Abstract
The deregulation of B cell differentiation has been shown to contribute to autoimmune disorders, hematological cancers, and aging. We provide evidence that the retinoic acid-producing enzyme aldehyde dehydrogenase 1a1 (Aldh1a1) is an oncogene suppressor in specific splenic IgG1+/CD19− and IgG1+/CD19+ B cells populations. Aldh1a1 regulated transcription factors during B cell differentiation in a sequential manner: 1) retinoic acid receptor alpha (Rara) in IgG1+/CD19− and 2) zinc finger protein Zfp423 and peroxisome proliferator-activated receptor gamma (Pparg) in IgG1+/CD19+ splenocytes. In Aldh1a1−/− mice, splenic IgG1+/CD19− and IgG1+/CD19+ B cells acquired expression of proto-oncogenic genes c-Fos, c-Jun, and Hoxa10 that resulted in splenomegaly. Human multiple myeloma B cell lines also lack Aldh1a1 expression; however, ectopic Aldh1a1 expression rescued Rara and Znf423 expression in these cells. Our data highlight a mechanism by which an enzyme involved in vitamin A metabolism can improve B cell resistance to oncogenesis.
Keywords: homeobox transcription factor, nuclear receptor, retinaldehyde, Raldh1, vitamin A metabolism, multiple myeloma
1.1 Introduction
The deregulation of B cell differentiation has been shown to play a causal role in autoimmune disorders, carcinogenesis, and aging [1]. B cells differentiate from B-lymphoid progenitors in bone marrow and progress through many stages during differentiation to ultimately express B cell receptor (BCR). Lymphocytes expressing surface IgM migrate to the spleen [2], where self-reactive splenic B cells undergo apoptosis; others become responsive to T-cell-dependent and T-cell-independent antigens. The various B-cell populations are compartmentalized in different splenic zones, including red pulp, marginal zone, and white pulp. After pathogen exposure, they complete differentiation in germinal centers [2]. Specific populations of B cells can undergo alternative differentiation. For instance, purified mouse splenic B cells respond to stimulation with cytokines, anti-CD38, anti-CD40, anti µ, and retinoic acid (RA) by enriching IgG1+ and CD138+ B cell populations and genes involved in the regulation of Ig somatic hypermutation and class switching [3]. In these B cells, RA contributed to the suppression of activation-induced deaminase (Aid), transcriptional regulators of differentiation (Pax5), and neoplastic transformation t(9;14) [3,4]. Oncogenic processes further diversify in B lymphoma cells [5]. The physiological mechanisms responsible for the formation of specific B cell subsets have remained unexplored.
The studies with dietary vitamin A (retinol or retinyl esters) highlighted a possible role for this pathway in specific B cell responses. Dietary vitamin A content influenced IgA production against T-cell dependent and T-cell independent type 2 antigens at mucosal locations [6]. Vitamin A deficiency in the diet diminishes immune responses and increases mortality [7–10]. These responses may be partially improved by supplementation with either vitamin A or its metabolite RA, arguing for RA as a mediator of these responses. The function of RA in B cell studies in vitro revealed that multiple aspects of B cell biology are RA-sensitive [10,11]. RA accelerated differentiation of a subset of proliferating lymphoid progenitor cells into B cells by targeting the oncogenes c-myc and cyclin D3, cytokines, and NFκB, as well as kinase p38/CDK2 [12,13]. RA treatment also promoted differentiation of malignant B cells, alone or in combination with rosiglitazone, an agonist for the nuclear receptor PPARγ [14]. The understanding of RA in immune function is incomplete. For example, a recent trial in Guinea-Bissau revealed a paradoxically higher mortality in girls supplemented with vitamin A than in placebo group [15]. In this study we dissected the role of endogenous vitamin A metabolism on gene regulation in B cells.
An increase in the intracellular RA concentrations is generated in response to various hormonal, dietary, and inflammatory stimuli and may be considered as a factor in endogenous differentiation of B cell subsets [11,16]. RA is produced sequentially. Alcohol dehydrogenases (ADH and SDR/RDH) oxidize retinol to retinaldehyde, which is dehydrogenated into RA by the members aldehyde dehydrogenase-1 family of enzymes: ALDH1a1, ALDH1a2, and ALDH1a3 [17]. The principal mechanism of RA action is through the activation of RA receptors (RAR). RA binding to RAR induces its heterodimerization with retinoid X receptor, and binding to cognate response element (RARE) sequences in the promoters of target genes [18]. In addition, RA regulates a plethora of signaling and transcriptional pathways [19], including a Zfp423-dependent induction of Pparg expression [20]. Paracrine RA production by ALDH1 enzymes in dendritic cells plays an important role in B cell homing to the mucosa, and promotes IgA isotype class switching [21,22]. The role of RA-generating enzymes in B cells has remained unexplored. Here, we investigated the transcriptional function of ALDH1a1 in in murine B cell subsets and in human multiple myeloma B cell lines.
1.2 Materials and Methods
Reagents
We purchased reagents from Sigma-Aldrich (St. Louis, MO) and cell culture media from Invitrogen (Carlsbad, CA) unless otherwise indicated. Anti-mouse antibodies were: CD19 from BD Bioscences (San Jose, CA) and β-galactosidase from Abcam (Cambridge, MA).
Animal studies
All experimental protocols were approved by the Institutional Animal Care and User Committee. Water and regular chow (Harlan Laboratories, Indianapolis, IL) was available ad libitum in all mouse studies.
Study 1 employed Tg RARE-Hspa1b/lacZ (denoted as RARE-lacZ) reporter mice developed by Dr. J. Rossant using a transgenic construct containing 3 copies of the 32bp RARE placed upstream of the mouse heat shock protein 1B promoter and β-galactosidase gene (lacZ) [23]. Female mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Three RARE-lacZ and three wild-type C57BL/6J (WT) female mice (12–15 weeks old) were fed regular chow throughout this study.
Study 2: Aldh1a1−/− mice were previously generated in the laboratory of G. Duester [24] and characterized for their metabolic responses [20,25,26]. Aldh1a1−/− (n=10) and WT (n=9) 13–14 month old male and female mice were used for these studies. Mice were fed regular chow diet. Blood was collected by cardiac puncture in EDTA-containing tubes. The spleen from 3 randomly-selected females were used for IgG1+/CD19− and IgG1+/CD19+ B-cell separation, and the remaining spleens and other organs were used for other analyses.
Human cells
Leukopacks (American Red Cross, Columbus, OH) were obtained from healthy donors under an Institutional Review Board-approved procurement protocol. Peripheral blood mononuclear cells (PBMC) were cultured in RPMI 1640 Media (Invitrogen) supplemented with 10% fetal bovine serum (ICN Biomedicals, Irvine, CA) and kept at 37°C in a 5% CO2/air incubator.
Flow cytometry analysis (FACS)
Splenocyte suspension was obtained from whole spleens dissected from 4 WT and 4 Aldh1a1−/− mice (2 males and 2 females in each group). Briefly, spleens were collected and mononuclear cell suspensions were prepared by mechanical disruption with the aid of a cell strainer (BD Biosciences, San Jose, CA) followed by brief incubation in NH4Cl (0.08%) to remove red blood cells. Splenocytes were resuspended in fluorescence-activated cell sorting buffer (FACS; PBS containing 0.1% BSA and 0.1% sodium azide) at 5×105/100µL. Cells were stained with a panel of antibodies from AbdSerotec, (Bio-Rad Laboratories, Ins, Hercules, CA) and available isotype controls. All antibodies were primary, non-conjugated with the exception of MHC class II which were directly conjugated to fluorescein isothiocyanate (FITC) and Alexa Fluor 647 (BD Biosciences), respectively. Secondary antibodies of either phycoerythrin (PE; 5 uL) or fluorescein isothiocyanate (FITC; 1uL) were purchased from AbD Serotec and were used at various dilutions (1:10, 1:50, and 1:100) [27]. Samples were analyzed using BD Accuri flow cytometer and analyzed with BD Accuri CFlow analysis software (BD Biosciences).
Purification of IgG1/CD19− and IgG1/CD19+ B cells
Splenic B cell subsets were obtained from four WT and Aldh1a1−/− female and one male mice. They were purified by automated magnetic cell separation (Auto MACS, Miltenyi Biotec). The cell suspensions were incubated with microbeads-conjugated with anti-mouse CD19. The CD19− and CD19+ populations were separated by AutoMACS. The CD19− and CD19+ fraction was further incubated with a biotinylated rat anti-mouse γ1 (clone G1-7.3, BD Biosciences) and streptavidin-conjugated microbeads. Populations of IgG1+/CD19− and IgG1+/CD19+ were separated by Auto MACS. Throughout, we denoted these populations as CD19− and CD19+. The purity (>98%) of the cell population was confirmed by FACS.
RNA isolation and quantitative real time PCR (qRT-PCR)
Total RNA was prepared using the RNeasy kit (Qiagen, Valencia, CA). qRT-PCR was performed with predesigned assays (Applied Biosystems, Foster City, CA) using on a 7900HT Fast Real-Time PCR System, TaqMan detection system, and validated primers (Applied Biosystems, Foster City, CA) in triplicate as described [16]. The mRNA expression was calculated based on Tata-box binding protein (TBP) expression for normalization using the comparative Ct method.
NanoString Gene Expression Profiling
The digital multiplexed NanoString nCounter mouse inflammation expression assay (NanoString Technologies) was performed with 100ng of total RNA according to the manufacturer’s instructions. RNA was isolated from CD19− and CD19+ fraction isolated from 3 female WT and Aldh1a1−/− mice. NanoString’s nCounter technology is based on direct detection of target molecules using color-coded molecular barcodes, providing a digital quantification of the number of target molecules [28]. Total mRNA (5µl) was hybridized overnight with nCounter Reporter (20µl) probes in hybridization buffer and nCounter Capture probes (5µl). The hybridizations were incubated at 65°C for 16–20h in excess of probes to ensure that each target finds a probe pair. Excess probes were removed using two-step magnetic bead based purification on the nCounter Prep Station. The hybridization mixture containing target/probe complexes was allowed to bind to magnetic beads containing complementary sequences on the Capture Probe and washed followed by a sequential binding to sequences on the Reporter Probe. Biotinylated capture probe-bound samples were immobilized and recovered on a streptavidin-coated cartridge. The abundance of specific target molecules was then quantified using the nCounter Digital Analyzer to count the individual fluorescent barcodes and assess target molecules present in each sample with a CCD camera. For each assay, a high-density scan (600 fields of view) was performed at the highest standard data resolution, 600 fields of view (FOV) that is the dynamic range and level of sensitivity in the system. Images were processed internally into a digital format and were normalized using the NanoString nSolver software analysis tool. Counts were normalized for all target RNAs in all samples based on the positive control RNA to account for differences in hybridization efficiency and post-hybridization processing, including purification and immobilization of complexes. Subsequently, a normalization of mRNA content was performed using six internal reference housekeeping genes that were included within the mouse inflammatory panel: Cltc, Gapdh, Gusb, Hprt1, Pgk1, and Tubb. The average was normalized by background counts for each sample obtained from the average of the eight negative control counts. Counts were corrected by subtracting the mean and 2 times standard deviation value of the negative control from the counts obtained for each target RNA.
Immunohistochemistry
Spleens and kidney were embedded in paraffin. Immunohistochemical analysis of spleens from WT and RARE-lacZ mice was performed with rabbit polyclonal β-galactosidase antibody (1:1000 dilution). Images were obtained using Olympus M081 IX50 and Pixera Viewfinder 3.0 software.
Transfections
U266B1 (U266) and RPMI8226 were purchased from American Type Culture Collection (Manassas, VA), other B cell lines were provided by Dr. D.M Benson, Jr. All human B cells were maintained in 15% fetal bovine serum/RPMI 1640 medium as previously described [29]. Human full length Aldh1a1 cDNA expression vector was purchased from OriGene (Rockville, MD). U266 cells (6 × 106 per tube) were transfected with human full length Aldh1a1 (PCMV6-XL5, Origene) or empty vector, using the Amaxa Cell Line Nucleofector kit C (Lonza, NJ). Transient transfections were performed in NIH-3T3 fibroblasts lacking Pparg expression using Fugene (Roche, South San Francisco, CA) and the following vectors: HoxA10 luciferase reporter vector (Switchgear Genomics, Menlo Park, CA), control Renilla reporter vector, murine full length Rara, Pparg, and Rxra constructs according to a previous protocol [16,20].
Statistical analysis
Oncomine cancer transcriptome database (https://www.oncomine.org) was used as a publicly available platform for data-mining in mRNA-expression studies [30]. Used the search terms ‘Aldh1a1 and multiple myeloma’ to identify relevant studies with sufficient sample numbers to compare expression of Aldh1a1 in multiple myeloma and plasma cells in the same dataset. One such dataset was identified [31]. All other data group comparisons were performed using Mann Whitney U-test unless otherwise indicated, and correlations were examined by Pearson’s test.
1.3 Results
Vitamin A metabolism regulates immature B cell populations in the spleen
The topography of RAR activation in mouse spleen was assessed in RARE-lacZ mice treated with and without RA (Fig.1 A). RARE activation in the spleen was heterogeneous and was predominant in the red pulp compared to lymphoid follicles in both non-treated and RA-treated samples.
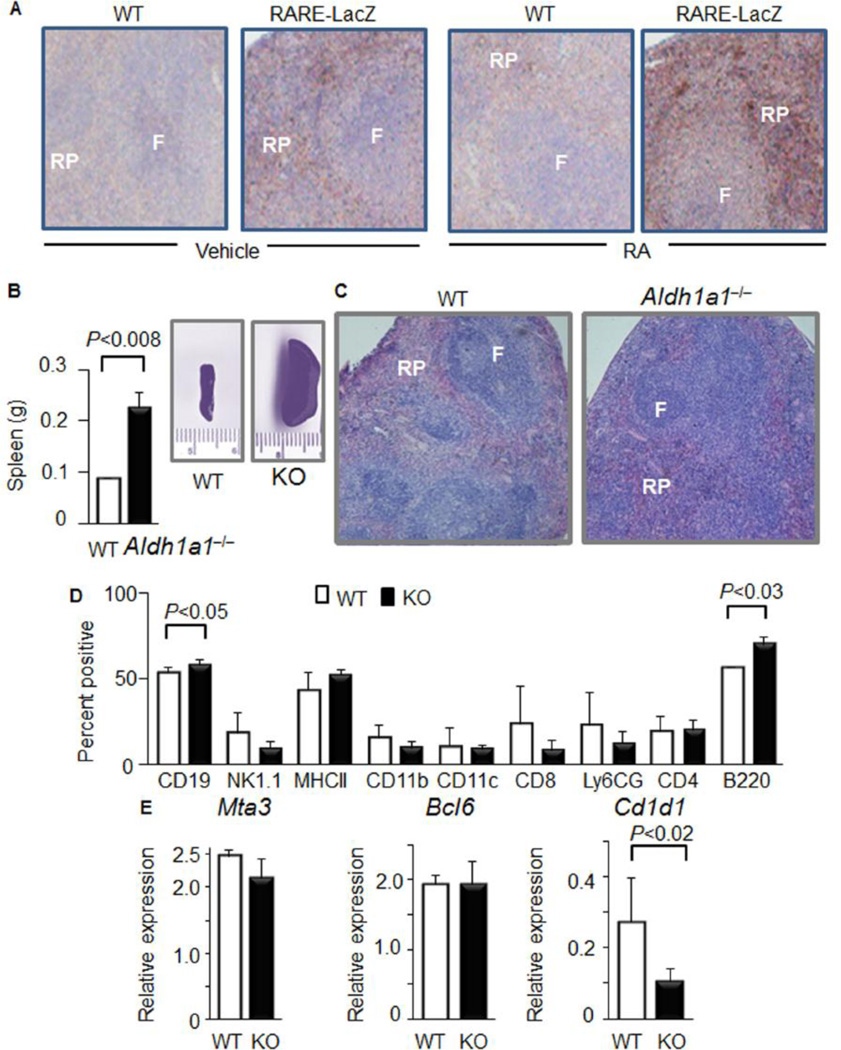
Figure 1. Activation of retinoic acid receptor response element (RARE) accompanies splenic red pulp development, which depends on Aldh1a1.
(A) RARE activation was studied in WT and RARE-LacZ mice (n=3 from each group) which were injected every 48h, up to a total of 3 injections with 1mL PBS (vehicle) without or with RA (500nM). RA was added into PBS from 500µM RA stock solution in ethanol immediately before injection. Vehicle PBS solution contained 1µL ethanol. All RA solutions were protected from light and stored under argon atmosphere. Total injected RA amount was 1.5nmol per mouse (0.15 microgram/dose). Immediately after the third injection, mice were harvested and their spleens were embedded in paraffin. Immunohistochemistry was performed with anti-β-galactosidase antibody. Heterogeneous brown β-galactosidase-positive areas were found in red pulp (RP) compared to follicular zone (F) (10× magnification). The RARE responses in adipose and hepatic tissues were described in [25].
(B) Weight of spleens (left panel, Study 2. Aldh1a1−/− (n=10) and WT (n=9)). Insert shows representative whole spleen images of WT and Aldh1a1−/− mice.
(C) Representative hematoxylin & eosin staining of paraffin embedded spleen section from WT and Aldh1a1−/− (KO) mice from the same study (n=3 per each group).
(D) FACS analysis of splenocytes suspension isolated from whole spleens of Aldh1a1−/− (n=4) and WT (n=4) mice. P, significance levels, Mann-Whithney U test.
(E) Expression of germinal center markers in the total spleen lysates isolated from WT (white bars) and Aldh1a1−/− mice was analyzed TaqMan assays (WT: n=3; Aldh1a1−/− n=5). Data were normalized by TBP. Significant difference was determined using Mann-Whitney U test
RA is produced by an ALDH1 family of enzymes ALDH1a1, ALDH1a2, and ALDH1a3. In Aldh1a1−/− mice, the spleen is enlarged (256%, Fig. 1B) compared to spleens in WT mice. Spleen architecture is altered in Aldh1a1−/− vs. WT mice (Fig. 1C) due to the increased proportion of CD19+ and B220+ B cell populations (Fig. 1D). Germinal center markers MTA3 and BCL6 [32] were expressed at similar levels in Aldh1a1−/− and WT spleens, indicating that they were not impaired by Aldh1a1 deficiency (Fig. 1E). The expression of Cd1d1 was 61% lower in Aldh1a1−/− than in WT splenocytes. The association of splenomegaly with the increase in CD19+ and B220+ B cell populations suggest that differentiation could be impaired in Aldh1a1−/− mice.
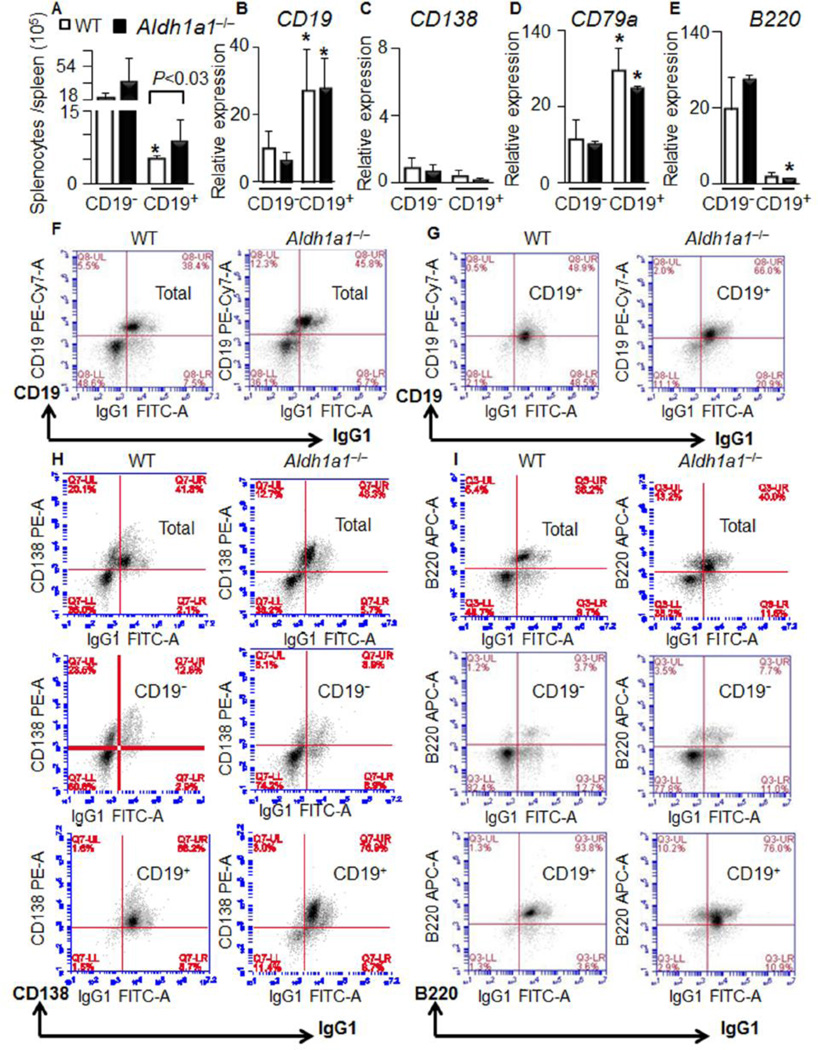
Differentiated CD19+ B cells are one of the major leukocyte populations in red pulp. Among them, the IgG1+ B cell population was sensitive to RA in pharmacological studies [3,4]. Therefore, we used magnetic cell separation technology to separate IgG1+/CD19− and IgG1+/CD19+ splenic B cell populations (Fig. 2) to test for effects of Aldh1a1 deficiency on transcriptional regulation of critical immune pathways in B cells. We termed IgG1+/CD19− as CD19− and IgG1+/CD19+ as CD19+ B cells throughout publication. The splenomegaly seen in Aldh1a1−/− mice (Fig. 1B) was associated with an increased number of CD19+ B cells (171%) compared to WT (Fig. 2A). The purity and characteristics of CD19+ B cell population was examined using CD19 expression (Fig. 2B) and FACS analysis (Fig. 2F–G). Although both cell populations expressed similar low levels of plasma cell marker CD138 (Fig. 2C), both CD19+ and CD19− B cell populations were CD138-positive in FACS analysis (Fig. 2H). CD19+ B cells also expressed a mature B cell marker CD79a (Fig. 2D). In contrast, an expression of pro-, mature and activated B cell marker B220 was lower in CD19+ than in CD19− B cells (Fig. 2E). Both groups were B220 positive in FACS analysis (Fig. 2I). Notably, the expression of all major studied B cell markers was similar in WT and Aldh1a1−/− mice. However, Aldh1a1 deficiency was associated with an increase in the CD19+ B cell population and splenomegaly.
Figure 2. Increased proportion of CD19+B cells contributes to splenomegaly in Aldh1a1−/− mice.
(A) CD19− and CD19+ B cells were isolated from whole spleens of WT and Aldh1a1−/− mice using automated magnetic cell separation and double selection with IgG1 and CD19 antibodies (n=5 per each group). Cell populations were quantified.
(B–E) Expression of CD19 (B) and plasma markers (CD138) (C) as well as differentiation (CD79a, D) and naive (B220, E) B cell markers were quantified in the isolated CD19− and CD19+ B cells using TaqMan assays (n=3 from each group). Data were normalized by TBP. Asterisk,- significant difference in expression between CD19− and CD19+ B cells of the same genetic background. Mann-Whitney U test.
(F–I) Representative immunostaining characteristics (from n=5 per each group) of total splenocytes (F) and isolated CD19− and CD19+ B cells (G) using FACS analysis. Total splenocytes and isolated CD19− and CD19+ B cells were simultaneously analyzed with CD19, CD138 (H), and B220 (I) antibodies conjugated with different secondary fluorescent antibodies. For all gates, the percent and total count of all cells staining positive for both antibodies was determined.
Dissimilar expression of Aldh1 in CD19− and CD19+ B cell populations
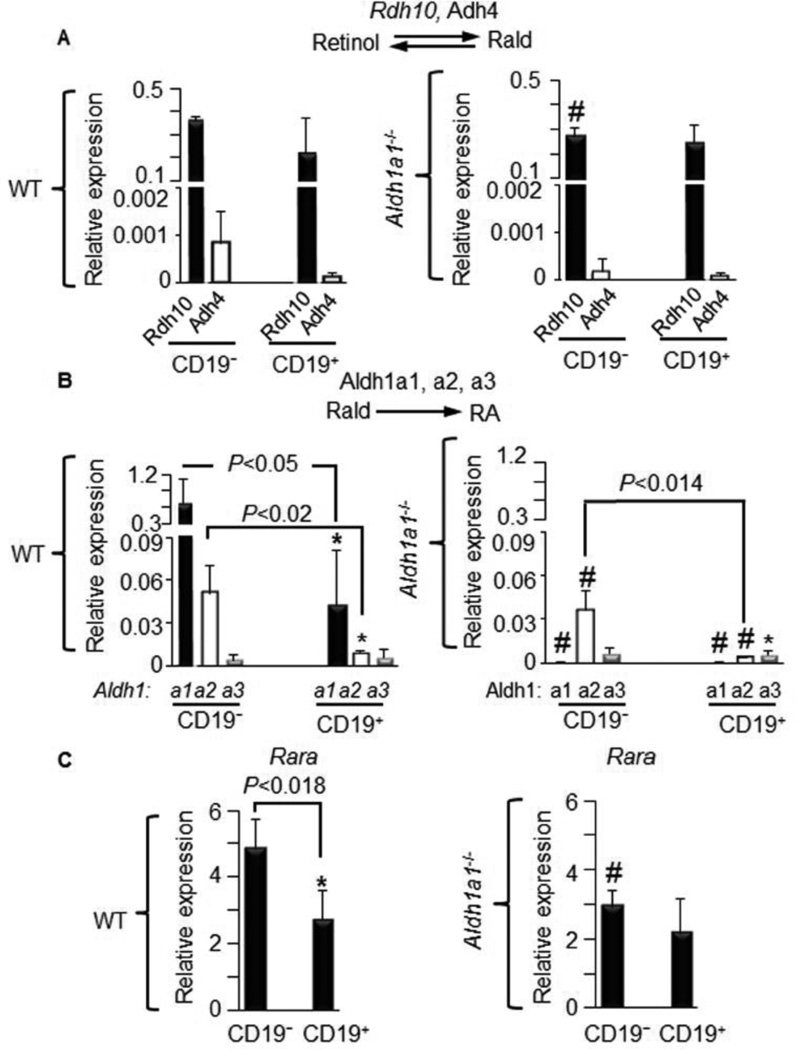
To investigate whether CD19− and CD19+ B cells metabolize vitamin A, we examined the expression of enzymes involved in synthesis of Rald (Fig. 3A) and RA (Fig. 3B). The expression of major Rald-generating enzymes (Rdh10, Adh4) was similar between CD19− and CD19+ B cells (Fig. 3A, left panel). Aldh1a1 deficiency moderately decreased Rdh10 levels (−24%, Fig. 3A, right panel). In contrast, expression of the RA-generating Aldh1a1 and Aldh1a2 enzymes was markedly reduced to 6.4% and 18%, respectively, in CD19+ compared to CD19− B cells (Fig. 3B left panel). Aldh1a1 was the predominantly expressed member of ALDH1 family of enzymes in both CD19− and CD19+ B cells (Fig. 3B, left panel). Aldh1a1 deficiency suppressed the expression of Aldh1a2 in CD19− and CD19+ B cells (Fig. 3B, right panel). Thus, CD19+ B cells in Aldh1a1−/− mice had reduced levels of all RA-producing enzymes. The change in Aldh1a1 expression also influenced expression of Rara, the primary transcription factor regulated by RA [18]. Rara was reduced to 44% in CD19+ vs. CD19− B cells in WT mice (Fig. 3C, left panel). In Aldh1a1−/− CD19− B cells, Rara expression was decreased to 60% compared to WT CD19− B cells and became similar to that seen in CD19+ B cells (Fig. 3C, right panel).
Figure 3. Diminished expression of RA-generating Aldh1 enzymes and Rara in B cells from Aldh1a1−/− vs. WT mice.
(A–C) CD19− and CD19+ B cells (same as in Fig. 2A, n=5 per each group) isolated from WT (left panels) and Aldh1a1−/− (right panels) mice were examined for the expression of (A) major retinaldehyde (Rald)-generating enzymes Rdh10 and Adh4, (B) RA-generating enzymes Aldh1a1, Aldh1a2, and Aldh1a3, and (C) Rara. Gene expression was analyzed by TaqMan assays and normalized with TBP. P, significant difference in expression between CD19− and CD19+ B cells. Mann-Whitney U test (throughout this Figure).
Aldh1a1 deficiency results in oncogene expression in CD19+ B cells
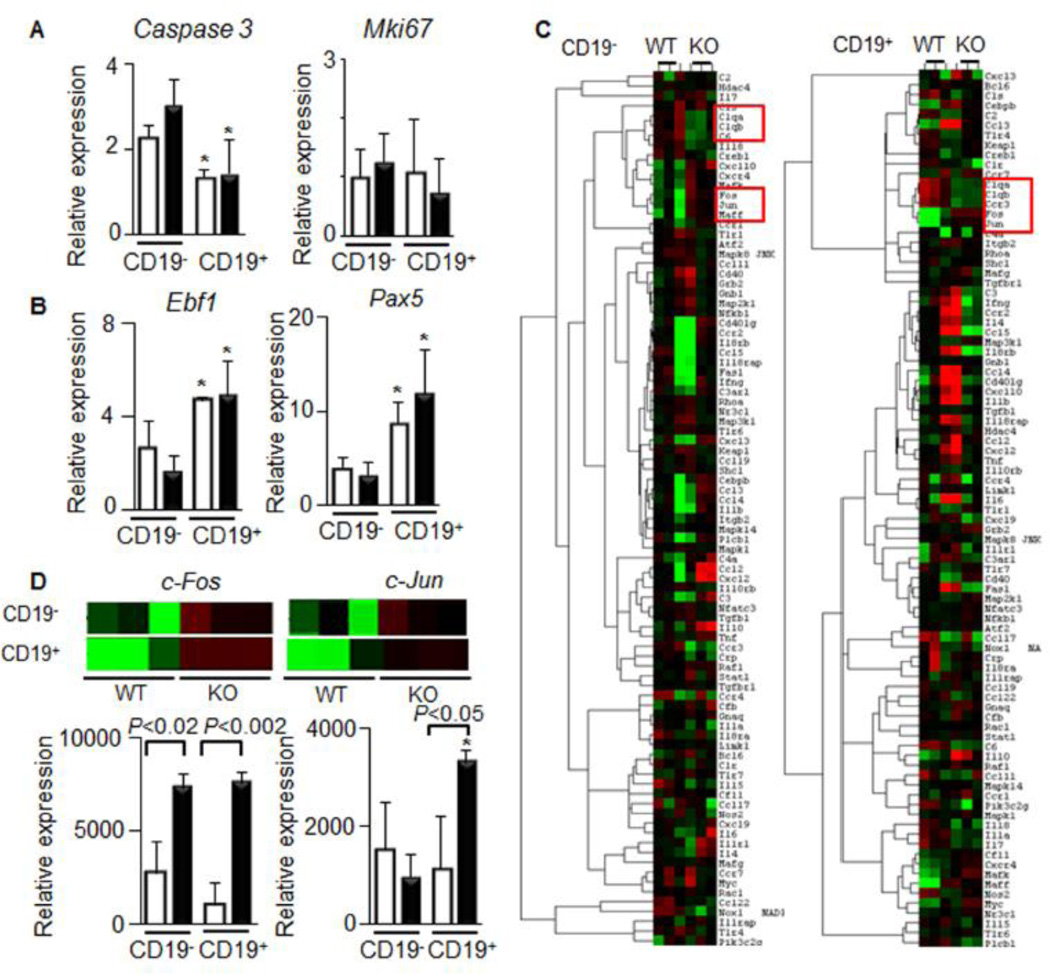
To identify mechanisms altering the number and properties of CD19+ B cells in Aldh1a1−/− mice, we analyzed classic markers of B cell apoptosis (caspase 3), proliferation (Mki67) (Fig. 4A), and differentiation (Ebf1, and Pax 5) in all groups (Fig. 4B). The expression of these genes was not altered between Aldh1a1−/− vs. WT genotype in the isolated CD19− and CD19+ B cells. Ebf1 and Pax5 expression were higher in differentiated CD19+ than in CD19− splenocytes; however, these levels were not influenced by WT and Aldh1a1−/− genotype. To identify genes increasing CD19+ B cells population in Aldh1a1−/− mice, we quantified expression of 250 inflammatory genes using nanoString Technologies’ nCounter System. Gene expression was normalized using six housekeeping genes. The gene cluster analysis revealed that Aldh1a1 influenced expression of proto-oncogenes (c-Fos, cJun, and Mafk kinase) and an opsonin (C1qb) (Fig. 4C). Aldh1a1−/− CD19− B cells expressed 267% higher levels of c-Fos than WT cells (Fig. 3D). These changes were in agreement with decreased expression of Rara, a known suppressor of c-Fos, in CD19− B cells (Fig. 3C) [33]. Aldh1a1 deficiency affected CD19+ B cells more than CD19− B cells. Specifically c-Fos, and c-Jun expression levels were 711% and 294% higher than those in WT cells (Fig. 4D). This finding was paradoxical because CD19− B cells expressed 15.5-times higher Aldh1a1 and 5.5-times higher Aldh1a2 levels than those seen in CD19+ B cells from WT mice (Fig. 3B). We hypothesized that another Aldh1a1-sensitive suppressor of c-Fos/c-Jun is active in CD19+ B cells.
Figure 4. Increased expression of proto-oncogenic genes in isolated splenic CD19− and CD19+ Aldh1a1−/− vs. WT B cells.
CD19− and CD19+ B cells were isolated from spleens of the same WT (white bars) and Aldh1a1−/− (black bars) group of mice (Fig. 2, n=5 per each group).
(A&B) Expression of apoptosis (Caspase3) and proliferation markers (Mki67) (A) as well as B cell differentiation markers (Ebf1 and Pax5) were semi-quantified using TaqMan assays. Data (n=3 per each group) were normalized by TBP. Asterisk, significant difference in expression between CD19− and CD19+ B cells of the same genetic background. Mann-Whitney U test.
(C) Selected expression heat maps (red and green colors represent high and low expression levels, respectively) obtained using nanoString Technologies’ nCounter mouse inflammation panel. The nCounter GX Mouse Inflammation Kit (NanoString Technologies) consists of 184 inflammation-related genes and six internal reference genes (www.nanostring.com). Red boxes showed statistically significant (n=3 per each group, P<0.05, Mann-Whitney U test) clusters of genes.
(D) Expression levels of c-Fos and c-Jun, using nanoString Technologies’ nCounter mouse inflammation panel Inserts show the extracted expression heat maps for c-Fos and c-Jun. P, significant difference between WT and Aldh1a1−/− B cells, Mann-Whitney U test (n=3 per each group).
Aldh1a1 limits oncogene expression CD19+ B cells by a sequential induction of Zfp423 and Pparg
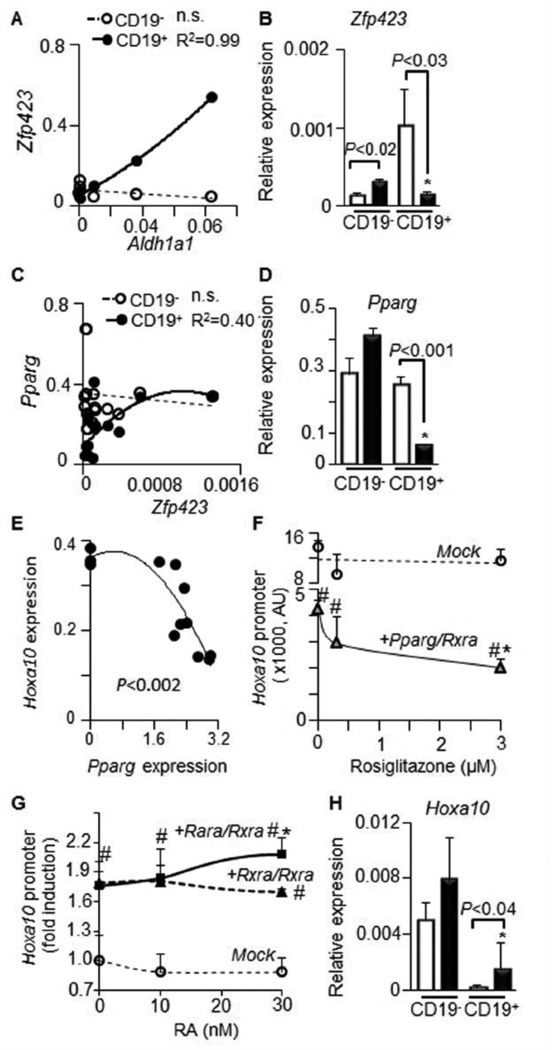
ALDH1 enzymes can induce the transcription factor Zfp423 which, in turn, controls expression of the anti-proliferative and anti-inflammatory transcription factor Pparg in adipocytes [20]. In splenic B cells, expression of Aldh1a1 positively correlated with Zfp423, specifically in CD19+ B cells (P<0.001) (Fig. 5A). Zfp423 was markedly up-regulated in CD19+ (625%) vs. CD19− WT B cells. However, this increase was abolished in Aldh1a1−/− CD19+ B cells (Fig. 5B), suggesting a regulatory role of Aldh1a1. The Zfp423 expression levels were also correlated with Pparg levels in CD19+ B cells (Fig. 5C). In agreement with Zfp423’s role in the induction of Pparg [20], only CD19+ B cells expressed less Pparg (−75%) in Aldh1a1−/− than WT splenocytes (Fig. 5D). PPAR response element (PPRE) has been identified in the promoter of transcription factor Hoxa10 [34], a key inducer of human lymphomyelopoiesis [35]. To examine a possible role for Pparg in the regulation of the Hoxa10 promoter, we performed transfection studies in NIH 3T3 fibroblasts, a cell line lacking endogenous Pparg expression. Forced expression of Pparg inhibited activation of Hoxa10 promoter in a gene dose-dependent manner (Fig. 5E). Pparg expression also markedly suppressed Hoxa10 promoter activation in a ligand (rosiglitazone)-dependent manner (Fig. 5F). In contrast, both Rxra and Rara only moderately activated Hoxa10 promoter reporter in the presence or absence of RA ligand (Fig. 5G). Consistent with a regulatory role of Pparg, CD19+ B cells expressed 400% higher levels of Hoxa10 in Aldh1a1−/− than in WT mice (Fig. 3H). Thus, elevated Hoxa10 expression in Aldh1a1−/− CD19+ B cells could be a direct effect of deficient Pparg expression. Other transcriptional mechanisms may also regulate Hoxa10 in CD19− B cells. Since elevated expression of proto-oncogenes AP1 (c-fos/c-jun) and Hoxa10 is involved in the development of multiple myeloma (MM), we examined the Aldh1a1 expression in B-cell related cancers.
Figure 5. Aldh1a1 influences expression of Zfp423 and Pparg genes in CD19+ B cells, suppressing promoter activity of Hoxa10.
(A&B). Correlation (A) between Aldh1a1 expression (Aldh1a1expression was shown in Fig.3 B) and expression levels of Zfp423 (B) in CD19− (white bars or circles) and CD19+ (black bars or circles) B cells. Gene expression was analyzed in triplicate by TaqMan assays (n=3 per each group).
(C&D) Correlation (C) between Zfp423 expression (B) and expression levels of Pparg (D) in CD19− (white bars or circles) and CD19+ (black bars or circles) B cells (n=6 in each correlation group). Gene expression was analyzed by TaqMan assays in triplicate. Asterisk, significant difference in expression between CD19− and CD19+ B cells of same genetic background; P, significant difference between WT and Aldh1a1−/− B cells, Mann-Whitney U test.
(E–G) Promoter analysis of Hoxa10 in NIH3-3T3 fibroblasts lacking Pparg (n=12). (E) NIH3-3T3 fibroblasts were transiently transfected with ful-length Pparg overexpression or empty (mock) vector (0). 48h after transfection the expression of Hoxa10 was measured by TagMan assay. P,-Pearson, correlation.
(F) Promoter analysis of Hoxa10 in NIH3-3T3 fibroblasts transiently transfected with mock or full-length Pparg overexpression vectors. 24h after transfection, cells were stimulated with different rosiglitazone concentrations for 15h (n=3 in each stimulated group). #, significantly different between cells expressing mock and Pparg vector; asterisk, significant difference between Pparg expressing cells stimulated with vehicle and rosiglitazone.
(G) Cell were transiently transfected with empty vector (Mock) or Rara and Rxra overexpression vectors (n=9). 24h after transfection, cells were stimulated with different retinoic acid (RA, n=3) concentrations for 27h. #, significantly different between cells expressing empty vector and Rara and/or Rxra; asterisk, significant difference between Rara/Rxra expressing cell stimulated with vehicle (ethanol/DMSO, 50/50%) and RA, Mann-Whitney U test.
(H) Expression levels of Hoxa10 in CD19− (white bars or circles) and CD19+ (black bars or circles) B cells measured by TaqMan assay (n=3 for each group). Significance was determined as in (D).
RA and/or Aldh1a1 rescues Rara and Zfp423 expression in myeloma B cell lines
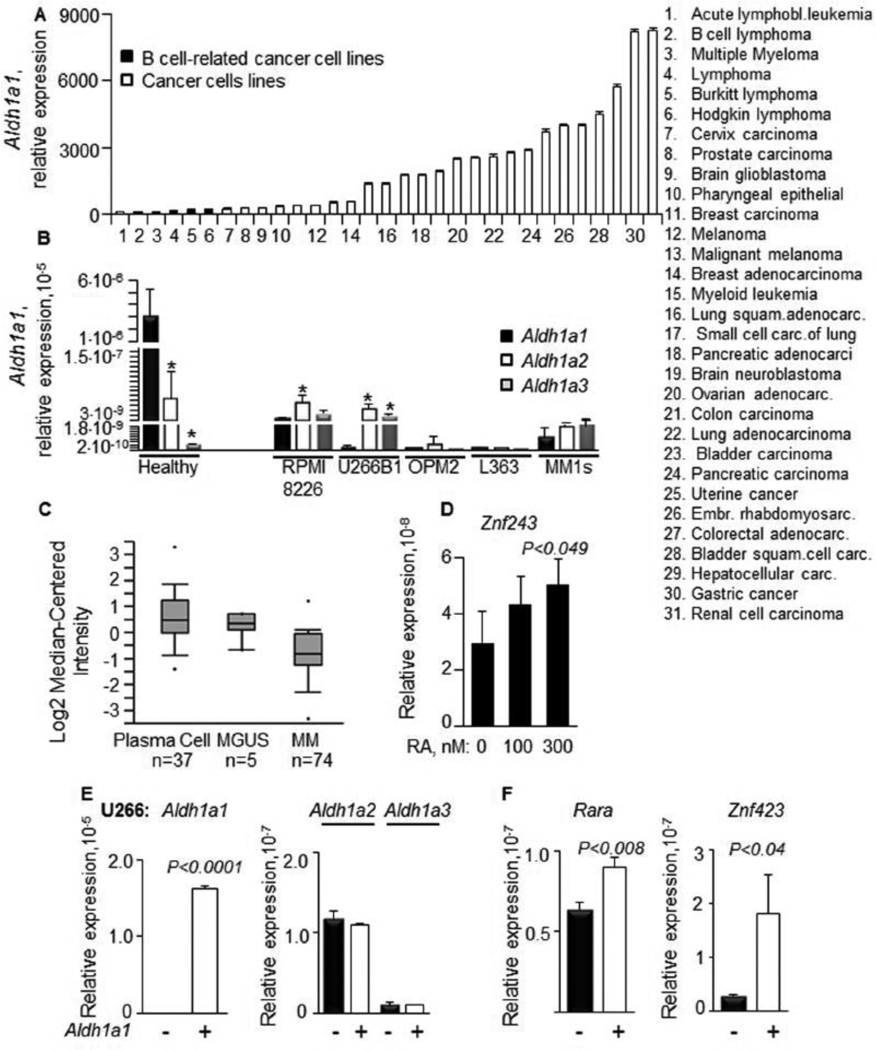
A search of publicly-available cancer gene expression data using Oncomine analysis revealed that B-cell-related cancer cell lines express low levels of Aldh1a1 compared to other cancer cell lines (Fig.6A). In our studies in peripheral blood mononuclear cells isolated from healthy donors (Fig.6B), Aldh1a1 was a predominantly expressed gene from the ALDH1 family. This pattern of expression was markedly changed in MM cells lines U266B1, RPMI8266, OPM2, L363, and MM1s (Fig. 6B). L363 MM cells expressed no Aldh1 genes. Aldh1a1 expression was lower compared to the expression of Aldh1a2 and Aldh1a3 in these MM cells. In agreement, the Oncomine analysis findings [31] showed reduced Aldh1a1 expression in 74 human MM patients, compared to healthy plasma cell controls (Fig. 6C). The RA stimulation of L363 MM cells increased expression of Znf423, a human analog of murine Zfp423 in MM cells (Fig. 6D). Aldh1a1 overexpression was even more effective in up-regulating suppressors of proto-oncogenes. The expression of a full-length human Aldh1a1 construct increased Aldh1a1 expression in U266 MM cells without altering expression of Aldh1a2 and Aldh1a3 (Fig. 6E). This overexpression of Aldh1a1 resulted in an increased expression of both Rara (30%) and Znf423 (500%) in U266 MM cells (Fig. 6F).
Figure 6. Impaired Aldh1a1 expression in human multiple myeloma B cell lines could be rescued by RA or Aldh1a1 overexpression that increased Rara and Znf423 levels.
(A) Relative expression of Aldh1a1 in hematological (n=3) and other cancer cell lines (n=28). Data were obtained from data mining in Oncomine database and based on the publication by Rhodes et al. [30].
(B) Expression levels of Aldh1 genes were measured in human PBMC cells (n=7 donors) and in myeloma cell lines (n=5): RPMI, U266B1, OPM2, L363, and MM1s (n=3 per a measurement). P, significant difference in expression between Aldh1a1, Aldh1a2, and Aldh1a3 enzymes within the same cell population.
(C) Analysis of Aldh1a1 gene expression in human multiple myeloma (n=74), plasma cells (n=37) and monoclonal gammopathy of undetermined significance (MGUS) (n=5). Gene expression database analysis (see Methods) identified the Zhan et al. 2002 [31] data shown here (adapted from Oncomine; Rhodes et al. 2007 [30]). The plot boxes are lined at lower, median and upper quartile score values; whiskers extend to 10th and 90th percentiles; dots mark minimum and maximum values.
(D) RA treatment of L363 myeloma cell lines increases Znf243 expression. L363 cells were maintained in RPMI medium containing 1% of UV treated FBS, which is depleted of retinoids. L363 cells were maintained in this medium 24h prior to RA stimulation and during treatment with RA. Znf243 expression was measured 48h after RA treatment (n=3). P, significant difference, Mann-Whitney U test.
(E) Expression levels of Aldh1a1 (left panel) and Aldh1a2 and Aldh1a3 (black and white bars in a right panel) in U266B1 cells transiently transfected with empty (−) or human full length Aldh1a1 overexpression plasmid (+) (n=3 independent experiments). Expression levels were measured in triplicate using TaqMan assays 24h after transfection. P, significant difference, Mann-Whitney U test.
(F) Expression of Rara (left panel) and Znf243 (human analog of mouse Znf423, right panel) in U266B1 (black bar) transfected with empty (−) and Aldh1a1 overexpression vector (+) (n=3 independent experiments). P, significant difference, Mann-Whitney U test.
1.4 Discussion
Humoral immune responses are mediated by mature follicular B cells with the help of T cells in splenic germinal centers and, to a minor extent (~10%), by marginal-zone B cells [2]. Cytokines/cytokine receptors, Ig recognition, and antigen presented by APC’s, dendritic cells, and/or macrophages can initiate differentiation of B cells and formation of germinal centers to achieve Ig production [11]. In these processes, dietary vitamin A or RA can facilitate differentiation by classic Pax5-dependent pathways in some splenic B cell population [11,12]. Our study revealed a key role for RA-generating ALDH1 enzymes in B cell biology. Aldh1a1 expression in immature CD19+ B cells and MM B cells is critical for the establishment of a transcriptional profile that prevents oncogene expression.
Aldh1a1 expression was consistently predominant over other RA-generating enzymes from the ALDH1 family in healthy B cells in mice and humans (Fig. 3B and 6B). Aldh1a1-dependent pathways in isolated B cell populations were different from those altered by administration of RA or manipulation of dietary vitamin A content. RA administration has two major sites of action related to B cell functions. It improves antigen presentation and IgA production at mucosal sites [15] and induces proliferation and differentiation of IgG1+ splenocytes in germinal centers [3,4,11]. In these scenarios, endogenous RA was produced by APC’s and stimulated final B cell differentiation in germinal centers [36]. We showed that under physiological conditions, intense endogenous RAR activity was associated with red pulp (Fig. 1A). In agreement, we found the highest Aldh1a1 and Rara expression levels in IgG1+CD19− (CD19−) B cell populations (Fig. 2, 3). CD19− is a potentially heterogeneous population comprised of naive and mature B220+ B cell populations. Both Rara and Aldh1 expression was decreased in the differentiated IgG1+CD19+ (CD19+) population. This loss of endogenous RA production in CD19+ B cells could later allow them to receive a paracrine signaling of RA-producing APC’s after they enter germinal centers. The physiological expression of Aldh1 genes in CD19+ vs. CD19− B cells in WT mice prevented increase in expression of oncogenes c-Fos/c-Jun, and Hoxa10 (Fig. 4D, Fig.5 H), due to the Aldh1a1-dependent induction of another oncogenes suppressors (Zfp423 and Pparg) in these cells (Fig. 5B,D). In contrast, the disruption of Aldh1a1 regulation in Aldh1a1−/− mice markedly compromised B cell oncogene profiles during differentiation (Fig. 4, 5), increased red pulp proportions, and reduced expression of Cd1d1 that is involved in interactions between T and B cells (Fig. 1D, right panel).
Classic CD19− and CD19+ B cell differentiation via Ebf1 and Pax5 [37] appear to be not impaired in the absence of Aldh1a1 (Fig. 4B). The breakthrough in the understanding of the mechanism increasing CD19+ population came from the NanoString analysis (Fig. 4C). Aldh1a1 deficient CD19+ B cells expressed proto-oncogenes c-Fos/c-Jun forming the transcription factor AP1. Elevated c-Fos/c-Jun and also Hoxa10 expression is a distinct feature of B cell-dependent hematological neoplasms, including lymphomas and multiple myeloma [38–40].
Hoxa10 overexpression in hematopoietic cells is sufficient to impair murine and human lymphomyelopoiesis and leads to acute myeloid leukemia [35,41]. Notably, all five human multiple myeloma (MM) cell lines expressed 100-times less Aldh1a1 than normal PBMC cells (Fig.6B). Similar findings in B cell cancers and in plasma cells from MM patients were available from other studies [31] that we identified through database analysis (Fig. 6A,C). The major RA-generating enzyme in these cancer cells was Aldh1a2 suggesting that the loss of Aldh1a1 contributed to B cell neoplastic pathology. Previous studies showed that only combined treatment of RA and rosiglitazone can induce U266 differentiation [14]. Our data provide a mechanism and rationale for the treatment of MM B cells lacking Aldh1a1 with RAR and PPARγ agonists.
Increased c-Fos expression in Aldh1a1−/− CD19− B cells appears to be consistent with the known competitive relation between AP1 and RARα activated by RA [33]. Indeed, Aldh1a1−/− CD19− B cells had reduced expression of both Aldh1a1 and Rara (Fig. 3). Forced expression of Aldh1a1 in U266 cells can readily increase Rara expression (Fig. 6). An unexpected result of our study was the finding of more profound transcriptional changes in CD19+ compared to CD19− B cells expressing markedly less Aldh1a1 in WT mice (Fig. 3B). CD19+ cells expressed higher levels of proto-oncogenes c-Fos, c-Jun, and Hoxa10 than CD19− B cells (c-Fos). Increased number of CD19+ cells contributed partially to the splenomegaly in Aldh1a1−/− mice. This phenomenon could be based on Aldh1a1-mediated changes on the transcriptome. During adipogenesis, Aldh1a1 induces expression of the transcription factor Zfp423, which in turn induces Pparg [20,42]. Previous investigations highlighted competition between Pparg and c-Fos/c-Jun without engaging PPRE [43,44]. PPRE response element was found in the Hoxa10 promoter [34]. Our studies connected Aldh1a1 to the regulation of all these transcription factors in B cells. Aldh1a1 did not support Zfp423 expression in CD19− B cells probably due to the specific transcriptional environment. It is possible that high Aldh1a1 levels in CD19− B cells produced RA for paracrine signaling that induced Zfp423 in CD19+ B cells. These regulatory mechanisms remain to be investigated in the future. We found that Aldh1a1 is required for the Zfp423 induction in differentiating CD19+ population (Fig. 5B). Zfp423 and Pparg levels were higher in CD19+ B cells in WT compared to Aldh1a1−/− mice. This link was suggested by a significant correlation in CD19+ B cells (Fig. 5C). The causative link between Aldh1a1 and Zfp423 expression was demonstrated in human U266 MM cells. Aldh1a1 expression rescued Znf423 (human analog of mouse Zfp423) in U266 MM cells (Fig. 6F). RA treatment of MM cells can also rescue Znf423 expression (Fig. 6D), suggesting that ALDH1a1 acts in part via autocrine RA generation. Since Pparg is induced by Znf423, the short (24h) transfection period was not sufficient to also observe significant increase in Pparg expression. However, the relationship between Zfp423 and Pparg has been widely documented [20,42]. Pparg induction by Aldh1a1 appears to be a critical event, because Pparg was an effective suppressor of Hoxa10, a key transcription factor perturbing myeloid and lymphoid differentiation in mice and humans [34,35,41]. Pparg inhibited the promoter of Hoxa10, while Rara and Rxra only modestly regulated this transcription factor (Fig. 5 F,G). Consequently, Hoxa10 was up-regulated in Aldh1a1−/− CD19+ B cells expressing less Pparg. Studies investigating effects of vitamin A-deficient diets reported splenomegaly and an increase in plasma IgG1 levels in mouse models of autoimmune disorders [45], whereas the production of specific IgG1 antibodies in the immunized mice was impaired [46]. Multiple mechanisms have been proposed to explain these phenomena, including RA-dependent production of IFNγ from T cells [47] and dendritic CD103+ cell subsets [48], but the role of B cells was unclear. Our data highlight an autocrine and/or paracrine ALDH1 function in differentiating B cells that regulates two key transcriptional oncogene suppressors Rara and Pparg. This suggests a gene-environment paradigm for early-stage deregulation of oncogene profiles in IgG1+ B cell subsets through compromised vitamin A metabolism.
1.5 Conclusion
Our findings showed the critical role of the retinoic acid-generating ALDH1a1 enzyme in the sequential induction of oncogene suppressors Rara in IgG1+/CD19− B cells and Zfp423/Pparg in IgG1+/CD19+ B cells during B cell differentiation. In the absence of these suppressors, B cells acquire oncogene Ap1 and Hoxa10 expression that leads to IgG1+/CD19+ B cell expansion and splenomegaly. Reduced expression of Aldh1a1 and oncogene suppressors Rara, Zfp423, and Pparg is a characteristic property of malignant human multiple myeloma B cells. Importantly, ectopic expression of Aldh1a1 or RA effectively rescues Rara and/or Zfp423 expression. The understanding of the role of ALDH1a1 in B cell differentiation can shed light on the early stages in the development of malignant hematological disorders and may lead to the development of novel therapeutics.
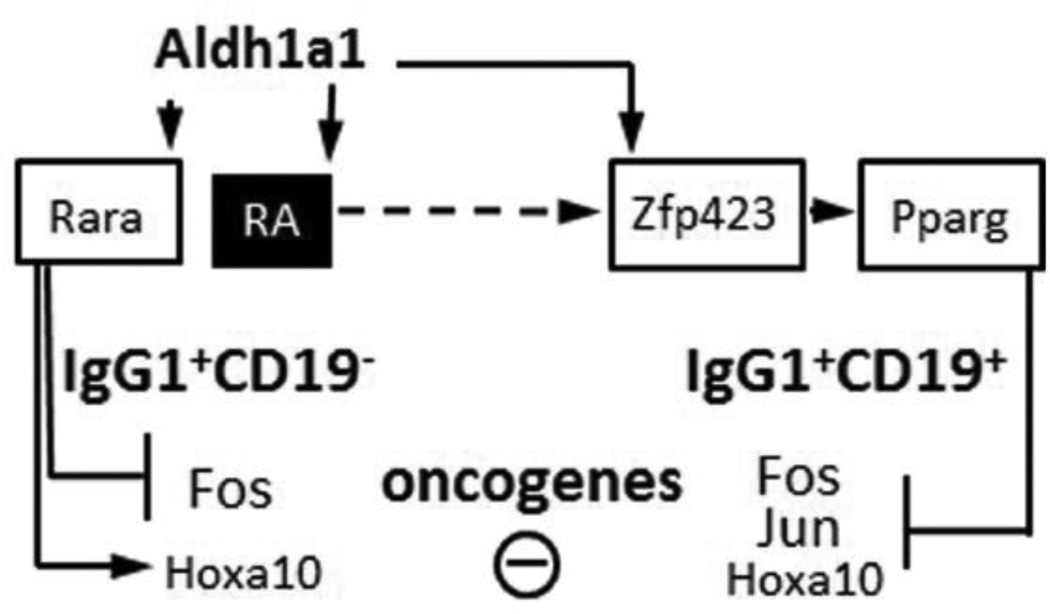
Figure 7.
Schematic diagram of the Aldh1a1 dependent pathways in CD19− and CD19+ IgG1+ B cell population in the spleen. Aldh1a1 is expressed at higher levels in CD19− vs. CD19+ B cells. Aldh1a1 also shapes the expression of transcription factors in CD19+ population, where it is responsible for the induction of Zfp423 and Pparg. Expression of Pparg suppressed proto-oncogenes c-Fos, c-Jun, and Hoxa10.
Highlights.
Aldh1a1 up-regulates oncogene suppressors Rar and Zfp423/Pparg in B cell populations
Aldh1a1−/− B cells acquire oncogene Ap1 and Hoxa10 expression causing splenomegaly
PPARγ suppresses promoter of multiple myeloma oncogene Hoxa1
Aldh1a1 rescues oncogene suppressors in human multiple myeloma cells
Acknowledgements
We thank Dr. C. Ross (Pennsylvania State University) for the helpful discussion. We express our gratitude to Kirsteen Maclean (NanoString Technologies) and the Nucleic Acid Shared Resource at The Ohio State University (OSU) for excellent technical and intellectual support.
Funding: We thank for helpful discussions. This research was supported by Food Innovation Center, Office for International Affairs and Center for Advanced Functional Foods Research and Entrepreneurship at OSU, Daskal Foundation (O.Z., R.Y.), and Mulitple Myeloma Opportunities for Research and Education (MMORE, DMB Jr). The project described was supported by Award Number Grant UL1TR000090 from the National Center For Advancing Translational Sciences and the Cancer Center Support Grant (CA016058) (O.Z). This study was also supported in part by a research Grant (CA100865) from the Department of Defense Congressionally Directed Medical Research Programs (C.E.A.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
Abbreviations
- Aldh1a1
aldehyde dehydrogenase 1a1
- AP1
Activator protein 1 a heterodimeric transcription factor formed by c-jun and c-fos
- c-Fos
transcription factor encoded by the FOS gene
- c-Jun
protein encoded by c-Jun gene
- Hoxa10
transcription factor homeobox protein a10
- Ig
immunoglobulin
- Pparg
peroxisome proliferator-activated receptor
- RA
retinoic acid
- Rara
retinoic acid receptor alpha
- RARE
retinoic acid receptor response element
- Zfp423
murine zinc finger protein
- Znf 423
human zinc finger protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dunn-Walters DK, Ademokun AA. B cell repertoire and ageing. Curr Opin Immunol. 2010;22:514–520. doi: 10.1016/j.coi.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Matthias P, Rolink AG. Transcriptional networks in developing and mature B cells. Nat Rev Immunol. 2005;5:497–508. doi: 10.1038/nri1633. [DOI] [PubMed] [Google Scholar]
- 3.Chen Q, Ross AC. Inaugural Article: Vitamin A and immune function: retinoic acid modulates population dynamics in antigen receptor and CD38-stimulated splenic B cells. Proc Natl Acad Sci U S A. 2005;102:14142–14149. doi: 10.1073/pnas.0505018102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Q, Ross AC. Retinoic acid promotes mouse splenic B cell surface IgG expression and maturation stimulated by CD40 and IL-4. Cell Immunol. 2007;249:37–45. doi: 10.1016/j.cellimm.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phillips-Quagliata JM, Faria AM, Han J, Spencer DH, Haughton G, Casali P. IgG2a and igA co-expression by the natural autoantibody-producing murine B lymphoma T560. Autoimmunity. 2001;33:181–197. doi: 10.3109/08916930109008046. [DOI] [PubMed] [Google Scholar]
- 6.Kim CH. Roles of retinoic acid in induction of immunity and immune tolerance. Endocr Metab Immune Disord Drug Targets. 2008;8:289–294. doi: 10.2174/187153008786848312. [DOI] [PubMed] [Google Scholar]
- 7.Amouzou A, Habi O, Bensaid K. Reduction in child mortality in Niger: a Countdown to 2015 country case study. Lancet. 2012;380:1169–1178. doi: 10.1016/S0140-6736(12)61376-2. [DOI] [PubMed] [Google Scholar]
- 8.NMWR Morb Mortal Wkly Rep. 2007;56:1237–1241. [PubMed] [Google Scholar]
- 9.Hussey GD, Klein M. A randomized, controlled trial of vitamin A in children with severe measles. N Engl J Med. 1990;323:160–164. doi: 10.1056/NEJM199007193230304. [DOI] [PubMed] [Google Scholar]
- 10.Ertesvag A, Naderi S, Blomhoff HK. Regulation of B cell proliferation and differentiation by retinoic acid. Semin Immunol. 2009;21:36–41. doi: 10.1016/j.smim.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 11.Ross AC, Chen Q, Ma Y. Vitamin A and retinoic acid in the regulation of B-cell development and antibody production. Vitam Horm. 2011;86:103–126. doi: 10.1016/B978-0-12-386960-9.00005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Esplin BL, Garrett KP, Welner RS, Webb CF, Kincade PW. Retinoids accelerate B lineage lymphoid differentiation. J Immunol. 2008;180:138–145. doi: 10.4049/jimmunol.180.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donjerkovic D, Mueller CM, Scott DW. Steroid- and retinoid-mediated growth arrest and apoptosis in WEHI-231 cells: role of NF-kappaB, c-Myc and CKI p27(Kip1) Eur J Immunol. 2000;30:1154–1161. doi: 10.1002/(SICI)1521-4141(200004)30:4<1154::AID-IMMU1154>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 14.Huang H, Wu D, Fu J, Chen G, Chang W, Chow HC, Leung AY, Liang R. All-trans retinoic acid can intensify the growth inhibition and differentiation induction effect of rosiglitazone on multiple myeloma cells. Eur J Haematol. 2009;83:191–202. doi: 10.1111/j.1600-0609.2009.01277.x. [DOI] [PubMed] [Google Scholar]
- 15.Jorgensen MJ, Fisker AB, Sartono E, Andersen A, Erikstrup C, Lisse IM, Yazdanbakhsh M, Aaby P, Benn CS. The effect of at-birth vitamin A supplementation on differential leucocyte counts and in vitro cytokine production: an immunological study nested within a randomised trial in Guinea-Bissau. Br J Nutr. 2012:1–11. doi: 10.1017/S0007114512001304. [DOI] [PubMed] [Google Scholar]
- 16.Yasmeen R, Jeyakumar SM, Reichert B, Yang F, Ziouzenkova O. The contribution of vitamin A to autocrine regulation of fat depots. Biochim Biophys Acta. 2012;1821:190–197. doi: 10.1016/j.bbalip.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Germain P, Chambon P, Eichele G, Evans RM, Lazar MA, Leid M, De Lera AR, Lotan R, Mangelsdorf DJ, Gronemeyer H. International Union of Pharmacology. LX. Retinoic acid receptors. Pharmacol Rev. 2006;58:712–725. doi: 10.1124/pr.58.4.4. [DOI] [PubMed] [Google Scholar]
- 19.Ziouzenkova O. Vitamin A metabolism: challenges and perspectives. Vitamins & Minerals. Element. 2012;1:E106. [Google Scholar]
- 20.Reichert B, Yasmeen R, Jeyakumar SM, Yang F, Thomou T, et al. Concerted action of aldehyde dehydrogenases influences depot-specific fat formation. Mol Endocrinol. 2011;25:799–809. doi: 10.1210/me.2010-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duriancik DM, Hoag KA. Vitamin A deficiency alters splenic dendritic cell subsets and increases CD8(+)Gr-1(+) memory T lymphocytes in C57BL/6J mice. Cell Immunol. 2010;265:156–163. doi: 10.1016/j.cellimm.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mora JR, Iwata M, Eksteen B, Song SY, Junt T, et al. Generation of gut-homing IgA-secreting B cells by intestinal dendritic cells. Science. 2006;314:1157–1160. doi: 10.1126/science.1132742. [DOI] [PubMed] [Google Scholar]
- 23.Rossant J, Zirngibl R, Cado D, Shago M, Giguere V. Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 1991;5:1333–1344. doi: 10.1101/gad.5.8.1333. [DOI] [PubMed] [Google Scholar]
- 24.Fan X, Molotkov A, Manabe S, Donmoyer CM, Deltour L, Foglio MH, Cuenca AE, Blaner WS, Lipton SA, Duester G. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol Cell Biol. 2003;23:4637–4648. doi: 10.1128/MCB.23.13.4637-4648.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yasmeen R, Reichert B, Deiuliis J, Yang F, Lynch A, et al. Autocrine function of aldehyde dehydrogenase 1 as a determinant of diet- and sex-specific differences in visceral adiposity. Diabetes. 2013;62:124–136. doi: 10.2337/db11-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziouzenkova O, Orasanu G, Sharlach M, Akiyama TE, Berger JP, et al. Retinaldehyde represses adipogenesis and diet-induced obesity. Nat Med. 2007;13:695–702. doi: 10.1038/nm1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherger M, Kisseberth W, London C, Olivo-Marston S, Papenfuss TL. Identification of myeloid derived suppressor cells in the peripheral blood of tumor bearing dogs. BMC Vet Res. 2012;8:209. doi: 10.1186/1746-6148-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 29.Dai Y, Landowski TH, Rosen ST, Dent P, Grant S. Combined treatment with the checkpoint abrogator UCN-01 and MEK1/2 inhibitors potently induces apoptosis in drug-sensitive and -resistant myeloma cells through an IL-6-independent mechanism. Blood. 2002;100:3333–3343. doi: 10.1182/blood-2002-03-0940. [DOI] [PubMed] [Google Scholar]
- 30.Rhodes DR, Kalyana-Sundaram S, Tomlins SA, Mahavisno V, Kasper N, Varambally R, Barrette TR, Ghosh D, Varambally S, Chinnaiyan AM. Molecular concepts analysis links tumors, pathways, mechanisms, and drugs. Neoplasia. 2007;9:443–454. doi: 10.1593/neo.07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhan F, Hardin J, Kordsmeier B, Bumm K, Zheng M, et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood. 2002;99:1745–1757. doi: 10.1182/blood.v99.5.1745. [DOI] [PubMed] [Google Scholar]
- 32.Okada T, Moriyama S, Kitano M. Differentiation of germinal center B cells and follicular helper T cells as viewed by tracking Bcl6 expression dynamics. Immunol Rev. 2012;247:120–132. doi: 10.1111/j.1600-065X.2012.01120.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhou XF, Shen XQ, Shemshedini L. Ligand-activated retinoic acid receptor inhibits AP-1 transactivation by disrupting c-Jun/c-Fos dimerization. Mol Endocrinol. 1999;13:276–285. doi: 10.1210/mend.13.2.0237. [DOI] [PubMed] [Google Scholar]
- 34.Gosiengfiao Y, Horvat R, Thompson A. Transcription factors GATA-1 and Fli-1 regulate human HOXA10 expression in megakaryocytic cells. DNA Cell Biol. 2007;26:577–587. doi: 10.1089/dna.2007.0575. [DOI] [PubMed] [Google Scholar]
- 35.Buske C, Feuring-Buske M, Antonchuk J, Rosten P, Hogge DE, Eaves CJ, Humphries RK. Overexpression of HOXA10 perturbs human lymphomyelopoiesis in vitro and in vivo. Blood. 2001;97:2286–2292. doi: 10.1182/blood.v97.8.2286. [DOI] [PubMed] [Google Scholar]
- 36.Iwata M. Retinoic acid production by intestinal dendritic cells and its role in T-cell trafficking. Semin Immunol. 2009;21:8–13. doi: 10.1016/j.smim.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanaitsuka T, Namba Y, Zu YL, Ishii K, Ashihara T, Hanaoka M, Suchi T. Detection of fos oncogene products by monoclonal antibody FO-120 in lymphoproliferative disorders. Leuk Res. 1989;13:1025–1033. doi: 10.1016/0145-2126(89)90010-6. [DOI] [PubMed] [Google Scholar]
- 39.Troen G, Nygaard V, Jenssen TK, Ikonomou IM, Tierens A, et al. Constitutive expression of the AP-1 transcription factors c-jun, junD, junB, and c-fos and the marginal zone B-cell transcription factor Notch2 in splenic marginal zone lymphoma. J Mol Diagn. 2004;6:297–307. doi: 10.1016/S1525-1578(10)60525-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Annunziata CM, Hernandez L, Davis RE, Zingone A, Lamy L, Lam LT, Hurt EM, Shaffer AL, Kuehl WM, Staudt LM. A mechanistic rationale for MEK inhibitor therapy in myeloma based on blockade of MAF oncogene expression. Blood. 2011;117:2396–2404. doi: 10.1182/blood-2010-04-278788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thorsteinsdottir U, Sauvageau G, Hough MR, Dragowska W, Lansdorp PM, Lawrence HJ, Largman C, Humphries RK. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol Cell Biol. 1997;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupta RK, Arany Z, Seale P, Mepani RJ, Ye L, Conroe HM, Roby YA, Kulaga H, Reed RR, Spiegelman BM. Transcriptional control of preadipocyte determination by Zfp423. Nature. 2010;464:619–623. doi: 10.1038/nature08816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Francois M, Richette P, Tsagris L, Raymondjean M, Fulchignoni-Lataud MC, Forest C, Savouret JF, Corvol MT. Peroxisome proliferator-activated receptor-gamma down-regulates chondrocyte matrix metalloproteinase-1 via a novel composite element. J Biol Chem. 2004;279:28411–28418. doi: 10.1074/jbc.M312708200. [DOI] [PubMed] [Google Scholar]
- 44.Delerive P, Martin-Nizard F, Chinetti G, Trottein F, Fruchart JC, Najib J, Duriez P, Staels B. Peroxisome proliferator-activated receptor activators inhibit thrombin-induced endothelin-1 production in human vascular endothelial cells by inhibiting the activator protein-1 signaling pathway. Circ Res. 1999;85:394–402. doi: 10.1161/01.res.85.5.394. [DOI] [PubMed] [Google Scholar]
- 45.Gershwin ME, Lentz DR, Beach RS, Hurley LS. Nutritional factors and autoimmunity. IV. Dietary vitamin A deprivation induces a selective increase in IgM autoantibodies and hypergammaglobulinemia in New Zealand Black mice. J Immunol. 1984;133:222–226. [PubMed] [Google Scholar]
- 46.Smith SM, Hayes CE. Contrasting impairments in IgM and IgG responses of vitamin A-deficient mice. Proc Natl Acad Sci U S A. 1987;84:5878–5882. doi: 10.1073/pnas.84.16.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carman JA, Hayes CE. Abnormal regulation of IFN-gamma secretion in vitamin A deficiency. J Immunol. 1991;147:1247–1252. [PubMed] [Google Scholar]
- 48.Chang JH, Cha HR, Chang SY, Ko HJ, Seo SU, Kweon MN. IFN-gamma secreted by CD103+ dendritic cells leads to IgG generation in the mesenteric lymph node in the absence of vitamin A. J Immunol. 2011;186:6999–7005. doi: 10.4049/jimmunol.1003484. [DOI] [PubMed] [Google Scholar]