., Kawai A., Tsukita S. (1994) ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J. Cell Biol. 126, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Takahashi K., Sasaki T., Mammoto A., Hotta I., Takaishi K., Imamura H., Nakano K., Kodama A., Takai Y. (1998) Interaction of radixin with Rho small G protein GDP/GTP exchange protein Dbl. Oncogene 16, 3279–3284 [DOI] [PubMed] [Google Scholar]
- 64. Tran Quang C., Gautreau A., Arpin M., Treisman R. (2000) Ezrin function is required for ROCK-mediated fibroblast transformation by the Net and Dbl oncogenes. EMBO J. 19, 4565–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Prag S., Parsons M., Keppler M. D., Ameer-Beg S. M., Barber P., Hunt J., Beavil A. J., Calvert R., Arpin M., Vojnovic B., Ng T. (2007) Activated ezrin promotes cell migration through recruitment of the GEF Dbl to lipid rafts and preferential downstream activation of Cdc42. Mol. Biol. Cell 18, 2935–2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yang H. S., Hinds P. W. (2006) Phosphorylation of ezrin by cyclin-dependent kinase 5 induces the release of Rho GDP dissociation inhibitor to inhibit Rac1 activity in senescent cells. Cancer Res. 66, 2708–2715 [DOI] [PubMed] [Google Scholar]
- 67. Ognibene M., Vanni C., Segalerba D., Mancini P., Merello E., Torrisi M. R., Bosco M. C., Varesio L., Eva A. (2011) The tumor suppressor hamartin enhances Dbl protein transforming activity through interaction with ezrin. J. Biol. Chem. 286, 29973–29983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Martín M., Simon-Assmann P., Kedinger M., Martin M., Mangeat P., Real F. X., Fabre M. (2006) DCC regulates cell adhesion in human colon cancer derived HT-29 cells and associates with ezrin. Eur. J. Cell Biol. 85, 769–783 [DOI] [PubMed] [Google Scholar]
- 69. Antoine-Bertrand J., Ghogha A., Luangrath V., Bedford F. K., Lamarche-Vane N. (2011) The activation of ezrin-radixin-moesin proteins is regulated by netrin-1 through Src kinase and RhoA/Rho kinase activities and mediates netrin-1-induced axon outgrowth. Mol. Biol. Cell 22, 3734–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lue R. A., Brandin E., Chan E. P., Branton D. (1996) Two independent domains of hDlg are sufficient for subcellular targeting: the PDZ1–2 conformational unit and an alternatively spliced domain. J. Cell Biol. 135, 1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lasserre R., Charrin S., Cuche C., Danckaert A., Thoulouze M. I., de Chaumont F., Duong T., Perrault N., Varin-Blank N., Olivo-Marin J. C., Etienne-Manneville S., Arpin M., Di Bartolo V., Alcover A. (2010) Ezrin tunes T-cell activation by controlling Dlg1 and microtubule positioning at the immunological synapse. EMBO J. 29, 2301–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. LaLonde D. P., Bretscher A. (2009) The scaffold protein PDZK1 undergoes a head-to-tail intramolecular association that negatively regulates its interaction with EBP50. Biochemistry 48, 2261–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zwaenepoel I., Naba A., Da Cunha M. M., Del Maestro L., Formstecher E., Louvard D., Arpin M. (2012) Ezrin regulates microvillus morphogenesis by promoting distinct activities of Eps8 proteins. Mol. Biol. Cell 23, 1080–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Naba A., Reverdy C., Louvard D., Arpin M. (2008) Spatial recruitment and activation of the Fes kinase by ezrin promotes HGF-induced cell scattering. EMBO J. 27, 38–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Helander T. S., Carpén O., Turunen O., Kovanen P. E., Vaheri A., Timonen T. (1996) ICAM-2 redistributed by ezrin as a target for killer cells. Nature 382, 265–268 [DOI] [PubMed] [Google Scholar]
- 76. Heiska L., Alfthan K., Grönholm M., Vilja P., Vaheri A., Carpén O. (1998) Association of ezrin with intercellular adhesion molecule-1 and -2 (ICAM-1 and ICAM-2). Regulation by phosphatidylinositol 4, 5-bisphosphate. J. Biol. Chem. 273, 21893–21900 [DOI] [PubMed] [Google Scholar]
- 77. Serrador J. M., Alonso-Lebrero J. L., del Pozo M. A., Furthmayr H., Schwartz-Albiez R., Calvo J., Lozano F., Sánchez-Madrid F. (1997) Moesin interacts with the cytoplasmic region of intercellular adhesion molecule-3 and is redistributed to the uropod of T lymphocytes during cell polarization. J. Cell Biol. 138, 1409–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Alonso-Lebrero J. L., Serrador J. M., Domínguez-Jiménez C., Barreiro O., Luque A., del Pozo M. A., Snapp K., Kansas G., Schwartz-Albiez R., Furthmayr H., Lozano F., Sánchez-Madrid F. (2000) Polarization and interaction of adhesion molecules P-selectin glycoprotein ligand 1 and intercellular adhesion molecule 3 with moesin and ezrin in myeloid cells. Blood 95, 2413–2419 [PubMed] [Google Scholar]
- 79. Oh H. M., Lee S., Na B. R., Wee H., Kim S. H., Choi S. C., Lee K. M., Jun C. D. (2007) RKIKK motif in the intracellular domain is critical for spatial and dynamic organization of ICAM-1: functional implication for the leukocyte adhesion and transmigration. Mol. Biol. Cell 18, 2322–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dickson T. C., Mintz C. D., Benson D. L., Salton S. R. (2002) Functional binding interaction identified between the axonal CAM L1 and members of the ERM family. J. Cell Biol. 157, 1105–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sakurai T., Gil O. D., Whittard J. D., Gazdoiu M., Joseph T., Wu J., Waksman A., Benson D. L., Salton S. R., Felsenfeld D. P. (2008) Interactions between the L1 cell adhesion molecule and ezrin support traction-force generation and can be regulated by tyrosine phosphorylation. J. Neurosci. Res. 86, 2602–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Matsui K., Parameswaran N., Bagheri N., Willard B., Gupta N. (2011) Proteomics analysis of the ezrin interactome in B cells reveals a novel association with Myo18aα. J. Proteome Res. 10, 3983–3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Denker S. P., Huang D. C., Orlowski J., Furthmayr H., Barber D. L. (2000) Direct binding of the Na-H exchanger NHE1 to ERM proteins regulates the cortical cytoskeleton and cell shape independently of H+ translocation. Mol. Cell 6, 1425–1436 [DOI] [PubMed] [Google Scholar]
- 84. Denker S. P., Barber D. L. (2002) Cell migration requires both ion translocation and cytoskeletal anchoring by the Na-H exchanger NHE1. J. Cell Biol. 159, 1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cha B., Tse M., Yun C., Kovbasnjuk O., Mohan S., Hubbard A., Arpin M., Donowitz M. (2006) The NHE3 juxtamembrane cytoplasmic domain directly binds ezrin: dual role in NHE3 trafficking and mobility in the brush border. Mol. Biol. Cell 17, 2661–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mykkänen O. M., Grönholm M., Rönty M., Lalowski M., Salmikangas P., Suila H., Carpén O. (2001) Characterization of human palladin, a microfilament-associated protein. Mol. Biol. Cell 12, 3060–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Rachlin A. S., Otey C. A. (2006) Identification of palladin isoforms and characterization of an isoform-specific interaction between Lasp-1 and palladin. J. Cell Sci. 119, 995–1004 [DOI] [PubMed] [Google Scholar]
- 88. Henning M. S., Stiedl P., Barry D. S., McMahon R., Morham S. G., Walsh D., Naghavi M. H. (2011) PDZD8 is a novel moesin-interacting cytoskeletal regulatory protein that suppresses infection by herpes simplex virus type 1. Virology 415, 114–121 [DOI] [PubMed] [Google Scholar]
- 89. D'Angelo R., Aresta S., Blangy A., Del Maestro L., Louvard D., Arpin M. (2007) Interaction of ezrin with the novel guanine nucleotide exchange factor PLEKHG6 promotes RhoG-dependent apical cytoskeleton rearrangements in epithelial cells. Mol. Biol. Cell 18, 4780–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Orlando R. A., Takeda T., Zak B., Schmieder S., Benoit V. M., McQuistan T., Furthmayr H., Farquhar M. G. (2001) The glomerular epithelial cell anti-adhesin podocalyxin associates with the actin cytoskeleton through interactions with ezrin. J. Am. Soc. Nephrol. 12, 1589–1598 [DOI] [PubMed] [Google Scholar]
- 91. Snapp K. R., Heitzig C. E., Kansas G. S. (2002) Attachment of the PSGL-1 cytoplasmic domain to the actin cytoskeleton is essential for leukocyte rolling on P-selectin. Blood 99, 4494–4502 [DOI] [PubMed] [Google Scholar]
- 92. Serrador J. M., Urzainqui A., Alonso-Lebrero J. L., Cabrero J. R., Montoya M. C., Vicente-Manzanares M., Yáñez-Mó M., Sánchez-Madrid F. (2002) A juxta-membrane amino acid sequence of P-selectin glycoprotein ligand-1 is involved in moesin binding and ezrin/radixin/moesin-directed targeting at the trailing edge of migrating lymphocytes. Eur. J. Immunol. 32, 1560–1566 [DOI] [PubMed] [Google Scholar]
- 93. Takai Y., Kitano K., Terawaki S., Maesaki R., Hakoshima T. (2007) Structural basis of PSGL-1 binding to ERM proteins. Genes Cells 12, 1329–1338 [DOI] [PubMed] [Google Scholar]
- 94. Domínguez-Luis M., Lamana A., Vazquez J., García-Navas R., Mollinedo F., Sánchez-Madrid F., Díaz-González F., Urzainqui A. (2011) The metalloprotease ADAM8 is associated with and regulates the function of the adhesion receptor PSGL-1 through ERM proteins. Eur. J. Immunol. 41, 3436–3442 [DOI] [PubMed] [Google Scholar]
- 95. Spertini C., Baïsse B., Spertini O. (2012) Ezrin-radixin-moesin-binding sequence of PSGL-1 glycoprotein regulates leukocyte rolling on selectins and activation of extracellular signal-regulated kinases. J. Biol. Chem. 287, 10693–10702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ramel D., Wang X., Laflamme C., Montell D. J., Emery G. (2013) Rab11 regulates cell-cell communication during collective cell movements. Nat. Cell Biol. 15, 317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Sperka T., Geissler K. J., Merkel U., Scholl I., Rubio I., Herrlich P., Morrison H. L. (2011) Activation of Ras requires the ERM-dependent link of actin to the plasma membrane. PLoS One 6, e27511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Takahashi K., Sasaki T., Mammoto A., Takaishi K., Kameyama T., Tsukita S., Takai Y. (1997) Direct interaction of the Rho GDP dissociation inhibitor with ezrin/radixin/moesin initiates the activation of the Rho small G protein. J. Biol. Chem. 272, 23371–23375 [DOI] [PubMed] [Google Scholar]
- 99. Koltzscher M., Neumann C., König S., Gerke V. (2003) Ca2+-dependent binding and activation of dormant ezrin by dimeric S100P. Mol. Biol. Cell 14, 2372–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Austermann J., Nazmi A. R., Müller-Tidow C., Gerke V. (2008) Characterization of the Ca2+-regulated ezrin-S100P interaction and its role in tumor cell migration. J. Biol. Chem. 283, 29331–29340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Bonilha V. L., Rodriguez-Boulan E. (2001) Polarity and developmental regulation of two PDZ proteins in the retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 42, 3274–3282 [PubMed] [Google Scholar]
- 102. Chirivino D., Del Maestro L., Formstecher E., Hupé P., Raposo G., Louvard D., Arpin M. (2011) The ERM proteins interact with the HOPS complex to regulate the maturation of endosomes. Mol. Biol. Cell 22, 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Jin C., Ge L., Ding X., Chen Y., Zhu H., Ward T., Wu F., Cao X., Wang Q., Yao X. (2006) PKA-mediated protein phosphorylation regulates ezrin-WWOX interaction. Biochem. Biophys. Res. Commun. 341, 784–791 [DOI] [PubMed] [Google Scholar]
- 104. Zaarour R. F., Chirivino D., Del Maestro L., Daviet L., Atfi A., Louvard D., Arpin M. (2012) Ezrin ubiquitylation by the E3 ubiquitin ligase, WWP1, and consequent regulation of hepatocyte growth factor receptor activity. PLoS One 7, e37490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wan X., Kim S. Y., Guenther L. M., Mendoza A., Briggs J., Yeung C., Currier D., Zhang H., Mackall C., Li W. J., Tuan R. S., Deyrup A. T., Khanna C., Helman L. (2009) β4 integrin promotes osteosarcoma metastasis and interacts with ezrin. Oncogene 28, 3401–3411 [DOI] [PMC free article] [PubMed] [Google Scholar]