Background: Yersinia enterocolitica yopD, lcrH, and yscM1 control the expression of yopQ encoding a secretion substrate.
Results: YopD associates with 30 S ribosomal particles, and YopD, LcrH, and YscM1 block yopQ mRNA translation.
Conclusion: In response to environmental signals, Yersinia prevent yopQ expression by blocking the translation of its transcripts.
Significance: These results demonstrate translational regulation for the Yersinia type III secretion pathway.
Keywords: Bacterial Pathogenesis, mRNA, Ribosomes, Secretion, Translation Regulation
Abstract
Yersinia enterocolitica type III secretion machines transport YopQ and other Yop effectors into host immune cells. YopD and its chaperone LcrH are essential components of the Yersinia type III pathway, enabling effector translocation into host cells. YopD, LcrH, and YscM1 also regulate yop expression post-transcriptionally in response to environmental signals; however, the molecular mechanisms for this regulation and Yop secretion are unknown. We show here that YopD associates with 30 S ribosomal particles in a manner requiring LcrH. When added to ribosomes, YopD, LcrH, and YscM1 block the translation of yopQ mRNA. We propose a model whereby LcrH-dependent association of YopD with 30 S ribosomal particles enables YscM1 to block yopQ translation unless type III machines are induced to secrete the effector.
Introduction
Yersinia enterocolitica and other pathogenic Yersinia species (Yersinia pestis and Yersinia pseudotuberculosis) require the 70-kb virulence plasmid-encoded type III secretion pathway to cause disease (1). During infection, Yersinia deploy type III secretion machines to inject Yop effectors (YopE, YopH, YopM, YopO, YopP, YopQ, and YopT) into host immune cells (2, 3). The type III pathway is assembled from 25 Ysc (Yop secretion) factors that transport secretion substrates across the bacterial envelope and through needle complexes composed of YscF needle protein, LcrV cap protein, and YopD translocator (4, 5). In addition to LcrV and YopD, Yersinia translocation of effectors also requires YopB, which assembles with LcrV and YopD to form a membrane pore complex for effector translocation into host cells (6, 7).
Yersinia type III secretion is regulated in response to environmental signals; specifically a low calcium signal that bacteria perceive as assembled type III machines encounter either the cytoplasm of host cells (<1 μm Ca2+) or extracellular body fluids (>1.2 mm Ca2+) (8, 9). Under high calcium conditions (>70 μm Ca2+), Y. enterocolitica assembles type III machines with needle complexes but without active translocation pores (10). Although Yop effector genes are transcribed under these conditions, gene expression is blocked at a post-transcriptional step (11). This regulatory mechanism requires two mRNA sequence elements, AUAAA sequences in the 5′-UTR and coding sequence of yop mRNA as well as AU-rich elements immediately adjacent to the AUG start codon (12, 13). Using genetic approaches, yopD, lcrH, and yscM1/yscM2 were identified as factors required for post-transcriptional control of yop effector expression under high calcium conditions (11, 12). Nevertheless, earlier work left the mechanism by which Yersinia control the expression yop effector genes unresolved.
Yersinia Yop effectors lack canonical signal peptide or amino acid sequence motifs that mark these polypeptides as substrates for the type III secretion pathway (14). Earlier work identified features of primary amino acid sequence, for example the attribute of some effectors to bind cytoplasmic chaperones, as contributing to their entry into the type III secretion pathway (15–19). Other studies characterized features of 5′ mRNA coding sequence that were shown to be essential for substrate recognition (20–22).
Genetic approaches designed to identify factors involved in the control of Yop effector gene expression, Yop secretion, or machine assembly identified three classes of genes. Class I genes (yopN-sycN-tyeA-yscB) control the secretion of Yop effectors in the absence of the low-calcium signal (23–25). Class II genes (yopD, lcrH, yscM1, yscM2) regulate the expression of effector Yop genes, whereas class III genes encode components of the type III secretion machine including YscF (26–28). For example, wild-type Y. enterocolitica do not express the yopQ gene unless the bacteria are provided with a low calcium signal, which couples yopQ expression and type III secretion of YopQ product (21). Class III mutants fail to express yopQ, irrespective of the presence or absence of the low calcium signal (11). Class II mutants express yopQ when grown under high calcium conditions; however, YopQ secretion is still regulated by a calcium signal (11). Class I mutants, however, express yopQ and secrete YopQ polypeptide under both low and high calcium conditions.
Mutants with defects in both class II and class III genes express yopQ, yet these variants are unable to promote type III secretion (11). Thus, class II regulation of Yop effector gene expression is epistatic over class III genes, in agreement with a general model whereby class II regulation of Yop effector expression precedes the type III secretion of Yops, for example YopQ. The genetic relationships between class I and class II or III genes have not yet been revealed. Here we investigated the epistatic relationships of class I, II, and III genes and sought to identify the mechanism whereby class II gene products regulate yopQ expression.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids
Escherichia coli strain BL21(DE3) (29), wild-type Y. enterocolitica W22703 (30), and its yopN, yopD, yscN, yopN/yopD, and yopD/yscN mutants have been described elsewhere (11, 31). Plasmids pyopQ, plcrH, pyopD, pGST-YscM1, and pKR6 have been reported earlier (11, 31). To generate pT7-yopQ (pKR10), yopQ was amplified using primers YopQUTR1 (5′-AATCTAGATCATATAAACAATGAGCAACGT-3′) and YopQCod2 (5′-AAGGATCCTCATCCCATAATACATTTTTGAT-3′). PCR products were digested with XbaI and BamHI and ligated into pET9a. To generate pT7-npt (pKR11), npt was amplified using Npt1 (5′-AATCTAGAATCAAGAGACAGGATGAGGAT-3′) and Npt2 (5′-AACATATGTCAGAAGAACTCGTCAAGAA-3′). PCR products were digested with XbaI and BamHI and ligated into pET9a. The yopN/yscN strain was generated using allelic replacement with pCT120 (ΔyscN allele) (32). Plasmid pCT120 was constructed with the replacement vector pLC28 (33) in which the yscN allele retains codons 1–50 and 391–439 joined by an intervening BglII site (32). Y. enterocolitica yopN mutant VTL1 (34) was used as parent to yield the double mutant strain. E. coli strains DH5α and S17.1 were used to generate plasmids or conjugate pLC28 derivatives into Y. enterocolitica W22703 (33).
Secretion Assay
Yersinia strains were grown in tryptic soy broth (TSB)4 Yersinia strains were grown overnight at overnight at 26 °C with shaking. Cultures were diluted 1:40 into 4 ml of TSB supplemented with either 5 mm CaCl2 (type III repressive) or 5 mm ethylene glycol tetraacetic acid (EGTA), type III inducing condition) and incubated at 26 °C for 2 h with shaking. Cultures were then shifted to 37 °C and incubated for 3 h with shaking. Where indicated, 1 mm IPTG was added at the time of temperature shift. Cultures of strains carrying plasmids with chloramphenicol resistance were supplemented with 30 μg/ml chloramphenicol. Culture aliquots (4 ml) were centrifuged at 8000 × g for 10 min. A 1-ml supernatant was removed, and proteins were precipitated with 14% trichloroacetic acid (TCA). The bacterial sediment was suspended in 4 ml of water, and a 1-ml suspension was removed for protein precipitation with 14% TCA. After incubation of samples on ice, proteins were sedimented by centrifugation at 15,000 × g for 15 min. Protein sediments were washed with ice-cold acetone, dried, and solubilized in 5 parts of 4% SDS, 0.5 m Tris-HCl (pH 8.0), and one part sample buffer (10% SDS, 0.35 m Tris-HCl (pH 6.8), 30% glycerol, 0.97 m β-mercaptoethanol, 180 μm bromphenol blue). Proteins were analyzed by SDS-PAGE. Gels were analyzed either by Coomassie Brilliant Blue staining or subjected to immunoblotting with specific antisera.
Ribosome Purification
Ribosomes were isolated using a previously developed protocol for E. coli ribosomes (35). Yersinia strains were grown overnight at 26 °C in TSB with shaking. Cultures were diluted 1:20 into 1 liter of tryptic soy broth supplemented with either 5 mm CaCl2 or 5 mm EGTA and incubated with rotation at 26 °C for 2 h and at 37 °C for 3 h. Where necessary, 1 mm IPTG was added at the time of temperature shift to induce the expression of genes cloned under control of the lac promoter. Cultures of strains carrying plasmids with chloramphenicol resistance were supplemented with 30 μg/ml chloramphenicol. Cells were sedimented by centrifugation at 8000 × g for 10 min. Cells were flash-frozen in liquid nitrogen and thawed on ice before use. Bacteria were suspended in 25 ml of ribosome lysis buffer (10 mm HEPES-KOH (pH 7.6), 50 mm KCl, 10 mm Mg(OAc)2, 7 mm β-mercaptoethanol) and broken with two cycles of French press lysis at 14,000 pounds/square inch. Cell debris was sedimented by centrifugation at 20,000 × g for 30 min. Supernatant was transferred to a 50-ml conical tube, and ammonium sulfate was added to a final concentration of 1.5 m (NH4)2·SO4. Samples were incubated for 5 min on ice and again centrifuged at 20,000 × g for 30 min. The supernatant was removed, filtered through a 0.45-μm surfactant-free cellulose acetate filter, and subjected to chromatography on 16 × 25 mm Hi-Trap Butyl FF (GE Healthcare) column pre-equilibrated with 100% Buffer A (20 mm HEPES·KOH (pH 7.6), 1.5 m (NH4)2·SO4, 10 mm Mg(OAc)2, 7 mm β-mercaptoethanol). Ribosomes were eluted during hydrophobic interaction chromatography by stepwise increases of 20, 50, and 100% Buffer B (20 mm HEPES-KOH (pH 7.6), 10 mm Mg(OAc)2, 7 mm β-mercaptoethanol) (35). For fractionation analysis, a flow-through sample (200-μl aliquot) was collected after 8 min. Sequential 10-ml fractions were collected comprising the 50 S wash and 70 S, and 30 S particle fractions. Aliquots of 200 μl from each fraction were analyzed by SDS-PAGE. For cell free translation and sucrose density ultracentrifugation, 10-ml 70, 30, or 50 S fractions were loaded onto 10 ml of a 30% sucrose cushion in 20 mm HEPES·KOH (pH 7.6), 30 mm NH4Cl, 10 mm Mg(OAc)2, and 7 mm β-mercaptoethanol and subjected to ultracentrifugation at 36,000 rpm for 16 h in a Ti-70 fixed angle rotor (Beckman). The ribosomal sediment was suspended in ribosome storage buffer (20 mm HEPES·KOH (pH 7.6), 30 mm KCl, 6 mm Mg(OAc)2, 7 mm β-mercaptoethanol) and stored at −80 °C. For SDS-PAGE analysis, ribosome preparations were mixed with sample buffer before electrophoresis. Gels were analyzed either by Coomassie Brilliant Blue staining or immunoblotting with specific antisera.
Immunoblotting
Proteins were resolved by electrophoresis on 15% SDS-PAGE gels. Proteins were transferred to polyvinylidene difluoride membranes (Millipore) and probed with either rabbit polyclonal antisera raised against Yersinia proteins or mouse monoclonal antibodies against E. coli S3 (mAb 373C9C3A1-s, Developmental Studies Hybridoma Bank, Iowa City). Immunoreactive signal was visualized by chemiluminescence using either rabbit or mouse IgG-secondary antibody conjugated to horseradish peroxidase. For quantifications listed in Table 1, chemiluminescence data were analyzed using Adobe Photoshop©. Signal intensities from three independent experiments were averaged, and a standard deviation was calculated. Differences between wild-type and mutant samples were analyzed for significance using the unpaired student's t test. For quantifications performed in Table 2, secondary antibody coupled to IRDye© 680 was used. Quantification of immunoblots was conducted using a Li-Cor Biosciences Odyssey imager. Signal intensities were normalized to control samples generated in the absence of any additional purified protein by generating a ratio between experimental and control samples. These ratios were analyzed with the paired Student's t test to assess statistical significance.
TABLE 1.
Association of YopD and YscM1 with ribosomal 30 S and 70 S particles
| Straina | Fractionb | YopDc |
YscM1c |
||||||
|---|---|---|---|---|---|---|---|---|---|
| CaCl2d | Pe | EGTAd | P | CaCl2 | P | EGTA | P | ||
| Wild type | 30 S | 12.3 (0.8)f | 15.1 (2.7) | 6.77 (3.9) | 5.7 (6.9) | ||||
| 70 S | 0.5 (0.4) | 1.7 (1.5) | 2.40 (2.4) | 10.1 (17.) | |||||
| lcrH | 30 S | 4.6 (3.2) | <0.05 | 0.5 (2.2) | 0.004 | 9.78 (11.2) | 0.7 | 1.8 (4.1) | 0.5 |
| 70 S | 0.6 (0.9) | 0.9 | 0.5 (2.4) | 0.3 | 17.3 (19.) | 0.3 | 21.0 (11.) | 0.4 | |
| lcrH (plcrH) | 30 S | 17.0 (6.6) | 0.7 | 14.3 (1.2) | 0.7 | 1.90 (1.5) | 0.2 | 0.7 (2.2) | 0.3 |
| 70 S | 3.2 (4.3) | 0.4 | 0.2 (0.2) | 0.2 | 2.45 (3.9) | 1.0 | 8.2 (7.8) | 0.9 | |
| yopD | 30 S | NDg | ND | 1.35 (0.4) | 0.1 | 3.9 (3.7) | 0.7 | ||
| 70 S | ND | ND | 2.71 (1.6) | 0.9 | 23.0 (21.) | 0.5 | |||
| yopD (pyopD) | 30 S | 12.1 (1.0) | 0.9 | 16.9 (2.6) | 0.086 | 1.11 (1.0) | 0.1 | 1.0 (1.1) | 0.3 |
| 70 S | 0 (0.3) | 0.9 | 0.5 (1.2) | 0.3 | 1.90 (0.7) | 0.8 | 0.8 (0.8) | 0.4 | |
| yscM1/yscM2 | 30 S | 13.7 (9.4) | 0.8 | 15.4 (2.6) | 0.9 | ND | ND | ||
| 70 S | 4.5 (6.2) | 0.4 | 0.00 (6.0) | 0.6 | ND | ND | |||
a Identifies Y. enterocolitica strain used to generate ribosomal fractions, including complementing plasmids in parentheses.
b Fractions comprising the 70 S and 30 S ribosomal particles were isolated by hydrophobic interaction chromatography.
c Fractions were subjected to quantitative immunoblotting using chemiluminescence to detect YopD or YscM1.
d Cells were grown in TSB with 5 mm CaCl2 (secretion non-permissive condition or with 5 mm EGTA (secretion-permissive).
e Statistical significance of YopD or YscM1 present in the indicated fractions compared to wild-type samples was evaluated with the unpaired Student's t test and p values recorded.
f Abundance of YopD or YscM1 was measured with quantitative immunoblotting in three independent determinations, averaged with S.D. calculated in parentheses.
g ND, not detected.
TABLE 2.
Inhibition of yopQ mRNA translation in the presence of H6-YopD/LcrH (added to Yersinia or E. coli ribosomes at a concentration of 13.3 μm) and GST-YscM
| Factora |
Y. enterocolitica ribosomesb |
E. coli ribosomesc |
||||||
|---|---|---|---|---|---|---|---|---|
| yopQd | Pe | nptf | Pg | yopQ | P | npt | P | |
| GST-YscM1 (13.3 μm) | 0.68 | 0.023 | 0.97 | 0.906 | 1.00 | 0.725 | 0.93 | 0.786 |
| GST (13.3 μm) | 1.09 | 0.95 | 1.04 | 1.25 | ||||
| GST-YscM1 (26.6 μm) | 0.37 | 0.005 | 0.83 | 0.357 | 0.95 | 0.227 | 1.02 | 0.544 |
| GST (26.6 μm) | 1.04 | 0.96 | 1.00 | 1.08 | ||||
| GST-YscM1 (39.9 μm) | 0.33 | 0.013 | 1.12 | 0.511 | 0.88 | 0.918 | 1.29 | 0.190 |
| GST (39.9 μm) | 0.98 | 0.98 | 0.85 | 1.58 | ||||
a Purified GST-YscM1 or GST were added to Yersinia or E. coli ribosomes at indicated concentrations.
b Ribosomes were isolated via hydrophobic interaction chromatography from Y. enterocolitica W22703 grown in TSB at 37 °C with 5 mm EGTA and added at a concentration of 13.3 μm.
c Ribosomes were isolated via hydrophobic interaction chromatography from E. coli BL21 (DE3) grown in LB at 37 °C and added at a concentration of 13.3 μm.
d Translation of yopQ was measured with quantitative immunoblotting using LI-COR in three or more independent determinations, normalized to control samples and averaged.
e Statistical significance of YopQ abundance between GST-YscM1 vs. GST samples was evaluated with the paired Student's t test and p values recorded.
f Translation of npt was measured with quantitative immunoblotting using LI-COR in three or more independent determinations, normalized to control samples, and averaged.
g Statistical significance of Npt abundance between GST-YscM1 vs. GST samples was evaluated with the paired Student's t test and p values recorded.
Purification of Class II Factors
E. coli BL21 (DE3) harboring plasmid pKR6 (H6-YopD/LcrH), pKR4 (H6-LcrH), pEC346 (GST-YscM1), or pGEX2TK (GST) was grown at 37 °C to A600 0.7 (11, 12). IPTG (1 mm) was added to induce expression of genes under T7p promoters, and cultures were incubated an additional 3 h at 37 °C. Bacteria were sedimented by centrifugation at 8000 × g and lysed with 2 cycles in French pressure cell at 14,000 p.s.i. Lysates were cleared with centrifugation at 13,000 × g for 30 min, and supernatant was applied to chromatography. For His-tagged constructs, supernatant was applied to nickel-nitrilotriacetic acid-agarose equilibrated with 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 20 mm imidazole. Proteins were eluted with 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, and 500 mm imidazole. For GST-tagged constructs, supernatant was applied to glutathione-Sepharose equilibrated with 50 mm Tris-HCl (pH 7.5), 150 mm NaCl. Proteins were eluted with 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, and 10 mm glutathione. Eluates were dialyzed at 4 °C in Slide-A-Lyzer dialysis cassettes (3500 molecular weight cutoff; Thermo Scientific) against Column Buffer 50 mm Tris-HCl (pH 7.5), 150 mm NaCl. Proteins were concentrated using Amicon® Ultra centrifugal filters (10,000 molecular weight cutoff; Millipore).
In Vitro Translation
Translation reactions were performed with the PURExpress ΔRibosome Kits (NEB) following the manufacturer's specifications with the following exceptions. Ribosomes were isolated as described above and added a final concentration of 13.3 μm. Purified class II factors were added before the addition of mRNA. Murine RNase inhibitor was added to each reaction (NEB). For experiments with translation inhibition, 3 μg of mRNA were used rather than 250 ng of plasmid DNA. mRNA was obtained using pKR10 or pKR11 as template for MEGAscript T7 in vitro transcription reactions.
Transmission Electron Microscopy
Ribosomal samples were pipetted on a carbon-coated copper grid and stained with 1% uranyl acetate before viewing with a Tecnai F30 electron microscope at 300 kV.
Sucrose Density Ultracentrifugation
Class II factors and ribosome particles were purified as described above. Twenty μg of H6-YopD/LcrH or GST-YscM1 were added alone or were mixed with the indicated concentrations of 30, 50, or 70 S particles in ribosome storage buffer and incubated for 15 min at 37 °C. The mixture was layered over 6 ml of 30% sucrose cushion and centrifuged for 4 h at 36,000 × g in a fixed angle rotor. Twelve 0.5-ml fractions were collected from the bottom of the tube using a peristaltic pump; sediments were suspended in 0.5 ml of ribosome storage buffer. For SDS-PAGE analysis, collected samples were heated with sample buffer before electrophoresis. To determine the dissociation constant, secondary antibody coupled to IRDye© 680 was used. Quantification of immunoblots was conducted using a Li-Cor Biosciences Odyssey imager. Data were used to calculate the dissociation constant Kd = [30 S] × ([YopD]total − [YopD]bound)/[YopD]bound.
RESULTS
Genetic Relationships for Three Classes of Yersinia Type III Regulators
Wild-type Y. enterocolitica W22703 and its yopN (class I), yopD (class II), and yscN (class III) mutants were analyzed for the production and the type III secretion of YopQ. As reported earlier, growth of Y. enterocolitica W22703 at 37 °C in the presence of calcium leads to the expression of type III machine components, and the assembly of the secretion machine (36). Under these conditions the yopQ effector gene is not expressed (Fig. 1).
FIGURE 1.
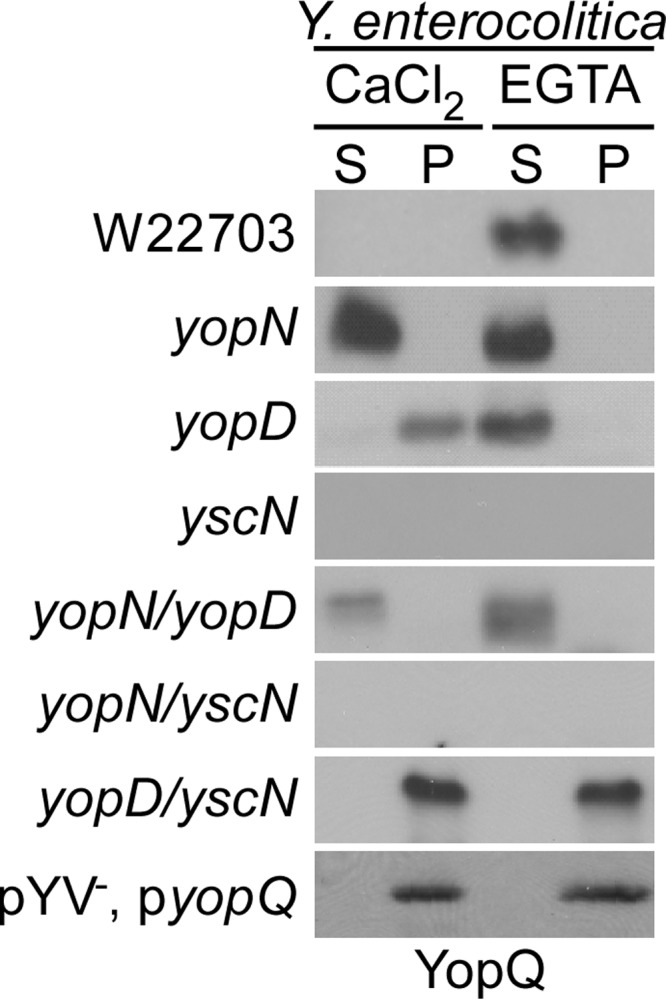
Control of Y. enterocolitica YopQ production and type III secretion by three classes of genes. Y. enterocolitica W22703 and its variants yopN (class I), yopD (class II), yscN (class III), yopN/yopD (class I/II), yopN/yscN (class I/III), and yopD/yscN (class II/III) and as well as Y. enterocolitica W22703 (pYV−) with plasmid pyopQ were analyzed for the production of YopQ and the type III secretion of YopQ. Cultures were grown in the presence of 5 mm CaCl2 (secretion non-permissive) or 5 mm EGTA (secretion permissive) and centrifuged. Proteins in the supernatant (S) and Yersinia sediment (P) fractions were analyzed by immunoblotting for the presence of YopQ.
Chelation of calcium ions with EGTA in growth media induces Yersinia YopQ production and secretion of YopQ polypeptide (Fig. 1). Mutations in class I genes (yopN) trigger Yersinia YopQ production and type III secretion of YopQ even in the presence of calcium ions (Fig. 1). In the presence of calcium ions, mutations in class II genes (yopD) cause Yersinia to synthesize YopQ without promoting YopQ secretion (Fig. 1). Finally, mutations in class III genes (yscN) abolish Yersinia YopQ production in the presence or absence of calcium (Fig. 1).
Mutants with defects in both class I (yopN) and II (yopD) genes trigger YopQ synthesis and secretion in the presence and absence of calcium, the same phenotype as class I (yopN) mutants (Fig. 1). Mutants with defects in class I (yopN) and III (yscN) genes cannot produce YopQ, similar to class III (yscN) mutants. Finally, Yersinia with mutational lesions in class II (yopD) and III (yscN) genes synthesize YopQ in the presence or absence of calcium ions but cannot secrete YopQ polypeptide (Fig. 1). Taken together, these data suggest effector translation is negatively regulated by class II genes, whereas secretion is negatively regulated by class I genes. Class I genes are only necessary to repress Yop effector synthesis in the presence of a secretion-competent type III machine, suggesting these gene products serve as a mediator between the machine and class II genes. We conclude that the class II genes perceive a translation signal originating with class III genes, and class I genes negatively regulate signal transmission.
Expression of yopQ mRNA in the Absence of Type III Secretion Factors
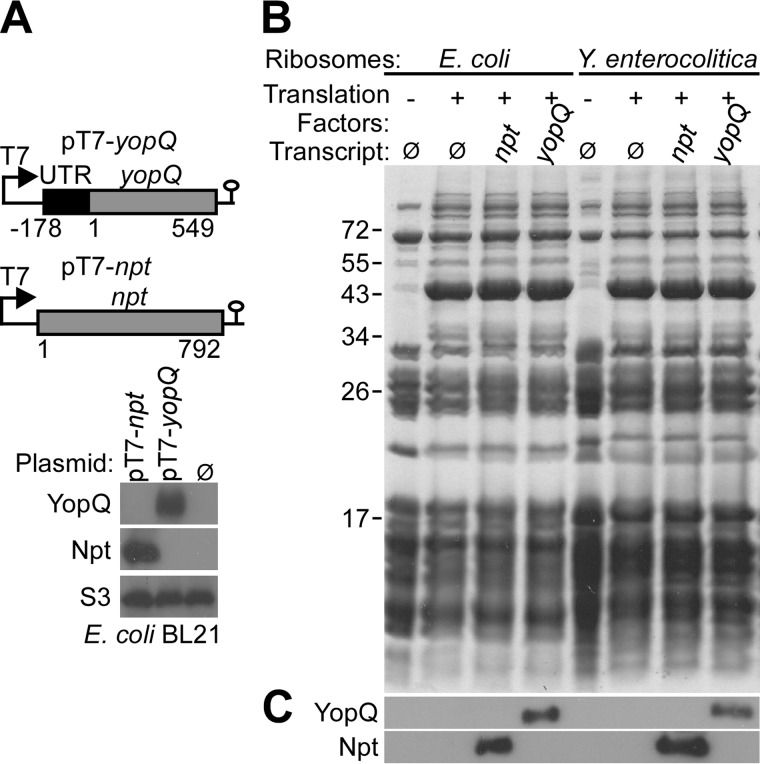
Previous studies entertained models whereby effector mRNA may assume structures that prevent their translation, which could subsequently be relieved by a regulatory activator (20, 37). However, genetic experiments did not identify an activator specially required for the expression of yopQ. In contrast, the observation that yopD/yopN, yopD/yscN or any other yopD-class III mutants express yopQ suggests to us that yopD and other class II genes may function as negative regulators of yopQ expression. As a further test for the existence of a positive regulatory factor, we expressed yopQ from an IPTG-inducible promoter in the Y. enterocolitica W22703 (pYV−) strain, which lacks genes of the type III secretion pathway. Immunoblotting experiments revealed yopQ expression in Y. enterocolitica W22703 (pYV−) independent of extracellular calcium ions (Fig. 1). Plasmid-encoded yopQ, placed under control of the IPTG-inducible T7 polymerase promoter, was expressed in E. coli BL21 (DE3) (pT7-yopQ) (Fig. 2A). Similarly, the neomycin-phosphotransferase gene (npt) was also expressed in E. coli BL21 (DE3) (pT7-npt) (Fig. 2A). To test whether yopQ and npt transcript are also effectively translated in vitro, we added pT7-yopQ or pT7-npt to purified E. coli or Y. enterocolitica ribosomes (Fig. 2B). In the presence of purified transcription and translation factors, both yopQ and npt were effectively translated by E. coli and Y. enterocolitica ribosomes and generated YopQ and Npt products, respectively (Fig. 2C). Together these results suggest that neither Yersinia nor E. coli require a specific activator for yopQ expression.
FIGURE 2.
In vivo and in vitro expression of yopQ. A, top panel, schematic of plasmids expressing yopQ or npt under control of the T7 promoter. Bottom panel, plasmids were transformed into E. coli BL21(DE3), and expression was induced by the addition of IPTG to induce T7 polymerase expression. Plasmid- or mock (Ø)-transformed bacteria were lysed, and cell extracts were analyzed by immunoblotting with polyclonal antisera raised against YopQ or Npt as well as monoclonal antibody against ribosomal protein S3. B, ribosomes were isolated from E. coli BL21(DE3) and Y. enterocolitica W22703 lysates with hydrophobic interaction chromatography and sedimentation through 30% sucrose cushion. T7 polymerase derived yopQ and npt mRNA from pT7-yopQ and pT7-npt and in vitro translation system were used to measure translation by E. coli BL21(DE3) and Y. enterocolitica W22703 ribosomes. Translation reactions were analyzed by Coomassie-stained SDS-PAGE. C, translation products were identified by immunoblotting with polyclonal antibodies against YopQ and Npt.
YopD Associates with Yersinia 30 S Ribosomal Particles
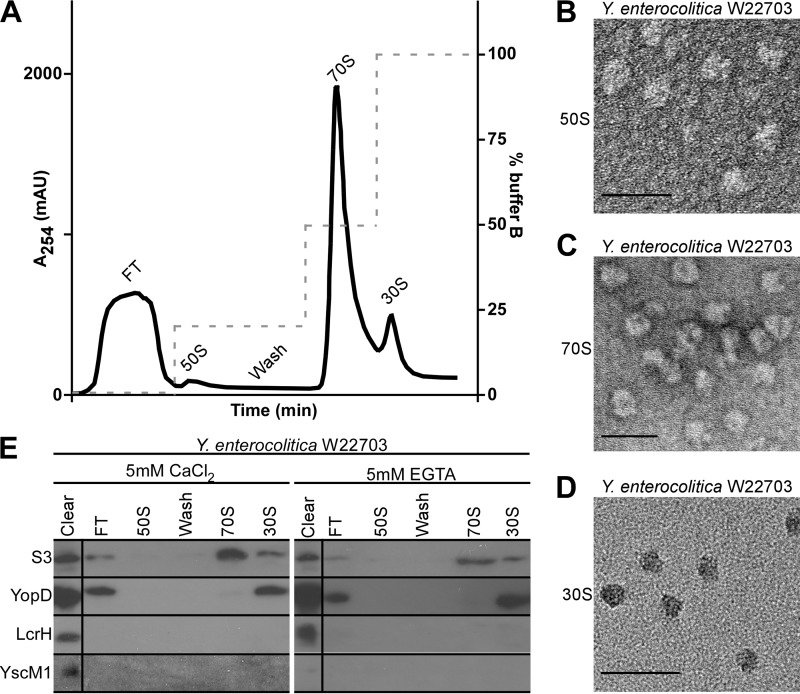
If class II gene products function as negative regulators of yopQ expression, this could occur via their specific association with yopQ mRNA or by implementing a translational block at the ribosome. Our initial experiments measured binding of YopD/LcrH or YscM1 to yopQ mRNA (11). However, we and others failed to detect a specific association between class II proteins and yopQ mRNA (11, 13). We, therefore, wondered whether class II gene products block yopQ translation at the ribosome and explored a possible association between these regulatory factors and ribosomal particles. Y. enterocolitica W22703 was grown in either secretion-permissive (chelation of calcium ions with 5 mm EGTA) or non-permissive (5 mm CaCl2) conditions. Bacteria were broken in a French pressure cell, insoluble material was removed by centrifugation and ammonium sulfate precipitation, and ribosomal particles were purified by hydrophobic interaction chromatography (Fig. 3A). Fractions that represent 50, 30, and 70 S ribosomal particles were subjected to transmission electron microscopy, which confirmed the presence of these particles, as previously described for E. coli and Thermus thermophilus ribosomes (38) (Fig. 3, B–D).
FIGURE 3.
YopD associates with Y. enterocolitica 30 S ribosomal particles. A, lysate of Y. enterocolitica W22703 were subjected to hydrophobic interaction chromatography with stepwise increases of ionic strength (buffer B with 1.5 m (NH4)2·SO4). Left, the y axis denotes absorbance at 254 nm to detect ribosomal RNA. Right, the y axis denotes the percentage of buffer B. mAU, milliabsorbance units. The indicated fractions were collected and subjected to transmission electron microscopy, which revealed the purification of 50 S (B), 70 S (C), and 30 S (D) ribosomal particles; scale bars represent 50 nm. E, immunoblot analysis of ribosomal particles purified in A. Polyclonal antisera raised against class II regulatory factors YopD, LcrH, or YscM1 as well as monoclonal antibodies directed against ribosomal protein S3 were used to probe collected fractions. FT = flow-through fractions. Average percentages of YopD or YscM1 in the cleared lysate that are associated with either 30 or 70 S ribosomal particles are listed in Table 1. Images and data analyses are representative examples from three independent experiments.
Ribosomal fractions were subjected to immunoblot analysis using a monoclonal antibody against the small ribosomal subunit protein S3 as well as polyclonal antibodies directed against YopD, LcrH, or YscM1. As expected, S3 was detected in the cleared lysate, in the flow-through (S3 molecules that are not associated with ribosomes), and in 70 S and 30 S particle fractions but not in the 50 S particle fraction (Fig. 3E). YopD was detected in the cleared lysate, the flow-through, and the 30 S particle fraction but not in 50 S and 70 S ribosomal particle fractions (Fig. 3E). LcrH and YscM1 were detected in lysates of wild-type Y. enterocolitica W22703 cells; however, these proteins sedimented during ammonium sulfate precipitation and were not found associated with ribosomal particles (Fig. 3E). We used quantitative immunoblotting of ribosomal particle fractions to measure YopD association with 30 S particles. In three independent experiments, 12% of YopD molecules in cleared lysates of Y. enterocolitica W22703 grown in 5 mm CaCl2 were associated with 30 S ribosomal particles (Table 1). Similarly, 15% of YopD from Y. enterocolitica W22703 grown in 5 mm EGTA were associated with 30 S particles. These data suggest that a significant portion of cytoplasmic YopD associates with 30 S ribosomal particles but not with assembled (70 S) ribosomes (Table 1).
LcrH-dependent Association of YopD with Yersinia Ribosomes
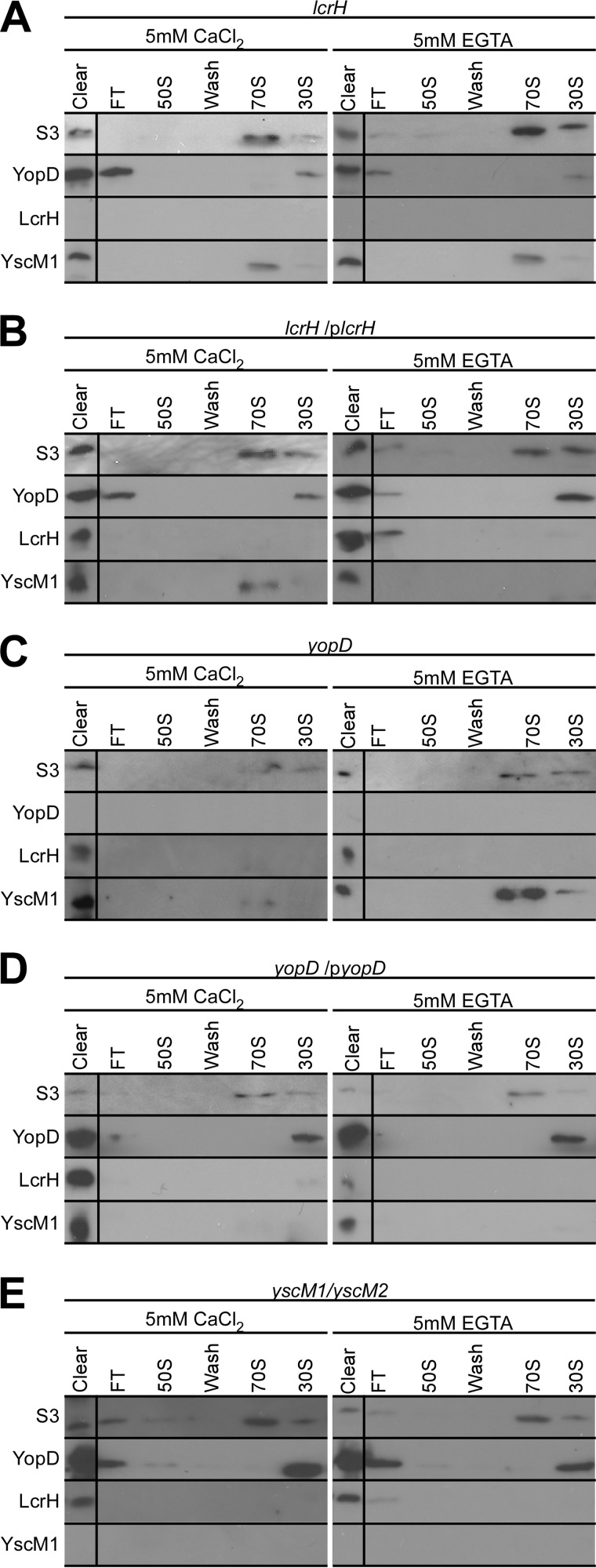
LcrH is the cytoplasmic-binding protein of YopD, which functions as a secretion chaperone promoting type III secretion of YopD and completion of the type III pathway for delivery of effectors into host cells (16, 39, 40). LcrH harbors tetratricopeptide-like repeats and, in addition to binding YopD and YopB at discrete sites, also associates with the class III machine component YscY (41–43). To explore a possible requirement of LcrH binding for the association of YopD with 30 S particles, we isolated ribosomal particles from the lcrH mutant strain CT133 (11) via hydrophobic interaction chromatography and analyzed fractions by immunoblotting for the presence or absence of S3, YopD, LcrH, and YscM1. When grown in the presence of 5 mm CaCl2, lcrH mutant Yersinia positioned 4.6% of YopD with 30 S particles (Fig. 4A and Table 1). When grown in the presence of 5 mm EGTA, lcrH mutant Yersinia positioned only 0.48% of YopD on 30 S particles (Table 1). The defect in YopD association with 30 S ribosomal particles was restored when lcrH mutant Yersinia was transformed with plasmid encoded wild-type lcrH (Fig. 4B and Table 1). As expected, YopD was not detected in the lysates and ribosomal fractions of the yopD mutant (Fig. 4C); however, yopD expression and association with 30 S ribosomal particles were restored after transformation with plasmid-encoded wild-type yopD (Fig. 4D and Table 1).
FIGURE 4.
LcrH is required for YopD association with 30 S ribosomal particles. A–E, immunoblot analysis of hydrophobic interaction chromatography fractions collected during isolation of ribosomal particles from Y. enterocolitica strains lcrH (A), lcrH (plcrH) (B), yopD (C), yopD (pyopD) (D), and yscM1/yscM2 (E). Each strain was grown in the presence of either 5 mm CaCl2 (secretion non-permissive) or 5 mm EGTA (secretion permissive). Polyclonal antisera raised against class II factors YopD, LcrH, and YscM1 as well as monoclonal antibodies against ribosomal protein S3 were used to probe collected fractions. FT = flow-through fractions. Average percentages of YopD or YscM1 in the cleared lysate that are associated with either 30 or 70 S ribosomal particles are listed in Table 1. Images and data analyses are representative examples from three independent experiments.
YscM1 was detected by immunoblotting in the 70 S ribosomal particle fractions of the lcrH mutant grown in the presence or absence of calcium ions (Fig. 4A). The percent amount of YscM1 on ribosomes of the lcrH mutant grown in the presence of 5 mm EGTA was 21% of YscM1 with 70 S and 1.8% on 30 S particles (Table 1). YscM1 associations with ribosomal fractions were reduced in the complemented lcrH (plcrH) mutant strain (Fig. 4B). Similar results were obtained with the yopD mutant and its complementing plasmid, pyopD (Fig. 4, C and D). As expected, YscM1 was not detected in the cleared lysate of the yscM1/yscM2 class II mutant strain (44) (Fig. 4E). Association of YopD with 30 S ribosomal particles was not affected in yscM1/yscM2 mutant Yersinia (Fig. 4E, Table 1). Taken together these data indicate that the association of YopD with 30 S ribosomal particles of Y. enterocolitica is dependent on LcrH (wild-type versus lcrH (5 mm CaCl2), p = 0.048; wild-type versus lcrH (5 mm EGTA), p = 0.004). Although we observed an association of YscM1 with 70 S ribosomes in both the presence and absence of calcium ions, the overall variability of this association did not allow us to reach a statistically firm conclusion whether or not YscM1 interacts with Yersinia 70 S ribosomes (Table 1).
Co-sedimentation of Purified YopD with Yersinia 30 S Particles
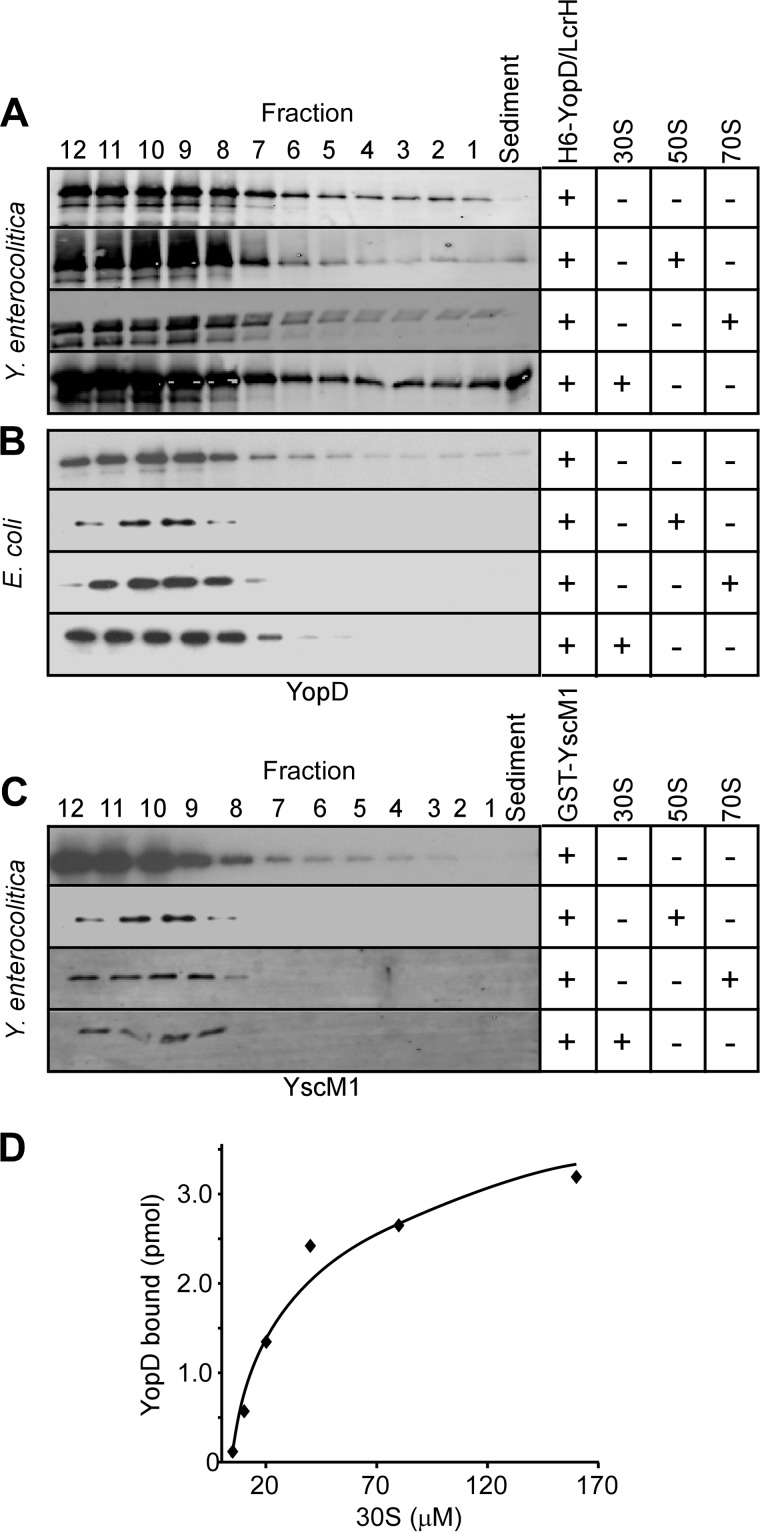
The class II regulatory functions of Y. enterocolitica yopD mutants can be complemented with plasmid encoded wild-type and six-histidyl (H6)-tagged yopD (11). Affinity purification of H6-YopD is, however, only successful from Yersinia strains expressing lcrH, as the production of soluble YopD is diminished in the absence of its LcrH chaperone (11, 43). For these reasons we used nickel-nitrilotriacetic acid affinity chromatography experiments to isolate soluble H6-YopD/LcrH complexes for the study of class II function. Plasmid-encoded yscM1 complements the class II regulatory defects of Y. enterocolitica yscM1 and yscM1/yscM2 mutants (44). Of note, overexpression of yscM1, but not overexpression of yopD or lcrH, abolishes the expression of Yersinia yop effector genes under low calcium conditions. Similar class II regulatory phenotypes are observed with GST-YscM1, which can be purified by affinity chromatography on glutathione-Sepharose (12, 45).
The data presented above suggest that YopD may associate with 30 S ribosomes. We sought to assess whether purified YopD can associate with isolated ribosomal particles. Purified H6-YopD/LcrH or GST-YscM1 were mixed with 30, 50, or 70 S ribosomal particles isolated from Y. enterocolitica W22703 (pYV−) and layered over a 30% sucrose cushion. Co-sedimentation of H6-YopD or GST-YscM1 with ribosomal particles was assessed by ultracentrifugation, which sedimented 30, 50, and 70 S particles. Fractions (0.5 ml) were collected from the bottom of each tube and analyzed by immunoblotting for the presence or absence of H6-YopD and GST-YscM1. Without the addition of ribosomal particles, neither H6-YopD nor GST-YscM1 sedimented through the sucrose cushion during ultracentrifugation (Fig. 5, A and C). When GST-YscM1 was mixed with 30, 50, or 70 S particles, the protein remained near the top of the gradient, indicating that GST-YscM1 does not sediment with purified ribosomes (Fig. 5C). H6-YopD mixed with either 50 or 70 S particles also did not associate with ribosomes. However, when mixed with 30 S particles, H6-YopD sedimented across the sucrose cushion (Fig. 5A). This attribute was unique to 30 S particles from Y. enterocolitica, as control experiments with 30 S particles from E. coli did not reveal H6-YopD co-sedimentation (Fig. 5B). These data suggest that purified H6-YopD can associate with Yersinia 30 S ribosomal particles. By varying the concentration of 30 S particles in the presence of fixed amounts of H6-YopD, the dissociation constant Kd 2.1 × 104 μm (±0.5 × 104) was calculated. The association curve for H6-YopD and 30 S particles is indicative of a weak association (Fig. 5D). We note that the association between YopD and 30 S ribosomal particles must be transient, as YopD is not found on fully assembled 70 S ribosomes. Presumably, the association between YopD and 30 S particles may function as a block for the translation of effector yop transcripts.
FIGURE 5.
Purified H6-YopD associates with 30 S ribosomal particles. A–C, immunoblot analysis of sucrose density ultracentrifugation fractions (12 indicates top of supernatant, 1 indicates bottom of supernatant) collected after mixing class II factors with the indicated ribosomal particles and analyzing mobility through 30% sucrose. Either purified H6-YopD/LcrH (A and B) or GST-YscM1 (C) were mixed with 30, 50, or 70 S ribosomal particles for 15 min at 37 °C before loading on 30% sucrose cushion and ultracentrifugation. YopD was found in sediment after mixing with 30 S ribosomes from Y. enterocolitica (A), not from E. coli (B). Using quantitative immunoblotting, the dissociation constant for the YopD/30 S ribosomal particle complex was determined: Kd 2.1 × 104 μm (±0.5 × 104). D, amount of YopD bound for a given concentration of 30 S particles as determined in A. Data are representative of four independent determinations.
Translational Control of yopQ Expression with Purified Ribosomes and Class II Proteins
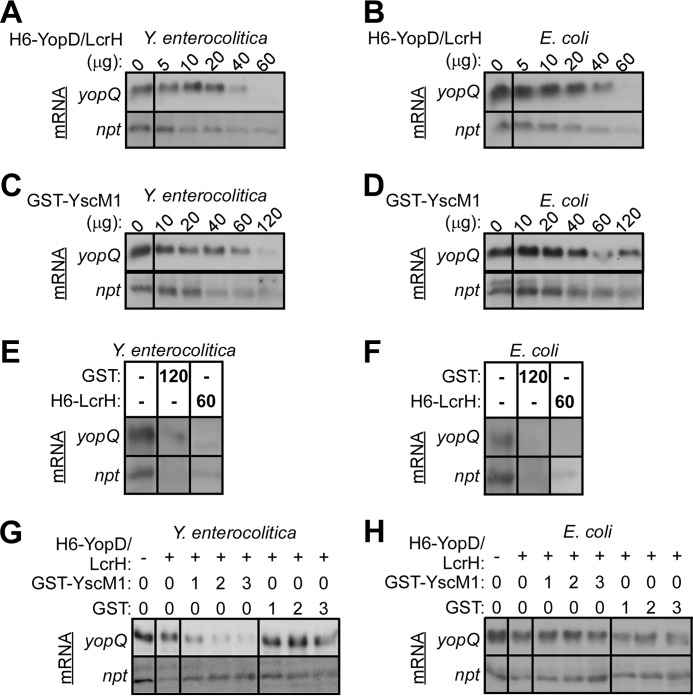
To explore whether class II regulatory factors affect the in vitro translation of yop transcripts, we measured translation of yopQ mRNA by Y. enterocolitica and E. coli ribosomes in the presence of H6-YopD/LcrH or GST-YscM1 (Fig. 6). In the presence of equimolar amounts (20 μg) of H6-YopD/LcrH (13.3 μm H6-YopD/LcrH and ribosome), Y. enterocolitica W22703 ribosomes translated yopQ and npt control transcripts similar to ribosomes without H6-YopD/LcrH (Fig. 6A). When the molar concentration of H6-YopD/LcrH exceeded that of ribosomes (3:1, 60 μg), the translation of yopQ and npt was quenched (Fig. 6A). The addition of equimolar amounts (20 μg) of GST-YscM1 to Y. enterocolitica W22703 ribosomes also did not affect the translation of yopQ or npt (Fig. 6C). At greater than a 6-fold molar excess (120 μg), GST-YscM1 quenched ribosomal translation of yopQ and npt (Fig. 6C). This effect was not specific, as the addition of excess purified GST or H6-LcrH alone also affected the translation of yopQ and npt (Fig. 6E). When yopQ and npt translation was measured using E. coli ribosomes, we obtained similar results. Equimolar amounts of H6-YopD/LcrH (Fig. 6B) or GST-YscM1 (Fig. 6D) did not affect translation, whereas large molar excesses of H6-YopD/LcrH, GST-YscM1, GST, or H6-LcrH reduced the production of YopQ and Npt (Fig. 6F).
FIGURE 6.
Class II factors control yopQ translation on Yersinia ribosomes. Ribosomes (70 S) were isolated from Y. enterocolitica W22703 (A, C, E, and G) and E. coli BL21 (B, D, F, and H) using hydrophobic interaction chromatography and mixed at a concentration of 13.3 μm ribosomes with 3 μg of in vitro transcribed yopQ or npt mRNAs (A–H). Purified class II factors H6-YopD/LcrH (A and B) or GST-YscM1 (C and D) were added at increasing concentrations (indicated as μg purified protein), and after 2 h of incubation, mRNA translation was monitored by immunoblotting of samples for the abundance of YopQ and Npt with specific antibodies. Yersinia (A) or E. coli (B) ribosomes were incubated with yopQ or npt mRNA in the presence of increasing concentrations (0–60 μg) of H6-YopD/LcrH complex. Yersinia (C) or E. coli (D) ribosomes were incubated with yopQ or npt mRNA in the presence of increasing concentrations (0–120 μg) of GST-YscM1. Yersinia (E) or E. coli (F) ribosomes were incubated with yopQ or npt mRNA in the presence of control proteins GST (120 μg) or H6-LcrH (60 μg). Yersinia (G) or E. coli (H) ribosomes were incubated with yopQ or npt mRNA in the presence of 13.3 μm H6-YopD/LcrH in either the absence (0) or the presence of equal (1 = 13.3 μm), or two- (2 = 26.6 μm) as well as 3-fold (3 = 39.9 μm) excess of GST-YscM1 or GST.
Increasing concentrations of GST-YscM1 were added to equimolar amounts of H6-YopD/LcrH and Y. enterocolitica W22703 ribosomes, which caused decreasing translation of yopQ transcripts without affecting the translation of npt control mRNA (Fig. 6G and Table 2). This effect was specific for GST-YscM1, as the addition of GST did not affect the translation of yopQ and npt transcripts (Fig. 6G and Table 2). The class II regulatory effects of H6-YopD/LcrH and GST-YscM1 were not observed with E. coli ribosomes, as increasing concentrations of GST-YscM1 incubated with equimolar amounts of H6-YopD/LcrH and E. coli ribosomes did not affect the translation of yopQ and npt transcripts (Fig. 6H and Table 2). Together these results suggest that the class II regulatory factors YopD/LcrH and YscM1 control the expression of yopQ by blocking the translation of its mRNA at Yersinia ribosomes.
DISCUSSION
Type III secretion machines can be thought of as bacterial weapons for close combat with host immune cells (26). A striking feature of the Yersinia type III pathway is its specificity of delivering effector Yops into the cytoplasm of host cells without secreting these polypeptides into the extracellular milieu (31, 46, 47). Yersinia accomplish this by distinguishing three stages of type III secretion and assembly reactions beginning with the assembly of the needle as an extension of the membrane embedded type III machine, which requires the secretion of YscH (YopR), YscI, and YscF (48–50). During the following stage, the needle cap/translocator proteins LcrV, YopD, and YopB are secreted, and these factors complete the type III pathway to form a conduit from bacterial cells into the cytoplasm of host cells (4, 10, 46). Finally, large amounts of effectors travel the type III pathway to block the motility and signaling functions of immune cells (2, 51).
Earlier work explored the secretion signals of type III substrates for entry into the pathway (52, 53). These studies identified discrete types of signals in different substrates, in agreement with a general model whereby type III secretion can be viewed as a developmental program that switches substrate specificity as different stages of pathway assembly are being completed (14). This concept also explains why fusions of some secretion substrates with impassable reporter proteins can block type III machines at one stage but not another. Of particular interest are the Yop effectors. Surprisingly, Yop fusions with impassable reporters are rejected from the type III pathway and cannot block the secretion of other effectors (54, 55). In all cases examined, the secretion signals of effectors have been mapped to the first 10–15 codons of yop genes, which when fused to passable reporter molecules promote the secretion of hybrid polypeptides (56–58). In several cases, the function of these secretion signals is not perturbed by mutations that frameshift the first 15 codons but can be abolished by mutations that change the nucleotide sequence without altering the amino acid sequence of Yops (20, 21, 59, 60). These signals are not restricted to Y. enterocolitica but have been described for other Yersinia spp. (61, 62) as well as Pseudomonas (37), Salmonella (22), and Xanthomonas spp. (63). Nevertheless, the molecular mechanisms whereby nucleic acid signals in yop mRNAs may be decoded for their entry into the secretion pathway have remained elusive.
The expression of yop effector genes is controlled by morphogenetic events associated with assembly of the type III pathway (64). Before assembly of the membrane-embedded machine, the formation of capped needle complexes and the completion of the type III conduit, yop expression is quenched by class II factors YopD, LcrH, and YscM1/YscM2 (65). Once regulators are secreted (YopD) and translocated into host cells (YscM1/YscM2), yop genes are expressed, and their products travel along the type III pathway (45). Genetic experiments reported here provide evidence that the regulation of yop expression and their secretion represent coupled events that involve class II genes (to regulate expression) and class I genes (to regulate secretion). Although earlier studies explored the possibility that class II factors YopD/LcrH and YscM1 interact with mRNA to control translational initiation (11), we report here that the class II factor YopD modifies the ribosome and blocks the translation of yop mRNA but not of other transcripts. The specificity for this block in translation can be found in the nucleic acid sequences of effector transcripts that harbor multiple repeats of the sequence AUAAA in addition to AU-rich elements in the vicinity of the Shine-Dalgarno sequence and AUG start codon (12, 13). The specific mechanism(s) of blocking translation is currently not appreciated; however, they must require the transient, LcrH-dependent interaction of YopD with 30 S particles in addition to the quenching action of YscM1 (LcrQ) and probably affect translational initiation, presumably perturbing the formation of the 30 S complex before 50 S ribosome engagement, as the interaction of YopD and the ribosome is lost once the 70 S particle is assembled.
How can Yersinia couple a class II factor-mediated block of yop mRNA translation to the low calcium signal for effector secretion? We hypothesize that class I gene products, for example YopN/TyeA, may accomplish this task by blocking transduction of the inducing signal to the class II regulatory factor YopD (23, 66). In view of the finding that YopD/LcrH and YscM1 block yopQ translation by Yersinia ribosomes, the purpose of specific class I and class III gene products may be to relieve the translational block and to guide effector polypeptides into the type III secretion pathway. Taking advantage of the experimental system developed here, the identity of the Yersinia factor(s) required for the YopD/LcrH/YscM1-mediated translational block, which is not operational with E. coli ribosomes, may be revealed by a combination of genetic and biochemical approaches. If so, one may be able to recapitulate the post-transcriptional control of yop expression and substrate recognition of effectors by type III machine components by measuring both the in vitro blockade and the relief of yop mRNA translation with purified proteins.
Acknowledgments
We thank Antoni P.A. Hendrickx for training in transmission electron microscopy and members of our laboratory for discussion and critical comments on the manuscript. The S3 monoclonal antibody developed by L. Kahan was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD, National Institutes of Health, and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242. O. S. acknowledges membership within and support from the Region V “Great Lakes” Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (National Institutes of Health Award 1-U54-AI-057153).
This work was supported, in whole or in part, by National Institutes of Health Grant AI42797 (NIAID, Infectious Diseases Branch; to O. S.).
- TSB
- tryptic soy broth
- EGTA
- ethylene glycol tetraacetic acid
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside.
REFERENCES
- 1. Cornelis G. R., Boland A., Boyd A. P., Geuijen C., Iriarte M., Neyt C., Sory M.-P., Stainier I. (1998) The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62, 1315–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rosqvist R., Magnusson K.-E., Wolf-Watz H. (1994) Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 13, 964–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marketon M. M., DePaolo R. W., DeBord K. L., Jabri B., Schneewind O. (2005) Plague bacteria target immune cells during infection. Science 309, 1739–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mueller C. A., Broz P., Müller S. A., Ringler P., Erne-Brand F., Sorg I., Kuhn M., Engel A., Cornelis G. R. (2005) The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science 310, 674–676 [DOI] [PubMed] [Google Scholar]
- 5. Broz P., Mueller C. A., Müller S. A., Philippsen A., Sorg I., Engel A., Cornelis G. R. (2007) Function and molecular architecture of the Yersinia injectisome tip complex. Mol. Microbiol. 65, 1311–1320 [DOI] [PubMed] [Google Scholar]
- 6. Håkansson S., Bergman T., Vanooteghem J.-C., Cornelis G., Wolf-Watz H. (1993) YopB and YopD constitute a novel class of Yersinia Yop proteins. Infect. Immun. 61, 71–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Costa T. R., Edqvist P. J., Bröms J. E., Ahlund M. K., Forsberg A., Francis M. S. (2010) YopD self-assembly and binding to LcrV facilitate type III secretion activity by Yersinia pseudotuberculosis. J. Biol. Chem. 285, 25269–25284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ferracci F., Schubot F. D., Waugh D. S., Plano G. V. (2005) Selection and characterization of Yersinia pestis YopN mutants that constitutively block Yop secretion. Mol. Microbiol. 57, 970–987 [DOI] [PubMed] [Google Scholar]
- 9. Ligtenberg K. G., Miller N. C., Mitchell A., Plano G. V., Schneewind O. (2013) LcrV mutants that abolish Yersinia type III injectisome function. J. Bacteriol. 195, 777–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mueller C. A., Broz P., Cornelis G. R. (2008) The type III secretion system tip complex and translocon. Mol. Microbiol. 68, 1085–1095 [DOI] [PubMed] [Google Scholar]
- 11. Anderson D. M., Ramamurthi K. S., Tam C., Schneewind O. (2002) YopD and LcrH regulate the expression of Yersinia enterocolitica YopQ at a post-transcriptional step and bind to yopQ mRNA. J. Bacteriol. 184, 1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cambronne E. D., Schneewind O. (2002) Yersinia enterocolitica type III secretion. yscM1 and yscM2 regulate yop gene expression by a post-transcriptional mechanism that targets the 5′-untranslated region of yop mRNA. J. Bacteriol. 184, 5880–5893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Y., Anderson D. M. (2011) Expression hierarchy in the Yersinia type III secretion system established through YopD recognition of RNA. Mol. Microbiol. 80, 966–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sorg J. A., Miller N. C., Schneewind O. (2005) Substrate recognition of type III secretion machines. Testing the RNA signal hypothesis. Cell. Microbiol. 7, 1217–1225 [DOI] [PubMed] [Google Scholar]
- 15. Sory M.-P., Boland A., Lambermont I., Cornelis G. R. (1995) Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. U.S.A. 92, 11998–12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wattiau P., Bernier B., Deslée P., Michiels T., Cornelis G. R. (1994) Individual chaperones required for Yop secretion by Yersinia. Proc. Natl. Acad. Sci. U.S.A. 91, 10493–10497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lloyd S. A., Sjöström M., Andersson S., Wolf-Watz H. (2002) Molecular characterization of the type III secretion signals via analysis of synthetic N-terminal amino acid sequences. Mol. Microbiol. 43, 51–59 [DOI] [PubMed] [Google Scholar]
- 18. Birtalan S. C., Phillips R. M., Ghosh P. (2002) Three-dimensional secretion signals in chaperone-effector complexes of bacterial pathogens. Mol. Cell 9, 971–980 [DOI] [PubMed] [Google Scholar]
- 19. Miao E. A., Miller S. I. (2000) A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. U.S.A. 97, 7539–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Anderson D. M., Schneewind O. (1997) A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science 278, 1140–1143 [DOI] [PubMed] [Google Scholar]
- 21. Anderson D. M., Schneewind O. (1999) Yersinia enterocolitica type III secretion. An mRNA signal that couples translation and secretion of YopQ. Mol. Microbiol. 31, 1139–1148 [DOI] [PubMed] [Google Scholar]
- 22. Niemann G. S., Brown R. N., Mushamiri I. T., Nguyen N. T., Taiwo R., Stufkens A., Smith R. D., Adkins J. N., McDermott J. E., Heffron F. (2013) RNA type III secretion signals that require Hfq. J. Bacteriol. 195, 2119–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheng L. W., Kay O., Schneewind O. (2001) Regulated secretion of YopN by the type III machinery of Yersinia enterocolitica. J. Bacteriol. 183, 5293–5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jackson M. W., Day J. B., Plano G. V. (1998) YscB of Yersinia pestis functions as a specific chaperone for YopN. J. Bacteriol. 180, 4912–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Day J. B., Plano G. V. (1998) A complex composed of SycN and YscB functions as a specific chaperone for YopN in Yersinia pestis. Mol. Microbiol. 30, 777–788 [DOI] [PubMed] [Google Scholar]
- 26. Cornelis G. R. (2006) The type III injectisome. Nat. Rev. Microbiol. 4, 811–825 [DOI] [PubMed] [Google Scholar]
- 27. Francis M. S., Lloyd S. A., Wolf-Watz H. (2001) The type III secretion chaperone LcrH co-operates with YopD to establish a negative, regulatory loop for control of Yop synthesis in Yersinia pseudotuberculosis. Mol. Microbiol. 42, 1075–1093 [DOI] [PubMed] [Google Scholar]
- 28. Williams A. W., Straley S. C. (1998) YopD of Yersinia pestis plays a role in negative regulation of the low-calcium response in addition to its role in translocation of Yops. J. Bacteriol. 180, 350–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Studier F. W. (1991) Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 219, 37–44 [DOI] [PubMed] [Google Scholar]
- 30. Cornelis G., Colson C. (1975) Restriction of DNA in Yersinia enterocolitica detected by the recipient ability for a derepressed R factor from Escherichia coli. J. Gen. Microbiol. 87, 285–291 [DOI] [PubMed] [Google Scholar]
- 31. Lee V. T., Anderson D. M., Schneewind O. (1998) Targeting of Yersinia Yop proteins into the cytosol of HeLa cells. one-step translocation of YopE across bacterial and eukaryotic membranes is dependent on SycE chaperone. Mol. Microbiol. 28, 593–601 [DOI] [PubMed] [Google Scholar]
- 32. Blaylock B., Riordan K. E., Missiakas D. M., Schneewind O. (2006) Characterization of the Yersinia enterocolitica type III secretion ATPase YscN and its regulator, YscL. J. Bacteriol. 188, 3525–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cheng L. W., Anderson D. M., Schneewind O. (1997) Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol. Microbiol. 24, 757–765 [DOI] [PubMed] [Google Scholar]
- 34. Lee V. T., Schneewind O. (1999) Type III machines of pathogenic Yersiniae secrete virulence factors into the extracellular milieu. Mol. Microbiol. 31, 1619–1629 [DOI] [PubMed] [Google Scholar]
- 35. Ohashi H., Shimizu Y., Ying B. W., Ueda T. (2007) Efficient protein selection based on ribosome display system with purified components. Biochem. Biophys. Res. Commun. 352, 270–276 [DOI] [PubMed] [Google Scholar]
- 36. Lee V. T., Tam C., Schneewind O. (2000) LcrV, a substrate for Yersinia enterocolitica type III secretion, is required for toxin targeting into the cytosol of HeLa cells. J. Biol. Chem. 275, 36869–36875 [DOI] [PubMed] [Google Scholar]
- 37. Anderson D. M., Fouts D. E., Collmer A., Schneewind O. (1999) Reciprocal secretion of proteins by the bacterial type III machines of plant and animal pathogens suggests universal recognition of mRNA targeting signals. Proc. Natl. Acad. Sci. U.S.A. 96, 12839–12843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Clemons W. M., Jr., Brodersen D. E., McCutcheon J. P., May J. L., Carter A. P., Morgan-Warren R. J., Wimberly B. T., Ramakrishnan V. (2001) Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus. Purification, crystallization, and structure determination. J. Mol. Biol. 310, 827–843 [DOI] [PubMed] [Google Scholar]
- 39. Neyt C., Cornelis G. R. (1999) Role of SycD, the chaperone of the Yersinia Yop translocators YopB and YopD. Mol. Microbiol. 31, 143–156 [DOI] [PubMed] [Google Scholar]
- 40. Francis M. S., Aili M., Wiklund M. L., Wolf-Watz H. (2000) A study of thr YopD-LcrH interaction from Yersinia pseudotuberculosis reveals a role for hydrophobic residues within the amphipathic domain of YopD. Mol. Microbiol. 38, 85–102 [DOI] [PubMed] [Google Scholar]
- 41. Pallen M. J., Francis M. S., Fütterer K. (2003) Tetratricopeptide-like repeats in type-III-secretion chaperones and regulators. FEMS Microbiol. Lett. 223, 53–60 [DOI] [PubMed] [Google Scholar]
- 42. Bröms J. E., Edqvist P. J., Carlsson K. E., Forsberg A., Francis M. S. (2005) Mapping of a YscY binding domain within the LcrH chaperone that is required for regulation of Yersinia type III secretion. J. Bacteriol. 187, 7738–7752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Edqvist P. J., Bröms J. E., Betts H. J., Forsberg A., Pallen M. J., Francis M. S. (2006) Tetratricopeptide repeats in the type III secretion chaperone, LcrH. Their role in substrate binding and secretion. Mol Microbiol. 59, 31–44 [DOI] [PubMed] [Google Scholar]
- 44. Cambronne E. D., Cheng L. W., Schneewind O. (2000) LcrQ/YscM1, regulators of the Yersinia yop virulon, are injected into host cells by a chaperone dependent mechanism. Mol. Microbiol. 37, 263–273 [DOI] [PubMed] [Google Scholar]
- 45. Cambronne E. D., Sorg J. A., Schneewind O. (2004) Binding of SycH chaperone to YscM1 and YscM2 activates effector yop expression in Yersinia enterocolitica. J. Bacteriol. 186, 829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Olsson J., Edqvist P. J., Bröms J. E., Forsberg A., Wolf-Watz H., Francis M. S. (2004) The YopD translocator of Yersinia pseudotuberculosis is a multifunctional protein comprised of discrete domains. J. Bacteriol. 186, 4110–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Garcia J. T., Ferracci F., Jackson M. W., Joseph S. S., Pattis I., Plano L. R., Fischer W., Plano G. V. (2006) Measurement of effector protein injection by type III and type IV secretion systems by using a 13-residue phosphorylatable glycogen synthase kinase tag. Infect. Immun. 74, 5645–5657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hoiczyk E., Blobel G. (2001) Polymerization of a single protein of the pathogen Yersinia enterocolitica into needles punctures eukaryotic cells. Proc. Natl. Acad. Sci. U.S.A. 98, 4669–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Blaylock B., Sorg J. A., Schneewind O. (2008) Yersinia enterocolitica type III secretion of YopR requires a structure in its mRNA. Mol. Microbiol. 70, 1210–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wood S. E., Jin J., Lloyd S. A. (2008) YscP and YscU switch the substrate specificity of the Yersinia type III secretion system by regulating export of the inner rod protein YscI. J. Bacteriol. 190, 4252–4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cornelis G. R. (2002) Yersinia type III secretion. Send in the effectors. J. Cell Biol. 158, 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ramamurthi K. S., Schneewind O. (2003) Substrate recognition by the Yersinia type III protein secretion machinery. Mol. Microbiol. 50, 1095–1102 [DOI] [PubMed] [Google Scholar]
- 53. Cornelis G. R. (2003) How Yop proteins find their way out of Yersinia. Mol. Microbiol. 50, 1091–1094 [DOI] [PubMed] [Google Scholar]
- 54. Sorg J. A., Miller N. C., Marketon M. M., Schneewind O. (2005) Rejection of impassable substrates by Yersinia type III secretion machines. J. Bacteriol. 187, 7090–7102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee V. T., Schneewind O. (2002) Yop fusions to tightly folded protein domains and their effects on Yersinia enterocolitica type III secretion. J. Bacteriol. 184, 3740–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sory M.-P., Cornelis G. R. (1994) Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14, 583–594 [DOI] [PubMed] [Google Scholar]
- 57. Michiels T., Cornelis G. R. (1991) Secretion of hybrid proteins by the Yersinia Yop export system. J. Bacteriol. 173, 1677–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schesser K., Frithz-Lindsten E., Wolf-Watz H. (1996) Delineation and mutational analysis of the Yersinia pseudotuberculosis YopE domains which mediate translocation across bacterial and eukaryotic cellular membranes. J. Bacteriol. 178, 7227–7233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ramamurthi K. S., Schneewind O. (2003) Yersinia yopQ mRNA encodes a bipartite type III secretion signal in the first fifteen codons. Mol. Microbiol. 50, 1189–1198 [DOI] [PubMed] [Google Scholar]
- 60. Ramamurthi K. S., Schneewind O. (2005) A synonymous mutation in Yersinia enterocolitica yopE affects the function of the YopE type III secretion signal. J. Bacteriol. 187, 707–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bröms J. E., Francis M. S., Forsberg A. (2007) Diminished LcrV secretion attenuates Yersinia pseudotuberculosis virulence. J. Bacteriol. 189, 8417–8429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Amer A. A., Åhlund M. K., Bröms J. E., Forsberg Å., Francis M. S. (2011) Impact of the N-terminal secretor domain on YopD translocator function in Yersinia pseudotuberculosis type III secretion. J. Bacteriol. 193, 6683–6700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mudgett M. B., Chesnokova O., Dahlbeck D., Clark E. T., Rossier O., Bonas U., Staskawicz B. J. (2000) Molecular signal required for type III secretion and translocation of the Xanthomonas campestris AvrBs2 protein to pepper plants. Proc. Natl. Acad. Sci. U.S.A. 97, 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pettersson J., Nordfelth R., Dubinina E., Bergman T., Gustafsson M., Magnusson K. E., Wolf-Watz H. (1996) Modulation of virulence factor expression by pathogen target cell contact. Science 273, 1231–1233 [DOI] [PubMed] [Google Scholar]
- 65. Ramamurthi K. S., Schneewind O. (2002) Type III protein secretion in Yersinia species. Annu. Rev. Cell Dev. Biol. 18, 107–133 [DOI] [PubMed] [Google Scholar]
- 66. Cheng L. W., Schneewind O. (2000) Yersinia enterocolitica TyeA, an intracellular regulator of the type III machinery, is required for the specific targeting of YopE, YopH, YopM, and YopN into the cytosol of eukaryotic cells. J. Bacteriol. 182, 3183–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]