Abstract
Transglutaminase 4 has been shown to enhance various biological properties of prostate cancer cells, e.g., cell–matrix adhesion, invasiveness and the epithelial–mesenchymal transition. The objectives of this study were to investigate the associations between transglutaminase 4 expression and the established features and biochemical recurrence of prostate cancer. Transglutaminase 4 immunostaining was performed on a tissue microarray. The expression of transglutaminase 4 was evaluated by a scoring method based on the intensity and extent of staining. The clinical and pathological information was obtained through a review of medical records. Follow-up data were obtained by consulting the hospital medical records and the prostate cancer database of our department and by contacting patients or family members. We then compared the transglutaminase 4 expression levels between the prostate cancer tissues and the paracarcinoma tissues and evaluated the correlation of transglutaminase 4 expression with the clinical parameters and biochemical recurrence of prostate cancer. Our results indicated that the transglutaminase 4 staining was significantly higher in tumour tissue than in paracarcinoma tissue (P<0.001) and was positively associated with higher Gleason score (P<0.001) and higher prostate-specific antigen level (P=0.005). Patients with transglutaminase 4 overexpression experienced shorter biochemical recurrence-free survival after surgery (P=0.042) in the univariate analysis but not in the multivariate analysis (P=0.139), which indicated that transglutaminase 4 may serve as a potential predictor of biochemical recurrence of prostate cancer.
Keywords: biochemical recurrence, prostate cancer, prostatectomy, tissue microarray, transglutaminase 4
Introduction
Prostate cancer is still a major challenge in the United States and throughout the world. It is estimated that 241 740 new cases of prostate cancer and 28 170 deaths related to this disease were recorded in 2012, making prostate cancer the second-leading cause of cancer death in men.1 In China, 33 802 new cases of prostate cancer and 14 297 deaths related to prostate cancer were recorded in 2008. It is worth noting that the incidence rate of prostate cancer increased from 1.6/100 000 in 2002 to 4.3/100 000 in 2008.2 In Shanghai, the incidence of and mortality due to prostate cancer were much higher (21.33/100 000 and 8.03/100 000) during 2003–2007.3 Radical prostatectomy is the standard treatment for organ-confined tumours; however, 20%–30% of patients experience recurrence after surgery.4 It is important to identify patients with poor prognosis as early as possible, especially those who need adjuvant treatment. The present clinical prognostic parameters are unable to accurately estimate the aggressiveness and recurrence of prostate cancer due to the biological variability of the tumours. Therefore, molecular markers are urgently needed to help identify and select patients who will benefit from adjuvant therapy. With the increased focus on this field, several markers, e.g., syndecan-1, syndecan-2, cysteine-rich angiogenic inducer 61 and the serum prostate-specific antigen (PSA) value adjusted for the non-cancerous prostate tissue volume,5,6,7 have been investigated to determine their prognostic significance for clinically localized diseases.
Transglutaminase 4 (TGM4), which is predominantly expressed in the prostate gland,8,9 is a member of the transglutaminase family that catalyses the post-translational modification of proteins by the formation of ε-(γ-glutamyl) lysine isopeptide bonds.10 Even though the biological effects of TGM4 on prostate cancer cells and other cells remain unclear, the existing data provide important indications. Dubbink et al.9 reported that TGM4 is androgen responsive, that is, androgen can upregulate the expression of TGM4. In prostate cancer cells, TGM4 has been shown to exert its influence by interacting with MDA-7/IL-24 and the HGFL/MSP-RON system.11 Further studies have indicated that TGM4 increases the invasiveness and migration of prostate cancer cells.12,13 In addition, TGM4 is linked to membrane micromotion, cell–matrix adhesion, the epithelial–mesenchymal transition and tumour–endothelium interactions.11
The expression pattern of TGM4 in prostate cancer remains controversial due to the limited amount of available literature. An et al.8 observed reduced expression of TGM4 in prostate tumour tissue and particularly in prostate cancer tissues with higher Gleason scores. Quantitative RT-PCR analysis of laser-microdissected prostate cancer tissues revealed that TGM4 was downregulated in moderately or poorly differentiated carcinoma glands compared with normal tissue.14 In contrast, Jiang et al.15 demonstrated that TGM4 appeared to be more highly expressed in human prostate cancer tissue and in high-Gleason-score tumours (>7) than in normal tissue.
However, due to inadequate sample sizes, the correlations between the expression level of TGM4 and relevant pathological data and follow-up information are yet to be clarified. To determine the association of TGM4 with the progression of prostate cancer and to assess the diagnostic and prognostic values of TGM4 for prostate cancer, immunohistochemistry (IHC) analysis using tissue microarray (TMA) slides was used to determine the expression level of TGM4 in this study.
Materials and methods
Patient population
This study was approved by our institutional medical ethics review committee. This study included 159 patients who had undergone radical prostatectomy and bilateral lymphadenectomy at the Department of Urology, Changhai Hospital, between October 2002 and December 2008 and for whom archival tissues were available. No patient was managed preoperatively with either hormonal or radiation therapy, and no secondary cancers were observed. The prostatectomy specimens were examined by two staff pathologists who were blinded to the clinical outcome and follow-up data. The evaluation of the specimen was performed according to the guidelines of the College of American Pathologists.16 The clinical and pathological information was obtained through a review of the patients' medical records. The stages of cancer for all patients were determined by the American Joint Committee on Cancer (AJCC) 2002 system.
Follow-up
All patients were scheduled to have their serum PSA level evaluated postoperatively every 3 months for the first year, every 6 months from the second to the fifth year and annually thereafter. Follow-up data were obtained by consulting the hospital medical records and the prostate cancer database of our department and by contacting the patients or their family members. Biochemical recurrence (BCR) was defined as the sustained elevation of the serum total PSA level above 0.2 ng ml−1 on two or more occasions, and the date of BCR was assigned as the date of the first value >0.2 ng ml−1. Censoring occurred if no BCR was observed prior to 30 November 2012.
TMA construction
The tissue microarray was constructed using archived formalin-fixed paraffin-embedded tissue samples from prostate cancer patients. Briefly, the original slides were reviewed by two pathologists, and tumour tissue and paracarcinoma tissue without inflammatory zones were selected and marked with different coloured inks. For each patient, two cores of the sample (1 mm in diameter) were transferred from the selected areas to the recipient block. Serial 3 µm sections of the TMA block were cut and stained with haematoxylin–eosin to verify that the cores adequately represented the diagnostic areas. The analysis included 159 patients, among whom there were 125 patients with paired tumour and paracarcinoma tissues, 18 patients with tumour tissue only and 16 patients with normal tissue only.
IHC and scoring
IHC analysis of TGM4 was performed on 3 µm TMA sections using a mouse monoclonal antihuman antibody (dilution 1∶50; Abcam, Cambridge, UK). After deparaffinisation, the samples were subjected to a heat antigen retrieval process using citrate buffer (1 mmol l−1, pH 6.0). The slides were then incubated overnight at 4 °C with the primary antibody after incubation with 2.5% blocking serum. Diaminobenzidine was used as the chromogen, and haematoxylin was used for counterstaining. The primary antibody was omitted for the negative control. All IHC staining procedures were performed by Dr Zhi Cao. The slides were interpreted by two independent observers who were blinded to the clinical data, and the slides were rescored to reach a consensus if the interobserver variability exceeded 10%. The expression of TGM4 was evaluated using a scoring method based on the intensity of staining (ranked from 0 to 3) and the extent of staining (percentage of tumour cells, ranked from 0 to 4). The results were recorded as the IE score (intensity score (I)×extent score (E) under ×200 visual field magnification), which varied from 0 to 12 and was divided into four grades, representing negative (a score of 0–2), weak (3–5), moderate (6–8) and strong staining (9–12). For some comparisons described later in the paper, low expression (score 0–5) comprised the negative and weak staining groups, whereas high expression comprised the moderate and strong staining groups. The Gleason score was also determined for each core.
Statistical analysis
Statistical analysis was performed with SAS 8.0 and SPSS 18.0 software. Biochemical recurrence-free survival was evaluated from the date of surgery to the date of the last follow-up visit or the point in time when BCR was observed. Comparisons of the IE score between the tumour tissue and the paracarcinoma tissue were performed using the Wilcoxon signed-rank test. The Kruskal–Wallis H test was used to evaluate the associations between the PSA level, the Gleason score, the pathological tumour stage and the IE score, and the Wilcoxon rank sum test was used to evaluate the associations between the IE score and both the surgical margin and the pathological lymph node category. The association between the IE score and biochemical recurrence-free survival was assessed by the log-rank test and Cox proportional hazard regression analysis. The Gleason score, the pathological category of the tumour and the PSA level were input into the multivariate analysis. The Gleason score mentioned above was the one scored by us, not the one recorded in the medical records. The level of statistical significance was set at least at P<0.05 (two-sided).
Results
Patient description
A total of 159 patients, aged 49–79 years (mean±s.d.: 66.8±6.6 years), were enrolled in the study. The clinical and pathological findings are reported in Table 1. The most common reason for missing information (36, 22.6%) was the inadequate description of the pathological reports in the medical records.
Table 1. Pathological and clinical data for the arrayed prostate cancers.
| Variable | n | Percentage (%) |
|---|---|---|
| Age (year) | ||
| <50 | 1 | 0.63 |
| 50–60 | 30 | 18.87 |
| 60–70 | 73 | 45.91 |
| >70 | 55 | 34.60 |
| Preoperative PSA level (ng ml−1) | ||
| <4 | 33 | 20.75 |
| 4–10 | 38 | 23.90 |
| 10–20 | 35 | 22.01 |
| >20 | 32 | 20.13 |
| N/A | 21 | 13.21 |
| pT category (AJCC 2002) | ||
| PIN | 1 | 0.63 |
| pT2a–2b | 9 | 5.67 |
| pT2c | 33 | 20.75 |
| pT3a | 42 | 26.42 |
| pT3b | 39 | 24.53 |
| N/A | 35 | 21.01 |
| Gleason score | ||
| ≤3+3 | 38 | 23.90 |
| 3+4 | 44 | 27.67 |
| 4+3 | 30 | 18.87 |
| ≤4+4 | 20 | 12.58 |
| N/A | 27 | 16.98 |
| pN category | ||
| pN0 | 127 | 79.87 |
| pN+ | 10 | 6.29 |
| NA | 23 | 14.47 |
| Surgical margin | ||
| Negative | 60 | 37.74 |
| Positive | 64 | 40.25 |
| N/A | 36 | 22.64 |
Abbreviations: AJCC, American Joint Committee on Cancer; N/A, not available; PIN, prostatic intraepithelial neoplasia; pN category, pathological lymph node category; PSA, prostate-specific antigen; pT category, pathological tumour category.
TGM4 expression pattern
In our study, TGM4 was commonly expressed in the prostate epithelial cells, and the lack of expression was observed only in the paracarcinoma tissue in nine patients. In addition, TGM4 staining was observed in the basal cells and a few stromal cells. Representative TGM4 staining patterns are shown in Figure 1. The staining for TGM4 was significantly stronger in the tumour tissues than in the paracarcinoma tissues (P<0.001) (Table 2).
Figure 1.
Examples of TGM4 immunostaining in prostate cancer and normal tissues. (a) Lack of staining, paracarcinoma tissue; (b) weak staining, T2aN0 M0, Gleason score: 3+3; (c) moderate staining, T3aN0 M0, Gleason score: 3+4; (d) strong staining, T3bN0 M0, Gleason score: 5+5. TGM4, transglutaminase 4.
Table 2. TGM4 expression and tumour phenotype.
| Variable | No. of successfully analysed samples | TGM4 immunohistochemistry result | P | |||
|---|---|---|---|---|---|---|
| Negative | Weak | Moderate | Strong | |||
| Tissue type | 125 | <0.001 | ||||
| Prostate cancer | 125 | 0 | 35 | 81 | 9 | |
| Paracancerous tissue | 125 | 9 | 58 | 55 | 3 | |
| pT stage | 115 | 0.299 | ||||
| PIN | 1 | 0 | 0 | 1 | 0 | |
| pT2a–2b | 7 | 0 | 2 | 3 | 2 | |
| pT2c | 30 | 0 | 10 | 20 | 0 | |
| pT3a | 39 | 0 | 11 | 26 | 2 | |
| pT3b | 38 | 0 | 7 | 26 | 5 | |
| Gleason score | 143 | <0.001 | ||||
| ≤3+3 | 57 | 0 | 24 | 31 | 2 | |
| 3+4 | 27 | 0 | 2 | 23 | 2 | |
| 4+3 | 34 | 0 | 9 | 23 | 2 | |
| ≥4+4 | 25 | 0 | 2 | 16 | 7 | |
| Pre-operative PSA level (ng ml−1) | 127 | 0.005 | ||||
| <4 | 30 | 0 | 8 | 19 | 3 | |
| 4–10 | 33 | 0 | 14 | 18 | 1 | |
| 10–20 | 35 | 0 | 8 | 24 | 3 | |
| >20 | 29 | 0 | 1 | 24 | 4 | |
| pN category | 125 | 0.297 | ||||
| pN0 | 116 | 0 | 30 | 76 | 10 | |
| pN+ | 9 | 0 | 0 | 9 | 0 | |
| Surgical margin | 114 | 0.760 | ||||
| Negative | 53 | 0 | 15 | 33 | 5 | |
| Positive | 61 | 0 | 14 | 43 | 4 | |
Abbreviations: N/A, not available; pN category, pathological lymph node category; PSA, prostate-specific antigen; pT category, pathological tumour category; TGM4, transglutaminase 4.
TGM4 expression was not associated with the pathological tumour stage, the pathological lymph node category or the surgical margin (P>0.05), but was positively associated with higher Gleason score (P<0.001) and higher PSA level (P=0.005) (Table 2).
Association between TGM4 expression and BCR
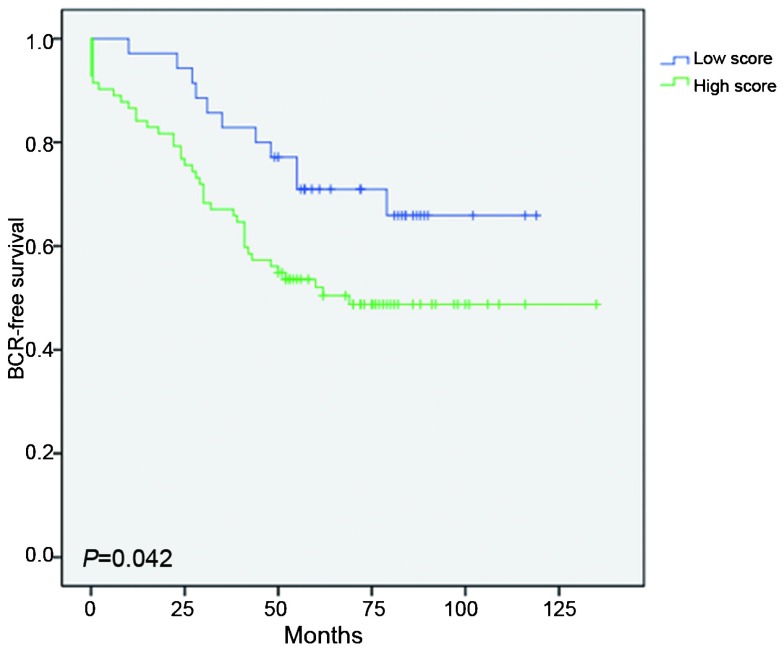
Adequate follow-up data were available for 117 of the 159 patients (73.6%), and 52 of these 117 patients (44.4%) experienced BCR after a median postoperative follow-up duration of 55.9±31.1 months. Log-rank test indicated that patients with high TGM4 expression levels experienced shorter BCR-free survival than those patients with low TGM4 expression levels (P=0.042). (Figure 2) In the univariate Cox proportional hazards regression analyses, TGM4 expression was significantly associated with BCR after radical prostatectomy (P=0.048). In the multivariate Cox proportional hazards regression analyses that was adjusted for the effects of standard clinicopathological features, TGM4 expression was not correlated with BCR (P=0.139) (Table 3).
Figure 2.
Influence of TGM4 expression on biochemical recurrence. Low score: IE scores 0–5, High score: IE scores 6–12. Adequate follow-up data were available for 117 patients; 52 patients experienced BCR, and BCR was not observed in 65 patients. BCR, biochemical recurrence; IE score, intensity score (I)×extent (E) score under ×200 visual field magnification; IHC, immunohistochemistry; RP, radical prostatectomy; PSA, prostate-specific antigen; TGM4, transglutaminase 4; TMA, tissue microarray.
Table 3. Cox's univariate and multivariate analyses of biochemical recurrence-free survival according to the IE score.
| BCR-free time | ||
|---|---|---|
| HR (95% CI) | P | |
| Univariate | ||
| Low IE score (score 0–5) | 1 | |
| High IE score (score 6–12) | 0.048 (1.006–3.816) | 0.048 |
| Multivariate | ||
| IE score (high/low) | 2.096 (0.787–5.581) | 0.139 |
| Gleason score (≥4+3/≤3+4) | 2.726(1.329–5.590) | 0.006 |
| PSA level (>10/≤10) | 1.391(0.712–2.716) | 0.334 |
| pT stage (T3/T2+PIN) | 1.558(0.644–3.772) | 0.325 |
Abbreviations: IE score=intensity score (I)×extent (E) score; PIN, prostatic intraepithelial neoplasia; PSA, prostate-specific antigen; pT category, pathological tumour category.
Discussion
Using TMA and IHC technologies, our study revealed that the expression level of TGM4 differentiates prostate cancer tissue from paracarcinoma tissue and is correlated with the Gleason score and the PSA level. Moreover, TGM4 overexpression was found to be associated with shorter BCR-free survival after radical prostatectomy in the univariate analysis. Unfortunately, this association did not remain statistically significant after controlling for the preoperative PSA level, the Gleason score and the pathological tumour category.
The availability of PSA testing has revolutionized the management of prostate cancer, which has greatly increased the likelihood of receiving appropriate treatment. However, up to a quarter of patients experience tumour recurrence after radical prostatectomy and received salvage therapy instead, such as endocrine therapy.17 When the tumour recurs, disease-free survival decreases, and morbidity increases.
If patients with a high risk of recurrence are detected and receive appropriate adjuvant therapy before BCR, the success rate of radical prostatectomy can be improved. Molecules that are differentially expressed between tumour tissues and normal tissues and those that reflect crucial biological functions involved in oncogenesis, such as adhesion, motility and cell cycle progression, can be regarded as candidates for prognostic biomarkers.
Even though TGM4 was discovered by Grant et al.18 using a human prostate cDNA library in the early 1990s, the interest in this protein experienced a relatively quiescent period. Currently, the interest in the role of TGM4 in prostate cancer is much higher, and several studies have focused on the expression pattern of TGM4, but no consensus has been reached. Although TGM4 is a protein, most studies using tissue samples were performed primarily at the RNA level. An et al.8 observed that the positivity for TGM4 expression was reduced in prostate tumour tissues, particularly in prostate tumour tissues with higher Gleason scores, and they further reported that the expression of TGM4 was very significantly inhibited in metastatic prostate cancer. Using a quantitative analysis of gene transcripts, Jiang et al.15 demonstrated that the expression level of TGM4 in human prostate cancer appeared to be higher than that in normal tissue and that it was also higher in high-Gleason-score tumours. Data from quantitative RT-PCR assays using RNA isolated from laser microdissected prostate tissue revealed that TGM4 was downregulated in moderately or poorly differentiated carcinoma glands compared with normal peripheral zone glands.14
The expression pattern of TGM4 in prostatic secretions is also under debate. It has been reported that selected reaction monitoring mass spectrometry indicated that TGM4 was downregulated in the organ-confined tumour group, whereas Western blot analysis demonstrated that the expression of TGM4 was reduced in the extracapsular tumour group.19 In another study, Western blot analysis indicated that TGM4 was downregulated in prostate cancer compared with normal tissue, a result that was confirmed by IHC.20 It is worth noting that the primary antibody for TGM4 used in the above two studies was a goat polyclonal IgG; thus, cross-reactivity may exist. Moreover, the sample sizes of these studies were relatively small.
Our study indicates that TGM4 is overexpressed in tumour tissue compared with paracarcinoma tissue and that the TGM4 expression level is positively associated with the Gleason score and the PSA level. These data indicate that TGM4 may prompt the development and progression of prostate cancer, which is consistent with the results from functional studies of TGM4. In the TGM4-positive prostate cancer cell line CA-HPV-10, the knockdown of TGM4 resulted in a marked reduction of in vitro invasiveness.12 By contrast, Jiang reported that TGM4 is colocalized with RON, the receptor for HGF-L/MSP, in prostate cancer cells. PC-3 cells overexpressing TGM4 exhibited increased cell motility, which is directly affected by the enhanced response of RON to HGF-L/MSP.13 In PC-3 cells, MDA-7 can inhibit cell adhesion, growth and migration. However, this phenomenon disappeared after a TGM4 expression vector was transfected into these cells.21 In another study, the ability of PC-3 cells to adhere to endothelial cells increased when TGM4 was overexpressed, and this ability was reduced after TGM4 expression was knocked down in CA-HPV-10 cells.22 Moreover, a recent study demonstrated that TGM4 is involved in the epithelial–mesenchymal transition, which has a critical role in cancer development and progression.23,24
Following these encouraging studies, we further explored the association between TGM4 expression and BCR. TGM4 overexpression was associated with shorter BCR-free survival after radical prostatectomy in the univariate analysis, but this association was not statistically significant after controlling for the preoperative PSA level, the Gleason score and the pathological category. Although the result is not striking, it indicates that TGM4 may be a potential predictor of BCR. Confirming the predictive power of TGM4 expression requires further investigation by studies with larger sample sizes and long-term follow-up. If this result is confirmed, TGM4 can be applied in clinical practice to improve the prognostic evaluation of patients with prostate cancer and can help identify patients with a higher likelihood of BCR, who thus require adjuvant therapies. At the same time, the use of TGM4 expression to predict BCR may help patients with a lower chance of BCR to avoid unnecessary adjuvant therapies and the associated complications.
Several limitations of this study should be considered. The results of the study may have been more persuasive if the sample size or follow-up time had been increased. Due to the good prognosis of prostate cancer and the short follow-up time, the numbers of patients with metastases and deaths were relatively small; therefore, it was difficult to evaluate the associations between TGM4 and both the development of metastases and the disease-specific survival of patients treated with radical prostatectomy.
Conclusion
The expression of TGM4 was upregulated in tumour tissue and was found to be associated with higher Gleason score and higher PSA level. In addition, the overexpression of TGM4 may serve as a potential predictor of BCR.
Author contributions
ZC, ZSZ and SCR designed the experiments, and ZC, BZ and MQ carried out the experiments. YW and YWY evaluated the IHC results. ZC and ZYL analysed the data. YHS and ZYL supervised the project. ZC, YW and ZSZ wrote the manuscript. All authors reviewed and approved the final manuscript.
Acknowledgments
This study was supported by the National Basic Research Program of China (No. 2012CB518300), the National Natural Science Foundation of China (No. 81101946), the Prostate Cancer Foundation Young Investigator Award, the Shanghai Pujiang Program (No. 12PJD008) and the Key Project of Science and Technology Commission of Shanghai Municipality (No. 09DJ1400400).
All authors declare that there are no competing financial interests.
References
- American Cancer Society Cancer Facts & Figures, 2012. http://www.cancer.org/research/cancerfactsfigures/cancerfactsfigures/ cancer-facts-figures-2012
- Zhang L, Yang BX, Zhang HT, Wang JG, Wang HL, et al. Prostate cancer: an emerging threat to the health of aging men in Asia. Asian J Androl. 2011;13:574–8. doi: 10.1038/aja.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renqiang H, Ming W, Wanqing C, Siwei Z, Rongshou Z. Analysis on incidence and mortality of prostate cancer in China during 2003∼2007. China Cancer. 2012;21:54–7. [Google Scholar]
- Pontes-Júnior J, Reis ST, de Oliveira LC, Sant'anna AC, Dall'oglio MF, et al. Association between integrin expression and prognosis in localized prostate cancer. Prostate. 2010;70:1189–95. doi: 10.1002/pros.21153. [DOI] [PubMed] [Google Scholar]
- Ledezma R, Cifuentes F, Gallegos I, Fullá J, Ossandon E, et al. Altered expression patterns of syndecan-1 and -2 predict biochemical recurrence in prostate cancer. Asian J Androl. 2011;13:476–80. doi: 10.1038/aja.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada N, Kulkarni P, Getzenberg RH. Cyr61 is a potential prognostic marker for prostate cancer. Asian J Androl. 2012;14:405–8. doi: 10.1038/aja.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku JH, Moon KC, Cho SY, Kwak C, Kim HH. Serum prostate-specific antigen value adjusted for non-cancerous prostate tissue volume in patients undergoing radical prostatectomy: a new predictor of biochemical recurrence in localized or locally advanced prostate cancer. Asian J Androl. 2011;13:248–53. doi: 10.1038/aja.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G, Meka CS, Bright SP, Veltri RW. Human prostate-specific transglutaminase gene: promoter cloning, tissue-specific expression, and down-regulation in metastatic prostate cancer. Urology. 1999;54:1105–11. doi: 10.1016/s0090-4295(99)00298-8. [DOI] [PubMed] [Google Scholar]
- Dubbink HJ, Verkaik NS, Faber PW, Trapman J, Schroder FH, et al. Tissue-specific and androgen-regulated expression of human prostate-specific transglutaminase. Biochem J. 1996;315:901–8. doi: 10.1042/bj3150901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folk JE, Chung SI. Molecular and catalytic properties of transglutaminases. Adv Enzymol. 1973;38:109–91. doi: 10.1002/9780470122839.ch3. [DOI] [PubMed] [Google Scholar]
- Jiang WG, Ablin RJ. Prostate transglutaminase: a unique transglutaminase and its role in prostate cancer. Biomark Med. 2011;5:285–91. doi: 10.2217/bmm.11.36. [DOI] [PubMed] [Google Scholar]
- Davies G, Ablin RJ, Mason MD, Jiang WG. Expression of the prostate transglutaminase (TGase-4) in prostate cancer cells and its impact on the invasiveness of prostate cancer. J Exp Therapeut Oncol. 2007;6:257–64. [PubMed] [Google Scholar]
- Jiang WG, Ye L, Ablin RJ, Kynaston HG, Mason MD. The prostate transglutaminase, TGase-4, coordinates with the HGFL/MSP-RON system in stimulating the migration of prostate cancer cells. Int J Oncol. 2010;37:413–8. doi: 10.3892/ijo_00000689. [DOI] [PubMed] [Google Scholar]
- Shaikhibrahim Z, Lindstrot A, Buettner R, Wernert N. Analysis of laser-microdissected prostate cancer tissues reveals potential tumor markers. Int J Mol Med. 2011;28:605–11. doi: 10.3892/ijmm.2011.746. [DOI] [PubMed] [Google Scholar]
- Jiang WG, Ablin RJ, Kynaston HG, Mason MD.Expression of the prostate type transglutaminase (TGase-4) in clinical prostate cancer (Abstract)In: Proceedings of the 98th Annual Meeting of the American Association for Cancer Research; 14–18 April 2007; Los Angeles, CA, USA. American Association for Cancer Research: Washington, DC, USA, 2007.
- Henson DE, Hutter RV, Farrow G. Practice protocol f or the examination of specimens removed from patients with carcinoma of the prostate gland. A publication of the Cancer Committee, College of American Pathologists. Task Force on the Examination of Specimens Removed from Patients with Prostate Cancer. Arch Pathol Lab Med. 1994;118:779–83. [PubMed] [Google Scholar]
- Potters L, Klein EA, Kattan MW, Reddy CA, Ciezki JP, et al. Monotherapy for stage T1–T2 prostate cancer: radical prostatectomy, external beam radiotherapy, or perma-nent seed implantation. Radiother Oncol. 2004;71:29–31. doi: 10.1016/j.radonc.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Grant FJ, Taylor DA, Sheppard PO, Mathewes SL, Lint W, et al. Molecular cloning and characterization of a novel transglutaminase cDNA from a human prostate cDNA library. Biochem Biophys Res Commun. 1994;203:1117–23. doi: 10.1006/bbrc.1994.2298. [DOI] [PubMed] [Google Scholar]
- Kim Y, Ignatchenko V, Yao CQ, Kalatskaya I, Nyalwidhe JO, et al. Identification of differentially expressed proteins in direct expressed prostatic secretions of men with organ-confined versus extracapsular prostate cancer. Mol Cell Proteomics. 2012;11:1870–84. doi: 10.1074/mcp.M112.017889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Principe S, Kim Y, Fontana S, Ignatchenko V, Nyalwidhe JO, et al. Identification of prostate-enriched proteins by in-depth proteomic analyses of expressed prostatic secretions in urine. J Proteome Res. 2012;11:2386–96. doi: 10.1021/pr2011236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablin RJ, Kynaston HG, Mason MD, Jiang WG. Prostate type transglutaminase (TGase-4) antagonizes the anti-tumor action of MDA-7/IL-24 in prostate cancer. J Transl Med. 2011;9:49. doi: 10.1186/1479-5876-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang WG, Ablin RJ, Kynaston HG, Mason MD. The prostate transglutaminase (TGase-4, TGaseP) regulates the interaction of prostate cancer and vascular endothelial cells, a potential role for the ROCK pathway. Microvasc Res. 2009;77:150–7. doi: 10.1016/j.mvr.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Iwatsuki M, Mimori K, Yokobori T, Ishi H, Beppu T, et al. Epithelial–mesenchymal transition in cancer development and its clinical significance. Cancer Sci. 2009;101:293–9. doi: 10.1111/j.1349-7006.2009.01419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablin RJ, Jiang WG.Prostate transglutaminase (TGase-4) induces epithelial–mesenchymal transition (EMT) in prostate cancer cellsIn: Proceedings of the 4th International Conference on Epithelial–Mesenchymal Transition; 23–28 September 2008; Tucson, AZ, USA.