Abstract
Chemical investigation of the tunicate Trididemnum solidum resulted in the isolation of two new chlorinated compounds belonging to the didemnin class, along with two known compounds didemnin A and didemnin B. The structural determination of the compounds was based on extensive NMR and mass spectroscopic analysis. The isolated compounds 1–4 were tested for their anti-inflammatory activity using in vitro assays for inhibition of inducible nitric oxide synthase (iNOS) and nuclear factor-kappa B (NF-κB) activity. The anti-cell proliferative activity of the above compounds was also evaluated against four solid tumor cell lines.
Keywords: chlorinated didemnins, anti-inflammatory activity, Trididemnum solidum
1. Introduction
Didemnins belonging to the class of depsipeptides were first isolated from the Caribbean tunicate Trididemnum solidum (Family Didemnidae) [1,2]. Didemnin B, a potent anticancer agent was the first marine natural product that entered into phase I clinical trials. Didemnin B exhibited a variety of biological activities which includes inhibition of the growth of both RNA and DNA viruses [3], in vitro and in vivo activity against B16 melanoma, in vivo activity against P388 leukemia and in vitro activity against L1210 lymphocytic leukemia [4,5]. In the course of our search to identify new drug candidates and targets, the tunicate Trididemnum solidum (KY10508001) was collected from the Little Cayman Island and the freeze dried sample was extracted with CH2Cl2/MeOH (1:1 v/v). The extract was found to exhibit strong anti-inflammatory activity using in vitro assays for inhibition of inducible nitric oxide synthase (iNOS) and nuclear factor-kappa B (NF-κB) activity. The IC50 values were 0.2 µg/mL and 0.4 µg/mL for inhibition of iNOS and NF-κB, respectively. The crude extract was further subjected to a series of chromatographic separations to yield the didemnin class of pure compounds (1–4), and the isolated compounds were tested for their in vitro anti-inflammatory and anticancer activities.
2. Results and Discussion
2.1. Bioassay-Guided Isolation
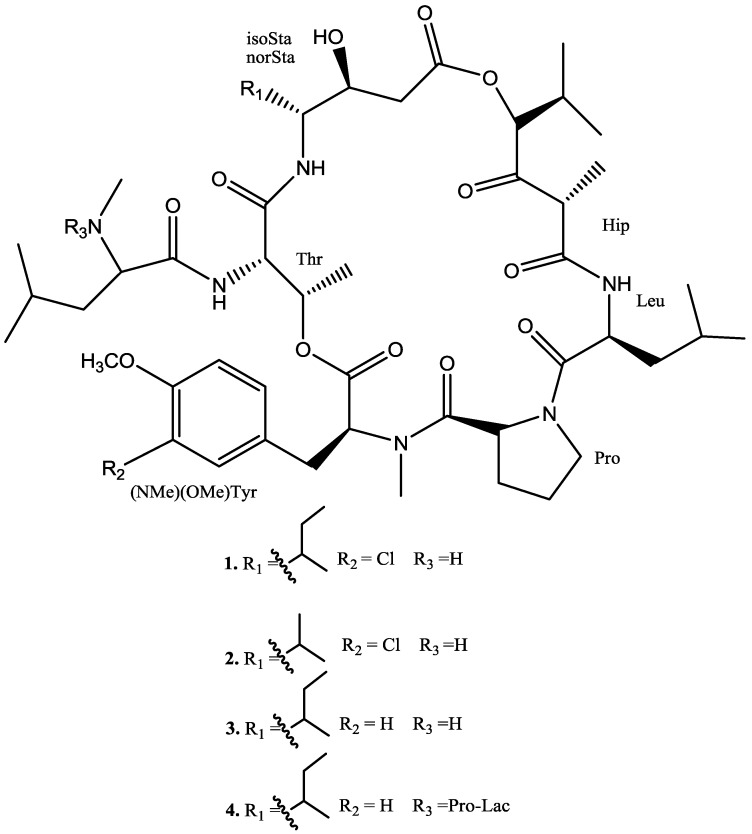
The crude DCM extract of the tunicate (5.5 g) was subjected to C18 flash column chromatography using water and methanol mixtures. Based on the anti-inflammatory activity, fraction D (IC50 of 0.14 µg/mL and 0.028 µg/mL for NF-κB and iNOS respectively) was further subjected to reversed phase HPLC (Phenomenex, Luna C18 (2)), using a gradient mixture (60:40 MeOH: H2O to 100% MeOH with 0.05% TFA over 65 min) to afford four pure compounds (1–4) (Figure 1).
Figure 1.
Structure of compounds 1–4.
2.2. Structural Elucidation of the New Compounds
Compound 1 was isolated as white solid with molecular formula of C49H77ClN6O12 as deduced by HRESIMS m/z 977.53505/979.53655 (actual 977.53662) corresponding to [M + H]+ ion. 49 resonances were observed in the 13C NMR spectrum (Supplementary Figure S1), which were identified to be 11× C (of which 8 were carbonyls); 17× CH; 8× CH2; 13× CH3 based on DEPT spectral data. Analysis of the 1H NMR data (Supplementary Figure S2) revealed the structural similarities of compound 1 to didemnin A [6]. Mass spectral data (Supplementary Figure S3) in combination with the 1H NMR spectrum (Table 1) indicated the presence of N,O-diMe-o-chlorotyrosine with signals at δ 7.02 (d), 112.7, 7.26 (d), 129.3, 7.43 (s), 131.5. HMBC correlations were observed from H-8 to C-6 & C-4; H-9 to C-7 & C-5; H-5 to C-7& C-9 indicating the position of chlorine to be ortho to the methoxy group on the tyrosine moiety. The structure of chlorinated tyrosine moiety was also confirmed by comparison of the data to the literature [7,8]. Analysis of the 1D (1H, 13C, DEPT) and 2D (COSY, HMQC, HMBC and ROESY) identified the rest of the amino acids as proline, leucine, threonine and Me-leucine. The stereochemistries of all the standard amino acids except for N-Me-d-leucine were determined to be l by LC-MS analysis (Supplementary Figure S4) of Marfey’s derivatives [9]. The stereochemistry of chloro N,O-di Me Tyr was determined by synthesis of the same using N,O-di Me-l-Tyr and sulfuryl chloride [10]. Comparison of spectral data of 1 with the literature [11,12] revealed the stereochemistry of isoSta and Hip moieties to be similar to that in didemninA. Thus, the structure of compound 1 was established as N,O-diMe-o-chlorotyrosine derivative of didemnin A.
Table 1.
13C NMR and 1H NMR data of compound 1 & 2 in d5-pyridine.
| 1 a | 2 b | ||||
|---|---|---|---|---|---|
| Subunit | C# | δC | δH (J in Hz) | δC | δH (J in Hz) |
| IsoSta or norSta | NH | - | 7.62 (d, 9.5) | - | 7.8 (d, 9.8) |
| CO | 169.1 | - | 169.7 | - | |
| C2H2 | 40.7 | 2.9 (m)/4.06 (d, 17.0) | 40.8 | 3.02 (m)/4.03 (d, 18.0) | |
| C3H | 67.1 | 4.68 (t, 10) | 67.6 | 4.69 (t, 10.2) | |
| C4H | 56.3 | 4.66 (brt, 9.5) | 57.7 | 4.51 (brt, 9.7) | |
| C5H | 34.5 | 2.51 (m) | 27.7 | 2.5 (m) | |
| C6H2/C6H3 | 28.1 | 1.45/1.73 (m) | 17.2 | 1.16 (d, 6.6) | |
| C7H3 | 12.1 | 1.07 (t, 7.5) | - | - | |
| C5CH3 | 14.6 | 1.18 (d, 7.0) | 22.1 | 1.22 (d, 6.3) | |
| Hip | CO | 172.2 | - | 172.5 | - |
| C2H | 49.4 | 4.27 * | 49.9 | 4.34 (q, 6.9) | |
| C3 | 204.6 | - | 208.8 | - | |
| C4H | 80.2 | 5.63 (d, 4.0) | 80.4 | 5.61 (d, 4.0) | |
| C5H | 29.6 | 2.39 m | 29.8 | 2.43 (m) | |
| C6H3 | 18.9 | 0.86 (d, 6.0) | 19.2 | 0.83 * | |
| C5CH3 | 16.9 | 0.84 (d, 6.0) | 17.1 | 0.85 * | |
| C2CH3 | 14.8 | 1.49 (d, 7.0) | 15.2 | 1.53 (d, 6.7) | |
| Leu | NH | - | 8.31 (d, 9.0) | - | 8.35 (d, 9.0) |
| CO | 171.2 | - | 171.4 | - | |
| CαH | 49.4 | 5.21 (brt, 9.5) | 49.8 | 5.22 (brt, 9.4) | |
| CβH2 | 41.8 | 1.96/1.68 (m) | 41.9 | 1.79/1.48 (m) | |
| CγH | 24.7 | 1.70 (m) * | 24.9 | 1.9 (m) | |
| CδH3 | 20.9 | 0.83 (d, 6.0) | 21.3 | 0.94 * | |
| Cδ′H3 | 22.0 | 1.03 (d, 6.0) | 21.5 | 1.03 * | |
| Pro | CO | 170.6 | - | 169.3 | - |
| CαH | 57.8 | 4.83 (m) | 58.8 | 4.83 (m) | |
| CβH2 | 27.7 | 1.96 (m),1.71 (m) | 27.9 | 2.04 (m), 1.78 (m) | |
| CγH2 | 24.8 | 1.63 (m), 1.91 (m) | 24.9 | 1.58 (m), 1.90 (m) | |
| CδH2 | 47.3 | 3.50 (m), 3.60 (m) | 47.5 | 3.50 (m), 3.58 (m) | |
| Cl-N,O-Me2Tyr | CO | 170.6 | - | 170.9 | - |
| NCH3 | 38.4 | 2.67 (s) | 38.8 | 2.66 (s) | |
| CαH | 65.3 | 4.28 * | 65.7 | 4.26 (dd, 9.5, 5.2) | |
| CβH2 | 33.9 | 3.50 (m) | 33.9 | 3.55 (m) | |
| Cγ | 131.6 | - | 131.2 | - | |
| CσH | 131.5 | 7.43 (s) | 131.8 | 7.44 (s) | |
| Cε | 122.1 | - | 122.4 | - | |
| Cξ | 154.1 | - | 154.5 | - | |
| CρH | 112.7 | 7.02 (d, 8.0) | 113.2 | 7.02 (d, 8.0) | |
| CψH | 129.3 | 7.26 (d, 8.0) | 129.7 | 7.28 (d, 8.0) | |
| OCH3 | 55.9 | 3.78 (s) | 56.3 | 3.80 (s) | |
| Thr | CO | 168.9 | - | 169.3 | - |
| NH | - | 10.3 (d, 6.0) | - | 10.3 (d, 6.1) | |
| CαH | 57.8 | 5.37 (brd, 6.5) | 58.1 | 5.38 (brd, 6.5) | |
| CβH | 70.4 | 5.74 (brd, 5.5) | 70.5 | 5.77 (brd, 6.3) | |
| Cβ-CH3 | 15.9 | 1.64 (d, 6.0) | 16.2 | 1.69 (d, 6.0) | |
| MeLeu | CO | 169.6 | - | 170.6 | - |
| NCH3 | 31.2 | 3.14 (s) | 31.8 | 3.15(s) | |
| CαH | 59.6 | 4.8 * | 59.1 | 4.84 (m) | |
| CβH2 | 39.5 | 2.15 (m) | 39.5 | 2.6 (m) | |
| CγH | 25.0 | 1.68 (m) | 25.2 | 1.8 (m) | |
| CδH3 | 23.4 | 0.84 (d, 6.0) | 23.7 | 0.94 (d, 5.6) | |
| Cδ′H3 | 21.8 | 0.92 (d, 6.0) | 22.9 | 1.03 * | |
a 500 MHz for 1H NMR and 125 MHz for 13C NMR; b 600 MHz for 1H NMR and 150 MHz for 13C NMR; * Multiplicity assignment not possible due to overlapping signals.
Compound 2 was also isolated as a white solid and had a molecular formula of C48H76ClN6O12 deduced by HERSIMS (Supplementary Figure S5) at m/z 963.52073/965.52101 (actual, 963.52097) corresponding [M + H]+ ion. Analysis of 1H and 13C NMR data revealed structural features similar to compound 1. The difference in mass by 14 amu’s compared to compound 1 indicated that compound 2 differed from compound 1 by a –CH2 unit. The absence of triplet methyl group (C7H3) from isoSta unit and the presence of a methyl doublet at δH 1.16 d indicated that the ethyl group in compound 1 was replaced by a methyl group. The stereochemistries of all the amino acids were same as in compound 1. Several nordidemnins were reported previously [11] and analysis of 2D NMR data indicated the presence of norStatine unit in compound 2. Thus compound 2 was identified as nor-N,O-diMe-o-chlorotyrosine derivative of didemnin A.
Compounds 3 & 4 were isolated as white solids and identified as didemnin A and didemnin B by comparison of spectral data to the literature [11,12,13].
The isolated compounds 1–4 were tested in a series of in vitro cell-based assays for their effects on selected targets involved in the process of inflammation and cancer. The results are presented in Table 2. Inflammation and oxidative stress are known to be linked to the development of many disorders such as cancer, organ damage, and neurodegenerative conditions. The NF-κB family of transcription factors plays a key role in inflammation, cell cycle regulation, apoptosis, and oncogenesis by controlling gene network expression [14,15]. The activation of NF-κB involves many cellular processes leading to inflammation and development of cancer [16,17]. In the assay for NF-κB activity, a luciferase construct with binding sites for specificity protein (SP-1) was used as a control because this transcription factor is relatively unresponsive to inflammatory mediators such as phorbol myristate acetate (PMA). Inducible nitric oxide synthase (iNOS) plays a key role in regulation of blood pressure, the immune system, infection, and inflammation [18]. Overproduction of nitric oxide (NO) by iNOS has been implicated in various pathological processes such as septic shock, inflammation, rheumatoid arthritis, cancer, and tissue damage [19,20]. The increase in NO response caused mainly by endotoxins and proinflammatory mediators such as lipopolysaccharide (LPS) can be reduced by anti-inflammatory agents acting as iNOS inhibitors. The isolated metabolites were also evaluated for their cytotoxicity towards four human solid tumor cell lines (SK-MEL: melanoma; KB: epidermal carcinoma; BT549: breast carcinoma and SK-OV-3: ovarian carcinoma) and non-cancer kidney cells (Vero: monkey kidney fibroblasts). The IC50 values for cell growth inhibition of compounds (1–4) are presented in Table 3.
Table 2.
Inhibition of Inducible Nitric Oxide Synthase (iNOS) and Nuclear Factor-Kappa B (NF-κB) activities by compounds (1–4) *.
| Compound | iNOS | NF-κB |
|---|---|---|
| 1 | 0.4 | 0.26 |
| 2 | 0.42 | 0.62 |
| 3 | 0.19 | 0.19 |
| 4 | 0.002 | 0.03 |
| Parthenolide α | 2.8 | 2.8 |
* IC50 values expressed in µM; α positive control.
Table 3.
Anti-cell proliferative activity of isolated compounds (1–4) *.
| Compound | SK-MEL | KB | BT-549 | SK-OV-3 | VERO |
|---|---|---|---|---|---|
| 1 | 0.12 | 0.26 | 0.16 | 0.26 | 4.8 |
| 2 | 0.06 | 0.42 | 0.16 | 0.38 | 2.08 |
| 3 | 0.055 | 0.16 | 0.07 | 0.16 | 4.78 |
| 4 | 0.022 | 0.09 | 0.02 | 0.1 | 0.15 |
| Doxorubicin α | 1.1 | 1.66 | 1.01 | 1.66 | 14 |
* IC50 values expressed in µM; α positive control.
All the compounds (1–4) showed strong activity against iNOS and NF-κB in LPS induced macrophages and PMA induced chondrocytes, respectively. The inhibition of NF-κB was not associated with the inhibition of SP-1 indicating that inhibition of NF-κB mediated transcription by these compounds was specific. Compound 4 was the most potent in inhibiting iNOS activity compared to 1–3, but its effect could be related to strong cytotoxicity towards all cell lines. All four compounds strongly inhibited cell proliferation of all four cancer cell lines in comparison to doxorubicin as the control drug (Table 3). Similar to the effect of doxorubicin, the compounds (1–3) were less toxic towards noncancerous cells (Vero cells) which were included for comparison. The strong anticancer activity can be partly explained due to a strong inhibition of NF-κB mediated transcription that may cause enhanced apoptosis and cell death [21].
2.3. Acid Hydrolysis and Marfey Analysis
Hydrolysis of 1 & 2 was achieved by dissolving 0.4 mg of each compound in 200 µL of 6 N HCl in a round bottom flask and stirred for 18 h on sand bath at 120 °C. The sample was dried and 1 N NaHCO3 (200 µL) and N-(3-fluoro-4,6-dinitrophyenyl)-l-alaninamide [FDAA, 50 µL (10 mg/mL solution in acetone)] were added and the mixture heated at 80 °C for 10 min. The reaction mixture was cooled to room temperature and neutralized with 2 N HCl (100 µL) and diluted with 1:1 CH3CN:H2O (300 µL). 10 µL of the FDAA derivative was analyzed by LCMS using a C18(2) column (Luna, 5 µm, 4.6 × 150 mm) and gradient (30:70 CH3CN: 20 mM ammonium formate both containing 0.1% formic acid to 70:30 over 25 min). The peaks were identified by injecting dl-mixture of Marfey’s derivatives of standard amino acids except for N,O-dimethyl tyrosine where only l amino acid was used. Retention times (min) for the amino acids were as follows: l-Leu (13.5), N-Me-d-Leu (15.1), l-Thr (6.27), l-Pro (9.9), o-Cl-N,O-dimethyl l-(Tyr) (14.1).
3. Experimental Section
3.1. General
Optical rotations were measured using a JASCO DIP-370 digital polarimeter. UV spectra were recorded on a Hewlett-Packard 8452A diode array spectrometer. NMR spectra were measured on Bruker Avance DRX-500 and Varian 600MHz spectrometer. 1H and 13C NMR spectra were measured and reported in ppm using d5-pyridine solvent peak (δH 8.74 and δC 150.4) as an internal standard. ESI-FTMS analyses were measured on a Bruker Magnex BioAPEX 30es ion cyclotron HR HPLC-FT spectrometer by direct injection into an electrospray interface. HPLC purifications were carried out on a Waters 2695 model system equipped with dual absorbance UV detector.
3.2. Tunicate Material
The tunicate (ID: KY10508001) was collected from Little Cayman Island at a depth of 10 m. A voucher specimen of the sample was deposited at the NOAA Ocean Biotechnology Center and Repository, Oxford, MS, USA.
3.3. Extraction and Isolation
Exactly 53.4 g of freeze-dried and finely ground tunicate material was exhaustively extracted with CH2Cl2/MeOH (1:1) to yield 5.5 g of the crude extract after concentration under reduced pressure. The crude extract was subjected to vacuum liquid chromatography on C18 column using gradient (100:0 to 0:100 H2O/MeOH). The 75% MeOH in water fraction (fraction D, 678.7 mg) which showed anti-inflammatory activity was further subjected to repeated reverse phased semi-preparative HPLC purification on Phenomenex, Luna, C18(2), 10 µL, 250 × 21.2 mm; flow rate, 10 mL/min, using a gradient of 40:60 (H2O:MeOH with 0.05% TFA) to 100% MeOH in 65 min. Peak 2 was identified as 3 (8.3 mg), peak 3 as 2 (4.5 mg), peak 4 as 1 ( 5.6 mg) and peak 10 as 4 (6.5 mg).
Compound 1: Colorless solid; 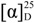 : −99.3 (c, 0.35, CHCl3); UV (MeOH) λmax (log ε) 208 (2.45) 232 (2.63); 1H- and 13C-NMR data, see Table 1; HRESIMS m/z [M + H]+ 977.53505/979.53655 (actual 977.53662) (calcd for C49H78ClN6O12+ 977.53662).
: −99.3 (c, 0.35, CHCl3); UV (MeOH) λmax (log ε) 208 (2.45) 232 (2.63); 1H- and 13C-NMR data, see Table 1; HRESIMS m/z [M + H]+ 977.53505/979.53655 (actual 977.53662) (calcd for C49H78ClN6O12+ 977.53662).
Compound 2: Colorless solid: 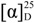 : −86.3 (c, 0.40, CHCl3); UV (MeOH) λmax (log ε) 206 (2.39) 230 (2.49); 1H- and 13C-NMR data, see Table 1; HERSIMS at m/z [M + H]+ 963.52073/965.52101 (calcd for C48H77 ClN6O12+ 963.52097).
: −86.3 (c, 0.40, CHCl3); UV (MeOH) λmax (log ε) 206 (2.39) 230 (2.49); 1H- and 13C-NMR data, see Table 1; HERSIMS at m/z [M + H]+ 963.52073/965.52101 (calcd for C48H77 ClN6O12+ 963.52097).
Compound 3: Colorless solid; 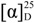 : −89.8 (c, 0.55, CHCl3); HRESIMS m/z 943.5461 [M + H]+ (calcd for C49H79N6O12+ 943.5755); 1H- and 13C-NMR data, see [12,13,14,15].
: −89.8 (c, 0.55, CHCl3); HRESIMS m/z 943.5461 [M + H]+ (calcd for C49H79N6O12+ 943.5755); 1H- and 13C-NMR data, see [12,13,14,15].
Compound 4: Colorless solid; 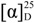 : −77.6 (c, 0.35, CHCl3); HRESIMS m/z 1112.6458 [M + H]+ (calcd for C57H90N7O15+ 1112.6494); 1H- and 13C-NMR data, see [12,13,14,15].
: −77.6 (c, 0.35, CHCl3); HRESIMS m/z 1112.6458 [M + H]+ (calcd for C57H90N7O15+ 1112.6494); 1H- and 13C-NMR data, see [12,13,14,15].
3.4. Biological Assays
Inhibition of NF-κB mediated transcription was determined in SW1353 cells by a reporter gene assay as described by Ma et al. [21]. Inhibition of iNOS activity was determined in RAW264.7 cells as described earlier [22]. In vitro cytotoxicity was determined against a panel of four solid tumor cell lines and compared with noncancerous kidney cells (Vero) [23]. Parthenolide and doxorubicin were used as positive controls for anti-inflammatory and cytotoxic activity, respectively.
4. Conclusions
The investigation of the tunicate Trididemnum solidum for bioactive natural products resulted in the isolation of two new chlorinated didemnin classes of compounds in addition to two known compounds. The recent reports of didemnin B from the α-proteobacterium Tistrella mobilis by Tsukimoto et al. [13], didemnin B and nordidemnin B from several species of Tistrella by Xu et al. [24] suggests that the chlorinated derivatives reported in this paper could also be of bacterial origin. Compound (1–4) exhibited significant inhibition of NF-κB and iNOS activity, as well as strong anticancer activity towards four cancer cell lines. Compound 1, the chlorinated derivative was less active compared to 3 in anti-inflammatory and cytotoxicity assays.
Acknowledgments
This work was supported by the National Institute for Undersea Science and Technology [NOAA, NIUST (NA16RU1496)], the United States Department of Agriculture (ARS Cooperative Agreement No. 58-6408-2-0009) and NIH COBRE grant number: P20GM104932. The authors thank Frank Wiggers of the National Center for Natural Products Research for providing NMR spectral data, Rama Sarma for his help in the synthesis of the chlorinated tyrosine derivative, Katherine Martin for technical assistance in bioassays. Samples were collected under a permit from the Cayman Islands Marine Conservation Board.
Supplementary Files
Supplementary Materials (PDF, 376 KB)
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Rinehart K.L., Jr., Gloer J.B., Hughes R.G., Jr., Renis H.E., McGovren J.P., Swynenberg E.B., Stringfellow D.A., Kuentzel S.L., Liv L.H. Didemnins: Antiviral and antitumor depsipeptides from a Caribbean tunicate. Science. 1981;212:933–935. doi: 10.1126/science.7233187. [DOI] [PubMed] [Google Scholar]
- 2.Rinehart K.L., Jr., Gloer J.B., Cook J.C., Jr., Mizsak S.A., Scahill T.A. Structures of the didemnins, antiviral and cytotoxic depsipeptides from a Caribbean tunicate. J. Am. Chem. Soc. 1981;103:1857–1859. doi: 10.1021/ja00397a055. [DOI] [Google Scholar]
- 3.Rinehart K.L., Jr., Gloer J.B., Wilson G.R., Hughes R.G., Jr., Li L.H., Renis H.E., McGovren J.P. Antiviral and antitumor compounds from tunicates. Fed. Proc. 1983;42:87–90. [PubMed] [Google Scholar]
- 4.Rinehart K.L., Jr. Peptides, Chemistry and Biology. ESCOM; Leiden, The Netherlands: 1988. Didemnin and Its Biological Properties; pp. 626–631. [Google Scholar]
- 5.Jouin P., Poncet J., Dufour M.N., Aumelas A., Pantaloni A., Cros S., Francois G. Antineoplastic activity of didemnin congeners: Nordidemnin and modified chain analogues. J. Med. Chem. 1991;34:486–491. doi: 10.1021/jm00106a002. [DOI] [PubMed] [Google Scholar]
- 6.Rinehart K.L., Kishore V., Bible K.C., Sakai R., Sullins D.W., Li K.M. Didemnins and tunichlorin: Novel natural products from the marine tunicate Trididemnum solidum. J. Nat. Prod. 1988;51:1–21. doi: 10.1021/np50055a001. [DOI] [PubMed] [Google Scholar]
- 7.Ishida K., Matsuda H., Murakami M., Yamaguchi K. Micropeptins 478-A and -B, plasmin inhibitors from the cyanobacterium Microcystis aeruginaosa. J. Nat. Prod. 1997;60:184–187. doi: 10.1021/np9606815. [DOI] [PubMed] [Google Scholar]
- 8.Apter S.G., Carmeli S. Protease inhibitors from a water bloom of the cyanobacterium Microcystis aeruginosa. J. Nat. Prod. 2009;72:1429–1436. doi: 10.1021/np900340t. [DOI] [PubMed] [Google Scholar]
- 9.Marfey P. Determination of d-amino acids. II. Use of a bifunctional reagent, 1,5-difluoro-2,4-dinitrobenzene. Carlsberg Re. Commun. 1984;49:591–596. doi: 10.1007/BF02908688. [DOI] [Google Scholar]
- 10.Gnaim J.M., Sheldon R.A. Selective ortho-chlorination of phenol using sulfuryl chloride in the presence of t-butylaminomethyl polystyrene as a heterogenous amine catalyst. Tetrahedron Lett. 2004;45:8471–8473. doi: 10.1016/j.tetlet.2004.09.102. [DOI] [Google Scholar]
- 11.Sakai R., Stroh J.G., Sullins D.W., Rinehart K. Seven new didemnins from the marine tunicate Trididemnum solidum. J. Am. Chem. Soc. 1995;117:3734–3748. doi: 10.1021/ja00118a010. [DOI] [Google Scholar]
- 12.Li W.R., Ewing W.R., Harris B.D., Joullie M.M. Total synthesis and structural investigations of Didemnins A, B and C. J. Am. Chem. Soc. 1990;112:7569–7672. [Google Scholar]
- 13.Tsukimoto M., Nagaoka M., Shishido Y., Fujimoto J., Nishisaka F., Matsumoto S., Harunari E., Imada C., Matsuzaki T. Bacterial production of the tunicate-derived antitumor cyclic depsipeptides didemnin B. J. Nat. Prod. 2011;74:2329–2331. doi: 10.1021/np200543z. [DOI] [PubMed] [Google Scholar]
- 14.Aggarwal B.B., Sethi G., Nair A., Ichikawa H. Nuclear factor-κB: A holy grail in cancer prevention and therapy. Curr. Signal Transduct. Ther. 2006;1:25–52. doi: 10.2174/157436206775269235. [DOI] [Google Scholar]
- 15.Melisi D., Chiao P.J. NF-kappa B as a target for cancer therapy. Expert Opin. Ther. Targets. 2007;11:133–144. doi: 10.1517/14728222.11.2.133. [DOI] [PubMed] [Google Scholar]
- 16.Brasier A.R. The NF-kappaB regulatory network. Cardiovasc. Toxicol. 2006;6:111–130. doi: 10.1385/CT:6:2:111. [DOI] [PubMed] [Google Scholar]
- 17.Navab M., Gharavi N., Watson A.D. Inflammation and metabolic disorders. Curr. Opin. Clin. Nutr. Metab. Care. 2008;11:459–464. doi: 10.1097/MCO.0b013e32830460c2. [DOI] [PubMed] [Google Scholar]
- 18.Yoo H.J., Kang H.J., Jung H.J., Kim K., Lim C.J., Park E.H. Anti-Inflammatory, anti-angiogenic and anti-nociceptive activities of Saururus chinensis extract. J. Ethnopharmacol. 2008;120:282–286. doi: 10.1016/j.jep.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 19.Lirk P., Hoffmann G., Rieder J. Inducible nitric oxide synthase—Time for reappraisal. Curr. Drug Targets Inflamm. Allergy. 2002;1:89–108. doi: 10.2174/1568010023344913. [DOI] [PubMed] [Google Scholar]
- 20.Kroncke K.D., Fehsel K., Kolb-Bachofen V. Inducible nitric oxide synthase in human diseases. Clin. Exp. Immunol. 1998;113:147–156. doi: 10.1046/j.1365-2249.1998.00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma G., Khan S.I., Benavides G., Schuhly W., Fischer N.H., Khan I.A., Pasco D.S. Inhibition of NF-kappaB-mediated transcription and induction of apoptosis by melampolides and repandolides. Cancer Chemother. Pharmacol. 2007;60:35–43. doi: 10.1007/s00280-006-0344-0. [DOI] [PubMed] [Google Scholar]
- 22.Quang D.N., Harinantenaina L., Nishizawa T., Hashimoto T., Kohchi C., Soma G., Asakawa Y. Inhibition of nitric oxide production in RAW 264.7 cells by azaphilones from xylariaceous fungi. Biol. Pharm. Bull. 2006;29:34–37. doi: 10.1248/bpb.29.34. [DOI] [PubMed] [Google Scholar]
- 23.Mustafa J., Khan S.I., Ma G., Walker L.A., Khan I.A. Synthesis and anticancer activities of fatty acid analogs of podophyllotoxin. Lipids. 2004;39:167–172. doi: 10.1007/s11745-004-1215-5. [DOI] [PubMed] [Google Scholar]
- 24.Xu Y., Kerten R.D., Nam S.-J., Lu L., Al-Wuwailem A.M., Zheng H., Fenical W., Dorrestein P.C., Moore S.B. Bacterial biosynthesis and maturation of the didemnin anti-cancer agents. J. Am. Chem. Soc. 2012;134:8625–8632. doi: 10.1021/ja301735a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials (PDF, 376 KB)