Abstract
Dendritic cells (DCs) form a remarkable cellular network that shapes adaptive immune responses according to peripheral cues. After four decades of research, we now know that DCs arise from a hematopoietic lineage distinct from other leukocytes, establishing the DC system as a unique hematopoietic branch. Recent work has also established that tissue DCs consist of developmentally and functionally distinct subsets that differentially regulate T lymphocyte function. This review discusses major advances in our understanding of the regulation of DC lineage commitment, differentiation, diversification, and function in situ.
Keywords: dendritic cells, plasmacytoid dendritic cells, inflammation, innate immunity, adaptive immunity
A BRIEF HISTORY OF THE DENDRITIC CELL LINEAGE
Ralph Steinman and Zanvil Cohn (1, 2) discovered the dendritic cell (DC) in the late 1970s, but the notion that DCs have a unique role in the immune system was met with decades of skepticism. Forty years later, the DC’s exquisite ability to mount adaptive immune responses to foreign antigens is indisputable, and its contribution to the induction of tolerance to self-antigens is also becoming increasingly evident. Consequently, the potential therapeutic benefits of modulating DCs for vaccines and suppressive therapies against pathogens, tumors, and/or autoimmune diseases are now being pursued in the academic and industrial worlds. In recognition of the importance of the discovery of the DC system, Ralph Steinman was awarded several prestigious prizes, including the Gairdner Foundation Award in 2003, the Albert Lasker Award in 2007 (3, 4), and the Nobel Prize in Physiology or Medicine in 2011 (5–8).
Soon after the identification of DCs in lymphoid organs, epidermal Langerhans cells (LCs) (first described by Paul Langerhans in 1868) were recognized as sharing several immunogenic properties with DCs (9), which led to the idea that more than one branch to the DC family might exist. This hypothesis was followed by studies that revealed the presence of cells with a similar phenotype in most non-lymphoid tissues that, upon antigen encounter, traffic through the lymphatics to lymphoid organs, where they localize in the T cell zone and present antigens to T lymphocytes. Such studies, taken together, define the concept of DCs as the sentinels of the immune system, whose main goal is to survey the tissues and instruct the adaptive immune system in response to peripheral cues (10).
In the mid-1990s, the full scope and significance of DC diversity first began to be acknowledged. The finding that murine lymphoid organ DCs consist of two subsets defined by the presence or absence of CD8 expression, with distinct immune functions, substantially broadened our consideration of the roles of DCs in the induction of tissue immunity (11). However, it took another 20 years to extend these findings to nonlymphoid tissues. This was complicated by the fact that although the nonlymphoid CD8+ DC equivalents shared several phenotypic features with their lymphoid counterparts, they did not, in fact, express CD8. Nevertheless, they were eventually characterized by the expression of the integrin CD103 (12).
More recently, a further major division in the DC family was uncovered with the identification of a population of cells that morphologically resemble plasma cells but, upon exposure to viral stimuli, produce enormous amounts of interferon (IFN)-α. Importantly, these cells also differentiate upon stimulation into immunogenic DCs that can prime T cells against viral antigens. The cells were named plasmacytoid DCs (pDCs) (13). To distinguish pDCs from Steinman’s DCs, the latter were renamed classical DCs (cDCs), and remain so today.
Despite strong evidence that DCs in different tissues share phenotypic and functional features that distinguish them from other leukocytes, the difficulties in identifying a dedicated DC precursor in the bone marrow (BM), together with the absence of DC-specific phenotypic markers, generated a level of skepticism that a unified DC lineage exists. In the past decade, different groups have identified a clonogenic DC progenitor that specifically gives rise to DCs and lacks other leukocyte potential (14–16). Several key cytokines and transcription factors that control DC development and diversification in mice have also been discovered. This review discusses major advances in our understanding of DC lineage regulation.
CLASSICAL AND PLASMACYTOID DENDRITIC CELLS
Plasmacytoid Dendritic Cells
pDCs represent a small subset of DCs that share a similar origin with, but a different life cycle than, cDCs (see Table 1). They accumulate mainly in the blood and lymphoid tissues and enter the lymph nodes (LNs) through the blood circulation. pDCs express low levels of major histocompatibility complex class II (MHC-II) and costimulatory molecules and low levels of the integrin CD11c in the steady state. They also express a narrow range of pattern-recognition receptors (PRRs) that include Toll-like receptors (TLRs) 7 and 9. Upon recognition of foreign nucleic acids, they produce massive amounts of type I IFN and acquire the capacity to present foreign antigens. The regulation of pDC development and function was recently reviewed in References 17 and 18 and is not discussed here.
Table 1.
Phenotype of mouse lymphoid and nonlymphoid tissue DCs
| Phenotypical marker | pDC | Lymphoid tissue cDC | Nonlymphoid tissue cDC | Langerhans cell | |||
|---|---|---|---|---|---|---|---|
| CD8+ cDC | CD11b+ cDC | CD103+ CD11b− cDC | CD103+ CD11b+ intestinal cDC | CD103− CD11b+ cDC | |||
| CD45 | + | + | + | + | + | + | + |
| CD11c | + | +++ | +++ | ++ | ++ | ++ | ++ |
| MHC class II | + | ++ | ++ | ++ | ++ | ++ | ++ |
| CD8 | subset | + | − | − | − | − | − |
| CD4 | + | − | +/− | − | − | − | ND |
| CD11b | − | − | + | − | + | + | + |
| CD103 | − | subset | − | ++ | ++ | − | − |
| Langerin | − | subset | − | + | − | − | ++ |
| EpCAM | − | − | − | − | − | − | ++ |
| B220 | + | − | − | − | − | − | − |
| CD24 | ND | ++ | + | ++ | ++ | +/− | ++ |
| Btla | + | ++ | + | ++ | + | +/− | ND |
| c-kit | − | + | + | + | + | +/− | ND |
| CD26 | + | + | + | + | + | +/− | ND |
| Xcr1 | − | + | − | + | − | − | − |
| CD36 | − | + | − | + | ND | − | − |
| Cystatin C | + | ++ | + | ND | ND | ND | ND |
| Clec9a (DNGR1) | + | ++ | − | ++ | − | − | − |
| Cadm1 (Necl2) | − | + | − | ND | ND | ND | ND |
| CD205 | − | ++ | + | ++ | ND | ND | ++ |
| CX3CR1 | − | subset | − | − | − | ++ | + |
| CD209 (dc-sign) | ++ | − | + | − | + | +/− | − |
| F4/80 | − | − | + | − | − | + | + |
| CD172a (Sirpa) | + | − | ++ | − | − | ++ | + |
| CD64 (Fc r1) | − | − | − | − | − | ++ | ND |
| Ly6C | ++ | − | − | − | − | +/− | − |
ND, not determined.
Classical Dendritic Cells
cDCs refer to all DCs other than pDCs. cDCs form a small subset of tissue hematopoietic cells that populate most lymphoid and nonlymphoid tissues. cDCs have an enhanced ability to sense tissue injuries, capture environmental- and cell-associated antigens, and process and present phagocytosed antigens to T lymphocytes. Through these processes, cDCs induce immunity to any foreign antigens that breach the tissues and enforce tolerance to self-antigens. cDCs have a unique potential to perform these functions because of a few key attributes:
Their critical location in nonlymphoid tissues and in the spleen marginal zone in the steady state, where they constantly acquire tissue and blood antigens.
Their superior antigen processing and presentation machinery (19–21).
A superior ability to migrate loaded with tissue antigens to the T cell zone of LNs in the steady state and inflamed state (22).
A superior ability to prime naive T cell responses (10).
PHENOTYPE AND TRANSCRIPTIONAL DEFINITION OF CLASSICAL DENDRITIC CELLS
Dendritic Cell Phenotype
The phenotypic definition of cDCs is evolving and will continue to do so with investigators’ widespread access to new technology such as polychromatic flow cytometry, mass spectrometry, and transcriptional profiling. On the cell surface, DCs constitutively express the hematopoietic markers CD45, MHC-II, and CD11c and lack T cell, natural killer (NK) cell, B cell, granulocyte, and erythrocyte lineage markers (see Figure 1 and Table 1). This definition is much too limited, however, and should not be used in isolation to define cDCs, especially in nonlymphoid tissues. Indeed, CD11c expression is evident on several macrophage populations—particularly lung and intestinal macrophages—but also on cDC precursors and other leukocytes. Additional markers have now been shown to facilitate the delineation of cDCs and macrophages. For example, expression of the tyrosine kinase receptor fms-like tyrosine kinase 3 [Flt3, also termed fetal liver kinase 2 (Flk2) or CD135] is an excellent marker by which to discern cDCs from macrophages. However, its expression is affected by collagenase digestion, which is required for analysis of nonlymphoid tissue DC populations (23). Below, we discuss useful markers to distinguish cDCs from macrophages in the context of each DC subset.
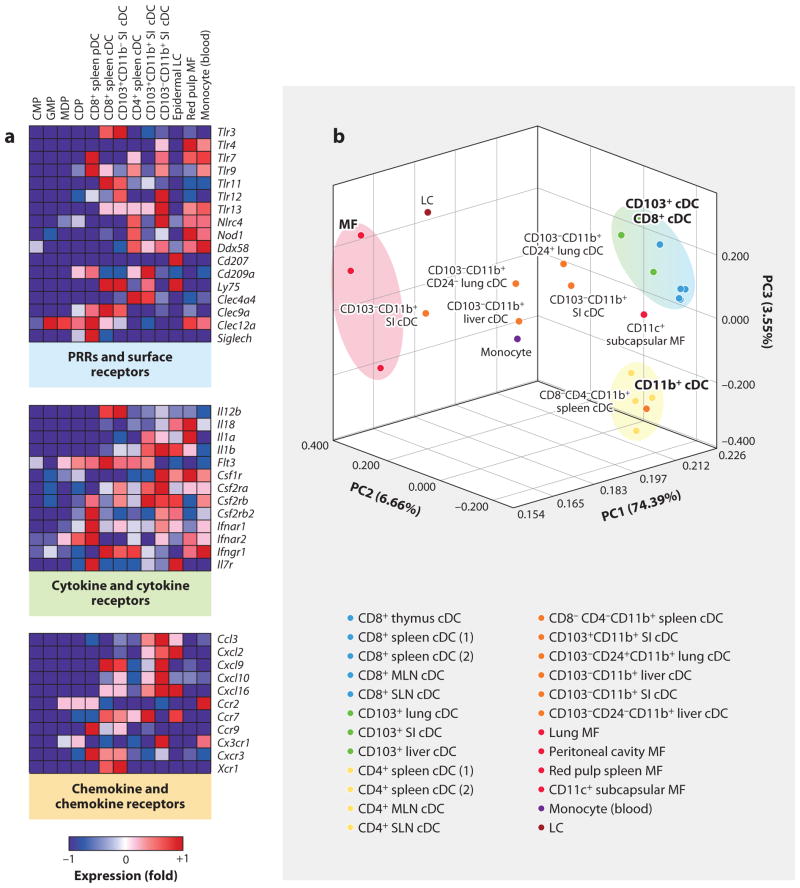
Figure 1.
Heat map representation of transcripts differentially expressed in progenitor and differentiated DCs. (a) Heat map representation of pathogen-recognition receptors (PRRs) and antigen receptors, cytokine and cytokine receptors, and chemokines and chemokine receptors in common myeloid progenitors (CMPs), granulocyte macrophage progenitors (GMPs), macrophage DC progenitors (MDPs), common DC progenitors (CDPs), and CD8+ spleen pDCs, CD8+ spleen cDCs, CD103+CD11b− lamina propria cDCs, CD4+ spleen cDCs, CD103+CD11b+ lamina propria DCs, CD103− CD11b+ lamina propria cDCs, epidermal Langerhans cells (LCs), red pulp macrophages (MFs), and blood monocytes. Red represents high and blue represents low relative expression. (b) Principal components analysis (PCA) of 15% of the most variable transcripts expressed by lymphoid tissue CD8+ cDCs, lymphoid tissue CD8− cDCs, nonlymphoid tissue CD103+ cDCs, nonlymphoid tissue CD11b+ cDCs, epidermal LCs, monocytes, and MF populations provides a visual representation of the heterogeneity of the mononuclear phagocytic lineage. cDC and MF populations cluster distinctly on opposite sides of the PCA, whereas the CD11b+ cDC distribution throughout the PCA suggests that these cells are more heterogeneous. Intriguingly, the CD11c subcapsular MF population clusters near DCs, suggesting that this population is more closely related to DCs than to macrophages (which is further suggested by the expression of zbtb46). (Additional abbreviations used in figure: MLN, mesenteric lymph node; SI, small intestine; SLN, skin-draining lymph node.)
Dendritic Cell Transcriptome
Analysis of the cDC transcriptome within the framework of the entire hematopoietic lineage suggests that cDCs form a unique transcriptional entity that segregates away from other leukocytes including macrophages (see Figure 1). Although few genes are uniquely expressed by cDCs in the context of all hematopoietic cells (24), a group of transcripts discriminates cDCs from macrophages and includes Flt3 (25, 26), c-kit (the receptor for stem cell factor), the chemokine receptor CCR7 (27), and the zinc finger transcription factor zbtb46 (28, 29).
Dendritic Cell Subsets
The role of cDCs as sentinels requires them to constantly sense and respond to environmental stimuli. Responses to environmental stimuli are manifested in the form of phenotypic changes. Thus, it is critical to distinguish between phenotypic plasticity within a cDC population and discrete cDC subsets. The definition of a cDC subset must be based on developmental specificity and functional specialization and not simply on a set of phenotypic differences (see sidebar). Below, we discuss ontogenically and functionally distinct DC subsets that have been identified in lymphoid and nonlymphoid tissues.
Dendritic Cells in Nonlymphoid Tissues
cDCs represent 1–5% of tissue cells depending on the organ and consist of two major subsets: CD103+CD11b− and CD11b+ cDCs.
HANDY HINTS TO CHARACTERIZE MOUSE TISSUE DENDRITIC CELLS IN VIVO.
-
1
The use of CD45 and autofluorescence gating in nonlymphoid tissue: cDCs can often be difficult to find using flow cytometry due not only to their rarity, but also to the propensity of autofluorescent macrophages to contaminate the cDC gate. In nonhematopoietic tissues, we recommend staining for CD45 to visualize hematopoietic cells and thus enrich for cDCs. We further recommend using empty channels to visualize autofluorescent cells before gating the cDC population.
-
2
Collagenase: Most protocols for isolating cDCs from tissues will call for enzymatic digestion of the tissues to create single-cell suspensions. This approach maximizes the cDC yield, but it can result in the destruction of certain cell surface markers. cDC culture in medium and serum for 24 h can help restore surface marker expression.
-
3
MHC-II and CD11c expression on their own are not sufficient: CD11c expression is neither specific nor uniform within the DC family. pDCs express lower levels of CD11c than cDCs, and cDCs downregulate CD11c upon migration or activation. Lung macrophages express CD11c at levels as high as those found on cDCs. Some CD11c expression has been found on spleen macrophages, NK cells, activated T cells, and monocytes. The expression of high levels of MHC-II in addition to CD11c is not sufficient to distinguish DCs in certain contexts. In the gut and lungs, macrophages express MHC-II levels that are as high as those found on cDCs. Furthermore, upon inflammation, both macrophages and pDCs upregulate MHC-II expression. Thus, to accurately visualize cDCs using flow cytometry, we recommend using a panel of markers that includes those highly expressed on macrophages, such as F4/80 Csf-1R and CD64, as well as those specific to cDCs such as Flt3.
-
5
Lineage: Be careful. The term lineage is often used as shorthand for a population of cells that do not express a certain combination of cell surface markers indicative of various lineages. Although this is most often found in the context of hematopoietic progenitors, some studies refer to the exclusion of lineages that are irrelevant to the study as “lin-.” Understandably, the excluded lineages (and thus the definition of lin-) differ according to the purpose of the study, rendering the common use of the term lin- slightly misleading. We recommend caution and careful attention to detail both when interpreting published data and when designing a lineage cocktail for use at home!
-
6
Migratory cDCs can be distinguished from resident cDCs in the steady state but not in the inflamed state: In LNs, migratory cDCs are often distinguished from resident cDCs by some hallmarks of maturation, namely the expression of higher levels of MHC-II and lower levels of CD11c. However, in an inflammatory setting, when resident cDCs are also activated, these markers are no longer sufficient to distinguish between migratory and resident cDCs. The Batf3-dependent cDC subsets can be distinguished by the selective expression of CD8 on resident cells and CD103 on migratory cells. Unfortunately, no equivalent marker(s) currently exist to distinguish between the CD11b+ resident and migratory subsets.
The CD103+CD11b− classical dendritic cell subset
The CD103+CD11b− cDC subset (referred to here as CD103+ cDCs) shares its origin and function with lymphoid tissue CD8+ cDCs (12, 30). CD103+ cDCs populate most connective tissues. The proportion of CD103+ cDCs among total cDCs rarely exceeds 20–30%. In the intestine, CD103+ cDCs are enriched in the Peyer’s patches, and they coexpress the marker CD8 on the cell surface, express low levels of MHC-II, and represent lymphoid tissue–resident CD8+ cDCs; in contrast, most lamina propria CD103+ cDCs express CD11b and thus are discussed in the CD11b+ cDC section below (31, 32) (Table 1). CD103+ cDCs lack the macrophage markers CD11b, CD115, CD172a, F4/80, and CX3CR1 (Table 1). They express higher Flt3 levels compared with CD11b+ cDCs, proliferate in response to Flt3 ligand (Flt3L), and are strongly reduced in Flt3L−/− mice (33). CD103+ cDCs commonly express the C-type lectin receptor langerin, except for intestinal and pancreatic CD103+ cDCs (33, 34) (Table 1). CD103 expression is dependent on the tissue environment and regulated by local production of the cytokine Csf-2 (35–38). CD103−/− mice do not have major defects in DC development (J. Helft & M. Merad, unpublished data), and CD103 is also expressed at high levels on epithelial T cells (39) and naive T cells in mice.
The CD11b+ classical dendritic cell subset
The CD11b+ cDC subset consists of a mixture of tissue cDCs and macrophages, which contributed to the confusion that still exists on the exact contribution of DCs and macrophages to tissue immunity. CD11b+ cDCs most often lack the integrin CD103 and express the integrin CD11b (Table 1). In the muscle and lamina propria, CD11b+ cDCs arise from cDC-restricted precursors and monocytes, but markers to distinguish the two ontogenically distinct subsets differ between tissues. For instance, in the muscle, expression of FcγRI helps distinguish between these two subpopulations (40), whereas expression of CD103 helps distinguish between the two CD11b+ cDC subsets in the lamina propria (31, 41).
Epidermal Langerhans Cells
Epidermal LCs are the DCs that populate the epidermal layer of the skin (34). LCs account for 3–5% of epidermal cells, with approximately 700 LCs/mm2 (34). LCs stand apart from other tissue cDCs through their unique ontogeny and homeostatic properties (42). Compared with dermal cDCs, LCs are characterized phenotypically by lower MHC-II levels, intermediate CD11c levels, and very high levels of the C-type lectin langerin (CD207). Langerin is involved in the formation of the intracytoplasmic Birbeck granules, which for a long time represented a conspicuous but pathognomonic marker for LCs. Murine LCs are uniformly CD11b+F4/80+ and lack CX3CR1 expression (34) (Table 1). In contrast to most DCs, LCs develop independently of Flt3 and Flt3L (33), and similar to many tissue macrophages, they require Csf-1R for their development (43).
Tissue-Migratory Dendritic Cells
Tissue-migratory cDCs are located in the peripheral LNs. They refer to nonlymphoid tissue DCs that have migrated to the tissue-draining LNs through the lymphatics as opposed to the blood-borne lymphoid-resident DCs (44). The nature of migratory cDCs depends on the site of LN drainage (45, 46). Although most tissue-migratory cDCs die in LNs, some exit through efferent lymphatics to access the blood and play a role in tissue immune responses and tolerance (for a complete review, see References 44, 47).
Nonlymphoid tissue cDCs constantly migrate through afferent lymphatics to the T cell areas of LNs charged with antigens (44) in the steady state, and this process increases manyfold in response to inflammatory signals (reviewed in 44, 47, 48). DC migration to the draining LNs is controlled by CCR7 (48, 49). CCR7−/− mice that lack migratory cDCs (49) have helped characterize tissue-migratory DCs. In the steady state, migratory cDCs are distinguished from resident DCs based on higher MHC-II expression and low-intermediate expression of CD11c. However, these features become obsolete during the inflamed setting (see sidebar entitled Handy Hints to Characterize Mouse Tissue Dendritic Cells In Vivo).
The migration process leads to a dramatic transformation of the tissue cDC phenotype, including the upregulation of MHC-II complexes and costimulatory molecules at the cell surface. This process, called DC maturation, occurs in the steady state and upon inflammation (10, 50), but in contrast to steady-state migratory cDCs, those that migrate in response to inflammation also produce inflammatory cytokines and upregulate costimulatory molecules that drive adaptive immunity (for a complete review, see Reference 50). Steady-state factors that trigger cDC migration remain unknown. Loss of E-cadherin-mediated cell adhesion by BM-derived DCs in vitro leads to phenotypical maturation and upregulation of CCR7 expression without production of inflammatory cytokines (51, 52). Whether a similar process controls steady-state cDC migration in vivo remains to be assessed.
Lymphoid Organ–Resident Dendritic Cells
Lymphoid tissue–resident cDCs differentiate in, and spend their entire lives within, lymphoid tissues. LNs also include nonlymphoid tissue–migratory cDCs, whereas lymphoid tissue–resident cDCs make up the entirety of the splenic cDC compartment (47). cDCs are also enriched in mucosa-associated lymphoid tissues in the nasopharynx, Peyer’s patches, and isolated lymphoid follicles in the intestine, which are mostly populated by lymphoid tissue–resident cDCs (53). Lymphoid tissue–resident cDCs consist mainly of two subsets, CD8+ and CD11b+ cDCs (11).
The CD8+ dendritic cell subset
CD8+ DCs represent 20–40% of spleen and LN cDCs. In contrast, most thymic cDCs consist of CD8+ cDCs and are generated locally from early thymocyte progenitors (54). CD8+ cDCs express the CD8α transcript and protein, but not CD8αβ, which is most commonly expressed by CD8+ T cells (11). CD8+ cDCs express no or low levels of the integrin CD11b and other macrophage markers (Table 1). CD8+ cDCs express high Flt3 levels, proliferate in response to Flt3L, and are strongly reduced in Flt3L−/− mice (25, 55). CD8 expression can also be detected on a subset of pDCs (56). In contrast to tissue-migratory cDCs that arrive in the LNs in a mature state, lymphoid tissue CD8+ cDCs are phenotypically immature in the steady state (11). Activation to a phenotypically mature state occurs upon stimulation with microbial products or when cDCs are isolated from the lymphoid tissue and cultured in vitro (57). The CD8+ cDC transcriptome is closely related to that of nonlymphoid tissue CD103+ cDCs and differs from that of CD11b+ cDCs (Figure 1). Accordingly, CD8+ cDCs express distinct lectin and TLRs compared with CD11b+ cDCs. Differentially expressed lectin receptors include CD205, Clec9A, and langerin, which are expressed mostly by CD8+ cDCs, whereas DCIR2 (also called 33D1) is expressed exclusively by CD11b+ DCs (58–60). These differences have been exploited to deliver antigens to specific DC subsets in vivo (60).
The CD11b+ dendritic cell subset
The CD11b+ cDC subset lacks the marker CD8 and most often predominates the lymphoid-resident cDC population in all organs except the thymus. Similar to lymphoid tissue CD8+ cDCs, CD11b+ cDCs proliferate in situ in response to Flt3L, and are reduced in Flt3-and Flt3L-deficient mice, albeit to a lesser extent than CD8+ cDCs (25, 55). In the spleen, CD11b+ cDCs are heterogeneous and are thought to consist of two populations that differentially express the endothelial cell–specific adhesion molecule (ESAM). ESAMhiCD11b+ DCs express higher CD4, CD11c, and Flt3 levels and lower Csf-1R, Csf-3R, and CCR2 levels than do ESAMloCD11b+ DCs. ESAMhiCD11b+ splenic DCs derive from DC-restricted precursors and are dependent on Notch2 signaling, whereas ESAMloCD11b+ DCs are thought to derive from circulating monocytes (61). Thymic CD11b+ DCs are best characterized by the expression of CD172a, express a more mature phenotype, and produce higher levels of the CD4+ T cell attractant chemokines CCL17 and CCL22 than do their CD8+ counterparts (62).
DENDRITIC CELL–RESTRICTED PROGENITORS
With the exception of epidermal LCs, most cDCs are short-lived hematopoietic cells that are constantly replaced by blood-derived precursors. After more than four decades of research, there is now evidence that cDCs arise from a hematopoietic lineage distinct from other leukocytes, establishing the cDC lineage as a distinct hematopoietic branch. Below we describe the successive steps that give rise to DC-restricted progenitors in the BM.
Early Progenitors
Early committed progenitors, including clonal common lymphoid progenitors (CLPs) and clonal common myeloid progenitors (CMPs), have been identified in mice and humans (see Figures 2 and 3) (63). In addition to these committed progenitors, overlapping and alternative graded stages of early lineage commitment have been revealed by recent studies (e.g., in Reference 64). Similar to other hematopoietic lineages, the identification of DC-restricted progenitor has relied on adoptive transfer studies of irradiated animals that carry abnormal levels of circulating cytokines, which will need to be validated using genetically based fate-mapping studies of clonogenic progenitors in the steady state (65). Nonetheless, adoptive transfer of CLPs and CMPs into irradiated animals produces cDCs and pDCs in mice, and similar potential has been found in human CLPs and CMPs cultured in vitro (66–70). CMPs on a per cell basis are much more efficient at generating splenic and LN cDCs, whereas CLPs are more potent at producing thymic cDCs (66, 71). Importantly, maintenance of cDC developmental potential in hematopoietic progenitors is linked to Flt3 expression and to their ability to respond to Flt3L (72–74).
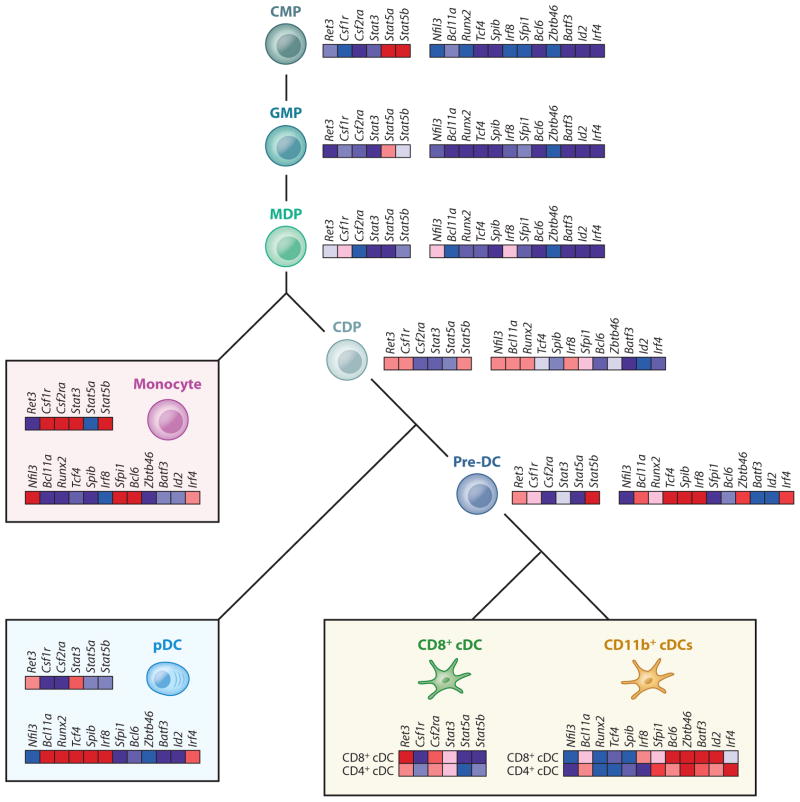
Figure 2.
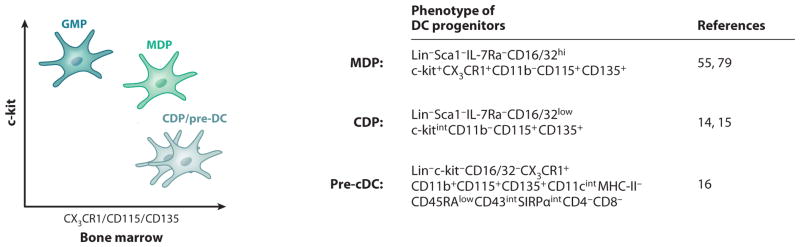
Phenotypes of murine DC progenitors. The illustration on the left suggests that the granulocyte macrophage progenitor (GMP) expresses high levels of c-kit and low levels of CX3CR1, Csf-1R (CD115), and Flt3 (CD135), whereas the macrophage DC progenitor (MDP) is positive for c-kit, CX3CR1, Csf-1R (CD115), and Flt3 (CD135). The common DC progenitor (CDP) expresses intermediate levels of c-kit and is positive for CX3CR1, Csf-1R (CD115), and Flt3 (CD135); pre-cDCs express no or low levels of c-kit but express CD115 and high levels of CD135. The table at the right summarizes the detailed phenotype of MDPs, CDPs, and pre-cDCs.
Figure 3.
Transcriptional control of DC commitment and differentiation. The illustration depicts a heat map representation of cytokines, TLRs, and some transcription factors expressed along the myeloid lineage, starting from the common myeloid progenitors (CMPs), to the granulocyte macrophage progenitors (GMPs), macrophage DC progenitors (MDPs), common DC progenitors (CDPs), pre-cDCs (circulating cDC progenitors), monocytes, plasmacytoid DCs (pDCs), and lymphoid tissue–resident CD8+ and CD11b+ cDCs.
Early Thymic Progenitors
CD8+ thymic cDCs derive predominantly from thymic precursors that take residence in the thymus (66–68, 75, 76). Early results have also shown that, in addition to producing thymic CD8+ cDCs, early thymic progenitors also produce spleen CD8+ DCs upon injection into irradiated animals. This finding led to the suggestion that CD8+ cDCs are lymphoid derived. However, the subsequent realization that upon adoptive transfer into irradiated animals CMPs and CLPs showed a similar bias toward CD8+ cDC generation has refuted this hypothesis, and thus the terms lymphoid and myeloid cDCs are no longer used in this context, with the exception of thymic cDCs (66–68, 75). pDCs and some thymic cDCs also show lymphoid characteristics such as immunoglobulin heavy chain gene rearrangements and are thought to differentiate from lymphoid progenitors (77, 78), although the use of this marker to denote a lymphoid developmental history has been questioned (69).
Intermediate Progenitors
Macrophage dendritic cell progenitors (MDPs)
The commitment of myeloid precursors to the mononuclear phagocyte lineage is thought to occur at the MDP stage (79). MDPs were initially described phenotypically as Lin− c-kit+CX3CR1+ (79) (Figure 2). Upon adoptive transfer, MDPs derive from CMPs or granulocyte macrophage progenitors (GMPs) (16) and produce spleen macrophages, lymphoid-resident cDCs (79), nonlymphoid tissue–resident cDCs (31–33), and some pDCs (80), but not granulocytes (79). A similarly restricted potential was subsequently described in the Lin− Csf-1R+ BM fraction (55). Thus, MDPs appear to be a more restricted developmental intermediate upon the pathway from early myeloid progenitors to macrophages and DCs (Figure 3).
Common dendritic cell progenitors (CDPs)
CDPs were originally isolated as Lin− c-kitintFlt3+Csf-1R+ (14, 15) (Figure 2) and shown to give rise at clonal levels to cDCs and pDCs in Flt3L-supplemented cultures (14, 15). Adoptive transfer of CDPs into irradiated and nonirradiated animals produces spleen and LN CD8+ cDCs, CD11b+ cDCs, and pDCs, with no developmental potential for macrophages (14). CDPs also produce liver and kidney CD103+CD11b− and CD11b+ cDCs (33) and intestinal CD103+CD11b− and CD103+CD11b+ cDCs (31, 32). CDPs were found to be immediately downstream of MDPs (16). Together, these results establish the CDP as the first dedicated DC progenitor in the BM that contributes to both pDCs and to the lymphoid and nonlymphoid tissue cDC pool. These results suggest a pathway in which MDPs differentiate into monocytes and CDPs, which then further give rise to pDCs and downstream cDC progenitors (Figure 3).
Downstream Progenitors
Pre-classical dendritic cells
A CD11c+MHC-II− proliferative precursor (pre-cDC) with a high clonal efficiency in mouse BM and lymphoid tissues, able to differentiate into CD8+ and CD11b+ cDCs but not into pDCs or macrophages in vivo, was identified by two separate groups (81, 82). A recent study defined a similar pre-DC population in the BM, blood, and lymphoid tissues and established the relationship of pre-cDCs to other progenitors by taking advantage of the persistent expression of Flt3 on cDCs throughout their development. This study revealed that pre-cDCs derive from CDPs and migrate from the BM through the blood to home to lymphoid and nonlymphoid tissues (16) (Figure 2). Upon adoptive transfer, pre-cDCs produce spleen CD8+ and CD11b+ cDCs but not pDCs (16). Pre-cDCs also produce CD103+ cDCs and some CD11b+ cDCs in the kidney and liver, along with CD103+CD11b− and CD103+CD11b+ cDCs in the intestine (31, 33). Thus, the pre-cDC is a cDC-restricted progenitor that constantly leaves the BM for the periphery to differentiate locally into cDCs (16, 31, 33) (Figure 3).
Monocytes
Monocytes were originally considered the immediate upstream precursors of cDCs. This hypothesis stemmed from studies showing that human monocytes can differentiate in vitro into cDCs (83). Subsequent studies in mice revealed that monocytes differentiate into cDCs in vivo mainly in infected or inflamed tissues, leading to the concept that monocytes are a precursor of inflammatory DCs (82). More recent studies, however, have established that monocytes contribute to intestinal CD103− CD11b+ DCs (31, 32), splenic CD11b+ ESAMlo DCs (61), and muscular FcγRI+ DCs in the steady state (40).
The Langerhans Cell Paradigm
LCs exhibit specific differentiation and homeostatic features that distinguish them from other cDC and pDC populations. In steady-state conditions, LCs self-renew in situ throughout life independently from the BM (84) and derive mostly from precursors that take residence in the skin prior to birth (85–87). A recent study has suggested that LCs may derive from yolk sac progenitors (88), although we found that the yolk sac contribution to adult LCs does not exceed 10% of the total LC pool and that most LCs derive from fetal liver–derived monocytes (89). Importantly, in inflammatory conditions that lead to severe LC depletion and damage of the epidermal-dermal basal membrane, LCs are repopulated by blood-borne monocytes (43).
CYTOKINE CONTROL OF THE DENDRITIC CELL LINEAGE
The differentiation and expansion of specific lineages are largely regulated extrinsically by different hematopoietic cytokines. Here we review the cytokines that control DC lineage commitment and differentiation in the BM and the maintenance of DC homeostasis in the periphery.
Flt3 Ligand
The cytokine Flt3L is a key regulator of DC commitment in hematopoiesis (90, 91). Flt3L is ubiquitously produced by multiple tissue stroma, endothelial cells, and activated T cells (92). The receptor for Flt3L, Flt3 (also termed CD135 and Flk2), is expressed on short-term repopulating hematopoietic stem cells (HSCs) (93), CLPs, and a subset of CMPs (73). Flt3 is also maintained on DC precursors including MDPs (55, 79), CDPs (14, 15), and pre-cDCs (16), whereas Flt3 expression is lost as progenitors become committed to non-DC lineages (73) (Figure 3). Flt3 is absent from most circulating and tissue leukocytes, with the exception of pre-cDCs and pDCs, and it is maintained on all tissue cDCs (73) (Figure 3), with the exception of LCs, which are unaffected by the loss of Flt3 or Flt3L (31, 33). Flt3 is also expressed on BM-derived DCs cultured in the presence of Flt3L, whereas it is absent from monocyte-derived DCs (73). Loss of Flt3 expression in hematopoietic progenitors correlates with the loss of DC differentiation potential (73), whereas enforcement of Flt3 expression in progenitors which lack DC potential restores some DC developmental potential (94). Mice that lack either Flt3 or Flt3L have reduced numbers of MDPs, CDPs, and tissue cDCs and pDCs (25, 55, 95). Interestingly, Flt3 inhibition and cDC depletion (96, 97) lead to increased Flt3L sera levels, suggesting the presence of a tightly regulated feedback loop in which low numbers of cDCs result in an upregulation of Flt3L production, which in turn induces the generation of DCs. Conversely, injection or overexpression of Flt3L in mice (26, 98) and humans (99, 100) leads to a dramatic expansion of cDCs and pDCs in the blood and in lymphoid and nonlymphoid tissues. In addition to its role in DC differentiation, Flt3L also regulates the proliferation of peripheral DCs to maintain homeostatic DC numbers (55).
Csf-1 (M-CSF)
Csf-1, also known as macrophage colony stimulating factor (M-CSF), is a hematopoietic factor that regulates the survival, proliferation, and differentiation of macrophages (101). The Csf-1 receptor (Csf-1R, also termed CD115) is expressed on GMPs, MDPs, monocytes, and macrophages (Figure 3). Csf-1R is expressed on CDPs, reduced in pre-cDCs, and lost on CD8+ and CD103+ cDCs, but is maintained on a subset of CD11b+ cDCs (Figure 3). Thus, the strength of Flt3 versus Csf-1R signals likely determines MDP progression to CDPs instead of monocytes (92). Csf-1R partly regulates the differentiation or survival of nonlymphoid tissue CD11b+ cDCs, potentially reflecting a monocytic origin of this subset or the heterogeneity of this population (31, 33). Epidermal LCs are totally absent in Csf-1R−/− mice (43) but develop normally in Flt3−/− or Flt3L−/− mice (31, 33). In contrast to Csf-1R−/− mice, mice bearing the natural null osteopetrotic mutation for Csf-1 (Csf-1op/op) mice can form LCs (102), which suggests that a Csf-1R ligand distinct from Csf-1 drives LC homeostasis. Strikingly, IL-34, a newly described high-affinity ligand for Csf-1R (103), is produced at high levels by keratinocytes in the epidermis, whereas epidermal Csf-1 levels are barely detectable (104, 105). Accordingly, IL-34−/− mice lack epidermal LCs but do not have any defects in dermal cDCs or dermal macrophages (104, 105).
Csf-2 (GM-CSF)
Csf-2 is a hematopoietic growth factor that controls the differentiation of the myeloid lineage (106). Csf-2 binds specifically to the Csf-2R, a heterodimer composed of a cytokine-specific alpha chain (Csf-2Rα) and a common signaling beta chain (Csf-2Rβ1) that is shared with the receptors for IL-3 (IL-3R) and IL-5 (IL-5R) (107). Csf-2Rα and Csf-2Rβ are expressed on GMPs, MDPs, and CDPs (Figure 3). Csf-2Rα and Csf-2Rβ are also expressed on CD8+, CD103+, and CD11b+ cDCs (38, 95) (Figure 3).
Csf-2 is a cytokine that is critical for promoting the differentiation of mouse and human hematopoietic progenitors and monocytes into cells that resemble mouse splenic cDCs (83, 108, 109) and remains to date a key cytokine for generating DC-based vaccines for clinical use (110). Therefore, it came as a surprise that mice lacking Csf-2 or its receptor displayed only minor impairment in the development of spleen and LN cDCs (111). More recent studies confirmed that the absence of Csf-2 does not impair the development of lymphoid tissue cDCs; however, they also revealed a reduction in the number of CD103+ cDCs and CD11b+ cDCs in the intestine, dermis, and lung of Csf-2−/− mice, consistent with high Csf-2 production in these tissues in wild-type mice (38, 95, 112). The tissue cDCs that remained in Csf-2−/− mice showed proapoptotic defects, suggesting that Csf-2 is a critical regulator of cDC survival in nonlymphoid tissues, but not in lymphoid organs (38). Importantly, Csf-2 also controls CD103 expression on tissue cDCs (35, 37, 38) and has been implicated in the final stages of cDC maturation and in the acquisition of the capacity to cross-present antigens (35, 36).
Lymphotoxin β
Mice deficient in the tumor necrosis factor (TNF) receptor family member lymphotoxin β receptor (LTβR) or its membrane-associated ligand LTα1β2 have reduced numbers of splenic cDCs (113–115), specifically of CD11b+ cDCs, whose proliferation is impaired (114, 115). B cells are a key source of LTα1β2 for CD11b+ cDC homeostasis, and transgenic overexpression of LTα1β2 on B cells leads to expansion of the CD11b+ cDC compartment (114). Furthermore, mice that lack RelB, through which the LTβR signals, have dramatically reduced numbers of splenic CD11b+ cDCs (116). The contribution of LTβR to the homeostasis of CD11b+ nonlymphoid tissue cDCs has yet to be explored.
TGF-β1
TGF-β1 is required for LC differentiation in mice and humans (117–120). Although TGF-β1 is expressed by both keratinocytes and LCs, an autocrine source of TGF-β1 is required for LCs to develop (121). TGF-β1 also acts directly on LCs in an autocrine and paracrine manner to inhibit steady-state and inflammation-induced migration (122, 123). Mice deficient in the inhibitor of DNA 2 (Id2), a TGF-β1-induced inhibitor of helix-loop-helix (HLH) transcription factors, also lack LCs (124), although the exact role of Id2 in LC development remains unclear. The transcription factor Runx3 mediates cDC responses to TGF-β1 and is required for LC development (125).
TRANSCRIPTIONAL CONTROL OF THE DENDRITIC CELL LINEAGE
Interferon Regulatory Factors
A number of IFN regulatory factors (IRFs) have been implicated in the development of DC subsets in mice and humans (see Table 2). IRF8 (also known as IFN consensus sequence–binding protein) plays a critical role in myeloid cell differentiation, promoting macrophage differentiation while inhibiting the development of granulocytes (126, 127). Irf8−/− animals develop a myeloproliferative disease distinguished by excessive granulocyte production; failure to generate adequate monocyte numbers (128); and lack of pDCs, spleen-resident CD8+ cDCs, and nonlymphoid tissue CD103+ cDCs (Figure 4) (33, 129–132). A spontaneous point mutation (R294C) of IRF8 in BHX2 mice also causes myeloproliferative disease (133) and, interestingly, impairs the development of CD8+ and CD103+ cDCs and IL-12 production without impairing pDC generation (33, 134). In addition to its role in DC development, IRF8 also plays a critical role in DC function. IRF8 controls CD8+ cDC maturation and IL-12 production (129), controls the migration of LCs to the draining LNs (135), and plays a role in the tolerogenic functions of DCs, positively regulating the expression of Indo, the gene that encodes the enzyme indoleamine 2,3-dioxygenase (136). In contrast to IRF8-deficient mice, mice deficient in IRF4 have reduced numbers of splenic CD4+ cDCs (137) but have no defects in CD8+ cDC development. IRF4 and IRF8 are differently regulated by Csf-2, which suppresses IRF8 while promoting IRF4 expression (138). IRF2-deficient mice have reduced numbers of splenic CD4+ cDCs and epidermal LCs (139). Interestingly, this defect was largely reversed in mice that lacked the IFN-α receptor IFNAR1, suggesting that IRF2 mediates its effect on DC development through its ability to attenuate type I IFN signaling. How this relates to the changes in hematopoiesis that occur upon inflammation will be an intriguing area for further investigation.
Table 2.
DC-deficient mice modelsa
| Phenotype of mice lacking regulator | pDC | Lymphoid tissue cDC | Nonlymphoid tissue cDC | Langerhans cell | |||
|---|---|---|---|---|---|---|---|
| CD8+ cDC | CD8− cDC | CD103+ cDC | CD103+ CD11b+ intestinal cDC | CD11b+ cDC | |||
| Batf3 | ↔ | ↓ ↓ ↓ | ↔ | ↓ ↓ ↓ | ↔ | ↔ | ↔ |
| Bcl6 | ↔ | ↓ ↓ ↓ | ↓ ↓ ↓ | ||||
| Gfi1 | ↓ | ↓ | ↓ | ↑ | |||
| ID2 | ↑ | ↓ ↓ ↓ | ↔ | ↓ ↓ ↓ | ↔ | ↔ | ↓ ↓ ↓ |
| Ikaros C | ↓ | ↓ ↓ ↓ | |||||
| Ikaros DN* | ↓ ↓ ↓ | ↓ ↓ ↓ | |||||
| Ikaros L/L** | ↓ ↓ ↓ | ↑ | ↓ | ||||
| IRF2 | ↔ | ↓ ↓ | ↓ | ||||
| IRF4 | ↓ | ↔ | ↓ ↓ | ||||
| IRF8 | ↓ ↓ ↓ | ↓ ↓ ↓ | ↔ | ↓ ↓ ↓ | ↔ | ↔ | ↓ |
| Nfil3 | ↔ | ↓ ↓ ↓ | ↔ | ||||
| Notch-RBPJ | ↔ | ↔ | ↓ ↓ ↓ | ||||
| Notch2 | ↔ | ↔ | ↓ ↓ | ↔ | ↓ ↓ ↓ | ↔ | ↔ |
| Pten | ↔ | ↑ ↑ | ↔ | ↑ ↑ | ↔ | ↔ | |
| PU.1 (Sfpi) | ↓ ↓ ↓ | ↓ ↓ ↓ | |||||
| RelB | ↔ | ↓ ↓ ↓ | ↔ | ||||
| Runx3 | ↑ ↑ | ↓ | ↓ ↓ ↓ | ||||
| SpiB | ↓ ↓ ↓ | ↔ | ↔ | ||||
| Stat3 | ↓ ↓ | ↓ ↓ ↓ | ↓ ↓ ↓ | ||||
| Stat5 | ↓ ↓ | ↓ ↓ | ↓ ↓ | ||||
| Tcf4 (E2-2) | ↓ ↓ ↓ | ↔ | ↔ | ||||
| Xbp1 | ↓ ↓ | ↓ | ↓ | ||||
| Zbtb46 | ↔ | ↔ | ↔ | ↔ | ↔ | ↔ | |
| Flt3 | ↓ ↓ ↓ | ↓ ↓ ↓ | ↓ ↓ ↓ | ↓ ↓ ↓ | ↓ ↓ ↓ | ↓ ↓ | ↔ |
| Csf-2R | ↔ | ↔ | ↓ ↓ ↓ | ↓ ↓ | ↓ | ↔ | |
| Csf-1R | ↔ | ↔ | ↔ | ↔ | ↔ | ↓ ↓ ↓ | ↓ ↓ ↓ |
| TGF-β | ↔ | ↔ | ↔ | ↔ | ↔ | ↔ | ↓ ↓ ↓ |
The table describes the phenotype of transcription factor knockout mice lacking specific DC subsets. ↑ indicates a reduction in cell numbers; ↑ indicates an increase in cell numbers; and ↔ means no change in cell numbers. The number of arrows is representative of the severity of the phenotype, with ↓ ↓ ↓ suggesting an almost complete loss of the population.
Ikaros DN = dominant negative allele.
Ikaros L/L = low-level expression.
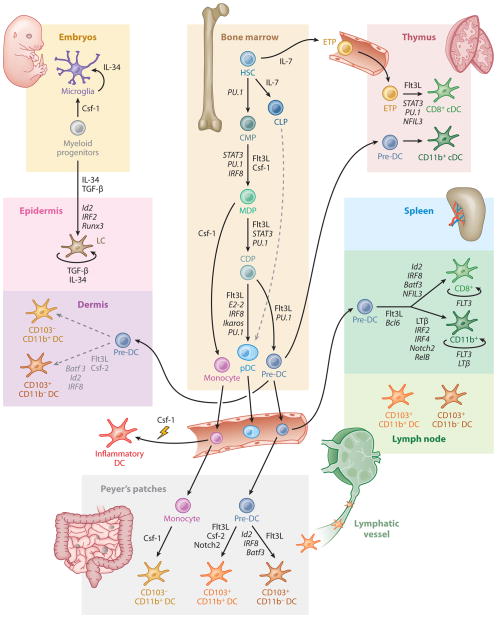
Figure 4.
Regulation of DC development and homeostasis in mice. This illustration summarizes the current model of the developmental pathways of both lymphoid tissue–resident and nonlymphoid tissue–resident murine DCs. Dashed lines indicate pathways that are likely but not yet definitively shown to operate in DC development. Cytokines and transcription factors that are important in each transition are indicated (HSC, hematopoietic stem cell; CMP, common myeloid progenitor; CLP, common lymphoid progenitor; MDP, macrophage DC progenitor; CDP, common DC progenitor; ETP, early thymic progenitor; mono, monocyte; LC, Langerhans cell).
Inhibitor of DNA Binding Protein 2
Id proteins, members of the HLH transcription factor family, act to inhibit the binding of other HLH proteins to DNA. Id2 and HLH family E-protein E2-2 mutually antagonize one another. E2-2 directly controls the expression of pDC genes, while also antagonizing several DC genes, including Id2 (140). Accordingly, E2-2 is required for pDC development (141), and the constitutive loss of E2-2 in peripheral pDCs induces the upregulation of cDC genes (140). In contrast, mice lacking Id2 have dramatic reductions in CD8+ and CD103+ cDCs (33, 124, 142); furthermore, enforced Id2 expression in early human hematopoietic progenitors inhibits pDC development but leaves cDC development unaffected (143).
Batf3
The basic leucine zipper transcription factor ATF-like 3 (Batf3) has a selective, nonre-dundant role in DC development. Although Batf3 is expressed in all cDCs including the CD8+ and CD103+ cDCs and the CD11b+ cDCs, mice lacking Batf3 have a selective de-ficiency in CD8+ and CD103+ cDCs in the 129S6/SvEv strain (132, 144). Batf3−/− mice on the C57BL/6 background lack CD103+ cDCs and have reduced spleen CD8+ cDCs, but retain normal numbers of CD8+ LN cDCs (37). Molecular compensation for Batf3 was recently observed in Batf3-deficient mice infected by Toxoplasma gondii and was shown to be provided by the induced cytokines that are related to the AP1 factors Batf and Batf2. Compensation among BATF factors was based on the shared capacity of their leucine zipper domains to interact with non-AP1 factors such as IRF8 to promote DC differentiation (145).
Zbtb46
The zinc finger transcription factor zbtb46 is expressed on endothelial cells and erythroid progenitors, but its expression within the immune system is restricted to the cDC lineage (28, 29). Specifically, zbtb46 starts to be expressed at the pre-cDC stage and remains expressed on spleen CD8+ and CD11b+ cDCs, nonlymphoid tissue CD103+ cDCs, and some CD11b+ cDCs, whereas it is absent in pDC, monocytes, and macrophages (28, 29). Deletion of zbtb46 does not alter cDC development in vivo (29, 146) but skews cDC composition in favor of CD8+ cDCs and results in partial activation of cDCs, establishing zbtb46 as a negative regulator of cDC activation (146). Diphtheria toxin (DT) administration to transgenic mice expressing DT under the zbtb46 promoter (zbtb46-DTR mice) is fatal within 24–48 h, suggesting that zbtb46 is expressed on radioresistant cells (28). Administration of DT to lethally irradiated mice reconstituted with zbtb46-DTR BM results in depletion of cDCs while sparing monocytes, macrophages, and NK cells, all of which are reduced upon DT treatment in CD11c-DTR mice (28). Thus, the identification of zbtb46 as a marker of the cDC lineage presents the field of DC biology with the exciting prospect of identifying and manipulating DC populations with a new specificity.
STATs
STAT3, a key component of the Flt3 signaling pathway, plays a nonredundant role in DC development (147). Mice lacking STAT3 have profound reductions in DCs and pDCs that cannot be rescued by Flt3L administration (147), whereas enforced expression of STAT3 in Flt3 negative progenitors restores some DC potential (94). STAT5 mediates Csf-2 suppression of pDC generation (148) via inhibition of IRF8 transcription (138). It also plays a role in the latter stages of human DC development in vitro in the presence of Csf-2 (149).
NF-κB Pathway Transcription Factors
The transcription factors RelB and TNF-associated factor 6 (TRAF6), which are involved in the NF-κB signaling pathway, have been implicated in the development of CD11b+ splenic cDCs. Mice deficient in either of these molecules show reduced levels of splenic CD11b+ cDCs (116, 150), their phenotype mimicking that seen in the LTβ−/− spleen (114). Both TRAF6 and RelB are involved in mediating signaling through the LTβ receptor, suggesting that activation of these transcription factors underlies the role of LTβ in CD11b+ cDC development.
Ikaros
The transcription factor Ikaros plays a role in the development of multiple hematopoietic lineages, including DCs; in two separate Ikaros mutant models, mice deficient in functional Ikaros lack thymic and splenic cDCs. Ikaros mutant BM failed to generate cDCs in mixed BM chimeric animals, indicating a cell-intrinsic requirement for Ikaros in DC generation (151).
Notch RBP-J
The transcription factor Notch RBP-J, which mediates signaling from the Notch receptor, plays an important role in the maintenance of the splenic CD11b+ cDC compartment; mice that lack Notch RBP-J in the CD11c+ compartment have a selective survival disadvantage in CD11b+ splenic cDCs (152). Interestingly, a more recent report found that Notch2 signaling is critical for the development of a subset of CD11b+ cDCs characterized by high ESAM expression (61) and for the development of CD103+CD11b+ lamina propria cDCs of the small intestine (61).
PU.1
PU.1, a member of the ETS family of transcription factors, has multiple roles in hematopoiesis and the process of lineage specification. Mice reconstituted with PU.1-deficient hematopoietic cells, in addition to other hematopoietic defects, have far fewer CD11b+ and CD8+ cDCs than those reconstituted with wild-type BM, and PU.1-deficient progenitors cultured in the presence of Csf-2 cannot differentiate into DCs in vitro (153, 154). Furthermore, PU.1−/− CMPs, CLPs, and CDPs showed defective DC differentiation potential both in vitro in response to Flt3L or Csf-2 and upon in vivo transfer, although in the latter case, the generation of most other immune cells was also diminished (155). Enforced expression of PU.1 in megakaryocyte erythroid progenitors (MEPs) that lack DC developmental potential restored both their Flt3 expression and DC potential (94). Conversely, enforced expression of Flt3 in MEPs led to PU.1 expression, suggesting a positive feedback loop between Flt3 and PU.1 that works to reinforce DC potential (94). However, restoration of Flt3 expression in PU.1-deficient progenitors did not restore their potential to give rise to DCs (155), indicating that in addition to Flt3, other targets of PU.1 are vital for DC differentiation.
REGULATION OF DENDRITIC CELL HOMEOSTASIS IN VIVO
In contrast to the long-term dogma that tissue cDCs are end-differentiated cells incapable of cell division, short-term bromodeoxyuridine (BrdU) labeling studies in mice revealed that 5% of lymphoid organ cDCs or their immediate progenitors are actively cycling at any given time (33, 84, 114, 156, 157). Thus, the rapid labeling kinetics and loss of BrdU labeling observed in experiments designed to measure DC half-life (158, 159) were due to cell division and not to cDC replacement or turnover. Parabiotic mice that share the same blood circulation for prolonged periods provide a model to follow the physiological turnover of blood-borne cells. Three weeks after parabiosis, 10–30% of cDCs derive from parabiont partners (33, 157) except for epidermal LCs (84). Upon separation, partner-derived DCs are entirely replaced by endogenously derived cells in 10–14 days, except for lung cDCs, which require more than 25 days to be replaced (33, 157). These data suggest that although cDC progenitors proliferate locally with small burst size, they do not self-renew and are continuously replaced by blood-borne precursors. In the steady state, cDC division is regulated by LTβR and Flt3. Flt3 is essential for maintaining DC homeostasis, in part by regulating the division of mature CD8+ and CD11b+ cDCs (Figure 4) (55) in situ, whereas the effects of LTβR ligands appear to be limited to CD11b+ spleen DCs (114, 115).
FUNCTIONAL SPECIALIZATION OF TISSUE-RESIDENT CLASSICAL DENDRITIC CELL SUBSETS
The development of conditional depletion models of CD11c+ cells in transgenic mice expressing DTR under the CD11c promoter (CD11c-DTR mice) has for the first time allowed the field to probe the role of DCs in a more physiological setting (Figure 5) (160). Although the use of CD11c-DTR mice has dramatically fostered DC biology, DT injection in CD11c-DTR mice also leads to the depletion of tissue macrophages, monocyte-derived DCs, and some NK cells. Results obtained with this model may have overestimated DC contributions to tissue immune responses (28). Thus, the use of conditional depletion models that specifically target DCs should help clarify the exact contributions of the DC subsets and macrophages to tissue immune responses.
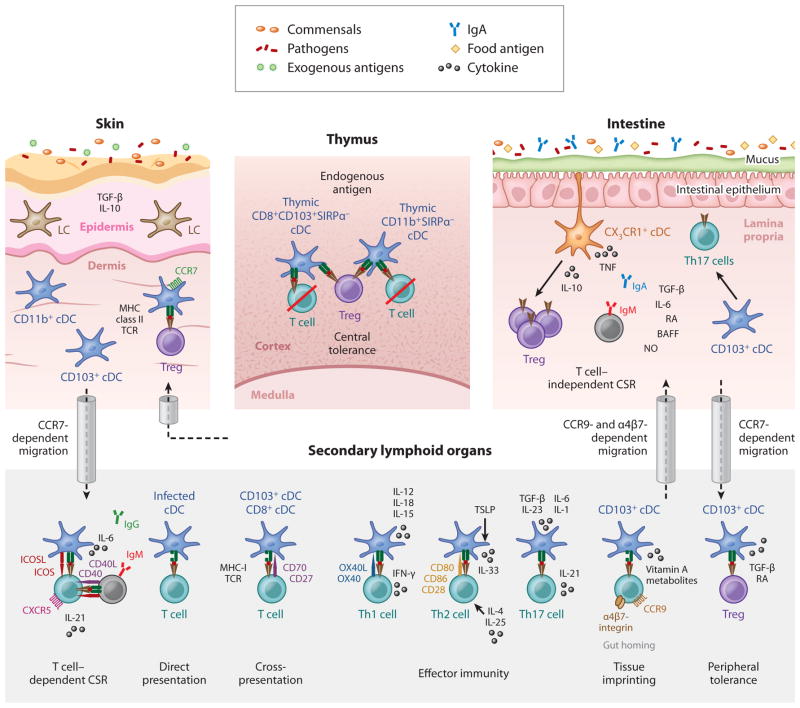
Figure 5.
DC controllers of adaptive immunity. This illustration summarizes key DC functions, highlighting their importance as regulators of adaptive immune functions in lymphoid and nonlymphoid tissues. DC subsets that populate peripheral tissues capture commensals, food antigens, or exogenous antigens and migrate in a CCR7-dependent manner to the draining lymph node, where they present tissue-derived antigens to CD8+ T cells (cross-presentation) and CD4+ T cells (direct presentation), thereby inducing peripheral tolerance in the steady state or effector immunity in the injured state. Gut tissue CD103+ cDCs that migrate to the draining lymph node can promote the induction of gut-homing molecules (α4β7 and CCR9) on naive T cells (tissue imprinting), thereby promoting T cell migration to gut tissue. cDCs also promote T cell–dependent class switch recombination (CSR).
Functional Specialization of the CD8+ and CD103+ Dendritic Cell Subsets
Lymphoid tissue CD8+ DCs and nonlymphoid tissue CD103+ DCs have the same origin and share a similar phenotype and transcriptional profile, as discussed above. In this section, we review data suggesting that these two DCs also share similar functional attributes and discuss the molecular cues that control the functional specialization of the CD8+ and CD103+ DC subsets.
Sensing pathogens and tissue damage
CD103+ cDCs are specifically enriched in nonlymphoid tissues at the interface with the environment and efficiently migrate charged with tissue antigens to the T cell zone of the draining LN (12). In the spleen, CD8+ cDCs are located in the marginal zone, an ideal location to filter blood antigens, whereas in the LNs CD8+ cDCs are located in the subcapsular sinus, the site of entry of afferent lymphatic vessels that drain nonlymphoid tissues (161, 162). From these strategic locations, CD8+ cDCs migrate to the T cell zone to present blood or tissue antigens to T lymphocytes (161–163). Splenic CD8+ cDCs express a specific pattern of recognition receptors (164–166). Recent studies have also found that CD103+ and CD8+ cDCs express a similar TLR, C-type lectin receptor, and chemokine receptor profile (23) (Figure 1). In particular, CD8+ and CD103+ cDCs are the only cDCs that express the double-stranded viral RNA sensor, TLR3 (164, 167), and the Toxoplasma gondii protein sensor TLR11 (168). Both subsets express high levels of the scavenger receptor CD36—which binds to dead cells (169)—and high levels of the C-type lectin Clec9A—which senses necrotic bodies (172). They also express DEC205 (CD205), and langerin (CD207) but lack DCIR2 and dectin 1 (60, 170–174).
Activation of naive CD8+ T cells
Ex vivo studies using cDCs purified after antigen inoculation in vivo revealed the superior ability of CD8+ and CD103+ cDCs over other cDCs to present microbial antigens (175–180) and cell-associated antigens (144, 176, 181–183) to CD8+ T cells. In Langerin-DTR mice, lung CD103+ DCs and lymphoid tissue CD8+ DCs are eliminated, along with LCs. This mouse model revealed for the first time in vivo the nonredundant role of langerin+ DCs in the induction of influenza virus–specific CD8+ T cells (184) and Leishmania major–specific CD8+ T cells (185). Similarly, Batf3-deficient mice that specifically lack CD8+ and CD103+ cDCs are unable to mount an efficient virus-specific cytotoxic T cell response upon subcutaneous injection of West Nile virus or pulmonary infection with an influenza virus; they are also unable to reject immunogenic fibrosarcoma tumors (144, 186). Together, these models have established the critical and nonredundant role of CD8+ and CD103+ DCs in the induction of CD8+ T cell immunity in vivo (132, 144).
The molecular cues that mediate the enhanced ability of CD8+ and CD103+ cDCs to mount CD8+ T cell immune responses are only starting to be unraveled.
Increased cross-presentation potential
Most of the studies on the molecular control of cross-presentation have used BM-derived DCs and are reviewed extensively elsewhere (21, 187–189). In this section, we highlight mainly the studies that assessed the cross-presentation potential of primary CD8+ and CD103+ cDCs. The ability of CD8+ cDCs to efficiently process and load exogenously acquired antigens on MHC-I molecules was established more than a decade ago (181). Similar to CD8+ cDCs, lung and dermal CD103+ cDCs have a superior ability compared with CD11b+ DCs to cross-present cell-associated antigens (190). Cross-presentation depends on two critical factors that include (a) an endocytic pathway with low degradative potential (191, 192) and (b) a phagosome-to-cytosol transport step that allows both the transfer of antigens from the phagosome to the cytosol and their loading onto MHC-I molecules (193). CD8+ cDCs demonstrate both of these attributes. Splenic CD8+ cDCs are enriched in Rac2, a GTPase that helps maintain an alkaline phagosome that limits protease activity, thereby favoring cross-presentation of exogenous antigens (194). Injection of cytochrome c, which induces apoptosis in cells equipped with the potential for cytosolic transfer, induces apoptosis selectively in CD8+ cDCs, indicating that these cells more efficiently transfer exogenous protein to the cytosol than do CD11b+ cDCs (195). Mechanisms leading to the disruption of phagosomal membranes have also been implicated in cytosolic transfer (196), and CD8+ cDCs were found to overexpress adipose differentiation–related protein, a molecule involved in lipid body formation that provides a source of oxidative stress that destabilizes phagosomal membranes and leads to the release of antigens to the cytosol (197).
Increased CD8+ T cell priming potential
CD8+ and CD103+ cDCs may also have a superior ability to prime CD8+ T cells independently of their cross-presentation potential. CD8+ cDCs express more genes related to MHC-I presentation than do CD11b+ cDCs (60). They are also the main source of IL-12 (198–200, 201) and IL-15 (202), two cytokines involved in the differentiation of cytotoxic CD8+ T cells (203). CD8+ and CD103+ cDCs are also the only hematopoietic cells that express the chemokine receptor XCR1; its ligand (XCL1) is rapidly produced by CD8+ T cells upon antigen presentation by CD8+ cDCs and promotes the differentiation of CD8+ cytotoxic T cells (204).
Activation of CD4+ T cells
The role of CD8+ cDCs in the activation of CD4+ T cells is not as clear as their role in the activation of CD8+ T cells. CD8+ splenic cDCs are the main producers of the Th1-polarizing cytokine IL-12 (198, 199). Dermal CD103+ cDCs control the induction of pathogen-specific CD4+IFN-γ+ T cells upon cutaneous infection with Candida albicans (205), and the ablation of dermal CD103+ DCs in langerin-DTR transgenic mice abrogates the induction of encephalitogenic CD4+ Th1 responses and the development of experimental autoimmune encephalomyelitis (EAE) (112). In addition, induction of CD8+ T cell effector and memory responses to herpes simplex virus 1 (HSV-1) depends on cognate licensing of CD4+ T cells by CD8+ cDCs in an antigen-specific manner, suggesting that the efficiency of CD8+ DCs to prime CD8+ T effector cells requires CD4+ T cell priming potential (206). However, Batf3-deficient mice that lack CD103+ and CD8+ cDCs can mount efficient CD4+ T cell responses to immunogenic fibrosarcoma tumors and to West Nile virus (144) as well as myelin oligodendrocyte glycoprotein–specific Th1 cells upon subcutaneous immunization, leading to severe EAE (37). Similarly, ablation of dermal CD103+ cDCs in Langerin-DTR mice does not affect the development of a CD4+ T cell response to Leishmania major infection (185). The discrepancy between these studies may suggest that the predominant role of cDCs in the induction of Th1 varies according to ligands encountered by each subset.
Central and peripheral tolerance
In the thymus, endogenous CD8+ cDCs and blood-borne CD11b+ cDCs contribute to central tolerance through the negative selection of developing thymocytes and the induction of T regulatory cells (Tregs) (207, 208), whereas in the periphery, CD8+ and CD103+ cDCs are thought to participate in deletional tolerance of self-reactive T cells and the induction of antigen-specific Tregs. A study done in rats revealed that the intestinal migratory OX41− DC subset constantly transports dying intestinal cells to the draining LNs (209), the first suggestion of functional specialization in the transport of dying cell–associated antigens. More recent studies showed that cutaneous migratory CD103+ DCs are the only cutaneous DCs able to cross-present keratinocyte-associated antigens to LN CD8+ T cells in the steady state (176, 183). Spleen CD8+ DCs capture dying cells and promote the deletional tolerance of antigen-specific CD8+ T cells (162, 210, 211). These results are consistent with the finding that constitutive deletion of CD11c+ DCs leads to autoimmune disorders (212), although autoimmune reactions were not reported in another strain of mice that constitutively lacks CD11c+ cells (97) or in Batf3−/− mice that lack CD8+ and CD103+ DCs (132, 144). The discrepancy between these studies remains to be clarified. However, it is worth noting that if CD8+ and CD103+ DCs are critical in the induction of tolerance as well as of autoimmune responses, deletion of this subset will subsequently not lead to autoimmune reactions despite the fact that these cells are critical for the maintenance of self-tolerance.
Distinct contribution of CD8+ and CD103+ dendritic cells to immune responses
Conditional depletion and knockout mouse models that specifically lack CD8+ or CD103+ cDCs are unavailable to researchers. This critical lack is due to the difficulty of identifying molecules expressed by one subset but not the other, such that assessing the exact contribution of each subset to tissue immunity is difficult and has relied mostly on ex vivo assays. Earlier studies have suggested that LN CD8+ cDCs are the main contributors to antiviral CD8+ T cell immune responses through their ability to cross-present tissue-migratory cDCs charged with viral antigens (213). Although this hypothesis has been proven true in some settings (214, 215), it is now clear that tissue CD103+ DCs are sufficient to drive T cell tolerance or immunity upon maturation (12, 186).
Functional Specialization of the CD11b+ Classical Dendritic Cell Subset
Similar to all cDCs, CD11b+ cDCs can sense pathogens and migrate from nonlymphoid tissues to regional LNs charged with self and foreign antigens (Figure 5) (165, 175, 216, 217). However, conditional depletion models of the CD11b+ cDC subset are lacking, limiting our current understanding of the in vivo contribution of this specific subset to tissue immunity. In addition, blood-derived cDCs that infiltrate inflamed lymphoid and nonlymphoid tissues share many phenotypical characteristics with the CD11b+ classical cDC subset, which further complicates the analysis of the contribution of tissue-resident CD11b+ cDCs to tissue immunity.
Sensing
CD11b+ cDCs express distinct PRRs compared with CD8+ and CD103+ cDCs (Figure 1), although the exact receptor profile of CD11b+ cDCs will need to be revisited when markers of CD11b+ cDC heterogeneity are described. In the gut, for example, the PRR expression profile of CD103+CD11b+ cDCs is more similar to that of CD103+CD11b− cDCs than to that of CD103− CD11b+ cDCs (Figure 1). Splenic CD11b+ cDCs express high cytoplasmic viral sensor levels (165) and are potent cytokine producers in the steady state and upon stimulation (218).
Activation of CD8+ T cells
Earlier studies suggested that although cross-presentation is constitutively active in CD8+ splenic cDCs, it can be induced in CD11b+ splenic cDCs via, for example, ligation of the Fcγ receptor (219). Ex vivo assays in viral infection models suggest that CD11b+ cDCs can also present viral antigens to CD8+ T cells (216, 220, 221), although it remains unclear whether MHC-I presentation results from cross-presentation of infected cell–associated antigens or direct presentation of viral antigens. We recently found that in influenza virus–infected mice, lung CD11b+ cDCs were protected from viral infection; they exclusively stimulated virus-specific CD4+ T cells but were unable to efficiently cross-present virally infected cells (186).
Activation of CD4+ T cells
CD11b+ cDCs are thought to have a predominant role in MHC-II presentation. CD4+CD11b+ splenic cDCs express higher levels of genes coding for proteins involved in the MHC-II antigenic pathway compared with CD8+ splenic cDCs (60). In vivo antigen delivery to CD11b+ splenic cDCs revealed that the CD4+CD11b+ cDC subset (which overlaps significantly with the ESAMhi cDC subset) is more efficient than CD8+ cDCs in MHC-II presentation to CD4+ T cells in the steady state (60, 222). Upon infection with HSV-1 or influenza virus, migratory CD11b+ cDCs present viral antigens predominantly to CD4+ T cells ex vivo (176, 216). CD11b+ dermal cDCs are the predominant subset to drive the accumulation of antigen-specific CD4+ T effector cells and Tregs upon subcutaneous vaccination with the relevant antigens combined with adjuvant (221, 223). The design of a mouse model allowing the specific depletion of CD11b+ cDCs is crucial to determining the exact contribution of CD11b+ cDCs to CD4+ T cell priming in vivo.
Central and peripheral tolerance
CD11b+ cDCs constantly migrate from the blood to the thymus (224) and can induce clonal deletion of autoreactive T cell or Treg differentiation (62, 225). However, the exact contribution of blood-borne CD11b+ cDCs to central tolerance remains unclear. Migratory lamina propria CD103+CD11b+ cDCs have a superior ability to induce peripheral Treg differentiation in vivo in part because of their superior ability to express aldehyde dehydrogenase (ALDH), an enzyme that metabolizes dietary vitamin A into retinoic acid (226, 227). In the dermis, CD11b+ cDCs also specifically express ALDH (228) and are potent at inducing the differentiation of antigen-specific Tregs in vitro (223, 228, 229).
Functional Specialization of the Langerhans Cells
Epidermal LCs were considered the prototype tissue cDC. However, the finding that LCs were unable to generate CD8+ T cell immunity to HSV-1 (214) and the realization that dermal cDCs are highly functional and diverse have together led to the suggestion that LCs may not be the ubiquitous T cell primers that had long been depicted in the literature. Mouse depletion models (230) have shown that LCs are alternately dispensable, required, or redundant with dermal CD103+ cDCs for induction of contact hypersentivity (CHS) (for a complete review, see Reference 231). The latter is consistent with results in the Batf3−/− mice that lack dermal CD103+ cDCs but not LCs and that develop normal CHS responses (132). Other studies have shown that CHS responses are increased in the absence of LCs, suggesting that LCs can have a tolerogenic role in vivo (205, 232, 233). In humans, LCs drive the proliferation of epidermal-resident Tregs in the steady state but limit Treg activation in the inflamed state (234). In the context of allogeneic BM transplantation, we found that host LCs are sufficient to prime allogeneic T cell responses to induce graft-versus-host disease (235). Recent results have also shown that upon infection with Candida albicans, LCs promote Th17 cell differentiation but are unable to induce Th1 responses or to cross-present antigens to CD8+ T cells, whereas dermal CD103+ cDCs are strong cross-presenters and drive Th1 cell differentiation (205). Together, these results suggest that LC function in vivo depends on the type of inflammatory signal to which they are subjected. Thus, results obtained with one specific model do not reveal information about the cell-intrinsic immunogenic properties of LCs.
DEVELOPMENT OF INFLAMMATORY DENDRITIC CELLS
Inflammatory DCs refer to a population of DCs that are transiently formed in response to microbial or inflammatory stimuli and disappear once the inflammation resolves. The regulation of inflammatory DC development and function remains poorly understood, in part because of the lack of comprehensive studies comparing results from different inflammation models used in inflammatory DC biology studies.
Definition of Inflammatory Dendritic Cells
The phenotype of inflammatory DCs is likely influenced by the nature of the stimuli, the tissues in which they arise, and the kinetics of analysis. Although most inflammatory DCs are characterized by the expression of Ly6C, CD11b, MHC-II, and intermediate CD11c levels (236), Ly6C, for example, is quickly downregulated upon tissue entry, making it difficult to distinguish inflammatory DCs from tissue-resident CD11b+ cDCs. Inflammatory DCs that accumulate in the LN in response to lipopolysaccharide (LPS) injection express the lectin DC-SIGN/CD209 and the mannose receptor CD206 (237, 238). LPS-induced DCs fail to accumulate in Flt3L−/− animals, express the DC-specific transcription factor zbtb46 (239), and are eliminated in zbtb46-DTR mice treated with DT (28), suggesting that these cells correspond to bona fide DCs. A subset of inflammatory DCs initially identified in animals infected with Listeria monocytogenes was termed TNF-α/iNOS-producing DCs (TipDCs) because of their ability to produce high amounts of TNF-α and iNOS (240). However, in contrast to LPS-induced inflammatory DCs, TipDCs lack zbtb46 (239) and are spared in zbtb46-DTR mice treated with DT (28), suggesting that they are more related to activated monocytes or macrophages than are bona fide DCs. The use of the zbtb46 marker should help classify inflammatory DCs and lead researchers to revisit their contribution to tissue immune responses.
Precursors of Inflammatory Dendritic Cells
Circulating monocytes consist of two main subsets, Ly6Chi and Ly6Clow monocytes, with distinct homing and functional properties (80). Among monocytes, circulating Ly6Chi monocytes are considered to be the direct precursors of inflammatory DCs (80, 236, 241, 242).
Recent studies have also established that in situations of stress, early hematopoietic precursors are also able to differentiate directly into DCs, bypassing normal growth and differentiation requirements (243). HSCs, CMPs, GMPs, and CLPs express TLRs (243, 244) (Figure 3). Upon TLR engagement, murine CLPs differentiate into DCs in vitro (244) and in vivo (245) at the cost of B cell development. Similarly, human CD34+ hematopoietic progenitors also express TLRs, and TLR engagement in vitro biases their lineage commitment to myelopoiesis over lymphopoiesis (246, 247). HSCs also reside in nonlymphoid tissues and differentiate into inflammatory DCs in response to TLR stimuli (248). Upon LPS injection in the periphery, BM CDPs downregulate CXCR4, allowing them to leave the BM, up-regulate CCR7, and home to the LNs, where they differentiate into DCs (249). These data indicate that direct pattern recognition via TLR ligation by early hematopoietic cells fosters the generation of innate immune cells and, in particular, DCs at the expense of other lineages. Whether the contributions of monocytes and DC-restricted precursors to the generation of inflammatory DCs differ quantitatively or qualitatively in vivo remains to be explored.
Cytokine and Chemokine Control of Inflammatory Dendritic Cells
The chemokine receptor CCR2 controls monocyte exit from the BM (240) and recruitment to the site of inflammation and infection (250, 251); accordingly, monocyte-derived DCs are greatly reduced in CCR2−/− mice (240). The cytokines and factors that control the differentiation of monocytes into inflammatory DCs are less well defined, but key requirements appear to be the recognition of bacterial products through TLRs and MyD88 (252, 253) or T cell activation signals (254). Csf-2 was thought to control the differentiation of inflammatory DCs (255). However, recent data from our laboratory revealed that the absence of Csf-2R does not affect the accumulation of inflammatory spleen DCs during the first few days following LPS injection, during infection with Listeria monocytogenes, or in influenza virus–infected lungs, although it remains possible that Csf-2 plays a role in the accumulation of DCs in chronic injuries, given its prosurvival role on nonlymphoid steady-state tissue cDCs (38).
THE HUMAN DENDRITIC CELL LINEAGE
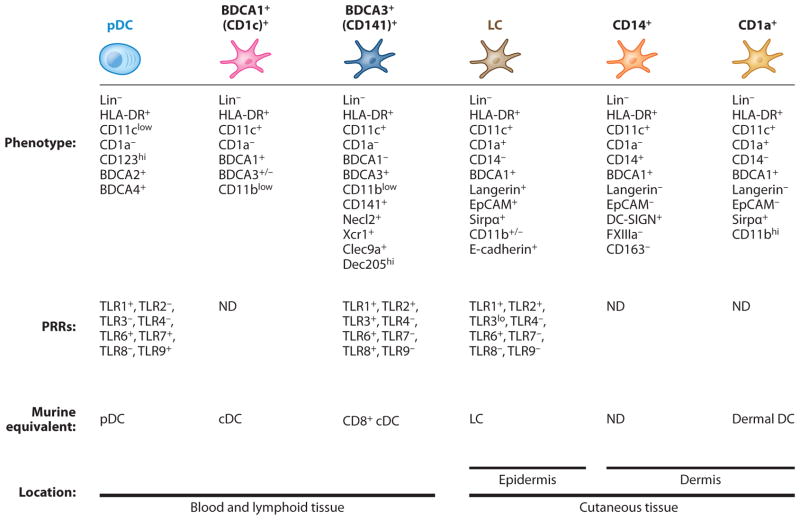
Most of our understanding of human DC heterogeneity stems from studies of skin and blood DCs. However, increased refinement of flow cytometric approaches allowing cross-correlation of a large number of surface markers as well as the development of novel genomic profiling methods have recently fostered our understanding of human DC heterogeneity (Figure 6).
Figure 6.
Phenotype of human DC subsets. This figure summarizes the phenotype and pathogen receptor expression profile location of human DC subsets known so far as well as the putative mouse DC subset equivalent. Abbreviations: LC, Langerhans cells; ND, not determined; PRRs, pattern-recognition receptors.
Blood Dendritic Cells
Human DCs are defined as cells that lack lineage (Lin) markers (CD3, CD19, CD14, CD20, CD56, glycophorin A) (see side-bar entitled Handy Hints to Characterize Mouse Tissue Dendritic Cells In Vivo) and constitutively express MHC-II. Human pDCs are characterized as Lin− MHC-II+CD303(BDCA2)+CD304(BDCA4)+ (17), whereas cDCs are characterized as Lin− MHC-II+CD11c+ (258–260), although in humans, CD11c is also expressed on most monocytes and macrophages. In contrast to mice, in which circulating pre-cDCs differentiate into mature cDC subsets once they reach peripheral tissues, in humans two cDC subsets expressing the nonoverlapping markers CD1c (BDCA1) or CD141 (BDCA3) (261–263) are present in the blood circulation (Figure 6). CD1c+ DCs represent by far the most predominant cDC subset in human blood, whereas CD141+ DCs form a minute blood population. Another population of Lin− MHC-II+ cells expressing FcγRIII (CD16), also termed Slan-DCs, has been identified in human blood (264). However, unlike CD1c+ and CD141+ DCs, they are absent from tissues and are thought to represent a monocyte subset (265–267).
Nonlymphoid Tissue Dendritic Cells
Analysis of nonlymphoid tissue DC compartments in humans has relied on tissue explants obtained from patients that undergo surgery for an underlying disease, which could affect tissue DC composition. Earlier studies have identified two subsets of DCs in the human dermis, including CD1a+CD14− DCs and CD1a− CD14+ DCs (268, 269). Subsequent studies found that the autofluorescence markers FXIII and CD163 can identify the dermal macrophages that contaminate dermal CD1a− CD14+ DCs (270, 271). Recent studies also identified a discrete subset of CD141+ DCs in the human dermis (270, 272). CD141+ DCs are the only dermal DCs to express XCR1, TLR3, CLEC9A, and Necl2 (273); they express the highest levels of Flt3 among dermal DCs and derive from blood CD141+ DCs (273). Similar populations were found in the lungs of humanized immunodeficient mice reconstituted with human hematopoietic progenitors (174) and in the kidney and lamina propria of humanized mice and human intestinal tissues (174).
Epidermal Langerhans Cells
LCs form the predominant hematopoietic cells in the human epidermis, which, in contrast to murine epidermis, contains few γδ T cells (274). Human LCs are easily identified in the epidermis based on the expression of the hematopoietic markers CD45, MHC-II, the epithelial cell adhesion molecule (EpCAM), and the lectin langerin (275). Human but not mouse LCs express high levels of CD1a (previously named OKT6) (276), a member of the group 1 CD1 proteins (CD1a, CD1b, and CD1c) that share the capacity to present lipid antigens to T cells (277).
Lymphoid Tissue Dendritic Cells
CD1c+ and CD141+ DC subsets that resemble blood DCs were found in the human spleen (278, 279) and tonsils (264, 280) and likely correspond to lymphoid tissue–resident DCs. Guided by the phenotypical definition of LN-resident and migratory DCs in mice, a recent study characterized noncancerous skin-draining LN DCs isolated from breast cancer patients. LN DCs were shown to consist of CD1c+ and CD141+ cDCs that phenotypically resembled blood cDCs (281) and were classified as LN-resident cDCs. LN cells also included MHC-IIhiCD11cintEpCAM+CD1a+ cells, EpCAM− CD1a+ cells, and CD206+ cells, classified as migratory LCs, migratory dermal CD1a+ DCs, and dermal CD14+ DCs, respectively (281). A recent analysis of dermopathic LNs identified the presence of CD141+ and CD1c+ cDCs in the resident and migratory cDC fractions of LN DCs, suggesting that cutaneous CD141+ DCs and CD1c+ cDCs also migrate to the draining LNs (273).
Earlier studies found that most human thymic DCs are CD11c+CD11b− CD45ROlow and lack myeloid markers resembling human CD141+ DCs and mouse CD8+ DCs (282). In contrast, a minority of thymic DCs are CD11chiCD11b+CD45ROhi and express myeloid markers, resembling mouse CD11b+CD172a+ thymic DCs (282).
Mouse-Human Dendritic Cell Relationship
One of the main barriers to comparing human DCs directly with mouse DCs has been the lack of CD8 expression on human DCs. Thus, it was assumed that this subset did not exist in humans until the recent identification of the human equivalent of the CD8+/CD103+ cDCs (283). Gene chip (meta)analysis of the transcriptome of multiple mouse and human DC subsets revealed that human CD141+ DCs are related to mouse CD8+ DCs, whereas human CD1c+ DCs are more related to mouse CD11b+ DCs (267).
Human CD141+ DCs uniquely express the lectin Clec9A (173, 284, 285) and the chemokine XCR1 (286, 287), an important finding considering that CD8+ DCs appear to be the only cells in the mouse that express this molecule (204, 288). Like mouse CD8+ DCs, human CD141+ DCs express Batf3 and IRF8 (267, 278, 280) and lack expression of IRF4 (267, 278, 280). Blood CD141+ DCs express TLR3, but not TLR7, and produce high amounts of IL-12 (278, 280) and type I IFN when activated with a TLR3 agonist (289). Further, they are capable of phagocytosing dead cells and of cross-presenting cell-associated and soluble antigens upon activation with the TLR3 ligand, poly(I:C) (278, 280, 287), with more efficiency than other dermal DC subsets (273). Whether the superior (compared with CD1c+ DCs) cross-presentation potential of CD141+ DCs is maintained upon stimulation with other triggers remains unclear (273). Of note, human CD1c+ DCs can also produce high IL-12 levels and cross-prime CD8+ T cells (290). The differentiation of human hematopoietic progenitors into CD141+ DCs occurs only when Flt3L is added to the cultures, and inhibition of Batf3 in these cultures abolishes the differentiation of CD141+ DCs but not of CD1c+ DCs (174), suggesting that CD141+ DCs are indeed developmentally related to mouse CD8+ and CD103+ DCs. In contrast to mice, however, IRF8 mutations in two immunodeficient patients were associated either with a complete absence of DCs and circulating monocytes or with the specific depletion of circulating CD1c+ DCs but not CD141+ DCs (291).
IN VITRO–DERIVED DENDRITIC CELLS IN MICE AND HUMANS.
Two in vitro models for DC generation predominate the DC biology landscape; these models use either the Flt3 ligand (Flt3L) or GM-CSF as a stimulus. However, although the DCs generated by both models share phenotypic, functional, and developmental characteristics with their in vivo counterparts, they are not identical. For example, the cDC subsets generated in Flt3L-stimulated cultures largely phenotypically resemble lymphoid tissue–resident cDCs, but they do not express the markers CD4 and CD8 in vitro, but do express these markers when transferred in vivo in mice. Although this may be a superficial difference, it serves as an important reminder that in vitro models are not identical to in vivo conditions and that in vitro findings must be validated in vivo. This sidebar summarizes both the benefits and disadvantages of the Flt3L- and Csf-2-stimulated systems, as well as models for the generation of human DCs.
Csf-2 (GM-CSF) DCs: Culturing BM in the presence of Csf-2 generates large numbers of DCs (108), which comprise a largely homogenous population of CD11bhighB220− cells that resemble inflammatory DCs (293) and do not give rise to pDCs (148). Csf-2 is often used in conjunction with IL-4, which is thought to further reduce macrophage differentiation in vitro.
Ftl3L DCs: In contrast to Csf-2-supplemented cultures, the culture of BM or hematopoietic progenitors in the presence of Flt3L generates, in 3–5 days, DC precursors that both phenotypically and developmentally resemble CDPs and pre-cDCs (15) and, in 7–10 days, gives rise to three DC subsets, including B220−CD11bhighCD172ahighCD24lowClec9A− cDCs, CD11blowCD172alowCD24highClec9A+ cDCs, and B220+ pDCs (294–296). These cells phenotypically and functionally resemble their splenic counterparts in their patterns of antigen presentation, cytokine production, and TLR expression (296).
Human in vitro–derived DCs: The dominant model for the generation of human DCs has involved (a) culturing CD34+ hematopoietic progenitors with Csf-2 supplemented with TNF-α, which gives rise to LC-like cells and dermal DC-like cells (297); or (b) culturing monocytes with Csf-2 and IL-4, which produces dermal-like CD1a+ cDCs (83, 298). In addition, the addition of TGF-β to CD34+ hematopoietic progenitor cultures (119) or monocyte cultures (120) promotes their differentiation into LC-like cells. More recently, culturing CD34+ progenitors with the Flt3L and thrombopoietin was shown to generate both pDCs and BDCA3+ and BDCA1+ cDCs in vitro (278, 299). Choosing which model to use should be dictated by the needs of the experiment. Csf-2-stimulated cultures reliably produce large numbers of cDCs, which may prove invaluable when designing interventions for human disease, whereas Flt3L-stimulated cultures more accurately mimic DC development in vivo and have proven instrumental in recent years, both in delineating the stages of the pathway by which DCs enter the steady state and in studies on the functional specialization of DC subsets.
Human Dendritic Cell Deficiency
Three genetically defined syndromes of DC deficiency have recently been described in humans. The first, DCML deficiency syndrome, is caused by a mutation of GATA-binding factor 2 (GATA2) that is associated with a complete absence of blood DCs, pDCs, tissue cDCs, circulating monocytes, and B and NK lymphoid cells (hence the name, DCML deficiency), but not with defects in granulocytes or platelets. This syndrome is also associated with an absence of multi-lymphoid progenitors and reduced GMPs, suggesting that GATA2 deficiency prevents the differentiation of these progenitors into DCs in vivo. DCML patients also have markedly elevated serum levels of FLT3L (approximately 100-fold) with no sign of myeloproliferation.
A second syndrome comes from the identification of patients with IRF8 mutations and helped establish the role of IRF8 in human DC development (291). One patient with an IRF8 null mutation had an increased number of granulocytes; a total absence of circulating monocytes, pDC, cDCs, and dermal DCs; and defective IL-12 production (291), resembling the phenotype of mice with a loss of function of IRF8 (129–131). Interestingly, epidermal LCs were present in normal numbers in this patient, suggesting that, similar to mice, epidermal LCs and cDCs have different origins. A distinct IRF8 mutation observed in two individuals was associated with reduced CD1c+ cDC numbers in the blood and defects in IL-12 production but no defects in granulocytes, monocytes, or pDCs, and resembled the hypomorphic mutation observed in the BXH-2 IRF8 mutant mice (134, 291). There were no defects in CD141+ cDCs in these patients, which is surprising given that CD8+ cDCs are the cDC subset most affected by the BXH-2 IRF8 mutation (134).
The third syndrome is associated with a mutation of adenylate kinase 2, a phosphotransferase required for nucleotide homeostasis. It causes a form of severe combined immunodeficiency known as reticular dysgenesis. This syndrome is characterized by impaired formation of all nucleated blood cells, including neutrophils, lymphocytes, monocytes, and cDCs as well as LCs (292).
CONCLUSIONS
Major progress has been made in our understanding of the regulation of the development, differentiation, and function of the cDC lineage. The major challenge of the years to come is to transport our emerging understanding of the molecular control of mouse cDC development and function onto the human stage; only then might we be able to harness our knowledge of the DC lineage to enable the development of better vaccines for the prevention or treatment of human disease.
Acknowledgments
We thank Drs. Bill Heath, Juliana Idoyaga, Elodie Segura, and Shalin Naik for discussions and critical input in the manuscript. We apologize to those colleagues whose work we have not cited, either through ignorance or space restrictions.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Steinman RM, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. I Morphology, quantitation, tissue distribution. J Exp Med. 1973;137:1142–62. doi: 10.1084/jem.137.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steinman RM, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. II Functional properties in vitro. J Exp Med. 1974;139:380–97. doi: 10.1084/jem.139.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashyam H. Ralph Steinman: dendritic cells bring home the Lasker. J Exp Med. 2007;204:2245–48. doi: 10.1084/jem.20071995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinman RM. Lasker Basic Medical Research Award. Dendritic cells: versatile controllers of the immune system. Nat Med. 2007;13:1155–59. doi: 10.1038/nm1643. [DOI] [PubMed] [Google Scholar]
- 5.Banchereau J, Cohn F, Inaba K, Muller B, Mellman I, et al. Remembering Ralph Steinman. J Exp Med. 2011;208:2343–47. doi: 10.1084/jem.20112295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mellman I, Nussenzweig M. Retrospective. Ralph M Steinman (1943–2011) Science. 2011;334:466. doi: 10.1126/science.1215136. [DOI] [PubMed] [Google Scholar]
- 7.Nussenzweig MC, Mellman I. Ralph Steinman (1943–2011) Nature. 2011;478:460. doi: 10.1038/478460a. [DOI] [PubMed] [Google Scholar]
- 8.Shortman K. Ralph Steinman and dendritic cells. Immunol Cell Biol. 2012;90:1–2. doi: 10.1038/icb.2011.91. [DOI] [PubMed] [Google Scholar]
- 9.Schuler G, Romani N, Steinman RM. A comparison of murine epidermal Langerhans cells with spleen dendritic cells. J Investig Dermatol. 1985;85:99s–106. doi: 10.1111/1523-1747.ep12275566. [DOI] [PubMed] [Google Scholar]
- 10.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–52. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 11.Shortman K, Heath WR. The CD8+ dendritic cell subset. Immunol Rev. 2010;234:18–31. doi: 10.1111/j.0105-2896.2009.00870.x. [DOI] [PubMed] [Google Scholar]
- 12.Helft J, Ginhoux F, Bogunovic M, Merad M. Origin and functional heterogeneity of non-lymphoid tissue dendritic cells in mice. Immunol Rev. 2010;234:55–75. doi: 10.1111/j.0105-2896.2009.00885.x. [DOI] [PubMed] [Google Scholar]
- 13.Colonna M, Trinchieri G, Liu YJ. Plasmacytoid dendritic cells in immunity. Nat Immunol. 2004;5:1219–26. doi: 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- 14.Onai N, Obata-Onai A, Schmid MA, Ohteki T, Jarrossay D, Manz MG. Identification of clono-genic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat Immunol. 2007;8:1207–16. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- 15.Naik SH, Sathe P, Park HY, Metcalf D, Proietto AI, et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat Immunol. 2007;8:1217–26. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- 16.Liu K, Victora GD, Schwickert TA, Guermonprez P, Meredith MM, et al. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324:392–97. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reizis B, Bunin A, Ghosh HS, Lewis KL, Sisirak V. Plasmacytoid dendritic cells: recent progress and open questions. Annu Rev Immunol. 2011;29:163–83. doi: 10.1146/annurev-immunol-031210-101345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reizis B. Regulation of plasmacytoid dendritic cell development. Curr Opin Immunol. 2010;22:206–11. doi: 10.1016/j.coi.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villadangos JA, Schnorrer P. Intrinsic and cooperative antigen-presenting functions of dendritic-cell subsets in vivo. Nat Rev Immunol. 2007;7:543–55. doi: 10.1038/nri2103. [DOI] [PubMed] [Google Scholar]
- 20.Segura E, Villadangos JA. Antigen presentation by dendritic cells in vivo. Curr Opin Immunol. 2009;21:105–10. doi: 10.1016/j.coi.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 21.Joffre OP, Segura E, Savina A, Amigorena S. Cross-presentation by dendritic cells. Nat Rev Immunol. 2012;12:557–69. doi: 10.1038/nri3254. [DOI] [PubMed] [Google Scholar]
- 22.Forster R, Braun A, Worbs T. Lymph node homing of T cells and dendritic cells via afferent lymphatics. Trends Immunol. 2012;33:271–80. doi: 10.1016/j.it.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Miller JC, Brown BD, Shay T, Gautier EL, Jojic V, et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol. 2012;13:888–99. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKenna HJ, Stocking KL, Miller RE, Brasel K, De Smedt T, et al. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95:3489–97. [PubMed] [Google Scholar]
- 26.Maraskovsky E, Brasel K, Teepe M, Roux ER, Lyman SD, et al. Dramatic increase in the numbers of functionally mature dendritic cells in Flt3 ligand-treated mice: multiple dendritic cell subpopulations identified. J Exp Med. 1996;184:1953–62. doi: 10.1084/jem.184.5.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forster R, Schubel A, Breitfeld D, Kremmer E, Renner-Muller I, et al. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99:23–33. doi: 10.1016/s0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- 28.Meredith MM, Liu K, Darrasse-Jeze G, Kamphorst AO, Schreiber HA, et al. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J Exp Med. 2012;209:1153–65. doi: 10.1084/jem.20112675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satpathy AT, Wumesh KC, Albring JC, Edelson BT, Kretzer NM, et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J Exp Med. 2012;209:1135–52. doi: 10.1084/jem.20120030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.del Rio ML, Bernhardt G, Rodriguez-Barbosa JI, Forster R. Development and functional specialization of CD103+ dendritic cells. Immunol Rev. 2010;234:268–81. doi: 10.1111/j.0105-2896.2009.00874.x. [DOI] [PubMed] [Google Scholar]
- 31.Bogunovic M, Ginhoux F, Helft J, Shang L, Hashimoto D, et al. Origin of the lamina propria dendritic cell network. Immunity. 2009;31:513–25. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varol C, Vallon-Eberhard A, Elinav E, Aychek T, Shapira Y, et al. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity. 2009;31:502–12. doi: 10.1016/j.immuni.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 33.Ginhoux F, Liu K, Helft J, Bogunovic M, Greter M, et al. The origin and development of non-lymphoid tissue CD103+ DCs. J Exp Med. 2009;206:3115–30. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merad M, Ginhoux F, Collin M. Origin, homeostasis and function of Langerhans cells and other langerin-expressing dendritic cells. Nat Rev Immunol. 2008;8:935–47. doi: 10.1038/nri2455. [DOI] [PubMed] [Google Scholar]
- 35.Zhan Y, Carrington EM, van Nieuwenhuijze A, Bedoui S, Seah S, et al. GM-CSF increases cross-presentation and CD103 expression by mouse CD8+ spleen dendritic cells. Eur J Immunol. 2011;41:2585–95. doi: 10.1002/eji.201141540. [DOI] [PubMed] [Google Scholar]
- 36.Sathe P, Pooley J, Vremec D, Mintern J, Jin JO, et al. The acquisition of antigen cross-presentation function by newly formed dendritic cells. J Immunol. 2011;186:5184–92. doi: 10.4049/jimmunol.1002683. [DOI] [PubMed] [Google Scholar]
- 37.Edelson BT, Bradstreet TR, Wumesh KC, Hildner K, Herzog JW, et al. Batf3-dependent CD11blow/− peripheral dendritic cells are GM-CSF-independent and are not required for Th cell priming after subcutaneous immunization. PLoS ONE. 2011;6:e25660. doi: 10.1371/journal.pone.0025660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greter M, Helft J, Chow A, Hashimoto D, Mortha A, et al. GM-CSF controls nonlymphoid tissue dendritic cell homeostasis but is dispensable for the differentiation of inflammatory dendritic cells. Immunity. 2012;36:1031–46. doi: 10.1016/j.immuni.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cepek KL, Shaw SK, Parker CM, Russell GJ, Morrow JS, et al. Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and the αEβ7 integrin. Nature. 1994;372:190–93. doi: 10.1038/372190a0. [DOI] [PubMed] [Google Scholar]
- 40.Langlet C, Tamoutounour S, Henri S, Luche H, Ardouin L, et al. CD64 expression distinguishes monocyte-derived and conventional dendritic cells and reveals their distinct role during intramuscular immunization. J Immunol. 2012;188:1751–60. doi: 10.4049/jimmunol.1102744. [DOI] [PubMed] [Google Scholar]
- 41.Schulz O, Jaensson E, Persson EK, Liu X, Worbs T, et al. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J Exp Med. 2009;206:3101–14. doi: 10.1084/jem.20091925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ginhoux F, Merad M. Ontogeny and homeostasis of Langerhans cells. Immunol Cell Biol. 2010;88:387–92. doi: 10.1038/icb.2010.38. [DOI] [PubMed] [Google Scholar]
- 43.Ginhoux F, Tacke F, Angeli V, Bogunovic M, Loubeau M, et al. Langerhans cells arise from monocytes in vivo. Nat Immunol. 2006;7:265–73. doi: 10.1038/ni1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Randolph GJ, Angeli V, Swartz MA. Dendritic-cell trafficking to lymph nodes through lymphatic vessels. Nat Rev Immunol. 2005;5:617–28. doi: 10.1038/nri1670. [DOI] [PubMed] [Google Scholar]
- 45.Henri S, Guilliams M, Poulin LF, Tamoutounour S, Ardouin L, et al. Disentangling the complexity of the skin dendritic cell network. Immunol Cell Biol. 2010;88:366–75. doi: 10.1038/icb.2010.34. [DOI] [PubMed] [Google Scholar]
- 46.Ginhoux F, Collin MP, Bogunovic M, Abel M, Leboeuf M, et al. Blood-derived dermal langerin+ dendritic cells survey the skin in the steady state. J Exp Med. 2007;204:3133–46. doi: 10.1084/jem.20071733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Randolph GJ, Ochando J, Partida-Sanchez S. Migration of dendritic cell subsets and their precursors. Annu Rev Immunol. 2008;26:293–316. doi: 10.1146/annurev.immunol.26.021607.090254. [DOI] [PubMed] [Google Scholar]
- 48.Forster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol. 2008;8:362–71. doi: 10.1038/nri2297. [DOI] [PubMed] [Google Scholar]
- 49.Ohl L, Mohaupt M, Czeloth N, Hintzen G, Kiafard Z, et al. CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity. 2004;21:279–88. doi: 10.1016/j.immuni.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 50.Reis e Sousa C. Dendritic cells in a mature stage. Nat Rev Immunol. 2006;6:476–83. doi: 10.1038/nri1845. [DOI] [PubMed] [Google Scholar]
- 51.Jiang A, Bloom O, Ono S, Cui W, Unternaehrer J, et al. Disruption of E-cadherin-mediated adhesion induces a functionally distinct pathway of dendritic cell maturation. Immunity. 2007;27:610–24. doi: 10.1016/j.immuni.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van den Bossche J, Malissen B, Mantovani A, De Baetselier P, Van Ginderachter JA. Regulation and function of the E-cadherin/catenin complex in cells of the monocyte-macrophage lineage and DCs. Blood. 2012;119:1623–33. doi: 10.1182/blood-2011-10-384289. [DOI] [PubMed] [Google Scholar]
- 53.Iwasaki A. Mucosal dendritic cells. Annu Rev Immunol. 2007;25:381–418. doi: 10.1146/annurev.immunol.25.022106.141634. [DOI] [PubMed] [Google Scholar]
- 54.Wu L, Shortman K. Heterogeneity of thymic dendritic cells. Semin Immunol. 2005;17:304–12. doi: 10.1016/j.smim.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 55.Waskow C, Liu K, Darrasse-Jeze G, Guermonprez P, Ginhoux F, et al. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat Immunol. 2008;9:676–83. doi: 10.1038/ni.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bar-On L, Birnberg T, Lewis KL, Edelson BT, Bruder D, et al. CX3CR1+ CD8α+ dendritic cells are a steady-state population related to plasmacytoid dendritic cells. Proc Natl Acad Sci USA. 2010;107:14745–50. doi: 10.1073/pnas.1001562107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vremec D, O’Keeffe M, Wilson A, Ferrero I, Koch U, et al. Factors determining the spontaneous activation of splenic dendritic cells in culture. Innate Immunol. 2011;17:338–52. doi: 10.1177/1753425910371396. [DOI] [PubMed] [Google Scholar]
- 58.Crowley M, Inaba K, Witmer-Pack M, Steinman RM. The cell surface of mouse dendritic cells: FACS analyses of dendritic cells from different tissues including thymus. Cell Immunol. 1989;118:108–25. doi: 10.1016/0008-8749(89)90361-4. [DOI] [PubMed] [Google Scholar]
- 59.Jiang W, Swiggard WJ, Heufler C, Peng M, Mirza A, et al. The receptor DEC-205 expressed by dendritic cells and thymic epithelial cells is involved in antigen processing. Nature. 1995;375:151–55. doi: 10.1038/375151a0. [DOI] [PubMed] [Google Scholar]
- 60.Dudziak D, Kamphorst AO, Heidkamp GF, Buchholz VR, Trumpfheller C, et al. Differential antigen processing by dendritic cell subsets in vivo. Science. 2007;315:107–11. doi: 10.1126/science.1136080. [DOI] [PubMed] [Google Scholar]
- 61.Lewis KL, Caton ML, Bogunovic M, Greter M, Grajkowska LT, et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 2011;35:780–91. doi: 10.1016/j.immuni.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Proietto AI, van Dommelen S, Zhou P, Rizzitelli A, D’Amico A, et al. Dendritic cells in the thymus contribute to T-regulatory cell induction. Proc Natl Acad Sci USA. 2008;105:19869–74. doi: 10.1073/pnas.0810268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kondo M, Wagers AJ, Manz MG, Prohaska SS, Scherer DC, et al. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol. 2003;21:759–806. doi: 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- 64.Buza-Vidas N, Luc S, Jacobsen SE. Delineation of the earliest lineage commitment steps of haematopoietic stem cells: new developments, controversies and major challenges. Curr Opin Hematol. 2007;14:315–21. doi: 10.1097/MOH.0b013e3281de72bb. [DOI] [PubMed] [Google Scholar]
- 65.Chow A, Brown BD, Merad M. Studying the mononuclear phagocyte system in the molecular age. Nat Rev Immunol. 2011;11:788–98. doi: 10.1038/nri3087. [DOI] [PubMed] [Google Scholar]
- 66.Traver D, Akashi K, Manz M, Merad M, Miyamoto T, et al. Development of CD8α-positive dendritic cells from a common myeloid progenitor. Science. 2000;290:2152–54. doi: 10.1126/science.290.5499.2152. [DOI] [PubMed] [Google Scholar]
- 67.Manz MG, Traver D, Miyamoto T, Weissman IL, Akashi K. Dendritic cell potentials of early lymphoid and myeloid progenitors. Blood. 2001;97:3333–41. doi: 10.1182/blood.v97.11.3333. [DOI] [PubMed] [Google Scholar]
- 68.Wu L, D’Amico A, Hochrein H, O’Keeffe M, Shortman K, Lucas K. Development of thymic and splenic dendritic cell populations from different hemopoietic precursors. Blood. 2001;98:3376–82. doi: 10.1182/blood.v98.12.3376. [DOI] [PubMed] [Google Scholar]
- 69.Shigematsu H, Reizis B, Iwasaki H, Mizuno S, Hu D, et al. Plasmacytoid dendritic cells activate lymphoid-specific genetic programs irrespective of their cellular origin. Immunity. 2004;21:43–53. doi: 10.1016/j.immuni.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 70.Chicha L, Jarrossay D, Manz MG. Clonal type I interferon-producing and dendritic cell precursors are contained in both human lymphoid and myeloid progenitor populations. J Exp Med. 2004;200:1519–24. doi: 10.1084/jem.20040809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Manz MG, Traver D, Akashi K, Merad M, Miyamoto T, et al. Dendritic cell development from common myeloid progenitors. Ann N Y Acad Sci. 2001;938:167–74. doi: 10.1111/j.1749-6632.2001.tb03586.x. [DOI] [PubMed] [Google Scholar]
- 72.D’Amico A, Wu L. The early progenitors of mouse dendritic cells and plasmacytoid predendritic cells are within the bone marrow hemopoietic precursors expressing Flt3. J Exp Med. 2003;198:293–303. doi: 10.1084/jem.20030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karsunky H, Merad M, Cozzio A, Weissman IL, Manz MG. Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J Exp Med. 2003;198:305–13. doi: 10.1084/jem.20030323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mende I, Karsunky H, Weissman IL, Engleman EG, Merad M. Flk2+ myeloid progenitors are the main source of Langerhans cells. Blood. 2006;107:1383–90. doi: 10.1182/blood-2005-05-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ardavin C, Wu L, Li CL, Shortman K. Thymic dendritic cells and T cells develop simultaneously in the thymus from a common precursor population. Nature. 1993;362:761–63. doi: 10.1038/362761a0. [DOI] [PubMed] [Google Scholar]
- 76.Luche H, Ardouin L, Teo P, See P, Henri S, et al. The earliest intrathymic precursors of CD8α+ thymic dendritic cells correspond to myeloid-type double-negative 1c cells. Eur J Immunol. 2011;41:2165–75. doi: 10.1002/eji.201141728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Corcoran L, Ferrero I, Vremec D, Lucas K, Waithman J, et al. The lymphoid past of mouse plasmacytoid cells and thymic dendritic cells. J Immunol. 2003;170:4926–32. doi: 10.4049/jimmunol.170.10.4926. [DOI] [PubMed] [Google Scholar]
- 78.Pelayo R, Welner R, Perry SS, Huang J, Baba Y, et al. Lymphoid progenitors and primary routes to becoming cells of the immune system. Curr Opin Immunol. 2005;17:100–7. doi: 10.1016/j.coi.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 79.Fogg DK, Sibon C, Miled C, Jung S, Aucouturier P, et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 2006;311:83–87. doi: 10.1126/science.1117729. [DOI] [PubMed] [Google Scholar]
- 80.Auffray C, Sieweke MH, Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu Rev Immunol. 2009;27:669–92. doi: 10.1146/annurev.immunol.021908.132557. [DOI] [PubMed] [Google Scholar]
- 81.Diao J, Winter E, Cantin C, Chen W, Xu L, et al. In situ replication of immediate dendritic cell (DC) precursors contributes to conventional DC homeostasis in lymphoid tissue. J Immunol. 2006;176:7196–206. doi: 10.4049/jimmunol.176.12.7196. [DOI] [PubMed] [Google Scholar]
- 82.Naik SH, Metcalf D, van Nieuwenhuijze A, Wicks I, Wu L, et al. Intrasplenic steady-state dendritic cell precursors that are distinct from monocytes. Nat Immunol. 2006;7:663–71. doi: 10.1038/ni1340. [DOI] [PubMed] [Google Scholar]
- 83.Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor α. J Exp Med. 1994;179:1109–18. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Merad M, Manz MG, Karsunky H, Wagers A, Peters W, et al. Langerhans cells renew in the skin throughout life under steady-state conditions. Nat Immunol. 2002;3:1135–41. doi: 10.1038/ni852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Romani N, Schuler G, Fritsch P. Ontogeny of Ia-positive and Thy-1-positive leukocytes of murine epidermis. J Investig Dermatol. 1986;86:129–33. doi: 10.1111/1523-1747.ep12284135. [DOI] [PubMed] [Google Scholar]
- 86.Chang-Rodriguez S, Hoetzenecker W, Schwarzler C, Biedermann T, Saeland S, Elbe-Burger A. Fetal and neonatal murine skin harbors Langerhans cell precursors. J Leukoc Biol. 2005;77:352–60. doi: 10.1189/jlb.1004584. [DOI] [PubMed] [Google Scholar]
- 87.Chorro L, Sarde A, Li M, Woollard KJ, Chambon P, et al. Langerhans cell (LC) proliferation mediates neonatal development, homeostasis, and inflammation-associated expansion of the epidermal LC network. J Exp Med. 2009;206:3089–100. doi: 10.1084/jem.20091586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 89.Hoeffel G, Wang Y, Greter M, See P, Teo P, et al. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J Exp Med. 2012;209:1167–81. doi: 10.1084/jem.20120340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Onai N, Obata-Onai A, Schmid MA, Manz MG. Flt3 in regulation of type I interferon-producing cell and dendritic cell development. Ann N Y Acad Sci. 2007;1106:253–61. doi: 10.1196/annals.1392.015. [DOI] [PubMed] [Google Scholar]
- 91.Merad M, Manz MG. Dendritic cell homeostasis. Blood. 2009;113:3418–27. doi: 10.1182/blood-2008-12-180646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schmid MA, Kingston D, Boddupalli S, Manz MG. Instructive cytokine signals in dendritic cell lineage commitment. Immunol Rev. 2010;234:32–44. doi: 10.1111/j.0105-2896.2009.00877.x. [DOI] [PubMed] [Google Scholar]
- 93.Adolfsson J, Borge OJ, Bryder D, Theilgaard-Monch K, Astrand-Grundstrom I, et al. Upregulation of Flt3 expression within the bone marrow Lin−Sca1+c-kit+ stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15:659–69. doi: 10.1016/s1074-7613(01)00220-5. [DOI] [PubMed] [Google Scholar]
- 94.Onai N, Obata-Onai A, Tussiwand R, Lanzavecchia A, Manz MG. Activation of the Flt3 signal transduction cascade rescues and enhances type I interferon-producing and dendritic cell development. J Exp Med. 2006;203:227–38. doi: 10.1084/jem.20051645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kingston D, Schmid MA, Onai N, Obata-Onai A, Baumjohann D, Manz MG. The concerted action of GM-CSF and Flt3-ligand on in vivo dendritic cell homeostasis. Blood. 2009;114:835–43. doi: 10.1182/blood-2009-02-206318. [DOI] [PubMed] [Google Scholar]
- 96.Tussiwand R, Onai N, Mazzucchelli L, Manz MG. Inhibition of natural type I IFN-producing and dendritic cell development by a small molecule receptor tyrosine kinase inhibitor with Flt3 affinity. J Immunol. 2005;175:3674–80. doi: 10.4049/jimmunol.175.6.3674. [DOI] [PubMed] [Google Scholar]
- 97.Birnberg T, Bar-On L, Sapoznikov A, Caton ML, Cervantes-Barragan L, et al. Lack of conventional dendritic cells is compatible with normal development and T cell homeostasis, but causes myeloid proliferative syndrome. Immunity. 2008;29:986–97. doi: 10.1016/j.immuni.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 98.Manfra DJ, Chen SC, Jensen KK, Fine JS, Wiekowski MT, Lira SA. Conditional expression of murine Flt3 ligand leads to expansion of multiple dendritic cell subsets in peripheral blood and tissues of transgenic mice. J Immunol. 2003;170:2843–52. doi: 10.4049/jimmunol.170.6.2843. [DOI] [PubMed] [Google Scholar]
- 99.Maraskovsky E, Daro E, Roux E, Teepe M, Maliszewski CR, et al. In vivo generation of human dendritic cell subsets by Flt3 ligand. Blood. 2000;96:878–84. [PubMed] [Google Scholar]
- 100.Fong L, Hou Y, Rivas A, Benike C, Yuen A, et al. Altered peptide ligand vaccination with Flt3 ligand expanded dendritic cells for tumor immunotherapy. Proc Natl Acad Sci USA. 2001;98:8809–14. doi: 10.1073/pnas.141226398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pixley FJ, Stanley ER. CSF-1 regulation of the wandering macrophage: complexity in action. Trends Cell Biol. 2004;14:628–38. doi: 10.1016/j.tcb.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 102.Witmer-Pack MD, Hughes DA, Schuler G, Lawson L, McWilliam A, et al. Identification of macrophages and dendritic cells in the osteopetrotic (op/op) mouse. J Cell Sci. 1993;104(Pt. 4):1021–29. doi: 10.1242/jcs.104.4.1021. [DOI] [PubMed] [Google Scholar]
- 103.Lin H, Lee E, Hestir K, Leo C, Huang M, et al. Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science. 2008;320:807–11. doi: 10.1126/science.1154370. [DOI] [PubMed] [Google Scholar]
- 104.Wang Y, Szretter KJ, Vermi W, Gilfillan S, Rossini C, et al. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol. 2012;13:753–60. doi: 10.1038/ni.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Greter M, Lelios I, Pelczar P, Hoeffel G, Price J, et al. Stroma-derived interleukin-34 controls the development and maintenance of Langerhans cells and the maintenance of microglia. Immunity. 2012;37:1050–60. doi: 10.1016/j.immuni.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Metcalf D. Hematopoietic cytokines. Blood. 2008;111:485–91. doi: 10.1182/blood-2007-03-079681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Martinez-Moczygemba M, Huston DP. Biology of common βreceptor-signaling cytokines: IL-3, IL-5, and GM-CSF. J Allergy Clin Immunol. 2003;112:653–65. doi: 10.1016/S0091. [DOI] [PubMed] [Google Scholar]
- 108.Inaba K, Inaba M, Romani N, Aya H, Deguchi M, et al. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp Med. 1992;176:1693–702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Caux C, Vanbervliet B, Massacrier C, Dezutter-Dambuyant C, de Saint-Vis B, et al. CD34+ hematopoietic progenitors from human cord blood differentiate along two independent dendritic cell pathways in response to GM-CSF+TNFα. J Exp Med. 1996;184:695–706. doi: 10.1084/jem.184.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Banchereau J, Palucka AK. Dendritic cells as therapeutic vaccines against cancer. Nat Rev Immunol. 2005;5:296–306. doi: 10.1038/nri1592. [DOI] [PubMed] [Google Scholar]
- 111.Vremec D, Lieschke GJ, Dunn AR, Robb L, Metcalf D, Shortman K. The influence of granulocyte/macrophage colony-stimulating factor on dendritic cell levels in mouse lymphoid organs. Eur J Immunol. 1997;27:40–44. doi: 10.1002/eji.1830270107. [DOI] [PubMed] [Google Scholar]
- 112.King IL, Kroenke MA, Segal BM. GM-CSF-dependent, CD103+ dermal dendritic cells play a critical role in Th effector cell differentiation after subcutaneous immunization. J Exp Med. 2010;207:953–61. doi: 10.1084/jem.20091844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu Q, Wang Y, Wang J, Hedgeman EO, Browning JL, Fu YX. The requirement of membrane lymphotoxin for the presence of dendritic cells in lymphoid tissues. J Exp Med. 1999;190:629–38. doi: 10.1084/jem.190.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kabashima K, Banks TA, Ansel KM, Lu TT, Ware CF, Cyster JG. Intrinsic lymphotoxin-β receptor requirement for homeostasis of lymphoid tissue dendritic cells. Immunity. 2005;22:439–50. doi: 10.1016/j.immuni.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 115.Wang YG, Kim KD, Wang J, Yu P, Fu YX. Stimulating lymphotoxin βreceptor on the dendritic cells is critical for their homeostasis and expansion. J Immunol. 2005;175:6997–7002. doi: 10.4049/jimmunol.175.10.6997. [DOI] [PubMed] [Google Scholar]
- 116.Wu L, D’Amico A, Winkel KD, Suter M, Lo D, Shortman K. RelB is essential for the development of myeloid-related CD8α− dendritic cells but not of lymphoid-related CD8α+ dendritic cells. Immunity. 1998;9:839–47. doi: 10.1016/s1074-7613(00)80649-4. [DOI] [PubMed] [Google Scholar]
- 117.Borkowski TA, Letterio JJ, Farr AG, Udey MC. A role for endogenous transforming growth factor β1 in Langerhans cell biology: the skin of transforming growth factor β1 null mice is devoid of epidermal Langerhans cells. J Exp Med. 1996;184:2417–22. doi: 10.1084/jem.184.6.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Strobl H, Bello-Fernandez C, Riedl E, Pickl WF, Majdic O, et al. flt3 ligand in cooperation with transforming growth factor-β1 potentiates in vitro development of Langerhans-type dendritic cells and allows single-cell dendritic cell cluster formation under serum-free conditions. Blood. 1997;90:1425–34. [PubMed] [Google Scholar]
- 119.Strobl H, Riedl E, Scheinecker C, Bello-Fernandez C, Pickl WF, et al. TGF-β1 promotes in vitro development of dendritic cells from CD34+ hemopoietic progenitors. J Immunol. 1996;157:1499–507. [PubMed] [Google Scholar]
- 120.Geissmann F, Prost C, Monnet JP, Dy M, Brousse N, Hermine O. Transforming growth factor β1, in the presence of granulocyte/macrophage colony-stimulating factor and interleukin 4, induces differentiation of human peripheral blood monocytes into dendritic Langerhans cells. J Exp Med. 1998;187:961–66. doi: 10.1084/jem.187.6.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kaplan DH, Li MO, Jenison MC, Shlomchik WD, Flavell RA, Shlomchik MJ. Autocrine/paracrine TGFβ1 is required for the development of epidermal Langerhans cells. J Exp Med. 2007;204:2545–52. doi: 10.1084/jem.20071401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bobr A, Igyarto BZ, Haley KM, Li MO, Flavell RA, Kaplan DH. Autocrine/paracrine TGF-β1 inhibits Langerhans cell migration. Proc Natl Acad Sci USA. 2012;109:10492–97. doi: 10.1073/pnas.1119178109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kel JM, Girard-Madoux MJ, Reizis B, Clausen BE. TGF-β is required to maintain the pool of immature Langerhans cells in the epidermis. J Immunol. 2010;185:3248–55. doi: 10.4049/jimmunol.1000981. [DOI] [PubMed] [Google Scholar]
- 124.Hacker C, Kirsch RD, Ju XS, Hieronymus T, Gust TC, et al. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat Immunol. 2003;4:380–86. doi: 10.1038/ni903. [DOI] [PubMed] [Google Scholar]
- 125.Fainaru O, Woolf E, Lotem J, Yarmus M, Brenner O, et al. Runx3 regulates mouse TGF-β-mediated dendritic cell function and its absence results in airway inflammation. EMBO J. 2004;23:969–79. doi: 10.1038/sj.emboj.7600085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Becker AM, Michael DG, Satpathy AT, Sciammas R, Singh H, Bhattacharya D. IRF-8 extinguishes neutrophil production and promotes dendritic cell lineage commitment in both myeloid and lymphoid mouse progenitors. Blood. 2012;119:2003–12. doi: 10.1182/blood-2011-06-364976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li L, Jin H, Xu J, Shi Y, Wen Z. Irf8 regulates macrophage versus neutrophil fate during zebrafish primitive myelopoiesis. Blood. 2010;117:1359–69. doi: 10.1182/blood-2010-06-290700. [DOI] [PubMed] [Google Scholar]
- 128.Holtschke T, Lohler J, Kanno Y, Fehr T, Giese N, et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307–17. doi: 10.1016/s0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 129.Schiavoni G, Mattei F, Sestili P, Borghi P, Venditti M, et al. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8α+ dendritic cells. J Exp Med. 2002;196:1415–25. doi: 10.1084/jem.20021263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tsujimura H, Tamura T, Ozato K. Cutting edge: IFN consensus sequence binding protein/IFN regulatory factor 8 drives the development of type I IFN-producing plasmacytoid dendritic cells. J Immunol. 2003;170:1131–35. doi: 10.4049/jimmunol.170.3.1131. [DOI] [PubMed] [Google Scholar]
- 131.Tamura T, Tailor P, Yamaoka K, Kong HJ, Tsujimura H, et al. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J Immunol. 2005;174:2573–81. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- 132.Edelson BT, Kc W, Juang R, Kohyama M, Benoit LA, et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8α+ conventional dendritic cells. J Exp Med. 2010;207:823–36. doi: 10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Turcotte K, Gauthier S, Tuite A, Mullick A, Malo D, Gros P. A mutation in the Icsbp1 gene causes susceptibility to infection and a chronic myeloid leukemia-like syndrome in BXH-2 mice. J Exp Med. 2005;201:881–90. doi: 10.1084/jem.20042170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tailor P, Tamura T, Morse HC, 3rd, Ozato K. The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood. 2008;111:1942–45. doi: 10.1182/blood-2007-07-100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schiavoni G, Mattei F, Borghi P, Sestili P, Venditti M, et al. ICSBP is critically involved in the normal development and trafficking of Langerhans cells and dermal dendritic cells. Blood. 2004;103:2221–28. doi: 10.1182/blood-2003-09-3007. [DOI] [PubMed] [Google Scholar]
- 136.Orabona C, Puccetti P, Vacca C, Bicciato S, Luchini A, et al. Toward the identification of a tolerogenic signature in IDO-competent dendritic cells. Blood. 2006;107:2846–54. doi: 10.1182/blood-2005-10-4077. [DOI] [PubMed] [Google Scholar]
- 137.Suzuki S, Honma K, Matsuyama T, Suzuki K, Toriyama K, et al. Critical roles of interferon regulatory factor 4 in CD11bhighCD8α− dendritic cell development. Proc Natl Acad Sci USA. 2004;101:8981–86. doi: 10.1073/pnas.0402139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Esashi E, Wang YH, Perng O, Qin XF, Liu YJ, Watowich SS. The signal transducer STAT5 inhibits plasmacytoid dendritic cell development by suppressing transcription factor IRF8. Immunity. 2008;28:509–20. doi: 10.1016/j.immuni.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ichikawa E, Hida S, Omatsu Y, Shimoyama S, Takahara K, et al. Defective development of splenic and epidermal CD4+ dendritic cells in mice deficient for IFN regulatory factor-2. Proc Natl Acad Sci USA. 2004;101:3909–14. doi: 10.1073/pnas.0400610101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ghosh HS, Cisse B, Bunin A, Lewis KL, Reizis B. Continuous expression of the transcription factor E2-2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity. 2010;33:905–16. doi: 10.1016/j.immuni.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cisse B, Caton ML, Lehner M, Maeda T, Scheu S, et al. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008;135:37–48. doi: 10.1016/j.cell.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kusunoki T, Sugai M, Katakai T, Omatsu Y, Iyoda T, et al. TH2 dominance and defective development of a CD8+ dendritic cell subset in Id2-deficient mice. J Allergy Clin Immunol. 2003;111:136–42. doi: 10.1067/mai.2003.29. [DOI] [PubMed] [Google Scholar]
- 143.Spits H, Couwenberg F, Bakker AQ, Weijer K, Uittenbogaart CH. Id2 and Id3 inhibit development of CD34+ stem cells into predendritic cell (pre-DC)2 but not into pre-DC1. Evidence for a lymphoid origin of pre-DC2. J Exp Med. 2000;192:1775–84. doi: 10.1084/jem.192.12.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, et al. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tussiwand R, Lee WL, Murphy TL, Mashayekhi M, Wumesh KC, et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature. 2012;490:502–7. doi: 10.1038/nature11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Meredith MM, Liu K, Kamphorst AO, Idoyaga J, Yamane A, et al. Zinc finger transcription factor zDC is a negative regulator required to prevent activation of classical dendritic cells in the steady state. J Exp Med. 2012;209:1583–93. doi: 10.1084/jem.20121003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Laouar Y, Welte T, Fu XY, Flavell RA. STAT3 is required for Flt3L-dependent dendritic cell differentiation. Immunity. 2003;19:903–12. doi: 10.1016/s1074-7613(03)00332-7. [DOI] [PubMed] [Google Scholar]
- 148.Gilliet M, Boonstra A, Paturel C, Antonenko S, Xu XL, et al. The development of murine plasma-cytoid dendritic cell precursors is differentially regulated by FLT3-ligand and granulocyte/macrophage colony-stimulating factor. J Exp Med. 2002;195:953–58. doi: 10.1084/jem.20020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.van de Laar L, van den Bosch A, Wierenga AT, Janssen HL, Coffer PJ, Woltman AM. Tight control of STAT5 activity determines human CD34-derived interstitial dendritic cell and Langerhans cell development. J Immunol. 2011;186:7016–24. doi: 10.4049/jimmunol.1003977. [DOI] [PubMed] [Google Scholar]
- 150.Kobayashi T, Walsh PT, Walsh MC, Speirs KM, Chiffoleau E, et al. TRAF6 is a critical factor for dendritic cell maturation and development. Immunity. 2003;19:353–63. doi: 10.1016/s1074-7613(03)00230-9. [DOI] [PubMed] [Google Scholar]
- 151.Wu L, Nichogiannopoulou A, Shortman K, Georgopoulos K. Cell-autonomous defects in dendritic cell populations of Ikaros mutant mice point to a developmental relationship with the lymphoid lineage. Immunity. 1997;7:483–92. doi: 10.1016/s1074-7613(00)80370-2. [DOI] [PubMed] [Google Scholar]
- 152.Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8− dendritic cells in the spleen. J Exp Med. 2007;204:1653–64. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Anderson KL, Perkin H, Surh CD, Venturini S, Maki RA, Torbett BE. Transcription factor PU. 1 is necessary for development of thymic and myeloid progenitor-derived dendritic cells. J Immunol. 2000;164:1855–61. doi: 10.4049/jimmunol.164.4.1855. [DOI] [PubMed] [Google Scholar]
- 154.Guerriero A, Langmuir PB, Spain LM, Scott EW. PU. 1 is required for myeloid-derived but not lymphoid-derived dendritic cells. Blood. 2000;95:879–85. [PubMed] [Google Scholar]
- 155.Carotta S, Dakic A, D’Amico A, Pang SHM, Greig KT, et al. The transcription factor PU. 1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity. 2010;32:628–41. doi: 10.1016/j.immuni.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 156.Bogunovic M, Ginhoux F, Wagers A, Loubeau M, Isola LM, et al. Identification of a radio-resistant and cycling dermal dendritic cell population in mice and men. J Exp Med. 2006;203:2627–38. doi: 10.1084/jem.20060667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liu K, Waskow C, Liu X, Yao K, Hoh J, Nussenzweig M. Origin of dendritic cells in peripheral lymphoid organs of mice. Nat Immunol. 2007;8:578–83. doi: 10.1038/ni1462. [DOI] [PubMed] [Google Scholar]
- 158.Kamath AT, Henri S, Battye F, Tough DF, Shortman K. Developmental kinetics and lifespan of dendritic cells in mouse lymphoid organs. Blood. 2002;100:1734–41. [PubMed] [Google Scholar]
- 159.Kamath AT, Pooley J, O’Keeffe MA, Vremec D, Zhan Y, et al. The development, maturation, and turnover rate of mouse spleen dendritic cell populations. J Immunol. 2000;165:6762–70. doi: 10.4049/jimmunol.165.12.6762. [DOI] [PubMed] [Google Scholar]
- 160.Jung S, Unutmaz D, Wong P, Sano G, De los Santos K, et al. In vivo depletion of CD11c+ dendritic cells abrogates priming of CD8+ T cells by exogenous cell-associated antigens. Immunity. 2002;17:211–20. doi: 10.1016/s1074-7613(02)00365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Idoyaga J, Suda N, Suda K, Park CG, Steinman RM. Antibody to Langerin/CD207 localizes large numbers of CD8α+ dendritic cells to the marginal zone of mouse spleen. Proc Natl Acad Sci USA. 2009;106:1524–29. doi: 10.1073/pnas.0812247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Qiu CH, Miyake Y, Kaise H, Kitamura H, Ohara O, Tanaka M. Novel subset of CD8α+ dendritic cells localized in the marginal zone is responsible for tolerance to cell-associated antigens. J Immunol. 2009;182:4127–36. doi: 10.4049/jimmunol.0803364. [DOI] [PubMed] [Google Scholar]
- 163.Edelson BT, Bradstreet TR, Hildner K, Carrero JA, Frederick KE, et al. CD8α+ dendritic cells are an obligate cellular entry point for productive infection by Listeria monocytogenes. Immunity. 2011;35:236–48. doi: 10.1016/j.immuni.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Edwards AD, Diebold SS, Slack EM, Tomizawa H, Hemmi H, et al. Toll-like receptor expression in murine DC subsets: lack of TLR7 expression by CD8 α+ DC correlates with unresponsiveness to imidazoquinolines. Eur J Immunol. 2003;33:827–33. doi: 10.1002/eji.200323797. [DOI] [PubMed] [Google Scholar]
- 165.Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, et al. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 2010;32:279–89. doi: 10.1016/j.immuni.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 166.Segura E, Kapp E, Gupta N, Wong J, Lim J, et al. Differential expression of pathogen-recognition molecules between dendritic cell subsets revealed by plasma membrane proteomic analysis. Mol Immunol. 2010;47:1765–73. doi: 10.1016/j.molimm.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 167.Davey GM, Wojtasiak M, Proietto AI, Carbone FR, Heath WR, Bedoui S. Cutting edge: priming of CD8 T cell immunity to herpes simplex virus type 1 requires cognate TLR3 expression in vivo. J Immunol. 2010;184:2243–46. doi: 10.4049/jimmunol.0903013. [DOI] [PubMed] [Google Scholar]
- 168.Yarovinsky F, Zhang D, Andersen JF, Bannenberg GL, Serhan CN, et al. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science. 2005;308:1626–29. doi: 10.1126/science.1109893. [DOI] [PubMed] [Google Scholar]
- 169.Ren Y, Silverstein RL, Allen J, Savill J. CD36 gene transfer confers capacity for phagocytosis of cells undergoing apoptosis. J Exp Med. 1995;181:1857–62. doi: 10.1084/jem.181.5.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cheong C, Idoyaga J, Do Y, Pack M, Park SH, et al. Production of monoclonal antibodies that recognize the extracellular domain of mouse langerin/CD207. J Immunol Methods. 2007;324:48–62. doi: 10.1016/j.jim.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Flacher V, Douillard P, Ait-Yahia S, Stoitzner P, Clair-Moninot V, et al. Expression of langerin/CD207 reveals dendritic cell heterogeneity between inbred mouse strains. Immunology. 2008;123:339–47. doi: 10.1111/j.1365-2567.2007.02785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sancho D, Joffre OP, Keller AM, Rogers NC, Martinez D, et al. Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature. 2009;458:899–903. doi: 10.1038/nature07750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sancho D, Mourao-Sa D, Joffre OP, Schulz O, Rogers NC, et al. Tumor therapy in mice via antigen targeting to a novel, DC-restricted C-type lectin. J Clin Investig. 2008;118:2098–110. doi: 10.1172/JCI34584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Poulin LF, Reyal Y, Uronen-Hansson H, Schraml B, Sancho D, et al. DNGR-1 is a specific and universal marker of mouse and human Batf3-dependent dendritic cells in lymphoid and nonlymphoid tissues. Blood. 2012;119:6052–62. doi: 10.1182/blood-2012-01-406967. [DOI] [PubMed] [Google Scholar]
- 175.Lukens MV, Kruijsen D, Coenjaerts FE, Kimpen JL, van Bleek GM. Respiratory syncytial virus-induced activation and migration of respiratory dendritic cells and subsequent antigen presentation in the lung-draining lymph node. J Virol. 2009;83:7235–43. doi: 10.1128/JVI.00452-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bedoui S, Whitney PG, Waithman J, Eidsmo L, Wakim L, et al. Cross-presentation of viral and self antigens by skin-derived CD103+ dendritic cells. Nat Immunol. 2009;10:488–95. doi: 10.1038/ni.1724. [DOI] [PubMed] [Google Scholar]
- 177.Lundie RJ, de Koning-Ward TF, Davey GM, Nie CQ, Hansen DS, et al. Blood-stage Plasmodium infection induces CD8+ T lymphocytes to parasite-expressed antigens, largely regulated by CD8α+ dendritic cells. Proc Natl Acad Sci USA. 2008;105:14509–14. doi: 10.1073/pnas.0806727105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Belz GT, Shortman K, Bevan MJ, Heath WR. CD8α+ dendritic cells selectively present MHC class I-restricted noncytolytic viral and intracellular bacterial antigens in vivo. J Immunol. 2005;175:196–200. doi: 10.4049/jimmunol.175.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Belz GT, Smith CM, Kleinert L, Reading P, Brooks A, et al. Distinct migrating and nonmigrating dendritic cell populations are involved in MHC class I-restricted antigen presentation after lung infection with virus. Proc Natl Acad Sci USA. 2004;101:8670–75. doi: 10.1073/pnas.0402644101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Belz GT, Bedoui S, Kupresanin F, Carbone FR, Heath WR. Minimal activation of memory CD8+ T cell by tissue-derived dendritic cells favors the stimulation of naive CD8+ T cells. Nat Immunol. 2007;8:1060–66. doi: 10.1038/ni1505. [DOI] [PubMed] [Google Scholar]
- 181.den Haan JM, Lehar SM, Bevan MJ. CD8+ but not CD8− dendritic cells cross-prime cytotoxic T cells in vivo. J Exp Med. 2000;192:1685–96. doi: 10.1084/jem.192.12.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Iyoda T, Shimoyama S, Liu K, Omatsu Y, Akiyama Y, et al. The CD8+ dendritic cell subset selectively endocytoses dying cells in culture and in vivo. J Exp Med. 2002;195:1289–302. doi: 10.1084/jem.20020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Henri S, Poulin LF, Tamoutounour S, Ardouin L, Guilliams M, et al. CD207+ CD103+ dermal dendritic cells cross-present keratinocyte-derived antigens irrespective of the presence of Langerhans cells. J Exp Med. 2010;207:189–206. doi: 10.1084/jem.20091964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.GeurtsvanKessel CH, Willart MA, van Rijt LS, Muskens F, Kool M, et al. Clearance of in-fluenza virus from the lung depends on migratory langerin+CD11b− but not plasmacytoid dendritic cells. J Exp Med. 2008;205:1621–34. doi: 10.1084/jem.20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Brewig N, Kissenpfennig A, Malissen B, Veit A, Bickert T, et al. Priming of CD8+ and CD4+ T cells in experimental leishmaniasis is initiated by different dendritic cell subtypes. J Immunol. 2009;182:774–83. doi: 10.4049/jimmunol.182.2.774. [DOI] [PubMed] [Google Scholar]
- 186.Helft J, Manicassamy B, Guermonprez P, Hashimoto D, Silvin A, et al. Cross-presenting CD103+ dendritic cells are protected from influenza virus infection. J Clin Investig. 2012;122(11):4037–47. doi: 10.1172/JCI60659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Amigorena S, Savina A. Intracellular mechanisms of antigen cross presentation in dendritic cells. Curr Opin Immunol. 2010;22:109–17. doi: 10.1016/j.coi.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 188.Trombetta ES, Mellman I. Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol. 2005;23:975–1028. doi: 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- 189.Guermonprez P, Valladeau J, Zitvogel L, Thery C, Amigorena S. Antigen presentation and T cell stimulation by dendritic cells. Annu Rev Immunol. 2002;20:621–67. doi: 10.1146/annurev.immunol.20.100301.064828. [DOI] [PubMed] [Google Scholar]
- 190.Heath WR, Carbone FR. Dendritic cell subsets in primary and secondary T cell responses at body surfaces. Nat Immunol. 2009;10:1237–44. doi: 10.1038/ni.1822. [DOI] [PubMed] [Google Scholar]
- 191.Delamarre L, Pack M, Chang H, Mellman I, Trombetta ES. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science. 2005;307:1630–34. doi: 10.1126/science.1108003. [DOI] [PubMed] [Google Scholar]
- 192.Lennon-Dumenil AM, Bakker AH, Maehr R, Fiebiger E, Overkleeft HS, et al. Analysis of protease activity in live antigen-presenting cells shows regulation of the phagosomal proteolytic contents during dendritic cell activation. J Exp Med. 2002;196:529–40. doi: 10.1084/jem.20020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kovacsovics-Bankowski M, Rock KL. A phagosome-to-cytosol pathway for exogenous antigens presented on MHC class I molecules. Science. 1995;267:243–46. doi: 10.1126/science.7809629. [DOI] [PubMed] [Google Scholar]
- 194.Savina A, Peres A, Cebrian I, Carmo N, Moita C, et al. The small GTPase Rac2 controls phagosomal alkalinization and antigen crosspresentation selectively in CD8+ dendritic cells. Immunity. 2009;30:544–55. doi: 10.1016/j.immuni.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 195.Lin ML, Zhan Y, Proietto AI, Prato S, Wu L, et al. Selective suicide of cross-presenting CD8+ dendritic cells by cytochrome c injection shows functional heterogeneity within this subset. Proc Natl Acad Sci USA. 2008;105:3029–34. doi: 10.1073/pnas.0712394105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ploegh HL. Bridging B cell and T cell recognition of antigen. J Immunol. 2007;179:7193. doi: 10.4049/jimmunol.179.11.7193. [DOI] [PubMed] [Google Scholar]
- 197.Bougneres L, Helft J, Tiwari S, Vargas P, Chang BH, et al. A role for lipid bodies in the cross-presentation of phagocytosed antigens by MHC class I in dendritic cells. Immunity. 2009;31:232–44. doi: 10.1016/j.immuni.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Maldonado-Lopez R, De Smedt T, Michel P, Godfroid J, Pajak B, et al. CD8α+ and CD8α− subclasses of dendritic cells direct the development of distinct T helper cells in vivo. J Exp Med. 1999;189:587–92. doi: 10.1084/jem.189.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Farrand KJ, Dickgreber N, Stoitzner P, Ronchese F, Petersen TR, Hermans IF. Langerin+ CD8α+ dendritic cells are critical for cross-priming and IL-12 production in response to systemic antigens. J Immunol. 2009;183:7732–42. doi: 10.4049/jimmunol.0902707. [DOI] [PubMed] [Google Scholar]
- 200.Hochrein H, Shortman K, Vremec D, Scott B, Hertzog P, O’Keeffe M. Differential production of IL-12, IFN-α, and IFN-γ by mouse dendritic cell subsets. J Immunol. 2001;166:5448–55. doi: 10.4049/jimmunol.166.9.5448. [DOI] [PubMed] [Google Scholar]
- 201.Mashayekhi M, Sandau MM, Dunay IR, Frickel EM, Khan A, et al. CD8α+ dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites. Immunity. 2011;35:249–59. doi: 10.1016/j.immuni.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mattei F, Schiavoni G, Belardelli F, Tough DF. IL-15 is expressed by dendritic cells in response to type I IFN, double-stranded RNA, or lipopolysaccharide and promotes dendritic cell activation. J Immunol. 2001;167:1179–87. doi: 10.4049/jimmunol.167.3.1179. [DOI] [PubMed] [Google Scholar]
- 203.Pulendran B. Immune activation: death, danger and dendritic cells. Curr Biol. 2004;14:R30–32. doi: 10.1016/j.cub.2003.12.020. [DOI] [PubMed] [Google Scholar]
- 204.Dorner BG, Dorner MB, Zhou X, Opitz C, Mora A, et al. Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity. 2009;31:823–33. doi: 10.1016/j.immuni.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 205.Igyarto BZ, Haley K, Ortner D, Bobr A, Gerami-Nejad M, et al. Skin-resident murine dendritic cell subsets promote distinct and opposing antigen-specific T helper cell responses. Immunity. 2011;35:260–72. doi: 10.1016/j.immuni.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Smith CM, Wilson NS, Waithman J, Villadangos JA, Carbone FR, et al. Cognate CD4+ T cell licensing of dendritic cells in CD8+ T cell immunity. Nat Immunol. 2004;5:1143–48. doi: 10.1038/ni1129. [DOI] [PubMed] [Google Scholar]
- 207.Klein L, Hinterberger M, Wirnsberger G, Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol. 2009;9:833–44. doi: 10.1038/nri2669. [DOI] [PubMed] [Google Scholar]
- 208.Hsieh CS, Lee HM, Lio CW. Selection of regulatory T cells in the thymus. Nat Rev Immunol. 2012;12:157–67. doi: 10.1038/nri3155. [DOI] [PubMed] [Google Scholar]
- 209.Huang FP, Platt N, Wykes M, Major JR, Powell TJ, et al. A discrete subpopulation of dendritic cells transports apoptotic intestinal epithelial cells to T cell areas of mesenteric lymph nodes. J Exp Med. 2000;191:435–44. doi: 10.1084/jem.191.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Liu K, Idoyaga J, Charalambous A, Fujii S, Bonito A, et al. Innate NKT lymphocytes confer superior adaptive immunity via tumor-capturing dendritic cells. J Exp Med. 2005;202:1507–16. doi: 10.1084/jem.20050956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Liu K, Iyoda T, Saternus M, Kimura Y, Inaba K, Steinman RM. Immune tolerance after delivery of dying cells to dendritic cells in situ. J Exp Med. 2002;196:1091–97. doi: 10.1084/jem.20021215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Ohnmacht C, Pullner A, King SB, Drexler I, Meier S, et al. Constitutive ablation of dendritic cells breaks self-tolerance of CD4 T cells and results in spontaneous fatal autoimmunity. J Exp Med. 2009;206:549–59. doi: 10.1084/jem.20082394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Heath WR, Belz GT, Behrens GM, Smith CM, Forehan SP, et al. Cross-presentation, dendritic cell subsets, and the generation of immunity to cellular antigens. Immunol Rev. 2004;199:9–26. doi: 10.1111/j.0105-2896.2004.00142.x. [DOI] [PubMed] [Google Scholar]
- 214.Allan RS, Smith CM, Belz GT, van Lint AL, Wakim LM, et al. Epidermal viral immunity induced by CD8α+ dendritic cells but not by Langerhans cells. Science. 2003;301:1925–28. doi: 10.1126/science.1087576. [DOI] [PubMed] [Google Scholar]
- 215.Allan RS, Waithman J, Bedoui S, Jones CM, Villadangos JA, et al. Migratory dendritic cells transfer antigen to a lymph node-resident dendritic cell population for efficient CTL priming. Immunity. 2006;25:153–62. doi: 10.1016/j.immuni.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 216.Kim TS, Braciale TJ. Respiratory dendritic cell subsets differ in their capacity to support the induction of virus-specific cytotoxic CD8+ T cell responses. PLoS ONE. 2009;4:e4204. doi: 10.1371/journal.pone.0004204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Bursch LS, Wang L, Igyarto B, Kissenpfennig A, Malissen B, et al. Identification of a novel population of Langerin+ dendritic cells. J Exp Med. 2007;204:3147–56. doi: 10.1084/jem.20071966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Beaty SR, Rose CE, Jr, Sung SS. Diverse and potent chemokine production by lung CD11bhigh dendritic cells in homeostasis and in allergic lung inflammation. J Immunol. 2007;178:1882–95. doi: 10.4049/jimmunol.178.3.1882. [DOI] [PubMed] [Google Scholar]
- 219.den Haan JM, Bevan MJ. Constitutive versus activation-dependent cross-presentation of immune complexes by CD8+ and CD8− dendritic cells in vivo. J Exp Med. 2002;196:817–27. doi: 10.1084/jem.20020295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ballesteros-Tato A, Leon B, Lund FE, Randall TD. Temporal changes in dendritic cell subsets, cross-priming and costimulation via CD70 control CD8+ T cell responses to influenza. Nat Immunol. 2010;11:216–24. doi: 10.1038/ni.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kastenmuller K, Wille-Reece U, Lindsay RW, Trager LR, Darrah PA, et al. Protective T cell immunity in mice following protein-TLR7/8 agonist-conjugate immunization requires aggregation, type I IFN, and multiple DC subsets. J Clin Investig. 2011;121:1782–96. doi: 10.1172/JCI45416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Idoyaga J, Cheong C, Suda K, Suda N, Kim JY, et al. Cutting edge: Langerin/CD207 receptor on dendritic cells mediates efficient antigen presentation on MHC I and II products in vivo. J Immunol. 2008;180:3647–50. doi: 10.4049/jimmunol.180.6.3647. [DOI] [PubMed] [Google Scholar]
- 223.McLachlan JB, Catron DM, Moon JJ, Jenkins MK. Dendritic cell antigen presentation drives simultaneous cytokine production by effector and regulatory T cells in inflamed skin. Immunity. 2009;30:277–88. doi: 10.1016/j.immuni.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Foss DL, Donskoy E, Goldschneider I. The importation of hematogenous precursors by the thymus is a gated phenomenon in normal adult mice. J Exp Med. 2001;193:365–74. doi: 10.1084/jem.193.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Bonasio R, Scimone ML, Schaerli P, Grabie N, Lichtman AH, von Andrian UH. Clonal deletion of thymocytes by circulating dendritic cells homing to the thymus. Nat Immunol. 2006;7:1092–100. doi: 10.1038/ni1385. [DOI] [PubMed] [Google Scholar]
- 226.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–85. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–64. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Guilliams M, Crozat K, Henri S, Tamoutounour S, Grenot P, et al. Skin-draining lymph nodes contain dermis-derived CD103− dendritic cells that constitutively produce retinoic acid and induce Foxp3+ regulatory T cells. Blood. 2010;115:1958–68. doi: 10.1182/blood-2009-09-245274. [DOI] [PubMed] [Google Scholar]
- 229.Vitali C, Mingozzi F, Broggi A, Barresi S, Zolezzi F, et al. Migratory, and not lymphoid-resident, dendritic cells maintain peripheral self-tolerance and prevent autoimmunity via induction of iTreg cells. Blood. 2012;120:1237–45. doi: 10.1182/blood-2011-09-379776. [DOI] [PubMed] [Google Scholar]
- 230.Kaplan DH, Kissenpfennig A, Clausen BE. Insights into Langerhans cell function from Langerhans cell ablation models. Eur J Immunol. 2008;38:2369–76. doi: 10.1002/eji.200838397. [DOI] [PubMed] [Google Scholar]
- 231.Kaplan DH. In vivo function of Langerhans cells and dermal dendritic cells. Trends Immunol. 2010;31:446–51. doi: 10.1016/j.it.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kaplan DH, Jenison MC, Saeland S, Shlomchik WD, Shlomchik MJ. Epidermal Langerhans cell-deficient mice develop enhanced contact hypersensitivity. Immunity. 2005;23:611–20. doi: 10.1016/j.immuni.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 233.Bobr A, Olvera-Gomez I, Igyarto BZ, Haley KM, Hogquist KA, Kaplan DH. Acute ablation of Langerhans cells enhances skin immune responses. J Immunol. 2010;185:4724–28. doi: 10.4049/jimmunol.1001802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Seneschal J, Clark RA, Gehad A, Baecher-Allan CM, Kupper TS. Human epidermal Langerhans cells maintain immune homeostasis in skin by activating skin resident regulatory T cells. Immunity. 2012;36:873–84. doi: 10.1016/j.immuni.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Merad M, Hoffmann P, Ranheim E, Slaymaker S, Manz MG, et al. Depletion of host Langerhans cells before transplantation of donor alloreactive T cells prevents skin graft-versus-host disease. Nat Med. 2004;10:510–17. doi: 10.1038/nm1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Dominguez PM, Ardavin C. Differentiation and function of mouse monocyte-derived dendritic cells in steady state and inflammation. Immunol Rev. 2010;234:90–104. doi: 10.1111/j.0105-2896.2009.00876.x. [DOI] [PubMed] [Google Scholar]
- 237.Cheong C, Matos I, Choi JH, Dandamudi DB, Shrestha E, et al. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209+ dendritic cells for immune T cell areas. Cell. 2010;143:416–29. doi: 10.1016/j.cell.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Segura E, Albiston AL, Wicks IP, Chai SY, Villadangos JA. Different cross-presentation pathways in steady-state and inflammatory dendritic cells. Proc Natl Acad Sci USA. 2009;106:20377–81. doi: 10.1073/pnas.0910295106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Satpathy AT, Wumesh KC, Albring JC, Edelson BT, Kretzer NM, et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J Exp Med. 2012;209:1135–52. doi: 10.1084/jem.20120030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Serbina NV, Salazar-Mather TP, Biron CA, Kuziel WA, Pamer EG. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 2003;19:59–70. doi: 10.1016/s1074-7613(03)00171-7. [DOI] [PubMed] [Google Scholar]
- 241.Randolph GJ, Inaba K, Robbiani DF, Steinman RM, Muller WA. Differentiation of phagocytic monocytes into lymph node dendritic cells in vivo. Immunity. 1999;11:753–61. doi: 10.1016/s1074-7613(00)80149-1. [DOI] [PubMed] [Google Scholar]
- 242.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19:71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- 243.Takizawa H, Boettcher S, Manz MG. Demand-adapted regulation of early hematopoiesis in infection and inflammation. Blood. 2012;119:2991–3002. doi: 10.1182/blood-2011-12-380113. [DOI] [PubMed] [Google Scholar]
- 244.Nagai Y, Garrett KP, Ohta S, Bahrun U, Kouro T, et al. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 2006;24:801–12. doi: 10.1016/j.immuni.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Welner RS, Pelayo R, Nagai Y, Garrett KP, Wuest TR, et al. Lymphoid precursors are directed to produce dendritic cells as a result of TLR9 ligation during herpes infection. Blood. 2008;112:3753–61. doi: 10.1182/blood-2008-04-151506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Sioud M, Floisand Y. TLR agonists induce the differentiation of human bone marrow CD34+ progenitors into CD11c+ CD80/86+ DC capable of inducing a Th1-type response. Eur J Immunol. 2007;37:2834–46. doi: 10.1002/eji.200737112. [DOI] [PubMed] [Google Scholar]
- 247.Sioud M, Floisand Y, Forfang L, Lund-Johansen F. Signaling through Toll-like receptor 7/8 induces the differentiation of human bone marrow CD34+ progenitor cells along the myeloid lineage. J Mol Biol. 2006;364:945–54. doi: 10.1016/j.jmb.2006.09.054. [DOI] [PubMed] [Google Scholar]
- 248.Massberg S, Schaerli P, Knezevic-Maramica I, Kollnberger M, Tubo N, et al. Immunosurveillance by hematopoietic progenitor cells trafficking through blood, lymph, and peripheral tissues. Cell. 2007;131:994–1008. doi: 10.1016/j.cell.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Schmid MA, Takizawa H, Baumjohann DR, Saito Y, Manz MG. Bone marrow dendritic cell progenitors sense pathogens via Toll-like receptors and subsequently migrate to inflamed lymph nodes. Blood. 2011;118:4829–40. doi: 10.1182/blood-2011-03-344960. [DOI] [PubMed] [Google Scholar]
- 250.Tsou CL, Peters W, Si Y, Slaymaker S, Aslanian AM, et al. Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J Clin Investig. 2007;117:902–9. doi: 10.1172/JCI29919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Serbina NV, Pamer EG. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat Immunol. 2006;7:311–17. doi: 10.1038/ni1309. [DOI] [PubMed] [Google Scholar]
- 252.Serbina NV, Kuziel W, Flavell R, Akira S, Rollins B, Pamer EG. Sequential MyD88-independent and -dependent activation of innate immune responses to intracellular bacterial infection. Immunity. 2003;19:891–901. doi: 10.1016/s1074-7613(03)00330-3. [DOI] [PubMed] [Google Scholar]
- 253.Copin R, De Baetselier P, Carlier Y, Letesson JJ, Muraille E. MyD88-dependent activation of B220− CD11b+LY-6C+ dendritic cells during Brucella melitensis infection. J Immunol. 2007;178:5182–91. doi: 10.4049/jimmunol.178.8.5182. [DOI] [PubMed] [Google Scholar]
- 254.De Trez C, Magez S, Akira S, Ryffel B, Carlier Y, Muraille E. iNOS-producing inflammatory dendritic cells constitute the major infected cell type during the chronic Leishmania major infection phase of C57BL/6 resistant mice. PLoS Pathog. 2009;5:e1000494. doi: 10.1371/journal.ppat.1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Shortman K, Naik SH. Steady-state and inflammatory dendritic-cell development. Nat Rev Immunol. 2007;7:19–30. doi: 10.1038/nri1996. [DOI] [PubMed] [Google Scholar]
- 256.Siegal FP, Kadowaki N, Shodell M, Fitzgerald-Bocarsly PA, Shah K, et al. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–37. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- 257.Rissoan MC, Soumelis V, Kadowaki N, Grouard G, Briere F, et al. Reciprocal control of T helper cell and dendritic cell differentiation [see comments] Science. 1999;283:1183–86. doi: 10.1126/science.283.5405.1183. [DOI] [PubMed] [Google Scholar]
- 258.O’Doherty U, Steinman RM, Peng M, Cameron PU, Gezelter S, et al. Dendritic cells freshly isolated from human blood express CD4 and mature into typical immunostimulatory dendritic cells after culture in monocyte-conditioned medium. J Exp Med. 1993;178:1067–76. doi: 10.1084/jem.178.3.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.O’Doherty U, Peng M, Gezelter S, Swiggard WJ, Betjes M, et al. Human blood contains two subsets of dendritic cells, one immunologically mature and the other immature. Immunology. 1994;82:487. [PMC free article] [PubMed] [Google Scholar]
- 260.Thomas R, Davis LS, Lipsky PE. Isolation and characterization of human peripheral blood dendritic cells. J Immunol. 1993;150:821–34. [PubMed] [Google Scholar]
- 261.Dzionek A, Fuchs A, Schmidt P, Cremer S, Zysk M, et al. BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J Immunol. 2000;165:6037–46. doi: 10.4049/jimmunol.165.11.6037. [DOI] [PubMed] [Google Scholar]
- 262.MacDonald KP, Munster DJ, Clark GJ, Dzionek A, Schmitz J, Hart DN. Characterization of human blood dendritic cell subsets. Blood. 2002;100:4512–20. doi: 10.1182/blood-2001-11-0097. [DOI] [PubMed] [Google Scholar]
- 263.Ito T, Inaba M, Inaba K, Toki J, Sogo S, et al. A CD1a+/CD11c+ subset of human blood dendritic cells is a direct precursor of Langerhans cells. J Immunol. 1999;163:1409–19. [PubMed] [Google Scholar]
- 264.Lindstedt M, Lundberg K, Borrebaeck CA. Gene family clustering identifies functionally associated subsets of human in vivo blood and tonsillar dendritic cells. J Immunol. 2005;175:4839–46. doi: 10.4049/jimmunol.175.8.4839. [DOI] [PubMed] [Google Scholar]
- 265.Schakel K, Kannagi R, Kniep B, Goto Y, Mitsuoka C, et al. 6-Sulfo LacNAc, a novel carbohydrate modification of PSGL-1, defines an inflammatory type of human dendritic cells. Immunity. 2002;17:289–301. doi: 10.1016/s1074-7613(02)00393-x. [DOI] [PubMed] [Google Scholar]
- 266.Randolph GJ, Sanchez-Schmitz G, Liebman RM, Schakel K. The CD16+ (FcγRIII+) subset of human monocytes preferentially becomes migratory dendritic cells in a model tissue setting. J Exp Med. 2002;196:517–27. doi: 10.1084/jem.20011608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Robbins SH, Walzer T, Dembele D, Thibault C, Defays A, et al. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 2008;9:R17. doi: 10.1186/gb-2008-9-1-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Nestle FO, Zheng XG, Thompson CB, Turka LA, Nickoloff BJ. Characterization of dermal dendritic cells obtained from normal human skin reveals phenotypic and functionally distinctive subsets. J Immunol. 1993;151:6535–45. [PubMed] [Google Scholar]
- 269.Lenz A, Heine M, Schuler G, Romani N. Human and murine dermis contain dendritic cells. Isolation by means of a novel method and phenotypical and functional characterization. J Clin Investig. 1993;92:2587–96. doi: 10.1172/JCI116873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Zaba LC, Fuentes-Duculan J, Steinman RM, Krueger JG, Lowes MA. Normal human dermis contains distinct populations of CD11c+BDCA-1+ dendritic cells and CD163+FXIIIA+ macrophages. J Clin Investig. 2007;117:2517–25. doi: 10.1172/JCI32282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Haniffa M, Ginhoux F, Wang XN, Bigley V, Abel M, et al. Differential rates of replacement of human dermal dendritic cells and macrophages during hematopoietic stem cell transplantation. J Exp Med. 2009;206:371–85. doi: 10.1084/jem.20081633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Zaba LC, Krueger JG, Lowes MA. Resident and “inflammatory” dendritic cells in human skin. J Investig Dermatol. 2009;129:302–8. doi: 10.1038/jid.2008.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Haniffa M, Shin A, Bigley V, McGovern N, Teo P, et al. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity. 2012;37:60–73. doi: 10.1016/j.immuni.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Tigelaar RE, Lewis JM, Bergstresser PR. TCR γ/δ+ dendritic epidermal T cells as constituents of skin-associated lymphoid tissue. J Investig Dermatol. 1990;94:58S–63S. doi: 10.1111/1523-1747.ep12875138. [DOI] [PubMed] [Google Scholar]
- 275.Valladeau J, Duvert-Frances V, Pin JJ, Dezutter-Dambuyant C, Vincent C, et al. The monoclonal antibody DCGM4 recognizes Langerin, a protein specific of Langerhans cells, and is rapidly internalized from the cell surface. Eur J Immunol. 1999;29:2695–704. doi: 10.1002/(SICI)1521-4141(199909)29:09<2695::AID-IMMU2695>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 276.Fithian E, Kung P, Goldstein G, Rubenfeld M, Fenoglio C, Edelson R. Reactivity of Langerhans cells with hybridoma antibody. Proc Natl Acad Sci USA. 1981;78:2541–44. doi: 10.1073/pnas.78.4.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Hunger RE, Sieling PA, Ochoa MT, Sugaya M, Burdick AE, et al. Langerhans cells utilize CD1a and langerin to efficiently present nonpeptide antigens to T cells. J Clin Investig. 2004;113:701–8. doi: 10.1172/JCI19655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Poulin LF, Salio M, Griessinger E, Anjos-Afonso F, Craciun L, et al. Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8α+ dendritic cells. J Exp Med. 2010;207:1261–71. doi: 10.1084/jem.20092618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.McIlroy D, Troadec C, Grassi F, Samri A, Barrou B, et al. Investigation of human spleen dendritic cell phenotype and distribution reveals evidence of in vivo activation in a subset of organ donors. Blood. 2001;97:3470–77. doi: 10.1182/blood.v97.11.3470. [DOI] [PubMed] [Google Scholar]
- 280.Jongbloed SL, Kassianos AJ, McDonald KJ, Clark GJ, Ju X, et al. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J Exp Med. 2010;207:1247–60. doi: 10.1084/jem.20092140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Segura E, Valladeau-Guilemond J, Donnadieu MH, Sastre-Garau X, Soumelis V, Amigorena S. Characterization of resident and migratory dendritic cells in human lymph nodes. J Exp Med. 2012;209:653–60. doi: 10.1084/jem.20111457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Shortman K, Liu YJ. Mouse and human dendritic cell subtypes. Nat Rev Immunol. 2002;2002:151–61. doi: 10.1038/nri746. [DOI] [PubMed] [Google Scholar]
- 283.Villadangos JA, Shortman K. Found in translation: the human equivalent of mouse CD8+ dendritic cells. J Exp Med. 2010;207:1131–34. doi: 10.1084/jem.20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Huysamen C, Willment JA, Dennehy KM, Brown GD. CLEC9A is a novel activation C-type lectin-like receptor expressed on BDCA3+ dendritic cells and a subset of monocytes. J Biol Chem. 2008;283:16693–701. doi: 10.1074/jbc.M709923200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Caminschi I, Proietto AI, Ahmet F, Kitsoulis S, Shin Teh J, et al. The dendritic cell subtype-restricted C-type lectin Clec9A is a target for vaccine enhancement. Blood. 2008;112:3264–73. doi: 10.1182/blood-2008-05-155176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Bachem A, Guttler S, Hartung E, Ebstein F, Schaefer M, et al. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J Exp Med. 2010;207:1273–81. doi: 10.1084/jem.20100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Crozat K, Guiton R, Contreras V, Feuillet V, Dutertre CA, et al. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8α+ dendritic cells. J Exp Med. 2010;207:1283–92. doi: 10.1084/jem.20100223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Crozat K, Tamoutounour S, Vu Manh TP, Fossum E, Luche H, et al. Cutting edge: expression of XCR1 defines mouse lymphoid-tissue resident and migratory dendritic cells of the CD8α+ type. J Immunol. 2011;187:4411–15. doi: 10.4049/jimmunol.1101717. [DOI] [PubMed] [Google Scholar]
- 289.Lauterbach H, Bathke B, Gilles S, Traidl-Hoffmann C, Luber CA, et al. Mouse CD8α+ DCs and human BDCA3+ DCs are major producers of IFN-λ in response to poly IC. J Exp Med. 2010;207:2703–17. doi: 10.1084/jem.20092720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Mittag D, Proietto AI, Loudovaris T, Mannering SI, Vremec D, et al. Human dendritic cell subsets from spleen and blood are similar in phenotype and function but modified by donor health status. J Immunol. 2011;186:6207–17. doi: 10.4049/jimmunol.1002632. [DOI] [PubMed] [Google Scholar]
- 291.Hambleton S, Salem S, Bustamante J, Bigley V, Boisson-Dupuis S, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med. 2011;365:127–38. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Emile JF, Geissmann F, Martin OC, Radford-Weiss I, Lepelletier Y, et al. Langerhans cell deficiency in reticular dysgenesis. Blood. 2000;96:58–62. [PubMed] [Google Scholar]
- 293.Xu Y, Zhan Y, Lew AM, Naik SH, Kershaw MH. Differential development of murine dendritic cells by GM-CSF versus Flt3 ligand has implications for inflammation and trafficking. J Immunol. 2007;179:7577–84. doi: 10.4049/jimmunol.179.11.7577. [DOI] [PubMed] [Google Scholar]
- 294.Brasel K, De Smedt T, Smith JL, Maliszewski CR. Generation of murine dendritic cells from flt3-ligand-supplemented bone marrow cultures. Blood. 2000;96:3029–39. [PubMed] [Google Scholar]
- 295.Brawand P, Fitzpatrick DR, Greenfield BW, Brasel K, Maliszewski CR, De Smedt T. Murine plasmacytoid pre-dendritic cells generated from Flt3 ligand-supplemented bone marrow cultures are immature APCs. J Immunol. 2002;169:6711–19. doi: 10.4049/jimmunol.169.12.6711. [DOI] [PubMed] [Google Scholar]
- 296.Naik SH, Proietto AI, Wilson NS, Dakic A, Schnorrer P, et al. Cutting edge: generation of splenic CD8+ and CD8− dendritic cell equivalents in Fms-like tyrosine kinase 3 ligand bone marrow cultures. J Immunol. 2005;174:6592–97. doi: 10.4049/jimmunol.174.11.6592. [DOI] [PubMed] [Google Scholar]
- 297.Caux C, Dezutter-Dambuyant C, Schmitt D, Banchereau J. GM-CSF and TNF-α cooperate in the generation of dendritic Langerhans cells. Nature. 1992;360:258–61. doi: 10.1038/360258a0. [DOI] [PubMed] [Google Scholar]
- 298.Palucka KA, Taquet N, Sanchez-Chapuis F, Gluckman JC. Dendritic cells as the terminal stage of monocyte differentiation. J Immunol. 1998;160:4587–95. [PubMed] [Google Scholar]
- 299.Chen W, Antonenko S, Sederstrom JM, Liang X, Chan AS, et al. Thrombopoietin cooperates with FLT3-ligand in the generation of plasmacytoid dendritic cell precursors from human hematopoietic progenitors. Blood. 2004;103:2547–53. doi: 10.1182/blood-2003-09-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]