Abstract
Significant efforts have been made to identify HIV-1 neutralizing antibodies because they are considered to be critical to the design of an effective HIV-1 vaccine. Although soluble HIV-1 envelope proteins can be used for this purpose, these reagents differ from membrane-anchored HIV-1 envelope spike in a number of important ways and display only a subset of its native epitopes. Consistent with this, some broadly neutralizing antibodies preferentially bind cell surface-expressed HIV-1 envelope, but not the soluble protein. Here we report the details of a new method for isolating anti-HIV-1 specific B cells based on capturing cells that produce antibodies to cell surface-expressed gp160ΔcBaL. While this method is far less efficient than sorting with soluble envelope proteins, it isolated broadly neutralizing anti-HIV-1 antibodies that bind cell surface expressed gp160ΔcBaL but not soluble envelope proteins.
Keywords: HIV-1, Surface-expressed envelope trimer, Single B cell sort, HIV-1 neutralizing Antibodies
1 Introduction
Broadly neutralizing antibodies (bNAbs) targeting HIV-1 can prevent infection in non-human primates, and control HIV-1 replication in humanized mice (Mascola et al., 2000; Hessell et al., 2009; Klein et al., 2012b). These antibodies are therefore of significant interest for vaccine design and as agents for novel therapeutic approaches (McCoy and Weiss, 2013).
Given their potential importance, substantial efforts have been made to dissect the human anti-HIV-1 antibody response in individuals who display broad and potent HIV-1 serum neutralizing activity (McCoy and Weiss, 2013). An essential component to this effort has been the development of new methods for antibody cloning from single B cells (Wardemann et al., 2003; Tiller et al., 2008). B cells expressing these antibodies were identified by staining them using labeled soluble HIV-1 envelope proteins (Scheid et al., 2009a; Scheid et al., 2009b; Wu et al., 2010; Scheid et al., 2011; Mouquet et al., 2012; Liao et al., 2013) or by screening for HIV-1 neutralization activity in culture supernatants (Walker et al., 2009; Walker et al., 2011).
All bNAbs target the HIV-1 envelope (Env) spike, a glycoprotein complex consisting of three gp120/gp41 heterodimers that are non-covalently associated. Several regions on the HIV-1 spike have been identified as targets of bNAbs, including the CD4 binding site (Wu et al., 2010; Diskin et al., 2011; Scheid et al., 2011), the base of the V3 loop (Walker et al., 2009; Pejchal et al., 2011; Walker et al., 2011; Mouquet et al., 2012), the V1/V2 loops (Walker et al., 2009; Bonsignori et al., 2011; Walker et al., 2011), the membrane-proximal external region (MPER) of gp41 (Morris et al., 2011; Huang et al., 2012), and an epitope recognized by the antibody 8ANC195 (Scheid et al., 2011) that harbors N-linked glycosylation sites at positions 234 and 276 (HXB2c numbering)(West et al., 2013).
Some bNAbs (e.g. PGTs 141–145, PG9/PG16, CH01-CH043) have been shown to bind to an epitope that is preferentially displayed on the surface-expressed HIV-1 spike (Walker et al., 2009; Bonsignori et al., 2011; Walker et al., 2011). These epitopes are potentially important because they are frequent targets of neutralizing antibodies (Gorny et al., 2005; Robinson et al., 2010; Walker et al., 2010; Bonsignori et al., 2011; Moore et al., 2011; Georgiev et al., 2013) and they are promising candidates for HIV-1 antibody-based therapy (Klein et al., 2012b). However, none of the soluble Env proteins (e.g. gp140YU2) developed to date fully mimic the complex antigenic nature of the surface-expressed HIV-1 envelope (Burton et al., 2012). Accordingly, when used as bait for single B cell isolation, soluble HIV-1 envelope proteins identify only a subset of anti-HIV-1 antibody expressing B cells and fail to capture B cells expressing antibodies to some conformational HIV-1 Env epitopes.
In order to overcome this limitation, we set out to develop a method to capture B cells producing anti-HIV-1 antibodies that preferentially react with HIV-1BaL gp160Δc trimer (gp160ΔcBaL) expressed on the surface of transfected cells. Here we report a detailed protocol for this new technique that was used to identify several new antibodies including the bNAbs 3BC176 and 3BC315 (Klein et al., 2012a).
2 Materials and methods
2.1 Human samples and cell lines
Human peripheral blood mononuclear cells (PBMCs) were obtained from HIV-1-infected subjects that were selected based on broad neutralizing serum activity. Subjects 3, 7, and 8 were selected from a cohort of elite controllers (International HIV Controllers Study) from the Ragon Institute (Scheid et al., 2009a; Scheid et al., 2011), and subject C69 from the University of Cologne, Germany (Klein et al., 2012a). Whole blood and leukapheresis samples were collected after signed informed consent in accordance with the Institutional Review Board at The Rockefeller University (protocol MNU-0628). PBMCs were isolated by Ficoll-Paque (GE Healthcare) density gradient centrifugation.
HEK293T/17 (293T; American Type Culture Collection; CRL-11268) cells and BOSC.23 cells (American Type Culture Collection; CRL-11270) were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco) supplemented with 1mM sodium pyruvate (Gibco), 1% antibiotic-antimycotic (Gibco) and 10% fetal bovine serum (FBS; HyClone, Thermo Scientific) at 37°C and 5% CO2. The recombinant murine B cell leukemia cell line 70z/3 (Paige et al., 1978) was grown in RPMI 1640 (Gibco) supplemented with 1mM sodium pyruvate, 2mM L-glutamine (Gibco), 10mM HEPES buffer solution (Gibco), 0.055mM β-Mercaptoethanol (Gibco), 1% antibiotic-antimycotic and 10% FBS (HyClone, Thermo Scientific). After retroviral infection, puromycin (Gibco) was added at a concentration of 1μg/ml to select for puromycin-resistant 70z/3 B cells.
2.2 Expression vectors
pMX-IRES-GFP encoding HIV-1BaL gp160Δc (pMX-gp160ΔcBaL-IRES-GFP) was used for gp160ΔcBaL expression on 293T cells (Fig. 1) (Pietzsch et al., 2010). pMX-IRES-GFP served as the no_insert control (Pietzsch et al., 2010).
Fig. 1. GFP-293TBaL cell viability and expression levels of GFP and gp160ΔcBaL.
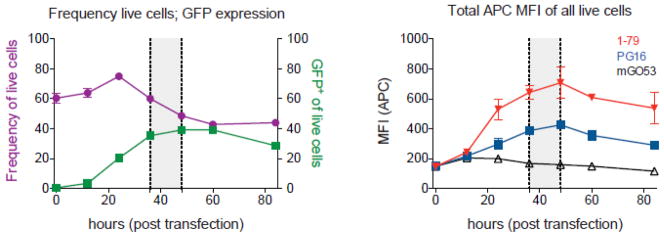
In order to determine the optimal time point for using GFP-293TBaL cells in B cell capturing experiments, cells were evaluated for viability, GFP and gp160ΔcBaL expression between 0 and 84 hours after transfection with pMX-gp160ΔcBaL-IRES-GFP. Frequencies of live cells and live 293T cells expressing GFP are illustrated in purple and green, respectively (left panel). gp160ΔcBaL expression (right panel) on total live cells was measured by using the V3 loop-binding antibody 1–79 (red) and the V1/V2 loop-binding antibody PG16 (blue). mGO53 was used as a negative control (black). Based on these data, GFP-293TBaL cells were collected between 36–48 h (grey field) post transfection to be used for B cell capturing experiments.
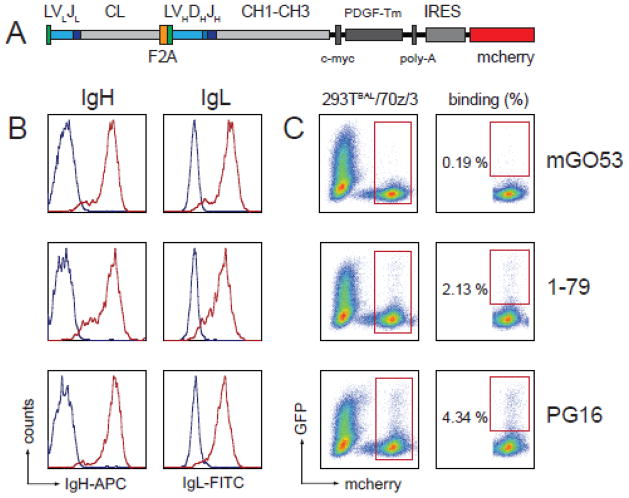
To express the antibodies 1–79 (V3 loop-specific (Scheid et al., 2009a)), PG16 (V1/V2-loop-specific (Walker et al., 2009)) or mGO53 (non HIV-1-reactive (Wardemann et al., 2003)) on the surface of 70z/3 B cells, respective heavy and light chains were cloned into pDisplay (Invitrogen) separated by a 2A self-processing signal sequence from the foot-and–mouth disease virus (F2A; VKQTLNFDLLKLAGDVESNPGP; Fig. 2A) (Szymczak et al., 2004). Antibody sequences included a 5′ Igκ leader and the PDGFR transmembrane domain (PDGFR-TM) in frame with the IgH constant domains enabling cell surface expression of the IgGs. The entire sequence (Leader-Igκ-F2A-IgH-PDGFR-TM) was subcloned into pMX-IRES-mCherry (Fig. 2A)(Bothmer et al., 2013).
Fig. 2. Binding of GFP-293TBaL cells to mGO53-, 1–79-, and PG16-expressing 70z/3 B cells.
(A) Schematic representation of the construct used for mGO53, 1–79, and PG16 surface expression on 70z/3 B cells. L, leader; V, variable gene segment; J, joining gene segment; D, D gene segment; CL, constant domain light chain; CH, constant domain heavy chain; IRES, internal ribosome entry site. F2A, 2A self-processing signal sequence from the foot-and–mouth disease virus (B) Illustration of human antibody heavy and light chain surface expression on 70z3 B cells after infection with the construct displayed in (A). Red and blue lines represent the expression levels measured on mCherryhigh+ and mCherry− B cells, respectively. Histograms on the left and right depict binding of anti-human IgG-APC and FITC-conjugated anti-human IgL mAbs (PG16 and 1–79, lambda; mGO53, kappa), respectively. (C) 70z/3 B cells expressing mGO53, 1–79 or PG16 were co-incubated with GFP-293TBaL cells and analyzed by flow cytometry. Frequencies of double positive (mCherry+ and GFP+) events of all live B cells are indicated in the right panels.
2.3 Antibody-expressing 70z/3 cells
To produce retrovirus BOSC.23 cells were cultured as described above and co-transfected at 80% confluency using FuGENE6 (Roche) and equal amounts of pCL-Eco (IMGENEX) and pMX-IRES-mCherry carrying PG16, 1–79, mGO, or no insert. The supernatant was taken off after 48h (McBride et al., 2006). 70z/3 B cells were infected by resuspending them in retroviral soup in the presence of polybrene (4–10 μg/ml, Sigma). Cells were spinoculated for 2 h at 1000 × g and medium was replaced after 6h. Infection was repeated after 24h following culturing of cells in the presence of 1μg/ml puromycin (Sigma) until the mCherry positive population was highly enriched.
2.4 Transfection of 293T cells
12.5 × 106 293T cells were seeded in 150mm tissue culture plates (Falcon) at a concentration of 0.5 × 106 cells/ml. After 12–24 hours, when the cells reached 60% cell confluency, medium was replaced and cells transfected with 40μg of pMX-gp160ΔcBaL-IRES-GFP or pMX-IRES-GFP using FuGENE6 (Roche) at a ratio of 1:2 according to the manufacturer’s instructions. gp160ΔcBaL-transfected 293T cells (GFP-293TBaL cells) were harvested at serial time points ranging from 0 to 84 hours using enzyme-free Cell Dissociation Buffer (Invitrogen) and resuspended in 1 × PBS supplemented with 2% FBS (FACS buffer) and 2–10mM EDTA. pMX-IRES-GFP-transfected 293T cells were harvested after 36–48h.
GFP expression was used as a surrogate indicator of gp160ΔcBaL cell surface expression using a BD FACSCalibur™ flow cytometer. Cells were subjected to B cell capture experiments only when GFP expression exceeded 40–50%.
2.5 Flow cytometry
GFP-293TBaL cells were incubated with primary antibodies (1–79, PG16, or mGO53) at a final concentration of 10μg/ml for 25min at 4°C. After washing, cells were incubated with anti-human IgG-APC (BD Pharmingen) at a 1:10 dilution for 25 min at 4°C. 4′,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI) at 50ng/ml (Invitrogen) was used for live/dead cells discrimination.
Antibody heavy and light chain surface expression on recombinant 70z/3 B cells was detected with heavy chain-specific anti-human IgG-APC and Ig lambda FITC (Abbiotec) for PG16 and 1–79 or anti-human Ig kappa FITC (Invitrogen) for mGO53 (Fig. 2B). To assess binding of recombinant antibody-expressing 70z/3 B cells to transfected GFP-293TBaL cells (Fig. 2C) both cell types were washed and re-suspended in FACS buffer containing 10 mM EDTA. 106 GFP-293TBaL and 105 70z/3 B cells were mixed in a 15 ml conical tube (BD Falcon) and centrifuged at 4°C for 30 minutes at 311 × g followed by 10 minutes at 76 × g. Supernatant was discarded and the cellular pellet gently re-suspended in FACS buffer (10mM EDTA). Cells were stained with DAPI and assayed for DAPI, GFP and mCherry. Double positive (GFP+ and mCherry+) events represent GFP-293TBaL – 70z/3 B cell doublets. Experiments were performed on an LSRFortessa (BD Biosciences) and data analysis carried out using FlowJo (Tree Star).
2.6 Flow sorting
B cells were isolated using B Cell Isolation Kit II (Miltenyi Biotec) according to the manufacturer’s instructions. Transfected GFP-293TBaL cells were prepared for flow cytometry as described above and mixed with B cells in FACS buffer supplemented with 10mM EDTA at a ratio of 2:1 in a 50ml conical tube (BD Falcon). The mixed cell suspension was centrifuged at 4°C for 10 min at 311 × g and then for additional 10 min at 76 × g (Fig. 3A). The mixed cell pellet was gently re-suspended in 800μl of FACS buffer (10mM EDTA) and incubated with 1:10 anti-human IgG-PE (BD), 1:20 CD20-APC_H7 (BD), 1:20 CD3-PerCP (Invitrogen) antibodies and 50ng/ml DAPI for 25 min at 4°C. After a final washing step cells were re-suspended in 4ml of FACS buffer and strained through a 35μm nylon mesh into a 5ml round-bottom tube (BD Falcon). The mixed cell suspension was kept at 4°C and briefly mixed before sample acquisition on a BD FACSAriaIII set up with a 100 μm nozzle. Healthy donor PBMCs and unlabeled 293T cells were used for single fluorochrome compensation. The gating hierarchy depicted in Fig. 3B was applied to select for live (DAPI−) CD20+ IgG+ GFP+ cell conjugates representing IgG+ B cells attached to GFP-293TBaL cells. CD20+ IgG+ GFP+ events were sorted into 96-well PCR plates (Eppendorf) containing 4μl of 4°C lysis buffer (0.5 × PBS, 10 mM DTT, 8 U RNAsin (Promega), and 0.4 U 5′-3′ Prime RNAse Inhibitor (Eppendorf)). Sorted plates were tightly sealed with Microseal ‘F’ Film (BioRad) and temporarily kept on dry ice (or −80°C freezer) before being subjected to the Ig cloning procedure. Controls sorts were performed under the same protocol, but with GFP-293Tno_insert cells or without any potential bait (i.e. CD20+ IgG+ only).
Fig. 3. Experimental layout of single B cell isolation using GFP-293TBaL cells.
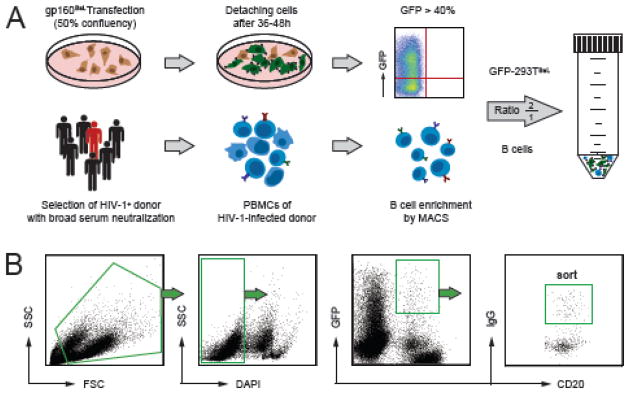
(A) Schematic diagram of the preparations for B cell capturing experiment. GFP-293TBaL cells are harvested between 36–48 h after transfection and are subjected to experiment when >40% of cells express GFP (upper panels). PMBCs obtained from HIV-1-infected individuals with broad serum neutralization are enriched for B cells (lower panels) and incubated with GFP-293TBaL cells in a 1:2 ratio. (B) Gating hierarchy for sorting GFP-293TBaL-B cell doublets represented by CD20+ GFP+ IgG+ events.
2.7 Ig gene amplification, sequence analysis and antibody cloning
Synthesis of B cell cDNA, nested PCR amplification of Ig variable heavy (VH), and light chain (VL) gene transcripts as well as antibody cloning and production were performed as previously described (Tiller et al., 2008; Scheid et al., 2011). Briefly, VH and VL gene transcripts were amplified from single B cell cDNA by two rounds of nested PCR using forward primer mixes specific for the leader region and reverse primers specific for the respective IgH and IgL constant regions (Scheid et al., 2011). All 2nd PCR amplicons were sequenced and analyzed using IgBLAST (http://www.ncbi.nlm.nih.gov/igblast/). After cloning heavy and light chain PCR products into their respective expression vectors antibody production in 293T cells was performed as previously described (Tiller et al., 2008; Mouquet et al., 2011).
3 Results
To determine the optimal conditions for using GFP-293TBaL cells we measured their viability and levels of GFP and HIV-1BaL expression over a 84 hour period after transfection with pMX-gp160ΔcBaL-IRES-GFP (Fig. 1). The average expression levels of GFP reached a maximum after 24h and remained stable thereafter while viability decreased after 24h (Fig. 1). Expression of HIV-1BaL envelope on live cells peaked between 40 and 50h after transfection as determined by staining with 1–79 and PG16 antibodies (Fig. 1). Therefore, cell-capture sorting experiments were performed with GFP-293TBaL cells harvested 36–48h (Fig. 1) after transfection.
To establish the feasibility of capturing HIV-1-specific B cells using GFP-293TBaL cells, we produced three 70z/3 B cell lines expressing the HIV-1-reactive antibodies 1–79 (Scheid et al., 2009a)) and PG16 (Walker et al., 2009)) as well as the non-reactive control antibody mGO53 (Wardemann et al., 2003)(Fig. 2A). Membrane-bound antibody heavy and light chain expression was confirmed by flow cytometry (Fig. 2B). mGO53, 1–79 and PG16 mCherry expressing B cells were incubated with GFP-293TBaL cells and subsequently analyzed for the presence of cell-cell conjugates (i.e. GFP-293TBaL - 70z/3 B cell doublets). 70z/3 B cells expressing surface IgG 1–79 and PG16 were captured by GFP-293TBaL cells at a frequency of 2.13% and 4.34%, respectively. In contrast, only 0.19% of mGO53 expressing 70z/3 B cells bound to GFP-293TBaL cells (Fig. 2C). We conclude that GFP-293TBaL cells are able to capture anti-HIV-1 antibody expressing B cells although at a low efficiency.
To determine whether GFP-293TBaL cells can be used to isolate HIV-1-specific B cells from HIV-1-infected individuals, we applied our sorting strategy (Fig. 3A) to four PBMC samples from HIV-1 clade B-infected donors as previously described (Klein et al., 2012a). Sample acquisition revealed DAPI- GFP+ CD20+ IgG+ cells corresponding to GFP-293TBaL–B cell conjugates (GFP-293TBaL/B cell doublets, Fig. 3B). Individual GFP-293TBaL/B cell doublets were sorted into 96-well PCR plates and cDNA libraries were produced to recover Ig variable heavy and light chain genes by PCR. In total, we retrieved 734 antibody sequences comprising 35 different clones (Klein et al., 2012a). Clonally related sequences within the single donors ranged from 1 – 26% (average 14%) (Klein et al., 2012a). Retrieving B cells carrying the same or related sequences (B cell clones) indicate capturing of B cells reactive to a specific antigen or bait. Thus, clonally related antibody sequences are less likely to be non-specific compared to single unrelated antibody sequences. Consequently, we mainly expressed clonally related antibody sequences for further characterization and tested them for binding to GFP-293TBaL cells by flow cytometry.
As shown in Klein et al., 2012, out of the 37 antibodies produced, 15 recognized GFP-293TBaL cells but not GFP-293Tno_insert cells, while only 4 out of these 15 recognized the soluble form of the HIV-1 envelope (i.e. gp140BaL) (Klein et al., 2012a). Most importantly, 6 of the 15 GFP-293TBaL binding antibodies showed neutralizing activity against two or more viruses (Klein et al., 2012a). The finding that only 15 of 37 cloned antibodies recognized GFP-293TBaL cells suggests that the cell-based HIV-1-specific B cell capture method is not entirely stringent and also captures non-HIV-1 specific B cells that bind to GFP-293TBaL cells.
To determine the level of HIV-1BaL gp160Δc-independent capturing we performed the cell-based capture experiment on the PBMCs from the same subject (#8) using GFP-293TBaL, GFP-293Tno_insert or no bait (Fig. 4). Heavy chain sequence analysis identified 13% and 14% clonally related sequences in the GFP-293Tno_insert and no bait sorts, respectively (Fig. 4). In contrast, 26% of the B cells sorted with GFP-293TBaL were clonally related. Comparison of antibody sequences among the groups identified two B cell clones that could be detected in control sorts as well as the original experiment using GFP-293TBaL as the bait (Fig. 4). As expected, these antibodies did not bind to cell surface expressed HIV-1 envelope. We conclude that GFP-293TBaL cells can be used, to identify HIV-1-specific antibodies that recognize surface HIV-1 envelope proteins but at a low sorting efficiency when compared to soluble protein baits (Scheid et al., 2009a; Scheid et al., 2011).
Fig. 4. Efficacy of B cell isolation using GFP-293TBaL cells.
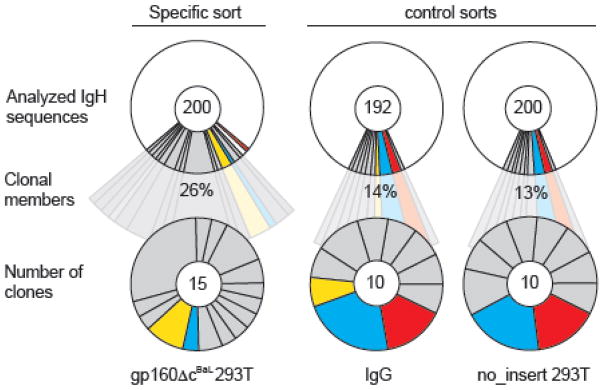
IgH sequences were retrieved from single B cell sorts of the same sample (subject 8) using GFP-293TBaL cells (left) and compared to IGH sequences obtained from control sorts (GFP-293Tno_insert, right; IgG, middle). Control sorts were performed with 293T empty-vector-transfected cells (pMX-no-insert-IRES-GFP) and staining for just IgG+ B cells. Between 192 and 200 IgH sequences were analyzed (upper panel) and the frequency of clonally related sequences (middle row) and number of clones (bottom row) examined. Antibody clones that were detected in more than one sort are highlighted in the same color.
4 Discussion
Single cell sorting techniques using soluble HIV-1 Env proteins are effective tools for studying the antibody response to HIV-1 and have been used to uncover a large number of potent broadly neutralizing antibodies (Scheid et al., 2009b; Wu et al., 2010; Scheid et al., 2011). However, none of the soluble Env trimers developed to date is a precise structural mimic of the functional HIV-1 envelope spike (Davenport et al., 2011; Burton et al., 2012). For example, bNAb PG9 (Walker et al., 2009) recognizes HIV-1 by contacting two of the three gp120 protomers in the native trimeric Env expressed by the virus (Julien et al., 2013), but fails to recognize most soluble protein trimers (Davenport et al., 2011). Single B cells sorting using soluble proteins would therefore fail to identify HIV-1-reactive antibodies like PG9 that preferentially recognize native epitopes on cell or virus surface-expressed HIV-1 trimers.
In order to overcome this limitation, we used 293T cells expressing gp160ΔcBaL as a bait for HIV-1-reactive B cells and were able to identify 15 HIV-1-reactive mAbs, some of which showed HIV-1 neutralizing or even broad neutralizing (i.e. 3BC176 and 3BC315) activity (Klein et al., 2012a).
Potential pitfalls in this approach include the fact that commercial cell sorters are designed to discriminate cells in close proximity ensuring that a single sort event does not result in sorting multiple cells (Battye et al., 2000). Moreover, incorporation of two cells in a droplet likely increases droplet size, which affects aiming accuracy of sorting into 96-well plates due to aerodynamic effects. Therefore, precise positioning of the 96-well plate is required to ensure accurate sorting into small volumes of lysis buffer at the bottom of single wells. (Battye et al., 2000)
Control experiments and the finding that only 40% of the cloned antibodies bound to GFP-293TBaL cells suggests that HIV-1BaL gp160Δc-independent interactions between B and GFP-293TBaL cells were a common occurrence. Consistent with our observations, sorting HIV-1-specific B cells using fluorescent Gag-Env virus-like-particles (VLPs) instead of GFP-293TBaL cells yielded only 0.7 – 10% of virus binding antibodies (Hicar et al., 2010).
The yield of specific conjugates might be increased by more stringent selection for GFP-293TBaL cells that express higher levels of Env and by reducing sheer forces during sample preparation. For example, the stability of antigen-specific conjugates between cytotoxic T lymphocytes (CTL) and fibroblasts was increased at higher temperatures while non-specific association was unaffected (Hubbard et al., 1990). In addition, the role of centrifugation and shear forces during FACS acquisition has been shown to affect conjugate stability (Segal and Stephany, 1984; Callewaert et al., 1991; Hauss et al., 1996). Finally, different cell ratios, densities and incubation kinetics could also enhance conjugate formation (Cavarec et al., 1990).
Binding to functional HIV-1 Env trimer is thought to be essential for activity of neutralizing antibodies (Fouts et al., 1997), whereas HIV-1-specific, but non-neutralizing antibodies seem to preferentially recognize non-functional forms of the envelope (i.e. uncleaved gp160 precursor protein, gp41 stumps, or gp120/gp41 monomers) (Poignard et al., 2003; Moore et al., 2006). The fact that only 6 out of 15 (40%) HIV-1-specific antibodies showed neutralizing activity suggests the existence of non-functional, possibly un-cleaved Env molecules on the cell surface of GFP-293TBaL cells leading to the capture of Env-specific B cells without neutralizing activity. gp160 cleavage efficiency differs among viral strains and HIV-1JR-FL, in particular, appeared to be efficiently cleaved in the context of surface expression on transfected cells (Pancera and Wyatt, 2005). Moreover cleavage efficiency of membrane bound Env proteins could be enhanced by glycosidase-protease digests (Crooks et al., 2011). Therefore, using gp160ΔcJR-FL instead of gp160ΔcBaL as well as improving cleavage efficiency might reduce non-native Env exposure and improve the frequency of bNAb isolation. In addition, the level of Env surface expression, the celltype on which the Envs are expressed as well as the stability of the Env trimers before and after antibody binding can influence the ability of the bait to capture bNAbs carrying B cells. Finally, soluble Env proteins that closely mimic the membrane-bound configuration (Binley et al., 2000; Harris et al., 2011) might be of high value to isolate bNAbs that are not detected by the currently used proteins.
5 Conclusions
We conclude that GFP-293TBaL cells can be used to isolate B cells expressing HIV-1-reactive antibodies. Cell-associated HIV-1 Env trimers are particularly useful for the identification of antibodies that recognize conformational epitopes (Klein et al., 2012a) and to define new neutralizing epitopes on the HIV-1 envelope. This method is far less efficient than methods that use soluble trimer as baits (Wu et al., 2010; Morris et al., 2011; Scheid et al., 2011; Huang et al., 2012), but might be improved by a variety of means to increase specific yield.
Highlights.
Single B cell sorts using surface-expressed HIV-1 envelope trimer
Isolation of HIV-1 neutralizing antibodies
Acknowledgments
We thank all HIV-1-infected volunteers who participated in this study. We also thank Yelena Shatalina for cell sorting support and all members of the Nussenzweig laboratory for helpful discussions. This research was supported in part by grant # UL1 TR000043 from the National Center for Advancing Translational Sciences (NCATS), the National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program, and NIH grants to M. C. N. (AI081677 and UM1AI100663) as well as the Bill and Melinda Gates Foundation (OPP1033115). C.G. and H.G. were supported by The German National Academic Foundation. M.C.N. is a Howard Hughes Medical Institute investigator. F.K. was supported by the German Research Foundation (DFG, KL 2389/1-1), the Stavros Niarchos Foundation and the Robert Mapplethorpe Foundation. The authors declare no conflicting financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Battye FL, Light A, Tarlinton DM. Single cell sorting and cloning. Journal of immunological methods. 2000;243:25–32. doi: 10.1016/s0022-1759(00)00225-8. [DOI] [PubMed] [Google Scholar]
- Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74:627–43. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, Sinangil F, Pancera M, Yongping Y, Zhang B, Zhu J, Kwong PD, O’Dell S, Mascola JR, Wu L, Nabel GJ, Phogat S, Seaman MS, Whitesides JF, Moody MA, Kelsoe G, Yang X, Sodroski J, Shaw GM, Montefiori DC, Kepler TB, Tomaras GD, Alam SM, Liao HX, Haynes BF. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Rommel PC, Gazumyan A, Polato F, Reczek CR, Muellenbeck MF, Schaetzlein S, Edelmann W, Chen PL, Brosh RM, Jr, Casellas R, Ludwig T, Baer R, Nussenzweig A, Nussenzweig MC, Robbiani DF. Mechanism of DNA resection during intrachromosomal recombination and immunoglobulin class switching. J Exp Med. 2013;210:115–23. doi: 10.1084/jem.20121975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, Kaufmann DE, McElrath MJ, Nussenzweig MC, Pulendran B, Scanlan CN, Schief WR, Silvestri G, Streeck H, Walker BD, Walker LM, Ward AB, Wilson IA, Wyatt R. A Blueprint for HIV Vaccine Discovery. Cell Host and Microbe. 2012;12:396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callewaert D, Radcliff G, Waite R, Lefevre J, Poulik M. Characterization of Effector-Target Conjugates for Cloned Human Natural-Killer and Human Lymphokine Activated Killer-Cells by Flow-Cytometry. Cytometry. 1991;12:666–676. doi: 10.1002/cyto.990120711. [DOI] [PubMed] [Google Scholar]
- Cavarec L, Quillet-Mary A, Fradelizi D, Conjeaud H. An improved double fluorescence flow cytometry method for the quantification of killer cell/target cell conjugate formation. Journal of immunological methods. 1990;130:251–261. doi: 10.1016/0022-1759(90)90055-z. [DOI] [PubMed] [Google Scholar]
- Crooks ET, Tong T, Osawa K, Binley JM. Enzyme digests eliminate nonfunctional Env from HIV-1 particle surfaces, leaving native Env trimers intact and viral infectivity unaffected. J Virol. 2011;85:5825–39. doi: 10.1128/JVI.00154-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport TM, Friend D, Ellingson K, Xu H, Caldwell Z, Sellhorn G, Kraft Z, Strong RK, Stamatatos L. Binding Interactions between Soluble HIV Envelope Glycoproteins and Quaternary-Structure-Specific Monoclonal Antibodies PG9 and PG16. Journal of Virology. 2011;85:7095–7107. doi: 10.1128/JVI.00411-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin R, Scheid JF, Marcovecchio PM, West AP, Klein F, Gao H, Gnanapragasam PNP, Abadir A, Seaman MS, Nussenzweig MC, Bjorkman PJ. Increasing the Potency and Breadth of an HIV Antibody by Using Structure-Based Rational Design. Science (New York, NY) 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts TR, Binley JM, Trkola A, Robinson JE, Moore JP. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. Journal of Virology. 1997;71:2779–2785. doi: 10.1128/jvi.71.4.2779-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev IS, Doria-Rose NA, Zhou T, Kwon YD, Staupe RP, Moquin S, Chuang GY, Louder MK, Schmidt SD, Altae-Tran HR, Bailer RT, McKee K, Nason M, O’Dell S, Ofek G, Pancera M, Srivatsan S, Shapiro L, Connors M, Migueles SA, Morris L, Nishimura Y, Martin MA, Mascola JR, Kwong PD. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science. 2013;340:751–6. doi: 10.1126/science.1233989. [DOI] [PubMed] [Google Scholar]
- Gorny MK, Stamatatos L, Volsky B, Revesz K, Williams C, Wang XH, Cohen S, Staudinger R, Zolla-Pazner S. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. Journal of Virology. 2005;79:5232–5237. doi: 10.1128/JVI.79.8.5232-5237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse PJ, Milne JLS, Wilson IA, Olson WC, Moore JP, Subramaniam S. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11440–11445. doi: 10.1073/pnas.1101414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauss P, Selz F, Fischer A. Comparative analysis of CD4-mediated down-regulation of T cell adhesion to B cells by flow cytometry and fluorescence microscopy. Cytometry. 1996;23:39–47. doi: 10.1002/(SICI)1097-0320(19960101)23:1<39::AID-CYTO6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Hessell AJ, Poignard P, Hunter M, Hangartner L, Tehrani DM, Bleeker WK, Parren PW, Marx PA, Burton DR. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nature medicine. 2009;15:951–4. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicar MD, Chen X, Briney B, Hammonds J, Wang JJ, Kalams S, Spearman PW, Crowe JE. Pseudovirion particles bearing native HIV envelope trimers facilitate a novel method for generating human neutralizing monoclonal antibodies against HIV. Journal of acquired immune deficiency syndromes (1999) 2010;54:223–235. doi: 10.1097/QAI.0b013e3181dc98a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard B, Glacken M, Rodgers J, Rich R. The role of physical forces on cytotoxic T cell-target cell conjugate stability. The Journal of Immunology. 1990;144:4129–4138. [PubMed] [Google Scholar]
- Julien JP, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, Sanders RW, Moore JP, Wilson IA, Ward AB. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci U S A. 2013;110:4351–6. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Gaebler C, Mouquet H, Sather DN, Lehmann C, Scheid JF, Kraft Z, Liu Y, Pietzsch J, Hurley A, Poignard P, Feizi T, Morris L, Walker BD, Fatkenheuer G, Seaman MS, Stamatatos L, Nussenzweig MC. Broad neutralization by a combination of antibodies recognizing the CD4 binding site and a new conformational epitope on the HIV-1 envelope protein. Journal of Experimental Medicine. 2012a;209:1469–1479. doi: 10.1084/jem.20120423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, Zang T, Dorner M, Billerbeck E, Labitt RN, Gaebler C, Marcovecchio PM, Incesu RB, Eisenreich TR, Bieniasz PD, Seaman MS, Bjorkman PJ, Ravetch JV, Ploss A, Nussenzweig MC. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 2012b;492:118–22. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Program NCS, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–76. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, Beary H, Hayes D, Frankel SS, Birx DL, Lewis MG. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6:207–10. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- McBride KM, Gazumyan A, Woo EM, Barreto VM, Robbiani DF, Chait BT, Nussenzweig MC. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc Natl Acad Sci U S A. 2006;103:8798–803. doi: 10.1073/pnas.0603272103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy LE, Weiss RA. Neutralizing antibodies to HIV-1 induced by immunization. J Exp Med. 2013;210:209–23. doi: 10.1084/jem.20121827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Crooks ET, Porter L, Zhu P, Cayanan CS, Grise H, Corcoran P, Zwick MB, Franti M, Morris L, Roux KH, Burton DR, Binley JM. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. Journal of Virology. 2006;80:2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Gray ES, Sheward D, Madiga M, Ranchobe N, Lai Z, Honnen WJ, Nonyane M, Tumba N, Hermanus T, Sibeko S, Mlisana K, Abdool Karim SS, Williamson C, Pinter A, Morris L, Study C. Potent and broad neutralization of HIV-1 subtype C by plasma antibodies targeting a quaternary epitope including residues in the V2 loop. Journal of Virology. 2011;85:3128–3141. doi: 10.1128/JVI.02658-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris L, Chen X, Alam M, Tomaras G, Zhang R, Marshall DJ, Chen B, Parks R, Foulger A, Jaeger F, Donathan M, Bilska M, Gray ES, Abdool Karim SS, Kepler TB, Whitesides J, Montefiori D, Moody MA, Liao HX, Haynes BF. Isolation of a human anti-HIV gp41 membrane proximal region neutralizing antibody by antigen-specific single B cell sorting. PLoS ONE. 2011;6:e23532. doi: 10.1371/journal.pone.0023532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H, Klein F, Scheid JF, Warncke M, Pietzsch J, Oliveira TY, Velinzon K, Seaman MS, Nussenzweig MC. Memory B cell antibodies to HIV-1 gp140 cloned from individuals infected with clade A and B viruses. PLoS One. 2011;6:e24078. doi: 10.1371/journal.pone.0024078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PN, Spencer DI, Seaman MS, Schuitemaker H, Feizi T, Nussenzweig MC, Bjorkman PJ. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A. 2012;109:E3268–77. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige CJ, Kincade PW, Ralph P. Murine B cell leukemia line with inducible surface immunoglobulin expression. Journal of immunology (Baltimore, Md: 1950) 1978;121:641–647. [PubMed] [Google Scholar]
- Pancera M, Wyatt R. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology. 2005;332:145–156. doi: 10.1016/j.virol.2004.10.042. [DOI] [PubMed] [Google Scholar]
- Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. A Potent and Broad Neutralizing Antibody Recognizes and Penetrates the HIV Glycan Shield. Science (New York, NY) 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietzsch J, Scheid JF, Mouquet H, Klein F, Seaman MS, Jankovic M, Corti D, Lanzavecchia A, Nussenzweig MC. Human anti-HIV-neutralizing antibodies frequently target a conserved epitope essential for viral fitness. Journal of Experimental Medicine. 2010;207:1995–2002. doi: 10.1084/jem.20101176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poignard P, Moulard M, Golez E, Vivona V, Franti M, Venturini S, Wang M, Parren PWHI, Burton DR. Heterogeneity of Envelope Molecules Expressed on Primary Human Immunodeficiency Virus Type 1 Particles as Probed by the Binding of Neutralizing and Nonneutralizing Antibodies. Journal of Virology. 2003;77:353–365. doi: 10.1128/JVI.77.1.353-365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JE, Franco K, Elliott DH, Maher MJ, Reyna A, Montefiori DC, Zolla-Pazner S, Gorny MK, Kraft Z, Stamatatos L. Quaternary epitope specificities of anti-HIV-1 neutralizing antibodies generated in rhesus macaques infected by the simian/human immunodeficiency virus SHIVSF162P4. Journal of Virology. 2010;84:3443–3453. doi: 10.1128/JVI.02617-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, Phogat A, Chakrabarti B, Li Y, Connors M, Pereyra F, Walker BD, Wardemann H, Ho D, Wyatt RT, Mascola JR, Ravetch JV, Nussenzweig MC. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009a;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Feldhahn N, Walker BD, Pereyra F, Cutrell E, Seaman MS, Mascola JR, Wyatt RT, Wardemann H, Nussenzweig MC. A method for identification of HIV gp140 binding memory B cells in human blood. Journal of immunological methods. 2009b;343:65–67. doi: 10.1016/j.jim.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Olivera TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton D, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. Sequence and Structural Convergence of Broad and Potent HIV Antibodies That Mimic CD4 Binding. Science. 2011 doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal DM, Stephany DA. The measurement of specific cell: cell interactions by dual-parameter flow cytometry. Cytometry. 1984;5:169–181. doi: 10.1002/cyto.990050211. [DOI] [PubMed] [Google Scholar]
- Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, Vignali DA. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–94. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. Journal of immunological methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien J-P, Wang S-K, Ramos A, Chan-Hui P-Y, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong C-H, Phogat S, Wrin T, Simek MD, PGP, Koff WC, Wilson IA, Burton DR, Poignard P. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011:1–6. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–9. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. A Limited Number of Antibody Specificities Mediate Broad and Potent Serum Neutralization in Selected HIV-1 Infected Individuals. PLoS Pathogens. 2010;6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science (New York, NY) 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- West AP, Jr, Scharf L, Horwitz J, Klein F, Nussenzweig MC, Bjorkman PJ. Computational analysis of anti-HIV-1 antibody neutralization panel data to identify potential functional epitope residues. Proc Natl Acad Sci U S A. 2013;110:10598–603. doi: 10.1073/pnas.1309215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O’Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–61. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]