Abstract
In the current studies we generated transgenic mice that overexpress human Insig-1 in the liver under a constitutive promoter. In cultured cells Insig-1 and Insig-2 have been shown to block lipid synthesis in a cholesterol-dependent fashion by inhibiting proteolytic processing of sterol regulatory element–binding proteins (SREBPs), membrane-bound transcription factors that activate lipid synthesis. Insig’s exert this action in the ER by binding SREBP cleavage-activating protein (SCAP) and preventing it from escorting SREBPs to the Golgi apparatus where the SREBPs are processed to their active forms. In the livers of Insig-1 transgenic mice, the content of all nuclear SREBPs (nSREBPs) was reduced and declined further upon feeding of dietary cholesterol. The nuclear content of the insulin-induced SREBP isoform, SREBP-1c, failed to increase to a normal extent upon refeeding on a high-carbohydrate diet. The nSREBP deficiency produced a marked reduction in the levels of mRNAs encoding enzymes required for synthesis of cholesterol, fatty acids, and triglycerides. Plasma cholesterol levels were strongly reduced, and plasma triglycerides did not exhibit their normal rise after refeeding. These results provide in vivo support for the hypothesis that nSREBPs are essential for high levels of lipid synthesis in the liver and indicate that Insig’s modulate nSREBP levels by binding and retaining SCAP in the ER.
Introduction
Insig-1 and Insig-2 are polytopic membrane proteins of the ER that play a central role in the feedback control of lipid synthesis in animal cells. Insig’s regulate lipid synthesis by binding in a sterol-dependent fashion to two ER proteins: HMG CoA reductase, a rate-limiting enzyme in cholesterol biosynthesis, and sterol regulatory element–binding protein (SREBP) cleavage-activating protein (SCAP), an escort protein required for the cleavage and activation of the SREBP family of membrane-bound transcription factors. Sterol-stimulated binding of Insig’s to HMG CoA reductase leads to its ubiquitination and proteosomal degradation (1, 2). Sterol-stimulated binding of Insig’s to SCAP leads to ER retention of the complexes between SCAP and SREBPs, thereby preventing the SREBPs from entering the Golgi apparatus for proteolytic processing (3, 4). ER retention prevents the proteolytic generation of the transcriptionally active nuclear forms of SREBPs (nSREBPs), thereby limiting transcription of SREBP target genes, which include all of the known genes required for synthesis of cholesterol, fatty acids, triglycerides, and phospholipids (5). In cultured cells, the net result of Insig action is to decrease lipid synthesis whenever sterols accumulate to high levels within cells.
The two known human Insig isoforms, Insig-1 and -2, are 59% identical over their lengths of 277 and 225 amino acids, respectively. Both are deeply embedded in ER membranes through the presence of six membrane-spanning segments (6). Studies in cultured cells indicate that the two Insig’s have overlapping functions in binding both HMG CoA reductase and SCAP, but the two Insig genes are controlled differently. Insig-1 is itself a target of nSREBPs, and its mRNA rises and falls coordinately with nSREBP levels (3, 7). The Insig-2 gene has two promoters/enhancers that give rise to two transcripts with different noncoding first exons, but identical coding exons (8). In cultured cells one of these transcripts, designated Insig-2b, is unvarying and is not influenced by nSREBP levels. The other transcript, Insig-2a, is expressed nearly exclusively in the liver, and it reaches high levels during insulin deficiency, such as occurs in fasting. When insulin levels rise, the Insig-2a transcript is strongly suppressed (8).
All detailed studies of Insig function have been carried out in cultured fibroblast-like cells, predominantly CHO cells. In these cells, overexpression of Insig-1 or -2 by transfection causes a marked leftward shift in the sterol-response curves for degradation of HMG CoA reductase (1, 2) and inhibition of SREBP processing (3, 4), rendering both processes much more sensitive to sterols. When expressed at extremely high levels, Insig-1 can trap the SCAP/SREBP complex in the ER, even without the addition of exogenous sterols. The sterol-sensitizing function of Insig’s is also apparent in an in vitro assay that measures a conformational change in SCAP upon sterol addition (9). Expression of mammalian Insig-1 or -2 in Drosophila cells together with mammalian SCAP confers the ability of sterols to suppress SCAP transport and SREBP processing (10). Combined knockdown of Insig-1 (by 95%) and Insig-2 (by 50%) through RNA interference abolishes the ability of sterols to accelerate degradation of HMG CoA reductase (2), but it does not reduce the ability of sterols to block SREBP processing, perhaps owing to the failure to reduce Insig levels sufficiently.
In the present studies, we explore the role of Insig-1 in regulating synthesis of cholesterol, fatty acids, and triglycerides in the mammalian liver. For this purpose, we produced transgenic mice that overexpress human Insig-1 in the liver. The data show that Insig-1 overexpression renders the liver much more sensitive to sterol-mediated feedback suppression of lipid synthesis, and it largely prevents the acute increase in lipid synthesis that accompanies refeeding of previously fasted mice. The latter finding suggests that insulin-mediated suppression of Insig-2a is required in order for insulin to increase lipid synthesis in the liver and that increased lipid synthesis does not occur when Insig levels are kept high through overexpession of Insig-1.
Results
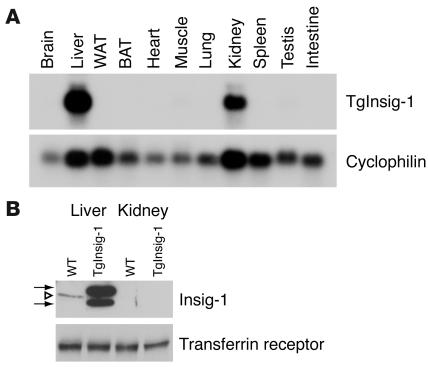
Figure 1 shows the expression in mouse tissues of the human Insig-1 transgene driven by a version of the apo E promoter/enhancer that is believed to be liver specific (11). Using Northern blotting, human Insig-1 mRNA was detectable only in the liver and kidney (Figure 1A). Despite expression of the mRNA in the kidney, the human protein was undetectable by immunoblotting in this organ (Figure 1B). In contrast, the liver expressed abundant amounts of human Insig-1 protein, which gives two bands, owing to the use of two start sites for translation (3). Mouse Insig-1 gives only a single band, owing to a 16–amino acid deletion that removes the downstream initiator methionine, which is at residue 37 in the human sequence. As a result, the single mouse Insig-1 protein is intermediate in size between the two human forms. In the livers of these ad libitum–fed mice, the level of human Insig-1 protein was many fold higher than the level of endogenous mouse Insig-1 (Figure 1B).
Figure 1.
Transgene expression in TgInsig-1 mice. (A) Tissue distribution of human Insig-1 transgene mRNA. Total RNA was extracted from tissues of transgenic mice consuming a chow diet. A total of 20 μg of RNA was subjected to electrophoresis and blot hybridization with 32P-labeled cDNA probes for human Insig-1 and mouse cyclophilin. After stringent washing, the membranes were exposed to Kodak X-Omat Blue XB-1 films for 4–12 hours at –80°C. (B) Immunoblot analysis of endogenous mouse Insig-1 and transgenic human Insig-1 from the livers and kidneys of WT and TgInsig-1 mice, respectively. Membrane fractions from five WT and five TgInsig-1 mice (same as those described in Table 1) were prepared and pooled as described in Methods. Aliquots of the pooled membrane fraction (45 μg protein) were subjected to SDS-PAGE and immunoblot analysis as described in Methods. Filters were exposed to Kodak X-Omat Blue XB-1 film for 15 seconds at room temperature. Open triangle denotes endogenous mouse Insig-1 (28 kDa); arrows denote transgenic human Insig-1 (doublet of 30 kDa and 26 kDa). Immunoblot of transferrin receptor served as a loading control. WAT, white adipose tissue; BAT, brown adipose tissue.
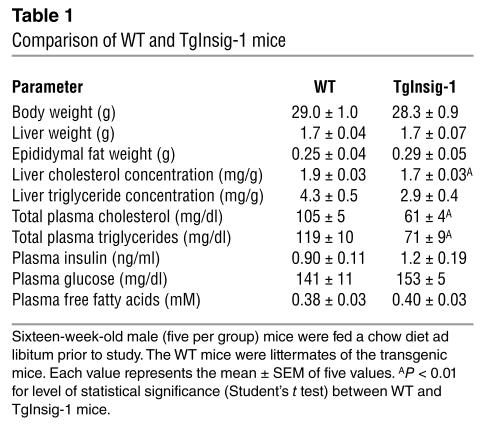
The transgenic mice appeared grossly normal. As shown in Table 1, at 16 weeks they had normal body weights, liver weights, and fat pad weights. The liver cholesterol content was significantly reduced. The triglyceride content was also reduced, but the data did not reach statistical significance. Plasma cholesterol and triglyceride levels were significantly reduced about 40%. Plasma glucose, insulin, and free fatty acids were unaffected.
Table 1.
Comparison of WT and TgInsig-1 mice
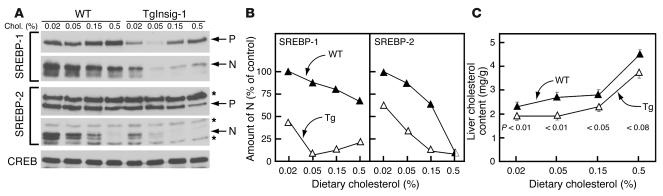
In cultured cells, Insig overexpression increases the sensitivity of SREBP processing to inhibition by sterols. To test for this phenomenon in vivo, WT and transgenic littermate mice were fed diets containing varying amounts of cholesterol for 2 days, after which the amounts of membrane-bound precursor and nSREBPs were measured by SDS-PAGE and immunoblotting (Figure 2A). When consuming the unsupplemented chow diet, which contains 0.02% cholesterol, the transgenic mice showed a lower content of nSREBP-1 and nSREBP-2 than did the WT mice (Figure 2B) These values declined further when small amounts of cholesterol were added to the diet. As dietary cholesterol increased, the decline in both nSREBPs was much more pronounced in transgenic than in WT mice. This decline occurred despite the fact that the cholesterol content of transgenic livers was lower than that in WT mice at all levels of dietary cholesterol (Figure 2C).
Figure 2.
Overexpression of Insig-1 increases sensitivity of SREBP processing to inhibition by dietary cholesterol. Eight-week old female mice (four per group) were fed an ad libitum chow diet supplemented with the indicated amount of cholesterol for 2 days prior to study. The WT mice were littermates of the transgenic mice. A detailed description of these mice is provided in Supplemental Table 1. (A) Immunoblot analysis. Livers from each group (four mice per group) were pooled, and 30-μg aliquots of the membrane and nuclear extract fractions were subjected to SDS-PAGE and immunoblotted with Ab’s against SREBP-1 and SREBP-2. Chol., cholesterol; N, nSREBP; P, precursor form of SREBP. Asterisks denote nonspecific bands. CREB (cAMP-responsive element binding) protein was used as a loading control for the nuclear extract fractions (9192; Ab from Cell Signaling Technology, Beverly, Massachusetts, USA). (B) The gels of nuclear extract fractions in A were scanned and quantified by densitometry. Intensities of the cleaved nuclear forms of SREBP-1 and SREBP-2 in lane 1 (WT mice fed with 0.02% cholesterol) were arbitrarily set at 100%. (C) Cholesterol content of the livers of WT and TgInsig-1 mice fed for 2 days with the indicated amount of cholesterol. Each value represents the mean ± SEM of data from four mice. The levels of statistical significance (Student’s t test) between the WT and TgInsig-1 groups are shown as P values.
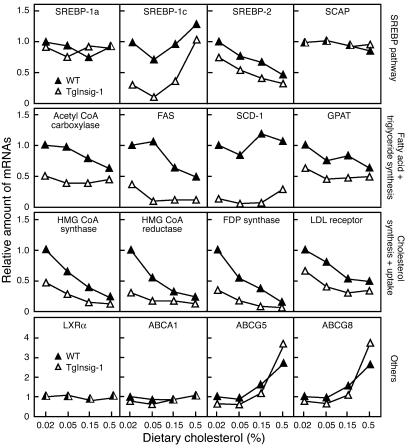
To determine the effects of Insig-1 overexpression on SREBP target genes, we measured the relevant mRNAs by quantitative real-time RT-PCR in the livers of cholesterol-fed mice (Figure 3). The liver produces two quantitatively major isoforms of SREBP, designated SREBP-1c and SREBP-2 (12). SREBP-1c activates predominantly the genes involved in fatty acid and triglyceride biosynthesis. SREBP-2 activates primarily the genes required for cholesterol synthesis. The mRNA for SREBP-1c is enhanced by nSREBPs in a feed-forward reaction. It is also induced by insulin and by the nuclear liver X receptor-α (LXRα) and LXRβ, whose activities are stimulated by oxygenated sterols, some of which can be derived from the cholesterol biosynthetic pathway or from dietary cholesterol (12–14). The mRNA for SREBP-2 is positively regulated by nSREBPs, but not by insulin or LXRs (12). The liver also produces small and unvarying amounts of another isoform, SREBP-1a, which can activate both fatty acid and cholesterol synthesis.
Figure 3.
Relative amount of various mRNAs in the livers of WT and TgInsig-1 mice fed with increasing amounts of cholesterol (0.02–0.5%). The mice used here are the same as those used in Figure 2 and Supplemental Table 1. Total RNA from four mouse livers was pooled and subjected to real-time PCR quantification as described in Methods. Each value represents the amount of mRNA relative to that in WT mice fed a chow diet (0.02% cholesterol), which is arbitrarily defined as 1. FAS, fatty acid synthase; FDP, farnesyl diphosphate; GPAT, glycerol-3-phosphate acyltransferase; SCD-1, stearoyl CoA desaturase-1.
The Insig-1 transgene had no effect on the small amount of SREBP-1a mRNA in the liver, and there was no response to cholesterol feeding (Figure 3, first row). In contrast, the mRNA for SREBP-1c was severely reduced in the transgenic livers, even on the 0.02% cholesterol chow diet. Cholesterol feeding induced a biphasic response. In WT littermates fed 0.05% cholesterol, the mRNA for SREBP-1c declined, perhaps owing to the reduction in nSREBPs. As dietary cholesterol increased, however, the amount of SREBP-1c mRNA rose, an effect that we attribute to the activation of LXRs (14). The effects of dietary cholesterol on SREBP-1c mRNA in the transgenic mice paralleled those in WT mice, but the absolute level in transgenic mice was always much lower than that in WT mice. The response of SREBP-2 mRNA was much less complex. This mRNA was reduced in the transgenic livers, and it declined in parallel in WT and transgenic animals as the dietary content of cholesterol rose. This decline is most likely caused by the loss of the feed-forward effect, owing to the cholesterol-mediated decline in nSREBPs (see Figure 2). SCAP mRNA was unaffected by the transgene or by cholesterol feeding.
All of the measured mRNAs in the fatty acid and triglyceride synthesis pathways were reduced in the Insig-1 transgenic mice fed the chow diet (0.02% cholesterol; second row in Figure 3). In WT mice cholesterol feeding partially reduced the mRNA for acetyl CoA carboxylase, but this was not observed in the Insig-1 transgenic mice. The mRNA for fatty acid synthase was reduced by cholesterol in both lines of mice. Cholesterol feeding had little effect on the mRNAs for stearoyl CoA desaturase or glycerol-3-phosphate acyltransferase. The mRNAs for three enzymes of cholesterol biosynthesis and the LDL receptor were reduced in the Insig-1 transgenic mice on the chow diet (third row in Figure 3), and they all declined further with cholesterol feeding. The mRNAs for LXRα and ABCA1 were not affected by the transgene or by cholesterol feeding. In contrast, the mRNAs for ABCG5 and ABCG8 were induced by cholesterol feeding, which reflects their known stimulation by LXRα and LXRβ. The Insig-1 transgene had little effect on these mRNAs, which encode transporters that secrete cholesterol and plant sterols into bile (15).
The data of Figures 2 and 3 indicate that overexpression of Insig-1 retards the processing of SREBP-1c and -2 and renders this process more sensitive to inhibition by cholesterol. The net effect is a reduction in the mRNAs for SREBP target genes. Increases in nSREBP-1c have also been invoked to explain the increase in hepatic fatty acid synthesis that occurs when fasted animals are refed with a high-carbohydrate diet, which stimulates insulin release and thereby increases the mRNA for SREBP-1c (12). To test the effect of the Insig-1 transgene on this response, we subjected the transgenic mice to a 12-hour fast followed by 12 hours of refeeding.
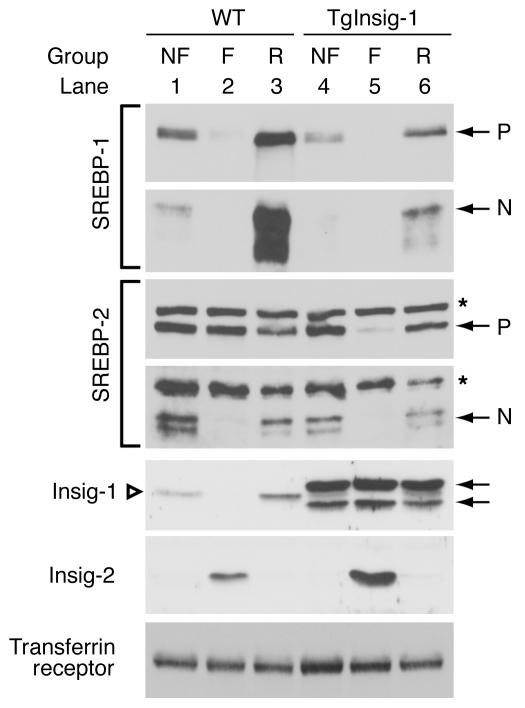
As shown by the immunoblots of Figure 4, in WT mice fasting reduced the amount of the precursor and nuclear forms of SREBP-1c, and refeeding induced a major “overshoot” in nSREBP-1c as well as in its precursor. In contrast, the transgenic mice showed a reduction in nSREBP-1c on an ad libitum diet, and there was no further reduction with fasting (although the precursor did fall in these mice). Refeeding restored the SREBP-1c precursor, but the amount of nSREBP-1c remained well below that in the WT littermates. In WT mice, fasting reduced nSREBP-2, and this was restored to control levels by refeeding without any overshoot. The response was similar in Insig-1 transgenic mice, except that the level of nSREBP-2 was reduced when compared with WT littermates. As previously reported (8), in the WT mice fasting caused the disappearance of Insig-1 (owing to the decrease in nSREBPs) and the appearance of Insig-2 (owing to the decrease in insulin). Refeeding reversed these changes. As expected, fasting and refeeding did not affect the elevated human Insig-1 in the transgenic mice. The induction of Insig-2 by fasting occurred normally in these animals. The modest increase of Insig-2 protein in the fasted transgenic mice compared with the WT controls was not observed in other studies.
Figure 4.
Effect of fasting and refeeding on SREBP and Insig proteins in the livers of WT and TgInsig-1 mice. Sixteen-week-old male mice (five per group) were subjected to fasting and refeeding as described in Methods. The WT mice were littermates of the transgenic mice. A detailed description of these mice is provided in Supplemental Table 2. The nonfasted group (NF) was fed a chow diet ad libitum, the fasted group (F) was fasted for 12 hours, and the refed group (R) was fasted for 12 hours and refed a high-carbohydrate/low-fat diet for 12 hours prior to study. Nuclear extract fractions were prepared from pooled livers (five mice per group); membrane fractions were prepared individually and pooled as described in Methods. Aliquots of membrane and nuclear extract fractions (45 μg) were subjected to SDS-PAGE and immunoblot analysis as described in Methods and Figures 1 and 2. Asterisks denote nonspecific bands. Open triangle denotes endogenous mouse Insig-1 (28 kDa) (lanes 1–3); arrows denote transgenic human Insig-1 (doublet of 30 kDa and 26 kDa) (lanes 4–6).
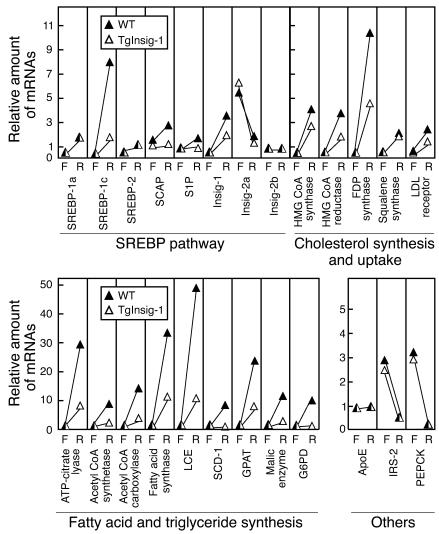
Figure 5 shows the levels of relevant mRNAs in the livers of the fasted and refed mice. With regard to genes of the SREBP pathway, refeeding induced a marked rise in SREBP-1c mRNA in WT but not transgenic mice (see Discussion for explanation). Refeeding also increased the mRNAs encoding three enzymes of cholesterol synthesis (HMG CoA synthase, HMG CoA reductase, and farnesyl diphosphate synthase). The response was blunted, but still detectable in the Insig-1 transgenic mice. Lesser increases were seen in squalene synthase and the LDL receptor. As expected, refeeding caused marked increases in all of the measured mRNAs encoding enzymes of fatty acid and triglyceride synthesis. The response was severely blunted in the Insig-1 transgenic mice. Refeeding suppressed the mRNAs for insulin receptor substrate-2 (IRS-2) and phosphoenolpyruvate carboxykinase (PEPCK), two genes known to be repressed by insulin (16). As a control, we measured the mRNA for apoE, which was not affected by refeeding in WT or transgenic mice.
Figure 5.
Relative amount of various mRNAs in the livers of WT and TgInsig-1 mice subjected to fasting and refeeding. The mice used here are the same as those used in Figure 4 and Supplemental Table 2. Total RNA from the livers of mice was pooled (five mice per group) and subjected to real-time PCR quantification as described in Methods. Each value represents the amount of mRNA relative to that in the nonfasted WT mice, which is arbitrarily defined as 1. For purposes of clarity, only the fasted and refed values are shown. FDP, farnesyl diphosphate; G6PD, glucose-6-phosphate dehydrogenase; LCE, long-chain fatty acyl elongase; S1P, site-1 protease. F, fasted group; R, refed group.
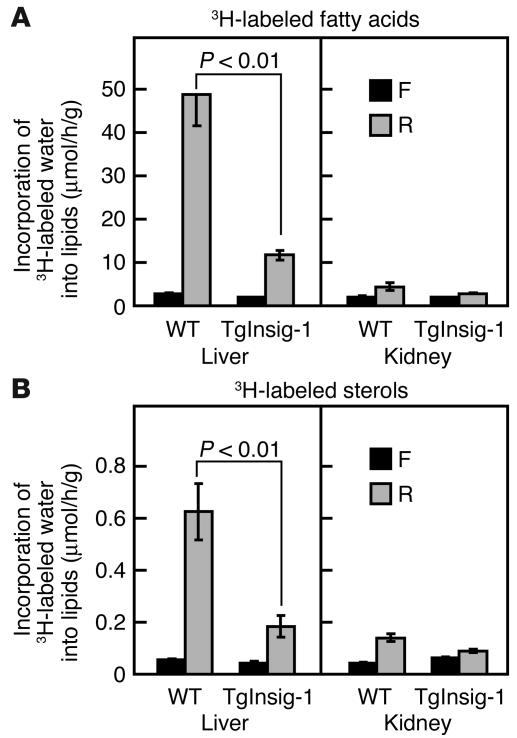
As expected from the changes in mRNA levels, refeeding caused a marked 18-fold increase in hepatic fatty acid synthesis in WT mice, as determined by measurement of the incorporation of intraperitoneally injected 3H-labeled water (Figure 6A). This response was severely blunted in the Insig-1 transgenic mice. Refeeding caused much less stimulation of fatty acid synthesis in the kidney, and this was similar in the WT and transgenic mice. Sterol synthesis was also markedly elevated in the livers of the refed WT mice, and this elevation was reduced threefold in the transgenic livers (Figure 6B). The kidneys of WT and transgenic mice showed a much smaller increase in sterol synthesis upon refeeding.
Figure 6.
In vivo synthesis rates of fatty acids (A) and sterols (B) in the livers and kidneys of WT and TgInsig-1 mice subjected to fasting and refeeding. Mice (16-week old male, five per group) were either fasted for 12 hours or fasted for 12 hours and then refed a high-carbohydrate/low-fat diet for 12 hours prior to study. Each mouse was then injected intraperitoneally with 3H-labeled water (50 mCi in 0.25 ml of isotonic saline). One hour later the tissues were removed for measurement of 3H-labeled fatty acids and digitonin-precipitable sterols. Each bar represents the mean ± SEM of values from five mice. The levels of statistical significance (Student’s t test) between the WT and TgInsig-1 groups are shown as P values. F, fasted group; R, refed group.
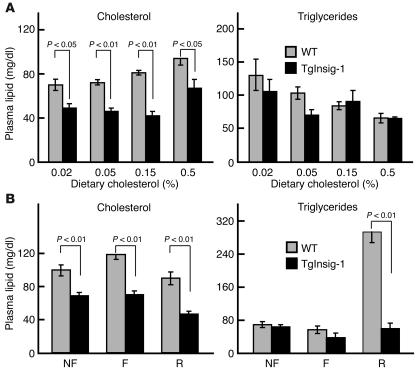
Figure 7 shows the plasma cholesterol and triglyceride levels in the animals that were used in the current studies. In the cholesterol-feeding experiment, plasma cholesterol levels were lower in the Insig-1 transgenic mice and remained lower as dietary cholesterol was increased to 0.5% (Figure 7A). Plasma triglyceride levels were not significantly lower in the transgenic mice as compared with their WT littermates. In the fasting-refeeding experiments, plasma cholesterol was also significantly reduced in the Insig-1 transgenic mice (Figure 7B). Refeeding the high-carbohydrate/low-fat diet caused a marked increase in plasma triglycerides in WT mice, reflecting the marked increase in hepatic fatty acid synthesis in these animals. This increase did not occur in the Insig-1 transgenic mice.
Figure 7.
Plasma lipid levels in WT and TgInsig-1 mice fed different amounts of cholesterol (A) or subjected to fasting and refeeding (B). Mice used in A are described in Figure 2; mice used in B are described in Figure 4. Each value is the mean ± SEM of data from four or five mice. The levels of statistical significance (Student’s t test) between the WT and TgInsig-1 groups are shown as P values. F, fasted group; R, refed group.
Discussion
The current results indicate that overexpressed Insig-1 traps the SCAP/SREBP complex in the ER of liver cells in living mice, thereby reducing the ability of SREBPs to activate transcription of genes encoding enzymes of cholesterol and fatty acid biosynthesis. In addition to reducing basal levels of nSREBPs, the overexpressed Insig-1 enhanced the ability of dietary cholesterol to further reduce these levels. Moreover, the overexpressed Insig-1 prevented the increase in nSREBP-1c that normally accompanies refeeding of previously fasted mice and thereby markedly reduced fatty acid synthesis in these animals. This latter observation is similar to the observation previously made in gene KO mice that lack SCAP or site-1 protease, both of which are required to process SREBPs (12, 17, 18). Considered together, these data establish the essential role of nSREBPs in mediating the insulin-stimulated increase in fatty acid synthesis that occurs upon feeding high-carbohydrate diets to mice.
The immunoblot data of Figure 4 illustrate the dramatic shifts in Insig isoforms that occur after fasting and refeeding in WT mice. Previous studies documented this shift at the mRNA level, but, to our knowledge, the data of Figure 4 are the first confirmation at the protein level, owing to the recent generation of Ab’s capable of recognizing endogenous Insig-1 and -2. Upon fasting, Insig-1 disappears and is replaced by Insig-2. Upon refeeding, Insig-2 markedly decreases and Insig-1 reappears. Inasmuch as SREBP-1c processing is blocked during fasting and returns after refeeding, these data suggest that Insig-2 may be responsible for this regulation.
In the Insig-1 transgenic mice, SREBP-1c processing was not enhanced to a normal extent during refeeding, even though Insig-2 levels declined (see lower panel of Figure 4). Apparently, the unphysiologically high levels of Insig-1 in the livers of the transgenic animals prevented SREBP-1c processing during refeeding, an action that might not occur when Insig-1 is present at its usual low level. To distinguish between the roles of Insig-1 and -2 in the liver, we are currently preparing transgenic mice that overexpress Insig-2. We are also creating gene KO animals that lack expression of the genes encoding Insig-1, Insig-2, and both.
The refed transgenic mice failed to show the normal rise in SREBP-1c mRNA that results from insulin action (Figure 5). This failure occurred even though plasma insulin rose to levels in the refed transgenic mice similar to those in the WT mice (see Supplemental Table 2; supplemental material available at http://www.jci.org/cgi/content/full/113/8/1168/DC1). Insulin had its normal effect on transcription of other hepatic genes, as reflected in the normal repression of IRS-2 and PEPCK mRNAs in the livers of the transgenic mice upon refeeding (see Figure 5). In studies to be published elsewhere, we have found that the promoter/enhancer of the SREBP-1c gene contains a functional sterol regulatory element, and this element is essential for the insulin-mediated enhancement of SREBP-1c mRNA levels. We believe that insulin fails to activate the SREBP-1c gene in the Insig-1 transgenic mice because of the deficiency of nSREBPs that results from the block in SREBP processing.
The current studies, considered together with studies in mice deficient in SCAP or site-1 protease, establish the role of SREBPs in mediating both cholesterol-repressible transcription and insulin-induced transcription of lipogenic genes in the mouse liver. Complex control mechanisms determine the nuclear content of SREBPs under any metabolic state. These mechanisms are a net result of metabolic and hormonal effects on transcription of the SREBP genes and on proteolytic processing of the SREBP proteins. Insig’s, along with SCAP, appear to play central roles in this regulation, which in turn is essential for normal homeostasis of carbohydrates and lipids in mice.
Methods
Materials and general methods.
Blood was drawn from the retro-orbital sinus; the plasma was separated immediately and stored at –70°C. Plasma glucose was measured using a glucose kit from Wako Chemicals USA Inc. (994-90902; Richmond, Virginia, USA). Plasma-free fatty acids were measured using a NEFA kit from Wako Chemicals USA Inc. (kit 994-75409). Plasma insulin was measured using a rat insulin RIA kit from Linco Research Inc. (SRI-13K; St. Charles, Missouri, USA). Cholesterol and triglycerides in plasma and the liver were measured as previously described (19). Immunoblot analyses of mouse SREBP-1 and -2 were carried out as previously described (20). Monoclonal antibody against transferrin receptor (13–6800) was obtained from Zymed Laboratories Inc. (San Francisco, California, USA).
Generation of transgenic mice overexpressing human Insig-1.
To generate transgenic mice overexpressing human Insig-1 (TgInsig-1) in the liver, we used a pLiv-11 vector that contains the constitutive human apoE gene promoter and its hepatic control region (a gift from John Taylor, Gladstone Institute, San Francisco, California, USA) (11, 21). The transgenic plasmid (pLiv-11-Insig-1) was generated by cloning a cDNA fragment encoding the open reading frame of human Insig-1 into MluI-ClaI sites of pLiv-11. The 11-kb SalI-SpeI fragment of pLiv-11-Insig-1 was then isolated and injected into fertilized eggs to generate transgenic mice as previously described (22). Tail DNA dot blot analyses were performed to identify founder mice that harbored the integrated transgene. Positive founders were then subjected to partial hepatectomy, and expression levels of the human Insig-1 transgene were determined by Northern blot analysis. Mice with high levels of transgene expression in the liver were bred to C57BL/6J × SJL/J F1 mice, and four lines of TgInsig-1 mice were established.
The transgenic mice were maintained as hemizygotes by breeding with WT C57BL/6J × SJL/J F1 mice. To genotype transgenic mice, tail DNA was prepared using a direct lysis kit from Viagen Biotech Inc. (102-T; Los Angeles, California, USA) and used for PCR with the following primers: 5′ primer, 5′-ATGGAGAGGAGGGGGCTGAGAATTGTGTGG-3′; 3’ primer, 5′-CTAAATTGGATTTTGCCAATAATGTCCAGC-3′. Tail DNA of TgInsig-1 mice produced a PCR product of 530 bp. All mice were housed in colony cages with a 12-hour light/12-hour dark cycle and fed Teklad Mouse/Rat Diet 7002 from Harlan Teklad (Madison, Wisconsin, USA). For animal experiments, nontransgenic littermates were used as controls for transgenic mice. All animal experiments were performed with the approval of the Institutional Animal Care and Research Advisory Committee at University of Texas Southwestern Medical Center at Dallas.
Diet studies.
For the cholesterol feeding study, mice were fed a chow diet (0.02% cholesterol) or chow diet supplemented with 0.05%, 0.15%, or 0.5% cholesterol for 2 days prior to the study. For the fasting and refeeding experiments, mice were divided into three groups: nonfasted, fasted, and refed. The nonfasted group was fed ad libitum, the fasted group was fasted for 12 hours, and the refed group was fasted for 12 hours and then refed a high-carbohydrate/low-fat diet (TD 88122; Harlan Teklad) for 12 hours prior to study. The starting times for the feeding regimens were staggered so that all mice were sacrificed at the same time, which was at the end of the dark cycle.
Real-time RT-PCR.
Total RNA was prepared from mouse tissues using an RNA STAT-60 kit from Tel-Test Inc. (Friendswood, Texas, USA). Equal amounts of RNA from four or five mice were then pooled and treated with DNase I (DNA-free; Ambion Inc., Austin, Texas, USA). First-strand cDNA was synthesized from 2 μg of DNase I–treated total RNA with random hexamer primers using the ABI cDNA synthesis kit (N808-0234; PE Biosystems, Foster City, California, USA). Specific primers for each gene were designed using Primer Express software (PE Biosystems). The real-time RT-PCR reaction was set up in a final volume of 20 μl containing 20 ng of reverse-transcribed total RNA, 167 nM of the forward and reverse primers, and 10 μl of 2× SYBR Green PCR Master Mix (4312704; PE Biosystems). PCR reactions were carried out in 384-well plates using the ABI PRISM 7900HT Sequence Detection System (PE Biosystems). All reactions were done in triplicate. The relative amount of all mRNAs was calculated using the comparative threshold cycle (CT) method. Mouse apoB or cyclophilin mRNAs were used as the invariant controls. The primers for real-time PCR were described previously (2, 8, 18, 23).
Ab’s against mouse Insig-1 and Insig-2.
Polyclonal Ab’s against mouse Insig-1 and Insig-2 were prepared by immunizing rabbits with (His)6-tagged mouse Insig-1 and Insig-2 proteins produced from Sf9 cells using the BAC-to-BAC Baculovirus Expression System from Invitrogen Corp. (Carlsbad, California, USA). Briefly, cDNA fragments encoding the Insig-1 and Insig-2 open reading frames were generated by PCR using mouse liver RNA as templates with a One-Step RT-PCR Kit from Clontech Laboratories Inc. (K1403-1; Palo Alto, California, USA). The PCR products were subcloned into the pFastBac HTA vector to generate pFastBac HTA-Insig-1 and pFastBac HTA-Insig-2, which were used to transform Escherichia coli strain DH10Bac to produce recombinant bacmids. The recombinant bacmids were then used to transfect Sf9 insect cells to produce recombinant baculoviruses. The recombinant baculoviruses were amplified and used to infect Sf9 insect cells to produce recombinant Insig-1 and Insig-2 proteins. The resultant (His)6-tagged proteins were solubilized with fos-choline-13 from Anatrace Inc. (Maumee, Ohio, USA), purified by Ni-NTA affinity columns, and injected into rabbits.
Immunoblot analyses of Insig-1 and Insig-2.
To prepare membrane fractions for immunoblot analyses, approximately 50 mg of frozen liver was homogenized in 1 ml buffer (20 mM Tris-HCl at pH 7.4, 2 mM MgCl2, 0.25 mM sucrose, 10 mM sodium EDTA, and 10 mM sodium EGTA) supplemented with protease inhibitor cocktail consisting of 5 mM DTT, 0.1 mM leupeptin, 1 mM PMSF, 0.5 mM Pefabloc, 10 μg/ml leupeptin, 5 μg/ml pepstatin A, 25 μg/ml N-acetyl-leu-leu-norleucinal (ALLN), and 10 μg/ml aprotinin. After centrifugation at 6,800 g for 5 minutes at 4°C in a microcentrifuge, the supernatant was centrifuged at 105 g for 30 minutes at 4°C in a Beckman TLA 100.2 rotor. The resulting membrane pellet was resuspended in 0.2 ml SDS-lysis buffer (10 mM Tris-HCl at pH 6.8, 1% wt/vol SDS, 100 mM NaCl, 1 mM sodium EDTA, and 1 mM sodium EGTA). After removal of an aliquot of the membrane fraction for measurement of protein concentration (BCA Kit; Pierce Biotechnology Inc., Rockford, Illinois, USA), 150 μl of the remaining fraction was mixed with an equal volume of buffer containing 62.5 mM Tris-HCl at pH 6.8, 15% SDS, 8 M urea, 10% (vol/vol) glycerol, and 100 mM DTT. To this mixture was added 100 μl of 4× SDS loading buffer (12% SDS, 0.02% bromophenol blue, 30% glycerol, 0.15 mM Tris-HCl at pH 6.8, and 6% β-mercaptoethanol). Equal amounts of protein from four to five mice per group were then pooled, and aliquots (45 μg) of the pooled membrane fraction were incubated at 37°C for 20 minutes and subjected to SDS-PAGE on 12% gels. After electrophoresis, the proteins were transferred to Hybond-C extra nitrocellulose filters from Amersham Biosciences Inc. (Piscataway, New Jersey, USA), and the filters were preincubated for 1 hour with blocking buffer (1× PBS with Tween 20) (P3563; Sigma-Aldrich, St. Louis, Missouri, USA) containing 5% (vol/vol) newborn calf serum (14-416Q; BioWhittaker Inc., Walkersville, Maryland, USA) and 5% (wt/vol) milk, followed by incubation for 3 hours at room temperature with blocking buffer containing a 1:1,000 dilution of either anti–Insig-1 or anti–Insig-2 antiserum. Bound Ab’s were visualized with peroxidase-conjugated, affinity-purified donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories Inc., West Grove, Pennsylvania, USA) using the SuperSignal CL-HRP substrate system (Pierce Biotechnology Inc.). Filters were exposed to Kodak X-Omat Blue XB-1 film at room temperature for 10–30 seconds.
Cholesterol and fatty acid synthesis
in vivo. Rates of cholesterol and fatty acid synthesis were measured in TgInsig-1 mice and their nontransgenic littermates using 3H-labeled water as previously described (22). The mice were either fasted for 12 hours or fasted for 12 hours and then refed a high-carbohydrate/low-fat diet (TD 88122; Harlan Teklad) for 12 hours prior to study. The rates of cholesterol and fatty acid synthesis were calculated as micromoles of 3H-radioactivity incorporated into fatty acids or digitonin-precipitable sterols per hour per gram of tissue.
Supplementary Material
Acknowledgments
We thank our colleagues Li-Ping Sun and Arun Radhakrishnan for help with Ab production; Monica Mendoza, Scott Clark, and Liz Lummus for excellent technical assistance; Samantha Castillo and Richard Gibson for invaluable help with animal studies; and Erin Friedman and Jeff Cormier for DNA sequencing and RT-PCR. This research was supported by grants from the NIH (HL-20948), the Perot Family Foundation, the Moss Heart Foundation, and the Keck Foundation.
Footnotes
See the related Commentary beginning on page 1112.
Nonstandard abbreviations used: insulin receptor substrate-2 (IRS-2); nuclear form of SREBP (nSREBP); phosphoenolpyruvate carboxykinase (PEPCK); SREBP cleavage-activating protein (SCAP); sterol regulatory element–binding protein (SREBP); transgenic mice overexpressing human Insig-1 (TgInsig-1 mice).
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Sever N, Yang T, Brown MS, Goldstein JL, DeBose-Boyd RA. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol. Cell. 2003;11:25–33. doi: 10.1016/s1097-2765(02)00822-5. [DOI] [PubMed] [Google Scholar]
- 2.Sever N, et al. Insig-dependent ubiquitination and degradation of mammalian 3-hydroxy-3-methylglutaryl-CoA reductase stimulated by sterols and geranylgeraniol. J. Biol. Chem. 2003;278:52479–52490. doi: 10.1074/jbc.M310053200. [DOI] [PubMed] [Google Scholar]
- 3.Yang T, et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 4.Yabe D, Brown MS, Goldstein JL. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12753–12758. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horton JD, et al. Combined analysis of olignucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feramisco JD, Goldstein JL, Brown MS. Membrane topology of human insig-1, a protein regulator of lipid synthesis. J. Biol. Chem. 2004;279:8487–8496. doi: 10.1074/jbc.M312623200. [DOI] [PubMed] [Google Scholar]
- 7.Janowski BA. The hypocholesterolemic agent LY295427 up-regulates INSIG-1, identifying the INSIG-1 protein as a mediator of cholesterol homeostasis through SREBP. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12675–12680. doi: 10.1073/pnas.202471599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yabe D, Komuro R, Liang G, Goldstein JL, Brown MS. Liver-specific mRNA for Insig-2 down-regulated by insulin: implications for fatty acid synthesis. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3155–3160. doi: 10.1073/pnas.0130116100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams CM, Goldstein JL, Brown MS. Cholesterol-induced conformational change in SCAP enhanced by Insig proteins and mimicked by cationic amphiphiles. Proc. Natl. Acad. Sci. U. S. A. 2003;100:10647–10652. doi: 10.1073/pnas.1534833100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobrosotskaya I, Goldstein JL, Brown MS, Rawson RB. Reconstitution of sterol-regulated endoplasmic reticulum-to-Golgi transport of SREBP-2 in insect cells by co-expression of mammalian SCAP and Insigs. J. Biol. Chem. 2003;278:35837–35843. doi: 10.1074/jbc.M306476200. [DOI] [PubMed] [Google Scholar]
- 11.Simonet WS, Bucay N, Lauer SJ, Taylor JM. A far-downstream hepatocyte-specific control region directs expression of the linked human apolipoprotein E and C-1 genes in transgenic mice. J. Biol. Chem. 1993;268:8221–8229. [PubMed] [Google Scholar]
- 12.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 2002;109:1125–1131. doi:10.1172/JCI200215593. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Repa JJ, et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeBose-Boyd RA, Ou J, Goldstein JL, Brown MS. Expression of sterol regulatory element-binding protein 1c (SREBP-1c) mRNA in rat hepatoma cells requires endogenous LXR ligands. Proc. Natl. Acad. Sci. U. S. A. 2001;98:1477–1482. doi: 10.1073/pnas.98.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berge KE, et al. Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science. 2000;290:1771–1775. doi: 10.1126/science.290.5497.1771. [DOI] [PubMed] [Google Scholar]
- 16.Shimomura I, et al. Decreased IRS-2 and increased SREBP-1c lead to mixed insulin resistance and sensitivity in livers of lipodystrophic and ob/ob mice. Mol. Cell. 2000;6:77–86. [PubMed] [Google Scholar]
- 17.Matsuda M, et al. SREBP cleavage-activating protein (SCAP) is required for increased lipid synthesis in liver induced by cholesterol deprivation and insulin elevation. Genes Dev. 2001;15:1206–1216. doi: 10.1101/gad.891301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J, et al. Decreased lipid synthesis in livers of mice with disrupted Site-1 protease gene. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13607–13612. doi: 10.1073/pnas.201524598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yokode M, Hammer RE, Ishibashi S, Brown MS, Goldstein JL. Diet-induced hypercholesterolemia in mice: prevention by overexpression of LDL receptors. Science. 1990;250:1273–1275. doi: 10.1126/science.2244210. [DOI] [PubMed] [Google Scholar]
- 20.Shimano H, et al. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J. Clin. Invest. 1997;100:2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allan CM, Taylor JM. Expression of a novel human apolipoprotein (apoC-IV) causes hypertriglyceridemia in transgenic mice. J. Lipid Res. 1996;37:1510–1518. [PubMed] [Google Scholar]
- 22.Shimano H, et al. Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J. Clin. Invest. 1996;98:1575–1584. doi: 10.1172/JCI118951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang G, et al. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J. Biol. Chem. 2002;277:9520–9528. doi: 10.1074/jbc.M111421200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.