Background: Botulinum HA is a component of the botulinum neurotoxin complex and dramatically increases the oral toxicity of the complex.
Results: The crystal structure of botulinum HA reveals that 12 subcomponents of HA constitute a huge triskelion-shaped molecule.
Conclusion: The complex is functionally and structurally separable into two parts.
Significance: This is the first crystal structure of the whole botulinum HA complex.
Keywords: Bacterial Toxins, Botulinum Toxin, Carbohydrate-binding Protein, Crystal Structure, E-cadherin, Epithelial Cell, Protein Complexes, Bacterial Pathogenesis, Hemagglutinin
Abstract
Clostridium botulinum HA is a component of the large botulinum neurotoxin complex and is critical for its oral toxicity. HA plays multiple roles in toxin penetration in the gastrointestinal tract, including protection from the digestive environment, binding to the intestinal mucosal surface, and disruption of the epithelial barrier. At least two properties of HA contribute to these roles: the sugar-binding activity and the barrier-disrupting activity that depends on E-cadherin binding of HA. HA consists of three different proteins, HA1, HA2, and HA3, whose structures have been partially solved and are made up mainly of β-strands. Here, we demonstrate structural and functional reconstitution of whole HA and present the complete structure of HA of serotype B determined by x-ray crystallography at 3.5 Å resolution. This structure reveals whole HA to be a huge triskelion-shaped molecule. Our results suggest that whole HA is functionally and structurally separable into two parts: HA1, involved in recognition of cell-surface carbohydrates, and HA2-HA3, involved in paracellular barrier disruption by E-cadherin binding.
Introduction
Botulinum neurotoxin (BoNT),4 the causative agent of the disease botulism, is a protein toxin produced by various strains of the spore-forming Gram-positive anaerobic bacteria Clostridium botulinum, Clostridium butyricum, and Clostridium baratii (1). BoNT is released by these bacteria in the form of a multimeric complex, along with a set of nontoxic proteins called neurotoxin-associated proteins (NAPs), encoded by genes adjacent to the BoNT gene (2–4). BoNT exists in seven distinct antigenic types (serotypes A–G), and the amino acid sequences of NAPs have diverged along with their associated BoNTs (3). Of the seven serotypes, types A, B, E, and F cause botulism in both humans and animals, whereas types C and D cause botulism mainly in animals but only very rarely in humans. The BoNT-NAP complex exists as 12 S, 16 S, and/or 19 S multimeric protein particles, depending on the serotype (2, 3). The 12 S toxin contains BoNT and a nontoxic component lacking HA activity, which is designated nontoxic non-HA (NTNH). The 16 S toxin consists of BoNT assembled with NTNH and three other proteins termed HA1, HA2, and HA3; together, these proteins exhibit HA activity and are designated the HA component. The 19 S toxin is presumed to be a dimer of the 16 S toxin.
BoNT targets peripheral nerve terminals, such as motor neuron nerve endings. In the cytosol of neurons, BoNT cleaves SNARE proteins involved in the fusion of synaptic vesicles with the presynaptic membrane, thereby blocking the release of acetylcholine and causing flaccid paralysis (5). Poisoning by BoNTs occurs mainly following oral ingestion of the toxin and subsequent absorption within the intestine (6). Therefore, it is clear that BoNTs in the intestinal lumen must cross the epithelial barrier. However, the detailed mechanisms by which BoNTs traverse this barrier and enter the circulation remain elusive. The presence of NAPs (HA and NTNH) in the toxin complex drastically increases BoNT toxicity following oral administration: the larger the molecular size of the toxin complex, the higher the oral toxicity (2). NAPs exert this effect by rendering BoNT resistant to low pH and proteases in the gastrointestinal tract (2). Recently, the structural basis for the protective effect of NTNH was elucidated by Gu et al. (7), who solved the crystal structure of the 12 S toxin. HA can bind intestinal epithelial cells via its carbohydrate-binding activity, which facilitates efficient transport of the toxin complex across the intestinal epithelial monolayers (8–18). In addition to this adhesive activity, we recently identified a novel activity of HA: type A HA and type B HA (HA/B) disrupt the epithelial paracellular barrier by directly binding to E-cadherin, thereby facilitating paracellular transport of macromolecules, including 12 S toxin (19–22). By contrast, type C HA does not bind E-cadherin but is capable of disrupting the epithelial barrier of certain cells (20). These studies indicate that HA plays an important role in toxin absorption and pathogenesis of botulism. Additionally, these properties of HA could be exploited to facilitate the transepithelial delivery of medicines. However, the molecular details of the mechanisms by which HA elicits these multiple biological activities remain unclear, in part because functional reconstitution studies have not been performed on the whole HA complex, and detailed structural information about the complex has not been previously available (Table 1).
TABLE 1.
Previously reported structures of HA subcomponents
| Structure | Serotype | PDBa ID | Ref. |
|---|---|---|---|
| Whole HA | B | 3WIN | This work |
| HA1-HA2 | D | 2E4M | 37 |
| HA1 | A | 1YBI | 35 |
| HA1 | C | 1QXM | 32 |
| HA1-sugars | C | 3AH1, 3AH2, 3AH4 | 33 |
| HA3 | C | 2ZS6 | 31 |
| HA3-sugars | C | 2ZOE, 4EN6, 4EN7, 4EN8, 4EN9 | 31, 40 |
a PDB, Protein Data Bank.
In this study, we achieved functional reconstitution of whole HA (HA1-HA2-HA3) using purified recombinant HA1, HA2, and HA3 of serotype B. Using this system, we obtained the first x-ray crystallographic structure of whole HA. The results of our structure-function studies suggest that HA1 proteins, which play a critical role in cell binding (9, 10, 13–18), are located in the most distal part of the complex, whereas the E-cadherin-binding site is in the inner part of the complex, the HA2-HA3 connecting region.
EXPERIMENTAL PROCEDURES
Plasmid Construction
DNA fragments encoding botulinum HA (HA1, amino acids (aa) 7–294; HA2, aa 2–146; and HA3, aa 19–626) (23) were amplified by PCR from C. botulinum type B strain Okra genomic DNA using gene-specific primers containing restriction sites at their 5′-ends. In the forward primer for HA1, an oligonucleotide encoding four consecutive aspartic acids was inserted between the restriction site and the gene-coding sequence. The HA1 and HA3 fragments were inserted into the KpnI-SalI site of the pET52b(+) expression vector (Merck Millipore). The HA2 fragment was inserted into the NheI-XhoI site of the pET28b(+) vector (Merck Millipore). The inserted regions were confirmed by DNA sequencing.
Protein Expression and Purification
To obtain high levels of the proteins at high purity, HA1, HA2, and HA3 were expressed as Strep-tag®-, His-, and Strep-tag-tagged proteins, respectively. Expression vectors harboring HA genes were transformed individually into Escherichia coli strain RosettaTM 2(DE3) (Merck Millipore). Protein expression was induced using Overnight ExpressTM Autoinduction System 1 (Merck Millipore). Expression of Strep-tag-HA1 and Strep-tag-HA3 was induced for 36 h at 30 °C. Expression of His-HA2 was induced for 40 h at 18 °C. The Strep-tag-tagged proteins and His-HA2 were purified using StrepTrap HP and HisTrap HP columns (GE Healthcare), respectively. Strep-tag was cut from Strep-tag-HA1 using human rhinovirus 3C protease (Merck Millipore), which was removed by passing the digest through a HisTrap HP column. The His tag was removed from His-HA2 using thrombin (GE Healthcare), and the HA2 protein was purified using a HiTrap DEAE FF column (GE Healthcare). Protein concentrations were determined using the BCA protein assay reagent (Thermo Scientific).
For reconstitution of the HA complex, recombinant HA1, HA2, and HA3 proteins were mixed at a molar ratio of 4:4:1 in PBS (pH 7.4) and incubated for 3 h at 37 °C. The HA complex was captured with a StrepTrap HP column, followed by a brief wash with PBS and incubation with human rhinovirus 3C protease on the column for 2 h at 20 °C to remove Strep-tag attached to the HA3 protein. The HA complex was eluted from the column and further purified with a Superdex 200 10/300 GL column (GE Healthcare). The protein was concentrated using Amicon Ultra-0.5 100K centrifugal filter units (Merck Millipore). The homogeneity of the purified preparation was confirmed by SDS-PAGE and native PAGE. The type B 16 S toxin (B16S toxin) was purified from a bacterial culture of C. botulinum type B strain Okra as described previously (20).
Analysis of Complex Formation by Gel Filtration
Purified HA proteins or mixtures of these proteins were incubated for 3 h at 37 °C prior to gel-filtration analysis. Proteins were individually loaded onto a Superdex 200 10/300 GL column previously equilibrated with PBS (pH 7.4) using an ÄKTA Pure system (GE Healthcare). The purified HA complex was analyzed using the same column equilibrated with 20 mm Tris-HCl (pH 7.4) and 200 mm NaCl. Absorbance was measured at 280 nm.
Measurement of Transepithelial Electrical Resistance (TER)
Measurement of TER was performed using a Millicell-ERS system (Merck Millipore) as described previously (20). Briefly, Caco-2 cells (derived from a human colon carcinoma) were plated onto filters (0.4-mm pore size) in Transwell chambers (Costar). After steady-state TER was achieved, HA proteins were added to the upper or lower side of the chamber. TER was measured at time points up to 24 h.
Crystallization
A 7 mg/ml solution of recombinant HA in 0.01 m Tris-HCl (pH 8.0) was employed for crystallization. Crystallization trials were set up at room temperature as sitting-drop vapor-diffusion experiments on Cryschem crystallization plates. The initial screening was performed at 295 K using the sparse matrix method (24) with commercial crystal-screening kits (Hampton Research). The crystallization droplets consisted of 1 μl of HA solution and 1 μl of reservoir solution containing 0.25 m ammonium chloride, 3% (w/v) polyethylene glycol 4000, 3% trehalose, 3% (w/v) benzamidine HCl, 3% (w/v) methylpentanediol, 3% (w/v) ethylene glycol, 0.1 m calcium chloride, and 0.1 m Tris-HCl (pH 8.0) and equilibrated with 500 μl of reservoir solution. Hexagon-shaped tabular crystals appeared within 1 week and grew to maximum dimensions of ∼0.3 × 0.3 × 0.2 mm.
Data Collection, Phasing, and Refinement
X-ray diffraction data from HA crystals were collected at 100 K in a nitrogen stream after the crystals were soaked in reservoir solution containing 23% ethylene glycol and 3% glycerol as a cryoprotectant. A native data set at 3.5 Å was collected from a single crystal using a Rayonix MX300-HE CCD detector on beamline BL44XU at SPring-8 (see Table 2). The x-ray wavelength was 0.9 Å, the angle oscillation range was 0.5°, and the crystal-to-detector distance was 500 mm.
TABLE 2.
Data collection, structure solution, and refinement statistics for HA
| Diffraction data | HA |
|---|---|
| X-ray source | SPring-8/BL44XU |
| Wavelength (Å) | 0.900 |
| Resolution (Å) | 3.5 |
| Space group | P6322 |
| Unique reflections | 45,595 |
| Rmerge (%)a | 8.6 (49.4) |
| Completeness (%)b | 99.3 (100) |
| Redundancy | 3.5 |
| Refinement | REFMAC5 |
| Resolution range (Å) | 50.0–3.5 |
| R-factorc/Rfreed (%) | 20.2/25.1 |
| No. of protein atoms | 10,522 |
| No. of solvent atoms | 50 |
| Ramachandran distribution (% favored, allowed, outlier) | 90.6, 8.3, 1.1 |
| r.m.s.d.e bonds (Å), angles | 0.011, 1.65° |
| Average B value (Å2) | 62.0 |
| PDB code | 3WIN |
a Rmerge = Σ|Ii − 〈Ii〉|/Σ〈Ii〉, where Ii is the observed intensity, and 〈Ii〉 is the average intensity over symmetry equivalent measurements.
b Completeness for all reflections and for the highest resolution shell is shown in parentheses.
c Rfactor = Σ‖Fobs| − |Fcalc‖/Σ|Fobs|.
d Rfree is calculated as R-factor but on 5% of all reflections that were never used in crystallographic refinement.
e r.m.s.d., root mean square deviation; PDB, Protein Data Bank.
Molecular replacement (Table 2) was performed using Phaser (25). The asymmetric unit contained one HA complex. The resulting electron density maps modified by Parrot (26) allowed us to automatically trace almost all of one independent complex. Model building and inspection were based on Coot/REFMAC5 (27, 28). The atomic coordinates and structure factors for HA/B were deposited in the Protein Data Bank with accession code 3WIN. All images of the molecular structure were prepared using PyMOL.
RESULTS
Reconstitution of the Functional HA Complex
To obtain a functional HA complex, we reconstituted the whole complex from the HA subcomponents in vitro. We expressed each subcomponent of HA separately in E. coli and purified them. When these three subcomponents were mixed and incubated at 37 °C, a large molecular complex formed spontaneously. When the three subcomponents were mixed pairwise and analyzed by gel filtration, only the HA2-HA3 complex was observed (Fig. 1A).
FIGURE 1.
Reconstitution of whole HA from recombinant HA subcomponents. A, each HA subcomponent, alone or in combination, was subjected to gel-filtration analysis as described under “Experimental Procedures.” Dashed lines on the chromatogram show the elution volume of the complexes or HA subcomponents, which are labeled at the top of the panel. Molecular masses were estimated from the elution volumes of the size marker proteins. B, Caco-2 cells were grown in Transwell chambers, and TER was measured in the presence of each HA subcomponent (HA1, 60 nm; HA2, 120 nm; and HA3, 30 nm), alone or in combination. The samples were applied from the basolateral (upper panel) or apical (lower panel) side of the chamber following a 3-h incubation at 37 °C. Values are means ± S.E. (n = 3). C, gel-filtration chromatogram of the whole HA complex used for crystallization. D, Caco-2 cells were treated with the indicated concentrations of B16S toxin (cyan) or HA complex (orange) from the basolateral side of the chamber, and TER was measured. Values are means ± S.E. (n = 3).
We reported previously that HA compromises the barrier function of epithelial cells and that E-cadherin binding of HA is sufficient for this effect (21). We assessed the barrier-disrupting activity of each HA subcomponent, alone or in combination, by measuring TER in Caco-2 cells. Neither the single subcomponents nor the pairwise combinations HA1-HA2 and HA1-HA3 exhibited any activity. By contrast, the HA2-HA3 complex and the full complex exhibited comparable activities when these proteins were applied from the basolateral side of the cells (Fig. 1B, upper panel). These results are consistent with the previous observation that the HA2-HA3 complex is sufficient for E-cadherin binding of HA (21). Because E-cadherins reside on the basolateral surface of epithelial cells, translocation of HA from the apical to the basolateral side of cells is a prerequisite for barrier disruption when the complex is applied from the apical side (19, 21). In this experimental setting, the full complex exhibited higher activity than the HA2-HA3 complex (Fig. 1B, lower panel), indicating that HA1 facilitates apical-to-basolateral translocation of the HA complex.
Our preliminary experiments showed that the stability of the whole HA complex was greatly affected by the tags attached to HA1. In particular, HA complexes readily aggregated when tag-free or His-tagged HA1 was used for reconstitution of the complex, whereas the presence of a FLAG tag (DYKDDDDK) on HA1 improved the solubility of the complex. However, the complex harboring FLAG-HA1 yielded two major peaks in gel-filtration analysis and appeared to aggregate to some extent. Therefore, we added four consecutive aspartic acid residues (a portion of the FLAG tag) to the N terminus of HA1. The resultant purified HA complex, which we used for crystallization, yielded a single peak in gel-filtration analysis, and exhibited barrier-disrupting activity comparable to that of the native B16S toxin (Fig. 1, C and D). In addition to the aspartic acid cluster in HA1, the N terminus of each HA subcomponent contains vector-derived sequences that are not identical to those in the native complex. However, electron density was not observed in these regions, and we assumed that these regions do not affect the overall structure of the complex (see below).
Structure Determination
The crystallographic data are shown in Table 2. Analysis of the diffraction pattern and systematic absences allowed us to assign HA crystals to the hexagonal space group P6322, with unit cell parameters a = b = 324.7 Å, c = 117.6 Å, α = β = 90°, and γ = 120°. A total of 45,592 unique reflections were obtained using the HKL2000 software package (29). The intensity data in the resolution range of 50.0 to 3.5 Å were processed with an Rmerge of 8.6%. Assuming a molecular mass of 148 kDa for the expressed HA (HA1-HA2-HA3 monomer), packing density calculations indicated the Vmax to be 6.06 Å3 Da−1, with one HA arm (HA1-HA2-HA3 monomer) per asymmetric unit. This corresponds to a solvent fraction of about 79.7%, an allowed value for protein crystals.
The crystal structure of whole HA was solved by the molecular replacement technique using the structures of type B HA35 and type D HA1-HA2 (Protein Data Bank ID 2E4M) and the Phaser program from CCP4. The electron density maps were of excellent quality, allowing unequivocal tracing of each subcomponent (HA1-HA2-HA3), and were subsequently refined at 3.5 Å resolution. The structure was refined using REFMAC (28); 5% of the unique reflections were used to monitor the free R-factor. The final values for general and free R-factors were 20.2 and 25.1%, respectively (all reflections in the 50.0–3.5 Å resolution range). The refined model consists of 10,554 atoms with 93 solvent molecules. Stereochemistry checks indicated that the refined model was in good agreement with expectations for models within this resolution range (Table 2). The asymmetric unit contains one HA1-HA2-HA3 complex (Fig. 2A); the structure of the trimer, which represents the natural form, was generated by crystallographic symmetry (Fig. 2B).
FIGURE 2.
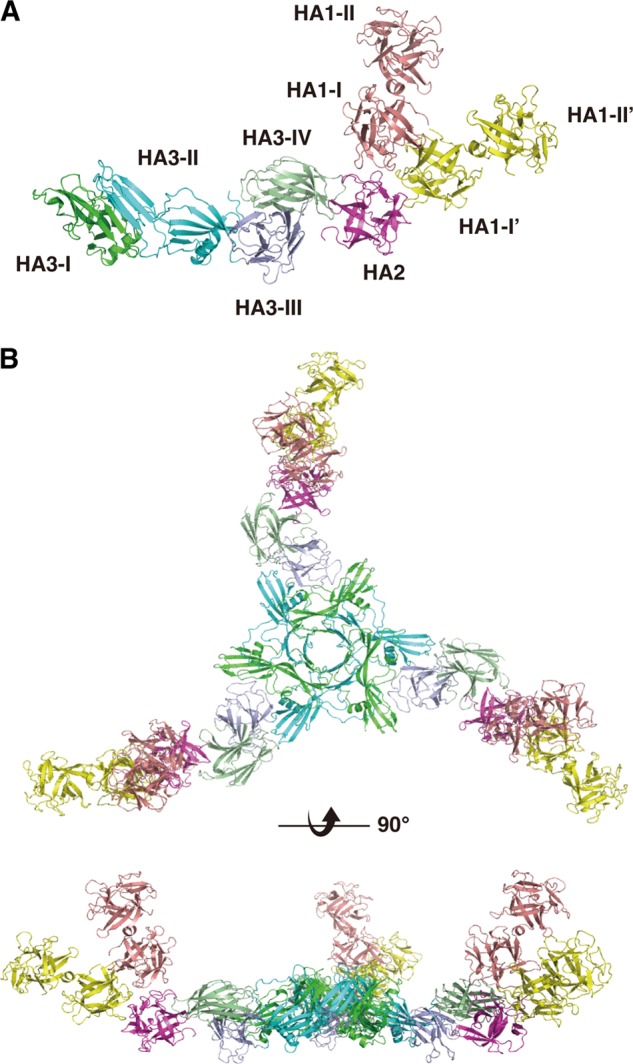
Structure of whole HA. A, ribbon diagram of the HA1-HA2-HA3 monomer. Pink, HA1-I and HA1-II; yellow, HA1-I′ and HA1-II′; magenta, HA2; dark green, HA3-I; cyan, HA3-II; purple, HA3-III; light green, HA3-IV. B, whole HA/B has a triskelion shape. Upper, view from the top of the trimer and parallel to the 3-fold axis. Lower, view perpendicular to the 3-fold axis.
Structure of Whole HA
Whole HA has a triskelion-like fold and forms a pore in the center. Measuring from the central pore, the HA complex has a radius of 160 Å. Notably, the central pore (diameter of ∼26 Å) is the same size as that of hemolysin, a pore-forming toxin produced by Staphylococcus aureus (30). The crystallographic trimeric fold is stabilized by a number of specific interactions between individual HA3 molecules, as observed in type C HA3 (31).
HA1-HA2-HA3 Monomer
Each arm of whole HA assumes a huge elongated Y shape (190 × 96 × 55 Å), which is composed of 91 β-strands, six helices, and seven 310 helices (Fig. 2A). Each HA1-HA2-HA3 monomer contains two molecules of HA1, one molecule of HA2, and one molecule of HA3. From the structural architecture, the two HA1 molecules can be distinguished as four domains, HA1-I, HA1-II, HA1-I′, and HA1-II′ (I and I′, aa 9–146; and II and II′, aa 147–294). Likewise, HA3 can be distinguished as four domains, HA3-I, HA3-II, HA3-III, and HA3-IV (I, aa 22–188; II, aa 209–379; III, aa 380–501; and IV, aa 502–626) (Fig. 2A). HA2 contains only one small domain.
Interactions between HA1 and HA2
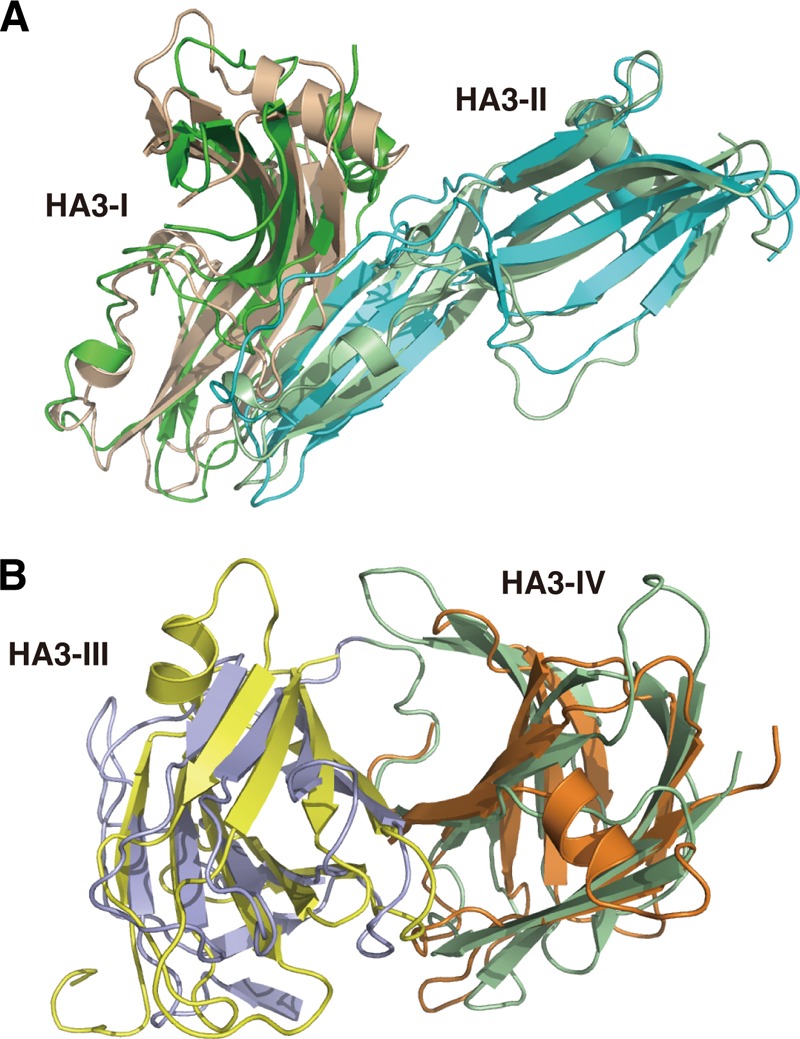
HA1 has a dumbbell-like fold consisting of two β-trefoil domains, the N-terminal (I and I′, aa 9–146) and C-terminal (II and II′, aa 147–294) domains, connected by a short α-helix (Tyr147–Phe152). According to co-crystal studies and mutation experiments using type A and C HA1 (32–34), the C-terminal β-trefoil domain harbors major sugar-binding site(s) that determine the spectrum of carbohydrate recognition of whole HA and the 16 S toxin. No density was observed for the N-terminal residues, including the aspartic acid cluster and residues 7 and 8, implying that these residues adopt a disordered structure. The HA2 molecule consists of a single polypeptide chain with residues 5–145; no electron density was visible for the three N-terminal residues or the C-terminal Ile146. Comparison of the interface residues of type B HA1-HA2 with those of type D HA1-HA2 (Protein Data Bank ID 2EM4) using PISA revealed that the interface hydrophobic residues are well conserved between the two serotypes (Fig. 3, A and B). Accordingly, when type B HA1-HA2 and type D HA1-HA2 were overlapped using SSM from Coot, the interfacing regions of HA2 and the HA1-I and HA1-I′ domains fit well with each other; however, HA1-II and HA1-II′ of types B and D do not overlap (Fig. 4A). This differential interdomain arrangement was observed between two crystal structures of type A HA1 and between type A and C HA1 (35). The arrangement of type B HA1 resembles one of the structures of type A HA1 (Fig. 4B).
FIGURE 3.
Views of the interface regions between HA1 and HA2. Residues directly involved in contacts between subcomponents are shown as stick models. The calculations of interfaces between subcomponents were performed using PISA. A, the interface between HA1 and HA2 of type B. HA1 and HA2 are shown in yellow and magenta, respectively. Residues directly involved in contacts are colored blue and green for HA1 and HA2, respectively. B, the interface between HA1 and HA2 of type D (37). HA1 and HA-2 are shown in blue and cyan, respectively. Residues directly involved in contacts are colored yellow and green for HA1 and HA2, respectively. The residues involved in the interface are conserved between type B and D HA1-HA2.
FIGURE 4.
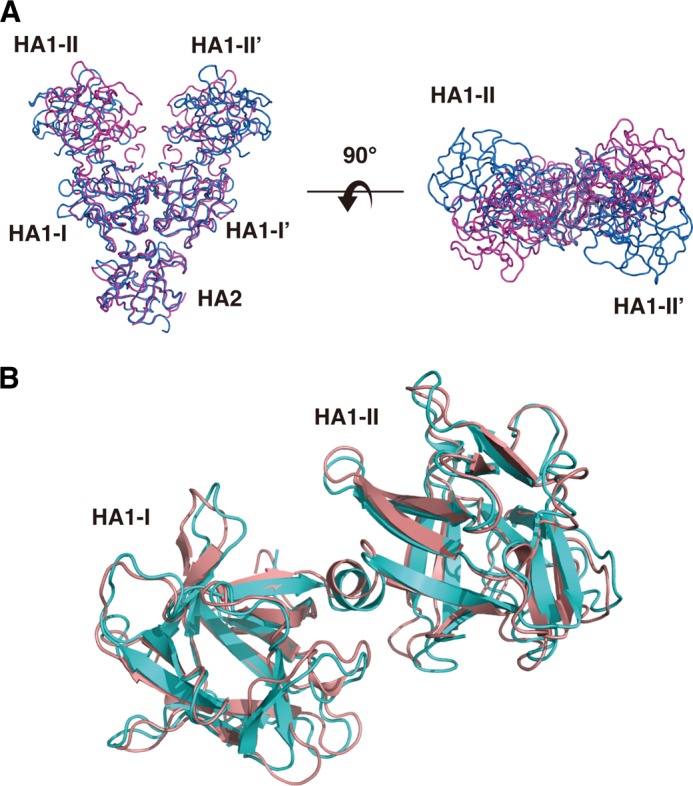
Comparison of the structure of HA1. A, structural comparison of type B HA1-HA2 (blue) with type D HA1-HA2 (magenta). The structures were superimposed using the SSM program in Coot. HA2, HA1-I, and HA1-I′ overlapped well with each other. The structural differences occur mainly on HA-II and HA1-II′, which are sugar-binding sites. B, structural comparison of type B HA1 (pink) with type A HA1 (cyan).
Interactions between HA2 and HA3
Among the key findings in the structure of whole HA are the details of the intramolecular interactions between HA2 and HA3. HA2 binds to the slightly lateral region of the apex of the triangle formed by the HA3 trimer. At the interface, one β-loop-β-region of HA2 is inserted into a crevice formed by two β-loop-β-regions of HA3-IV. Analyses of the scores of the interface performed using PISA revealed that the associations are mediated mainly by hydrophobic interactions, as well as by several salt bridges and hydrogen bonds. The HA3 β-loop-β-regions from Ile565 to Gln584 interact with the hydrophobic phenylalanine cluster of HA2 (Phe7, Phe56, and Phe88) (Fig. 5A). Ile575 of HA3-IV makes a hydrophobic contact with the phenylalanine cluster of HA2, and the next residue (Asp576) forms a hydrogen bond with Arg54 of HA2 (Fig. 5A). On another side, Phe547 of the HA3 β-loop-β-regions from Tyr536 to Thr559 forms a hydrophobic interaction with Ile18, Ile92, and Ala93 (Fig. 5B), and the outside Lys550 of HA3-IV forms a hydrogen bond with Glu119 of HA2. These two specific interactions determine the relative positions of HA2 and HA3.
FIGURE 5.
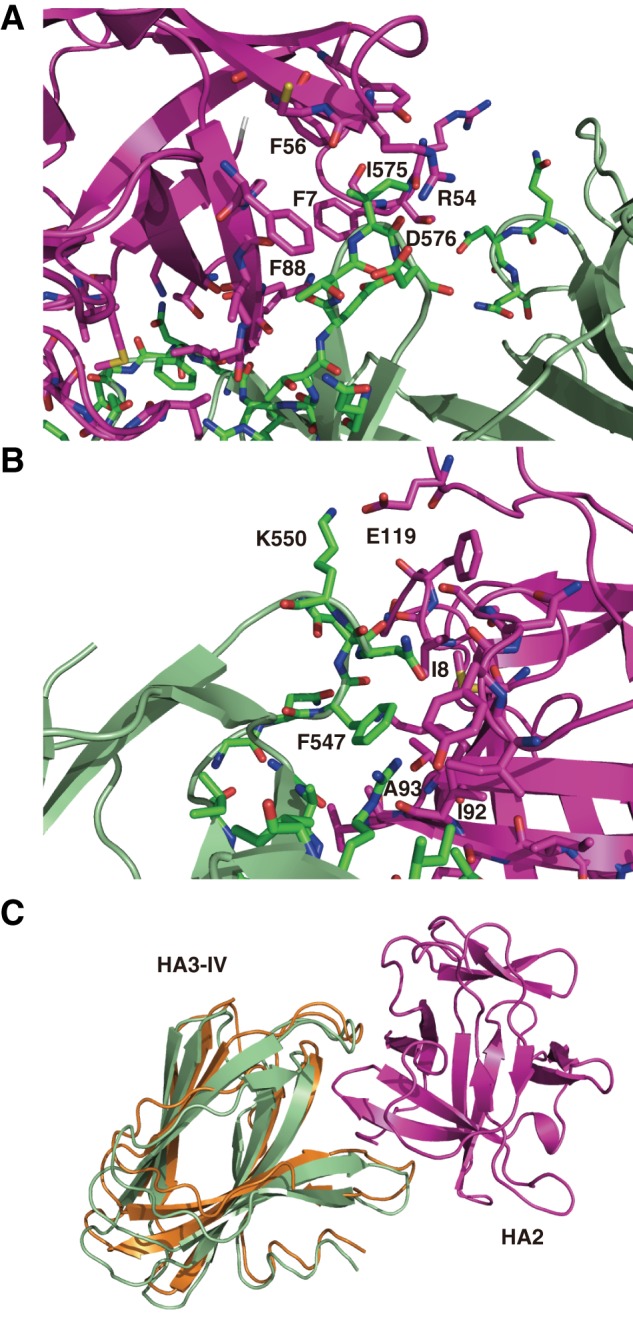
Interactions between HA2 and HA3 in whole HA/B. A and B, close-up view of the HA2-HA3 binding regions. Residues important for interactions between HA2 and HA3 are shown in stick representation. The oxygen, nitrogen, and sulfur atoms are colored red, blue, and yellow, respectively. Carbon atoms are colored magenta in HA2 and green in HA3. C, structural comparison of type B HA3-IV domains in the HA3 trimer (orange) and in the whole HA/B complex (green). The HA2 molecule is also shown in magenta.
Structure of the Type B HA3 Trimer
HA3 is the biggest subcomponent in the whole HA complex, containing residues 19–626; no electron density was visible for the three N-terminal residues 19–21 and aa 189–207. HA3 exists naturally as a trimer, and HA3 is stable for crystallization. We have already obtained three types of type B HA3 crystals: one symmetrically monomeric, one trimeric, and one containing four molecules in the asymmetric unit.5 Comparison of the type B and C HA3 structures (31) revealed that the overall structures are similar. The type C HA3 structure was determined in the monomeric form, but the authors of that study incorrectly traced against the asymmetric unit because they lacked maps of electron density from residues 185–203. The crystal structures that we have solved with four molecules in the asymmetric unit (lacking density maps for aa 189–195) clearly indicate that the natural HA3 monomer has an elongated shape (Fig. 2A).
There are no remarkable structural differences between type B HA3 in the trimeric form and that in the whole HA complex, except in domain HA3-IV, which contains the HA2-binding regions (Fig. 5C). The HA2-binding sites of HA3-IV in whole HA adopt a closed conformation, in contrast to the corresponding region of domain IV in the trimeric HA3 structures. In particular, the two loops of HA3-IV in whole HA have moved to accommodate the HA2 molecule.
Structural Homolog of HA
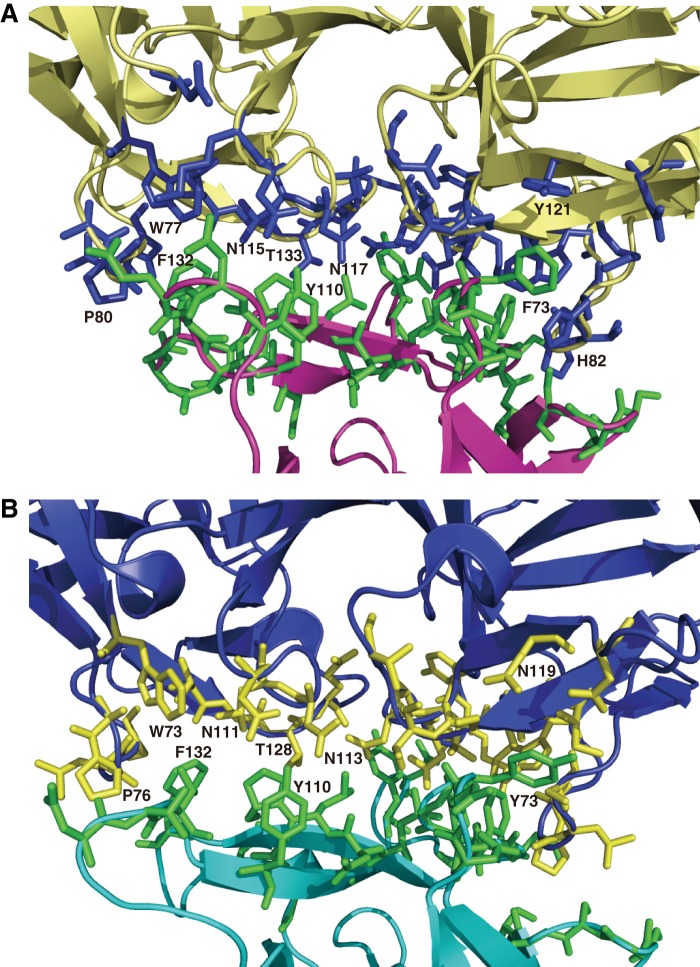
A DALI search was performed on whole HA. As predicted, the results revealed that type C HA1, type C HA3, and type D HA1-HA2 complexes are similar to type B HA1, HA3, and HA1-HA2, respectively. Another structural homolog proposed for type B HA3 is CPE, a β-pore-forming enterotoxin produced by Clostridium perfringens (Protein Data Bank ID 3AM2; Z = 9.7). (Fig. 6, A and B). As described above, HA3 monomers consist primarily of four domains, HA3-I, HA3-II, HA3-III, and HA3-IV. The overall shapes of HA3-I and HA3-II are quite similar to that of the N-terminal pore-forming domain of CPE (called N-CPE), whereas HA3-III and HA3-IV resemble the C-terminal receptor-binding domain of CPE (called C-CPE), which binds to certain members of the claudin family, components of tight junctions (36).
FIGURE 6.
Comparisons of the structures of type B HA1-HA2-HA3 against N-CPE and C-CPE. The structures were superimposed using the SSM program in Coot. HA/B is colored as described for Fig. 2A. HA3-I, HA3-II, HA3-III, and HA3-IV are shown in dark green, cyan, purple, and light green, respectively. A, two N-CPE molecules are overlapped on the HA3-I and HA3-II domains. The two N-CPE molecules are colored wheat and light green, respectively. B, two C-CPE molecules are superimposed on the HA3-III and HA3-IV domains. The two C-CPE molecules are colored yellow and orange, respectively.
DISCUSSION
In this study, we have provided the first crystal structure of whole HA, a multifunctional large protein complex that is critical for the high oral toxicity of the BoNT complex. Type B whole HA is a huge triskelion-shaped complex with an HA1:HA2:HA3 stoichiometry of 6:3:3. The crystal structure is consistent with the three-arm structure proposed previously for the 16 S toxin based on electron microscopy (37, 38).
Our reconstitution experiment showed that among all three pairwise combinations of the three proteins, only HA2 and HA3 form a complex. Although an HA1-HA2 complex was reported in type C HA (39), HA1 formed a stable complex with neither HA2 nor HA3 and was found in a complex only when all three subcomponents were present. These observations suggest that HA2-HA3 complex formation precedes HA1-HA2 association at least in type B. HA2 appears to play a key role in this process. Nevertheless, the detailed mechanism of HA complex formation remains to be elucidated.
As we described previously, HA/B disrupts the epithelial paracellular barrier by binding to E-cadherin via specific protein-protein interactions (21). Our TER data (Fig. 1B) show that basolaterally added HA3 alone does not possess barrier-disrupting activity; however, the HA2-HA3 complex disrupts the barrier almost as potently as whole HA and the 16 S toxin. These data indicate that HA3 has no ability to bind to E-cadherin, whereas the HA2-HA3 complex, whole HA, and the 16 S toxin possess comparable abilities to bind E-cadherin. The structural conformations of HA3 domains are highly conserved between the HA3 trimer and HA3 in whole HA, with the exception of domain HA3-IV, which contains HA2-binding regions. These results indicate that the parts of HA that are essential for E-cadherin binding are located at the connecting region between HA2 and HA3-IV.
A database search identified CPE as a structural homolog of HA3. HA3-I and HA3-II are homologous to N-CPE, whereas HA3-III and HA3-IV are homologous to C-CPE, which is the binding site for receptors (claudins). As described above, HA3-IV is presumed to be involved in E-cadherin binding. Furthermore, type C HA3-IV contains a sialic acid-binding site (31, 40), although the role of sialic acid binding is totally unclear. Type B HA3 has also been reported to bind sialic acid (15), and the amino acid residues involved in sialic acid binding in type C HA3 are conserved in type B. Therefore, the ligand-binding function appears to be conserved between C-CPE and HA3-IV. In contrast, the significance of the homology between the N-terminal parts of these proteins is unclear. The N terminus of CPE is responsible for β-pore formation on the host plasma membrane, in a manner analogous to that of S. aureus hemolysin (41, 42); however, the depth of the HA pore is insufficient to cross the membrane, and some of the amino acid residues conserved among β-pore-forming toxins are not present in HA3 (42). Consistent with this, HA3 has never been reported to form pores on the plasma membrane. In HA, this structure might be exploited for other purposes, e.g. NTNH binding. The N-terminally located long loop of NTNH, designated the nLoop, is involved in 16 S toxin formation (7, 38). This loop is located at the opposite side of the BoNT-interacting surface of NTNH, where it is fully accessible to HA (7). Taken together, these observations suggest that the pore might be used as the docking site for nLoop.
In nature, HA-containing BoNT complexes enter the host through the apical surface of intestinal epithelial cells. Our previous results show that HA disrupts the paracellular barrier of the epithelial monolayer via a two-step mechanism (6, 19, 21). First, HA on the apical surface translocates to the basolateral surface via transcytosis. Next, HA located on the basolateral surface binds to the extracellular domain of E-cadherin, thereby disrupting the paracellular barrier. The results of our functional reconstruction experiment show that the HA2-HA3 complex has attenuated barrier-disrupting activity relative to whole HA when these proteins are added apically, whereas it has the same potency as whole HA when applied basolaterally. These data provide evidence that HA1 plays an important role in the first step of HA action and that the HA2-HA3 complex is necessary and sufficient for the second step. Our results are consistent with the observation that the carbohydrate-binding activity of HA1 in the 16 S toxin plays an important role in the binding and transport of the toxin complex across intestinal epithelial monolayers (9, 10, 13–18). In the structure of whole HA, the orientation of the BoNT-NTNH binding side was easily inferred from studies of negative-stain electron microscopy of 16 S toxins (37, 38). On the basis of this, we propose a model for the interaction of the 16 S toxin with polarized epithelial cells (Fig. 7). First, HA1, which is located on the front of the 16 S toxin, binds to the apical cell surface via its carbohydrate-binding sites and translocates from the apical to the basolateral surface by transcytosis. Next, the HA2-HA3 connecting region interacts with the extracellular domain of E-cadherin at the basolateral surface. Our structure shows that the deduced E-cadherin-binding site at the HA2-HA3 connecting area is located inside whole HA; however, this site would be accessible from the opposite side of the NTNH-binding region even after a complex is formed between HA and cell-surface carbohydrates. In the future, crystallization of whole HA in complex with E-cadherin will facilitate the understanding of the molecular mechanisms by which HA disrupts the paracellular barrier.
FIGURE 7.
Model of the interaction of the B16S toxin with intestinal epithelial cells. A structural model of the B16S toxin was constructed based on the crystal structure of the type A 12 S toxin (7), HA/B (this study), and a model of the type A 12 S and B16S toxins obtained by electron microscopy (38). Left panel, on the apical side of the epithelial cells, the toxin complex binds to the cell surface via the most distally located HA1 (dotted cyan circles), which allows toxin translocation across the cell. Right panel, on the basolateral side, the complex interacts with E-cadherin through the inner region of HA (dotted red circles), thereby disrupting the epithelial barrier. Red, type A BoNT; green, type A NTNH; blue, HA.
In summary, our structure of whole HA in the botulinum neurotoxin complex provides a comprehensive view of this huge multifunctional protein, which plays important roles in intestinal absorption of BoNT. This new information will generate opportunities for understanding the pathogenicity of the botulinum neurotoxin complex, for development of therapeutics against botulinum intoxication, and also for development of novel methods for transepithelial delivery of various macromolecule-based medications.
Acknowledgments
We thank Dr. E. Yamashita and Prof. A. Nakagawa (SPring-8 beamline BL44XU) for help with the x-ray data collection. Diffraction data were collected at Osaka University beamline BL44XU at SPring-8 (equipped with a Rayonix MX300-HE detector), which is financially supported by Academia Sinica and the National Synchrotron Radiation Research Center (Taiwan). We also thank Asami Sano, Takayuki Yoshimura, and Kaoru Sasaki for technical assistance.
This work was supported by the Japan Society for the Promotion of Science (JSPS) through the Funding Program for Next Generation World-Leading Researchers (NEXT Program), initiated by the Council for Science and Technology Policy (CSTP).
The atomic coordinates and structure factors (code 3WIN) have been deposited in the Protein Data Bank (http://wwpdb.org/).
S. Amatsu and K. Kitadokoro, unpublished data.
- BoNT
- botulinum neurotoxin
- NAP
- neurotoxin-associated protein
- NTNH
- nontoxic non-hemagglutinin
- HA/B
- type B HA
- aa
- amino acids
- B16S toxin
- type B 16 S toxin
- TER
- transepithelial electrical resistance
- CPE
- C. perfringens enterotoxin.
REFERENCES
- 1. Poulain B., Popoff M. R., Molgo J. (2008) How do the botulinum neurotoxins block neurotransmitter release: from botulism to the molecular mechanism of action. Botulinum J. 1, 14–87 [Google Scholar]
- 2. Sakaguchi G. (1982) Clostridium botulinum toxins. Pharmacol. Ther. 19, 165–194 [DOI] [PubMed] [Google Scholar]
- 3. Oguma K., Inoue K., Fujinaga Y., Yokota K., Watanabe T., Ohyama T., Takeshi K., Inoue K. (1999) Structure and function of Clostridium botulinum progenitor toxin. J. Toxicol. Toxin Rev. 18, 17–34 [Google Scholar]
- 4. Hill K. K., Smith T. J. (2013) Genetic diversity within Clostridium botulinum serotypes, botulinum neurotoxin gene clusters and toxin subtypes. Curr. Top. Microbiol. Immunol. 364, 1–20 [DOI] [PubMed] [Google Scholar]
- 5. Schiavo G., Matteoli M., Montecucco C. (2000) Neurotoxins affecting neuroexocytosis. Physiol. Rev. 80, 717–766 [DOI] [PubMed] [Google Scholar]
- 6. Fujinaga Y., Sugawara Y., Matsumura T. (2013) Uptake of botulinum neurotoxin in the intestine. Curr. Top. Microbiol. Immunol. 364, 45–59 [DOI] [PubMed] [Google Scholar]
- 7. Gu S., Rumpel S., Zhou J., Strotmeier J., Bigalke H., Perry K., Shoemaker C. B., Rummel A., Jin R. (2012) Botulinum neurotoxin is shielded by NTNHA in an interlocked complex. Science 335, 977–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fujinaga Y., Inoue K., Watanabe S., Yokota K., Hirai Y., Nagamachi E., Oguma K. (1997) The haemagglutinin of Clostridium botulinum type C progenitor toxin plays an essential role in binding of toxin to the epithelial cells of guinea pig small intestine, leading to the efficient absorption of the toxin. Microbiology 143, 3841–3847 [DOI] [PubMed] [Google Scholar]
- 9. Fujinaga Y., Inoue K., Nomura T., Sasaki J., Marvaud J. C., Popoff M. R., Kozaki S., Oguma K. (2000) Identification and characterization of functional subunits of Clostridium botulinum type A progenitor toxin involved in binding to intestinal microvilli and erythrocytes. FEBS Lett. 467, 179–183 [DOI] [PubMed] [Google Scholar]
- 10. Fujinaga Y., Inoue K., Watarai S., Sakaguchi Y., Arimitsu H., Lee J. C., Jin Y., Matsumura T., Kabumoto Y., Watanabe T., Ohyama T., Nishikawa A., Oguma K. (2004) Molecular characterization of binding subcomponents of Clostridium botulinum type C progenitor toxin for intestinal epithelial cells and erythrocytes. Microbiology 150, 1529–1538 [DOI] [PubMed] [Google Scholar]
- 11. Nishikawa A., Uotsu N., Arimitsu H., Lee J. C., Miura Y., Fujinaga Y., Nakada H., Watanabe T., Ohyama T., Sakano Y., Oguma K. (2004) The receptor and transporter for internalization of Clostridium botulinum type C progenitor toxin into HT-29 cells. Biochem. Biophys. Res. Commun. 319, 327–333 [DOI] [PubMed] [Google Scholar]
- 12. Kojima S., Eguchi H., Ookawara T., Fujiwara N., Yasuda J., Nakagawa K., Yamamura T., Suzuki K. (2005) Clostridium botulinum type A progenitor toxin binds to Intestine-407 cells via N-acetyllactosamine moiety. Biochem. Biophys. Res. Commun. 331, 571–576 [DOI] [PubMed] [Google Scholar]
- 13. Uotsu N., Nishikawa A., Watanabe T., Ohyama T., Tonozuka T., Sakano Y., Oguma K. (2006) Cell internalization and traffic pathway of Clostridium botulinum type C neurotoxin in HT-29 cells. Biochim. Biophys. Acta 1763, 120–128 [DOI] [PubMed] [Google Scholar]
- 14. Niwa K., Koyama K., Inoue S., Suzuki T., Hasegawa K., Watanabe T., Ikeda T., Ohyama T. (2007) Role of nontoxic components of serotype D botulinum toxin complex in permeation through a Caco-2 cell monolayer, a model for intestinal epithelium. FEMS Immunol. Med. Microbiol. 49, 346–352 [DOI] [PubMed] [Google Scholar]
- 15. Arimitsu H., Sakaguchi Y., Lee J. C., Ochi S., Tsukamoto K., Yamamoto Y., Ma S., Tsuji T., Oguma K. (2008) Molecular properties of each subcomponent in Clostridium botulinum type B haemagglutinin complex. Microb. Pathog. 45, 142–149 [DOI] [PubMed] [Google Scholar]
- 16. Niwa K., Yoneyama T., Ito H., Taira M., Chikai T., Kouguchi H., Suzuki T., Hasegawa K., Miyata K., Inui K., Ikeda T., Watanabe T., Ohyama T. (2010) Sialic acid-dependent binding and transcytosis of serotype D botulinum neurotoxin and toxin complex in rat intestinal epithelial cells. Vet. Microbiol. 141, 312–320 [DOI] [PubMed] [Google Scholar]
- 17. Inui K., Ito H., Miyata K., Matsuo T., Horiuchi R., Ikeda T., Watanabe T., Ohyama T., Niwa K. (2010) Involvement of sialic acid in transport of serotype C1 botulinum toxins through rat intestinal epithelial cells. J. Vet. Med. Sci. 72, 1251–1255 [DOI] [PubMed] [Google Scholar]
- 18. Ito H., Sagane Y., Miyata K., Inui K., Matsuo T., Horiuchi R., Ikeda T., Suzuki T., Hasegawa K., Kouguchi H., Oguma K., Niwa K., Ohyama T., Watanabe T. (2011) HA-33 facilitates transport of the serotype D botulinum toxin across a rat intestinal epithelial cell monolayer. FEMS Immunol. Med. Microbiol. 61, 323–331 [DOI] [PubMed] [Google Scholar]
- 19. Matsumura T., Jin Y., Kabumoto Y., Takegahara Y., Oguma K., Lencer W. I., Fujinaga Y. (2008) The HA proteins of botulinum toxin disrupt intestinal epithelial intercellular junctions to increase toxin absorption. Cell. Microbiol. 10, 355–364 [DOI] [PubMed] [Google Scholar]
- 20. Jin Y., Takegahara Y., Sugawara Y., Matsumura T., Fujinaga Y. (2009) Disruption of the epithelial barrier by botulinum haemagglutinin (HA) proteins–differences in cell tropism and the mechanism of action between HA proteins of types A or B, and HA proteins of type C. Microbiology 155, 35–45 [DOI] [PubMed] [Google Scholar]
- 21. Sugawara Y., Matsumura T., Takegahara Y., Jin Y., Tsukasaki Y., Takeichi M., Fujinaga Y. (2010) Botulinum hemagglutinin disrupts the intercellular epithelial barrier by directly binding E-cadherin. J. Cell Biol. 189, 691–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sugawara Y., Fujinaga Y. (2011) The botulinum toxin complex meets E-cadherin on the way to its destination. Cell Adh. Migr. 5, 34–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arimitsu H., Inoue K., Sakaguchi Y., Lee J., Fujinaga Y., Watanabe T., Ohyama T., Hirst R., Oguma K. (2003) Purification of fully activated Clostridium botulinum serotype B toxin for treatment of patients with dystonia. Infect. Immun. 71, 1599–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jancarik J., Scott W. G., Milligan D. L., Koshland D. E., Jr., Kim S. H. (1991) Crystallization and preliminary X-ray diffraction study of the ligand-binding domain of the bacterial chemotaxis-mediating aspartate receptor of Salmonella typhimurium. J. Mol. Biol. 221, 31–34 [DOI] [PubMed] [Google Scholar]
- 25. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cowtan K. (2010) Recent developments in classical density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emsley J., Knight C. G., Farndale R. W., Barnes M. J. (2004) Structure of the integrin α2β1-binding collagen peptide. J. Mol. Biol. 335, 1019–1028 [DOI] [PubMed] [Google Scholar]
- 28. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 29. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 30. Song L., Hobaugh M. R., Shustak C., Cheley S., Bayley H., Gouaux J. E. (1996) Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science 274, 1859–1866 [DOI] [PubMed] [Google Scholar]
- 31. Nakamura T., Kotani M., Tonozuka T., Ide A., Oguma K., Nishikawa A. (2009) Crystal structure of the HA3 subcomponent of Clostridium botulinum type C progenitor toxin. J. Mol. Biol. 385, 1193–1206 [DOI] [PubMed] [Google Scholar]
- 32. Inoue K., Sobhany M., Transue T. R., Oguma K., Pedersen L. C., Negishi M. (2003) Structural analysis by X-ray crystallography and calorimetry of a haemagglutinin component (HA1) of the progenitor toxin from Clostridium botulinum. Microbiology 149, 3361–3370 [DOI] [PubMed] [Google Scholar]
- 33. Nakamura T., Tonozuka T., Ide A., Yuzawa T., Oguma K., Nishikawa A. (2008) Sugar-binding sites of the HA1 subcomponent of Clostridium botulinum type C progenitor toxin. J. Mol. Biol. 376, 854–867 [DOI] [PubMed] [Google Scholar]
- 34. Nakamura T., Tonozuka T., Ito S., Takeda Y., Sato R., Matsuo I., Ito Y., Oguma K., Nishikawa A. (2011) Molecular diversity of the two sugar-binding sites of the β-trefoil lectin HA33/C (HA1) from Clostridium botulinum type C neurotoxin. Arch. Biochem. Biophys. 512, 69–77 [DOI] [PubMed] [Google Scholar]
- 35. Arndt J. W., Gu J., Jaroszewski L., Schwarzenbacher R., Hanson M. A., Lebeda F. J., Stevens R. C. (2005) The structure of the neurotoxin-associated protein HA33/A from Clostridium botulinum suggests a reoccurring β-trefoil fold in the progenitor toxin complex. J. Mol. Biol. 346, 1083–1093 [DOI] [PubMed] [Google Scholar]
- 36. Sonoda N., Furuse M., Sasaki H., Yonemura S., Katahira J., Horiguchi Y., Tsukita S. (1999) Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: evidence for direct involvement of claudins in tight junction barrier. J. Cell Biol. 147, 195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hasegawa K., Watanabe T., Suzuki T., Yamano A., Oikawa T., Sato Y., Kouguchi H., Yoneyama T., Niwa K., Ikeda T., Ohyama T. (2007) A novel subunit structure of Clostridium botulinum serotype D toxin complex with three extended arms. J. Biol. Chem. 282, 24777–24783 [DOI] [PubMed] [Google Scholar]
- 38. Benefield D. A., Dessain S. K., Shine N., Ohi M. D., Lacy D. B. (2013) Molecular assembly of botulinum neurotoxin progenitor complexes. Proc. Natl. Acad. Sci. U.S.A. 110, 5630–5635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kouguchi H., Watanabe T., Sagane Y., Sunagawa H., Ohyama T. (2002) In vitro reconstitution of the Clostridium botulinum type D progenitor toxin. J. Biol. Chem. 277, 2650–2656 [DOI] [PubMed] [Google Scholar]
- 40. Yamashita S., Yoshida H., Uchiyama N., Nakakita Y., Nakakita S., Tonozuka T., Oguma K., Nishikawa A., Kamitori S. (2012) Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides. FEBS Lett. 586, 2404–2410 [DOI] [PubMed] [Google Scholar]
- 41. Kitadokoro K., Nishimura K., Kamitani S., Fukui-Miyazaki A., Toshima H., Abe H., Kamata Y., Sugita-Konishi Y., Yamamoto S., Karatani H., Horiguchi Y. (2011) Crystal structure of Clostridium perfringens enterotoxin displays features of β-pore-forming toxins. J. Biol. Chem. 286, 19549–19555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Briggs D. C., Naylor C. E., Smedley J. G., 3rd, Lukoyanova N., Robertson S., Moss D. S., McClane B. A., Basak A. K. (2011) Structure of the food-poisoning Clostridium perfringens enterotoxin reveals similarity to the aerolysin-like pore-forming toxins. J. Mol. Biol. 413, 138–149 [DOI] [PMC free article] [PubMed] [Google Scholar]