Abstract
In migratory birds, traits such as orientation and distance are known to have a strong genetic background, and they often exhibit considerable within-population variation. How this variation relates to evolutionary responses to ongoing selection is unknown because the underlying mechanisms that translate environmental changes into population genetic changes are unclear. We show that within-population genetic structure in southern German blackcaps (Sylvia atricapilla) is related to individual differences in migratory behavior. Our 3-year study revealed a positive correlation between individual migratory origins, denoted via isotope (δ2H) values, and genetic distances. Genetic diversity and admixture differed not only across a recently established migratory polymorphism with NW- and SW-migrating birds but also across δ2H clusters within the same migratory route. Our results suggest assortment based on individual migratory origins which would facilitate evolutionary responses. We scrutinized arrival times and microhabitat choice as potential mechanisms mediating between individual variation in migratory behavior and assortment. We found significant support that microhabitat choice, rather than timing of arrival, is associated with individual variation in migratory origins. Moreover, examining genetic diversity across the migratory divide, we found migrants following the NW route to be genetically more distinct from each other compared with migrants following the traditional SW route. Our study suggests that migratory behavior shapes population genetic structure in blackcaps not only across the migratory divide but also on an individual level independent of the divide. Thus, within-population variation in migratory behavior might play an important role in translating environmental change into genetic change.
Keywords: Blackcap, bottleneck, micro-evolution, microhabitat choice, migratory connectivity, migratory divide, spatial isolation, stable isotopes
Introduction
In several migratory species, hybrid zones between recently diverged taxa are located on migratory divides, which are areas where populations with different migratory behavior meet (Bensch et al. 2009; Brelsford and Irwin 2009; Ruegg et al. 2012). Thus, migratory behavior has been considered an important factor influencing assortative mating and, consequently, population divergence and even speciation (Price 2007; Liedvogel et al. 2011). However, mechanisms how migratory behavior can lead to divergence are not well understood, mainly because the genetic structure of migratory populations is typically unknown. It is thus important to link within-population variation in migratory traits with population genetic structure.
In particular, it has been hypothesized that migratory connectivity (i.e., the linkage between breeding, stopover, and wintering areas) can affect the genetic structure of populations and the ability of migratory species to evolve in response to changing selective pressures (Webster et al. 2002). For example, if individuals from a breeding population overwinter in different regions, there will be substantial genetic variation for migratory traits (such as distance and orientation) in that population. Nonrandom aggregation on the breeding grounds based on similarities in individual wintering origins would then facilitate ongoing population divergence, whereas random structuring would prevent divergence (Helbig 1996; Pulido et al. 1996). While previous studies analyzed whether migratory behavior contributed to population divergence (Bensch et al. 2009; Rolshausen et al. 2009), to our knowledge, no study has attempted to ask how individual migratory routes, and distances contribute to within-population structuring.
In this study, we examine whether individual variation in wintering origins predicts population genetic structure of sympatric blackcaps (Sylvia atricapilla) in southern Germany. Contingent upon our findings, we then analyse whether the observed genetic structure of the population is explicable by temporal and spatial isolation in arrival times and microhabitat choice of territorial blackcaps. Earlier work showed that migratory orientation and distance in blackcaps, a species that migrates alone, are not learned but have a strong genetic component (Berthold 2000). Furthermore, rapid micro-evolutionary changes of its migratory phenology have been observed (Berthold et al. 1992; Pulido and Berthold 2010). In particular, the recently established migratory polymorphism in southern German blackcaps (Berthold et al. 1992) is known to facilitate reproductive isolation and drive population divergence in sympatry (Bearhop et al. 2005; Rolshausen et al. 2009). Thus, because it is known that individual differences in migratory traits influence mating behavior (Bearhop et al. 2005), we do not focus on this topic but rather specifically ask how within-population variation in individual claw tip stable isotope (δ2H) values, a proxy for the birds' wintering area (Bearhop et al. 2005), relates to within-population genetic structuring, arrival times, and microhabitat choice. We further scrutinize within-population variation in δ2H and microsatellites to ask how the migratory polymorphism in our study population affects genetic variability on either side of the migratory divide. Uncovering this link between individual migratory behavior and population genetic structure will help the evaluation of the potential for micro-evolutionary responses to contemporary changes in selective regimes, for instance advancing spring phenology due to climate change.
Previous studies used stable-hydrogen isotope values (δ2H) to directionally assign blackcaps to either one of the two main wintering origins of southern German blackcaps. Those origins were northwestern (NW) migrants wintering in Great Britain or south-western (SW) migrants wintering in the Mediterranean (Bearhop et al. 2005; Rolshausen et al. 2009). Assignments were based on the fact that δ2H values of foodwebs determining blackcap claw isotope values were driven primarily by latitudinal gradients in amount-weighted long-term patterns of average δ2H in precipitation for Europe (Bowen et al. 2005). On average, more southern wintering populations were expected to arrive on German breeding grounds with higher δ2H values than more northern populations such as those wintering in the U.K. (Bearhop et al. 2005). As these probability assignments were a priori referenced with tissue δ2H distributions from prespecified wintering grounds, they necessarily resulted in a dichotomous “either-or” outcome and therefore discounted individual variation in wintering origins within each of the regions. However, the wintering range of blackcaps is large and ringing recoveries reveal that it is continuous rather than dichotomous covering substantial areas throughout Western Europe, including Belgium, northern Germany, France, southern and northern Spain (Snow and Perrins 1997; Mokwa 2009). Furthermore, contemporary shifts in wintering ranges and a reduction in migratory activity in response to changing climate are well documented for central European migrants, including the blackcap (Fiedler 2003). In a recent study, Pulido and Berthold (2010) showed that migratory activity within a south-west migrating population of southern German blackcaps decreased significantly over 14 years arguing that these changes presumably involve a micro-evolutionary response to climate-induced directional selection for shorter migration routes favoring earlier arrival and earlier breeding.
We suggest that such particularly rapid genetic adjustment would be facilitated by a link between individual wintering origin and genetic structuring within breeding populations mediated by nonrandom pairing according to individual migratory strategies. This hypothesis has not yet been investigated.
To examine the hypothesis that the population genetic structure of southern German blackcaps is affected by differences in individual wintering origins within a population, we used δ2H values in the birds' claw tips as a continuous proxy for wintering area. While the pattern of δ2H values in keratin of songbirds in Europe is expected to primarily reflect a northeast–southwest axis for origins of individuals (Bowen et al. 2005; Hobson 2011), populations differing in δ2H values can be assumed to derive from different, albeit unknown, wintering origins (Hobson 2005; Studds et al. 2012). Also, within-population variance in δ2H values among songbirds from the same location is of the order of 9–12‰ (Hobson et al. 2012). With these caveats in mind, we used blackcap tissue δ2H values as a conservative means of exploring linkages between genetic structure and potentially different wintering origins. We considered detectable trends in any relationship between blackcap tissue δ2H values and population genetic structure as strong evidence linking isolation on wintering grounds to nonrandom mating on sympatric breeding grounds.
Unlike previous studies (Bearhop et al. 2005; Rolshausen et al. 2009, 2010), we do not use a priori information to assign individuals to prespecified wintering origins but calculate pairwise similarities in δ2H values across the whole population (to account for a continuous wintering range) and perform an uninformed cluster analysis to portion individuals based on δ2H similarities (δ2H clusters). We then partitioned the overall genetic diversity in our dataset on the basis of variation in blackcap tissue δ2H values. Assuming that individual differences in migratory behavior contribute to population genetic structuring, we predicted (1) pairwise genetic distances to increase with pairwise geographical distances between wintering areas and (2) pairwise genetic distances within δ2H clusters to be lower than the mean pairwise genetic distance across the whole study population. We (3) examined whether spatio-temporal isolation of blackcaps contributed to the observed genetic structure of sympatric blackcaps. To our knowledge, this is the first study that investigates population genetic structure in relation to migratory behavior on an individual level.
Methods
Field procedures and sampling
We caught blackcaps with mist nets upon their spring arrival on southern German breeding grounds in Radolfzell (47°45′N 08°59′E) in 2006 and in Freiburg (48°00′N 07°51′E) in 2007 and 2008. The Freiburg site was chosen because of logistical reasons and was also surveyed for habitat analyses in 2010 and 2011 (see below). In each year, we captured birds from mid-March on, when the first migrants arrived from their wintering quarters to mate on their breeding grounds, and caught birds every day from early morning to noon until the end of April. Blackcaps were caught within an area of 50 ha in a deciduous forest with mist nets using tape recordings of their song as a decoy. Each morning we intensively patrolled for singing activity and captured all newly singing males as well as nonsinging individuals within this area. Because of this procedure, we considered the first day a bird was caught as a proxy for arrival date and we calculated a dayscore as the difference in arrival relative to the start of the field season for statistical analysis (days from 15th March). Our proxy for arrival date reflects accurately arrival in the study area. However, we cannot exclude that an individual had already arrived a few days before in an area outside the study area. All individuals were marked with a standard aluminum ring (additional color coded rings in case of habitat examination, see below), sexed, aged (Shirihai et al. 2001), and weighed (digital balance ±0.1 g precision) before obtaining blood samples (50–75 μL) and claw tip samples. Claw tips were then analyzed for their individual stable isotope signature (2H/1H ratio, denoted as δ2H) using the comparative equilibration method described in detail by Wassenaar and Hobson (2003) and through the use of calibrated keratin isotope reference materials (see: Rolshausen et al. 2010 for methodological details).
Signals based on tissue δ2H measurements have been used successfully to infer wintering origins of migratory passerines, and blackcaps in particular, based on a large-scale latitudinal stable isotope gradient across central Europe (Bearhop et al. 2005; Bowen et al. 2005; Rolshausen et al. 2010). Along this gradient δ2H values are higher in the north-northwest and lower in the south (Bowen et al. 2005, see also: http://www.waterisotopes.org). Individual δ2H values entered our analyses as a continuous variable and were used as a proxy for (north–south) geographical distance between wintering origins (Webster et al. 2002; Hobson 2005). As our dataset potentially includes both locally breeding migrants and migrants caught “en route,” we used wing morphology to assign individuals as either local southern German breeding birds or long-distance migrants on their migration to more northerly breeding grounds. The assignment was based on probability density functions (see Rolshausen et al. 2010 for details) using Fiedler (2005) as well as our own measurements as reference data. From the 195 individuals that were analyzed in this study, 142 were considered as locally breeding middle-distance migrants based on their wing morphology. In the result section, we provide test statistics for this specific subset along with our main results for the whole dataset.
Nuclear DNA was extracted from blood samples using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). We used the following microsatellites for the genetic analysis: Syl1, Syl2, Syl3, Syl4, Syl5, Syl6, Syl7, Syl8, Syl9, Syl10 (for details on markers and PCR conditions see: Segelbacher et al. 2008; Rolshausen et al. 2009). In total, we analyzed isotope signatures and genomic data for 195 birds from the three consecutive study years.
Habitat analyses
To investigate whether microhabitat choice of blackcaps on the breeding grounds is related to individual variation in wintering origins, we analyzed the habitat structure for 36 breeding territories observed in 2010 and 2011 at our field sites in Freiburg. At the beginning of the respective breeding season, a male was considered territorial for a specific area if he was repeatedly spotted in that area for a minimum of 10 consecutive days, displayed territorial behavior (i.e., calls and singing), and/or was accompanied by a female. The margins of a territory were either defined as a 20 m radius from the center where the male was spotted most of the time or they were inferred from male's behavioral patterns (i.e., antagonistic interactions with other males). The territory size estimated by both measures did not differ ( C. Hermes, G. Segelbacher and H. Martin Schaefer, unpubl. data).
In mid June, each territory was surveyed for its vegetation structure by recording the variables described in Table 1; some of them were previously identified as being important for the establishment of territories in blackcaps (Hoi-Leitner et al. 1995). The data were then analyzed with a principal component analysis (PCA) to examine the distribution of individuals in an ordination diagram (i.e., habitat space). To examine whether microhabitat choice is associated with wintering origins, we (1) use the individual scores on the first two habitat principal components (PC1 and PC2) to test for a linear relationship with the respective individual δ2H values and (2) use a generalized additive model (GAM) to fit a smooth response surface of δ2H values over the limits of the ordination biplot. We include the latter procedure to additionally test for a combined nonlinear association between the habitat ordination axes (“habitat space”) and individual δ2H values.
Table 1.
List of the variables recorded for the microhabitat analysis along with a brief description of how they were recorded. Loadings correspond to correlation coefficients between principal components and variables, respectively (importance cutoff limit: 0.2). Numbers 1–9 correspond to the arrows 1–9 in Fig. 5
| Loadings | ||||
|---|---|---|---|---|
| Habitat variable | Description | PC1 | PC2 | PC3 |
| 1. Lower shrubbery layer (1.5–3 m) | % of vegetation in this layer | 0.559 | −0.038 | 0.272 |
| 2. Upper shrubbery layer (3–5 m) | % of vegetation in this layer | 0.516 | 0.277 | 0.253 |
| 3. Crown layer (above 5 m) | % of vegetation in this layer | 0.035 | 0.593 | −0.341 |
| 4. Nettle (Urtica dioica) covering | % of vegetation covered by nettles | −0.054 | −0.324 | −0.339 |
| 5. Bramble (Rubus sp.) covering | % of vegetation covered by brambles | −0.059 | −0.010 | 0.116 |
| 6. Number of ivy (Hedera helix) trees | Number of trees covered in ivy | 0.189 | 0.528 | −0.318 |
| 7. Blooming shrubs | % of blooming individuals in shrubbery layer | 0.390 | −0.209 | 0.121 |
| 8. Blooming herbs | % of blooming individuals in herbaceous layer | 0.424 | −0.323 | −0.315 |
| 9. Disturbance | % of the territory that is disturbed (e.g., proximity to highways, noise, walking paths) | 0.214 | −0.197 | −0.634 |
Individual genetic distance, diversity, and admixture
Individual genetic distance matrices based on the 10 microsatellites were calculated using the software package microsat (http://hpgl.stanford.edu/projects/microsat). The following genetic distance measures were calculated in our study: Reynolds Theta (Reynolds et al. 1983), Slatkins Rst (Slatkin 1995), and Goldsteins deltaMu ([δμ]2, Goldstein et al. 1995). Furthermore, we calculated a pairwise distance matrix based on individual δ2H values to infer individual differences in wintering origin within the studied population. Correlations between pairwise geographical distances and pairwise genetic distances were then analyzed with Mantel's nonparametric test (Mantel 1967). In addition, to correct for pseudo-replication due to pairwise comparisons, we analyzed the relation between genetic distance and geographical distances using a linear mixed model with geographical distance as fixed effect and individuals as a random structure. Significance for the fixed effect was based on (1) the posterior distribution of the model parameter obtained via MCMC simulations (n = 10,000) and (2) loglikelihood comparisons between the full model and the reduced model (excluding δ2H as a fixed effect).
To examine the relation between individual wintering origins and population genetic structuring, we applied a nonhierarchical cluster analysis to our δ2H variable that separates migrants based primarily on the north–south geographical distance between individual wintering origins. The appropriate number of “δ2H clusters” was objectively evaluated on the basis of the cumulative explained variance among clusters. This method recommends choosing that number of clusters so that further splitting does not provide more relevant information (i.e., “elbow criterion”). Respective clusters were then analyzed for deviations of their mean within-cluster genetic distance from the overall mean genetic distance to ask whether the cluster-intern genetic structure was more homogenous than that of the overall population structure of the dataset (i.e., indicating assortative pairing). This was tested by permutating the composition of respective clusters, while keeping cluster sizes constant. We analyzed between-cluster differences in individual genetic diversity using two different measures: mean individual heterozygosity and mean d² (Coulson et al. 1998). Inbreeding coefficients (FIS) between δ2H clusters were calculated with FSTAT (Goudet 1995).
The software BOTTLENECK (Piry et al. 1999) was used to scrutinize δ2H clusters for deviations from expected heterozygote excess relative to allelic diversity across all microsatellite loci. These tests within the δ2H clusters were performed on the basis of the proposed mutation models for microsatellites: the infinite alleles model (IAM), the stepwise mutation model (SMM), and the two-phase model (TPM: 70% SMM, 30% IAM). Studies on avian microsatellite evolution suggest that the SMM/TPM models are most appropriate (Primmer and Ellegren 1998; Beck et al. 2003). However, a proportion of our markers are compound markers with imperfect sequence motifs (Segelbacher et al. 2008), more likely to evolve under the IAM (Estoup et al. 1995). We therefore included the SMM, TPM, and IAM in our analysis. Note that the heterozygosity excess compares observed and expected heterozygosity and should not be confused with an excess of heterozygotes (Cornuet and Luikart 1996).
A Bayesian admixture model analysis was conducted on population structure using the software STRUCTURE 2.3 (Pritchard et al. 2000) and the therein implemented model of informative priors (LOCPRIOR, Hubisz et al. 2009). The LOCPRIOR model included the δ2H clustering and was run for k = 2 and k = 3 population clusters. These k-values were chosen according to (1) an evaluation of the most likely k for our dataset (without a priori information) using STRUCTURE and the Δk criterion (Evanno et al. 2005; yielded k = 2, for details see: Rolshausen et al. 2009) and (2) the most informative clustering of the δ2H variable (yielded k = 3, Fig. 2). STRUCTURE was run for 10 separate MCMC simulations over 50,000 burn-ins with 100,000 repeats for each k, and the different runs were then merged using CLUMPP (Jakobsson and Rosenberg 2007). Based on the likelihood assignment of the admixture model, we calculated (1) individual confidence coefficients (Δpr) as the residuals of the highest assignment score to the random assignment value (pr,2clusters = ½ and pr,3clusters = ⅓) and (2) the overall percentage of each simulated genetic cluster within the respective δ2H clusters. These two simple measures allow a proportionate comparison of the genetic clusters within the δ2H clusters.
Figure 2.
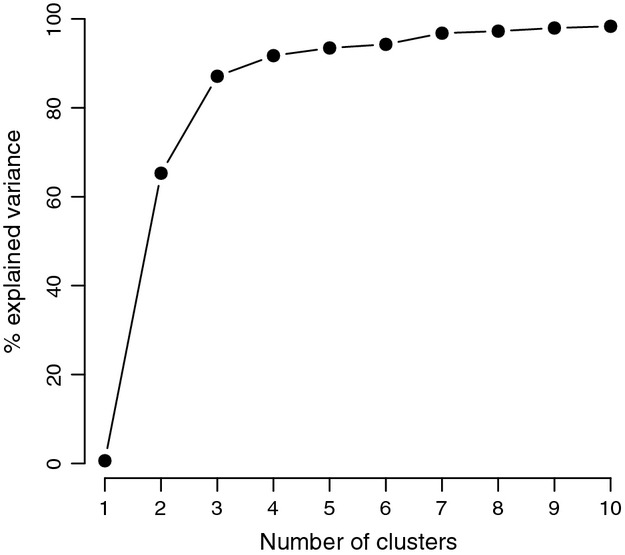
Evaluation of best clustering based on partitioning around medoids (pam) applied to the δ2H variable. The graph depicts the course of the ratio of within-group variances by the total variance along an increasing number of clusters. We chose three clusters to be the optimal representation of the data with ∼90% of the total variance explained.
All results reported for the genetic distance are based on Reynolds Theta distance but do not change qualitatively if Slatkins Rst (Slatkin 1995) or Goldsteins deltaMu (Goldstein et al. 1995) are employed. We provide the alternative test statistics along with our results. All basic statistical procedures were performed using R (R Development Core Team 2010).
Results
We found a significant positive correlation between the individual pairwise genetic distance (Theta) and the individual pairwise δ2H distances in claw stable isotope values (r = 0.13, P = 0.001, n = 195). The mixed model incorporating individuals as a random structure also revealed a significant effect of δ2H distance on genetic distance (Model estimates: Intercept = 0.760, δ2H = 0.005, MCMC simulation: P-valueδ2H = 0.001; reduced vs. full model: ΔAIC = 510.29, P-value<0.0001). The relatively low correlation coefficient is typical for individual pairwise comparisons illustrating substantial genetic and geographical variation in overwintering locations in our blackcap population. Similar results were found when we (1) based the test on alternative genetic distance measures (Slatkins Rst: r = 0.10, P = 0.003; Goldsteins [δμ]2: r = 0.10, P = 0.001, Mantel's test with 1000 permutations, n = 195), (2) excluded potential long-distance migrants from our dataset (r = 0.15, P = 0.002, n = 142), and (3) partitioned our dataset according to study year and/or site (Fig. 1). To account for a possible bias from an underlying population structure caused by the NW–SW migratory divide in our data (Rolshausen et al. 2009), we additionally ran Mantel's test on the SW migrants only. This test yielded comparable population structuring within SW migrants (r = 0.10, P = 0.033). We (2) repeated the test on 500 randomly chosen subsets comprising 100 individuals from our dataset, respectively. Here, we found that 80% of all 500 Mantel tests were significant (P < 0.05) and 93% were marginally significant (P < 0.1) with a mean test statistic comparable to our original results (r ± SE=0.13 ± 0.002). Isotope distances were not related to individual distances in arrival times of birds on their breeding grounds (r = 0.03, P > 0.7, Mantel's test with 1000 permutations) suggesting that individual birds from distinct wintering regions did not differ strongly in arrival times.
Figure 1.
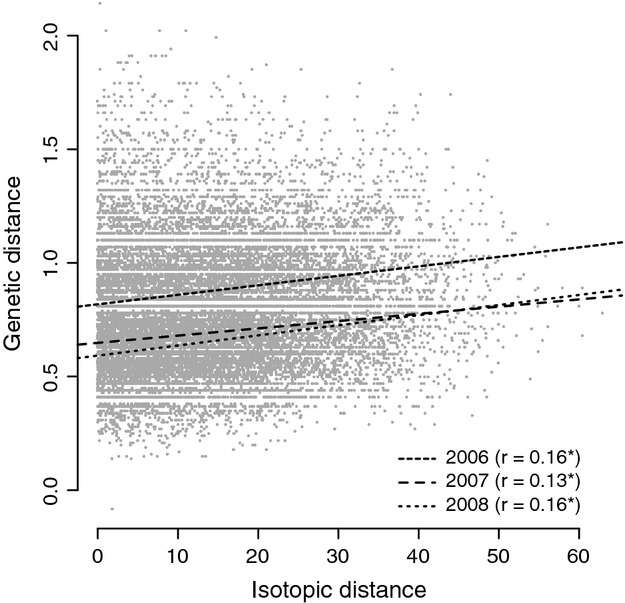
Correlation between individual genetic distance and delta distance (based on δ2H values). The scatterplot shows individual pairwise distances from all three study years (2006–2008, note that 2006 birds and 2007/2008 birds were not caught at the same site), and the lines denote the positive correlations for each year separately. Numbers in parentheses depict the respective Pearson correlation coefficients (*: all P≪0.01).
To analyse the relation of migratory origin and the underlying population structure, we partitioned the δ2H variable into separate clusters. The evaluation of the most informative partition according to the cumulative explained variance yielded three wintering origins illustrated as k = 3 δ2H clusters as the fitting selection (Fig. 2): one δ2H cluster containing birds presumably from more northerly wintering areas (N migrants, δ2H cluster 3: n = 55, mean δ2H = −91.2‰) and two δ2H clusters containing birds presumably from more southerly wintering quarters (S-migrants), separated according to their approximate distance to the breeding grounds (farther: δ2H cluster 1: n = 52, mean δ2H = −59.6‰; nearer: δ2H cluster 2: n = 88, mean δ2H = −74.4‰).
A particularly interesting result was that the mean individual genetic distances among birds overwintering at northern latitudes were higher compared with birds that overwintered south of the breeding grounds (δ2H cluster 3 vs. δ2H cluster 1 and 2, Fig. 3). Further, δ2H clusters tended to differ in their mean within-genetic distance from the mean within-genetic distance obtained by permutation of the entire population. The northerly migrants were significantly more distant from each other than the southerly wintering migrants which were less distant from each other compared with the permutation mean (δ2H cluster 1: P = 0.015, δ2H cluster 2: P = 0.004, δ2H cluster 3: P = 0.000, permutation test, Fig. 3). Both measures of genetic diversity, mean heterozygosity, and mean d² were significantly lower in N migrants (δ2H cluster 3) compared with the other δ2H clusters (δ2H cluster 3 vs. δ2H cluster 1 and 2: all P < 0.01, comparisons between δ2H clusters 1 and 2: P > 0.1). The inbreeding coefficient (FIS) was higher for the N migrants and not different among the two S-migrating clusters (FIS δ2H cluster 3: 0.45; FIS δ2H cluster 2: 0.27; FIS δ2H cluster 1: 0.22; P ≤ 0.10, pairwise Wilcoxon tests with adjusted P-values). No heterozygote excess in either δ2H cluster was found when testing under the assumptions of the SMM (P > 0.80), whereas there was a significant heterozygote excess found under TPM and IAM assumptions (all P ≤ 0.05, Wilcoxon signed rank test), indicating no clear evidence for a recent bottleneck in any cluster.
Figure 3.
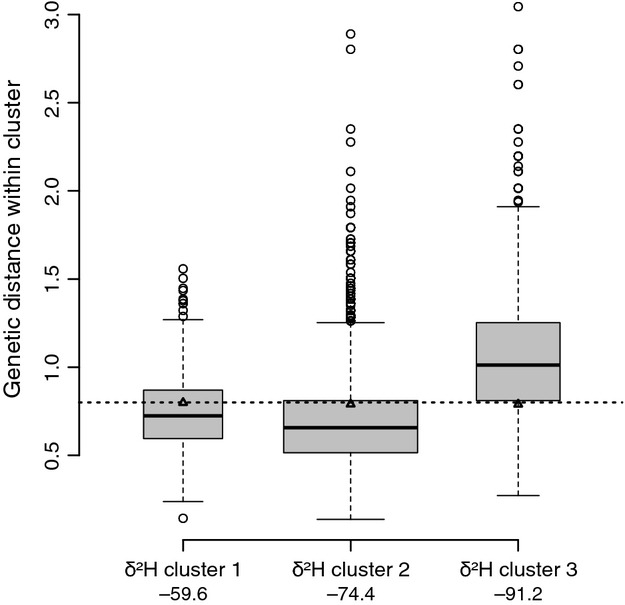
Individual pairwise genetic distances within δ2H clusters obtained from partitioning the δ2H variable. The x-axis denotes the respective clusters with their mean δ2H values (‰) and the y-axis denotes the genetic distance as Reynolds Theta. The dotted horizontal line marks the overall mean genetic distance for the dataset, and triangles denote the respective mean distances within clusters after randomized cluster assignment (1000 permutations). Mean genetic distances within clusters were significantly different from either the permutation means or the overall mean (cluster 1 [n = 52]: P = 0.015, cluster 2 [n = 88]: P = 0.004, cluster 3 [n = 55]: P = 0.000, overall mean: 0.785, P-values obtained over 1000 permutations).
STRUCTURE simulations assuming k = 2 genetic clusters yielded no significant differences in admixture among the three δ2H clusters (Fig. 4A). Individual confidence coefficients (Δpr) did not differ between the δ2H clusters (all P ≥ 0.10), nor did the overall proportions of each genetic cluster in the δ2H clusters (genetic cluster 1: 66% in δ2H cluster 1; 68% in δ2H cluster 2; 56% in δ2H cluster 3). However, when simulating k = 3, we found significant differences in Δpr and overall proportions of genetic clusters between δ2H clusters (Fig. 4B). Blackcaps wintering in more northern areas, represented by δ2H cluster 3, had significantly higher Δpr scores compared with δ2H clusters 1 and 2 (all P < 0.010), indicating a more confident assignment of individuals from that δ2H cluster to either of the three simulated populations. We also found marginal differences in Δpr scores between δ2H clusters 1 and 2 (P = 0.100, pairwise Wilcoxon test with adjusted P-values). The percentage represented by the third genetic cluster within each of the δ2H clusters increased from 14% in cluster 1 to 21% in cluster 2, to 36% in cluster 3 (Fig. 4B) and assignment scores to the third genetic cluster correlated significantly with individual δ2H values (r = 0.48, P < 0.001, n = 195, Fig 4C). Given the unbiased approach of the LOCPRIOR admixture model (Hubisz et al. 2009), the reported differences between δ2H clusters suggest asymmetric admixture in relation to overwintering origins.
Figure 4.
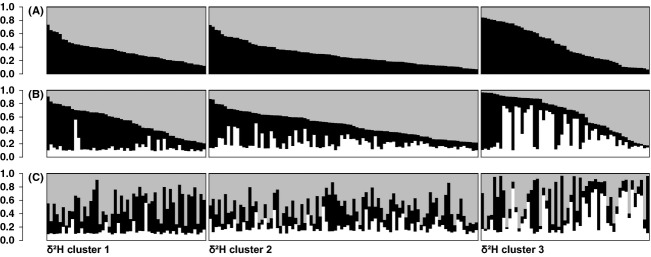
Coefficients of admixture for each individual estimated using STRUCTURE. Individuals in their respective δ2H cluster are represented by columns, and individual admixture coefficients are shown as proportions of different colors in each column. A: admixture model assuming two underlying populations; B and C: admixture model assuming three underlying populations. Barplots A and B align individuals within δ2H clusters according to their admixture coefficients, barplot C aligns individuals in ascending order of their δ2H values.
The analysis of the microhabitat choice of blackcaps resulted in three main principal components accounting for a total of 60% of variation (PC1:25%, PC2:18%, PC3:17%) in the birds' distribution (Fig. 5). Five of nine habitat variables were considered important loadings (correlation coefficient >0.2, Table 1) for PC1 basically describing vegetation density (in lower and upper shrubbery layers) and the amount of blooming shrubs and herbs. PC2 was mainly affected by vegetation density in the crown layer, the density of ivy (Hedera helix), and presence of nettles (Urtica dioica), and PC3 was mainly affected by disturbance and density in the crown layer and presence of nettles (Table 1).
Figure 5.
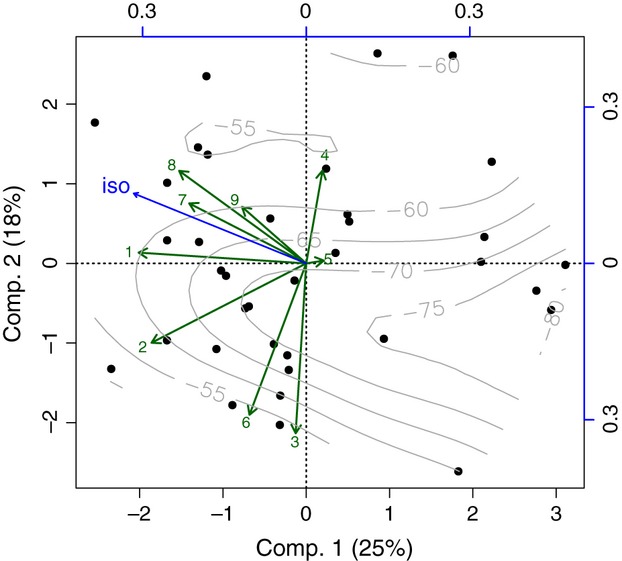
Habitat choice ordination biplot with δ2H variable response surface overlain in gray and the surface was fitted using a generalized additive model approach. The arrows 1–9 depict the respective habitat variables (see Table 1). The iso-arrow depicts the direct biplot projection for the δ2H variable onto the ordination and indicates the significant association with Comp. 1 but not with Comp. 2.
Using linear models, we found a significant correlation of individual PC1 scores with δ2H values (cor.r = −0.323, P = 0.050, Fig. 5) but no relation between δ2H values and PC2 (cor.r = 0.120, P = 0.467) and a marginal relation with PC3 (cor.r = −0.268, P = 0.102). However, assuming a nonlinear relation, the generalized additive model fitted onto the first two PCs indicated a significant association between individual δ2H values and the main individual variation in occupancy of the habitat space (GAM fit: F = 2.24, P = 0.047, Fig. 5). Taken together, both results suggest that – independent of the underlying assumption – blackcaps establish territories in different habitats depending on their individual wintering origins.
Discussion
Migratory connectivity and genetic structuring
Here, we show that individual migratory behavior within a population contributes to the genetic structure of sympatric southern German blackcaps. In particular, birds with more similar δ2H values were genetically more closely related to each other (pairwise differences at neutral loci) than to birds wintering at more distant locations. Initially, it seems that the overall correlation (r = 0.13) in the pairwise comparisons among 195 individuals was weak. However, this only shows that there is substantial genetic variation within blackcap populations which is consistent with other studies on migratory birds (Bensch et al. 2009; Prochazka et al. 2011). A comparable coefficient was recently reported for a population of Darwin's finches where individual beak morphology correlated with genetic distance at r = 0.13 (De León et al. 2012). Indeed, the pattern we report is likely to be biologically relevant because it was consistently found in three consecutive years, at two different study sites (Fig. 1), and also occurred for >80% of 500 randomly drawn subsets of our dataset and for the subset excluding potential long-distance migrants. Taken together, these results indicate that similarity in wintering origin influences genetic distances even on shared breeding grounds. This conclusion mirrors that from Bearhop et al. (2005). However, our results indicate that the influence of wintering origin on mating decisions not only occurs dichotomously along the migratory divide (Bearhop et al. 2005; Rolshausen et al. 2009) but also to a similar extent within the same migration route (SW route) on an individual level throughout the population. Hence, an important novel insight from our study is that the influence of wintering origin on population structuring is not solely attributable to the two migratory directions in Southern German blackcaps (NW and SW), but may be more prevalent in migratory birds in general. Thus, rapid translations of environmental change into micro-evolutionary changes, such as the genetic reduction in migratory activity in blackcaps due to climate-driven selection (Pulido and Berthold 2010), could be facilitated by genetic structuring within populations according to individual migratory routes and distances.
The proximate mechanisms mediating the effect of migratory connectivity on the genetic structure of sympatric populations are currently not well understood. Our habitat analyses reveal a significant relation between the characteristics of individual territories and the tissue δ2H values of males occupying these territories (Fig. 5). This link between breeding habitat choice and wintering origin suggests that where a bird overwinters may have profound influence on which habitat it chooses for breeding and that this in turn may affect reproductive output (see also Norris and Taylor 2006; Norris and Marra 2007). For example, blackcaps might colonize breeding territories that are more similar to their wintering quarters. While this conjecture has not yet been investigated, carry-over effects are more generally important at the individual level and can translate variation in migratory behavior into assortative mating on shared breeding grounds (Reudink et al. 2009). Our study showed no effect of temporal isolation because differences in arrival times were not significantly related to the respective wintering origins of individual blackcaps inferred from tissue δ2H values. This result supports previous analyses from our population (Rolshausen et al. 2010) and contrasts those of more eastern blackcap populations where assortative mating according to migratory route seems to be more prevalent (Bearhop et al. 2005). Spatial rather than temporal separation might therefore contribute to explain the effect of migratory origins on the genetic structure of the sympatric populations in south-western Germany. Furthermore, it is currently unknown whether postzygotic barriers additionally contribute to the observed genetic structure of blackcaps. In general, postzygotic barriers are deemed important in incipient divergence (Coyne and Orr 2004) and apparently maintain genetic divergence in willow warblers (Phylloscopus trochilus) and yellow-rumped warblers (Setophaga coronata) complex in the absence of strong assortative mating (Bensch et al. 2009; Brelsford and Irwin 2009). Therefore, future research on the incipient divergence blackcaps should also consider postzygotic isolation as a potential mechanism driving divergence.
Genetic diversity along the migratory divide
Our study on blackcaps differing in wintering origins found significant differences in genetic diversity along the migratory polymorphism in southern Germany. Migrants following the recently established northwestern route (Berthold et al. 1992) showed higher levels of inbreeding (FIS) and significantly lower individual genetic diversity, denoted as overall heterozygosity and mean d², compared with the two clusters of SW migrants that follow the traditional south-western route. These findings are consistent with the new route having evolved only recently, involving far fewer individuals than the SW route, and document thus for the first time that the genetic structure of the NW-migrating population deviates from that of SW-migrating populations.
At present, it remains unknown whether the lower heterozygosity we found among the NW-migrating blackcaps might affect their future adaptive potential. In general, low levels of heterozygosity are linked to lower population fitness (Reed and Frankham 2003) and also to the ability of populations to respond to selection (i.e., evolvability, Houle 1992). On the one hand, moderate inbreeding might constrain the evolvability of traits with strong additive genetic backgrounds, whereas on the other hand, it might assist the evolvability of traits with a nonadditive genetic background (Cheverud et al. 1999; Zhang et al. 2004; Van Buskirk and Willi 2006). Interestingly, life-history traits (e.g., timing of breeding) are known to have a strong nonadditive genetic background (Merilä and Sheldon 1999, Teplitsky et al. 2009) and are also important targets of selection in migratory birds (Both et al. 2006; Hedenström et al. 2007). Moreover, both the expression of the underlying genetic variance in life-history traits and the selection acting on them often depend on environmental conditions (Husby et al. 2011). However, our analyses are based on the variation in microsatellites and therefore on (neutral) genetic markers that generally (1) reflect only a very small portion of the genome and (2) are known to be poor indicators of adaptive genetic differences (Reed and Frankham 2001). Hence, further investigation of the evolutionary potential in our study population initially requires that quantitative genetic information of respective traits is measured directly.
Multiple founder effects following the migratory divide?
The new migratory behavior in southern German blackcaps probably evolved within only a few decades from preexisting variation for migratory directions in central European blackcaps (Berthold et al. 1992; Helbig 1996). The newly evolved migratory route entails restricted gene flow between sympatrically breeding populations using different wintering quarters (Rolshausen et al. 2009). Assuming that the population genetic differences along the migratory divide can be explained by a recent colonization event (i.e., “founder effect,” Nei et al. 1975), two nonmutually exclusive hypotheses can be applied: The founding event(s) could have arisen (1) from within the local breeding population but also (2) from multiple contributions of more distant populations of blackcaps that also adopted a northwestern orientated migration (Helbig 1996). In favor of the first hypothesis, we found elevated inbreeding (FIS) and significantly lower genetic diversity (heterozygosity) for the NW migrants. However, NW migrants were on average genetically more distinct from each other than SW migrants (i.e., higher genetic distances within the third δ2H cluster), which did not differ genetically between isotope clusters (Fig. 3). Moreover, the bottleneck analysis of heterozygosity excess corrected for allelic diversity (Cornuet and Luikart 1996; Piry et al. 1999) did not yield evidence for a recent severe population bottleneck in the NW migrants. As such, we hypothesize that multiple founder events occurred as this hypothesis is consistent with a more diverse genetic background of individual NW migrants.
Both scenarios, introgression from more distant populations of northwest migrating blackcaps, as well as a founder event from within the local breeding population require incipient reproductive isolation and therefore asymmetric admixture in sympatry along the migratory divide to maintain the genetically based differences in migratory behavior (Berthold et al. 1992; Helbig 1996). Our admixture model in STRUCTURE that assumed three underlying populations to match the number of δ2H clusters (Fig. 2) found significant asymmetric admixture on neutral genetic markers related to the wintering areas of blackcaps (Fig. 4). Migrants from δ2H cluster 3 (i.e., NW migrants) were on average assigned with higher confidence to one of the three simulated genetic populations, that is, they had a higher probability of pertaining to a specific genetic cluster, as denoted in higher residual assignment coefficients (Δpr). Moreover, the within-cluster fractions of the third genetic population decreased along the N-S gradient from 36% in δ2H cluster 3 to 14% in δ2H cluster 1, a result which is strongly confirmed by examining the extremes of the δ2H spectrum and the correlation between assignment scores and individual δ2H values (Fig. 4). These patterns suggest that the migratory divide in southern German blackcaps has a significant impact on population genetic dynamics in sympatry. Yet, given the low overall genetic differentiation in the studied population (FST, N vs. S ≤0.008, for details see Rolshausen et al. 2009) along with nonequilibrium population dynamics due to the newly established NW route, the STRUCTURE simulation might not be a powerful tool to resolve the actual population structure at this early stage (see Pritchard et al. 2000). Still, our analyses motivate the hypothesis that NW migrants from foreign populations are more successful in introgressing the δ2H cluster 3, already containing NW migrants, and that this introgression could therefore be facilitated via assortative mating based on wintering origins and migratory strategies.
Conclusions
Our analyses show a positive relationship between individual wintering origins and population genetic structure in sympatrically breeding blackcaps from southern Germany. The migratory divide of SW- and NW-migrating birds strongly contributes to this genetic structure, but it is also present within SW migrants. Moreover, individual differences in migratory behavior relate to differences in admixture among groups of blackcaps with distinct migratory behavior. We also found support for the hypothesis that the establishment of the migratory divide in southern German blackcaps may have involved multiple founder events that may have arisen not only from within the local breeding population but also from more distant blackcap populations also migrating in a northwestern direction. Taken together, our study indicates that individual migratory behavior influences genetic structuring in sympatric populations of migratory birds that show weak migratory connectivity. In general, this mechanism might be an important link between the genetic basis of migratory behavior and (1) the translation of environmental change into adaptive population genetic change (e.g., Pulido and Berthold 2010) as well as (2) the establishment and maintenance of migratory divides (e.g., Bensch et al. 2009; Brelsford and Irwin 2009). A potential proximate mechanism facilitating the genetic structuring in our study population is spatial divergence caused by differential microhabitat selection. We did not find evidence that individual arrival times varied consistently according to migration distances. Although differential arrival times of populations with distinct migratory routes have been discussed in the incipient population divergence of blackcaps in southern Germany (Bearhop et al. 2005; Rolshausen et al. 2010), the null model by Rolshausen et al. (2010) suggests that temporal isolation can maximally explain part of the incipient isolation and that other mechanisms are likely to also contribute to it. Hence, the results from our territory analyses are encouraging that future research on microhabitat selection in relation to migratory behavior in birds will provide more detailed insights into micro-evolutionary dynamics within populations responding to contemporary changes in selection.
Acknowledgments
The authors thank Carlo Catoni, Rebecca Bloch, Kalliope Stournaras, Nicola Moratscheck, Claudia Hermes, Raeann Mettler, and Frederick Wehrle for their help in collecting the field data. We also thank Andrew Hendry and Renauld Kaeuffer for valuable comments and discussion. Stable isotope analyses were performed by Len I. Wassenaar at the Environment Canada stable isotope facility in Saskatoon, Canada. This work was supported by DFG Grant (Scha 1008/6-1).
Conflict of Interest
None declared.
References
- Bearhop S, Fiedler W, Furness RW, Votier SC, Waldron S, Newton J, et al. Assortative mating as a mechanism for rapid evolution of a migratory divide. Science. 2005;310:502–504. doi: 10.1126/science.1115661. [DOI] [PubMed] [Google Scholar]
- Beck NR, Double MC, Cockburn A. Microsatellite evolution at two hypervariable loci revealed by extensive avian pedigrees. Mol. Biol. Evol. 2003;20:54–61. doi: 10.1093/molbev/msg005. [DOI] [PubMed] [Google Scholar]
- Bensch S, Grahn M, Müller N, Gay L, Akesson S. Genetic, morphological, and feather isotope variation of migratory willow warblers show gradual divergence in a ring. Mol. Ecol. 2009;18:3087–3096. doi: 10.1111/j.1365-294X.2009.04210.x. [DOI] [PubMed] [Google Scholar]
- Berthold P. 2000. Vogelzug. Eine aktuelle Gesamtübersicht. Edition 4. Wissenschaftliche Buchgesellschaft Darmstadt. ISBN-10: 353413656X, ISBN-13: 978-3534136568.
- Berthold P, Helbig A, Mohr G, Querner U. Rapid microevolution of migratory behaviour in a wild bird species. Nature. 1992;360:668–670. [Google Scholar]
- Both C, Bouwhuis S, Lessells CM, Visser ME. Climate change and population declines in a long-distance migratory bird. Nature. 2006;441:81–83. doi: 10.1038/nature04539. [DOI] [PubMed] [Google Scholar]
- Both C, Van Turnhout CA, Bijlsma RG, Siepel H, Van Strien AJ, Foppen RP. Avian population consequences of climate change are most severe for long-distance migrants in seasonal habitats. Proc. Biol. Sci. 2010;277:1259–1266. doi: 10.1098/rspb.2009.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen GJ, Wassenaar LI, Hobson KA. Global application of stable hydrogen and oxygen isotopes to wildlife forensics. Oecologia. 2005;143:337–348. doi: 10.1007/s00442-004-1813-y. [DOI] [PubMed] [Google Scholar]
- Brelsford A, Irwin DE. Incipient speciation despite little assortative mating: the yellow-rumped warbler hybrid zone. Evolution. 2009;63:3050–3060. doi: 10.1111/j.1558-5646.2009.00777.x. [DOI] [PubMed] [Google Scholar]
- Cheverud JM, Vaughn TT, Pletscher LS, King-Ellison K, Adams E, Erickson C, et al. Epistasis and the evolution of additive genetic variance in populations that pass through a bottleneck. Evolution. 1999;53:1009–1018. doi: 10.1111/j.1558-5646.1999.tb04516.x. [DOI] [PubMed] [Google Scholar]
- Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson TN, Pemberton JM, Albon SD, Beaumont M, Marshall TC, Slate J, et al. Microsatellites reveal heterosis in red deer. Proc. Biol. Sci. 1998;265:489–495. doi: 10.1098/rspb.1998.0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sinauer Associates. 2004;1:247–281. [Google Scholar]
- De León LF, Rolshausen G, Bermingham E, Podos J, Hendry AP. Individual specialization and the seeds of adaptive radiation in Darwin's finches. Evol. Ecol. Res. 2012;14:365–380. [Google Scholar]
- Estoup A, Tailliez C, Cornuet J, Solignac M. Size homoplasy and mutational processes of interrupted microsatellites in two bee species, Apis mellifera and Bombus terrestris (Apidae) Mol. Biol. Evol. 1995;12:1074–1084. doi: 10.1093/oxfordjournals.molbev.a040282. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Fiedler W. Recent changes in migratory behaviour of birds: a compilation of field observations and ringing data. In: Berthold P, Gwinner E, Sonnenschein E, editors. Avian migration. Berlin: Springer; 2003. [Google Scholar]
- Fiedler W. Ecomorphology of the external flight apparatus of blackcaps (Sylvia atricapilla) with different migration behavior. Annals of the NY Acad. Sci. 2005;1046:253–263. doi: 10.1196/annals.1343.022. [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Ruiz-Linares A, Cavalli-Sforza LL, Feldman MW. An evaluation of genetic distances for use with microsatellite loci. Genetics. 1995;139:167–178. doi: 10.1093/genetics/139.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. FSTAT (version 1.2): A computer program to calculate F-statistics. J. Hered. 1995;86:1994–1995. [Google Scholar]
- Hedenström A, Barta Z, Helm B, Houston A, McNamara J, Jonzén N. Migration speed and scheduling of annual events by migrating birds in relation to climate change. Clim. Res. 2007;35:79–91. [Google Scholar]
- Helbig A. Genetic basis, mode of inheritance and evolutionary changes of migratory directions in palaearctic warblers (Aves: Sylviidae) J. Exp. Biol. 1996;199:49–55. doi: 10.1242/jeb.199.1.49. [DOI] [PubMed] [Google Scholar]
- Hobson KA. Using stable isotopes to trace long-distance dispersal in birds and other taxa. Divers. Distrib. 2005;11:157–164. [Google Scholar]
- Hobson KA. Isotopic ornithology: a perspective. J. Ornithol. 2011;152:49–66. [Google Scholar]
- Hobson KA, Wassenaar SL, Van Wilgenburg LI, Larson K. Linking hydrogen (δ2H) isotopes in feathers and precipitation: sources of variance and consequences for assignment to isoscapes. PLoS ONE. 2012;7:1–9. doi: 10.1371/journal.pone.0035137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoi-Leitner M, Nechtelberger H, Hoi H. Song rate as a signal for nest-site quality in Blackcaps (Sylvia atricapilla. Behav. Ecol. Sociobiol. 1995;37:399–405. [Google Scholar]
- Houle D. Comparing evolvability and variability of quantitative traits. Genetics. 1992;130:195–204. doi: 10.1093/genetics/130.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Res. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husby A, Visser ME, Kruuk LEB. Speeding up microevolution: the effects of increasing temperature on selection and genetic variance in a wild bird population. PLoS Biol. 2011;9:1–9. doi: 10.1371/journal.pbio.1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Liedvogel M, Akesson S, Bensch S. The genetics of migration on the move. Trends Ecol. Evol. 2011;26:561–569. doi: 10.1016/j.tree.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Merilae J, Sheldon BC. Genetic architecture of fitness and nonfitness traits: empirical patterns and development of ideas. Heredity. 1999;83:103–109. doi: 10.1046/j.1365-2540.1999.00585.x. [DOI] [PubMed] [Google Scholar]
- Mokwa K. Wintering range of the Blackcap (Sylvia atricapilla) in Europe – stabilized or changing? Ring. 2009;31:45–58. [Google Scholar]
- Nei M, Maruyama T, Chakraborty R. The bottleneck effect and genetic variability in populations. Evolution. 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- Norris DR, Marra P. Seasonal interactions, habitat quality, and population dynamics in migratory birds. The Condor. 2007;109:535–547. [Google Scholar]
- Norris DR, Taylor CM. Predicting the consequences of carry-over effects for migratory populations. Biol. Lett. 2006;2:148–151. doi: 10.1098/rsbl.2005.0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piry S, Luikart G, Cornuet JM. BOTTLENECK: A computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 1999;90:502–503. [Google Scholar]
- Price T. Speciation in birds. Greenwood Village, Colorado: Roberts & Company Publishers; 2007. ISBN 0-9747077-8-3. [Google Scholar]
- Primmer CR, Ellegren H. Patterns of molecular evolution in avian microsatellites. Mol. Biol. Evol. 1998;15:997–1008. doi: 10.1093/oxfordjournals.molbev.a026015. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochazka P, Stokke BG, Jensen H, Fainová D, Bellinvia E, Fossøy F, et al. Low genetic differentiation among reed warbler Acrocephalus scirpaceus populations across Europe. J. Avian Biol. 2011;42:103–113. [Google Scholar]
- Pulido F, Berthold P. Current selection for lower migratory activity will drive the evolution of residency in a migratory bird population. Proc. Natl. Acad. Sci. U.S.A. 2010;107:7341–7346. doi: 10.1073/pnas.0910361107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido F, Berthold P, vanNoordwijk AJ. Frequency of migrants and migratory activity are genetically correlated in a bird population: Evolutionary implications. PNAS. 1996;93:14642–14647. doi: 10.1073/pnas.93.25.14642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2010. [Google Scholar]
- Reed DH, Frankham R. Correlation between fitness and genetic diversity. Conserv. Biol. 2003;17:230–237. [Google Scholar]
- Reudink MW, Marra PP, Kyser TK, Boag PT, Langin KM, Ratcliffe LM. Non-breeding season events influence sexual selection in a long-distance migratory bird. Proc. Biol. Sci. 2009;276:1619–1626. doi: 10.1098/rspb.2008.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolshausen G, Segelbacher G, Hobson KA, Schaefer HM. Contemporary evolution of reproductive isolation and phenotypic divergence in sympatry along a migratory divide. Curr. Biol. 2009;19:2097–2101. doi: 10.1016/j.cub.2009.10.061. [DOI] [PubMed] [Google Scholar]
- Rolshausen G, Hobson KA, Schaefer HM. Spring arrival along a migratory divide of sympatric blackcaps (Sylvia atricapilla. Oecologia. 2010;162:175–183. doi: 10.1007/s00442-009-1445-3. [DOI] [PubMed] [Google Scholar]
- Ruegg K, Anderson EC, Slaabekoorn H. Differences in timing of migration and response to sexual signalling drive asymmetric hybridization across a migratory divide. J. Evol. Biol. 2012;25:1741–1750. doi: 10.1111/j.1420-9101.2012.02554.x. [DOI] [PubMed] [Google Scholar]
- Segelbacher G, Rolshausen G, Weis-Dootz T, Serrano D, Schaefer HM. Isolation of 10 tetranucleotide microsatellite loci in the blackcap (Sylvia atricapilla. Mol. Ecol. Res. 2008;8:1108–1110. doi: 10.1111/j.1755-0998.2008.02171.x. [DOI] [PubMed] [Google Scholar]
- Shirihai H, Gargallo G, Helbig AJ. Sylvia Warblers. In: Kirwan G, Svensson L, editors. Identification, taxonomy, and phylogeography of the genus Sylvia. London: A&C Black; 2001. [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;462:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow DW, Perrins C. The birds of the Western Palaeartic. Oxford University Press; 1997. Abridged edition. ISBN-10: 019854099X, ISBN-13: 978-0198540991. [Google Scholar]
- Studds CE, McFarland KP, Aubury Y, Rimmer CC, Hobson KA, Marra PP, et al. Stable-hydrogen isotope measures of natal dispersal reflect observed population declines in a threatened migratory songbird. Divers. Distrib. 2012;18:919–930. [Google Scholar]
- Teplitsky C, Mills JA, Yarrall JW, Merilä J. Heritability of fitness components in a wild bird population. Evolution. 2009;63:716–726. doi: 10.1111/j.1558-5646.2008.00581.x. [DOI] [PubMed] [Google Scholar]
- Van Buskirk J, Willi Y. The change in quantitative genetic variation with inbreeding. Evolution. 2006;60:2428–2434. [PubMed] [Google Scholar]
- Wassenaar LI, Hobson KA. Comparative equilibration and online technique for determination of non-exchangeable hydrogen of keratins for use in animal migration studies. Isot. Environ. Health Stud. 2003;39:211–217. doi: 10.1080/1025601031000096781. [DOI] [PubMed] [Google Scholar]
- Webster MS, Marra PP, Haig SM, Bensch S, Holmes RT. Links between worlds: unraveling migratory connectivity. Trends Ecol. Evol. 2002;17:76–83. [Google Scholar]
- Zhang X-S, Wang J, Hill WG. Redistribution of gene frequency and changes of genetic variation following a bottleneck in population size. Genetics. 2004;167:1475–1492. doi: 10.1534/genetics.103.025874. [DOI] [PMC free article] [PubMed] [Google Scholar]


