Abstract
Background
Epidermal growth factor receptor (EGFR) signaling is one of the most promising targets for molecular-targeted therapies in esophageal squamous cell carcinoma (ESCC). Thus, the molecular diagnosis of KRAS and BRAF mutations is clinically important in therapeutic decision making. However, the frequency of KRAS and BRAF mutations in ESCCs remains inconclusive because of the limited sample sizes of previous studies (all N ≤ 80). Pyrosequencing is a nonelectrophoretic nucleotide extension sequencing technology that can be used for mutation testing.
Methods
The frequency of KRAS and BRAF mutations was examined using a nonbiased database of 203 resected ESCCs and a high-throughput pyrosequencing assay.
Results
The validity of the KRAS pyrosequencing method was initially demonstrated by detection of all 4 types of KRAS mutations [c.35G>T (codon 12 GGT>GTT), c.35G>A (codon 12 GGT>GAT), c.34G>T (codon 12 GGT>TGT), c.38G>A mutation (codon 13 GGC>GAC)], which had been previously diagnosed using Scorpion-ARMS technology, in 9 colon cancer tissues (9 of 9; 100 %). Similar results were demonstrated for BRAF mutational status in 3 colon cancer cell lines (HCT116, Colo201, and HT29), which were validated by Sanger dideoxy sequencing. Subsequently, the KRAS mutation was found to be extremely rare (1 of 203; 0.5 %), and the BRAF mutation was absent (0 of 203; 0 %), in the dataset of 203 ESCCs.
Conclusions
These results suggest that KRAS and BRAF mutations play a limited role in the development of ESCC and that mutation analysis is not useful as a screening test for sensitivity to anti-EGFR therapy in ESCC.
Esophageal squamous cell carcinoma (ESCC) is the major histological type of esophageal cancer in East Asian countries and is one of the most aggressive malignant tumors.1 Despite remarkable advances in multimodal therapies, patient prognosis remains poor, even for those whose carcinomas have been completely resected.2–5 The limited improvement in treatment outcomes by conventional therapies urged us to seek innovative strategies for treating ESCC, especially those that are molecularly targeted. One of the most promising targets is the inhibition of the epidermal growth factor receptor (EGFR) by monoclonal antibodies (e.g., cetuximab, panitumumab) or small molecule tyrosine kinase inhibitors (e.g., erlotinib, gefitinib).6–10 The EGFR signal transduction network plays a crucial role in multiple tumorigenic processes, contributing to cell-cycle progression, angiogenesis, metastasis, and protection of the cancer cell from apoptosis.11 Mutations in the Kirsten Ras 1 (KRAS) and V-Raf Murine Sarcoma Viral Oncogene Homolog B1 (BRAF) genes may be predictive of response to drugs directly linked to the EGFR pathway.12–14 Thus, molecular diagnosis of these mutations is increasingly important in making therapeutic decisions. Several previous studies have examined the frequency of KRAS and BRAF mutations in ESCCs; however, they were all limited by small sample sizes (all N ≤ 80) (Table 1), yielding inconclusive results.8,15–20
Table 1.
Studies on KRAS and BRAF mutations in ESCC
| Study | Sample size | Methods | Mutation detected N (%) | Codons examined |
|---|---|---|---|---|
| Studies on KRAS mutations in ESCC | ||||
| Ma et al.15 | 35 | Pyrosequencing | 2 (5.7 %) | Codons 12–13 |
| Liu et al.16 | 50 | Pyrosequencing | 6 (12 %) | Codons 12–13 |
| Lorenzen et al.8 | 37 | Direct sequencing | 0 (0 %) | Codons 12–13 |
| Hollstein et al.17 | 16 | Direct sequencing | 0 (0 %) | Codons 12–13 |
| Victor et al.18 | 27 | PCR and oligomer hybridization assay | 0 (0 %) | Codons 12–13 |
| Hollstein et al.19 | 25 | PCR and oligomer hybridization assay | 0 (0 %) | Codons 12–13 |
| Present study | 203 | Pyrosequencing | 1 (0.5 %) | Codons 12–13 |
Pyrosequencing is a nonelectrophoretic nucleotide extension sequencing technology for various applications, including mutation testing of tumors.21–24 This technology has several advantages. First, it has higher sensitivity than classical Sanger dideoxy sequencing.22 Sanger dideoxy sequencing needs more than 20 % of tumor load in a specimen to render a reliable result, while pyrosequencing can render a reliable result with a tumor load of 5 %. Second, pyrosequencing is faster than Sanger dideoxy sequencing. Third, pyrosequencing is more cost effective. Collectively, pyrosequencing is a useful method in molecular diagnostics and large-scale epidemiological studies.25,26
Therefore, in the present study, KRAS and BRAF mutations were screened using a nonbiased database of 203 ESCCs and pyrosequencing technology.
Patients and Methods
Study Subjects
A total of 217 consecutive patients with ESCC who were undergoing curative resection at Kumamoto University Hospital between April 2005 and December 2010 were enrolled in this study. There were 13 patients excluded because of the unavailability of adequate tissue samples. We initially quantified KRAS and BRAF mutation in 204 cancer specimens and obtained valid results in 203 cases (99.5 %). Thus, 203 ESCCs were finally included in this study. Tumor staging was done by the American Joint Committee on Cancer Staging Manual (7th edition).27 Written informed consent was obtained from each subject, and the institutional review board of Kumamoto University approved the study procedures.
A total of 9 patients with colon cancers harboring 4 different KRAS mutations [c.35G>T (codon 12 GGT>GTT; p.Gly12Val), c.35G>A (codon 12 GGT>GAT; p.Gly12Asp), c.34G>T (codon 12 GGT>TGT; p.Gly12Cys), and c.38G>A mutation (codon 13 GGC>GAC; p.Gly13Asp)], which had been already diagnosed by Scorpion-ARMS technology, were also included in this study to validate the pyrosequencing method for the detection of KRAS mutations.
Genomic DNA Extraction
One pathologist marked the tumor areas on slides stained with hematoxylin-eosin. Genomic DNA was extracted from tumor lesions enriched with neoplastic cells, without adjacent normal tissue, using an FFPE kit (Qiagen, Valencia, CA). DNA was also extracted from 3 cell lines: Colo201 and HT29 with the BRAF mutation c.1799T>A (p.V600E), and HCT116 with wild-type BRAF using a QIAmp DNA mini kit (Qiagen, Valencia, CA).28,29 DNA was stored at −20 °C before use.
Whole Genome Amplification
Whole genome amplification (WGA) is a useful technique for preserving original study material for many different assays and for future studies. In WGA, genomic DNA is amplified by polymerase chain reaction (PCR) using primers consisting of a random sequence of 15 nucleotides. Each PCR mix contained 40 pmol of the random primers, 1.0 nmol each of dNTP, 2.0 mmol/L MgCl2, 1× PCR buffer (Applied Biosystems, Foster City, CA), 0.25 U of AmpliTaq Gold 360 (Applied Biosystems), and 5 μl of template DNA solution in a total volume of 50 μl. PCR conditions consisted of initial denaturation at 95 °C for 10 min; 50 cycles of 95 °C for 60 s, annealing (37 °C for 2 min), ramping from 37 to 55 °C (0.1 °C/s), 55 °C for 2 min, and 68 °C for 30 s; and a final extension at 72 °C for 7 min.
Pyrosequencing for KRAS and BRAF Mutations
Pyrosequencing technology has been shown to reliably detect KRAS mutations with 100 % analytic sensitivity and specificity, even when the proportion of mutant alleles is as low as 10 %. PCR amplification primers for pyrosequencing targeted for KRAS (codons 12, 13), BRAF (codon 600) were: KRAS-F, forward, 5′-NNNGGCCTGCTGAAAATGACTGAA-3′; and KRAS-R, reverse biotinylated primer, 5′-TTAGCTGTATCGTCAAGGCACTCT-3′; and BRAF-F, forward biotinylated primer, 5′-CAGTAAAAATAGGTGATTTTG-3′; and BRAF-R reverse, 5′-TCCAGACAACTGTTCAAACTGA-3′. Each PCR mix contained the forward and reverse primers (20 pmol each), 1.0 nmol of each dNTP with dUTP, 2 mmol/L MgCl2, 1× PCR buffer, 1.25 U of AmpliTaq Gold 360, 0.5 U of AmpErase UNG and 5 μl of template WGA product in a total volume of 50 μl. PCR condition consisted of initial denaturation at 50 °C (10 min) for AmpErase UNG; initial denaturation at 94 °C (10 min) for AmpliTaq Gold 360; 50 cycles of 95 °C (30 s), annealing (30 s; 55 °C for BRAF, 57 °C for KRAS), and 72 °C (30 s); and a final extension at 72 °C (7 min). The PCR products were electrophoresed through an agarose gel to confirm successful amplification of the 82-bp (KRAS) and the 80-bp PCR (BRAF) product.
KRAS and BRAF pyrosequencing was performed using the PyroMark Q24 System (Qiagen, Valencia, CA) according to the manufacturer’s instructions (Fig. 1 for KRAS; Fig. 2 for BRAF). All forward sequencing results were confirmed by reverse sequencing. In the KRAS pyrosequencing assay, the presence of a mutation was routinely confirmed by 3 different sequencing primers and by the creation of the frameshifted open reading frame of the mutant sequence relative to a wild-type sequence in a program. The primer KRAS-PF1 (5′-TGTGGTAGTTGGAGCTG-3′; pyrosequencing nucleotide dispensation order, ACTGATCG ATCGATCGATCGATCGATCG) could detect the c.35G>T (codon 12 GTT) and c.35G>A (codon 12 GAT) mutations. The primer KRAS-PF2 (5′-TGTGGTAGTTGGAGCT-3′; pyrosequencing nucleotide dispensation order, ATCGATCGATCGATCGATCGATCATCG) could detect the c.34G>T (codon 12 TGT) mutation. The primer KRAS-PF3 (5′-TGGTAGTTGGAGCTGGT-3′; pyrosequencing nucleotide dispensation order, GATGCATGCATGCATGCATGCATGCATGC) could detect the c.34G>A (codon 13 GAC) mutation. In the BRAF pyrosequencing assay, the primer was 5′-CACTCCATCGAGATTTC-3′, and the pyrosequencing nucleotide dispensation order was CTGCATGCATGCTGCA.
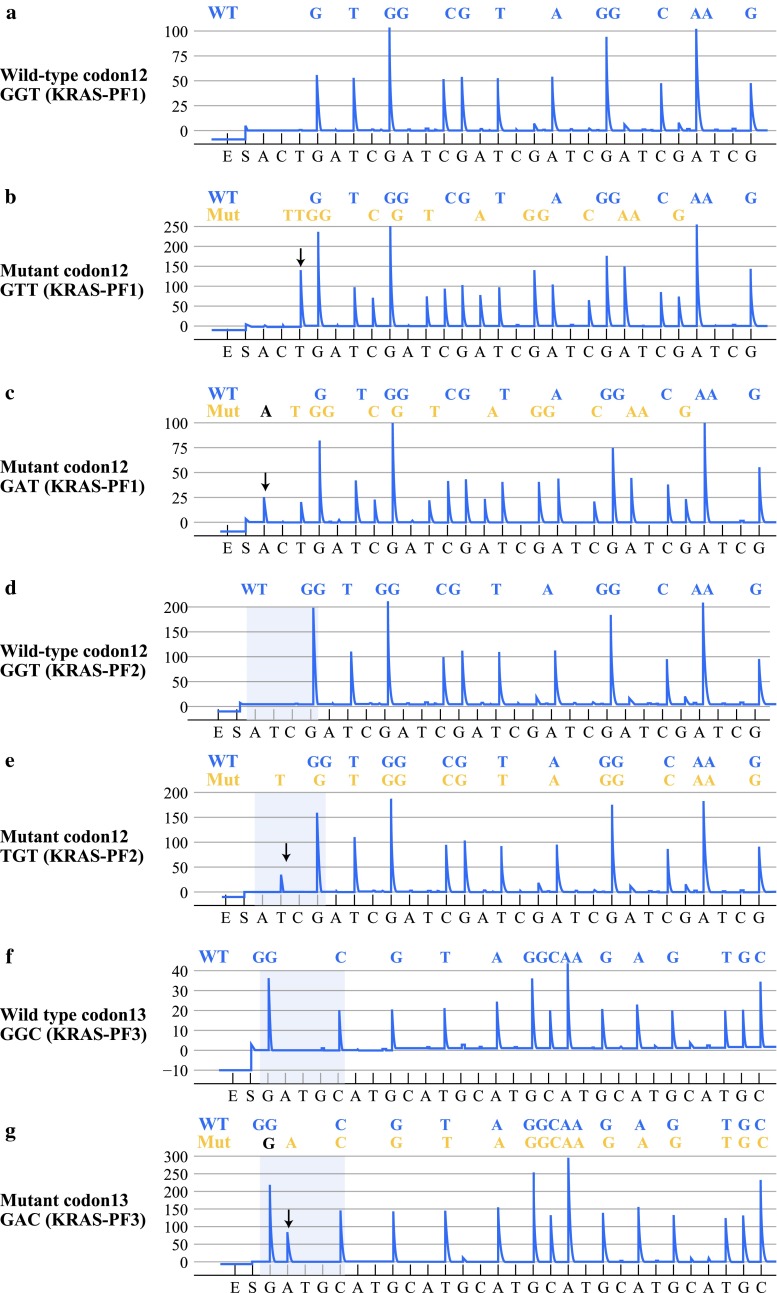
Fig. 1.
Pyrograms of wild-type and mutant KRAS in colorectal carcinoma. a Wild-type codon 12 detected by the KRAS-PF1 primer. b c.35GT (codon 12 GTT) mutation detected by the KRAS-PF1 primer. c c.35GA (codon 12 GAT) mutation detected by the KRAS-PF1 primer. d Wild-type codon 12 detected by the KRAS-PF2 primer. e c.34GT (codon 12 TGT) mutation detected by the KRAS-PF2 primer. f Wild-type codon 13 detected by the KRAS-PF3 primer. g c.38GA (codon 13 GAC) mutation detected by the KRAS-PF3 primer. Arrows indicate the presence of mutant alleles
Fig. 2.
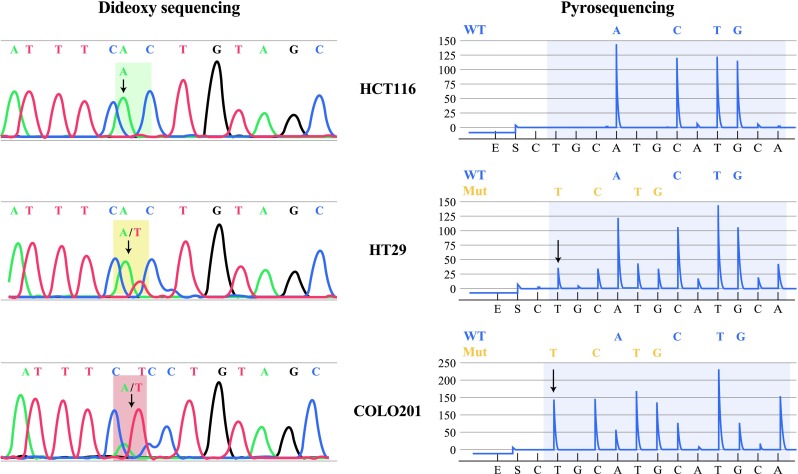
Detection of BRAF V600E in colon cancer cell lines by dideoxy sequencing and pyrosequencing; detection of homozygous wild type (HCT116), heterozygous mutant (HT29, COLO201). BRAF V600E variants identified by dideoxy sequencing (left panel) and pyrosequencing (right panel). The pyrosequencing nucleotide dispensation order is shown below each pyrogram. The numerical position for each nucleotide is indicated at the top. Arrows indicate the presence of mutant alleles
Results
Clinical Data
A summary of the clinical characteristics of the patients is given in Table 2. In this cohort, the 5-year overall survival rates of patients treated by esophagectomy were 83.9 % for stage I, 59.7 % for stage II, and 36.7 % for stage III. These rates are similar to those from the “Comprehensive Registry of Esophageal Cancer in Japan” (79.5 % for stage I, 58.9 % for stage II, and 39.8 % for stage III), which supports the absence of bias in our database.30
Table 2.
Patient characteristics
| Clinical characteristics | Total (N) |
|---|---|
| All cases | 203 |
| Mean age ± SD | 66.8 ± 9.24 |
| Sex | |
| Male | 178 (87.7 %) |
| Female | 25 (12.3 %) |
| Preoperative treatment | |
| Present | 68 (33.5 %) |
| Absent | 135 (66.5 %) |
| Cancer location | |
| Upper thoracic | 31 (15.3 %) |
| Middle thoracic | 105 (51.7 %) |
| Lower thoracic | 58 (28.6 %) |
| Esophagogastric junction | 9 (4.4 %) |
| Stage | |
| I (IA, IB) | 76 (37.4 %) |
| II (IIA, IIB) | 65 (32.0 %) |
| III (IIIA, IIIB, IIIC) | 62 (30.5 %) |
| Lymph node metastasis | |
| Positive | 94 (46.3 %) |
| Negative | 109 (53.7 %) |
| Histological grade | |
| G1 | 83 (40.9 %) |
| G2 | 82 (40.4 %) |
| G3–4 | 28 (13.8 %) |
Validation of the Pyrosequencing Assay for KRAS Mutation Detection
We first examined the validity of the pyrosequencing method using eleven colon cancer tissues harboring 4 different KRAS mutations [c.35G>T (codon 12 GGT>GTT; p.Gly12Val), c.35G>A (codon 12 GGT>GAT; p.Gly12Asp), c.34G>T (codon 12 GGT>TGT; p.Gly12Cys), and c.38G>A mutation (codon 13 GGC>GAC; p.Gly13Asp)]. These mutations had already been diagnosed by the sensitive Scorpion-ARMS technology [i.e., Amplification Refractory Mutation System incorporating a unique bifunctional florescent primer/probe molecule (Scorpion)]. As shown in Table 3, all 4 types of mutations could be detected using the pyrosequencing technology (11 of 11, 100 %) (Fig. 1), which demonstrated that the method was reliable for the detection of KRAS mutations in tumors.
Table 3.
Comparative analysis of Scorpion-ARMS and pyrosequencing for detection of KRAS mutation in 9 colorectal carcinoma tissues
| KRAS mutation type | Results of Scorpion-ARMS (n) | Results of pyrosequencing (n) | Concordance rate |
|---|---|---|---|
| Codon12 GAT | 3 | 3 | 100 % (3/3) |
| Codon12 GTG | 2 | 2 | 100 % (2/2) |
| Codon12 TGT | 2 | 2 | 100 % (2/2) |
| Codon13 GAC | 2 | 2 | 100 % (2/2) |
| Total | 9 | 9 | 100 % (9/9) |
KRAS Mutation in ESCC
Pyrosequence analysis of KRAS codon 12 and 13 mutations was successful for 203 of 204 (99.5 %) ESCC paraffin-embedded tissues. Among the 203 ESCCs, only 1 case harbored a KRAS mutation [c.34G>T (codon 12 GGT>TGT; p.Gly12Cys)]. This mutation was also diagnosed by the Scorpion-ARMS technology (data not shown). This result showed that the frequency of KRAS mutations in ESCC was extremely low.
Validation of Pyrosequencing Assay for BRAF Mutation Detection
To validate our BRAF pyrosequencing assay, both pyrosequencing and Sanger dideoxy sequencing were performed on the same set of DNA samples from 3 colon cancer cell lines (HCT116, Colo201, and HT29). HCT116 possesses wild-type BRAF, and Colo201 and HT29 harbors a BRAF mutation [c.1799T>A (p.V600E)]. In Sanger dideoxy sequencing, HCT116 showed was homozygous wild-type for the BRAF gene, and HT29 and COLO201 were shown to be heterozygous BRAF V600E mutants (Fig. 2). Among all 3 cell lines, pyrosequencing gave the same results as Sanger sequencing for BRAF mutational status (Fig. 2).
Furthermore, the BRAF mutation could be detected in paraffin-embedded tissues of colon cancer that had been previously diagnosed by Sanger dideoxy sequencing. Collectively, these preliminary experiments supported the validity of the pyrosequencing method for the detection of BRAF mutation in paraffin-embedded specimens.
BRAF Mutation in ESCC
The mutational status in BRAF exon 15 (V600E) was examined in 204 ESCCs, and valid results were obtained in 203 tissues (99.5 %). All 203 ESCCs were wild-type at codon 600 in BRAF exon 15.
Discussion
KRAS and BRAF mutational status could represent a predictive marker for anti-EGFR therapies; therefore, better understanding of the incidence of these mutations is important. However, the frequency of KRAS and BRAF mutations in ESCCs remains inconclusive because of the limited sample sizes of previous studies.8,15–20 The validity of the pyrosequencing method for detecting KRAS and BRAF mutations was initially demonstrated using colon cancer cell lines and colon cancer tissues harboring KRAS and BRAF mutations. Thereafter, KRAS mutations were shown to be extremely rare in a database of more than 200 ESCCs. In addition, the BRAF mutation was absent in ESCC tumors.
Pyrosequencing is a nonelectrophoretic nucleotide extension sequencing technology that can be used for mutation detection in tumors.21–24 Pyrosequencing offers a higher sensitivity than classical Sanger dideoxy sequencing for the detection of KRAS mutations.22 In addition, because of its simplicity and cost effectiveness, pyrosequencing represents a potentially useful method in molecular diagnostics and epidemiological studies, particularly in the setting of large-scale projects and clinical assays.25,26
Mutations in the KRAS gene occur early in the development of several types of cancers.31 Commonly restricted to codon 12 and 13 in exon 2, these mutations cause impaired GTPase activity and result in a continual stimulus for cellular proliferation. They have been found in more than 40 % of colorectal cancers, 90 % of pancreatic carcinomas, and 33 % of non-small-cell lung carcinomas.32 Importantly, KRAS mutation status has recently become an important biomarker when identifying resistance to anti-EGFR therapy. Several previous studies have examined the frequency of KRAS mutations in ESCCs; only 2 studies (N = 35 and 50) showed the presence of KRAS mutations in ESCCs; however, others have demonstrated the absence of KRAS mutation in ESCCs.8,15–19 Unfortunately, all previous studies were limited by small sample sizes (N ≤ 80). It should be noted that small studies are more prone to “publication bias” than large studies. The phenomenon of publication bias occurs because studies with null findings (e.g., absence of KRAS mutation in tumors) have a higher likelihood of being unwritten and unpublished compared with those with significant results (e.g., presence of KRAS mutation in tumors). Compared with small studies with null data, large studies with null data are more likely to be published. As a result, large studies are less prone to publication bias than small studies. Publishing null data from well-powered studies are important because publishing significant results from small underpowered studies also leads to publication bias. In this respect, our finding that KRAS mutations are extremely rare in a nonbiased database of more than 200 ESCCs may have considerable implications.
The BRAF gene encodes a serine/threonine kinase of the RAS RAF MEK MAPK signaling pathway and is mutated in a variety of cancer types.33 The V600E point mutation in exon 15 of the BRAF gene has been shown to be associated with insensitivity to antigrowth signals, cell-cycle dysregulation, tumor invasion and metastasis, escape from apoptosis, unlimited replicative potential and angiogenesis, and can be used as a predictive biomarker for BRAF-targeted therapy. In addition, the prognostic role of the BRAF mutation has been emphasized in several types of cancers.34–37 However, the incidence of the BRAF mutation in ESCC remains less clear. One study showed the absence of a BRAF mutation in 35 ESCCs, and the other showed that only 1 tumor harbored a BRAF mutation among 80 ESCC tumors. In this study, the BRAF mutation was absent in 203 ESCCs.
In summary, KRAS mutations were extremely rare, and the BRAF mutation was absent in a nonbiased database of 203 ESCCs. This suggests that KRAS and BRAF mutations play a limited role in the development of ESCC and that mutation analysis is not useful as a predictive marker for sensitivity to anti-EGFR therapy in ESCC.
Acknowledgment
This work was supported in part by the Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research, Grant No. 23591941.
References
- 1.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 2.Thallinger CM, Raderer M, Hejna M. Esophageal cancer: a critical evaluation of systemic second-line therapy. J Clin Oncol. 2011;29:4709–4714. doi: 10.1200/JCO.2011.36.7599. [DOI] [PubMed] [Google Scholar]
- 3.Stahl M. Is there any role for surgery in the multidisciplinary treatment of esophageal cancer? Ann Oncol. 2010;21(Suppl 7):vii283–vii285. doi: 10.1093/annonc/mdq294. [DOI] [PubMed] [Google Scholar]
- 4.Chen WH, Chao YK, Chang HK, Tseng CK, Wu YC, Liu YH, et al. Long-term outcomes following neoadjuvant chemoradiotherapy in patients with clinical T2N0 esophageal squamous cell carcinoma. Dis Esophagus. 2012;25:250–255. doi: 10.1111/j.1442-2050.2011.01243.x. [DOI] [PubMed] [Google Scholar]
- 5.Pantling AZ, Gossage JA, Mamidanna R, Newman G, Robinson A, Manifold DK, et al. Outcomes from chemoradiotherapy for patients with esophageal cancer. Dis Esophagus. 2011;24:172–176. doi: 10.1111/j.1442-2050.2010.01121.x. [DOI] [PubMed] [Google Scholar]
- 6.Martini M, Vecchione L, Siena S, Tejpar S, Bardelli A. Targeted therapies: how personal should we go? Nat Rev Clin Oncol. 2011;9:87–97. doi: 10.1038/nrclinonc.2011.164. [DOI] [PubMed] [Google Scholar]
- 7.McNamara MJ, Adelstein DJ. Current developments in the management of locally advanced esophageal cancer. Curr Oncol Rep. 2012;14:342–349. doi: 10.1007/s11912-012-0239-7. [DOI] [PubMed] [Google Scholar]
- 8.Lorenzen S, Schuster T, Porschen R, Al-Batran SE, Hofheinz R, Thuss-Patience P, et al. Cetuximab plus cisplatin-5-fluorouracil versus cisplatin-5-fluorouracil alone in first-line metastatic squamous cell carcinoma of the esophagus: a randomized phase II study of the Arbeitsgemeinschaft Internistische Onkologie. Ann Oncol. 2009;20:1667–1673. doi: 10.1093/annonc/mdp069. [DOI] [PubMed] [Google Scholar]
- 9.Ilson DH, Kelsen D, Shah M, Schwartz G, Levine DA, Boyd J, et al. A phase 2 trial of erlotinib in patients with previously treated squamous cell and adenocarcinoma of the esophagus. Cancer. 2011;117:1409–1414. doi: 10.1002/cncr.25602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adelstein DJ, Rodriguez CP, Rybicki LA, Ives DI, Rice TW. A phase II trial of gefitinib for recurrent or metastatic cancer of the esophagus or gastroesophageal junction. Invest New Drugs. 2010;30:1684–1689. doi: 10.1007/s10637-011-9736-z. [DOI] [PubMed] [Google Scholar]
- 11.Han W, Lo HW. Landscape of EGFR signaling network in human cancers: biology and therapeutic response in relation to receptor subcellular locations. Cancer Lett. 2012;318:124–134. doi: 10.1016/j.canlet.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 2009;9:489–499. doi: 10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
- 13.Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004;6:313–319. doi: 10.1016/j.ccr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 14.De Roock W, De Vriendt V, Normanno N, Ciardiello F, Tejpar S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011;12:594–603. doi: 10.1016/S1470-2045(10)70209-6. [DOI] [PubMed] [Google Scholar]
- 15.Ma H, Xue Y, Li C, Zhang J, Ren Z. A preliminary study on K-ras, EGFR, and B-raf mutations of esophageal squamous cell carcinoma. Chinese-German. J Clin Oncol. 2011;10:497–501. [Google Scholar]
- 16.Liu QW, Fu JH, Luo KJ, Yang HX, Wang JY, Hu Y, et al. Identification of EGFR and KRAS mutations in Chinese patients with esophageal squamous cell carcinoma. Dis Esophagus. 2011. doi:10.1111/j.1442-2050.2010.01155.x. [Epub ahead of print]. [DOI] [PubMed]
- 17.Hollstein MC, Peri L, Mandard AM, Welsh JA, Montesano R, Metcalf RA, et al. Genetic analysis of human esophageal tumors from two high incidence geographic areas: frequent p53 base substitutions and absence of ras mutations. Cancer Res. 1991;51:4102–4106. [PubMed] [Google Scholar]
- 18.Victor T, Du Toit R, Jordaan AM, Bester AJ, van Helden PD. No evidence for point mutations in codons 12, 13, and 61 of the ras gene in a high-incidence area for esophageal and gastric cancers. Cancer Res. 1990;50:4911–4914. [PubMed] [Google Scholar]
- 19.Hollstein MC, Smits AM, Galiana C, Yamasaki H, Bos JL, Mandard A, et al. Amplification of epidermal growth factor receptor gene but no evidence of ras mutations in primary human esophageal cancers. Cancer Res. 1988;48:5119–5123. [PubMed] [Google Scholar]
- 20.Maeng CH, Lee J, van Hummelen P, Park SH, Palescandolo E, Jang J, et al. High-throughput genotyping in metastatic esophageal squamous cell carcinoma identifies phosphoinositide-3-kinase and BRAF mutations. PLoS One. 2012;7:e41655. doi: 10.1371/journal.pone.0041655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fakhrai-Rad H, Pourmand N, Ronaghi M. Pyrosequencing: an accurate detection platform for single nucleotide polymorphisms. Hum Mutat. 2002;19:479–485. doi: 10.1002/humu.10078. [DOI] [PubMed] [Google Scholar]
- 22.Ogino S, Kawasaki T, Brahmandam M, Yan L, Cantor M, Namgyal C, et al. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen S, Qin D. Pyrosequencing data analysis software: a useful tool for EGFR, KRAS, and BRAF mutation analysis. Diagn Pathol. 2012;7:56. doi: 10.1186/1746-1596-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen DC, Saarela J, Nuotio I, Jokiaho A, Peltonen L, Palotie A. Comparison of GenFlex Tag array and Pyrosequencing in SNP genotyping. J Mol Diagn. 2003;5:243–249. doi: 10.1016/S1525-1578(10)60481-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imamura Y, Morikawa T, Liao X, Lochhead P, Kuchiba A, Yamauchi M, et al. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers. Clin Cancer Res. 2012;18:4753–4763. doi: 10.1158/1078-0432.CCR-11-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogino S, Meyerhardt JA, Irahara N, Niedzwiecki D, Hollis D, Saltz LB, et al. KRAS mutation in stage III colon cancer and clinical outcome following intergroup trial CALGB 89803. Clin Cancer Res. 2009;15:7322–7329. doi: 10.1158/1078-0432.CCR-09-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice TW, Blackstone EH, Rusch VW. 7th edition of the AJCC Cancer Staging Manual: esophagus and esophagogastric junction. Ann Surg Oncol. 2010;17:1721–1724. doi: 10.1245/s10434-010-1024-1. [DOI] [PubMed] [Google Scholar]
- 28.Brandi G, Tavolari S, De Rosa F, Di Girolamo S, Agostini V, Barbera MA, et al. Antitumoral efficacy of the protease inhibitor gabexate mesilate in colon cancer cells harbouring KRAS, BRAF and PIK3CA mutations. PLoS One. 2012;7:e41347. [DOI] [PMC free article] [PubMed]
- 29.Wang H, Daouti S, Li WH, Wen Y, Rizzo C, Higgins B, et al. Identification of the MEK1(F129L) activating mutation as a potential mechanism of acquired resistance to MEK inhibition in human cancers carrying the B-RafV600E mutation. Cancer Res. 2011;71:5535–5545. doi: 10.1158/0008-5472.CAN-10-4351. [DOI] [PubMed] [Google Scholar]
- 30.Ozawa S, Tachimori Y, Baba H, Fujishiro M, Matsubara H, Numasaki H, et al. Comprehensive registry of esophageal cancer in Japan, 2004. Esophagus. 2012;9:75–98. doi: 10.1007/s10388-012-0327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loriot Y, Mordant P, Deutsch E, Olaussen KA, Soria JC. Are RAS mutations predictive markers of resistance to standard chemotherapy? Nat Rev Clin Oncol. 2009;6:528–534. doi: 10.1038/nrclinonc.2009.106. [DOI] [PubMed] [Google Scholar]
- 32.Adjei AA. Blocking oncogenic Ras signaling for cancer therapy. J Natl Cancer Inst. 2001;93:1062–1074. doi: 10.1093/jnci/93.14.1062. [DOI] [PubMed] [Google Scholar]
- 33.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 34.Ogino S, Shima K, Meyerhardt JA, McCleary NJ, Ng K, Hollis D, et al. Predictive and prognostic roles of BRAF mutation in stage III colon cancer: results from intergroup trial CALGB 89803. Clin Cancer Res. 2012;18:890–900. doi: 10.1158/1078-0432.CCR-11-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grisham RN, Iyer G, Garg K, Delair D, Hyman DM, Zhou Q, et al. BRAF Mutation is associated with early stage disease and improved outcome in patients with low-grade serous ovarian cancer. Cancer. 2012. doi:10.1002/cncr.27782. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 37.Moreau S, Saiag P, Aegerter P, Bosset D, Longvert C, Hélias-Rodzewicz Z, et al. Prognostic value of BRAF (V600) mutations in melanoma patients after resection of metastatic lymph nodes. Ann Surg Oncol. 2012;19:4314–4321. doi: 10.1245/s10434-012-2457-5. [DOI] [PubMed] [Google Scholar]