Abstract
Accurate diagnosis of thyroid tumors is challenging. A particular problem is distinguishing between follicular thyroid carcinoma (FTC) and benign follicular thyroid adenoma (FTA), where histology of fine-needle aspirates is not conclusive. It is often necessary to remove healthy thyroid to rule out carcinoma. In order to find markers to improve diagnosis, we quantified gene transcript expression from FTC, FTA, and normal thyroid, revealing 73 differentially expressed transcripts (P ≤ 0.0001). Using an independent set of 23 FTCs, FTAs, and matched normal thyroids, 17 genes with large expression differences were tested by real-time RT-PCR. Four genes (DDIT3, ARG2, ITM1, and C1orf24) differed between the two classes FTC and FTA, and a linear combination of expression levels distinguished FTC from FTA with an estimated predictive accuracy of 0.83. Furthermore, immunohistochemistry for DDIT3 and ARG2 showed consistent staining for carcinoma in an independent set 59 follicular tumors (estimated concordance, 0.76; 95% confidence interval, [0.59, 0.93]). A simple test based on a combination of these markers might improve preoperative diagnosis of thyroid nodules, allowing better treatment decisions and reducing long-term health costs.
Introduction
The incidence of thyroid cancer is increasing, with a global estimate of one-half million new cases this year. Thyroid carcinoma is usually first suspected by the physician when a solitary nodule is palpated on physical examination. Thyroid nodules, however, can result from a wide spectrum of causes, and a major concern is to accurately differentiate between benign and malignant nodules.
Fine-needle aspiration (FNA) biopsy is the most widely used and cost-effective preoperative test for initial thyroid nodule diagnosis (1). When the FNA findings are diagnostic of papillary thyroid carcinoma, the specificity for malignancy approaches 95% (2). A common problem in clinical practice, however, is the evaluation and management of thyroid tumors with a follicular pattern. FNA cannot differentiate between follicular thyroid adenoma (FTA) and follicular thyroid carcinoma (FTC). Since cytology cannot distinguish FTA and FTC, they are often grouped together as indeterminate or follicular-patterned thyroid lesions, and surgical biopsy is needed to look for invasion through the tumor capsule or the blood vessels. Most guidelines recommend that a nodule diagnosed as having a follicular pattern should be surgically removed to provide an accurate diagnosis. Complete thyroid resection, and subsequent radioiodine therapy, is indicated for those patients for whom carcinoma is ultimately indicated. Overall, only 8–17% of these cytologically suspicious nodules are indeed malignant on histology (3). A large percentage of patients would, therefore, benefit greatly from improved diagnosis of FNA material, which could reduce the number or extent of surgeries, long-term health costs, and postsurgical complications. In particular, in many areas of the world where health care systems are overburdened, limited resources for surgery could be directed more rapidly toward those most likely to have carcinoma. Accurate molecular markers based on genes expressed differentially between FTC and FTA would be one means of improving the accuracy of diagnoses made from FNA.
Several genes are already reported to be associated with thyroid tumors. LGALS3 expression was proposed as a potential marker for preoperative diagnosis of thyroid carcinoma (4–6). Subsequent findings, however, showed LGALS3 expression in benign lesions such as multinodular goiter and FTA (7, 8). Recently, a chromosomal translocation, t(2;3)(q13;p25), was reported in five of eight cases of FTC, but not in 20 FTAs. The authors suggested that the resulting PAX8-PPARG fusion gene could be useful in the diagnosis and treatment of thyroid cancer (9). This rearrangement, however, was found in 13–30% of follicular adenomas (10–12). In addition, several molecular markers have been analyzed for their ability to discriminate between benign and malignant follicular tumors, including TPO, TP53, telomerase, and HMBE-1. Nonetheless, these candidate markers have not proved to be of practical value for preoperative FNA diagnosis of FTC (13–15). More recently, cDNA array technology has been used to identify potentially important thyroid cancer–associated genes (16). Although many of the genes or gene patterns expressed in thyroid tumors have been described, the clinical problem of distinguishing FTC from FTA remains.
In this project we sought to directly address the problem of finding diagnostic markers that would distinguish FTA from FTC. We first quantified gene expression in follicular tumors using serial analysis of gene expression (SAGE) (17). SAGE counts cDNA transcript tags in large numbers, making it possible to identify the restricted set of genes that are highly expressed in one tissue and not detectable in another. Transcript counts from FTC, FTA, and normal-thyroid libraries were generated and compared. Next, we evaluated independent FTA and FTC samples by real-time PCR for candidate genes identified by SAGE. Four of 17 genes differed between the two classes, FTA and FTC. In addition, we developed a predictor that proved to accurately distinguish FTA from FTC. The expression levels of two genes were further confirmed by immunohistochemistry. These markers may eventually improve diagnosis and, consequently, treatment of patients with thyroid follicular tumors and may play a role in the functional differences between adenoma and carcinoma.
Results
SAGE analysis.
SAGE was used to analyze the gene-expression profile in normal thyroid tissue, FTA, and FTC. A total of 359,478 tags were obtained, representing 116,037 unique transcript tags. Using a SAGE tag sequence error rate of 6.8% (18, 19), we estimated a total of 108,146 unique transcripts detected, of which 10,048 were detected at least five times and 32,748 were detected at least twice.
Two comparisons were performed using SAGE 2000 software version 4.12: one between normal thyroid and FTA and one between FTA and FTC. The expression levels of 305 genes were statistically significant (P value ≤ 0.0001), as analyzed using SAGE software to perform Monte Carlo simulations as previously described (18). Thirty-seven of these 305 transcripts were highly expressed in the FTC library, while 36 transcripts were not expressed in FTC but were highly expressed in FTA and normal-thyroid libraries. Among these 73 candidates genes with a P value ≤ 0.0001, those with the greatest fold induction or fold repression in FTC were considered first. Accordingly, we selected 17 transcripts for RT-PCR validation (Supplemental Table 1; supplemental material available at http://www.jci.org/cgi/content/full/113/8/1234/DC1). Twelve transcripts were highly expressed in the FTC library, and five were expressed only in FTA and normal-thyroid libraries. The expression levels of these genes in FTA and FTC libraries ranged from 43- to 10-fold. Table 1 lists the 17 genes and, for comparison, the transcript levels found for well-characterized genes for normal thyroid physiology.
Table 1.
Transcript expression of candidate FTA and FTC tumor markers with examples of thyroid function genes.
Quantitative RT-PCR.
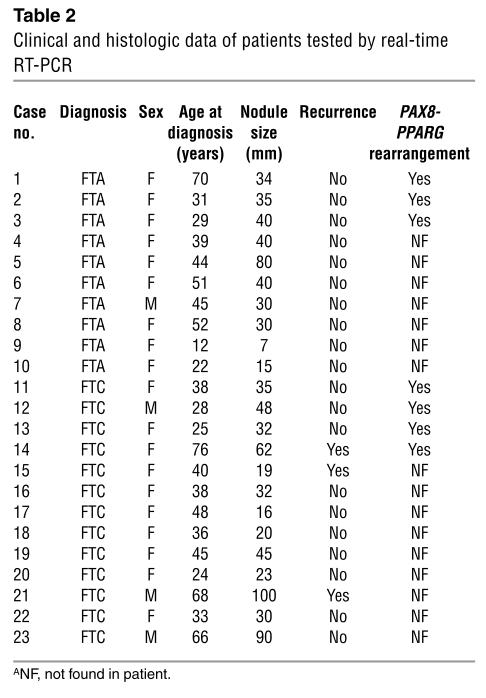
The gene-expression profiles in SAGE libraries for 17 transcripts showing the largest difference between adenoma and carcinoma were tested by quantitative RT-PCR in an independent set of ten FTAs, 13 FTCs, and eight normal patient-matched tissues (Table 2). We first compared the results obtained by SAGE with those obtained by RT-PCR analysis for the samples used to generate FTA and FTC libraries (cases 5 and 12, respectively). When RT-PCR and the original samples were used, 14 of 17 genes showed the predicted difference between FTA and FTC, and three did not.
Table 2.
Clinical and histologic data of patients tested by real-time RT-PCR
Next, using the full panel of samples, we observed that 9 of 12 transcripts that were overexpressed in FTCs maintained high expression in 50–100% of FTCs tested, compared with expression of the same transcripts in FTAs and patient-matched normal tissue. DDIT3 (DNA damage–inducible transcript 3) and ARG2 (arginase type II) were expressed at higher levels in FTCs. The increase in average of expression was at least fivefold in nearly all FTCs, and some exhibited at least 11-fold higher levels, as predicted by SAGE. The gene ITM1 (integral membrane protein 1) was expressed in all FTCs, with low levels of expression in six FTAs. The genes C1orf24 (niban) and ACO1 (aconitase 1) were expressed in 76% of FTCs, with low but detectable expression in 40% of FTAs. The hypothetical protein FLJ13576 was expressed in 67% of FTCs and in two cases of FTA (cases 6 and 8). Six genes did not distinguish well: ODZ1, PCSK2 (proprotein convertase subtilisin/kexin type 2), DNASE2 (deoxyribonuclease II, lysosomal), LOC92196 (similar to death-associated protein), PDK4 (pyruvate dehydrogenase kinase 4), and PPP1R14B (protein phosphatase 1, regulatory subunit 14B) were expressed in 30–69% of FTCs and in about 30–40% of FTAs.
Analysis also revealed that, of the five genes predicted to be adenoma specific, the TARSH gene (target of Nesh-SH3) was not expressed in most FTCs but was expressed at high levels in normal thyroid and FTAs. The genes putative Emu1, NID2, COL14A1, and endothelial receptor type B were expressed in about 60–80% of FTCs and were not discriminatory between FTA and FTC. The RT-PCR results from the six genes that appeared to discriminate between FTC and FTA are summarized in Figure 1.
Figure 1.
Relative levels of expression determined by quantitative RT-PCR in 23 samples of FTA and FTC (black bars) and in normal thyroid tissues (gray bars). Transcript levels were normalized to the average of ribosomal protein 8 and t-complex 1, which were uniformly expressed in all three thyroid SAGE libraries. Numbers 1–10 correspond to FTA and 11–23 to FTC cases described in Table 2. The statistical analysis of RT-PCR values revealed that expression of the genes DDIT3, ARG2, and ITM1 was significantly different at the 0.05 level, and expression of C1orf24 was significant at the 0.10 level.
In addition, we analyzed the expression levels of selected genes in three well-characterized thyroid cell lines from different types of thyroid tumors (20–22). All the transcripts elevated in FTCs (Table 1) were expressed in all thyroid cell lines. We suggest that the expression of the candidate markers in the pure populations of cultured carcinoma cells indicates that the expression was due to the malignant component of the tumor. Conversely, the genes downregulated in the FTC library were present at lower levels or absent in the cell lines (Figure 2).
Figure 2.
Quantitative RT-PCR products of three genes with statistically significant expression differences, showing adenoma (A), FTC (C), and normal thyroid (N) tissues and thyroid carcinoma cell lines (CL). The genes DDIT3, ARG2, and ITM1 are expressed in most of the FTCs, the thyroid follicular carcinoma cell line (CL1), the papillary thyroid carcinoma cell line (CL2), and the undifferentiated thyroid carcinoma cell line (CL3), but not in normal tissue and most FTAs. Case 6 (A6) expressed ARG2 and ITM1 and was misclassified according to our class-predicted genes. Universal Human Reference RNA (UHR) was used as a control. Ribosomal protein 8 was used as a calibrator gene. The 100-bp DNA ladder (M) is shown in the far left and far right lanes. The results are shown in triplicate, and the numbers correspond to cases analyzed (Table 2). The product sizes and PCR conditions are described in Methods and summarized in Supplemental Table 1. Negative control (NC) consists of PCR mix and primers, but no template.
Immunohistochemistry.
For two of the genes, DDIT3 and ARG2, antibodies were commercially available. Immunohistochemistry on paraffin-embedded sections was performed to determine protein expression and differential expression in FTA (n = 32) and FTC (n = 27), and the results are summarized in Table 3 (for details see Supplemental Table 2). Staining for DDIT3 expression (GADD153 antibody) showed a moderate to strong (++/+++) expression in 23 FTCs (85.2%). The staining was detected in both the nucleus and the cytoplasm of neoplastic follicular cells (Figure 3, D–F). Adjacent non-neoplastic thyroid tissue did not stain. Three of four FTCs negative for DDIT3 staining were classified as minimally invasive follicular tumors, and one was classified as moderately differentiated. No nuclear and cytoplasmic staining in epithelial cells was observed in 29 sections from FTAs (90.6%; Figure 3, A–C) and IgG-negative controls. A weak or moderate staining for DDIT3 was found in three FTAs (9.4%). Two of three were diagnosed as Hürthle cell adenoma (HCA), and one was an atypical adenoma (data not shown).
Table 3.
Immunoreactivity for DDIT3 and ARG2 in FTA and FTC
Figure 3.
Immunohistochemical analysis of DDIT3 (A–F) and ARG2 (G–L) in paraffin-embedded sections of FTAs and FTCs. FTCs exhibited strong brown immunostaining for DDIT3 (D–F) and ARG2 (J–L). In contrast, FTAs (A–C and G–I) exhibited no immunoreactivity. The arrows in D and F show the vascular invasion in FTCs and the follicular cells that are positive for DDIT3. The arrow in L shows the normal thyroid tissue that was negative for ARG2, adjacent to tumor area that was positive for ARG2. Hematoxylin was used as a nuclear counterstain. Original magnification, ×100 for A–E and G–L, ×40 for F.
ARG2 staining was consistently negative in 29 FTAs (90.6%) and adjacent non-neoplastic thyroid tissue, whereas specific staining was found in the cytoplasm of neoplastic follicular thyroid cells in 23 of the FTCs analyzed (85.2%; Figure 3, G–L). All four FTCs negative for ARG2 were diagnosed as minimally invasive.
Overall, a moderate to strong expression of ARG2 and DDIT3 was observed in 85.2% of FTCs, whereas 90.6% of FTAs were negative, indicating the utility of these antibodies to discriminate FTC from FTA. In addition, the immunoreactivity with both antibodies in FTCs was more often diffuse than focal and was stronger in intensity compared with that observed in the four FTAs.
Staining with von Willebrand factor VIII was used to distinguish endothelial cells in all tissues. Moderate staining with CA9 antibody was observed in two FTCs (cases 11 and 12), but not in FTAs and normal tissues (data not shown).
Statistical analysis.
We first used the SAGE-predicted differences between the global gene expressions of FTA and FTC to identify 17 candidate molecular markers in thyroid. Fourteen of the 17 genes showed some difference by RT-PCR on an independent set of patients and were used for statistical analysis. This analysis comparing FTC and FTA revealed that expression levels of four genes differed in this data set. Genes were declared different between the two groups if the P value was less than the family-wise error rate of 0.10. The Wilcoxon test showed that the difference in gene expression of DDIT3, ARG2, and ITM1 was statistically significant at the 0.05 level. Expression of an additional gene (C1orf24) was statistically significant at the 0.10 level. The Student’s t test showed that expression levels of DDIT3 and ITM1 were significant at the 0.05 level. No additional genes were significant at the 0.10 level. Thus, expression levels of four genes (DDIT3, ARG2, ITM1, and C1orf24) were declared significantly different between the two groups; expression levels of DDIT3 and ITM1 were declared significantly different by both analyses.
The class predictor used genes whose expression levels were declared significantly different at the 0.10 level using the t test. The sample t statistics were used as weights in the compound covariate predictor. To evaluate the predictor, we used leave-one-out cross-validation: for each sample, in turn, one sample was left out, and the predictor was developed on the remaining 22 samples. The left-out sample was predicted. We used all the steps of the prediction procedure, including selection of differentially expressed genes, as well as creation of the prediction rule (23, 24). Using leave-one-out cross-validation, 19 of the 23 samples (83%) were correctly predicted. To assess the significance of these prediction results, we implemented a permutation test (23, 24); the proportion of random permutations with four or fewer misclassifications was 0.007. Thus, we declared the results of the prediction analysis significant. Two of the genes, DDIT3 and ITM1, were always selected in each step of the cross-validation procedure (i.e., each time a sample was left out).
Figure 2 shows the final products for three genes, the differential expression of which was shown to be statistically significant at the 0.05 level, after 40 cycles of PCR were run using templates from FTC, FTA, normal thyroid, and cell lines.
The concordance between the results of the immunohistochemistry staining on an independent set of tumors and the diagnosis by histopathology was estimated by κ. The estimated κ was 0.76 with a 95% confidence interval of [0.59, 0.93]. The value of 0.76 corresponds to a substantial strength of agreement based on previously developed guidelines (25).
Clinical, pathologic, and molecular correlations.
All patients received postsurgical radioiodine ablation and suppressive thyroxine therapy. Tumor recurrence was observed in three cases of FTC (Table 2).
All patients tested by RT-PCR were also checked for PPARG-PAX8 rearrangement. Analysis revealed that the rearrangement of PPARG-PAX8, previously identified as an FTC marker (9), was found in about 33% of FTAs and in 33% of FTCs (C. Nakabashi et al., unpublished observations) and did not distinguish between adenoma and carcinoma (Table 2). We then compared the clinical and pathologic information with the results obtained from quantitative RT-PCR analysis. Using the cross-validation procedure, the prediction accuracy was estimated to be 83%. Four of the cases were misclassified (cases 6, 8, 13, and 21; Table 2). Case 6, which exhibited DDIT3, ARG2, and ITM1 expression, was re-evaluated by an experienced pathologist and showed no evidence of either capsule or blood vessel invasion. However, Hashimoto thyroiditis and positive staining for both ERBB2 and P53 were reported. In case 8, Hashimoto thyroiditis was also involved. A longer follow-up for both cases will reveal whether they are true FTAs. Case 13 was an FTC that was diagnosed as minimally invasive. Case 21, however, was an FTC in which both blood and capsule invasion were present.
Discussion
In this study, we sought molecular markers that could improve the preoperative diagnosis of follicular thyroid tumors. Two deeply sampled SAGE libraries, one of FTA and one of FTC, were compared, yielding 305 candidate genes that were differentially expressed (P value ≤ 0.0001 as determined by the SAGE 2000 software). By selecting for transcripts that were absent (from more than 100,000 tags sampled) in one of the two libraries compared, we narrowed the list to 73 candidate markers.
SAGE has been previously used for a shallower transcript sampling of thyroid tissue but has not been specifically used to distinguish between FTA and FTC (26, 27). Using deep sampling of representative FTC and FTA cases allowed us to apply selection criteria for candidate genes that were likely to have large differences in expression that could be easily detected by immunohistochemistry.
Seventeen transcripts with the greatest predicted expression differences were selected for testing by quantitative RT-PCR in an independent set of samples. Twelve transcripts were candidate FTC markers, whereas five transcripts were markers for FTA. Four genes, DDIT3, ARG2, ITM1, and C1orf24, were statistically the most consistent markers for FTC. A linear combination of expression levels accurately predicted tumor class in 19 of 23 samples, using leave-one-out cross-validation (estimated prediction accuracy, 0.83; P value from permutation test, 0.007). The genes DDIT3 and ITM1 were consistently selected in the cross-validation of the prediction procedure. In the Wilcoxon test, ARG2 was statistically significant at the 0.05 level. An additional gene, C1orf24, expressed in most of the FTCs, may be a potential predictor, but further analysis is needed. Although DDIT3 and ITM1 transcripts were elevated in most FTC cases, use of one of these independently to identify tumors may result in some misclassifications; for example, case 14 had low levels of DDIT3 but expressed ITM1 and ARG2 at higher levels (Figure 1).
These findings were also confirmed in immunohistochemical analysis. Even though 82.5% of FTCs were positive for both DDIT3 and ARG2, two FTCs exhibited immunoreactivity to only DDIT3 or ARG2, and two cases were negative for both. Use of one marker separately could, therefore, easily lead to both false-positive and false-negative classifications and should not be employed for this purpose. The use of ITM1 might be helpful to better classify the tumors as benign or malignant.
Immunohistochemical analysis also revealed the expression of DDIT3 in three FTAs, which were diagnosed as atypical adenoma and HCA (Supplemental Table 2). These results support the idea that some follicular Hürthle tumors should be considered as a separate thyroid cancer class and that a small percentage of tumors with the diagnosis of FTA are actually early in situ carcinomas with malignant potential. Longer follow-up will be needed to determine whether these tumors are a less benign variant. In addition, both follicular lesions coexisted with Hashimoto thyroiditis, which is a possible source of diagnostic error (9). Immunohistochemical analysis in a large set of HCAs would be necessary to understand whether the use of additional class predicted genes (such as ITMI and C1orf24) in combination with DDIT3 and ARG2 can better classify this type of follicular lesion or whether additional profiling is necessary to find new markers for the Hürthle subtype.
DDIT3 , also named GADD153 (growth arrest and DNA damage-inducible 153 gene), is a transcription factor shown to be induced in response to cellular stresses such as UV light, hypoxia, nutrient deprivation, environmental toxicants, and certain DNA-damaging agents (19, 28). When induced, DDIT3 inhibits cell proliferation and promotes repair and/or apoptosis. The induction of DDIT3 leads to distinct biologic effects, such as growth stimulation, differentiation, invasiveness, and migration (29). In this study, overexpression of DDIT3 transcript was found in FTCs and thyroid cancer cell lines. Immunohistochemistry showed that DDIT3 protein expression was moderate to strong in 23 FTCs (82.5%) and specific for the follicular cells of the tumor (Figure 3). No expression of DDIT3 was found in four FTCs, three of which were diagnosed as minimally invasive. This observation suggested a correlation between DDIT3 expression and capsular and vascular invasion. Interestingly, Nikiforova et al. (30) reported that 85% of FTCs were able to develop through nonoverlapping RAS or PAX8-PPARG pathways. The authors suggested that RAS activation by itself appears insufficient to determine malignant growth but may predispose to acquisition of additional genetic or epigenetic alteration that leads to a fully transformed phenotype. Brenner et al. showed a signaling cascade from the FAS receptor via the G proteins RAS and RAC to JNK/p38 kinase and the transcription factor DDIT3 (31). Expression of DDIT3 was also elevated after induction with thiazolidinedione via PPARG1 (32). It is therefore possible that either or both of these pathways activate DDIT3 expression.
ITM1 encodes a highly conserved protein that contains 10–14 membrane-spanning domains. The protein does not have any identifiable domains with enzymatic activity and is probably not involved in direct transmembrane signaling (33). In addition, the transmembrane domain of ITM1 does not present any features of a transporter protein, such as an ATP-binding cassette. Accordingly, it has been speculated that ITM1 is a novel type of permease/transporter membrane protein (33). The ITM1 gene was mapped to human chromosome 11q23.3 (34, 35), where loss of heterozygosity has been found in follicular adenomas (36, 37).
Another FTC-associated gene was ARG2, which catalyzes the hydrolysis of arginine to ornithine plus urea. At least two isoforms of mammalian arginase exist (ARG1 and ARG2), which differ in their tissue distribution, subcellular localization, immunologic cross-reactivity, and physiologic function (38). The type II isoform is located in the mitochondria and is expressed in extrahepatic tissues, especially in the kidney (39). ARG2 is thought to play a role in nitric oxide and polyamine metabolism (40). Since polyamines are vital for cell proliferation, it is possible that the increased level of ornithine, due to the elevated arginase activity, may be linked to carcinogenesis (41).
C1orf24 was described as a candidate marker for renal tumor, especially in early-stage renal carcinogenesis (42). The pattern of gene expression showed that C1orf24 is expressed in normal muscle, pancreas, colon, and prostate. The gene is very conserved in humans and rats, but the protein function is unknown. A similarity with the DNAJ-1 motif, part of a chaperone system, has been described (43).
TARSH, a candidate marker for FTAs, was downregulated in most FTCs and expressed in normal thyroid tissues but was just above the 0.05 significance level. TARSH encodes a protein containing an Src-homology 3–binding (SH3-binding) motif, a nuclear target sequence and no catalytic domain. Its biochemical and physiologic role has not been identified. TARSH is thought to be a binding partner of NESH-SH3, a member of the E3B1/ArgBP/Avi2/NESH family (44). Members of this family are involved in membrane ruffling and lamellipodia formation, which suggests that the loss of their expression could be involved in the mechanism of cell motility and metastasis. Re-expression of NESH suppresses motility and metastasis dissemination in the U-87 MG malignant glioma cell line (45). Although the binding activity between NESH and TARSH is yet to be confirmed, the loss of TARSH expression in FTCs might be a mechanism by which the follicular cells acquire motility and promote invasion. Another fact that supports this hypothesis is that the TARSH gene was mapped at 3q12, where loss of heterozygosity was found in FTC but not in FTA (46, 47). Loss of heterozygosity in 3q was also correlated with survival in FTC (48).
Since follicular cell interaction and differentiation is guided by a variety of factors, such as ECM glycoprotein and receptor and cell adhesion molecules, we also expected to find genes involved in this process to be differentially expressed between FTC and FTA. In fact, we found ODZ1 (tenascin M), ANXA1 (annexin 1), LAMB1 (laminin β1), MYL6 (myosin light polypeptide 6), MSN (moesin), CLU (clusterin), TMSB4X (thymosin β4), SPARC (osteonectin), CLDN1 (claudin 1), NID2 (nidogen 2), Emu1, CANX (calnexin), SDC2 (syndecan 2), FMOD (fibromodulin), CDH1 (cadherin 1), and COL14A1 (undulin) differentially expressed in thyroid SAGE libraries. Some of these genes were described previously as being involved in thyroid-tumor genesis, but they were not used to discriminate between FTA and FTC (27, 49).
While this manuscript was in preparation, Barden et al. (16), by oligonucleotide array, found the gene DDIT3 upregulated in FTC and the genes putative Emu1 and NID2 upregulated in FTA. Although the investigators did not validate the expression of these genes in a set of samples, their findings help to corroborate ours.
By applying SAGE to representative adenoma and carcinoma, we were able to determine the genes that had the largest quantitative differences in expression, and to confirm for the two genes tested that the protein levels differed as predicted. Definition of a small set of the best predictive antibodies would likely be necessary to improve FNA diagnosis — an important goal, since clinicians currently use FNA as the first preoperative diagnostic test. About 10–25% of nodules are classified as indeterminate or suspicious by FNA, and most patients with these nodules are referred for surgery. Unnecessary removal of benign follicular tumors increases long-term health costs. Additionally, sensitive markers that detect carcinoma by FNA might speed detection and subsequent treatment. In the group of patients analyzed in this study by immunohistochemistry, the cytologic reports of FNA revealed that 36 of 59 follicular tumors were diagnosed as “suspicious.” A total thyroidectomy was the treatment of choice, and a negative result was confirmed on permanent pathology in 20 cases (66%; Supplemental Table 2). The immunohistochemistry test using the two available antibodies against the two differentially expressed genes DDIT3 and ARG2 correctly classified 29 of 32 FTAs (90.6%) and 23 of 27 FTCs (85.2%) (Table 3). However, the sensitivity and specificity of this test should be evaluated further by analysis of other differentially expressed genes, starting with ITM1. Other subtypes, such as Hürthle tumors, may need to be evaluated by SAGE to generate markers specific for that molecular class of thyroid tumor.
This simple test, based on the tumor markers we describe, could improve both preoperative diagnosis of thyroid nodules by FNA and subsequent treatment, while reducing costs. A simple and cost-effective approach will be necessary, in particular, for regions of the world where health care systems are financially constrained. Although this small data set produced a robust prediction model, and high concordance between the results of pathology and immunohistochemistry, larger sample sizes and the testing of additional antibodies to find the optimum combination should improve this model.
Methods
Tissue samples.
For the RT-PCR analysis, 23 primary tumors were obtained from patients initially diagnosed with follicular thyroid tumor; the tumors were frozen immediately after surgical biopsy. All samples were obtained from patients followed at Hospital São Paulo, Federal University of São Paulo, and at Hospital Heliópolis (São Paulo, Brazil). The study was approved by the Ethics and Research Committees of the Federal University of São Paulo and Hospital Heliópolis and was in agreement with the World Medical Associations’ 1975 Declaration of Helsinki, revised in 1983. A signed letter of informed consent was obtained from each patient. Tissue histology confirmed the initial diagnoses, as summarized in Table 1. The samples included 10 FTAs and 13 FTCs. In addition, we analyzed eight patient-matched normal tissues obtained from patients with FTC (n = 5) and with FTA (n = 3). Total Universal Human Reference RNA (Stratagene, La Jolla, California, USA) was used as a control. For the immunohistochemical study, we retrieved pathologic materials from specimens diagnosed with FTC (n = 27) and FTA (n = 32) at Hospital São Paulo, Federal University of São Paulo, in an 8-year period from 1996 to 2003. H&E-stained sections were reviewed by an experienced pathologist.
Cell lines.
The human FTC cell line UCLA RO-82W-1, the papillary thyroid carcinoma line UCLA NPA-87-1, and an undifferentiated thyroid carcinoma cell line, UCLA RO-81A-1, were grown in DMEM (Invitrogen Corp., Carlsbad, California, USA) supplemented with 10% FCS (Invitrogen Corp.) in a 5% CO2 environment at 37°C, as previously described (20).
SAGE libraries.
One FTA, one FTC, and one normal thyroid were chosen for SAGE (17). The libraries were constructed using a microSAGE procedure (50) and were sequenced through the SAGE portion of the Cancer Genome Anatomy Project (51). Tags were extracted from automated sequence text files; and duplicate ditags, linker sequences, and repetitive tags were removed using SAGE 2000 software version 4.12 (available at http://www.sagenet.org). The Monte Carlo simulation function of this program was used to determine P values of differentially expressed genes. The full set of tag counts for all three libraries is available for downloading or analysis at the Cancer Genome Anatomy Project SAGE Genie website at http://cgap.nci.nih.gov/SAGE (52).
RNA isolation, cDNA synthesis, and quantitative RT-PCR.
To validate the differential gene profile predicted by SAGE, we tested 17 genes with the highest fold induction and analyzed them by quantitative real-time RT-PCR. Total RNA was isolated using RNAgents (Promega Corp., Madison, Wisconsin, USA), according to the manufacturer’s recommendation. One microgram of total RNA was treated with DNA-free (Ambion Inc., Austin, Texas, USA) and was reverse-transcribed to cDNA using the Omniscript Reverse Transcriptase kit (QIAGEN Inc., Germantown, Maryland, USA) with oligo-dT12-18 primer and 10 U of RNase inhibitor (Invitrogen Corp.). Reverse transcriptase–negative samples were prepared for each individual reaction and were used as controls for detection of assay contamination. The cDNA was then diluted fivefold, and 1.5-μl aliquots were used in 20-μl PCR reactions containing 10 μM of each specific primer, 1× iQ Supermix (Bio-Rad Laboratories Inc., Hercules, California, USA), and SYBR Green (Sigma-Aldrich, St. Louis, Missouri, USA). The PCR reaction was performed for 40 cycles of a four-step program: 94°C for 30 seconds, annealing temperature for 15 seconds, 72°C for 15 seconds, and a fluorescence-read step for 10 seconds. After PCR, a melting-curve analysis was performed, and the read temperature of each assay was set above the melting point of short primer-dimers and below that of the target PCR product. Quantitative PCR reactions were performed twice in triplicate; threshold cycles (Ct) were obtained using iCycler software version 3.0 (Bio-Rad Laboratories Inc.) and were averaged (SD ≤ 1). Gene expression was normalized to the average of two control genes, ribosomal protein S8 and t-complex 1, shown by SAGE to be at equivalent levels in all three SAGE libraries. A relative expression was calculated according to the formula 2(Rt – Et)/2(Rn – En), where Rt is the Ct number observed in the experimental sample for two control genes, Et is the Ct number observed in the experimental sample for the reference gene, Rn is the average Ct number observed in ten adenomas for two control genes, and En is the average Ct number observed in ten adenomas for the reference gene (53). The results obtained from 14 of the 17 relative-expression levels in 23 samples and normal tissue, shown in Figure 1, were used for statistical analysis. Only 14 were used because three genes showed no difference by PCR. The PCR-specific primers, annealing temperatures, and fluorescence-read temperatures are summarized in Supplemental Table 1. The PCR products were resolved by electrophoresis in a 3% agarose/ethidium gel.
Immunohistochemical analysis.
Immunohistochemical staining was performed on paraffin-embedded tissue sections (3 μm) placed on 0.1% polylysine–coated slides (Sigma-Aldrich), deparaffinized with xylene, and rehydrated through a series of graded alcohols. The endogenous alkaline phosphatase activity was blocked by 3% hydrogen peroxide. After pressure-cooking retrieval (10 mmol/l citrate buffer, pH 7.4, for 2 minutes), the sections were blocked in 1× PBS/0.1% BSA for 1 hour at room temperature and incubated with the first antibody for at least 16 hours at 4°C. The labeled complex of streptavidin and biotin reagents (DAKO LSAB+ kit and HRP; DAKO Corp., Carpinteria, California, USA) was used with 3,3-diaminobenzidine tetrahydrochloride (DAB) (Sigma-Aldrich) as a substrate. Hematoxylin was used as the nuclear counterstain. The slides were mounted in Faramount mounting medium (DAKO Corp.) and examined by light microscopy. The immunopositivity was evaluated by two independent observers in a semiquantitative fashion in which the relative abundance of each antigen was evaluated by counting of 1,000 cells in at least five randomly chosen fields of the tissue sections at ×400 magnification and scored as follows: –, negative; +, weak; ++, moderately abundant; +++, strong. Polyclonal antiserum GADD153, originated against a peptide mapping at the C-terminus of DDIT3 of human origin, was used at 1:200 dilution (R-20; Santa Cruz Biotechnology Inc., Santa Cruz, California, USA). Polyclonal antiserum arginase II, raised against a recombinant protein to amino acids 291–354 mapping at the C-terminus of arginase type II of human origin, was used at 1:100 dilution (H-64; Santa Cruz Biotechnology Inc.). Monoclonal von Willebrand factor VIII was used at 1:25 dilution (M0616; DAKO Corp.). CA9 monoclonal antibody (gift of E. Oosterwijk, University Medical Center, Nijmegen, The Netherlands) was used at 1:400 dilution. The control for antibody specificity included incubation with rat IgG, used in the same concentration as the first antibody (Vector Laboratories Inc., Burlingame, California, USA). Positive and negative controls were included in each run.
Statistical analysis.
To identify genes for which expression levels were significantly different between FTA and FTC, we used the relative-expression data obtained from RT-PCR analysis on 14 of 17 genes (Supplemental Table 1). The initial comparison of expression levels was carried out using rank-based (Wilcoxon rank sum) and mean-based (Student’s t) tests. Data were log-transformed before application of the Student’s t test. A comparison was designated as statistically significant if either the rank-sum statistic or the corresponding t statistic was found to be significant, using an α level that had been adjusted (using a Bonferroni adjustment) to keep the family-wise error rate at 0.10. We next investigated development of an expression-based model that could be used to predict class of diagnosis for the tumor (FTA or FTC). We followed the framework outlined by Radmacher et al. (24), and we used the compound covariate predictor for gene-expression data (24, 54). The performance of the predictor was tested using leave-one-out cross-validation for all steps of the prediction procedure (i.e., selection of differentially expressed genes as well as creation of the prediction rule) (23). We assessed the significance of the performance of the predictor using the permutation-based test outlined by Radmacher et al. (24), in which the class labels are randomly permuted and the proportion of data sets that have a cross-validated error rate as small as the error rate observed in the data set is calculated. Because it was prohibitive to compute all possible permutations, we used 2,000 random permutations to estimate the achieved significance level.
We estimated the concordance of the results of the immunohistochemical staining and the pathological identification of class (FTC vs. FTA) by using a κ statistic and constructing a 95% confidence interval (24). The use of κ corrects for agreements between the two methods (immunohistochemistry and pathology) that would be expected by chance. The maximum value of κ, corresponding to perfect agreement, is 1.0. We used previously suggested guidelines (25) to assess the significance of the magnitude of the statistic.
Supplementary Material
Acknowledgments
We thank Tracy-Ann Read, Anita Lal, Kathy Boon, Rita G. Coimbra, and Maria José Carregosa Pinheiro dos Santos for assistance. J.M. Cerutti is a scholar of the Coordenação de Aperfeicoamento de Pessoal de Nível Superior Brasília (Brazil) and the Federal University of São Paulo. Ann S. Tamariz edited the manuscript. This project was also supported in part by the USA National Cancer Institute’s Cancer Genome Anatomy Project (NCI contract S98-146), the Molecular Classification of Tumors Initiative (U01 CA88128), and the Ludwig Trust.
Footnotes
Nonstandard abbreviations used: fine-needle aspiration (FNA); follicular thyroid adenoma (FTA); follicular thyroid carcinoma (FTC); Hürthle cell adenoma (HCA); serial analysis of gene expression (SAGE); Src-homology 3 (SH3); threshold cycle (Ct).
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Gharib H. Fine-needle aspiration biopsy of thyroid nodules: advantages, limitations, and effect. Mayo Clin. Proc. 1994;69:44–49. doi: 10.1016/s0025-6196(12)61611-5. [DOI] [PubMed] [Google Scholar]
- 2.Mazzaferri EL. Management of a solitary thyroid nodule. N. Engl. J. Med. 1993;328:553–559. doi: 10.1056/NEJM199302253280807. [DOI] [PubMed] [Google Scholar]
- 3.Goellner JR, Gharib H, Grant CS, Johnson DA. Fine needle aspiration cytology of the thyroid, 1980 to 1986. Acta Cytol. 1987;31:587–590. [PubMed] [Google Scholar]
- 4.Inohara H, et al. Expression of galectin-3 in fine-needle aspirates as a diagnostic marker differentiating benign from malignant thyroid neoplasms. Cancer. 1999;85:2475–2484. [PubMed] [Google Scholar]
- 5.Bartolazzi A, et al. Application of an immunodiagnostic method for improving preoperative diagnosis of nodular thyroid lesions. Lancet. 2001;357:1644–1650. doi: 10.1016/s0140-6736(00)04817-0. [DOI] [PubMed] [Google Scholar]
- 6.Xu XC, el-Naggar AK, Lotan R. Differential expression of galectin-1 and galectin-3 in thyroid tumors. Potential diagnostic implications. Am. J. Pathol. 1995;147:815–822. [PMC free article] [PubMed] [Google Scholar]
- 7.Cvejic D, et al. Immunohistochemical localization of galectin-3 in malignant and benign human thyroid tissue. Anticancer Res. 1998;18:2637–2641. [PubMed] [Google Scholar]
- 8.Bernet VJ, et al. Determination of galectin-3 messenger ribonucleic acid overexpression in papillary thyroid cancer by quantitative reverse transcription-polymerase chain reaction. J. Clin. Endocrinol. Metab. 2002;87:4792–4796. doi: 10.1210/jc.2002-020390. [DOI] [PubMed] [Google Scholar]
- 9.Kroll TG, et al. PAX8-PPARγ1 fusion oncogene in human thyroid carcinoma [erratum 2000, 289:1474] Science. 2000;289:1357–1360. doi: 10.1126/science.289.5483.1357. [DOI] [PubMed] [Google Scholar]
- 10.Marques AR, et al. Expression of PAX8-PPAR γ 1 rearrangements in both follicular thyroid carcinomas and adenomas. J. Clin. Endocrinol. Metab. 2002;87:3947–3952. doi: 10.1210/jcem.87.8.8756. [DOI] [PubMed] [Google Scholar]
- 11.Nikiforova MN, Biddinger PW, Caudill CM, Kroll TG, Nikiforov YE. PAX8-PPARγ rearrangement in thyroid tumors: RT-PCR and immunohistochemical analyses. Am. J. Surg. Pathol. 2002;26:1016–1023. doi: 10.1097/00000478-200208000-00006. [DOI] [PubMed] [Google Scholar]
- 12.Cheung L, et al. Detection of the PAX8-PPAR gamma fusion oncogene in both follicular thyroid carcinomas and adenomas. J. Clin. Endocrinol. Metab. 2003;88:354–357. doi: 10.1210/jc.2002-021020. [DOI] [PubMed] [Google Scholar]
- 13.Fagin JA. Tumor suppressor genes in human thyroid neoplasms: p53 mutations are associated undifferentiated thyroid cancers. J. Endocrinol. Invest. 1995;18:140–142. doi: 10.1007/BF03349723. [DOI] [PubMed] [Google Scholar]
- 14.Haugen BR, et al. Telomerase activity in benign and malignant thyroid tumors. Thyroid. 1997;7:337–342. doi: 10.1089/thy.1997.7.337. [DOI] [PubMed] [Google Scholar]
- 15.Sack MJ, Astengo-Osuna C, Lin BT, Battifora H, LiVolsi VA. HBME-1 immunostaining in thyroid fine-needle aspirations: a useful marker in the diagnosis of carcinoma. Mod. Pathol. 1997;10:668–674. [PubMed] [Google Scholar]
- 16.Barden CB, et al. Classification of follicular thyroid tumors by molecular signature: results of gene profiling. Clin. Cancer Res. 2003;9:1792–1800. [PubMed] [Google Scholar]
- 17.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, et al. Gene expression profiles in normal and cancer cells. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 19.Velculescu VE, et al. Analysis of human transcriptomes. Nat. Genet. 1999;23:387–388. doi: 10.1038/70487. [DOI] [PubMed] [Google Scholar]
- 20.Pang XP, Hershman JM, Chung M, Pekary AE. Characterization of tumor necrosis factor-alpha receptors in human and rat thyroid cells and regulation of the receptors by thyrotropin. Endocrinology. 1989;125:1783–1788. doi: 10.1210/endo-125-4-1783. [DOI] [PubMed] [Google Scholar]
- 21.Cerutti J, et al. Block of c-myc expression by antisense oligonucleotides inhibits proliferation of human thyroid carcinoma cell lines. Clin. Cancer Res. 1996;2:119–126. [PubMed] [Google Scholar]
- 22.Visconti R, et al. Expression of the neoplastic phenotype by human thyroid carcinoma cell lines requires NFkappaβ p65 protein expression. Oncogene. 1997;15:1987–1994. doi: 10.1038/sj.onc.1201373. [DOI] [PubMed] [Google Scholar]
- 23.Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J. Natl. Cancer Inst. 2003;95:14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- 24.Radmacher MD, McShane LM, Simon R. A paradigm for class prediction using gene expression profiles. J. Comput. Biol. 2002;9:505–511. doi: 10.1089/106652702760138592. [DOI] [PubMed] [Google Scholar]
- 25.Kramer MS, Feinstein AR. Clinical biostatistics LIV: the biostatistics of concordance. Clin. Pharmacol. Ther. 1981;29:111–123. doi: 10.1038/clpt.1981.18. [DOI] [PubMed] [Google Scholar]
- 26.Pauws E, van Kampen AH, van de Graaf SA, de Vijlder JJ, Ris-Stalpers C. Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: implications for SAGE analysis. Nucleic Acids Res. 2001;29:1690–1694. doi: 10.1093/nar/29.8.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takano T, et al. Gene expression profiles in thyroid carcinomas. Br. J. Cancer. 2000;83:1495–1502. doi: 10.1054/bjoc.2000.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin K, et al. cDNA microarray analysis of changes in gene expression induced by neuronal hypoxia in vitro. Neurochem. Res. 2002;27:1105–1112. doi: 10.1023/a:1020913123054. [DOI] [PubMed] [Google Scholar]
- 29.Talukder AH, Wang RA, Kumar R. Expression and transactivating functions of the bZIP transcription factor GADD153 in mammary epithelial cells. Oncogene. 2002;21:4289–4300. doi: 10.1038/sj.onc.1205529. [DOI] [PubMed] [Google Scholar]
- 30.Nikiforova MN, et al. RAS point mutations and PAX8-PPARgamma rearrangement in thyroid tumors: evidence for distinct molecular pathways in thyroid follicular carcinoma. J. Clin. Endocrinol. Metab. 2003;88:2318–2326. doi: 10.1210/jc.2002-021907. [DOI] [PubMed] [Google Scholar]
- 31.Brenner B, et al. Fas- or ceramide-induced apoptosis is mediated by a Rac1-regulated activation of Jun N-terminal kinase/p38 kinases and GADD153. J. Biol. Chem. 1997;272:22173–22181. doi: 10.1074/jbc.272.35.22173. [DOI] [PubMed] [Google Scholar]
- 32.Satoh T, et al. Activation of peroxisome proliferator-activated receptor-gamma stimulates the growth arrest and DNA-damage inducible 153 gene in non-small cell lung carcinoma cells. Oncogene. 2002;21:2171–2180. doi: 10.1038/sj.onc.1205279. [DOI] [PubMed] [Google Scholar]
- 33.Hong G, et al. Molecular cloning of a highly conserved mouse and human integral membrane protein (Itm1) and genetic mapping to mouse chromosome 9. Genomics. 1996;31:295–300. doi: 10.1006/geno.1996.0051. [DOI] [PubMed] [Google Scholar]
- 34.Van Hul W, et al. Assignment of the human integral transmembrane protein 1 gene (ITM1) to human chromosome band 11q23.3 by in situ hybridization and YAC mapping. Cytogenet. Cell Genet. 1996;74:218–219. doi: 10.1159/000134417. [DOI] [PubMed] [Google Scholar]
- 35.Meerabux JM, et al. Molecular cloning of a novel 11q23 breakpoint associated with non-Hodgkin’s lymphoma. Oncogene. 1994;9:893–898. [PubMed] [Google Scholar]
- 36.Matsuo K, Tang SH, Fagin JA. Allelotype of human thyroid tumors: loss of chromosome 11q13 sequences in follicular neoplasms. Mol. Endocrinol. 1991;5:1873–1879. doi: 10.1210/mend-5-12-1873. [DOI] [PubMed] [Google Scholar]
- 37.Ward LS, Brenta G, Medvedovic M, Fagin JA. Studies of allelic loss in thyroid tumors reveal major differences in chromosomal instability between papillary and follicular carcinomas. J. Clin. Endocrinol. Metab. 1998;83:525–530. doi: 10.1210/jcem.83.2.4550. [DOI] [PubMed] [Google Scholar]
- 38.Gotoh T, Araki M, Mori M. Chromosomal localization of the human arginase II gene and tissue distribution of its mRNA. Biochem. Biophys. Res. Commun. 1997;233:487–491. doi: 10.1006/bbrc.1997.6473. [DOI] [PubMed] [Google Scholar]
- 39.Morris SM, Jr, Bhamidipati D, Kepka-Lenhart D. Human type II arginase: sequence analysis and tissue-specific expression. Gene. 1997;193:157–161. doi: 10.1016/s0378-1119(97)00099-1. [DOI] [PubMed] [Google Scholar]
- 40.Russell DH, McVicker TA. Polyamine biogenesis in the rat mammary gland during pregnancy and lactation. Biochem. J. 1972;130:71–76. doi: 10.1042/bj1300071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian W, Boss GR, Cohen DM. Ras signaling in the inner medullary cell response to urea and NaCl. Am. J. Physiol. Cell Physiol. 2000;278:C372–C380. doi: 10.1152/ajpcell.2000.278.2.C372. [DOI] [PubMed] [Google Scholar]
- 42.Majima S, Kajino K, Fukuda T, Otsuka F, Hino O. A novel gene “Niban” upregulated in renal carcinogenesis: cloning by the cDNA-amplified fragment length polymorphism approach. Jpn. J. Cancer Res. 2000;91:869–874. doi: 10.1111/j.1349-7006.2000.tb01027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sood R, et al. Cloning and characterization of 13 novel transcripts and the human RGS8 gene from the 1q25 region encompassing the hereditary prostate cancer (HPC1) locus. Genomics. 2001;73:211–222. doi: 10.1006/geno.2001.6500. [DOI] [PubMed] [Google Scholar]
- 44.Matsuda S, et al. Cloning and sequencing of a novel human gene that encodes a putative target protein of Nesh-SH3. J. Hum. Genet. 2001;46:483–486. doi: 10.1007/s100380170049. [DOI] [PubMed] [Google Scholar]
- 45.Ichigotani Y, Yokozaki S, Fukuda Y, Hamaguchi M, Matsuda S. Forced expression of NESH suppresses motility and metastatic dissemination of malignant cells. Cancer Res. 2002;62:2215–2219. [PubMed] [Google Scholar]
- 46.Zedenius J, et al. Allelotyping of follicular thyroid tumors. Hum. Genet. 1995;96:27–32. doi: 10.1007/BF00214182. [DOI] [PubMed] [Google Scholar]
- 47.Roque L, Rodrigues R, Pinto A, Moura-Nunes V, Soares J. Chromosome imbalances in thyroid follicular neoplasms: a comparison between follicular adenomas and carcinomas. Genes Chromosomes Cancer. 2003;36:292–302. doi: 10.1002/gcc.10146. [DOI] [PubMed] [Google Scholar]
- 48.Grebe SK, et al. Frequent loss of heterozygosity on chromosomes 3p and 17p without VHL or p53 mutations suggests involvement of unidentified tumor suppressor genes in follicular thyroid carcinoma. J. Clin. Endocrinol. Metab. 1997;82:3684–3691. doi: 10.1210/jcem.82.11.4352. [DOI] [PubMed] [Google Scholar]
- 49.Fonseca E, Soares P, Rossi S, Sobrinho-Simoes M. Prognostic factors in thyroid carcinomas. Verh. Dtsch. Ges. Pathol. 1997;81:82–96. [PubMed] [Google Scholar]
- 50.St Croix B, et al. Genes expressed in human tumor endothelium. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- 51.Lal A, et al. A public database for gene expression in human cancers. Cancer Res. 1999;59:5403–5407. [PubMed] [Google Scholar]
- 52.Boon K, et al. An anatomy of normal and malignant gene expression. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11287–11292. doi: 10.1073/pnas.152324199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buckhaults P, et al. Secreted and cell surface genes expressed in benign and malignant colorectal tumors. Cancer Res. 2001;61:6996–7001. [PubMed] [Google Scholar]
- 54.Tukey JW. Tightening the clinical trial. Control. Clin. Trials. 1993;14:266–285. doi: 10.1016/0197-2456(93)90225-3. [DOI] [PubMed] [Google Scholar]
- 55.The Cancer Genome Anatomy Project. All about the CGAP GO browser. http://cgap.nci.nih.gov/Genes/AllAboutGO.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.