Abstract
Background
Transforming growth factor-βs (TGF-βs), bone morphogenetic proteins (BMPs) and activins are important regulators of developmental cell growth and differentiation. Signaling by these factors is mediated chiefly by the Smad family of latent transcription factors.
Results
There are a large number of uncharacterized cDNA clones that code for novel proteins with homology to known signaling molecules. We have identified a novel molecule from the HUGE database that is related to a previously known molecule, AMSH (associated molecule with the SH3 domain of STAM), an adapter shown to be involved in BMP signaling. Both of these molecules contain a coiled-coil domain located within the amino-terminus region and a JAB (Domain in Jun kinase activation domain binding protein and proteasomal subunits) domain at the carboxy-terminus. We show that this novel molecule, which we have designated AMSH-2, is widely expressed and its overexpression potentiates activation of TGF-β-dependent promoters. Coimmunoprecipitation studies indicated that Smad7 and Smad2, but not Smad3 or 4, interact with AMSH-2. We show that overexpression of AMSH-2 decreases the inhibitory effect of Smad7 on TGF-β signaling. Finally, we demonstrate that knocking down AMSH-2 expression by RNA interference decreases the activation of 3TP-lux reporter in response to TGF-β.
Conclusions
This report implicates AMSH and AMSH-2 as a novel family of molecules that positively regulate the TGF-β signaling pathway. Our results suggest that this effect could be partially explained by AMSH-2 mediated decrease of the action of Smad7 on TGF-β signaling pathway.
Keywords: Signal transduction, bioinformatics, serine/threonine kinase, phosphorylation
Background
TGF-β ligands belong to a large and expanding family of multifunctional cytokines which include the activins, inhibins, bone morphogenetic proteins (BMPs), growth/differentiation factors, Mullerian inhibiting substance and Nodal. Members of this family regulate a broad range of physiological processes, including organogenesis, proliferation, differentiation, adhesion, motility and apoptosis [1,2]. Mutations in downstream components of the TGF-β signaling pathways have been implicated in several human diseases [3].
TGF-β superfamily members signal through a receptor complex formed by the association of two transmembrane proteins designated I and II, each possessing serine/threonine kinase activity (for reviews see [1,4,5]). TGF-β binding induces the assembly of the type I and type II receptors [6,7], thereby enabling the type II receptor to phosphorylate and activate the type I receptor [8,9]. Activated type I receptors then phosphorylate Smad2 and Smad3 receptor-regulated Smad proteins (R-Smads) at the carboxy terminal motif SSxS [10,11]. Phosphorylated R-Smads interact with Smad 4, the unique Co-Smad [11-13]. This complex accumulates in the nucleus where it regulates transcriptional activity of several genes in association with transcriptional activators and corepressors (for reviews see [14-17]).
The specific transcriptional outcome in response to TGF-β is achieved by the interplay of intracellular regulators at different levels. Within the Smad family of proteins, Smad6 and 7 (I-Smads) negatively regulate TGF-β signaling [18-20]. Smad6 also inhibits BMP signaling, while activin signaling is negatively regulated by Smad7 [21]. In addition to the Smad proteins, other regulatory molecules work as adapter and/or scaffolding proteins, such as SARA, that mediates the access of Smads to activated receptors [22]. Finally, the specific outcome of TGF-β signaling is modulated by several other signaling pathways (for review see [15,23]).
The human unidentified gene-encoded large proteins database (HUGE) contains long cDNAs and is a valuable tool to discover new proteins, and to predict structural features with a possible functional relevance [24,25]. Through the analysis of the cDNA clones contained in this database, we have identified a novel protein that is highly similar to AMSH (associated molecule with the SH3 domain of STAM) [26]. Systematic analysis using specific reporters for several pathways showed that this novel molecule, designated AMSH-2, positively regulates TGF-β signaling system but not the TNF signaling pathway.
Results and discussion
Identification of a novel signaling molecule, AMSH-2
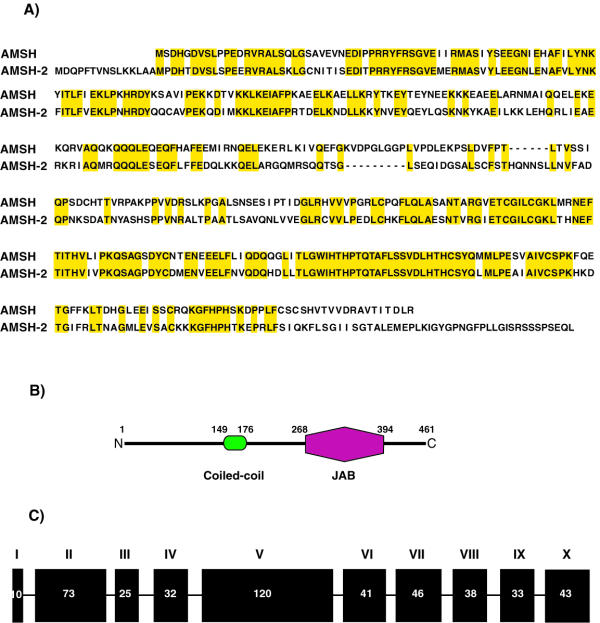
We have adopted a systematic strategy to identify and characterize cDNAs encoding novel proteins containing domains that have a role in signal transduction pathways [27-29]. We have used the HUGE database as a primary public resource as it contains mostly uncharacterized full-length cDNAs. By analyzing the domain composition of the predicted proteins, we identified a cDNA [Genbank AB037794] with an open reading frame of 461 amino acids (Figure 1A). The domain composition of the predicted protein was obtained by searching the SMART database [30] and revealed a coiled-coil domain spanning amino acids 149 to 176 in the N-terminus of the molecule and a JAB domain [31-33] (amino acids 268 to 394) located in the carboxy terminal region of the molecule (Figure 1B). A search of GenBank database revealed a similar molecule, AMSH, which had been identified on the basis of its ability to interact with STAM and showed a similar domain composition. Alignment of the novel protein against human AMSH revealed a 75% identity within the JAB domain, with an overall identity of 56% between the two molecules (Figure 1A). We have designated this novel protein AMSH-2 after its paralog AMSH. Upon searching the human genomic database, we found that AMSH-2 gene was located on 10q23.33. A detailed analysis of the genomic structure of the gene encoding for AMSH-2 revealed that the coding region spans 10 exons (Figure 1C). All the intron-exon boundaries followed the GT/AG rule for splice junctions [34].
Figure 1.
Sequence, domain architecture and genomic structure analysis of a novel protein AMSH-2. A) Alignment of AMSH family of proteins. Identical residues are shaded in yellow. B) The domain architecture of AMSH-2 was analyzed by SMART program. The coiled-coil region and the JAB domains are represented by green and red figures respectively. The numbers indicate the position of the domain within the protein sequence. C) Analysis of the genomic structure of the human gene encoding for AMSH-2. Only the coding exons are represented. Each exon is represented by a box with a length scaled to the size of the exon. The number of the amino acids encoded by each exon are indicated.
Expression of AMSH-2
In order to determine the tissue expression of AMSH-2, we probed a Northern blot containing poly A+ mRNA from several human tissues with a radiolabeled AMSH-2 probe. We found a single band of approximately 2.3 kb which was detected in almost all the tissues examined (Figure 2A, left panel). Probing a cancer cell line northern blot revealed a band of similar size in most of the cell lines, including HL60, Hela S3, Molt-4, Raji, SW480 and A549 (Figure 2A, right panel).
Figure 2.
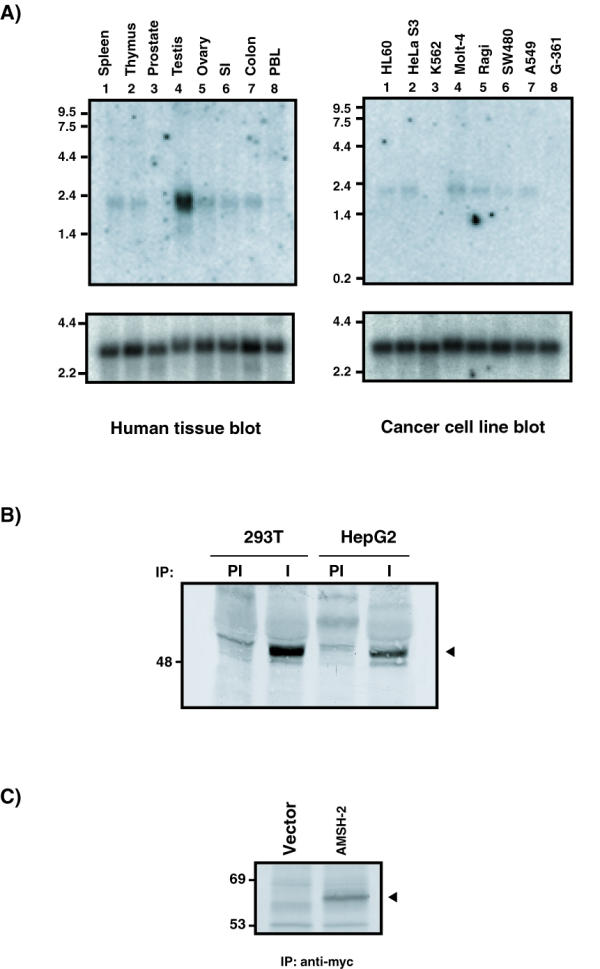
A) Northern blot analysis of AMSH-2 in different human tissues and cell lines. Each lane contains 2 μg of poly A+ mRNA. The top panels show the result of hybridization of the membranes with a specific probe for AMSH-2 mRNA. The lower panels show β-actin reprobing of the membranes. The sizes of the transcripts are indicated in kb on the left. B) Expression of AMSH-2 protein. Cell lysates from metabolically labeled 293T and HepG2 cells were immunoprecipitated with preimmune (PI) and AMSH-2 antiserum (I). A specific band for AMSH-2 can be observed in the lanes immunoprecipitated with AMSH-2 antiserum (indicated by an arrow). C) Expression of AMSH-2 Myc construct. 293T cells were transfected with the indicated constructs and metabolically labeled with S35. Cell lysates were immunoprecipitated with 9E10 monoclonal antibody and after SDS-polyacrylamide gel separation and blotting, the membrane was exposed. The arrow points a specific band corresponding to the Myc tagged AMSH-2 protein.
We corroborated the mRNA distribution of AMSH-2 by performing a BLAST search against the EST database. We found ESTs corresponding to AMSH-2 mRNA in brain, testes, bone, pancreas, fetal liver, kidney, colon, stomach, bone marrow, placenta, breast, pectoral muscle, hypothalamus, ovary as well as in a number of cell lines of different origins, including T cell leukemia, NT2 neuronal and B-cell CLL [35]. These data indicate that, like AMSH [26], AMSH-2 mRNA is widely expressed.
We raised an antibody against the carboxy terminus of AMSH-2 and measured the expression of AMSH-2 protein in different cell lines. For this purpose, HepG2 and 293T cells lines were metabolically labeled with 35S and cell lysates were immunoprecipitated with preimmune or anti-AMSH-2 antiserum. We observed a band with a molecular weight of approximately 56 kDa, which is in agreement with the predicted molecular weight of AMSH-2. This band was specific for AMSH-2 as it was absent in the samples immunoprecipitated with pre-immune serum (Figure 2B).
AMSH-2 enhances gene expression of TGF-β-dependent promoters
In order to study the functional role of AMSH-2, we constructed several epitope-tagged versions of AMSH-2. The entire open reading frame of AMSH-2 was subcloned by PCR into pEF mammalian expression vectors that provide a Myc or V5 epitopes at the C-terminus. Ectopic expression of AMSH-2/Myc was undertaken and confirmed by metabolic labeling of 293T cells, followed by immunoprecipitation with anti-Myc antibodies (Figure 2C).
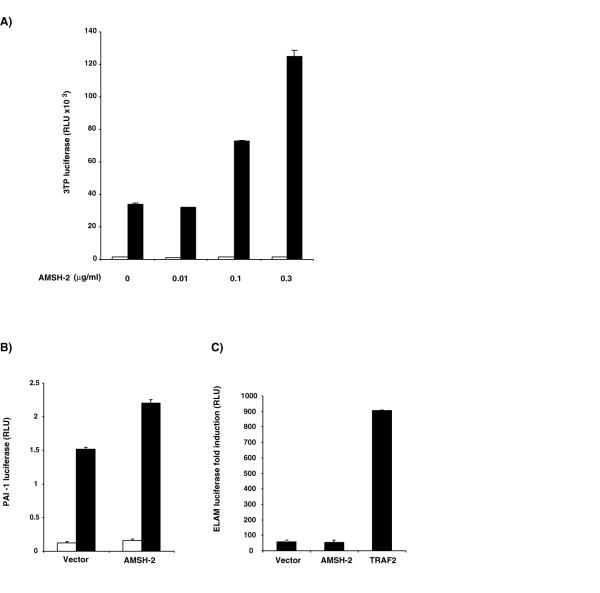
We next examined the effect of overexpression of AMSH-2 on a number of signaling pathways. Since AMSH-2 is expressed in HepG2 and 293T cells, we chose these cell lines for our assays. Given the similarity between AMSH-2 and AMSH, and the ability to AMSH to regulate BMP signaling, we hypothesized that AMSH-2 may affect signaling by TGF-β. We first tested the effect of AMSH-2 overexpression on TGF-β signaling pathway using p3TP-lux reporter plasmid which is a standard reporter to evaluate TGF-β signaling [6]. This reporter construct contains the luciferase gene under the control three consecutive TPA response elements and a 96 bp fragment of the PAI-1 promoter that is regulated by TGF-β and is also highly inducible in response to TGF-β. Overexpression of AMSH-2 in HepG2 cells increased the level of luciferase activity driven by the 3TP promoter upon TGF-β stimulation as compared to cells transfected with vector alone (Figure 3A). As this promoter is derived from the Plasminogen activator inhibitor-1 (PAI-1) promoter, we also tested the effect on PAI-1 luciferase activity [36]. When we cotransfected AMSH-2 into HepG2 cells together with this reporter, a similar increase in the luciferase levels was observed indicating that AMSH-2 is likely a positive regulator of TGF-β signaling (Figure 3B).
Figure 3.
A) Effect of AMSH-2 on TGF-β induced transcriptional activity of 3TP driven luciferase. HepG2 cells were cotransfected with 3TP-lux reporter together with the empty vector or increasing amounts of AMSH-2 Myc construct. Twenty-four hours posttransfection, cells were serum-starved and left untreated (white bars) or stimulated (black bars) with 5 ng/ml TGF-β1 for 16–20 h before the luciferase and β-galactosidase activities were measured. B) Effect of AMSH-2 on TGF-β induced transcriptional activity of PAI-1 luciferase. HepG2 cells were cotransfected with PAI-1 reporter together with the empty vector or 0.9 μg/ml of AMSH-2 Myc construct. A similar procedure as described in A was followed. C) AMSH-2 does not stimulate NF-kB signaling pathway. 293 cells were cotransfected with ELAM luciferase reporter construct together with an empty vector, 0.9 μg/ml of AMSH-2 Myc construct or 0.9 μg/ml of TRAF-2, used as a positive control. Forty-eight hours posttransfection, luciferase and β-galactosidase activities were measured.
To account for possible non-specific effects, we tested the effect of AMSH-2 overexpression on the TNF signaling pathway. We used an ELAM-luciferase reporter construct as a reporter for activation of this pathway. This reporter contains the ELAM-1 promoter (-730 to +52) and is well known to be induced by TNF stimulation [37]. We therefore cotransfected 293 cells with ELAM-luciferase and either AMSH-2, empty vector or TRAF2, a prototypical member of the TRAF family of adapter proteins known to potentiate TNF signaling pathway [38-40]. We did not observe any increase in the luciferase levels in AMSH-2 transfected cells as compared with the empty vector, although overexpression of TRAF2 led to a significant upregulation of the promoter (Figure 3C). Taken together, these results imply that the AMSH-2 regulates TGF-β signaling pathway.
AMSH-2 negates the inhibitory effect of Smad7 on TGF-β signaling
The ability of AMSH-2 to regulate TGF-β signaling could be explained by several mechanisms, including interaction with Smad2, Smad3, Smad4, or Smad7 [41]. To determine whether AMSH-2 interacts with these Smads, we coexpressed AMSH-2 with epitope tagged Smad constructs in 293T cells. As shown in Figure 4A, we observed an association between AMSH-2 and Smad7 but not with Smads 3 or 4 (Figure 4A). We also observed association between Smad2 and AMSH-2. These findings are similar to that observed for AMSH, since this molecule interacts strongly with Smad6 and 7; but differ in that AMSH does not with any R-Smads [41].
Figure 4.
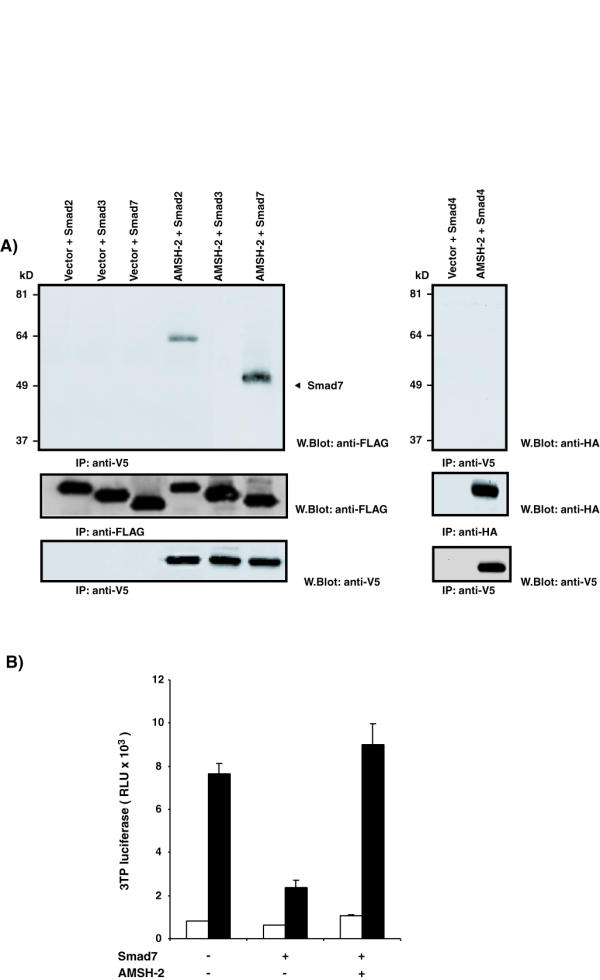
A) AMSH-2 interacts with Smad2 and Smad7, but not with Smad3 or Smad4. Vector or AMSH-2 V5 construct were coexpressed in 293T cells together with the Smad proteins as indicated on the top of the figure. Lysates were immunoprecipitated with anti-V5 antibody and western blotted with anti-Flag or anti-HA antibodies. To control for Smad and AMSH-2 protein expression part of the lysates were immunoprecipitated with anti-Flag, anti-HA or anti-V5 antibodies and western blotted with the same antibodies. B) Smad7 inhibitory effect on TGF-β signaling pathway is rescued by AMSH-2. HepG2 cells were cotransfected with 3TP-lux reporter together with the empty vector or 10 ng/ml of Smad7 with or without 0.3 μg/ml of AMSH-2 Myc construct. Twenty-four hours posttransfection, cells were serum-starved and left untreated (white bars) or stimulated (black bars) with 5 ng/ml TGF-β1 for 16–20 h before luciferase and β-galactosidase activities were measured.
Because AMSH-2 interacts with Smad7, we also analyzed the effect of AMSH-2 on the ability of Smad7 to inhibit TGF-β signaling in HepG2 cells. As expected, overexpression of Smad7 attenuated the induction of p3TP-luciferase activity by TGF-β (Figure 4B). Interestingly, overexpression of AMSH-2 negated the inhibitory effect of Smad7 on p3TP expression stimulated by TGF-β (Figure 4B). Therefore AMSH-2 can rescue the inhibitory effects of Smad7 on TGF-β signaling pathway. Given the sequence similarity between AMSH and AMSH-2, it is indeed possible that AMSH-2 also interacts with Smad6. Smad6 has been shown to inhibit the TGF-β pathway [42] and AMSH is able to rescue the inhibitory effect of Smad6 on BMP signaling pathway [41]. If AMSH-2 is able to interact with Smad6, the effect of AMSH-2 on TGF-β signaling pathway could be explained by its interaction with both Smad7 and Smad6.
AMSH-2 is a positive regulator of TGF-β-dependent transcriptional activation
The use of small interfering RNAs (siRNA) has been proven a powerful tool to suppress the expression of specific proteins in mammalian cells [43,44]. We have shown that overexpression of AMSH-2 positively regulates TGF-β-dependent transcriptional activation of 3TP-lux reporter. To assess the effect of depletion of endogenous AMSH-2, we designed a specific siRNA to silence AMSH-2. This siRNA was cotransfected in HepG2 cells and the effect on the transcriptional activity of the 3TP driven luciferase reporter was analyzed. As a positive control we used an siRNA targeted against Smad7. Cotransfection of siRNA specific for AMSH-2 cause a decrease in the luciferase activity upon stimulation with TGF-β when compared with cells cotransfected with a commercially available randomly ordered RNA scrambled RNA. As expected, we observed an increase in luciferase activity in the cells transfected with a siRNA specific for Smad7 (Figure 5). These knock down experiments confirm that AMSH-2 positively regulates TGF-β signaling pathway.
Figure 5.
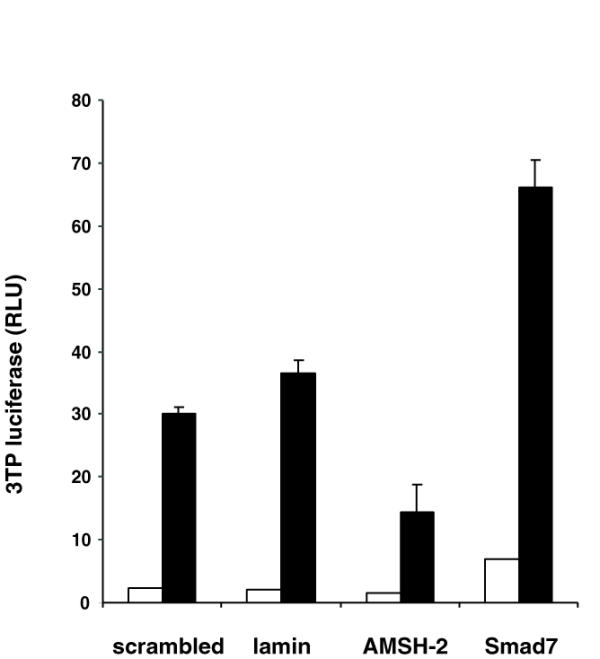
AMSH-2 RNA interference has a negative effect on TGF-β signaling pathway. HepG2 cells were transfected with 3TP-lux reporter and scramble, laminA/C, AMSH-2 or Smad7 siRNA duplexes as indicated in the bottom of the graph. Forty-eight hours posttransfection cells were serum-starved and left treated (black bars) or not (white bars) with 5 ng/ml TGF-β1.
Conclusions
Our results indicate that AMSH-2 is a positive regulator of TGF-β signaling. This effect might be mediated through the interaction with the inhibitory Smad7 as AMSH-2 is capable of negating the inhibitory effects of Smad7 in TGF-β signaling pathway. Further, use of siRNA to knock down the expression of AMSH-2 reduces signaling by TGF-β. Similar to our studies, it has been suggested that AMSH might be involved in TGF-β signaling. AMSH can interact with both I-Smads (i. e. Smads 6 and 7) and is a positive regulator of BMP signaling pathway [41]. The high degree of homology between these paralogs suggests that AMSH-2 might also interact with Smad6, and therefore might regulate BMP signaling. Although further research needs to be conducted to elucidate the exact mechanism of action of AMSH-2, it is clear that AMSH and AMSH-2 belong to a new family of molecules that positively regulate TGF-β signaling pathway.
Methods
cDNAs and constructs
A human cDNA clone [Genbank AB037794] was used as described [24,25]. To generate wild type AMSH-2 expression vector with a carboxy-terminal Myc epitope, we subcloned AMSH-2 open reading frame into the Nco I and Xho I sites of pEF/myc/cyto (Invitrogen, Carsbad, CA) by standard PCR procedures using the primers: AAAACCATGGATCAGCCTTTTACTGTGAATTC (5' primer) and AAACTCGAGCTGTTCAGATGGTGATGATGAC (3' primer). To generate AMSH-2 V5 carboxy-terminal-tagged expression vector, the open reading frame of AMSH-2 was subcloned into Nco I and Xho I sites of pEntr4 (Invitrogen) using the same 5' primer as described above and AAACTCGAGAGCTGTTCAGATGGTGATGATGACCTAG (3' primer), and transferred to pEF1/V5-HisB (Invitrogen), which has been previously made gateway compatible by inserting a cassette in the EcoRV site.
3TP-lux, PAI-1-luc, pELAMP-luc+, TRAF2, Flag-tagged Smad7, Smad3, have been previously described [6,20,36-38,45] while Flag-tagged Smad2 and HA-tagged Smad4 were generously provided by Joan Massague.
Northern blot analysis
Human tissue northern blot II and cancer cell line blot were obtained from CLONTECH (Palo Alto, CA). We isolated and radiolabeled an AMSH-2 cDNA (nucleotides 1088–2010) produced by digestion of AMSH-2 cDNA clone with HindIII and SphI. Hybridization signals were detected on a BAS-2000 bioimaging analyzer (Fuji film, Tokyo, Japan). After exposure, the blots were stripped and reprobed with beta-actin to control for equal amounts of PolyA+ RNA loading.
Cell culture and antibodies
293 and 293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mM L-glutamine and penicillin/streptomycin. HepG2 cells were grown in MEM supplemented with 10% fetal bovine serum, non essential amino-acids, sodium pyruvate and penicillin/streptomycin.
The peptide corresponding to the C-terminus of human AMSH-2 was synthesized by Boston Biomolecules, Inc (Woburn, MA). The specific AMSH-2 rabbit polyclonal antibody (anti-AMSH-2) was raised at Covance Research Products Inc. (Denver, PA). The following antibodies were obtained: (i) c-Myc mAb 9E10 (Covance Research Products Inc);(ii) anti-V5 and anti-V5 HRP antibodies (Invitrogen); (iii) anti-Flag HRP (UPSTATE, Lake Placid, NY);and (v) anti-HA 12CA5, anti-HA HRP (Roche Diagnostics Corp, Indianapolis, IN).
Metabolic labeling and immunoprecipitation
To test endogenous expression of AMSH-2, 293T and HepG2 cells were metabolically labeled with 35S methionine and 35S cysteine and lysed in modified RIPA buffer (150 mM NaCl, 50 mM Tris, pH 7.5, 1 mM EDTA, 1% NP-40, 0.25% sodium deoxycholate) with 1 mM sodium orthovanadate and protease inhibitors. Samples were incubated either with pre-immune serum or AMSH-2 antiserum. Immunoprecipitated proteins were resolved by SDS-PAGE. To test expression of the Myc-tagged AMSH-2 construct, cells were transfected with 15 μg of DNA using the standard calcium phosphate method. Twenty-four h postransfection, the cells were metabolically labeled for 16 h. and subsequently lysed and immunoprecipitated with anti-Myc antibody as above.
Luciferase assays
HepG2 cells were transfected with 0.5 μg of the corresponding luciferase reporter, 50 ng of β-galactosidase reporter and 4.5 μg of DNA. Twenty-four h postransfection, the cells were depleted from serum and stimulated for 16–20 h with 5 ng/ml of purified recombinant human TGF-β1 (R&Dsystems. Inc. Minneapolis, MN). Luciferase and β-galatosidase activities were measured according to manufacturer's instructions (Tropix, Bedford, MA). Measurements were corrected by β-galactosidase activity. The experiments were repeated at least three times.
Coimmunoprecipitation experiments
293T cells were cotransfected with 13 μg of AMSH-2 V5 construct and 2 μg of the corresponding Smad constructs. After 48 h, the cells were lysed in lysis buffer (150 mM NaCl, 50 mM Tris, pH 7.5, 1 mM EDTA, 1% NP-40) with 1 mM sodium orthovanadate and protease inhibitors. Lysates were incubated with anti-V5 antibody, and the resulting immunocomplexes were separated in SDS-PAGE. Blots were probed with the indicated HRP conjugated antibody.
siRNA experiments
To target AMSH-2 and Smad 7, we designed 21 nt siRNA duplexes: AACAATTCCTTGCTGAATGTA and AACCGCAGCAGTTACCCCATC, according to [44]. All siRNA duplexes including lamin A/C siRNA duplexes and scrambled siRNA were obtained from Dharmacon Research. Inc. (Lafayette, Colorado).
HepG2 cells grown in 6 well plates were transfected with 250 ng of 3TP-lux reporter, 50 ng of β-galactosidase reporter and 0.12 nmol of the corresponding siRNA duplex. After 48 h of transfection cells were depleted from serum and treated with 5 ng/ml TGF-β1. Luciferase and β-galactosidase activities were measured after 17 h stimulation.
Acknowledgments
Acknowledgments
We thank Harvey Lodish for support and encouragement. Akhilesh Pandey is supported by a Career Development Award from the Breast Cancer SPORE (CA 88843) at the Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins and by a Sidney Kimmel Scholar Award. Work at the Center of Experimental BioInformatics (CEBI) is supported by a generous grant from the Danish National Research Foundation. N.I was supported by a grant from Ministerio de Ciencia y Technologia (Spain).
Contributor Information
Nieves Ibarrola, Email: nibarro1@jhmi.edu.
Irina Kratchmarova, Email: ihk@bmb.sdu.dk.
Daisuke Nakajima, Email: nakajima@kazusa.or.jp.
William P Schiemann, Email: schiemannwp@njc.org.
Aristidis Moustakas, Email: aris.moustakas@licr.uu.se.
Akhilesh Pandey, Email: pandey@jhmi.edu.
Matthias Mann, Email: mann@bmb.sdu.dk.
References
- Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Piek E, Heldin CH, Ten Dijke P. Specificity, diversity, and regulation in TGF-beta superfamily signaling. Faseb J. 1999;13:2105–2124. [PubMed] [Google Scholar]
- Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- ten Dijke P, Miyazono K, Heldin CH. Signaling via hetero-oligomeric complexes of type I and type II serine/threonine kinase receptors. Curr Opin Cell Biol. 1996;8:139–145. doi: 10.1016/S0955-0674(96)80058-5. [DOI] [PubMed] [Google Scholar]
- Josso N, di Clemente N. Serine/threonine kinase receptors and ligands. Curr Opin Genet Dev. 1997;7:371–377. doi: 10.1016/S0959-437X(97)80151-7. [DOI] [PubMed] [Google Scholar]
- Wrana JL, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang XF, Massague J. TGF beta signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- Yamashita H, ten Dijke P, Franzen P, Miyazono K, Heldin CH. Formation of hetero-oligomeric complexes of type I and type II receptors for transforming growth factor-beta. J Biol Chem. 1994;269:20172–20178. [PubMed] [Google Scholar]
- Wrana JL, Attisano L, Wieser R, Ventura F, Massague J. Mechanism of activation of the TGF-beta receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- Weis-Garcia F, Massague J. Complementation between kinase-defective and activation-defective TGF-beta receptors reveals a novel form of receptor cooperativity essential for signaling. Embo J. 1996;15:276–289. [PMC free article] [PubMed] [Google Scholar]
- Macias-Silva M, Abdollah S, Hoodless PA, Pirone R, Attisano L, Wrana JL. MADR2 is a substrate of the TGFbeta receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Feng X, We R, Derynck R. Receptor-associated Mad homologues synergize as effectors of the TGF-beta response. Nature. 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Partnership between DPC4 and SMAD proteins in TGF-beta signalling pathways. Nature. 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin CH, Miyazono K, ten Dijke P. TGF-beta receptor-mediated signalling through Smad2, Smad3 and Smad4. Embo J. 1997;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attisano L, Wrana JL. Smads as transcriptional co-modulators. Curr Opin Cell Biol. 2000;12:235–243. doi: 10.1016/S0955-0674(99)00081-2. [DOI] [PubMed] [Google Scholar]
- Itoh S, Itoh F, Goumans MJ, Ten Dijke P. Signaling of transforming growth factor-beta family members through Smad proteins. Eur J Biochem. 2000;267:6954–6967. doi: 10.1046/j.1432-1327.2000.01828.x. [DOI] [PubMed] [Google Scholar]
- Miyazono K, Kusanagi K, Inoue H. Divergence and convergence of TGF-beta/BMP signaling. J Cell Physiol. 2001;187:265–276. doi: 10.1002/jcp.1080. [DOI] [PubMed] [Google Scholar]
- ten Dijke P, Miyazono K, Heldin CH. Signaling inputs converge on nuclear effectors in TGF-beta signaling. Trends Biochem Sci. 2000;25:64–70. doi: 10.1016/S0968-0004(99)01519-4. [DOI] [PubMed] [Google Scholar]
- Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Smad6 inhibits signalling by the TGF-beta superfamily. Nature. 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, Richardson MA, Topper JN, Gimbrone M. A., Jr., Wrana JL, Falb D. The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, ten Dijke P. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- Hata A, Lagna G, Massague J, Hemmati-Brivanlou A. Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev. 1998;12:186–197. doi: 10.1101/gad.12.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- Massague J, Chen YG. Controlling TGF-beta signaling. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- Ohara O, Nagase T, Ishikawa K, Nakajima D, Ohira M, Seki N, Nomura N. Construction and characterization of human brain cDNA libraries suitable for analysis of cDNA clones encoding relatively large proteins. DNA Res. 1997;4:53–59. doi: 10.1093/dnares/4.1.53. [DOI] [PubMed] [Google Scholar]
- Kikuno R, Nagase T, Waki M, Ohara O. HUGE: a database for human large proteins identified in the Kazusa cDNA sequencing project. Nucleic Acids Res. 2002;30:166–168. doi: 10.1093/nar/30.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Kaneko K, Asao H, Kasai H, Endo Y, Fujita T, Takeshita T, Sugamura K. Possible involvement of a novel STAM-associated molecule "AMSH" in intracellular signal transduction mediated by cytokines. J Biol Chem. 1999;274:19129–19135. doi: 10.1074/jbc.274.27.19129. [DOI] [PubMed] [Google Scholar]
- Dan I, Ong SE, Watanabe NM, Blagoev B, Nielsen MM, Kajikawa E, Kristiansen TZ, Mann M, Pandey A. Cloning of MASK, a novel member of the mammalian germinal center kinase III subfamily, with apoptosis-inducing properties. J Biol Chem. 2002;277:5929–5939. doi: 10.1074/jbc.M110882200. [DOI] [PubMed] [Google Scholar]
- Pandey A, Dan I, Kristiansen TZ, Watanabe NM, Voldby J, Kajikawa E, Khosravi-Far R, Blagoev B, Mann M. Cloning and characterization of PAK5, a novel member of mammalian p21-activated kinase-II subfamily that is predominantly expressed in brain. Oncogene. 2002;21:3939–3948. doi: 10.1038/sj.onc.1205478. [DOI] [PubMed] [Google Scholar]
- Pandey A, Blagoev B, Kratchmarova I, Fernandez M, Nielsen M, Kristiansen TZ, Ohara O, Podtelejnikov AV, Roche R, Lodish HF, Mann M. Cloning of a novel phosphotyrosine binding domain containing molecule, Odin, involved in signaling by receptor tyrosine kinases. Oncogene. 2002;21:8029–8036. doi: 10.1038/sj.onc.1205988. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. doi: 10.1016/0968-0004(96)10052-9. [DOI] [PubMed] [Google Scholar]
- Aravind L, Ponting CP. Homologues of 26S proteasome subunits are regulators of transcription and translation. Protein Sci. 1998;7:1250–1254. doi: 10.1002/pro.5560070521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Bucher P. The PCI domain: a common theme in three multiprotein complexes. Trends Biochem Sci. 1998;23:204–205. doi: 10.1016/S0968-0004(98)01217-1. [DOI] [PubMed] [Google Scholar]
- Padgett RA, Grabowski PJ, Konarska MM, Seiler S, Sharp PA. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Peri S, Ibarrola N, Blagoev B, Mann M, Pandey A. Common pitfalls in bioinformatics-based analyses: look before you leap. Trends Genet. 2001;17:541–545. doi: 10.1016/S0168-9525(01)02443-X. [DOI] [PubMed] [Google Scholar]
- Keeton MR, Curriden SA, van Zonneveld AJ, Loskutoff DJ. Identification of regulatory sequences in the type 1 plasminogen activator inhibitor gene responsive to transforming growth factor beta. J Biol Chem. 1991;266:23048–23052. [PubMed] [Google Scholar]
- Schindler U, Baichwal VR. Three NF-kappa B binding sites in the human E-selectin gene required for maximal tumor necrosis factor alpha-induced expression. Mol Cell Biol. 1994;14:5820–5831. doi: 10.1128/mcb.14.9.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe M, Wong SC, Henzel WJ, Goeddel DV. A novel family of putative signal transducers associated with the cytoplasmic domain of the 75 kDa tumor necrosis factor receptor. Cell. 1994;78:681–692. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- Rothe M, Sarma V, Dixit VM, Goeddel DV. TRAF2-mediated activation of NF-kappa B by TNF receptor 2 and CD40. Science. 1995;269:1424–1427. doi: 10.1126/science.7544915. [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Rothe M, Goeddel DV. Anatomy of TRAF2. Distinct domains for nuclear factor-kappaB activation and association with tumor necrosis factor signaling proteins. J Biol Chem. 1996;271:19935–19942. doi: 10.1074/jbc.271.33.19935. [DOI] [PubMed] [Google Scholar]
- Itoh F, Asao H, Sugamura K, Heldin CH, ten Dijke P, Itoh S. Promoting bone morphogenetic protein signaling through negative regulation of inhibitory Smads. Embo J. 2001;20:4132–4142. doi: 10.1093/emboj/20.15.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Smad6 inhibits signalling by the TGF-beta superfamily. Nature. 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- Liu X, Sun Y, Constantinescu SN, Karam E, Weinberg RA, Lodish HF. Transforming growth factor beta-induced phosphorylation of Smad3 is required for growth inhibition and transcriptional induction in epithelial cells. Proc Natl Acad Sci U S A. 1997;94:10669–10674. doi: 10.1073/pnas.94.20.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]