Abstract
Intratumour heterogeneity is a longstanding field of focus for both researchers and clinicians. It refers to the diversity amongst cells within the same tumour. Two major hypotheses have attempted to explain the existence of intratumour heterogeneity: (i) the clonal evolution (CE) theory and (ii) the cancer stem cell (CSC) model. CE theory emphasizes the evolutionary biological characteristics of the tumour, underscoring the initiation and progression of the disease. In contrast, the CSC model focuses on stem cell differentiation into distinct functions in order to stabilize the tumour microenvironment. Here we consider single-cell sequencing (SCS) as a newly developed technique for application to the investigation of intratumour heterogeneity and assess its relevance within research and clinical environments. Early detection of rare tumour cells, monitoring of circulating tumour cells (CTCs) and control of the occurrence of drug resistance are important goals in early diagnosis, prognosis prediction and individualized medicine.
Keywords: clinical applications, intratumour heterogeneity, single-cell sequencing
INTRODUCTION
Intratumour heterogeneity refers to a tumour composed of cancerous cells with alternative genotypes and phenotypes, which results in differentiated proliferation, aggressiveness and drug sensitivity, thereby affecting the patient's diagnosis and prognosis. With the advent of single-cell sequencing (SCS), researchers have gained a deeper and broader insight into the formation of heterogeneity. And applications of the technique, including early diagnosis, prognosis prediction and guided personalized treatment, are promising for future medical practice. Here, we present a review to summarize intratumour heterogeneity and SCS and suggest the integration of both into future research and clinical practice.
INTRATUMOUR HETEROGENEITY
Definition and history
Intratumour heterogeneity has been recognized for many years and is defined as the presence of different genotypes and phenotypes in different tumour cells within solid tumours. In the nineteen century, pathologists found the occurrence of cellular polymorphisms within a tumour using a microscope, indicating that the tissue was composed of various types of cells.1 In the 1970s, Heppner et al.2 and Fidler et al.3 demonstrated the existence of tumour subpopulations, which present different manifestations of the tumour, such as tumorigenicity, metastasis and drug resistance, puzzling clinicians. Later, with the development of biological techniques, intratumour heterogeneity was described as a solid tumour consisting of cells that have unique cellular surface biomarkers.4 Currently, with the advent of genetic sequencing, tumour heterogeneity considers different levels of genetic architecture, including the genetic heterogeneity and epigenetic heterogeneity that eventually induce the various phenotypes and functions in tumour cells.
The history of researching tumour heterogeneity is one of progressive levels of resolution, commensurate with the increasing sophistication of technology, moving from cellular morphology, tumour histology, karyotyping and cellular biomarkers (consisting of receptors and immunological characteristics) towards genetic alterations. Tumour heterogeneity still perplexes the researcher and clinician when exploring the mechanisms of tumorigenesis, development, metastasis and resistance to therapy.
Two hypotheses
The acquisition of genetic heterogeneity and epigenetic heterogeneity is based on two major theories: The cancer stem cell (CSC) model, which assumes an ability for self-renewal and differentiation, and the clonal evolution (CE) theory, which postulates that tumours may evolve under the stress of environmental intervention.
CSC model
Recently, the hypothesis of tumour stem cells has been highlighted in the tumour research field. Researchers assumed that CSCs, a small subpopulation of tumour, have the ability to retain their genetic composition via the process of duplication (self-renewal) and differentiation as seen in normal adult stem cells, thus producing the phenotypic and functional heterogeneity that are considered to be polymorphisms of tumour cells.5
CSC was first identified by Dick et al.6 in acute myelogenous leukaemia, which was subsequently proven to present in other solid tumours.7 The identification of cancer stem cells relies on the recognition of the cell surface marker expressions. In colorectal cancer, several studies found cancer stem cells with CD44 and CD133 expression as biomarkers.8,9 Eramo et al.10 discovered tumorigenic cells that express CD133 in lung cancer. These expression positive cells promoted proliferation, thus generating tumour xenografts in mice. And in other tumour types, such as pancreatic cancer, liver cancer and breast cancer, they all have their specific surface markers and also share the same ones. In prostate cancer, cancer stem cells are also regarded as the major reason and fundamental molecular signature for tumour recurrence and development, although the understanding of tumour heterogeneity is evolving. Prostate CSC was recently defined by TRA-1-60, CD151 and CD166 expression as immature AR-negative cells in prostate cancer xenografts.11
In all, the identification of CSCs in various types of cancers can help researchers to understand the origin of tumours, and thus develop approaches to locate and target the biomarkers of CSCs before their development and evolution.
Clonal evolution theory
However, CSC is not the only hypothesis proposed for intratumour heterogeneity; Nowell12 first suggested the CE theory as another possible mechanism of tumorigenicity. Multiple mutations occurring within a random single cell can provide a survival advantage over other normal cells. Along with treatment and medical intervention, these cells may grow insensitive to therapy (drug resistance) and gain the ability to migrate to other tissues (metastasis).13,14
The concept of ‘driver' and ‘passenger' mutations is the core of the CE theory: a driver mutation is defined as one that gives a selective advantage to a clone in its microenvironment, through either increasing its survival or reproduction. Passenger mutation refers to a mutation that has no effect on the fitness of a clone yet may be associated with a clonal expansion because it occurs in the same genome with a driver mutation. When considering tumour evolution, mutations can be separated into three types, including public mutation (in all cancer cells), semiprivate mutation (in a detectable fraction of cancer cells) and private mutation (in a single or few cancer cells).15 In a recent study, Gerlinger et al.16 performed exome sequencing on multiple spatially separated samples from primary renal carcinomas and several metastatic sites, revealing that 63%–69% of all somatic mutations were private ones. The results implied that tumour heterogeneity exists not only between primary and metastatic lesions but also within multiple regions of the same tumour bulk. Because there is no apparent functional advantage of private mutations for the fitness of a clone, we speculate that a newly emerged tumour cell population consists of a few private mutations. To some extent, the private mutation frequency can be considered to correspond to tumour age. The greater the frequency, the longer the transformation time the tumour has had. Therefore, the tumour evolves with the presentation of newly emerged private mutations, as described by the trunk-branch model, a theory confirmed by Gerlinger and his colleagues.16 An ancestral relationship and a phylogenetic tree of tumour regions was constructed using their genetic analysis.16
The CE theory has also been clearly identified in many studies. Sidransky et al.17 found that the histological progression of brain tumours was related to the p53 gene mutation in cells. Several somatic mutations that differ between primary and metastatic breast cancer have been discovered, indicating that evolution occurs with disease progression.18 Researchers suspect that there is at least a decade between the occurrence of the initiating mutation and the non-metastatic founder cell, and another 5 years before the acquisition of metastatic ability.19
Current understanding and research significance
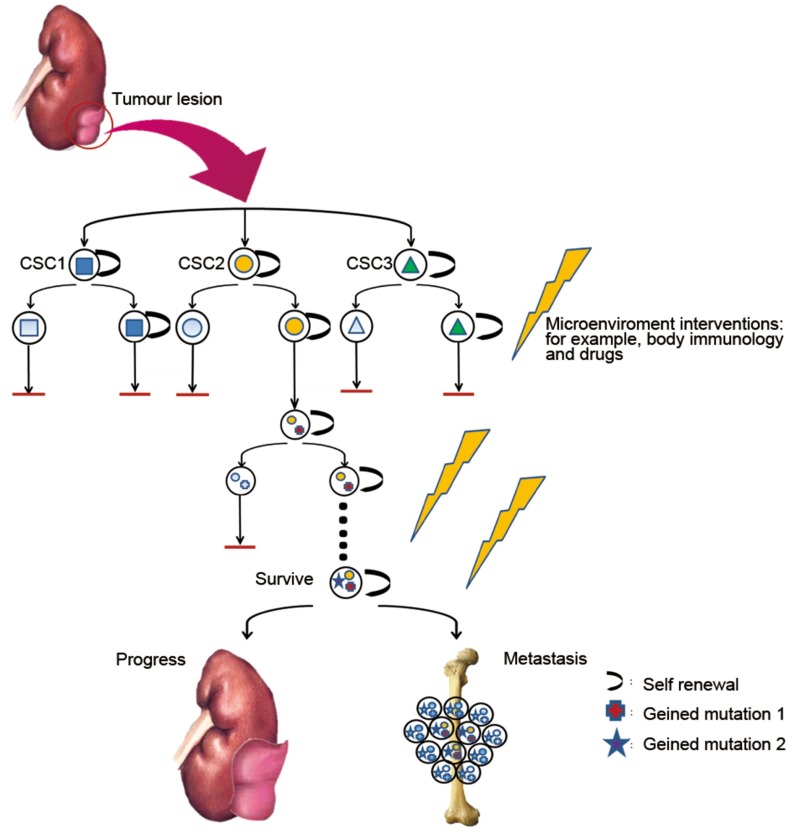
The models of cancer stem cell and clonal evolution are likely not mutually exclusive to one another but instead coexist in a dynamic state (Figure 1). Tumour cells are more likely to have spatial and temporal changes, which eventually lead to intratumour heterogeneity as dictated by the complexity of the tumour microenvironment. Tumour CE provides cancer cells with different genetic features. In contrast, in the process of evolution, newly emerged CSC populations with special features help cancers by creating a more stable state through specific differentiation. Thus, heterogeneous tumours should be regarded as complex societies where even a minor subpopulation may affect the growth and invasiveness of the entire tumour.
Figure 1.
Intratumour heterogeneity through space and time. In a tumour, genetic and epigenetic alterations may result from the emergence of a group of self-renewing CSCs. Through microenvironment interventions, including the immune response and drug treatment, some CSCs are inhibited. However, some genetic alterations occur in specific CSCs, helping the CSC to gain the ability to survive treatment, a process that is known as clonal evolution. After several cycles of intervention, the surviving CTC is capable of tumour progression and metastasis, affecting the patient's survival. CSC, cancer stem cell; CTC, circulating tumour cell.
A recent study sequenced epidermal growth factor receptor (EGFR) mutations in 264 patients with advanced non-small-cell lung cancer who received first-line chemotherapy before and after the treatment. They found that the frequency of EGFR mutation was 34.5% before the therapy and decreased to 23.1% after, indicating that heterogeneity occurs swiftly under selection pressure.20 Further, Martins et al.21 presented the analysis of BRCA1, PTEN and p53 in 55 BRCA-1-associated breast tumours, revealing that loss of PTEN is the most common event in the basal-like subtype, whereas TP53 mutation occurs primarily with a rare PIK3CA mutant in most luminal tumours. The results show the different mutations in different spatial tumours. Sottoriva's experiments with spatially distinct tumour specimens from 11 patients with glioblastoma discovered copy number alterations of EGFR and CDKN2A/B/p14ARF in the early stage and aberrations of PDGFRA and PTEN in later stages, revealing that the tumour evolves in a dynamic process with some mutational losses and acquisitions.22
A wealth of emerging evidence suggests that the temporal and spatial intratumour heterogeneity that branched evolution produces in linear models may support tumour survival in a hostile microenvironment by adaptation to the human body. The survival advantage of the tumour ‘society' may cause progression and metastasis, worsening the patient's condition. Thus, investigation and evaluation of heterogeneous tumour cells are of great value to diagnosis, prognosis prediction and early therapy interventions.
SCS AS A NEWLY EMERGED TECHNIQUE
The SCS landscape
Because intratumour heterogeneity is based on the CSC model and CE theory, which emphasize that a tumour is composed of complex mixtures of cells with various genomes and functions, it is suspected that the admixture might influence the results and analysis revealed by traditional next-generation sequencing (NGS). Therefore, single-cell analysis is a feasible approach to solving the problems caused by multiple mixtures of mutations within tumour cells (Table 1).
Table 1. Single-cell profiling in tumour research.
| Cancer | Level | Method |
|---|---|---|
| Melanoma27 | mRNA | NGS |
| Myeloproliferative neoplasm24 | Exome | NGS |
| Bladder45 | DNA | NGS |
| Kidney23 | Exome | NGS |
| Prostate25 | RNA | Microarray |
| Breast26,46 | RNA | Microarray, FISH |
| Colon47 | RNA | Microarray |
Abbreviations: FISH, fluorescence in situ hybridisation; NGS, next generation sequencing.
Generally, SCS can be divided into single-cell genomic sequencing and single-cell transcriptomic sequencing, which provide different genetic analyses for different objects. Single-cell genomic sequencing mainly focuses on the general landscape of mutations, such as single nucleotide variations and indels (insertions and deletions) in a single cell. Xu et al.23 presented a single-cell exome sequencing in renal cell carcinoma that revealed a small number of ‘mountain' genes and a significantly greater number of ‘hill' genes, providing the first detailed intratumoural genetic landscape at a single-cell level. A whole-exome SCS of 58 cells from a JAK2-negative myeloproliferative neoplasm patient, carried out by Hou et al.,24 found thrombocythemia-related candidate mutations, such as sSESN2 and NTRK1, in the neoplasm, indicating the ability of this sequencing approach to characterize the genetic architecture of the neoplasm, its clonal evolution and candidate driver genes.
Genetic network regulation is usually analysed by single-cell transcriptomic sequencing, especially in stem cells and early embryonic cell populations with a high degree of heterogeneity. It is helpful in analysing the process and regulatory networks of cell differentiation, reprogramming and transdifferentiation. Further, another aspect of single-cell transcriptomic sequencing is revealing information about transcriptomic alterations, including those related to mRNAs, microRNAs, retained introns, alternative splicings, long-noncoding RNAs and fusion genes, with a much higher detection rate. Welty et al.25 performed transcriptomic profiling in a single prostate cancer, showing a low false positive rate when compared with 5-cell pools and 10-cell pools. In another study, elevated transcript levels of epithelial mesenchymal transition-associated genes during metastasis were discovered for the first time by high dimensional gene expression analysis of CTCs from patients with metastatic breast cancer.26 However, existing methods of single-cell transcriptomic expression profiling are not sufficient. Ramskold et al.27 described a robust mRNA-seq protocol called Smart-Seq with higher sensitivity and quantitative accuracy to improve read coverage across transcripts, enhancing detailed analysis of alternative transcript isoforms and identification of single nucleotide polymorphisms.
The SCS procedure
SCS consists of two procedures: the isolation of single cells and their subsequent genomic profiling. In the first stage, there are three major methods for isolating single cells from solid tumour bulks. Micromanipulation is used to isolate cells from liquid samples, but this process is labour intensive. Another method is laser-capture microdissection, which acquires single cells directly isolated from tissues with their own characteristic histological structures, such as ducts or lobules. However, during the process of microdissection, some chromosome sections are inevitably sliced, potentially influencing the results. So far, flow cytometry, by means of fluorescence-activated cell sorting,28 is the most efficient method of single-cell isolation.
After the large-scale collection of various single cells, genomic profiling is performed. There are several methods to acquire the genomic characteristics of single cells, including cytological approaches to visualize chromosomes, microarray comparative genomic hybridisation coupled with whole-genome amplification, as well as NGS. Compared with the former two methods, NGS has the advantage of high throughput with several gigabytes of data produced from one cycle and at low cost. Thus, more and more scientists are choosing to use NGS to discover genomic and transcriptomic aberrations.
However, because of the tiny amount of DNA extracted from a single cell, if a copying error occurs during the replication cycle, the copies will be overrepresented in the final samples. The bias existing in the SCS can result in an uneven distribution of sequencing reads through the genome, affecting final sequential outcomes. Thus, simple SCS paired with whole-genome amplification is not suitable for quantifying copy number or allelic imbalances in single nucleotide polymorphisms or copy number variants. Multiple displacement amplification, which is thought to provide advantages over PCR-based methods by using priming and strand-displacing phi29 polymerase under isothermal conditions,29 still exhibits reckoned bias because of nonlinear amplification without optimisation. Zong et al.30 reported a new whole-genome amplification method, multiple annealing and looping-based amplification cycles (MALBAC), to reduce the bias caused by low genome coverage. This method employs special primers to start amplification, allowing DNA to be copied to form loops that cannot be used as templates for further replication. This process results in only the initial genomic DNA being copied in each cycle and thus lowering the sequencing bias. This technique solves the biggest problem in SCS and is used in both basic research science and individualized medicine. Recently, the genomes of 99 sperm cells from an Asian cohort were sequenced using MALBAC, determining that sperm DNA abnormalities, such as single nucleotide polymorphisms and copy number variants, exist even in healthy populations.31 These results offer insight into the relationship between genome instability and fertility.
CLINICAL APPLICATION
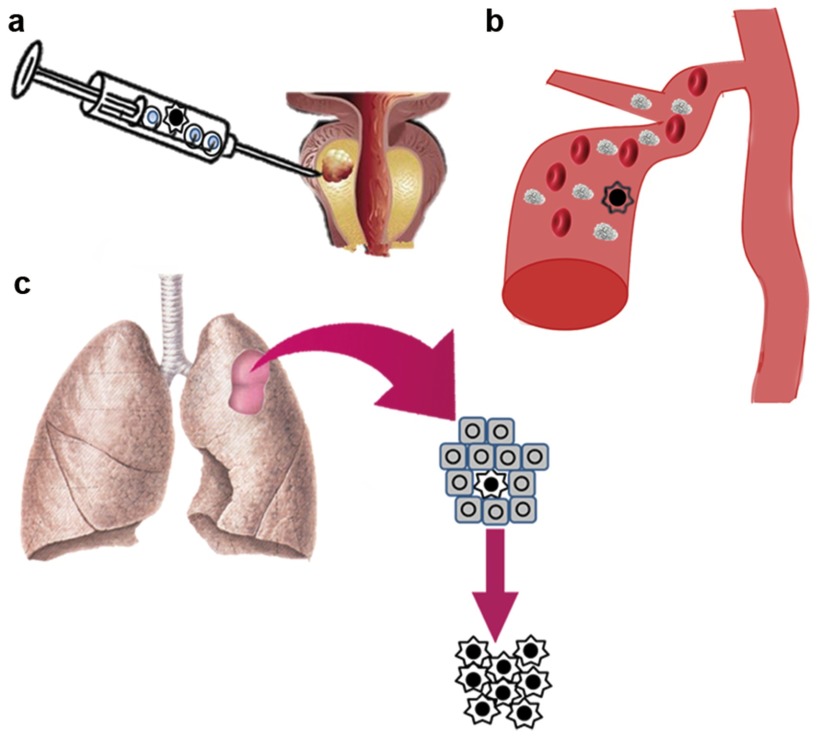
SCS has been applied in the cancer research field with fantastic results, and research indicates that it has the potential to solve clinical problems and eventually bring great benefits to patients (Figure 2).
Figure 2.
The application of SCS in clinics. (a) Profiling of rare tumour cells in a clinical sample, assisting in early diagnosis. (b) Isolation and profiling of CTCs for detection. (c) Identification and profiling of some drug resistant tumour cells for the purpose of individualized treatment. CTC, circulating tumour cell; SCS, single-cell sequencing.
Diagnosis in rare tumour cells
In clinics, some cancers may go undetected because of fewer tumour cells in the tissue during early stages, leading to missed diagnoses and poor prognoses. In breast cancer, low-grade ductal carcinoma in situ (a cancer with low aggressiveness) is challenging to pathologists, as only 5%–10% patients typically progress towards an invasive stage.32 Thus, we propose that SCS can detect the aggressive cancer stem cells and cells that have specific genetic aberrations, indicating the degree of malignancy in the tumour tissue without being hampered by a small amount of the specific cells that carry limited DNA or RNA. Allred and his colleagues supported the theory that many early stage breast cancer cells present specific characteristics during pathogenesis.33 Applying SCS to the early detection of cancer cells in small clinical samples might lead to earlier and more appropriate treatment decisions by clinicians.
Early detection of metastatic malignancies in circulating tumour cells (CTCs)
Another major application of SCS is the detection and genetic profiling of circulating tumour cells for disease evaluation and early intervention to inhibit the occurrence of metastasis.
CTCs are potentially invasive cells that are released into the circulatory system outside primary lesions. These cells are very rare and are estimated to exist in a ratio of 1∶100 000 000 relative to blood cells.34 Although the frequency of CTCs is extremely low, it is possible to recognize them in the blood because of their specific phenotypic markers of epithelial–mesenchymal transition, regarded as an important characteristic in the metastatic process.35
Systemically, CTCs offer a useful way to predict metastasis and corresponding prognosis of patients with tumours. Cristofanilli et al.36 demonstrated that monitoring CTCs is related to prognosis in patients with metastatic breast cancer and prediction of treatment response. In castration-resistant prostate cancer patients, CTC levels via immunomagnetic enrichment and fluorescence-based microscopy are better than PSA decrement algorithms in predicting overall survival.37
The value of investigating CTCs with SCS involves two aspects. First, it solves the problem of recognizing impure mixtures of the tumour bulk, simplifying certain complicated procedures such as retroperitoneal lymph node biopsy. And second, analysis can be carried out despite a limited amount of DNA or specimen such as a 7.5-ml blood sample, or a ‘liquid biopsy'.38 Moreover, another application of SCS in CTCs detection can be predictive of response or resistance to cancer therapy, which will be discussed in the following section.
Intratumour heterogeneity-guided personalized medicine
Some cancer cells are likely to miss detection during tumour biopsy because of the fact that some specific cancer cells are rare or that the specimen may not represent the population as a whole. Thus, the patient cannot be treated as precisely or as soon as possible, which may lead to tumour progression, affecting the prognosis of the patient. Therefore, personalized medicine based on Intratumour heterogeneity by SCS is a recent research trend in the tumour therapy field.
In a recent study, Navin et al.39 profiled genomic copy number variations in two primary breast carcinomas by applying SCS and found that one tumour was monogenomic, whereas the other was polygenomic. Gerlinger et al.16 discovered polygenetic reconstruction, revealing that 63%–69% of somatic mutations were not detectable across all regions within a solid tumour. These results indicate that even the same tumour is composed of cells with different genomic alterations, which cannot be tested by the pathological identification of the whole solid tumour tissue, and that these heterogeneities might lead to therapeutic failure. This type of treatment resistance can be regarded as primary, which is defined as a resistance existing prior to any given treatment due to primary intratumour heterogeneity. It has been reported that more than 50% of patients with adenocarcinoma will relapse after surgery and chemotherapy because of a chemoresistant phenotype.40 Another study revealed that approximately 20% of patients with acute lymphoblastic leukaemia experienced primary resistance to treatment.41 Thus, intratumour heterogeneity detection by appropriate methods such as SCS is of utmost importance in order to discover cells with rare genotypes that may cause primary drug resistance. In clinics, many target drugs are used in specific diseases associated with specific genomic alterations.
Moreover, in accordance with Darwin's theory of survival of the fittest, tumour cells adjust and adapt to changes in their microenvironment, including catastrophic pressures such as cytotoxic agents and target therapy. This type of accommodation or resistance is known as acquired resistance and follows initial therapy. Geioli et al.42 found resistance emergence during the medication process as a result of mutations in the case of BCR/ABL and EGFR inhibitors, which finally lost the ability to bind their targets.
In general, individualized medicine based on the phenomenon that tumours are composed of different cells with different genomic alterations suggests that detection by SCS is the best approach to solve the problem of therapeutic resistance, regardless of whether it is performed before or during treatment.
DISCUSSION
The advantages and limitations of SCS
Since the emergence of SCS, less DNA or RNA is needed for genetic analysis, providing earlier and more convenient opportunities for researchers to obtain comprehensive tumour information from patients. First and foremost, SCS can contribute to a fundamental understanding of single tumour cell biology, as single cells are the fundamental units of life. The underlying characterisation and features of a specific tumour cell cannot be found by other conventional methods, because of the complex nature of a bulk that is composed of different tumour cells. Furthermore, some specific amplification methods may increase results with less bias when compared with conventional sequencing methods, which makes analyses faster and more accurate. The most important aspect of SCS is its application in clinics. It helps patients and clinicians obtain complete information about a disease, monitor early cancer diagnosis, evaluate treatment effectiveness and predict prognosis, which cannot be achieved by other methods.
However, SCS is not perfect in all aspects. The first challenge is the high percentage of genome with minimal amplification bias. Although a new method presented by Zong et al.30 called MALBAC, as previously discussed, has less amplification bias than multiple displacement amplification, which is regarded as the most reliable method for amplifying a whole genome of a single cell, detailed studies are required to prove the effect of MALBAC on reducing amplification bias. Further, another limitation of MALBAC involves regions of the genome that are reproducibly underrepresented, as reported by Lasken.43 The ability to assay multiple properties in single-cell integrated analyses is still a long way down the road. The translation of SCS from basic research to clinical application challenges researchers because of the lack of cost–benefit analyses.44 New methods for isolating single cells and modifications of current methods are needed, which will eventually develop into a unified single-cell multiproperty analysis protocol. A novel strategy to profile single cells quickly for a reasonable price is necessary. Furthermore, methods for profiling paraffin-embedded tissues instead of frozen ones and for providing information security for sequenced patient genomes are also essential in clinics.
Further prospectives
Intratumour heterogeneity is a major hotspot in the field of tumorigenesis research. The tumour itself is like a family composed of members who have different functions and obligations in all aspects. Thus, understanding all the members within the tumour is a must for early detection and intervention in patients afflicted with the disease.
SCS can be applied in several capacities based on the existence of intratumour heterogeneity. It can be applied to the detection of rare tumour cells both in lesions and in the peripheral circulation for early diagnosis, evaluation of patient prognosis and determination of the efficacy of therapeutic approaches by profiling CTCs and monitoring primary tumour cells. Therefore, novel treatment strategies may be developed based on the results of these applications.
We believe that we can improve early diagnosis, prognosis and therapeutic effects in patients by applying SCS to tumours with intratumour heterogeneity.
AUTHOR CONTRIBUTIONS
YHS and SCR conceived the idea and made up the structure. MQ searched the literature reviews and drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgments
This study was supported by the National Basic Research Program of China (No. 2012CB518306), the National Natural Science Foundation of China (No. 81101946), the Prostate Cancer Foundation Young Investigator Award and the Shanghai Pujiang Program (12PJD008).
The authors declare no competing financial interests.
References
- Brown TM, Fee E. Rudolf Carl Virchow: medical scientist, social reformer, role model. Am J Public Health. 2006;96:2104–5. doi: 10.2105/AJPH.2005.078436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppner GH, Miller BE. Tumor heterogeneity: biological implications and therapeutic consequences. Cancer Metastasis Rev. 1983;2:5–23. doi: 10.1007/BF00046903. [DOI] [PubMed] [Google Scholar]
- Fidler IJ. Tumor heterogeneity and the biology of cancer invasion and metastasis. Cancer Res. 1978;38:2651–60. [PubMed] [Google Scholar]
- Dexter DL, Kowalski HM, Blazar BA, Fligiel Z, Vogel R, et al. Heterogeneity of tumor cells from a single mouse mammary tumor. Cancer Res. 1978;38:3174–81. [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Prosise GL, Bonnet MH, Berry RB, Dickel MJ. Effects of abstinence from smoking on sleep and daytime sleepiness. Chest. 1994;105:1136–41. doi: 10.1378/chest.105.4.1136. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–68. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–5. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158–63. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504–14. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- Rajasekhar VK, Studer L, Gerald W, Socci ND, Scher HI. Tumour-initiating stem-like cells in human prostate cancer exhibit increased NF-kappaB signalling. . Nat Commun. 2011;2:162. doi: 10.1038/ncomms1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- Merlo LMF, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–35. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- Crespi B, Summers K. Evolutionary biology of cancer. Trends Ecol Evol. 2005;20:545–52. doi: 10.1016/j.tree.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Shibata D. Cancer. Heterogeneity and tumor history. Science. 2012;336:304–5. doi: 10.1126/science.1222361. [DOI] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidransky D, Mikkelsen T, Schwechheimer K, Rosenblum ML, Cavanee W, et al. Clonal expansion of p53 mutant cells is associated with brain tumour progression. Nature. 1992;355:846–7. doi: 10.1038/355846a0. [DOI] [PubMed] [Google Scholar]
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–13. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- Yachida S, Jones S, Bozic I, Antal T, Leary R, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H, Wang Z, Chen K, Zhao J, Lee JJ, et al. Influence of chemotherapy on EGFR mutation status among patients with non-small-cell lung cancer. J Clin Oncol. 2012;30:3077–83. doi: 10.1200/JCO.2011.39.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins FC, De S, Almendro V, Gonen M, Park SY, et al. Evolutionary pathways in BRCA1-associated breast tumors. Cancer Discov. 2012;2:503–11. doi: 10.1158/2159-8290.CD-11-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA. 2013;110:4009–14. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Hou Y, Yin X, Bao L, Tang A, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–95. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Song L, Zhu P, Zhang B, Tao Y, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–85. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- Welty CJ, Coleman I, Coleman R, Lakely B, Xia J, et al. Single cell transcriptomic analysis of prostate cancer cells. BMC Mol Biol. 2013;14:6. doi: 10.1186/1471-2199-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, et al. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PloS ONE. 2012;7:e33788. doi: 10.1371/journal.pone.0033788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramskold D, Luo S, Wang YC, Li R, Deng Q, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–82. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N, Hicks J. Future medical applications of single-cell sequencing in cancer. Genome Med. 2011;3:31. doi: 10.1186/gm247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–9. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–6. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Zong C, Fan W, Yang M, Li J, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338:1627–30. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerlikowske K, Molinaro AM, Gauthier ML, Berman HK, Waldman F, et al. Biomarker expression and risk of subsequent tumors after initial ductal carcinoma in situ diagnosis. J Natl Cancer Inst. 2010;102:627–37. doi: 10.1093/jnci/djq101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allred DC, Wu Y, Mao S, Nagtegaal ID, Lee S, et al. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution. Clin Cancer Res. 2008;14:370–8. doi: 10.1158/1078-0432.CCR-07-1127. [DOI] [PubMed] [Google Scholar]
- Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol. 2011;192:373–82. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosse SA, Hannemann J, Spotter J, Bauche A, Andreas A, et al. Changes in keratin expression during metastatic progression of breast cancer: impact on the detection of circulating tumor cells. Clin Cancer Res. 2012;18:993–1003. doi: 10.1158/1078-0432.CCR-11-2100. [DOI] [PubMed] [Google Scholar]
- Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- Scher HI, Morris MJ, Larson S, Heller G. Validation and clinical utility of prostate cancer biomarkers. Nat Rev Clin Oncol. 2013;10:225–34. doi: 10.1038/nrclinonc.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoe M, Ogawa Y, Morita J, Omori K, Takeshita K, et al. Detection of circulating urothelial cancer cells in the blood using the CellSearch System. Cancer. 2007;109:1439–45. doi: 10.1002/cncr.22543. [DOI] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–4. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castells M, Thibault B, Delord JP, Couderc B. Implication of tumor microenvironment in chemoresistance: tumor-associated stromal cells protect tumor cells from cell death. Int J Mol Sci. 2012;13:9545–71. doi: 10.3390/ijms13089545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor D, Sibson K, Caswell M, Connor P, Cummins M, et al. Early UK experience in the use of clofarabine in the treatment of relapsed and refractory paediatric acute lymphoblastic leukaemia. Br J Haematol. 2011;154:482–5. doi: 10.1111/j.1365-2141.2011.08752.x. [DOI] [PubMed] [Google Scholar]
- Gajria D, Chandarlapaty S. HER2-amplified breast cancer: mechanisms of trastuzumab resistance and novel targeted therapies. Expert Rev Anticancer Ther. 2011;11:263–75. doi: 10.1586/era.10.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasken RS. Single-cell sequencing in its prime. Nat Biotechnol. 2013;31:211–2. doi: 10.1038/nbt.2523. [DOI] [PubMed] [Google Scholar]
- Soon WW, Hariharan M, Snyder MP. High-throughput sequencing for biology and medicine. Mol Syst Biol. 2013;9:640. doi: 10.1038/msb.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xu X, Song L, Hou Y, Li Z, et al. Single-cell sequencing analysis characterizes common and cell-lineage-specific mutations in a muscle-invasive bladder cancer. Gigascience. 2012;1:12. doi: 10.1186/2047-217X-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heselmeyer-Haddad K, Berroa Garcia LY, Bradley A, Ortiz-Melendez C, Lee WJ, et al. Single-cell genetic analysis of ductal carcinoma in situ and invasive breast cancer reveals enormous tumor heterogeneity yet conserved genomic imbalances and gain of MYC during progression. Am J Pathol. 2012;181:1807–22. doi: 10.1016/j.ajpath.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29:1120–7. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]