Abstract
We recently showed that antigen-nonspecific inflammatory cells are recruited into the liver when hepatitis B virus (HBV)-specific CTLs are injected into HBV transgenic mice, and that this process amplifies the severity of liver disease. We also showed that the severity of CTL-induced liver disease is ameliorated by the depletion of Gr-1+ cells (Gr-1 is an antigen highly expressed by neutrophils), which, secondarily, abolishes the intrahepatic recruitment of all antigen-nonspecific Gr-1– mononuclear cells (NK and NKT cells, T and B lymphocytes, monocytes, macrophages, dendritic cells) despite the strong induction of chemokine gene expression. Those results suggested that in addition to chemokine expression, CTL-induced functions are necessary for mononuclear cell recruitment to occur. We now report that MMPs known to be produced by Gr-1+ cells are rapidly induced in the livers of CTL-injected mice. The inhibition of MMP activity reduced the intrahepatic recruitment of antigen-nonspecific mononuclear cells and much of the attending liver disease without affecting the migration or antiviral potential of antigen-specific CTLs. The notion that the inhibition of MMP activity is associated with maintenance of antiviral effects but diminished tissue damage may be significant for the development of immunotherapeutic approaches for the treatment of chronic HBV infection.
Introduction
The hepatitis B virus (HBV) is a noncytopathic, enveloped DNA virus that causes acute and chronic liver disease characterized by a necroinflammatory cell infiltrate that is triggered by the cellular immune response to viral antigens (1). Using transgenic mice that sustain high-level HBV replication in the liver (2), we have shown that most of the antiviral potential of virus-specific CTLs is mediated by noncytolytic mechanisms that involve the intrahepatic production of IFN-γ by the CTLs following antigen recognition (3–5). We have also previously shown that the direct cytopathic potential of the CTLs is limited, involving the apoptotic death of a small number of hepatocytes (6). This process is followed by the recruitment into the liver of many host-derived, antigen-nonspecific, polymorphonuclear and mononuclear cells (i.e., NK and NKT cells, T and B lymphocytes, monocytes, macrophages, dendritic cells) that contribute to the formation of necroinflammatory foci that are scattered throughout the liver parenchyma and are typical of acute viral hepatitis in humans (6–8).
The recruitment process is chemokine-dependent, since we have shown that blocking the IFN-γ–inducible chemokines CXCL9 and CXCL10 reduces the migration of antigen-nonspecific mononuclear cells and the severity of liver disease after CTL injection (7). The association of reduced liver disease with reduced recruitment of antigen-nonspecific mononuclear cells implies that these cells can amplify the liver damage initiated by the antigen-specific CTLs. We recently showed that depletion of Gr-1+ cells also reduces the severity of liver disease in this model. Gr-1+ cells include polymorphonuclear neutrophils (PMNs) (9), plasmacytoid dendritic cells (pDCs) (10, 11), and a subset of monocytes/macrophages (12, 13). Interestingly, depletion of Gr-1+ cells completely blocks the recruitment of all Gr-1– intrahepatic mononuclear cells into the liver, even though the expression of many chemokines (including that of CXCL9 and CXCL10) is induced at high levels in the organ (8). These results indicate that Gr-1+ cells are necessary for the intrahepatic recruitment of antigen-nonspecific mononuclear cells, and they suggest that functions by Gr-1+ cells in addition to chemokine induction are necessary for the recruitment process to occur.
These functions may include the release of the matrix-degrading metalloproteinases by PMNs or other Gr-1+ cells. Although it is not known whether pDCs or Gr-1+ monocytes/macrophages produce MMPs, PMNs are known to produce high levels of collagenases (such as MMP-8, neutrophil collagenase) and gelatinases (such as MMP-9, gelatinase B) (14). MMP-8 and MMP-9 are stored within PMN granules, and they are rapidly liberated in response to inflammatory mediators (14). A major action of these enzymes involves the remodeling of the ECM, a process that is thought to facilitate leukocyte trafficking through endothelial barriers and solid organs (14). The capacity of MMP-8 and MMP-9 to cleave and modulate the function of various chemokines (including CXCL9 and CXCL10) is another potentially important biological action (15–17). Although the role of MMP-8 and MMP-9 in the pathogenesis of CTL-induced immunopathology is unknown, it has been previously shown that the administration of a broad-spectrum MMP inhibitor can reduce liver disease in a mouse model of hepatitis induced by TNF-α in combination with D-galactosamine (TNF/GalN) (18). Likewise, amelioration of TNF/GalN-induced liver disease was observed in mice deficient in MMP-2, MMP-3, and MMP-9 (18). Other studies involving MMP-9 KO mice have also indicated a role of MMP-9 in (a) leukocyte recruitment during allergen-induced airway inflammation (19), (b) resistance to cancer invasion (20), and (c) angiogenesis caused by ocular infection with herpes simplex virus (21).
Importantly, the physiological balance between degradation and deposition of the ECM is maintained by the tissue inhibitors of MMPs (TIMPs). Four mammalian TIMPs (TIMP-1, -2, -3, and -4) have been cloned and characterized, and they represent the major cellular inhibitors of MMPs (22). All TIMPs are secreted proteins that have the potential to reversibly inhibit active forms of all MMPs (22).
Based on these observations, the current study was undertaken to determine the role of MMPs in our system. First, we monitored the association between the recruitment of inflammatory cells into the liver and the intrahepatic expression or activity of MMPs. Second, we took advantage of the hydrodynamic injection technique (23) to transduce hepatocytes of HBV transgenic mice with an expression vector encoding murine TIMP-1 and to determine whether enhanced hepatic secretion of TIMP-1 would inhibit MMP activity and alter the intrahepatic recruitment and pathogenetic or antiviral effector functions of HBV-specific CTLs and other inflammatory cells.
Results
Recruitment of antigen-nonspecific inflammatory cells into the liver is associated with induced expression and activity of MMP-8 and MMP-9.
To monitor whether (a) MMP RNAs were induced in the liver of CTL-injected HBV transgenic mice, and (b) their induction was correlated with the recruitment of intrahepatic leukocytes (IHLs), total hepatic RNA isolated from the same mice described by Sitia et al. (8) was analyzed for MMP expression by RNase protection assay (RPA). As shown in Figure 1A, when compared to NaCl-injected controls, the total number of IHLs significantly increased in the livers of CTL-injected HBV transgenic mice that received irrelevant Ab’s and were sacrificed at day 1 after CTL injection. As previously reported, IHL recruitment was completely blocked in transgenic littermates that were depleted of Gr-1+ cells with anti–Gr-1 antibodies (α–Gr-1) prior to CTL injection (Figure 1A) (8).
Figure 1.
Recruitment of antigen-nonspecific inflammatory cells into the liver is associated with induced expression and activity of MMP-8 and MMP-9. (A) IHL analysis of transgenic mice from lineage 1.3.32 that were injected with α–Gr-1 and HBsAg-specific CTLs (black bar). The indicated numbers of total IHLs represent the numbers detected in the whole liver. The results were compared with additional groups of transgenic mice that were injected either with saline (NaCl) alone (gray bar) or with CTLs and irrelevant control antibodies (Irr. Ab) (white bar). (B) Total hepatic RNA derived from the same mice was analyzed by RPA for the expression of various MMPs, as indicated. The housekeeping mRNA encoding the ribosomal protein L32 was used to normalize the amount of RNA loaded in each lane. (C) Quantitative phosphor imaging analysis of the same MMP RNAs shown in B. The indicated numbers in the CTL-injected groups (Irr. Ab and α–Gr-1) are displayed as fold induction over NaCl-injected controls (set as 1). The numbers were obtained by dividing each MMP value by the amount of the corresponding housekeeping gene L32 RNA, and they represent the mean of three mice per group. (D) Gelatin-PAGE zymography was performed as described in the Methods on liver extracts from the same mice shown above. (E and F) Collagen type I and gelatin in situ zymography were performed on liver sections from the same mice described above. Cryostat sections were overlaid with a solution containing quenched fluorescent collagen type I (E) or quenched fluorescent gelatin (F), and sections were examined 18 hours later. Collagenase and gelatinase activities become apparent as fluorescence (E and F, middle panels). Note that no detection of collagenase and gelatinase activity was observed in either NaCl-injected controls (E and F, left panels) or CTL-injected mice depleted of Gr-1+ cells (E and F, right panels). Original magnification, ×400.
As shown in Figure 1B, when compared to NaCl-injected controls, the intrahepatic messages for MMP-8 (neutrophil collagenase), MMP-7 (matrilysin), MMP-12 (macrophage metalloelastase), MMP-9 (gelatinase B), MMP-13 (collagenase-3), MMP-14 (MT-1 membrane type-1 metalloproteinase), and MMP-2 (gelatinase A) were induced in the CTL-injected mice that received irrelevant control Ab’s (about sixfold for MMP-8, 18-fold for MMP-7, 28-fold for MMP-12, twofold for MMP-9, sevenfold for MMP-3, twofold for MMP-14, and twofold for MMP-2, respectively, as measured by phosphor imaging analysis; see Figure 1C). In contrast, no increase in gene expression was observed for MMP-11 (Figure 1B). Interestingly, treatment with α–Gr-1 completely inhibited the increase of MMP-8 and MMP-9 RNAs but not that of the other induced MMP RNAs (MMP-2, MMP-3, MMP-7, MMP-12, MMP-13, and MMP-14) (Figure 1, B and C). The association between induction of MMP-8 and MMP-9 RNA and IHL recruitment, along with the observation that no induction of these enzymes was observed in the livers of mice in which IHL recruitment did not occur may indicate that MMP-8 and MMP-9 participate in the recruitment process. The results herein described, coupled with the notion that PMNs are an abundant source of MMP-8 and MMP-9 (14), suggest that PMNs may be an important source of these enzymes in our system. In contrast to MMP-8 and MMP-9 RNAs, α–Gr-1 treatment either had no effect (MMP-2, MMP-3, and MMP-14) or even induced (MMP-7, MMP-12, and MMP-13) the expression of other MMPs in the liver (Figure 1, B and C), suggesting that these MMPs did not contribute to IHL recruitment.
To monitor whether the changes in MMP-8 and MMP-9 RNAs correlated with commensurate changes in collagenase and gelatinase activities, collagen type I and gelatin in situ zymography and gelatin-PAGE zymography were performed on liver sections from the same mice just described.
As shown in Figure 1, E and F (left panels), no activity was observed in the uninflamed livers of NaCl-injected control mice. In good correlation with the increase in MMP-8 and MMP-9 RNA, induction of collagenase and gelatinase activities was observed in the livers of CTL-injected mice that received irrelevant Ab’s (Figure 1, E and F, middle panels). Such activities were scattered throughout the liver parenchyma and confined within necroinflammatory foci (Figure 1, E and F, middle panels) containing apoptotic hepatocytes as well as mononuclear and polymorphonuclear cells (data not shown). This suggests that cleavage of ECM components was colocalized with recruited IHLs. Importantly, lack of MMP-8 and MMP-9 RNA induction in the livers of animals treated with α–Gr-1 (Figure 1, B and C) correlated with absence of collagenase and gelatinase activities (Figure 1, E and F, right panels), suggesting that Gr-1+ cells were responsible for this effect. These results were also confirmed by gelatin-PAGE zymography, in which an activity at 105 kDa (corresponding to the inactive form of MMP-9, gelatinase B) was observed in the livers of CTL-injected mice that received irrelevant Ab’s but not in the livers of mice that were injected either with NaCl or with CTL and α–Gr-1 (Figure 1D). Finally, in keeping with the minor changes of MMP-2 (gelatinase A) RNA levels detected in the different groups of animals shown in Figure 1, B and C, little or no change in gelatinase activity at 72 kDa (corresponding to MMP-2) was observed in the same livers (Figure 1D).
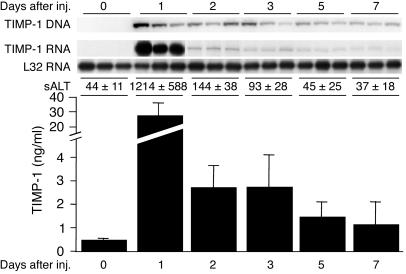
Hydrodynamic injection of a plasmid encoding murine TIMP-1 leads to enhanced expression of TIMP-1 RNA in the liver and TIMP-1 protein in the serum.
Based on the foregoing results, we sought to monitor the effect of the MMP inhibitor, TIMP-1, on the intrahepatic recruitment and pathogenetic effector functions of HBV-specific CTLs and other inflammatory cells in our system. Five groups (three mice per group) of age- and hepatitis B e antigen–matched (HBeAg-matched) female HBV transgenic mice were hydrodynamically injected with pcDNA3.1-TIMP-1, and they were sacrificed at various times after injection. The intrahepatic contents of the TIMP-1 expression plasmid and of TIMP-1 RNA were measured by Southern blot and RPA, respectively, and the serum levels of TIMP-1 protein were measured by ELISA.
As shown in Figure 2 (top), the TIMP-1 plasmid DNA was readily detectable in the liver by day 1 after injection, and it was still present, albeit at slightly lower levels, on day 7. By day 1, the content of hepatic TIMP-1 RNA (Figure 2, top) and serum TIMP-1 protein (Figure 2, bottom) had increased almost 65-fold over control levels before injection (day 0). Their induction decreased quite abruptly thereafter, so that TIMP-1 RNA and protein levels were four- to fivefold and two- to threefold higher than those of controls were by days 3 and 7, respectively (Figure 2). Importantly, the intrahepatic content of HBV RNAs, including the 2.1-kb RNA that encodes the target antigen (hepatitis B surface antigen; HBsAg) of the transferred CTLs, was identical to that of uninjected controls at all time points after hydrodynamic injection of pcDNA3.1-TIMP-1 (data not shown).
Figure 2.
Hydrodynamic injection (inj.) of a plasmid encoding murine TIMP-1 leads to enhanced expression of TIMP-1 RNA in the liver and TIMP-1 protein in the serum. Five groups of age- and HBeAg-matched female HBV transgenic mice (three mice per group) were hydrodynamically injected with pcDNA3.1-TIMP-1. Mice were bled and sacrificed, and the livers were harvested at days 1, 2, 3, 4, and 5 after hydrodynamic injection. The intrahepatic content of pcDNA3.1-TIMP-1 DNA and TIMP-1 RNA were measured by Southern blot and RPA, respectively (upper panels), and the serum levels of TIMP-1 protein were measured by ELISA (lower panel). The mean sALT activity (± SD), measured at the time of autopsy, is indicated for each group and is expressed in units/liter.
It is noteworthy that a sharp elevation in serum alanine aminotransferase (sALT) activity was consistently observed in these animals by day 1 after injection, returning to almost normal levels by days 2–3 (Figure 2). The transient surge in sALT activity completely resolved within a few days of the injection. These changes are probably due to the hydrodynamic injection technique, since similar findings were observed when empty vectors were injected (data not shown).
Enhanced expression of TIMP-1 ameliorates the severity of liver disease and reduces the recruitment of IHLs.
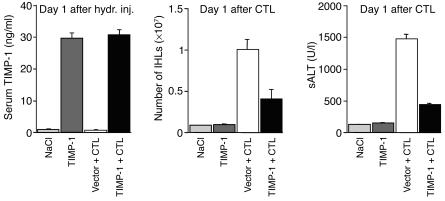
Based on the results just described, we injected HBsAg-specific CTLs 2 days after hydrodynamic injection of pcDNA3.1-TIMP-1, when sALT levels were returning to normal and when the concentration of circulating TIMP-1 was still significantly enhanced (four- to fivefold above baseline). Thus, two groups of age- and serum HBeAg-matched female transgenic mice from lineage 1.3.32 (six mice per group) were hydrodynamically injected with either pcDNA3.1-TIMP-1 or an empty vector, and 2 days later they received 1 × 107 HBsAg-specific CTLs. Mice were bled, sacrificed, and perfused; their livers were harvested on day 1 after CTL transfer. The results were also compared with two additional groups of transgenic littermates (six mice per group) that were hydrodynamically injected with pcDNA3.1-TIMP-1 or NaCl alone and sacrificed 3 days after injection.
As shown in Figure 3 (left panel), mice that were hydrodynamically injected with pcDNA3.1-TIMP-1 (dark gray and black bars) showed high levels of circulating TIMP-1 on day 1 after injection, whereas no elevation of TIMP-1 was observed at the same time point in mice that were injected either with NaCl (light gray bar) or the empty vector (white bar). Serum TIMP-1 levels remained elevated (fivefold above baseline) in animals injected with pcDNA3.1-TIMP-1 at the time of CTL transfer (data not shown). As also shown in Figure 3 (middle and right panels), when compared with mice injected either with NaCl (light gray bars) or pcDNA3.1-TIMP-1 alone (dark gray bars), the total number of IHLs and disease severity (monitored as sALT activity) significantly increased in the livers of CTL-injected HBV transgenic mice that received the empty vector and were sacrificed on day 1 after CTL injection (white bars). Importantly, both parameters were reduced approximately 3- and 2.4-fold, respectively, in transgenic littermates that received the pcDNA3.1-TIMP-1 prior to CTL injection (Figure 3, middle and right panels, black bars). These results indicate that the enhanced expression of TIMP-1 reduces the recruitment of IHLs and ameliorates the severity of liver disease.
Figure 3.
Enhanced expression of TIMP-1 reduces the recruitment of IHLs and ameliorates the severity of liver disease. Two groups of age- and serum HBeAg-matched female transgenic mice from lineage 1.3.32 (six mice per group) were hydrodynamically injected (hydr. inj.) with either pcDNA3.1-TIMP-1 (TIMP-1 + CTL, black bars) or an empty vector (Vector + CTL, white bars), and 2 days later they received 1 × 107 HBsAg-specific CTLs. Mice were bled, sacrificed, and perfused; the livers were harvested on day 1 after CTL transfer (day 3 after hydrodynamic injection). Serum TIMP-1 ELISA (left panel) measured 1 day after hydrodynamic injection and total IHLs (middle panel) or sALT (right panel) measured 1 day after CTL injection were also compared with two additional groups of transgenic littermates (six mice per group) that were hydrodynamically injected with pcDNA3.1-TIMP-1 (TIMP-1, dark gray bars) or saline (NaCl, light gray bars).
Enhanced expression of TIMP-1 inhibits the induction of collagenase and gelatinase activities within the intrahepatic necroinflammatory foci of CTL-injected mice.
To monitor whether the enhanced expression of TIMP-1 inhibited the CTL-induced activity of those MMPs (i.e., MMP-8 and MMP-9) that were associated with IHL recruitment (Figure 1), collagen type I and gelatin in situ zymography was performed on liver sections from the same mice just described.
As shown in Figure 4, collagenase and gelatinase activities were induced within necroinflammatory foci in the livers of CTL-injected mice treated with empty vector (left panels), and this effect was blocked in the intrahepatic foci of mice that received pcDNA3.1-TIMP-1 (right panels). These results indicate that the reduced IHL recruitment observed in animals overexpressing TIMP-1 (Figure 3) was associated with reduced MMP-8 and MMP-9 activity, suggesting that these enzymes contribute to IHL recruitment in this model.
Figure 4.
Induction of collagenase and gelatinase activities within the intrahepatic necroinflammatory foci of CTL-injected mice is inhibited by the enhanced TIMP-1 expression. Collagen type I (A, top panels) and gelatin (B, bottom panels) in situ zymography were performed on liver sections from the same mice described in the legend to Figure 3. Note that no collagenase and gelatinase activity was observed in CTL-injected mice that received the pcDNA3.1-TIMP-1 (right panels). Original magnification, ×400.
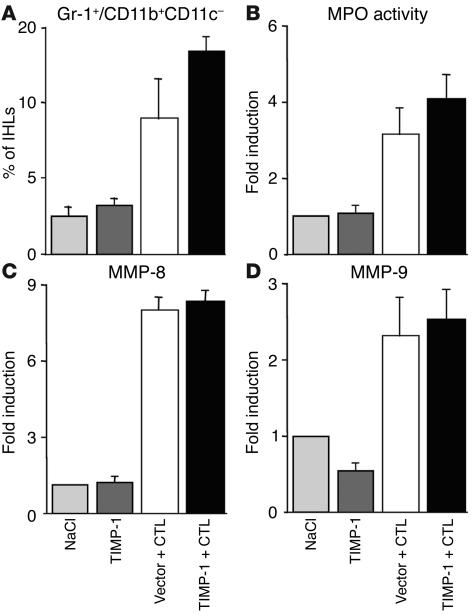
Enhanced expression of TIMP-1 does not affect the recruitment of PMNs but significantly diminishes the recruitment of antigen-nonspecific mononuclear cells into the liver.
Since the reduced MMP-8 and MMP-9 activity in the livers of animals overexpressing TIMP-1 could be due to impaired intrahepatic recruitment of PMNs, the recruitment of these cells was monitored in the same livers described already. PMNs were quantified by measuring (a) the percentage of Gr-1+/CD11b+/CD11c– cells (mostly PMNs) in the total IHL population; (b) total liver myeloperoxidase (MPO) activity, an enzymatic activity that is mostly present in PMNs and thus is used to quantitate PMN recruitment into the liver (24); (c) the intrahepatic content of MMP-8 and MMP-9 RNAs, whose induction is inhibited in the livers of animals depleted of Gr-1+ cells (Figure 1).
As shown in Figure 5A, when compared by FACS analysis to control mice that were injected with NaCl (light gray bars) or pcDNA3.1-TIMP-1 (dark gray bars) alone, the percentage of Gr-1+/CD11b+/CD11c– cells in the total IHL population increased more than threefold in CTL-injected mice that received the empty vector (Figure 5A, white bar). The percentage of Gr-1+/CD11b+/CD11c– cells increased at even higher levels (four- to fivefold over controls) in the livers of CTL-injected animals overexpressing TIMP-1 (Figure 5A, black bar). This suggests that PMNs were efficiently recruited into the liver of the latter group of mice, and their increased percentage in the total IHL population may be due to impaired intrahepatic recruitment of other IHLs (i.e., mononuclear cells, see later) in these animals. In keeping with these results and when compared to the same controls, liver MPO activity (Figure 5B) and MMP-8 or MMP-9 RNAs (Figure 5, C and D) were similarly induced in CTL-injected mice that received either the empty vector (white bar) or pcDNA3.1-TIMP-1 (black bar). All together, the data suggest that the reduced intrahepatic MMP-8 and MMP-9 activity in the livers of CTL-injected mice that overexpress TIMP-1 (Figure 4) was not due to impaired PMN recruitment but instead to impaired function of these cells.
Figure 5.
Enhanced expression of TIMP-1 does not affect the recruitment of PMNs into the liver. (A) Intrahepatic analysis of Gr-1+/CD11b+/CD11c– cells in the same animals described in the legend to Figure 3. Livers were weighed at the time of autopsy. Total IHLs were isolated from two liver lobes of a known weight and analyzed by flow cytometry. The indicated numbers represent the percentage of Gr-1+/CD11b+/CD11c– cells detected in the total IHL population isolated from the whole liver. (B) Total liver MPO activity was determined as described in the Methods in the same mice. The indicated numbers are displayed as fold induction over NaCl-injected controls (set as 1), and they represent the mean of six mice per group. Total hepatic RNA from the same mice was also analyzed by RPA to measure the intrahepatic content of MMP-8 (C) and MMP-9 (D) mRNAs. The indicated numbers, calculated by phosphor imaging analysis, are displayed as fold induction over NaCl-injected controls (set as 1). The numbers were obtained by dividing each MMP value by the amount of the corresponding housekeeping gene L32 RNA, and they represent the mean of six mice per group.
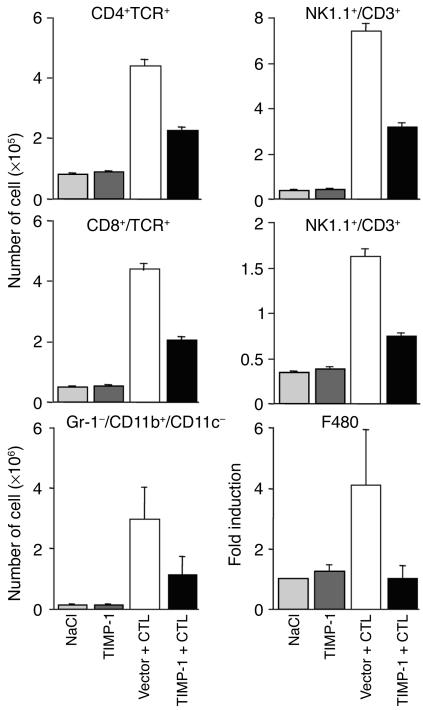
In contrast, CTL-injected mice that received pcDNA3.1-TIMP-1 showed a marked reduction in the number of intrahepatic antigen-nonspecific mononuclear cells. As shown in Figure 6, when compared by FACS analysis with mice injected either with NaCl (light gray bars) or pcDNA3.1-TIMP-1 alone (dark gray bars), the number of CD4+/T cell receptor–positive (CD4+/TCR+) cells (mostly Th cells), CD8+/TCR+ cells (mostly CTLs), Gr-1–/CD11b+/CD11c– cells (mostly macrophages), NK1.1+/CD3– cells (mostly NK cells), and NK1.1+/CD3+ cells (mostly NKT cells) significantly increased in the livers of CTL-treated mice that received the empty vector (Figure 6, white bars). As also shown in Figure 6, the enhanced expression of TIMP-1 (black bars) reduced the increase in the number of CD8+ and CD4+ cells, NK, NKT cells, and macrophages by about twofold. The extent of reduction in macrophage recruitment was also confirmed in this group of mice by using RPA to measure the intrahepatic content of F480 mRNA, a macrophage-specific marker (25, 26) (Figure 6, bottom right panel). All together, these results indicate that the enhanced expression of TIMP-1 and the inhibition of MMP activity diminish the recruitment of mononuclear cells into the liver without affecting the recruitment of PMNs. That the recruitment of mononuclear cells was not completely abolished (Figure 6) suggests that other influences in addition to MMP activity can contribute to this process.
Figure 6.
Enhanced expression of TIMP-1 diminishes the recruitment of antigen-nonspecific mononuclear cells into the liver. Intrahepatic mononuclear cells analysis was done in the same animals described in the legend to Figure 3. Livers were weighed at the time of autopsy. IHLs were isolated from two liver lobes of a known weight and analyzed by flow cytometry. The indicated numbers of the different mononuclear cell subsets represent the total numbers detected in the whole liver. Total hepatic RNA from the same mice was also analyzed by RPA to measure the intrahepatic content of F480 mRNA, a macrophage specific marker (bottom right panel). The indicated numbers are displayed as fold induction over NaCl-injected controls (set as 1), and they represent the mean of six mice per group.
Enhanced expression of TIMP-1 affects neither the intrahepatic recruitment of HBsAg-specific CTLs nor the intrahepatic expression of cytokines and chemokine
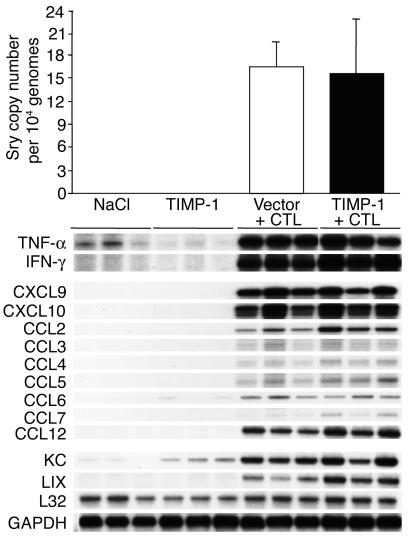
s. To determine whether the enhanced expression of TIMP-1 affected the recruitment process of passively transferred HBsAg-specific male CTLs, the intrahepatic CTL number was measured by quantifying the amount of Sry-specific sequences in the liver of the same HBV transgenic female mice just described. As shown in Figure 7 (top), upon normalization with the internal control, we calculated that the average copy number (per 10,000 liver cell genomes) of Sry-specific amplicons was very similar in the livers of mice that were sacrificed on day 1 after CTL transfer and were injected either with the empty vector (white bar) or pcDNA3.1-TIMP-1 (black bar). These results suggest that similar numbers of CTLs reached the liver at this time point and, therefore, the enhanced expression of TIMP-1 did not affect their intrahepatic recruitment. This conclusion was also supported by the finding that the levels of IFN-γ and TNF-α mRNAs (markers of antigen recognition by activated CTLs; refs. 3, 4, 7, 8) were virtually identical in the livers of the two groups of mice (Figure 7).
Figure 7.
Enhanced expression of TIMP-1 affects neither the intrahepatic recruitment of HBsAg-specific CTLs nor the intrahepatic expression of cytokines and chemokines. The recruitment of the passively transferred CTL lines was measured in the same livers described in the legend to Figure 3 by quantifying the amount of Sry-specific sequences and CCKAR-specific sequences by real-time PCR. The indicated numbers represent the average copy numbers of Sry-specific amplicons per 10,000 liver cell genomes (top). Total hepatic RNA from the same mice was also analyzed by RPA for the expression of various cytokines and chemokines as indicated (bottom). The housekeeping mRNAs encoding the ribosomal protein L32 or GAPDH were used to normalize the amount of RNA loaded in each lane.
We also monitored the effect of enhanced TIMP-1 expression on the intrahepatic induction of CXCL9, CXCL10, and other chemokines, since we have recently shown that blocking the chemokines CXCL9 and CXCL10 reduces the recruitment of host-derived mononuclear cells into the liver and the severity of liver disease in this model (7).
As shown in Figure 7, the messages for CXCL9 and CXCL10 (chemoattractants for mononuclear cells; refs. 27, 28) were induced at similar high levels in the livers of mice that were sacrificed on day 1 after CTL transfer, irrespective of the expression of pcDNA3.1-TIMP-1. Likewise, other chemoattractants for mononuclear cells such as CCL2 (monocyte chemotactic protein, MCP-1), CCL3 (macrophage inflammatory protein, MIP1-α), CCL4 (MIP1-β), CCL5 (RANTES), CCL6 (C10), CCL7 (MCP-3), and CCL12 (MCP-5), or chemoattractants for polymorphonuclear cells such as keratinocyte-derived chemokine (KC) and LPS-induced chemokine (LIX) (27, 28) were similarly upregulated in these animals (Figure 7). All together, these results suggest that the reduced IHL recruitment and the amelioration of disease severity observed in mice overexpressing TIMP-1 was not due to impaired CTL homing, CTL activation, or chemokine production.
Enhanced expression of TIMP-1 does not affect the antiviral potential of HBV-specific CTLs.
To determine whether the enhanced expression of TIMP-1 affected the potential of HBV-specific CTLs to induce a sustained inhibition of viral replication in the face of decreased liver damage, we monitored the content of HBV-replicative intermediates in the livers of transgenic mice from lineage 1.3.32 (three mice per group) that were hydrodynamically injected with either pcDNA3.1-TIMP-1 or an empty vector and then 2 days later received 1 × 107 HBsAg-specific CTLs. Mice were bled, sacrificed, and perfused; their livers were harvested on day 7 after CTL transfer. The results were also compared with two additional groups of transgenic littermates (three mice per group) that were hydrodynamically injected with pcDNA3.1-TIMP-1 or NaCl alone and sacrificed at the same time.
As shown in Figure 8, when compared with mice injected either with NaCl or pcDNA3.1-TIMP-1 alone in which HBV replication was not inhibited, the enhanced expression of TIMP-1 did not affect the CTL-induced inhibition of hepatic HBV replication that was also detected in control mice that received the empty vector plus CTLs. This is not surprising, since similar number of CTLs and similar transcript levels for IFN-γ and other cytokines (i.e., TNF-α) with known antiviral activity against HBV (3, 4) were detected in the livers of groups of animals that were similarly treated and sacrificed at day 1 after CTL transfer (Figure 7). Importantly, the enhanced expression of TIMP-1 reduced the severity of the CTL-induced liver disease (monitored as sALT activity) at the indicated time points after CTL injection (Figure 8, bottom). These results indicate that the enhanced expression of TIMP-1 does not affect the potential of HBV-specific CTLs to induce a sustained inhibition of viral replication in the face of decreased liver damage.
Figure 8.
Enhanced expression of TIMP-1 does not affect the antiviral potential of HBV-specific CTLs. Age- and serum HBeAg–matched female transgenic mice (three mice per group) were hydrodynamically injected (hydro) with either pcDNA3.1-TIMP-1 (TIMP-1 + CTL, black bars) or an empty vector (Vector + CTL, white bars), and 2 days later they received 1 × 107 HBsAg-specific CTLs. Mice were bled at the indicated time points, then sacrificed at day 7 after CTL transfer. Total hepatic DNA was then analyzed for HBV replication by Southern blot analysis. Bands corresponding to the integrated transgene (Trans.), relaxed-circular (RC), and single-stranded (SS) linear HBV DNA replicative forms are indicated. The integrated transgene can be used to normalize the amount of DNA bound to the membrane. The mean sALT activity (± SD), measured at the indicated time points, is indicated for each group and is expressed in units/liter. These results were also compared with two additional groups of transgenic littermates that were hydrodynamically injected with pcDNA3.1-TIMP-1 alone (TIMP-1, dark gray bars) or injected with saline (NaCl, light gray bars).
Discussion
In this study we showed that following the passive transfer of HBV-specific CTLs into transgenic mice that express and replicate HBV at high levels in their hepatocytes (2), various MMPs were rapidly induced in the liver. Interestingly, MMP-8 and MMP-9, which are known to be produced by PMNs (14), were not induced in mice treated with α–Gr-1, whereas MMP-2, MMP-3, MMP-7, MMP-12, MMP-13, and MMP-14, which are known to be produced by many other myeloid and nonmyeloid cell types (14), were induced, suggesting that Gr-1+ cells, especially PMNs, are the probable source of MMP-8 and MMP-9 in our system. Since depletion of Gr-1+ cells also inhibits CTL-induced recruitment of antigen-nonspecific inflammatory cells into the liver (8), the tight association between MMP-8 and MMP-9 activities and IHL recruitment is compatible with the hypothesis that these enzymes facilitate leukocyte trafficking through the endothelial barrier and entry into the liver parenchyma.
To test this hypothesis, we inhibited MMP activity in vivo and monitored whether this altered the intrahepatic recruitment and pathogenetic or antiviral effector functions of HBV-specific CTLs and other inflammatory cells in our system. Inhibition of MMP activity was achieved by enhancing hepatic TIMP-1 expression via the hydrodynamic injection (23) of a plasmid encoding murine TIMP-1 (pcDNA3.1-TIMP-1), which transduces hepatocytes in vivo, thereby increasing the concentration of TIMP-1 in the serum and, especially, in the extracellular compartment of the liver. Finally, TIMP-1 is a specific natural MMP inhibitor, in contrast to synthetic MMP inhibitors that have been previously used in cancer studies (29) and other models of liver disease (18), and that are known to interfere with the activity of many proteases other than MMPs (14).
The enhanced expression of TIMP-1 inhibited MMP function, including that of MMP-8 and MMP-9, which are associated with IHL recruitment and that are known to be produced by PMNs (14). The notion that PMNs were recruited in the livers of mice overexpressing TIMP-1 suggests that, rather than PMN recruitment, the TIMP-1–mediated repression of MMP function inhibited PMN-derived MMP activity.
Importantly, the enhanced expression of TIMP-1 reduced the CTL-induced recruitment of host-derived, antigen-nonspecific mononuclear cells into the liver and the attending liver disease, indicating that the recruitment of these cells requires MMP activity. This result, combined with the observation that the messages for many chemoattractants for mononuclear cells were strongly induced in the livers of mice overexpressing TIMP-1, suggests that chemokines alone are not sufficient to recruit those cells into the liver. This concept is also supported by previous work showing that blocking CXCL9 and CXCL10 reduces the intrahepatic recruitment of mononuclear cells (7), and depletion of Gr-1+ cells profoundly inhibits mononuclear cell recruitment in face of high intrahepatic levels of CXCL9 and CXCL10 and other chemokine RNAs (8).
It is also worth mentioning that MMP-8 and MMP-9 have the potential to cleave chemoattractants for mononuclear (such as CXCL9 and CXCL10) (15) and polymorphonuclear (such as LIX) (16) cells. This enzymatic activity is thought to decrease the biological function of CXCL9 and CXCL10 and slightly increase that of LIX. Although at first sight our results (i.e., that inhibition of MMP activity was associated with decreased recruitment of mononuclear cells and normal recruitment of PMNs) do not support this notion, future studies aimed at defining the role of MMPs in the processing of chemokines are certainly warranted in our system.
All mononuclear cell subsets measured were similarly affected, so that a two- to threefold reduction in the number of recruited T cells, NK cells, NKT cells, and macrophages was observed in mice overexpressing TIMP-1 when compared with controls. The observation that the TIMP-1–induced inhibition of mononuclear cell recruitment was significant but not complete suggests that other factors in addition to MMPs participate in the recruitment process. The association of reduced liver disease with reduced recruitment of antigen-nonspecific mononuclear cells confirms previous studies that showed that the intrahepatic accumulation of large numbers of these cells can amplify the liver damage initiated by the antigen-specific CTLs (7, 8). Similar mechanisms may contribute to the pathogenesis of viral hepatitis in humans, where, as in our system, the number of HBV-specific T cells detected in the liver is outnumbered by recruited non-virus-specific T cells (30, 31) and other inflammatory cells (32). The pathogenetic mechanisms whereby antigen-nonspecific mononuclear cells may induce liver damage are not understood. Future studies will attempt to address this important issue.
The results herein described also showed that inhibition of MMP activity does not affect the capacity of HBV-specific CTLs to abolish HBV replication. The notion that inhibition of MMP activity is associated with maintenance of antiviral effects but diminished tissue damage may be important for the development of immunotherapeutic approaches for the treatment of chronic HBV infection.
In conclusion, we found that MMPs are dispensable for the migratory and antiviral activity of virus-specific CTLs, but they are necessary for the intrahepatic recruitment of antigen-nonspecific cells that amplify the CTL-initiated liver damage. The data are compatible with the hypothesis that PMNs represent the first cell type to be recruited into the liver following antigen recognition by the CTLs. According to this hypothesis, the production of MMPs by PMNs, and perhaps by other cells, may cleave components of the ECM and/or perform as yet undefined functions that facilitate the recruitment of mononuclear cells into the liver parenchyma in response to their own chemoattractants. It is possible that similar events may play a role in the pathogenesis of other immune-mediated diseases that involve solid organs.
Methods
Mice.
HBV transgenic mouse lineage 1.3.32 used in this study has been previously described (2). Lineage 1.3.32 (inbred C57BL/6, H-2b) was bred one generation against B10D2 mice (H-2d) to produce H-2bxd F1 hybrids prior to injection of H-2d-restricted HBsAg-specific CTL lines. In all experiments, the mice were matched for age (8 weeks), sex (female), and HBeAg levels in their serum before experimental manipulations. All animals were housed in pathogen-free rooms under strict barrier conditions. These studies were approved by the Animal Research Committee of The Scripps Research Institute.
Injection of HBsAg-specific CTL lines.
HBsAg-specific CTL lines were derived from spleen cells of nontransgenic B10.D2 male mice immunized as previously described (33). Briefly, 8-week-old B10.D2 male mice received a single intramuscular injection of a plasmid that expresses the middle and major ENV proteins (preS2/S) of HBV (33). After 2 weeks, the animals were boosted with a single intravenous injection of vaccinia virus encoding preS2/S (33), and 3 weeks later they were sacrificed and spleen cells collected. The CTL lines were stimulated weekly as described (33). After 2 weeks of in vitro stimulation, the cells were tested for antigen specificity by flow cytometry using a recombinant soluble dimeric H-2 Ld:Ig Fusion Protein (BD Biosciences Pharmingen, San Diego, California, USA) that was incubated with the immunodominant peptide epitope (Env 28-39) of HBsAg (34), according to the manufacturer’s instructions. Over 95% of cells were CD8+ and specific for Env 28-39. At this time, the cells were washed, counted, suspended in HBSS containing 2% FCS, and injected intravenously (1 × 107 cells/mouse) into transgenic female mice from lineage 1.3.32. Groups of mice (three mice per group) were sacrificed on day 1 after CTL injection, and their livers were perfused and harvested for histological and flow cytometry analyses, or they were snap frozen in liquid nitrogen and stored at –80°C for subsequent molecular analyses (see later).
α–Gr-1.
Rat IgG2b mAb’s specific for mouse Ly-6G (Gr-1) (clone RB6-8C5) and rat IgG2b control Ab’s (clone A95-1) (BD Biosciences Pharmingen) were used as described (8). Mice were injected intraperitoneally twice with α–Gr-1 or control Ab’s (100 μg/mouse), initially 16 hours before and then simultaneously with the intravenous injection of the CTLs, and they were sacrificed on day 1 after CTL injection.
Tissue DNA and RNA analyses.
Total liver DNA was isolated from frozen livers (left lobe) and extracted as previously described (2). Thirty micrograms of DNA were digested with HindIII and analyzed for TIMP-1 and HBV DNA by Southern blot. Total liver RNA isolated from the same lobe was analyzed for HBV RNA by Northern blot and for cytokine, chemokine, macrophage markers, and for MMP and TIMP-1 mRNAs by RPA, exactly as previously described (2, 3, 7, 35). The MMP-8 (neutrophil collagenase) specific transcript was analyzed by RPA using a 322-nucleotide probe obtained by amplification of a cDNA fragment spanning nucleotides 827–1,148 of the murine Mmp8 gene (GenBank accession number NM_008611). The probe was validated by sequencing and cloned into pGEM-T Easy vector (Promega Corp., Madison, Wisconsin, USA) as described (35). The relative abundance of specific DNA and RNA molecules was determined by phosphor imaging analysis, using the Optiquant image analysis software (Packard Instrument Co., Meriden, Connecticut, USA).
Biochemical and histological analyses.
The extent of hepatocellular injury was monitored by measuring sALT activity at multiple time points after treatment. sALT activity was measured in a Paramax chemical analyzer (Baxter International Inc., McGaw Park, Illinois, USA) exactly as previously described (3).
Gelatin PAGE-Zymography.
Gelatinolytic protease activity was determined as previously described (36). Briefly, frozen livers were homogenized in lysis buffer (50 mmol/liter Tris-HCl pH 7.5, 150 mmol/liter NaCl, 100 mmol/liter L-arginine, 1.0% Triton X-100, and 0.005% Brij 35). The homogenate was centrifuged at 4°C for 20 minutes at 10,600 g. Supernatants were divided into aliquots and stored at –80°C until analyzed. The protein concentration of each sample was measured with a BCA protein assay kit (Pierce Chemical Co., Rockford, Illinois, USA) using a bovine gamma globulin standard. Total liver homogenate (10 μg) was mixed with an equal volume of sample buffer (80 mmol/liter Tris-HCl pH 6.8, 4% SDS, 10% glycerol, 0.01% Bromphenol Blue) and subjected to electrophoresis at 4°C on an 8% SDS-polyacrylamide resolving gel containing 1 mg/ml gelatin. After electrophoresis, gels were washed three times with 2.5% Triton X-100 solution to extract SDS from the gels, allowing gelatinases to renature within the gel. The gels were then incubated in reaction buffer (50 mmol/liter Tris-HCl, pH 7.5, containing 10 mmol/liter CaCl2 and 0.02% NaN3) at 37°C for 42 hours. After incubation, gels were washed in water and stained with Coomassie Brilliant Blue R-250 for 60 minutes. The gelatinolytic activity was detected as clear bands on a dark blue background, and molecular weight markers as well as standards of recombinant MMPs (Chemicon International, Temecula, California, USA) were included in the gels for identification. Quantitative analysis of activity was determined using a Personal Densitometer SI (Molecular Dynamics, Sunnyvale, California, USA).
In situ zymography.
In situ gelatin zymography was performed as described by Oh and coworkers (37). In situ collagen type I zymography was adapted from the same protocol (37). Briefly, cryostat sections (8 μm) of livers were overlaid with assay solution (50 mmol/liter Tris-HCl, 5 mmol/liter CaCl2, 0.2 mmol/liter NaN3, pH 7.5) containing either 40 μg/ml FITC-labeled DQ gelatin or 40 g/ml FITC-labeled DQ collagen type I (Molecular Probes Inc., Eugene, Oregon, USA). Sections were incubated at 37°C for 18 hours and examined by fluorescence microscopy. Endogenous gelatinolytic proteases or collagen type I proteases cleaved quenched DQ gelatin or DQ collagen type I, respectively, resulting in fluorescent breakdown products that allowed the localization of either gelatinase or collagenase type I activity.
Isolation and analysis of IHLs.
Mouse livers were weighed at the time of autopsy. Single-cell suspensions were prepared from two liver lobes of known weight, and analysis of the IHL population was performed by flow cytometry, exactly as described (7). The cells were surface-stained with FITC-labeled, phycoerythrin-labeled (PE-labeled), or allophycocyanin-labeled (APC-labeled) anti-CD4, anti-CD8, anti-CD3, anti-TCR, anti-NK1.1, α–Gr-1, anti-CD11b, and anti-CD11c Ab’s (BD PharMingen) for the detection of CD4+/TCR+ (mostly Th) cells, CD8+/TCR+ cells (mostly CTLs), NK1.1+/CD3– cells (NK cells), NK1.1+/CD3+ cells (NKT cells), Gr-1–/CD11b+/CD11c– (mostly macrophages), and Gr-1+/CD11b+/CD11c– cells (mostly PMNs). Samples were acquired on a FACSCalibur flow cytometer, and the data were analyzed using CELLQuest software (Becton-Dickinson Immunocytometry Systems, San Jose, California, USA).
Quantitative analyses of the intrahepatic content of the transferred CTLs.
Male CTLs were injected into female transgenic mice, and the animals were sacrificed 1 day later. Total liver DNA was extracted as previously described (2), and mouse Y chromosome sequences contained within the Sry gene were quantitated by real-time PCR upon normalization with an internal control (cholecystokinin type A receptor, CCKAR), exactly as described (8). The average copy number of Sry-specific amplicons in each group of mice (six mice per group) was calculated on the basis of 10,000 liver cell genomes (Figure 7, top).
Hydrodynamic injection of plasmids encoding TIMP-1.
A plasmid encoding murine TIMP-1 under the control of a CMV promoter (pcDNA3.1-TIMP-1) was generously provided by B.T. Rouse (The University of Tennessee, Knoxville, Tennessee, USA) (21). An empty vector pcDNA3.1 was used as a negative control. A total of 15 μg of pcDNA3.1-TIMP-1 were injected into the tail vein of groups of 8-week-old lineage 1.3.32 female mice in a volume of NaCl equivalent to 8.5% of the body mass of the mouse. The total volume was delivered within 5–8 seconds as previously described (23).
TIMP-1 ELISA.
Quantitation of serum TIMP-1 content was determined by a commercially available ELISA assay according to the manufacturer’s instructions (DuoSet IC murine TIMP-1 ELISA; R&D Systems Inc., Minneapolis, Minnesota, USA).
MPO assay.
Liver MPO activity is specific for PMNs and therefore quantifies their intrahepatic recruitment (24). Liver MPO content was determined as previously described (38). Briefly, frozen liver tissue was homogenized in 2 ml of buffer A (3.4 mmol/liter KH2HPO4, 16 mmol/liter Na2HPO4, pH 7.4). After centrifugation for 20 minutes at 10,000 g, the pellet was resuspended in 10 volumes of buffer B (43.2 mmol/liter KH2HPO4, 6.5 mmol/liter Na2HPO4, 10 mmol/liter EDTA, 0.5% hexadecyltrimethylammonium, pH 6.0) and sonicated for 10 seconds. After heating for 2 hours at 60°C, the supernatant was reacted with 3,3’,3,5′-tetramethylbenzidine (Sigma-Aldrich, St. Louis, Missouri, USA), and the optical density was determined at 655 nm.
Acknowledgments
We are grateful to Barry Rouse for providing the pcDNA3.1-TIMP-1 plasmid. We also thank Heike Mendez, Sadie Medrano, and Margie Chadwell for excellent technical assistance, and Gregory Del Zoppo for helpful discussions. This work was supported by grants NS36979 to I.L. Campbell, CA40489 to F.V. Chisari, and AI40696 to L.G. Guidotti from the NIH. This is manuscript number 16131-MEM from the Scripps Research Institute.
Footnotes
Giovanni Sitia and Masanori Isogawa contributed equally to this work.
Nonstandard abbreviations used: allophycocyanin (APC); anti–Gr-1 antibodies (α–Gr-1); cholecystokinin type A receptor (CCKAR); hepatitis B e antigen (HBeAg); hepatitis B surface antigen (HBsAg); hepatitis B virus (HBV); intrahepatic leukocyte (IHL); keratinocyte-derived chemokine (KC); lipopolysaccharide-induced chemokine (LIX); macrophage inflammatory protein (MIP); myeloperoxidase (MPO); phycoerythrin (PE); polymorphonuclear neutrophils (PMNs); RNase protection assay (RPA); serum alanine aminotransferase (sALT); T cell receptor (TCR); tissue inhibitor of matrix metalloproteinases (TIMP).
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Chisari FV, Ferrari C. Hepatitis B virus immunopathogenesis. Annu. Rev. Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 2.Guidotti LG, Matzke B, Schaller H, Chisari FV. High level hepatitis B virus replication in transgenic mice. J. Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guidotti LG, et al. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity. 1996;4:25–36. doi: 10.1016/s1074-7613(00)80295-2. [DOI] [PubMed] [Google Scholar]
- 4.McClary H, Koch R, Chisari FV, Guidotti LG. Relative sensitivity of hepatitis B virus and other hepatotropic viruses to the antiviral effects of cytokines. J. Virol. 2000;74:2255–2264. doi: 10.1128/jvi.74.5.2255-2264.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guidotti LG, Chisari FV. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 2001;19:65–91. doi: 10.1146/annurev.immunol.19.1.65. [DOI] [PubMed] [Google Scholar]
- 6.Ando K, et al. Class I restricted cytotoxic T lymphocytes are directly cytopathic for their target cells in vivo. J. Immunol. 1994;152:3245–3253. [PubMed] [Google Scholar]
- 7.Kakimi K, et al. Blocking chemokine responsive to γ-2/interferon (IFN)-γ inducible protein and monokine induced by IFN-γ activity in vivo reduces the pathogenetic but not the antiviral potential of hepatitis B virus-specific cytotoxic T lymphocytes. J. Exp. Med. 2001;194:1755–1766. doi: 10.1084/jem.194.12.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sitia G, et al. Depletion of neutrophils blocks the recruitment of antigen-nonspecific cells into the liver without affecting the antiviral activity of hepatitis B virus-specific cytotoxic T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 2002;99:13717–13722. doi: 10.1073/pnas.172521999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagasse E, Weissman IL. Flow cytometric identification of murine neutrophils and monocytes. J. Immunol. Methods. 1996;197:139–150. doi: 10.1016/0022-1759(96)00138-x. [DOI] [PubMed] [Google Scholar]
- 10.Nakano H, Yanagita M, Gunn MD. CD11c(+)B220(+)Gr-1(+) cells in mouse lymph nodes and spleen display characteristics of plasmacytoid dendritic cells. J. Exp. Med. 2001;194:1171–1178. doi: 10.1084/jem.194.8.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalod M, et al. Dendritic cell responses to early murine cytomegalovirus infection: subset functional specialization and differential regulation by interferon α/β. J. Exp. Med. 2003;197:885–898. doi: 10.1084/jem.20021522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bronte V, et al. Identification of a CD11b(+)/Gr-1(+)/CD31(+) myeloid progenitor capable of activating or suppressing CD8(+) T cells. Blood. 2000;96:3838–3846. [PMC free article] [PubMed] [Google Scholar]
- 13.Mordue DG, Sibley LD. A novel population of Gr-1+-activated macrophages induced during acute toxoplasmosis. J. Leukoc. Biol. 2003;74:1015–1025. doi: 10.1189/jlb.0403164. [DOI] [PubMed] [Google Scholar]
- 14.Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu. Rev. Cell Dev. Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van den Steen PE, Husson SJ, Proost P, Van Damme J, Opdenakker G. Carboxyterminal cleavage of the chemokines MIG and IP-10 by gelatinase B and neutrophil collagenase. Biochem. Biophys. Res. Commun. 2003;310:889–896. doi: 10.1016/j.bbrc.2003.09.098. [DOI] [PubMed] [Google Scholar]
- 16.Van Den Steen PE, et al. Gelatinase B/MMP-9 and neutrophil collagenase/MMP-8 process the chemokines human GCP-2/CXCL6, ENA-78/CXCL5 and mouse GCP-2/LIX and modulate their physiological activities. Eur. J. Biochem. 2003;270:3739–3749. doi: 10.1046/j.1432-1033.2003.03760.x. [DOI] [PubMed] [Google Scholar]
- 17.Opdenakker G, et al. Gelatinase B functions as regulator and effector in leukocyte biology. J. Leukoc. Biol. 2001;69:851–859. [PubMed] [Google Scholar]
- 18.Wielockx B, et al. Inhibition of matrix metalloproteinases blocks lethal hepatitis and apoptosis induced by tumor necrosis factor and allows safe antitumor therapy. Nat. Med. 2001;7:1202–1208. doi: 10.1038/nm1101-1202. [DOI] [PubMed] [Google Scholar]
- 19.Cataldo DD, et al. Matrix metalloproteinase-9 deficiency impairs cellular infiltration and bronchial hyperresponsiveness during allergen-induced airway inflammation. Am. J. Pathol. 2002;161:491–498. doi: 10.1016/S0002-9440(10)64205-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itoh T, et al. Experimental metastasis is suppressed in MMP-9-deficient mice. Clin. Exp. Metastasis. 1999;17:177–181. doi: 10.1023/a:1006603723759. [DOI] [PubMed] [Google Scholar]
- 21.Lee S, Zheng M, Kim B, Rouse BT. Role of matrix metalloproteinase-9 in angiogenesis caused by ocular infection with herpes simplex virus. J. Clin. Invest. 2002;110:1105–1111. doi:10.1172/JCI200215755. doi: 10.1172/JCI15755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker AH, Edwards DR, Murphy G. Metalloproteinase inhibitors: biological actions and therapeutic opportunities. J. Cell Sci. 2002;115:3719–3727. doi: 10.1242/jcs.00063. [DOI] [PubMed] [Google Scholar]
- 23.Yang PL, Althage A, Chung J, Chisari FV. Hydrodynamic injection of viral DNA: a mouse model of acute hepatitis B virus infection. Proc. Natl. Acad. Sci. U. S. A. 2002;99:13825–13830. doi: 10.1073/pnas.202398599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 25.Guidotti LG, et al. Viral cross talk: intracellular inactivation of the hepatitis B virus during an unrelated viral infection of the liver. Proc. Natl. Acad. Sci. U. S. A. 1996;93:4589–4594. doi: 10.1073/pnas.93.10.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hume DA, et al. The mononuclear phagocyte system revisited. J. Leukoc. Biol. 2002;72:621–627. [PubMed] [Google Scholar]
- 27.Rossi D, Zlotnik A. The biology of chemokines and their receptors. Annu. Rev. Immunol. 2000;18:217–242. doi: 10.1146/annurev.immunol.18.1.217. [DOI] [PubMed] [Google Scholar]
- 28.Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 29.Stamenkovic I. Matrix metalloproteinases in tumor invasion and metastasis. Semin. Cancer Biol. 2000;10:415–433. doi: 10.1006/scbi.2000.0379. [DOI] [PubMed] [Google Scholar]
- 30.Maini MK, et al. The role of virus-specific CD8(+) cells in liver damage and viral control during persistent hepatitis B virus infection. J. Exp. Med. 2000;191:1269–1280. doi: 10.1084/jem.191.8.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertoletti A, Maini MK. Protection or damage: a dual role for the virus-specific cytotoxic T lymphocyte response in hepatitis B and C infection? Curr. Opin. Immunol. 2000;12:403–408. doi: 10.1016/s0952-7915(00)00108-4. [DOI] [PubMed] [Google Scholar]
- 32.Ishak KG. Light microscopic morphology of viral hepatitis. Am. J. Clin. Pathol. 1976;65:787–827. [PubMed] [Google Scholar]
- 33.Kakimi K, Isogawa M, Chung J, Sette A, Chisari FV. Immunogenicity and tolerogenicity of hepatitis B virus structural and nonstructural proteins: implications for immunotherapy of persistent viral infections. J. Virol. 2002;76:8609–8620. doi: 10.1128/JVI.76.17.8609-8620.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ando K, et al. Mechanisms of class I restricted immunopathology. A transgenic mouse model of fulminant hepatitis. J. Exp. Med. 1993;178:1541–1554. doi: 10.1084/jem.178.5.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pagenstecher A, Stalder AK, Campbell IL. RNAse protection assays for the simultaneous and semiquantitative analysis of multiple murine matrix metalloproteinase (MMP) and MMP inhibitor mRNAs. J. Immunol. Methods. 1997;206:1–9. doi: 10.1016/s0022-1759(97)00077-x. [DOI] [PubMed] [Google Scholar]
- 36.Heo JH, et al. Matrix metalloproteinases increase very early during experimental focal cerebral ischemia. J. Cereb. Blood Flow Metab. 1999;19:624–633. doi: 10.1097/00004647-199906000-00005. [DOI] [PubMed] [Google Scholar]
- 37.Oh LY, et al. Matrix metalloproteinase-9/gelatinase B is required for process outgrowth by oligodendrocytes. J. Neurosci. 1999;19:8464–8475. doi: 10.1523/JNEUROSCI.19-19-08464.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kato A, Yoshidome H, Edwards MJ, Lentsch AB. Regulation of liver inflammatory injury by signal transducer and activator of transcription-6. Am. J. Pathol. 2000;157:297–302. doi: 10.1016/S0002-9440(10)64540-3. [DOI] [PMC free article] [PubMed] [Google Scholar]