Abstract
Lipogenesis is regulated by sterols and by insulin through the regulated expression and activation of the sterol regulatory element–binding proteins (SREBPs). A new study shows one way in which sterol and insulin regulation can be decoupled. In transgenic mice overexpressing a protein that regulates SREBP activation, lipogenesis is more sensitive to cholesterol and less sensitive to insulin.
Animal cells, like nations, have a domestic and a foreign policy. “Domestic policy” involves local control of metabolism, responses to energy charge, redox status, or the concentration of key allosteric regulators (e.g., malonyl-CoA and fructose 2,6-bisphosphate). The world of a multicellular organism imposes a “foreign policy” on its cells. They must respond to global signals in the interest of the whole organism.
Global and local signals invariably converge (as former Congressman Tip O’Neil famously quipped, “All politics is local.”). Lipogenesis is a case in point. Feeding, fasting, and refeeding cause large changes in the rates of fatty acid and cholesterol synthesis. These changes are evoked by both the carbohydrate and the lipid components of the diet. We now have a molecular window into some of the key molecules that integrate nutrient-derived signals.
A key breakthrough was the simultaneous discovery by Tontonoz et al. (1) and Yokoyama et al. (2) of the sterol regulatory element–binding protein/adipocyte differentiation factor-1 (SREBP/ADD1). The Brown and Goldstein laboratory went on to show that in the liver, two isoforms, SREBP-1c and SREBP-2, are master transcriptional regulators of the fatty acid and cholesterol synthesis pathways, respectively (3). The SREBPs reside as inactive precursors in the ER membrane. Upon vesicular transport to the Golgi, they are proteolytically cleaved to liberate a fragment that enters the nucleus and activates SRE-containing genes. The transport step is blocked by sterols. This defines the ER membrane as a locus wherein proteins sense lipids and act as the gatekeepers of SREBPs.
What are the protein gatekeepers? Through somatic cell genetic studies of mutant cell lines with defective feedback regulation of sterol synthesis, Brown and Goldstein discovered an SREBP escort protein, SREBP cleavage–activating protein (SCAP). SCAP has a sterol-sensing domain that is necessary for sterols to inhibit SREBP transport to the Golgi (4). Without SCAP, cells require exogenous cholesterol to survive.
Gene dosage studies of SCAP in hamster cells suggested that an additional factor could be titrated to abolish the sterol responsiveness of SREBP processing (5). Coimmunoprecipitation studies led to the discovery of a SCAP-interacting protein, Insig-1 (6). A homologous gene, Insig-2, was also identified (7). Careful gene dosage studies of Insig-1 showed that only within a narrow window of the Insig-1/SCAP stoichiometry does the system respond to sterol regulation. When SCAP is in molar excess relative to Insig-1, SREBP processing is insensitive to sterols. With a large excess of Insig-1, SREBP is retained in the ER, even in the absence of sterols. This has led researchers to ask whether these titration studies extend to the liver of an intact animal.
Insig-1 overexpression suppresses lipogenesis in vivo
In this issue of the JCI, Engelking and co-workers show that hepatic overexpression of Insig-1 in transgenic mice caused a marked reduction in nuclear SREBPs, which further decreased when mice were fed a high-cholesterol diet (8). Thus, the in vitro titration experiments correctly predicted the effect of Insig-1 on the lipogenesis rate in vivo. The in vivo effects of cholesterol on expression of SREBP are complicated by the fact that sterols activate SREBP-1c (not SREBP-2) gene expression through the liver X receptor transcription factors (9). Thus, at low levels, dietary cholesterol decreases the level of mature SREBP-1c protein, but at higher levels, it induces SREBP-1c gene expression (8).
Insig-1 overexpression reduces the sensitivity of lipogenesis to insulin
The first link between SREBP and insulin came from the discoveries that insulin increases the expression of SREBP-1c (10, 11), that transgenic mice overexpressing SREBP-1c do not suppress lipogenesis in response to fasting (12), and that lipogenesis is not induced in mice lacking SREBP-1c when they are fed a high-carbohydrate diet (13). Given that lipogenesis is regulated by insulin, inevitable questions arise: How do cells integrate regulation of lipogenesis by insulin with regulation by lipids? What molecules “sense” lipid and hormonal status? When they act together, which signals are dominant?
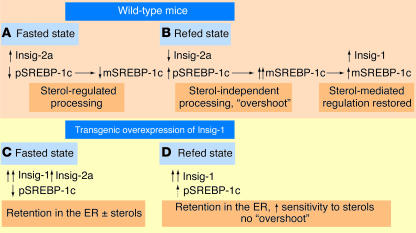
Normally, fasting causes a downregulation of SREBP-1c and thus a reduction in mature SREBP-1c. Refeeding leads to an induction of SREBP-1c and Insig-1 and greatly increased levels of mature SREBP-1c (Figure 1). Surprisingly, transgenic overexpression of Insig-1 blunted the insulin response; refeeding produced only a modest increase in SREBP-1c (8). In contrast, the sensitivity of SREBP-1c processing to cholesterol was enhanced.
Figure 1.
Integration of metabolic signals at the ER membrane. (A) During fasting, expression of the SREBP-1c precursor (pSREBP-1c) is reduced. Insig-2a is expressed and binds to SCAP, causing retention of a very low amount of pSREBP-1c in the ER in the presence of sterols. (B) Upon refeeding, insulin increases the expression of pSREBP-1c and decreases the expression of Insig-2a. The low abundance of both Insig-1 and Insig-2a leads to greatly increased processing of pSREBP-1c to mature SREBP-1c (mSREBP-1c), leading to a lipogenesis rate that is more than tenfold higher than that of the basal state, which represents an “overshoot.” The increased level of mSREBP-1c promotes expression of Insig-1. At a critical stoichiometry relative to SCAP, Insig-1 restores sterol-mediated regulation of pSREBP-1c processing. The lipogenesis rate then returns to the original basal level. (C) With transgenic overexpression of Insig-1, the cycling of lipogenesis between the fed and fasted states is dampened. The high ratio of Insig-1 to SCAP causes retention of pSREBP-1c in the ER. Fasted mice have a modest additional drop in lipogenesis due to the reduction of pSREBP-1c expression and increased Insig-2a levels. (D) With refeeding, the transgenic mice express extremely low levels of pSREBP-1c due to their inability to produce enough mSREBP-1c to drive the transcription of the pSREBP-1c gene. The persistently elevated level of Insig-1 abolishes the dramatic “overshoot” in lipogenesis seen in the wild-type refed mice but still mediates an enhanced sensitivity of pSREBP-1c processing to sterols.
Insulin cycling during the transition from feeding to fasting causes the liver to switch from glycolysis/lipogenesis to gluconeogenesis/ketogenesis. After refeeding, insulin suppresses the expression of insulin receptor substrate-2 (IRS-2) and phosphoenolpyruvate carboxykinase, resulting in decreased gluconeogenesis (14, 15). This pathway was intact in the Insig-1 transgenic mice (8).
Left undefined is the role of Insig-2a, a liver-specific isoform of Insig-2. Like Insig-1, Insig-2a causes retention of SREBP in the ER; however, it does so only in the presence of a concentration of sterols (7). Unlike Insig-1, Insig-2a is suppressed by insulin and induced by fasting. Early in the transition from fasting to feeding, Insig-1 levels are low and Insig-2a expression is repressed, allowing the insulin-induced SREBP-1c protein to be processed. Increased SREBP-1c induces Insig-1 expression and restores sterol sensing (Figure 1). Upon fasting, Insig-2a levels rise, suppressing lipogenesis in a sterol-dependent fashion. Insulin cycling might be critical for the prevention of excessive lipogenesis. It creates a brief window during refeeding when Insig levels begin to decline and there is insufficient Insig-1 to suppress lipogenesis. However, with induction of Insig-1, lipogenesis is held in check. Thus, a wild-type refed animal “overshoots” its rate of lipogenesis (7). In contrast, transgenic mice overexpressing Insig-1 effectively abolish the cycling of Insig-1 expression and therefore do not overshoot lipogenesis upon refeeding (Figure 1). This leads to a dramatic 80% reduction in plasma triglycerides in the refed mice (8).
Potential implications for insulin resistance
With chronic hyperinsulinemia, the normally converse relationship between lipogenesis and gluconeogenesis can be disrupted (16). This occurs in models of leptin deficiency (e.g., the leptinob mutation or congenital lipodystrophy) or leptin resistance. Deletion of IRS-2 leads to leptin resistance (17), suggesting a convergence between the leptin and insulin signaling pathways. Leptin resistance boosts lipogenesis in the liver through increased SREBP-1c. Deletion of SREBP-1c reduces the rate of lipogenesis of leptin-deficient animals but does not reverse insulin resistance, hence other aspects of leptin signaling influence insulin signaling (18).
The focus of the Insig story will likely turn to Insig-2a. Is Insig-2a affected by leptin deficiency or leptin resistance? What happens to Insig-2a under conditions of chronic hyperinsulinemia? We hope to learn why two closely related proteins are oppositely regulated by insulin.
Footnotes
See the related article beginning on page 1168.
Nonstandard abbreviations used: insulin receptor substrate-2 (IRS-2); SREBP cleavage–activating protein (SCAP); sterol regulatory element–binding protein/adipocyte differentiation factor-1 (SREBP/ADD1).
Conflict of interest: The author has declared that no conflict of interest exists.
References
- 1.Tontonoz P, Kim JB, Graves RA, Spiegelman BM. ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol. Cell. Biol. 1993;13:4753–4759. doi: 10.1128/mcb.13.8.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yokoyama C, et al. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
- 3.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 2002;109:1125–1131. doi:10.1172/JCI200215593. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hua X, Sakai J, Brown MS, Goldstein JL. Regulated cleavage of sterol regulatory element binding proteins requires sequences on both sides of the endoplasmic reticulum membrane. J. Biol. Chem. 1996;271:10379–10384. doi: 10.1074/jbc.271.17.10379. [DOI] [PubMed] [Google Scholar]
- 5.Yang T, Goldstein JL, Brown MS. Overexpression of membrane domain of SCAP prevents sterols from inhibiting SCAP.SREBP exit from endoplasmic reticulum. J. Biol. Chem. 2000;275:29881–29886. doi: 10.1074/jbc.M005439200. [DOI] [PubMed] [Google Scholar]
- 6.Yang T, et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 7.Yabe D, Brown MS, Goldstein JL. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12753–12758. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelking LJ, et al. Overexpression of Insig-1 in the livers of transgenic mice inhibits SREBP processing and reduces insulin-stimulated lipogenesis. J. Clin. Invest. 2004;113:1168–1175. doi:10.1172/JCI200420978. doi: 10.1172/JCI20978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Repa JJ, et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foretz M, et al. ADD1/SREBP-1c is required in the activation of hepatic lipogenic gene expression by glucose. Mol. Cell. Biol. 1999;19:3760–3768. doi: 10.1128/mcb.19.5.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimomura I, et al. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc. Natl. Acad. Sci. U. S. A. 1999;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimano H, et al. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J. Clin. Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang G, et al. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J. Biol. Chem. 2002;277:9520–9528. doi: 10.1074/jbc.M111421200. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, et al. Insulin inhibits transcription of IRS-2 gene in rat liver through an insulin response element (IRE) that resembles IREs of other insulin-repressed genes. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3756–3761. doi: 10.1073/pnas.071054598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puigserver P, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 16.Shimomura I, et al. Decreased IRS-2 and increased SREBP-1c lead to mixed insulin resistance and sensitivity in livers of lipodystrophic and ob/ob mice. Mol. Cell. 2000;6:77–86. [PubMed] [Google Scholar]
- 17.Tobe K, et al. Increased expression of the sterol regulatory element-binding protein-1 gene in insulin receptor substrate-2–/– mouse liver. J. Biol. Chem. 2001;276:38337–38340. doi: 10.1074/jbc.C100160200. [DOI] [PubMed] [Google Scholar]
- 18.Yahagi N, et al. Absence of sterol regulatory element-binding protein-1 (SREBP-1) ameliorates fatty livers but not obesity or insulin resistance in Lepob/Lepob mice. J. Biol. Chem. 2002;277:19353–19357. doi: 10.1074/jbc.M201584200. [DOI] [PubMed] [Google Scholar]