Abstract
Unexplained male infertility (UMI), the inability to reproduce despite having a normal sexual history, physical exam and semen analysis, can have a genetic origin. Currently, few diagnostic tools are available for detecting such genetic abnormalities. Karyotyping and fluorescence in situ hybridization (FISH) are respectively used for chromosomal alterations in somatic cells and sperm aneuploidy assessment. Gene sequencing and mutational analysis have been introduced for identifying specific mutations and polymorphisms. Other approaches to the molecular evaluation of spermatozoa are under investigation, including array comparative genomic hybridization and whole-genome sequencing and non-coding ribonucleic acid arrays. Although treating cytogenetic abnormalities and genetic aberrations is still out of reach, the integration of these novel techniques may unravel hidden genetic defects in UMI. Finally, a deeper understanding of the sperm epigenome might allow the development of therapies based on epigenome modifications. This review focuses on the genetic basis of UMI and highlights the current and future methods for the evaluation of genetic defects as they relate to UMI. Review of the literature was carried out using ScienceDirect, OVID, PubMed and MedLine search engines.
KEY WORDS: Diagnosis, epigenetics, genetics, genome, male infertility, unexplained infertility
INTRODUCTION
An estimated 15% of couples remain childless after 1 year of unprotected intercourse, which represents approximately 140 million people worldwide.[1,2] Concerning men in reproductive age, about 8% seek medical counseling for infertility-related problems.[3] Despite proper diagnostic work-up and as our knowledge of the events involved in normal conception is still limited, we fail to determine the cause of infertility in nearly half of these cases.[4] It is also difficult to determine a threshold to discriminate fertile from infertile ejaculates based on the results of the conventional semen analysis as approximately 40% of infertile men have normal parameters.[5,6] Likewise, it is impossible to make a final diagnosis of infertility based on the conventional approaches.[7,8]
When there is no identifiable cause and the results of the semen analysis are normal the patients are categorized as having unexplained male infertility (UMI).[9,10] Abnormalities are likely to be present, but are not detected by conventional approaches. Genetic defects may be partly or entirely involved in the true cause of infertility in such men. Since assisted reproductive technology (ART) is overall successful regardless of the underlying infertility cause, this is often the next step for many couples with unexplained infertility. However, genetically compromised spermatozoa used in ART have been associated with a wide range of adverse outcomes including abnormal embryo development that may either fail to implant or result in an increased risk of miscarriage and defects in the offspring.[11] Therefore, it is sound to determine the origin of the problem to allow appropriate counseling and management.
In this review, I first discuss the genetic disorders associated with UMI and then, I outline the testing performed to diagnose genetic conditions associated with UMI. Finally, I propose a workable plan for genetic evaluation of men with unexplained infertility and discuss the future perspective for diagnosis in line with the novel genomic platforms under investigation.
REVIEW CRITERIA
An extensive search of studies examining the relationship between genetics and UMI was performed using search engines such as ScienceDirect, OVID, PubMed and MedLine. The overall strategy for study identification and data extraction was based on the following key words: “Genetics,” “epigenetics,” “unexplained infertility,” “male infertility,” “diagnosis,” “infertile men,” “semen parameters,” “pregnancy rate” and the specific genetic tests. Articles published in English only were considered. Data that were solely published in conference or meeting proceedings, websites or books were not included. Websites and book-chapter citations providing conceptual content were used.
GENETIC DISORDERS IN UMI
Spermatogonial series are kept in a latent state inside the fetal testis until puberty when they increase in number by repeated mitotic divisions. Spermatogenesis starts in adolescence and is controlled by genetic factors. It has been estimated that 2000 genes are essential for the full process to be completed; of these, only 30 genes are present in the Y chromosome.[12,13]
Genetic abnormalities including chromosomal aberrations and monogenic diseases have been estimated to respond in 10-15% of infertility cases.[13] Infertile men with genetic alterations usually present with impaired spermatogenesis, genital structural abnormalities, reduced testicular size, hypogonadism and sperm dysfunction.[14] Although its prevalence is unknown, genetic abnormalities may also occur in males with UMI.[9,10,11,12,13,14,15]
Didactically, genetic abnormalities can be grouped into four main categories: (i) Chromosomal defects in the somatic cells; (ii) gene mutations and polymorphisms in the somatic cells; (iii) sperm chromosomal abnormalities; and (iv) epigenetic disorders. Although the first two categories affect men with abnormal genotypes in somatic cells, sperm chromosomal abnormalities can be originated from individuals with either abnormal or normal genotypes. Epigenetics, on the other hand, refers to the mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in deoxyribonucleic acid (DNA) sequence.
Chromosomal defects
Chromosomal defects are the most common genetic abnormalities in infertile males, accounting for 2.1-15.5% of cases.[16] Klinefelter syndrome, chromosome translocations, inversions and deletions fall in this category and the vast majority of affected individuals display severely compromised semen quality. Translocation carriers, however, may have varying sperm production phenotypes ranging from normal spermatogenesis to inability to produce spermatogonia.[17] Chromosomal translocations occur when non-homologous chromosomes exchange segments. Translocations involving the sex chromosomes and autosomes can be either balanced or unbalanced; most often they are either associated with severe sperm abnormalities or lethal for fetuses. Robertsonian translocation (RT), however, may account for few cases of unexplained infertility.[18]
RT represents a translocation category in which two acrocentric chromosomes fuse at the region next to the centromere causing loss of their short arms. The resulting balanced karyotype has only 45 chromosomes including the chromosome with the translocation, which is actually constituted by long arms of two chromosomes. As the short arms of the five acrocentric chromosomes (chromosomes 13, 14, 15, 21 and 22) harbor multiple copies of ribosomal ribonucleic acid (RNA), loss of their short arms is not harmful. RTs are the most frequent structural chromosome abnormalities in humans, affecting the fertility status of one in 1000 men.[19] Despite being estimated to 0.8% in subfertile males, its prevalence is nine times higher than the general population.[20] In heterozygous carriers, RT chromosomes and their acrocentric homologs may undergo either alternate segregation or adjacent segregation at meiosis. Alternate segregation results in the formation of balanced gametes carrying either RT or a normal karyotype, whereas adjacent segregation leads to the formation of aneuploidy gametes. Carriers of RT may exhibit normal phenotype but otherwise be infertile due to more or less severe sperm abnormalities.[16] There is also a risk of unbalanced gamete production that results in an increased risk of spontaneous abortion and unbalanced offspring. The most relevant clinical aspect involves the carriers of translocations in chromosome 21 as they are at risk of producing a child with Down Syndrome due to a 21q trisomy inheritance.
Gene mutations
Like chromosomal defects, gene mutations are usually related to severely abnormal sperm production phenotypes. Microdeletions in the Y chromosome Azoospermia factor region, mutations in the cystic fibrosis gene and mutations and polymorphisms of the androgen receptor gene are classic examples of genetic abnormalities in this disease category.[14,16] Abnormalities of interest in UMI include mutations of cation channel of sperm (CatSper) and sperm mitochondrial genes. The diagnosis of gene mutations can be made only by molecular genetic testing.
CatSper gene
Voltage-gated calcium channels (CatSper 1-4) and proton pumps located in the principal piece of the sperm flagellum regulates hyperactivation.[21,22] In humans, the extension of hyperactivated motility in a sperm population is positively correlated with the extent of zona pellucida binding, acrosome reaction, zona-free oocyte penetration and fertilization capacity in vitro.[23] The CatSper ion channel has been a recently discovered protein complex composed of 6 subunits. Among these, four are α subunits (CatSper 1-4) with calcium-selective pore while two are transmembrane proteins with large extracellular domains and unknown function (CatSper beta and CatSper gamma). CatSper 1, 2, 3 and 4 have been mapped to chromosomes 11q12.1, 15q13-q15, 5q31.2 and 1p35.3, respectively.[24] An abnormally low proportion of sperm exhibiting hyperactivation was found in UMI associated with CatSper1 gene mutations.[25] In mice, mutations in CatSper 1-4 ion channel proteins lead to infertility despite normal semen and testicular development.[26] CatSper-related male infertility is inherited in an autosomal recessive manner. CatSper1 mutations can be identified by sequencing analysis and testing is clinically available. In summary, mutations in the CatSper channel genes can be considered a potential cause in UMI.
Sperm mitochondrial deoxyribonucleic acid mutations
Sperm mitochondria are located around the mid-piece in a helical arrangement containing 1-2 mitochondrial deoxyribonucleic acid (mtDNA). Mitochondrial DNA encodes 37 genes that regulate oxidative phosphorylation. It differs from nuclear DNA in respect to replication, repair mechanism, genome packing and position. Unlike nuclear DNA, mtDNA is not protected by histones and physically associates with the inner mitochondrial membrane, where highly mutagenic oxygen radicals are generated by the respiratory chain.[27,28] The leakage of these free radicals makes mitochondria a major source of reactive oxygen species and may also explain why mtDNA is more prone to mutations than nuclear DNA.
Mitochondrial DNA polymerase gamma (POLG) is a key enzyme involved in the elongation and repair of mitochondrial DNA strands that encode for the POLG gene. Studies have shown an association between POLG gene polymorphisms and UMI.[29,30] Polymorphism of this gene might decrease sperm oocyte penetration and fertilization even when sperm parameters were normal. In one study, 14.3% of men with unexplained infertility had POLG gene polymorphisms compared with only 2.3% in the unselected control group and 0.9% in fertile controls.[30] Despite the yet undefined role of determining mtDNA mutations in UMI, this information may prove beneficial when recommending infertile couples for ART. POLG gene mutations can be detected using sequence analysis and testing is clinically available (www.transgenomic.com). Of note, prognosis for pregnancy is good in cases treated with ART since mtDNA is not transmitted to the offspring.[16]
Sperm chromosomal abnormalities
A threefold increase in the frequency of sperm aneuploidy is found in infertile men (around 3%) compared with fertile counterparts.[31] Sperm aneuploidy has been associated with severe sperm defects, unexplained infertility, recurrent miscarriage, in vitro fertilization (IVF) failure and increased risk of chromosome abnormalities in newborns.[31,32] In fact, sperm aneuploidy has been associated with paternally derived de novo chromosome abnormalities in embryos, fetuses and newborns conceived after intracytoplasmic sperm injection (ICSI). Such abnormalities occur in 2-3% of ICSI conceptions, which is three times greater than natural conceptions.[33,34] Although not a first-line investigation, measurement of chromosomal abnormalities in sperm may be indicative in selected cases of UMI.
Epigenetic disorders
Epigenetics applied to male infertility refers to all types of molecular information that are transmitted from the spermatozoa to the embryo. The epigenetic regulatory mechanisms required for proper embryogenesis include: (i) Functional role of centrosome; (ii) DNA methylation; (iii) histone modifications; (iv) chromatin remodeling; and (v) role of RNA transcripts and telomere length. Methylation is the best example of the sperm epigenetic contribution to the embryo as human embryos cannot develop unless paternal methylation is normal. DNA methylation is referred to as ‘imprinting’, and it determines, which genes from both parental and maternal genomes will be expressed in the embryo.[35] DNA imprinting regions are reset at every reproductive cycle thus allowing renewing of parental imprints in parental germ cells.[36] Activation of imprinting is a result of differential marking of DNA regions with histone modifications, methylation, or a combination of both, to allow only one allele to remain active.[36] Methylation occurs at the 5-carbon position of cytosines found in cytosine-phosphate-guanine dinucleotides (CpGs). CpGs are found in high concentrations near the gene promoter and are termed “CpGs islands”. Decreased methylation of the paternal IGF2/H19 imprinting control region 1 (ICR1) and GTL2 have been found in spermatozoa of men with disturbed spermatogenesis.[37] The degree of methylation in the IGF2/H19 ICR1 and mesoderm-specific transcript locus has been also associated with infertility.[38]
Histones also play an important role in the transmission of paternal epigenetic information. Histone covalent modifications are associated with several nuclear functions including transcriptional control, chromatin packaging and DNA methylation. During chromatin packaging, 85% of histones are replaced by protamines.[39] In an intermediate phase of the replacement process, transitional proteins (TP) are inserted into the chromatin structure. In mice, disruption of TP1 and TP2 encoding genes can produce infertile phenotypes, as well as unbalanced protamination ratios and early transcription of mRNA.[40,41] Histones are more easily extracted from DNA than protamines and histone-bound DNA is more susceptible to DNA-damaging agents than protamine counterparts.[42] If abnormally modified, histones are candidates for impeding normal embryogenesis.[43] Finally, telomeres have also been targeted as potential candidates to explain some infertile phenotypes. Its functions include protection of the genetic information encoded on the chromosome, localizing the chromosomes in the nucleus and supporting DNA replication.[35] Abnormal shortening of telomeres has been associated with male factor infertility.[44] Studies in mice have suggested that there is a protective mechanism which degrades spermatocytes with reduced telomere length to prevent their maturation.[45] If this process failed, spermatocytes with shortened telomeres would reach meiosis I, which is indicative they cleared this checkpoint without being degraded.[46] Despite these considerations, the role of telomere length in male infertility remains to be further investigated.
In summary, variations in the sperm epigenome may also contribute to male fertility by DNA protamination and methylation. It means that a series of epigenetic signals coming from the paternal chromatin are needed for the proper execution of the DNA-encoded genetic program. Genetic inheritance is thus much more complex than the transmission of the information provided exclusively by the paternal DNA.
GENETIC TESTING IN UMI
A series of tests can be used to identify genetic and epigenetic defects in UMI [Table 1]. At present the most common test used to evaluate the genetic status of such men is the cytogenetic analysis by karyotyping. Since the incidence of karyotype anomalies is inversely proportional to sperm concentration, abnormal results are found in less than 1% in UMI.[47] Karyotyping involves the collection of heparinized peripheral blood samples and the isolation of a plasma lymphocyte suspension. Lymphocytes are cultured to promote mitotic stimulation and then division is arrested at the metaphase.[16] A dye, often Giemsa (G-banding), is used to stain bands on the chromosomes. With the use of light microscopy to evaluate the appearance of chromosomes, karyotyping resolves variations in the DNA complement of ≥4 Mb.[48] Structural chromosome abnormalities such as translocations can be easily detected by cytogenetic techniques.[16]
Table 1.
Genetic tests in unexplained male infertility
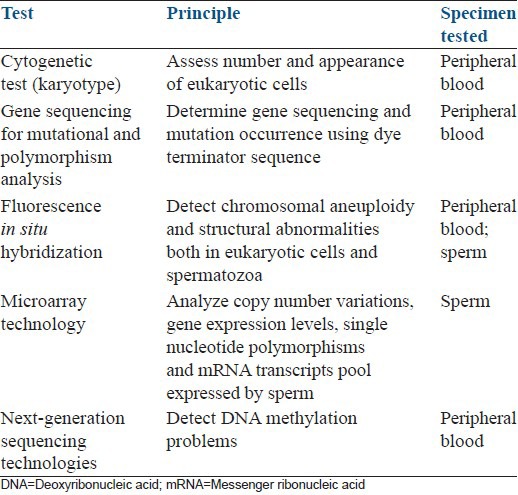
For specific gene sequencing and mutational analyses, the “dye terminator sequence” method is usually performed. It also involves the collection of peripheral blood. Its principle is the premature termination of four separate sequencing reactions that contain all standard deoxynucleotides (Desoxyadenosine triphosphate [dATP], Desoxyguanosine triphosphate [dGTP], Desoxycytidine triphosphate [dCTP] and Desoxythymidine triphosphate [dTTP]) and the DNA polymerase. Only one of the four dideoxynucleotides (ddATP, ddGTP, ddCTP, or ddTTP), which are chain-terminating nucleotides that lack the 3’-OH group required for the formation of a phosphodiester bond between two nucleotides, is added to each reaction. Thus, DNA strand extension is terminated, resulting in DNA fragments of varying lengths. Next, these labeled DNA fragments are separated by gel electrophoresis on a denaturing polyacrylamide-urea gel and read in a specific manner from the shortest to the longest.[14]
High resolution assessment of sperm genetic complement can be achieved using fluorescence in situ hybridization (FISH). It combines the classic karyotype method with molecular techniques using fluorescent DNA probes to bind selectively to a specific genetic sequence after denaturation. The fluorochromes are then visualized with the use of fluorescent microscopy. FISH can be used to detect chromosomal aneuploidy and structural abnormalities both in eukaryotic cells and sperm.[14,16] Unlike karyotype that requires metaphase cells, FISH can be applied to interphase nuclei. Studies from the last decade using FISH on sperm showed that the rate of sperm aneuploidy was inversely correlated to sperm quality.[49,50] The incidence of sperm disomy (two pairs of a single chromosome), diploidy (two pairs of all chromosomes) and polyploidy are also positively correlated to the occurrence of sperm morphology abnormalities including macrocephalic, multinucleate and multiflagellate sperm.[49,50] FISH of sperm is most often used in cases of recurrent miscarriage as it defines meiotic defects in the form of aneuploid sperm.[51]
The survey of sperm epigenome is carried out with the use of next-generation sequencing technologies. Bisulfite sequencing (Bi-seq), reduced-representation Bi-seq, methylated DNA immunoprecipitation sequencing, methylated DNA capture by affinity purification sequencing, methylated DNA binding domain sequencing and ethylation-sensitive restriction enzyme sequencing are the methods for detecting DNA methylation problems. Chromatin immunoprecipitation followed by sequencing is the standard approach used to detect histone-tail modifications while long non-coding RNAs can be detected using RNA-sequencing studies.[52]
CURRENT SCENARIO AND FUTURE PERSPECTIVES FOR THE GENETIC DIAGNOSIS IN UMI
Genetic testing not only allows clarifying an obscure infertility diagnosis, but also helps to prevent miscarriage and iatrogenic transmission of genetic defects to the offspring by ART. The most important strengths of genetic testing lie in its ability to identify men with genetically defective sperm thus aiding couples make informed reproductive decisions. In the past, only large structural chromosomal aberrations, defined by karyotype analysis, were detected. Using modern testing much smaller genomic regions has been found to be responsible for infertility. In fact, current estimates indicate that genetic abnormalities cause 15-30% of male factor infertility.[19]
Despite being limited by the widespread use of ART, a cost-effective genetic evaluation should be considered as an integral part of the work-up plan in UMI. Given the simplicity and low-cost of karyotype analysis, the test might be offered as a first-line investigation in cases of UMI, especially after ART failure or recurrent pregnancy loss. Although not a first line investigation, sperm aneuploidy assessment might also be considered in cases of UMI associated with IVF failures or recurrent pregnancy loss.
At present, the routine use of specific gene sequences, mutation analysis and sperm epigenome survey are limited by several factors including cost, availability, clinical relevance and endorsement by societies’ guidelines. In the near future, however, novel platforms that have the potential to redefine how male infertility is diagnosed are likely to become widely available. Microarray technology, which evaluates for copy number variations, gene expression levels and single nucleotide polymorphisms, has yielded several male fertility gene candidates with strong association with infertility and sperm quality and function.[53,54,55,56] An example of its application is the array comparative genomic hybridization to assess the relative quantities of DNA between samples, which allows to determine gene copy number as a function of chromosomal location.[48] In a recent study, the genetic sperm expression profile was used to classify the fertility status of men with normal sperm parameters based on the expression signature of four genes (EIF5A, RPLI3, RPL23A, RPS27A). The authors found that such analysis was able to predict the chances of pregnancy in intrauterine insemination with sensitivity and specificity of 82% and 90%, respectively.[57] Microarray technologies have also enabled evaluation of sperm messenger RNA that correlate with spermatogenesis, sperm motility, germ cell antiapoptotic processes, DNA repair, oxidative stress reduction and histone modification.[54,58,59,60,61] Recent investigations have shown that the mRNA profiles differ in sperm that succeed or fail to result in pregnancies in ART.[58,60,61]
Concerning sperm epigenetics, there is still a lot to be learned but this field bears the promise of reversing the effect due to its dynamic nature. Unlike genetic studies, a deeper understanding of the epigenetic processes could shed light on therapies based on epigenome modifications.
CONCLUSIONS
Male fertility, including spermatogenesis and sperm function, is regulated by thousands of genes. As men with unexplained infertility can harbor genetic abnormalities that may compromise their reproductive potential, efforts should be made to identify such conditions. Currently, few diagnostic tools are available for routine use and their usefulness is not yet completely clear. Chromosomal abnormalities in somatic cells can be detected by karyotyping. Sperm aneuploidy assessment in couples with unexplained infertility experiencing either repeated IVF failures or recurrent pregnancy loss can be performed by FISH while mutations and polymorphisms are identified by specific gene sequencing and mutational analysis methods. Many advances are currently being made and the use of novel genetic microarray technology may hold the key to more accurately diagnosing and treating men with unexplained infertility.
ACKNOWLEDGMENTS
The author is grateful to Dr. Ricardo Miyaoka for his help in the acquisition of data and Mrs. Fabiola C. Bento for language revision.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Cambridge: Cambridge University Press; 2000. World Health Organization. WHO Manual for Standardised Investigation and Diagnosis of the Infertile Couple. [Google Scholar]
- 2.Vital and Health Statistics, Series 23, No. 26, CDC. [Last accessed on 2013 Aug 30]. Available from: http://www.cdc.gov .
- 3.RightDiagnosis.com. Statistics by Country for Infertility. Health Grades Inc. [Last accessed on 2013 Aug 30]. Available from: http://www.righdiagnosis.com/i/infertility/stats-country.htm .
- 4.Kamel RM. Management of the infertile couple: An evidence-based protocol. Reprod Biol Endocrinol. 2010;8:21. doi: 10.1186/1477-7827-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esteves SC, Zini A, Aziz N, Alvarez JG, Sabanegh ES, Jr, Agarwal A. Critical appraisal of world health organization's new reference values for human semen characteristics and effect on diagnosis and treatment of subfertile men. Urology. 2012;79:16–22. doi: 10.1016/j.urology.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 6.van der Steeg JW, Steures P, Eijkemans MJ, F Habbema JD, Hompes PG, Kremer JA, et al. Role of semen analysis in subfertile couples. Fertil Steril. 2011;95:1013–9. doi: 10.1016/j.fertnstert.2010.02.024. [DOI] [PubMed] [Google Scholar]
- 7.Rybar R, Markova P, Veznik Z, Faldikova L, Kunetkova M, Zajicova A, et al. Sperm chromatin integrity in young men with no experiences of infertility and men from idiopathic infertility couples. Andrologia. 2009;41:141–9. doi: 10.1111/j.1439-0272.2008.00905.x. [DOI] [PubMed] [Google Scholar]
- 8.Guzick DS, Overstreet JW, Factor-Litvak P, Brazil CK, Nakajima ST, Coutifaris C, et al. Sperm morphology, motility, and concentration in fertile and infertile men. N Engl J Med. 2001;345:1388–93. doi: 10.1056/NEJMoa003005. [DOI] [PubMed] [Google Scholar]
- 9.Hamada A, Esteves SC, Nizza M, Agarwal A. Unexplained male infertility: Diagnosis and management. Int Braz J Urol. 2012;38:576–94. doi: 10.1590/s1677-55382012000500002. [DOI] [PubMed] [Google Scholar]
- 10.Sigman M, Lipshultz L, Howard S. Office evaluation of the subfertile male. In: Lipshultz LI, Howards SS, Niederberger CS, editors. Infertility in the Male. 4th ed. Cambridge: Cambridge University Press; 2009. pp. 153–76. [Google Scholar]
- 11.Aitken RJ, Koopman P, Lewis SE. Seeds of concern. Nature. 2004;432:48–52. doi: 10.1038/432048a. [DOI] [PubMed] [Google Scholar]
- 12.Hamada A, Esteves SC, Agarwal A. Genetics and male infertility. In: Dubey AK, editor. Infertility, Diagnosis, Management and IVF. 1st ed. New Delhi: Jaypee Medical Publishers Inc; 2012. pp. 113–57. [Google Scholar]
- 13.Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–7. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 14.Hargreave TB. Genetic basis of male fertility. Br Med Bull. 2000;56:650–71. doi: 10.1258/0007142001903454. [DOI] [PubMed] [Google Scholar]
- 15.Matzuk MM, Lamb DJ. Genetic dissection of mammalian fertility pathways. Nat Cell Biol. 2002;4(Suppl):s41–9. doi: 10.1038/ncb-nm-fertilityS41. [DOI] [PubMed] [Google Scholar]
- 16.Dada R, Thilagavathi J, Venkatesh S, Esteves SC, Agarwal A. Genetic testing in male infertility. Open Reprod Sci J. 2011;3:42–56. [Google Scholar]
- 17.Georgiou I, Syrrou M, Pardalidis N, Karakitsios K, Mantzavinos T, Giotitsas N, et al. Genetic and epigenetic risks of intracytoplasmic sperm injection method. Asian J Androl. 2006;8:643–73. doi: 10.1111/j.1745-7262.2006.00231.x. [DOI] [PubMed] [Google Scholar]
- 18.Conn CM, Cozzi J, Harper JC, Winston RM, Delhanty JD. Preimplantation genetic diagnosis for couples at high risk of Down syndrome pregnancy owing to parental translocation or mosaicism. J Med Genet. 1999;36:45–50. [PMC free article] [PubMed] [Google Scholar]
- 19.Ferlin A, Raicu F, Gatta V, Zuccarello D, Palka G, Foresta C. Male infertility: Role of genetic background. Reprod Biomed Online. 2007;14:734–45. doi: 10.1016/s1472-6483(10)60677-3. [DOI] [PubMed] [Google Scholar]
- 20.De Braekeleer M, Dao TN. Cytogenetic studies in male infertility: A review. Hum Reprod. 1991;6:245–50. [PubMed] [Google Scholar]
- 21.Lishko PV, Kirichok Y. The role of Hv1 and CatSper channels in sperm activation. J Physiol. 2010;588:4667–72. doi: 10.1113/jphysiol.2010.194142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlson AE, Burnett LA, del Camino D, Quill TA, Hille B, Chong JA, et al. Pharmacological targeting of native CatSper channels reveals a required role in maintenance of sperm hyperactivation. PLoS One. 2009;4:e6844. doi: 10.1371/journal.pone.0006844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esteves SC, Verza S., Jr Relationship of in vitro acrosome reaction to sperm function: An update. Open Reprod Sci J. 2011;3:72–84. [Google Scholar]
- 24.Wang H, Liu J, Cho KH, Ren D. A novel, single, transmembrane protein CATSPERG is associated with CATSPER1 channel protein. Biol Reprod. 2009;81:539–44. doi: 10.1095/biolreprod.109.077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hildebrand MS, Avenarius MR, Fellous M, Zhang Y, Meyer NC, Auer J, et al. Genetic male infertility and mutation of CATSPER ion channels. Eur J Hum Genet. 2010;18:1178–84. doi: 10.1038/ejhg.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi H, Moran MM, Navarro B, Chong JA, Krapivinsky G, Krapivinsky L, et al. All four CatSper ion channel proteins are required for male fertility and sperm cell hyperactivated motility. Proc Natl Acad Sci U S A. 2007;104:1219–23. doi: 10.1073/pnas.0610286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venkatesh S, Deecaraman M, Kumar R, Shamsi MB, Dada R. Role of reactive oxygen species in the pathogenesis of mitochondrial DNA (mtDNA) mutations in male infertility. Indian J Med Res. 2009;129:127–37. [PubMed] [Google Scholar]
- 28.Richter C, Suter M, Walter PB. Mitochondrial free radical damage and DNA repair. Biofactors. 1998;7:207–8. doi: 10.1002/biof.5520070308. [DOI] [PubMed] [Google Scholar]
- 29.Rovio AT, Marchington DR, Donat S, Schuppe HC, Abel J, Fritsche E, et al. Mutations at the mitochondrial DNA polymerase (POLG) locus associated with male infertility. Nat Genet. 2001;29:261–2. doi: 10.1038/ng759. [DOI] [PubMed] [Google Scholar]
- 30.Jensen M, Leffers H, Petersen JH, Nyboe Andersen A, Jørgensen N, Carlsen E, et al. Frequent polymorphism of the mitochondrial DNA polymerase gamma gene (POLG) in patients with normal spermiograms and unexplained subfertility. Hum Reprod. 2004;19:65–70. doi: 10.1093/humrep/deh038. [DOI] [PubMed] [Google Scholar]
- 31.Moosani N, Pattinson HA, Carter MD, Cox DM, Rademaker AW, Martin RH. Chromosomal analysis of sperm from men with idiopathic infertility using sperm karyotyping and fluorescence in situ hybridization. Fertil Steril. 1995;64:811–7. doi: 10.1016/s0015-0282(16)57859-5. [DOI] [PubMed] [Google Scholar]
- 32.Tempest HG, Martin RH. Cytogenetic risks in chromosomally normal infertile men. Curr Opin Obstet Gynecol. 2009;21:223–7. doi: 10.1097/GCO.0b013e32832947c2. [DOI] [PubMed] [Google Scholar]
- 33.Bonduelle M, Van Assche E, Joris H, Keymolen K, Devroey P, Van Steirteghem A, et al. Prenatal testing in ICSI pregnancies: Incidence of chromosomal anomalies in 1586 karyotypes and relation to sperm parameters. Hum Reprod. 2002;17:2600–14. doi: 10.1093/humrep/17.10.2600. [DOI] [PubMed] [Google Scholar]
- 34.Devroey P, Van Steirteghem A. A review of ten years experience of ICSI. Hum Reprod Update. 2004;10:19–28. doi: 10.1093/humupd/dmh004. [DOI] [PubMed] [Google Scholar]
- 35.Emery BR, Carrell DT. The effect of epigenetic sperm abnormalities on early embryogenesis. Asian J Androl. 2006;8:131–42. doi: 10.1111/j.1745-7262.2006.00127.x. [DOI] [PubMed] [Google Scholar]
- 36.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–93. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi H, Sato A, Otsu E, Hiura H, Tomatsu C, Utsunomiya T, et al. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet. 2007;16:2542–51. doi: 10.1093/hmg/ddm187. [DOI] [PubMed] [Google Scholar]
- 38.Poplinski A, Tüttelmann F, Kanber D, Horsthemke B, Gromoll J. Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/H19 ICR1. Int J Androl. 2010;33:642–9. doi: 10.1111/j.1365-2605.2009.01000.x. [DOI] [PubMed] [Google Scholar]
- 39.Carrell DT, Emery BR, Hammoud S. Altered protamine expression and diminished spermatogenesis: What is the link? Hum Reprod Update. 2007;13:313–27. doi: 10.1093/humupd/dml057. [DOI] [PubMed] [Google Scholar]
- 40.Yu YE, Zhang Y, Unni E, Shirley CR, Deng JM, Russell LD, et al. Abnormal spermatogenesis and reduced fertility in transition nuclear protein 1-deficient mice. Proc Natl Acad Sci U S A. 2000;97:4683–8. doi: 10.1073/pnas.97.9.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho C, Jung-Ha H, Willis WD, Goulding EH, Stein P, Xu Z, et al. Protamine 2 deficiency leads to sperm DNA damage and embryo death in mice. Biol Reprod. 2003;69:211–7. doi: 10.1095/biolreprod.102.015115. [DOI] [PubMed] [Google Scholar]
- 42.Yamauchi Y, Shaman JA, Ward WS. Non-genetic contributions of the sperm nucleus to embryonic development. Asian J Androl. 2011;13:31–5. doi: 10.1038/aja.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nanassy L, Carrell DT. Paternal effects on early embryogenesis. J Exp Clin Assist Reprod. 2008;5:2. doi: 10.1186/1743-1050-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zalenskaya IA, Zalensky AO. Telomeres in mammalian male germline cells. Int Rev Cytol. 2002;218:37–67. doi: 10.1016/s0074-7696(02)18011-9. [DOI] [PubMed] [Google Scholar]
- 45.Hemann MT, Rudolph KL, Strong MA, DePinho RA, Chin L, Greider CW. Telomere dysfunction triggers developmentally regulated germ cell apoptosis. Mol Biol Cell. 2001;12:2023–30. doi: 10.1091/mbc.12.7.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu L, Blasco M, Trimarchi J, Keefe D. An essential role for functional telomeres in mouse germ cells during fertilization and early development. Dev Biol. 2002;249:74–84. doi: 10.1006/dbio.2002.0735. [DOI] [PubMed] [Google Scholar]
- 47.Chandley AC, Edmond P, Christie S, Gowans L, Fletcher J, Frackiewicz A, et al. Cytogenetics and infertility in man. I. Karyotype and seminal analysis: Results of a five-year survey of men attending a subfertility clinic. Ann Hum Genet. 1975;39:231–54. doi: 10.1111/j.1469-1809.1975.tb00126.x. [DOI] [PubMed] [Google Scholar]
- 48.Kovac JR, Pastuszak AW, Lamb DJ. The use of genomics, proteomics, and metabolomics in identifying biomarkers of male infertility. Fertil Steril. 2013;99:998–1007. doi: 10.1016/j.fertnstert.2013.01.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tempest HG, Griffin DK. The relationship between male infertility and increased levels of sperm disomy. Cytogenet Genome Res. 2004;107:83–94. doi: 10.1159/000079575. [DOI] [PubMed] [Google Scholar]
- 50.Lewis-Jones I, Aziz N, Seshadri S, Douglas A, Howard P. Sperm chromosomal abnormalities are linked to sperm morphologic deformities. Fertil Steril. 2003;79:212–5. doi: 10.1016/s0015-0282(02)04411-4. [DOI] [PubMed] [Google Scholar]
- 51.Bernardini LM, Costa M, Bottazzi C, Gianaroli L, Magli MC, Venturini PL, et al. Sperm aneuploidy and recurrent pregnancy loss. Reprod Biomed Online. 2004;9:312–20. doi: 10.1016/s1472-6483(10)62147-5. [DOI] [PubMed] [Google Scholar]
- 52.Bell JT, Spector TD. A twin approach to unraveling epigenetics. Trends Genet. 2011;27:116–25. doi: 10.1016/j.tig.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park JH, Lee HC, Jeong YM, Chung TG, Kim HJ, Kim NK, et al. MTHFR C677T polymorphism associates with unexplained infertile male factors. J Assist Reprod Genet. 2005;22:361–8. doi: 10.1007/s10815-005-6795-0. [DOI] [PubMed] [Google Scholar]
- 54.Garrido N, Martínez-Conejero JA, Jauregui J, Horcajadas JA, Simón C, Remohí J, et al. Microarray analysis in sperm from fertile and infertile men without basic sperm analysis abnormalities reveals a significantly different transcriptome. Fertil Steril. 2009;91:1307–10. doi: 10.1016/j.fertnstert.2008.01.078. [DOI] [PubMed] [Google Scholar]
- 55.Matzuk MM, Lamb DJ. The biology of infertility: Research advances and clinical challenges. Nat Med. 2008;14:1197–213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang I, Emery BR, Christensen GL, Griffin J, Peterson CM, Carrell DT. Novel UBE2B-associated polymorphisms in an azoospermic/oligozoospermic population. Asian J Androl. 2008;10:461–6. doi: 10.1111/j.1745-7262.2008.00386.x. [DOI] [PubMed] [Google Scholar]
- 57.Bonache S, Mata A, Ramos MD, Bassas L, Larriba S. Sperm gene expression profile is related to pregnancy rate after insemination and is predictive of low fecundity in normozoospermic men. Hum Reprod. 2012;27:1556–67. doi: 10.1093/humrep/des074. [DOI] [PubMed] [Google Scholar]
- 58.García-Herrero S, Meseguer M, Martínez-Conejero JA, Remohí J, Pellicer A, Garrido N. The transcriptome of spermatozoa used in homologous intrauterine insemination varies considerably between samples that achieve pregnancy and those that do not. Fertil Steril. 2010;94:1360–73. doi: 10.1016/j.fertnstert.2009.07.1671. [DOI] [PubMed] [Google Scholar]
- 59.Montjean D, De La Grange P, Gentien D, Rapinat A, Belloc S, Cohen-Bacrie P, et al. Sperm transcriptome profiling in oligozoospermia. J Assist Reprod Genet. 2012;29:3–10. doi: 10.1007/s10815-011-9644-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garrido N, García-Herrero S, Meseguer M. Assessment of sperm using mRNA microarray technology. Fertil Steril. 2013;99:1008–22. doi: 10.1016/j.fertnstert.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 61.García-Herrero S, Garrido N, Martínez-Conejero JA, Remohí J, Pellicer A, Meseguer M. Differential transcriptomic profile in spermatozoa achieving pregnancy or not via ICSI. Reprod Biomed Online. 2011;22:25–36. doi: 10.1016/j.rbmo.2010.09.013. [DOI] [PubMed] [Google Scholar]


