Abstract
MicroRNAs (miRNAs) have gradually been recognized as regulators of embryonic development; however, relatively few miRNAs have been identified that regulate cardiac development. A series of recent papers have established an essential role for the miRNA-17-92 (miR-17-92) cluster of miRNAs in the development of the heart. Previous research has shown that the Friend of Gata-2 (FOG-2) is critical for cardiac development. To investigate the possibility that the miR-17-92 cluster regulates FOG-2 expression and inhibits proliferation in mouse embryonic cardiomyocytes we initially used bioinformatics to analyze 3′ untranslated regions (3′UTR) of FOG-2 to predict the potential of miR-17-92 to target it. We used luciferase assays to demonstrate that miR-17-5p and miR-20a of miR-17-92 interact with the predicted target sites in the 3′UTR of FOG-2. Furthermore, RT-PCR and Western blot were used to demonstrate the post-transcriptional regulation of FOG-2 by miR-17-92 in embryonic cardiomyocytes from E12.5-day pregnant C57BL/6J mice. Finally, EdU cell assays together with the FOG-2 rescue strategy were employed to evaluate the effect of proliferation on embryonic cardiomyocytes. We first found that the miR-17-5p and miR-20a of miR-17-92 directly target the 3′UTR of FOG-2 and post-transcriptionally repress the expression of FOG-2. Moreover, our findings demonstrated that over-expression of miR-17-92 may inhibit cell proliferation via post-transcriptional repression of FOG-2 in embryonic cardiomyocytes. These results indicate that the miR-17-92 cluster regulates the expression of FOG-2 protein and suggest that the miR-17-92 cluster might play an important role in heart development.
Keywords: MicroRNAs, miR-17-92, FOG-2, Proliferation
Introduction
MicroRNAs (miRNAs) are a class of small singlestranded (∼22 nucleotides; nt), non-coding RNA genes that, by binding complementary sequences in the 3′ untranslated region (3′UTR) of messenger RNAs (mRNAs), mediate translational repression or direct mRNA cleavage (1). miRNAs are usually copied by RNA polymerase α as primary miRNA transcripts (pri-miRNAs) (2,3). The pri-miRNAs are subsequently cleaved into approximate 70-nt stem-loop precursor hairpins, known as pre-miRNAs, by the nuclear RNase β enzyme Drosha (4). The pre-miRNAs are exported from the nucleus to the cytoplasm via the Exportin-5 pathway, where it is cleaved into mature miRNAs by the RNase β enzyme Dicer in partnership with another RNA-binding protein called transactivation-responsive RNA-binding protein (5-8). Finally, the mature miRNAs become singe-stranded RNAs with a complex of associated proteins known as the RNA-induced silencing complex bound to sites of complementarity in the 3′UTR of mRNAs. Subsequently, targeted mRNAs are translated less efficiently and undergo accelerated turnover (9).
Despite the large number of candidate targets, the definite cellular pathways, which are regulated by miRNAs, are not well understood. miRNAs have been implicated in development, differentiation, cell proliferation, apoptosis, and metabolism (10,11). In animals, miRNAs are frequently transcribed together as polycistronic primary transcripts, which are subsequently processed into multiple individual mature miRNAs (12). miR-17-92 (also called Oncomir-1) is a polycistronic cluster of highly conserved miRNAs that has been shown to contribute to tumorigenesis (13-17), lung development (18,19), angiogenesis (20), adipocyte differentiation (21), and heart development (9,22). The miR-17-92 cluster consists of seven individual miRNAs (miR-17-3p, miR-17-5p, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92-1) located in the third intron of a primary transcript of approximately 7 kb known as C13orf25 (23,24).
In humans, congenital heart disease usually refers to abnormalities in the heart's structure or function, which are generated before birth and occur in approximately 12 of 1000 live births (25). The development of the heart is a fascinating process. It involves the differentiation of cardiac precursors into many kinds of cell types including myocardium, cardiac mesenchyme, and endocardium. Each of these cell types contributes in its own specific way to form the cardiac chambers, conduction system, and a highly specialized valvuloseptal apparatus (26). It is clear that precise regulation of its development is critical for normal functioning of the heart.
Friend of Gata-2 (FOG-2, also called zfpm2) is a multizinc finger nuclear corepressor protein, which is necessary for cardiac development (27,28). FOG-2 interacts directly with GATA factors to modulate GATA-dependent transcriptional activation and can act as an activator or repressor, depending on the promoter and cell type (29). FOG-2−/− embryos die around day E13.5 of cardiac defects, including thin myocardium, large ventricular septal defect, severe atrioventricular endocardial cushion defect, overriding aorta, and severe underdevelopment of the coronary vascular plexus (27,28,30). In this study, we initially used bioinformatics to analyze the 3′UTR of FOG-2. A putative 8mer seed site of miR-17-5p, an 8mer seed site of miR-20a, a 7mer-m8 seed site of miR-19a, a 7mer-m8 seed site of miR-19b, and a 7mer-1A seed site of miR-92-1 are known to exist; furthermore, these targeted regions are highly conserved among mammals (www.targetscan.org).
Here, we demonstrated that miR-17-5p and miR-20a directly target the 3′UTR of FOG-2 and post-transcriptionally repress the expression of FOG-2, and we also showed that over-expression of miR-17-92 may inhibit cell proliferation via post-transcriptional repression of FOG-2 in embryonic cardiomyocytes. This was the first study to show that FOG-2 is targeted by the specific miR-17-92 cluster. Moreover, these results suggest that the miR-17-92 cluster might also play an important role in the regulation of cardiac development.
Material and Methods
Plasmid construction
The murine FOG-2 3′UTR was amplified from mouse genomic DNA with the forward primer: 5′-GCTCTAGAACTAACTGAGTTACTAAAG-3′ and the reverse primer: 5′-CGGGATCCAACGTCTCCAACAAT-3′. This 1016-bp fragment was cloned into the XbaI/BamHI site of the pGL3-promoter vector (Promega, USA) to create pGL3-FOG-2 after the SV40 polyadenylation sequence was removed. pRL-SV40 was obtained from Promega. The miR-17-92 cluster expression construct was generated by using PCR to amplify a 1102-bp fragment encoding the mir-17-92 cluster precursor from mouse genomic DNA with the forward primer: 5′-GTCAGAATTCCCCCTTGGGTATAAGCTGTAATT-3′ and the reverse primer: 5′-GTCACTCGAGACCAACGAAAGCAATAGAAATCA-3′. This fragment was cloned into the EcoRI/XhoI sites of pcDNA3.1(+) (Invitrogen, USA) to create pcDNA3.1-miR-17-92. The pcDNA3.1-miR-17-5p (forward primer: 5′-GCGAATTCCAAATTTAGCAGGAATAAAG-3′, reverse primer: 5′-CGCTCGAGGACTGGACGCAGCCAGTG-3′, EcoRI/XhoI), pcDNA3.1-miR-19a (forward primer: 5′-GCGAATTCATCCAATCCAAGTCAAGCAA-3′, reverse primer: 5′-CGCTCGAGTACATCGACACAATAAAAATACAA-3′, EcoRI/XhoI), pcDNA3.1-miR-19b (forward primer: 5′-GCGAATTCCCTCTAAAATACCCAACATG-3′, reverse primer: 5′-CGCTCGAGATGCTGCTCGGTTGGTGG-3′, EcoRI/XhoI), pcDNA3.1-miR-20a (forward primer: 5′-GCGAATTCTATTGTGTCGATGTAGAGCCT-3′, reverse primer: 5′-CGCTCGAGAACTAACCATAGACCAGTGCTCA-3′, EcoRI/XhoI) and pcDNA3.1-miR-92-1 (forward primer: 5′-GCGAATTCCCATGCAAAACTGACTGTGG-3′, reverse primer: 5′-CGCTCGAGAGTGGAAATCGGCATCTTCA-3′, EcoRI/XhoI) expression constructs were cloned in the same way. The FOG-2 expression construct was generated by using PCR to amplify a 3456-bp fragment open reading frame encoding the N- and C-terminal of FOG-2 (aa 1-1151) from mouse heart cDNA with the forward primer: 5′-CGGGATCCATGTCCCGGCGAAAGCAGAG-3′ and the reverse primer: 5′-CCGCTCGAGTCATTTGACATGTTCTGCTGCATGTGATG-3′. This fragment was cloned into the BamHI/XhoI sites of pcDNA3.1(+) to create pcDNA3.1-FOG-2. All constructs were verified by DNA sequencing.
Cell culture and transfection
Hearts of E12.5-day mouse embryos were removed and placed in 1X Dulbecoo's phosphate-buffered solution lacking magnesium and calcium (Gibco, USA). Ventricular tissue was minced and the cells were dissociated using 0.25% trypsin-EDTA (Gibco) at 37°C with gentle shaking. Trypsin digestion was halted after 10 min by transferring the cells to a trypsin neutralization solution [Dulbecco's modified eagle medium (DMEM) with 20% fetal bovine serum (FBS); Gibco]. The cells were then centrifuged for 3 min at 1500 rpm at 4°C and resuspended in nutritive medium. Cells were cultivated in DMEM, supplemented with 20% FBS, 100 U/mL penicillin and 100 µg/mL streptomycin. HEK 293T cells were cultivated in DMEM supplemented with 10% FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin. Cells were cultivated in a humidified atmosphere of 5% CO2 at 37°C. All transfections were carried out using Lipofectamine™ 2000 (Invitrogen) according to manufacturer instructions.
Luciferase assays
The pcDNA3.1-miR-17-92 activity was determined by co-transfection of the pGL3-FOG-2 reporter with pRL-SV40 into HEK 293T cells using Lipofectamine™ 2000. Forty-eight hours later, luciferase activity was determined with the Dual-Luciferase Reporter Assay System (Promega) according to manufacturer instructions. Activity was defined as Firefly/Renilla ratio, normalized to control vector transfection. All experiments were performed in triplicate and standard deviations calculated.
RT-PCR
Total RNA was isolated from cells using Trizol (Invitrogen) and cDNA was synthesized from this RNA using the RevertAid™ First-Strand cDNA Synthesis Kit (Fermentas Life Sciences, USA) according to manufacturer instructions. RT-PCR was performed using the sense primer: 5′-ACTGCACATCAGCGTAATGACC-3′ and anti-sense primer: 5′-AATATCGCATAGCCGGCAGTA-3′ to amplify a 741-bp fragment corresponding to the coding region. Data were normalized to results obtained with primers specific for GAPDH. Each experiment was performed in triplicate and standard deviations were calculated.
Western blotting
The cells were washed three times with ice-cold PBS buffer, lysed with RIPA Lysis Buffer (Boste, China) and placed on ice for 30 min. Proteins were separated on 10% SDS-PAGE and subsequently electrotransferred to a PVDF membrane (Millipore, USA). The membrane was blocked with 5% BSA in Tris-buffered saline and Tween 20 (10 mM Tris, pH 7.5, 140 mM NaCl, 0.05% Tween-20) for 1 h at room temperature. A rabbit polyclonal antibody against FOG-2 (1:1000; GeneTex, USA) was used as the primary antibody, and horseradish peroxidase-conjugated goat anti-rabbit IgG was used as a secondary antibody. Western Blot Luminol Reagent (Zhongshan Bio, China) was used for antibody detection with X-ray film.
Northern blotting
Total RNA was isolated from cells using Trizol (Invitrogen). Total RNA was separated by 15% polyacrylamide gel electrophoresis with 0.5X TBE. After 1-h electrotransfer to an Immobilon-Ny+ membrane (Millipore) the membrane was prehybridized and hybridized in hybridization buffer (0.5 M sodium phosphate, pH 7.1, 2 mM EDTA, 7% SDS, 0.1% sodium pyrophosphate, 50% formamide, 10 mg/mL salmon sperm DNA) at 42°C for DNA probes. Probe sequences were as follows: miR-17-5p, 5′-ACTACCTGCACTGTAAGCACTTTGCCAGAG-3′; miR-20a, 5′-CTACCTGCACTATAAGCACTTTACCAGAG-3′; U6, 5′-GCAGGGGCCATGCTAATCTTCTCTGTATCG-3′. Probes were labeled with [32P]-γ-ATP by T4 polynucleotide kinase (Ta-KaRa, Japan). The membrane was washed twice with 10X SSC at room temperature for autoradiography.
5-. Ethynyl-2′-deoxyuridine (EdU) assays
Cardiomyocyte proliferation was determined in vitro using the EdU DNA Proliferation in vitro Detection kit (RiboBio, China) according to manufacturer instructions.
Statistical analysis
Data are reported as means ± SEM for at least three separate experiments. Statistical analysis was performed by the Student t-test and ANOVA. P < 0.05 was considered to be statistically significant.
Results
Bioinformatics predicted miRNAs target FOG-2
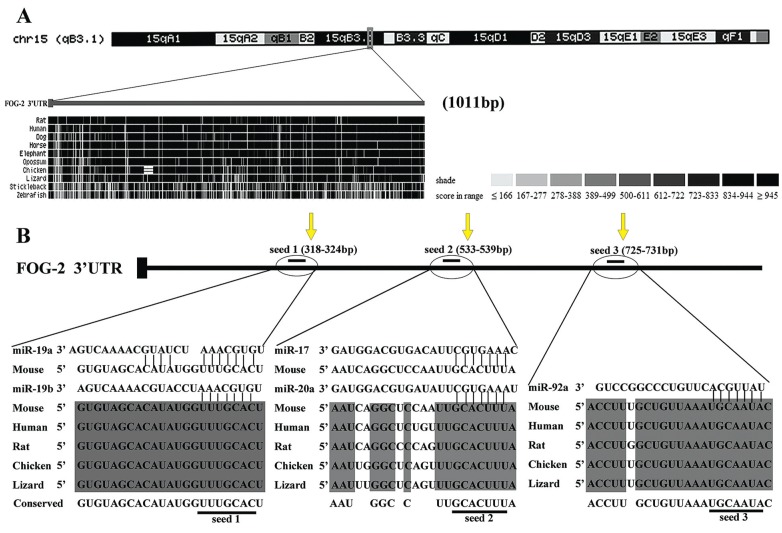
In order to determine if miRNA was associated with the FOG-2 gene, we analyzed the 3′UTR of FOG-2 via the UC Santa Cruz genome database (UCSC, http://genome.ucsc.edu). The data showed that the 3′UTR of FOG-2 was 1011 bp in length and highly conserved across mammals (Figure 1A). To identify the candidate miRNAs associated with FOG-2, we utilized the TargetScan release 5.1 (http://www.targetscan.org) to predict the miRNAs target FOG-2. As a result, the entire mouse FOG-2 3′UTR was found to contain 17 conservative target sites for 34 different miRNAs. Of the 34 miRNAs, the miR-17-92 cluster includes five miRNAs (miR-17-5p, miR-19a, miR-19b, miR-20a, and miR-92-1), and of the 17 target sites, the miR-17-92 cluster occupies three putative target sites. After retrieving the 3′UTR of human, mouse, rat, chicken, and lizard from the TargetScan (http://www.targetscan.org), the alignments with miR-17-5p, miR-19a, miR-19b, miR-20a, and miR-92-1 are illustrated in Figure 1B.
Figure 1. Bioinformatic analysis of the 3′ untranslated regions (3′UTR) of Friend of Gata-2 (FOG-2). A, The figure shows that the 3′UTR of the FOG-2 is highly conserved across mammals, as shown by the UC Santa Cruz genome database. The chromosome tracks show the location of the 3′UTR of FOG-2. In the dense display tracks, conservation is shown in grayscale using darker values to indicate higher levels of overall conservation as scored by phastCons. B, Schematic diagrams of the 3′UTR of FOG-2 and the predicted binding sites for the miR-17-92 cluster via the TargetScan release 5.1 database. The upper panel shows three putative target sites predicted on FOG-2 3′UTR and the lower panel shows multiple sequence alignment of miR-17-92 with the three binding sites on 3′UTR of FOG-2. The underlined bases represent the location of predicted miR-17-92 seed sites and spaces are added where needed to facilitate the alignment. The number in parentheses (below the yellow arrows) represents the location of miR-17-92 potential binding sites. Nucleotides in dark-shaded boxes indicate sequences that are present in all five species.
The miR-17-92 cluster directly targeted FOG-2 3′UTR
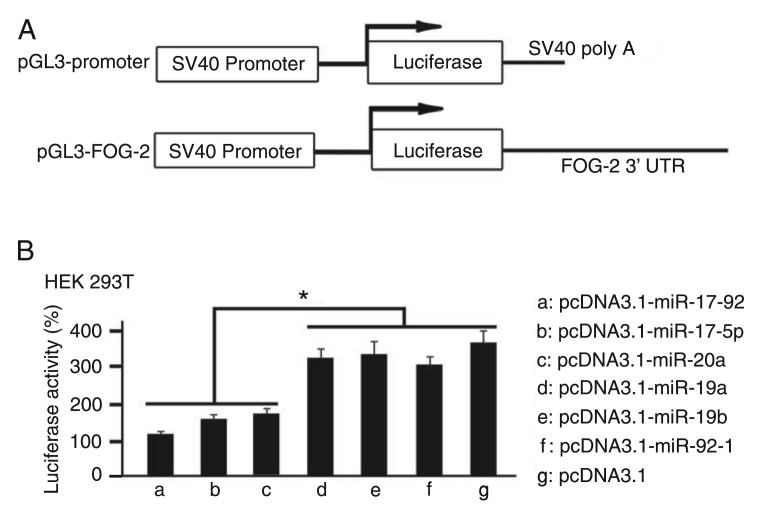
To explore whether FOG-2 is targeted directly by the miR-17-92 cluster in vitro, the full-length fragment of the 3′UTR of FOG-2 was inserted into a luciferase reporter gene (Figure 2A). The pcDNA3.1, pcDNA3.1-miR-17-92, pcDNA3.1-miR-17-5p, pcDNA3.1-miR-19a, pcDNA3.1-miR-19b, pcDNA3.1-miR-20a, pcDNA3.1-miR-92-1, pGL3-FOG-2, and pRL-SV40 were transfected into HEK 293T cell lines using Lipofectamine™ 2000. The luciferase assays were carried out in triplicate 48 h after transfection. For comparative purposes, activity was defined as firefly/renilla ratio, normalized to control vector transfection. As shown in Figure 2B, in HEK 293T cotransfected with the miR-17-92, miR-17-5p or miR-20a precursor and the pGL3-FOG-2 construct, luciferase reporter activities obviously decreased compared to HEK 293T cotransfected with the pcDNA3.1 and pGL3-FOG-2 construct. We also did not find any significant changes in the expression of luciferase reporter activities in HEK 293T cotransfected with the miR-19a, miR-19b or miR-92-1 precursor and pGL3-FOG-2. These data imply that miR-17-5p and miR-20a may regulate FOG-2 protein expression by directly binding to the 3′UTR region of FOG-2.
Figure 2. miR-17-5p and miR-20a directly targeted the 3′ untranslated regions (3′UTR) of Friend of Gata-2 (FOG-2). A, A schematic diagram of the constructs used to evaluate the function of the conserved region of the 3′UTR of FOG-2. B, HEK 293T cell lines were transfected with pRL-SV40 and a luciferase reporter containing the 3′UTR of FOG-2. Forty-eight hours after transfection, cell lysates were assayed for luciferase activity and normalized to Renilla luciferase activity. Data are reported as means ± SEM for three independent experiments. *P < 0.05 (ANOVA).
The miR-17-92 cluster post-transcriptionally regulated FOG-2 expression
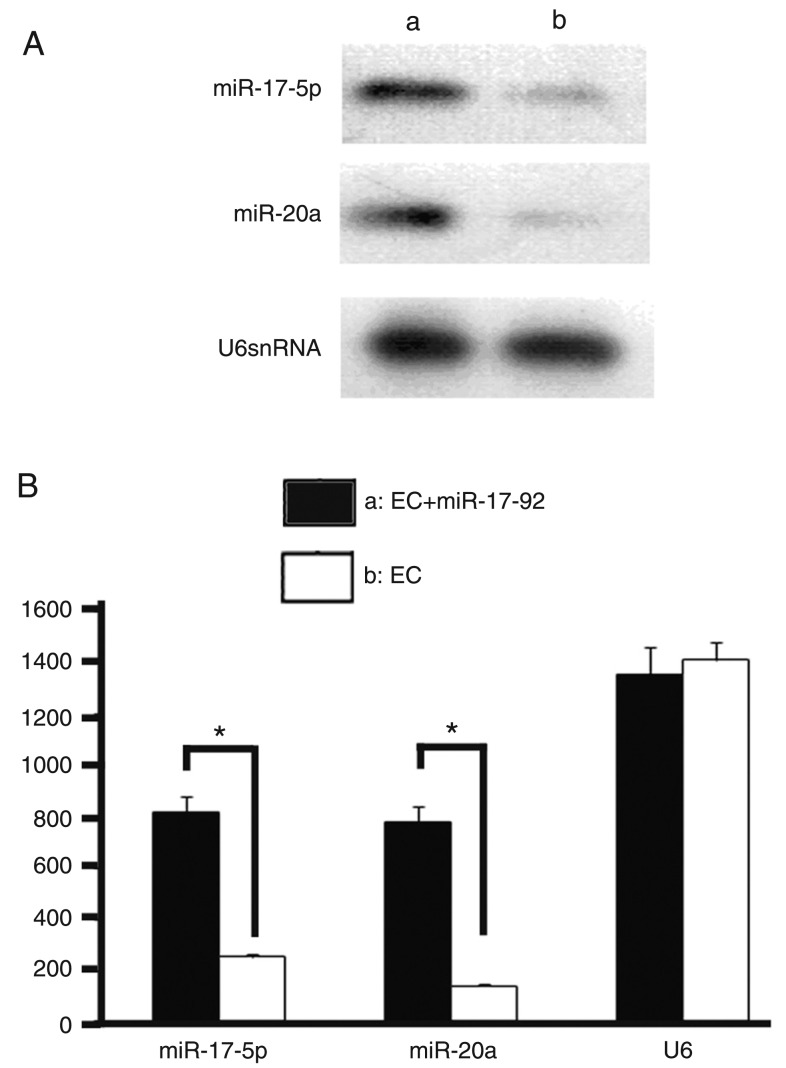
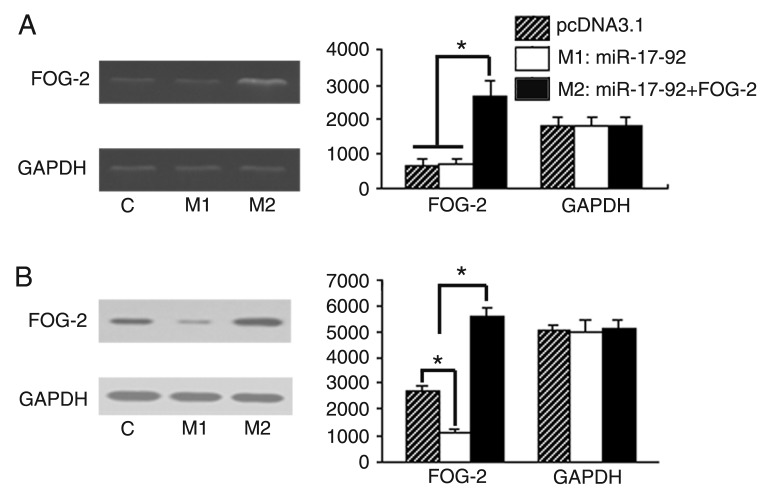
To further explore the roles of miR-17-92 in directly targeting the 3′UTR of FOG-2 in the expression of FOG-2, we constructed miR-17-92 and pcDNA3.1-FOG-2 overexpression vectors as described above. The pcDNA3.1, pcDNA3.1-miR-17-92 and pcDNA3.1-FOG-2&pcDNA3.1-miR-17-92 plasmids were transfected into E12.5 embryonic cardiomyocytes and RNA and protein were extracted from cardiomyocytes after transfection for 48 h. To confirm that miR-17-5p and miR-20a were indeed expressed in the embryonic cardiomyocytes, we performed Northern analysis on total RNA from mouse embryonic cardiomyocytes using radiolabeled probes specific for miR-17-5p and miR-20a. miR-17-5p and miR-20a were expressed in the embryonic cardiomyocytes and both were overexpressed with the transfected pcDNA3.1-miR-17-92 vector (Figure 3). Nevertheless, as anticipated, miR-17-92 did not decrease the mRNA expression in cardiomyocytes of FOG-2 compared with pcDNA3.1 and pcDNA3.1-miR-17-92 (Figure 4A). The protein expression in cardiomyocytes of FOG-2 was obviously decreased in pcDNA3.1-miR-17-92 compared with pcDNA3.1 (Figure 4B). The mRNA and protein expression of FOG-2 in cotransfected pcDNA3.1-FOG-2&pcDNA3.1-miR-17-92 cardiomyocytes were increased (Figure 4A,B), indicating that miR-17-92 might post-transcriptionally regulate FOG-2.
Figure 3. miR-17-5p and miR-20a were expressed in the embryonic cardiomyocytes (EC). miR-17-5p and miR-20a were overexpressed with the transfected pcDNA3.1-miR-17-92 vector in embryonic cardiomyocytes. *P < 0.05 (t-test).
Figure 4. miR-17-92 post-transcriptionally down-regulates FOG-2 protein in cardiomyocytes 48 h after transfection. A, Detection of Friend of Gata-2 (FOG-2) mRNA expression by RT-PCR. Overexpression of miR-17-92 did not reduce the level of endogenous FOG-2 mRNA. Overexpression of FOG-2 increased the level of exogenous FOG-2 mRNA. Data were normalized to the GAPDH level. B, Detection of FOG-2 protein expression by Western blotting. Overexpression of miR-17-92 reduced the levels of endogenous FOG-2 protein. Overexpression of FOG-2 increased the level of exogenous FOG-2 protein. Data were normalized to the level of GAPDH. Data are reported as means ± SEM for three independent experiments. *P < 0.05 (ANOVA).
Expression of miR-17-92 inhibited cell proliferation
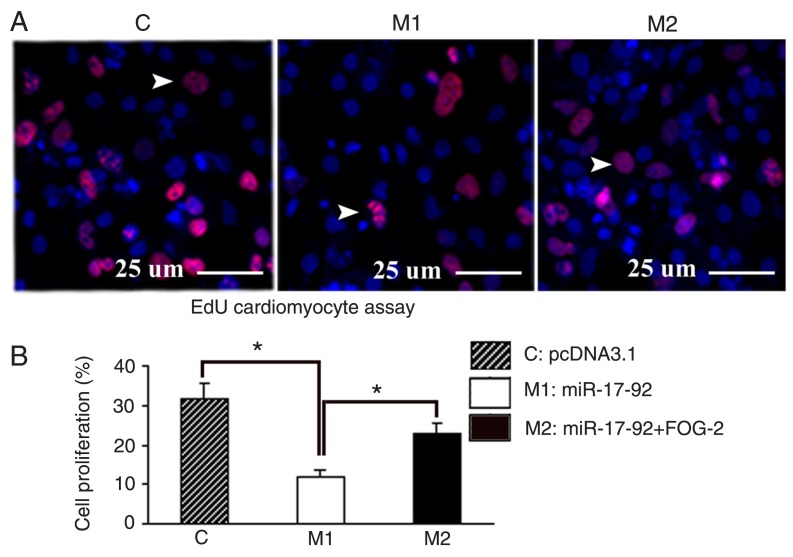
To estimate the effects of miR-17-92 on cardiomyocyte proliferation, pcDNA3.1 and pcDNA3.1-miR-17-92 plasmids were transfected into E12.5-day embryonic cardiomyocytes. Cardiomyocyte proliferation was determined using the EdU DNA Detection kit after transfection for 48 h (Figure 5A). The results of the EdU assay are illustrated in Figure 5B. The proliferation rate was lower in pcDNA3.1 than in pcDNA3.1-miR-17-92 (P < 0.05). These findings suggest that overexpression of miR-17-92 might inhibit cardiomyocyte proliferation.
Figure 5. Expression of miR-17-92 inhibited cardiomyocyte proliferation. A, Cardiomyocyte morphology was assessed by fluorescence microscopy (400X). Cardiomyocyte proliferation was determined by the 5-ethynyl-2′-deoxyuridine (EdU) assay. Forty-eight hours after transfection, cardiomyocytes were stained with EdU and DAPI. The arrowheads indicate cells undergoing proliferation. B, Cell proliferation rate was decreased by overexpression of miR-17-92, but might be rescued by overexpression of Friend of Gata-2 (FOG-2). Data are reported as means ± SEM for three independent experiments. *P < 0.05 (ANOVA).
FOG-2 partially rescued proliferation induced by miR-17-92
To further identify the effects of FOG-2 on miR-17-92-inhibited cell proliferation, we introduced the FOG-2 overexpression vector (pcDNA3.1-FOG-2) and miR-17-92 overexpression vector (pcDNA3.1-miR-17-92) simultaneously into cardiomyocytes by applying the EdU assays described above (Figure 5A). The results of the EdU assays are illustrated in Figure 5B. The proliferation rate was higher in pcDNA3.1-miR-17-92&pcDNA3.1-FOG-2 than in pcDNA3.1-miR-17-92 (P < 0.05), but was lower than in pcDNA3.1 (P < 0.05). These findings suggest that the miR-17-92 cluster might inhibit at least in part cardiomyocyte proliferation by post-transcriptionally regulating FOG-2 in embryonic cardiomyocytes.
Discussion
miRNAs are a class of small RNA molecules that participate in the developmental control of gene expression, and up to 30% of human protein coding genes may be regulated by miRNAs (31). miRNAs play substantial roles in development, differentiation, cell proliferation, apoptosis, and metabolism (10,11). miR-17-92 is a polycistronic cluster of highly conserved miRNAs that has been shown to contribute to tumorigenesis (13-17), lung development (18,19), angiogenesis (20), adipocyte differentiation (21), and heart development (9,22). However, the definite cellular pathways that are regulated by miR-17-92 in the heart are not known. Whether there is a relationship between gene n3′UTR target seeds and miR-17-92 in the heart and what its function may be are still unanswered questions. These speculations are worth exploring.
Congenital heart diseases are common in humans, and dysregulation of heart development is the origin of this disease. FOG-2 is a multi-zinc finger nuclear corepressor protein, which is necessary for cardiac development (27,28). However, it is unclear whether FOG-2 is regulated by miR-17-92. Since the functions of miRNAs are clearly defined as patterns of complementarity to the 3′UTR of their targeted transcripts (32), the target region, which is located in the 2-7 position of the miRNAs, is a crucial determinant of the specificity of binding sites, and is required for perfect complementarity (33). Thus, we applied these principles to predict the theoretical miR-17-92 targets of the 3′UTR of FOG-2, and analyzed the evolutionary conservation of these targeted sites among humans, mice, rats, chickens, and lizards. On the basis of the results of bioinformatic analysis, the 3′UTR of FOG-2 was 1011 bp in length and highly conserved across mammals. The miR-17-92 cluster includes five miRNAs (miR-17-5p, miR-19a, miR-19b, miR-20a, and miR-92-1) and occupies three putative target sites associated with the 3′UTR of FOG-2. Moreover, the targeted 3′UTR sites in FOG-2 are highly conserved among mammals. These findings suggest that miR-17-92 might target FOG-2 via the putative seed sites.
However, whether FOG-2 is directly targeted by the miR-17-92 cluster requires further experimental proof. According to the miRNA genesis process and transcription mechanisms, there are two main modes of miRNA overexpression: pri-miRNAs are driven by type α promoters or premiRNAs are driven by type β promoters (34). In the present study, we used type α CMV promoters to drive genomic pri-miRNAs to gain high expression of mature miRNAs (miR-17-5p, miR-19a, miR-19b, miR-20a, and miR-92-1), and the pri-miRNA was cloned from mouse genomic DNA by PCR. A luciferase activity assay then confirmed that miR-17-5p and miR-20a directly targeted the 3′UTR of FOG-2. This was consistent with our prediction by bioinformatic analysis. In most mammals, miRNAs directly targeting with the complementary 3′UTR of genes would post-transcriptionally inhibit the expression of the target gene (35). Our results initially showed that miR-17-92 post-transcriptionally suppressed the translation of FOG-2, which also means that the regulation of miR-17-92 may reduce the expression of FOG-2. Since the abnormal expression of FOG-2 would lead to heart dysplasia we inferred that miR-17-92 might be involved in heart dysplasia through FOG-2.
It is well known that FOG-2 interacts directly with GATA factors to modulate GATA-dependent transcriptional activation and can act as an activator or repressor, depending on the promoter and cell type (29). In embryos deficient in FOG-2 or carrying a mutation, there is ventricular hypoplasia GATA-4 independent of FOG-2 (28). Moreover, FOG-2 regulating the expression of FoxM1 via GATA-4 contributes to cardiomyocyte proliferation and ventricular heart development (36). In our research, we further examined the effect of miR-17-92 post-transcriptionally regulated by FOG-2 on cell proliferation. Our findings demonstrated that miR-17-92 might inhibit cardiomyocyte proliferation. In addition, EdU allied to FOG-2 rescue strategies confirmed that miR-17-92 might indeed regulate cardiomyocyte proliferation by post-transcriptionally regulating FOG-2 in heart development.
This study showed for the first time that the miR-17-5p and miR-20a of the miR-17-92 cluster directly target the 3′UTR of FOG-2 and post-transcriptionally repress the expression of FOG-2. Moreover, we found that overexpression of miR-17-92 may inhibit cell proliferation via post-transcriptional repression of FOG-2 in embryonic cardiomyocyte. These results demonstrated the importance of the miR-17-92 cluster in the regulation of FOG-2 protein expression and suggest that the miR-17-92 cluster may also play a role in the regulation of cardiac development. Of course, the miR-17-92 cluster has many other target genes and additional functions. On the other hand, FOG-2 has numerous unknown functions besides cell proliferation. It is worthwhile to explore novel cell functions for the FOG-2 and miR-17-92-FOG-2 target relationship as well as the phenotype when knocking down the miR-17-92 cluster. In a subsequent study, we plan to generate the overexpression and knockdown of miR-17-92 transgenic mice for further research on the functions of the miR-17-92 cluster and the miR-17-92-FOG-2 relationship in vivo.
Acknowledgments
We thank all members of the Core Facility of Gene Engineered Mice for technical assistance. Research supported by the National Key Basic Research Program of China to Qin Zhou (#2011CB944002).
References
- 1.Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 5.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 8.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of premicroRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 11.Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 12.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 13.Fontana L, Fiori ME, Albini S, Cifaldi L, Giovinazzi S, Forloni M, et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One. 2008;3:e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 15.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tagawa H, Seto M. A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia. 2005;19:2013–2016. doi: 10.1038/sj.leu.2403942. [DOI] [PubMed] [Google Scholar]
- 17.Uziel T, Karginov FV, Xie S, Parker JS, Wang YD, Gajjar A, et al. The miR-17∼92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci U S A. 2009;106:2812–2817. doi: 10.1073/pnas.0809579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carraro G, El-Hashash A, Guidolin D, Tiozzo C, Turcatel G, Young BM, et al. miR-17 family of microRNAs controls FGF10-mediated embryonic lung epithelial branching morphogenesis through MAPK14 and STAT3 regulation of E-Cadherin distribution. Dev Biol. 2009;333:238–250. doi: 10.1016/j.ydbio.2009.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suarez Y, Fernandez-Hernando C, Yu J, Gerber SA, Harrison KD, Pober JS, et al. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci U S A. 2008;105:14082–14087. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Li YC, Wang J, Kong J, Qi Y, Quigg RJ, et al. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Natl Acad Sci U S A. 2008;105:2889–2894. doi: 10.1073/pnas.0800178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, et al. Identification and characterization of a novel gene, C 13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 24.Mestdagh P, Bostrom AK, Impens F, Fredlund E, Van Peer G, De Antonellis P, et al. The miR-17-92 microRNA cluster regulates multiple components of the TGF-beta pathway in neuroblastoma. Mol Cell. 2010;40:762–773. doi: 10.1016/j.molcel.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffman JI, Kaplan S, Liberthson RR. Prevalence of congenital heart disease. Am Heart J. 2004;147:425–439. doi: 10.1016/j.ahj.2003.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Snarr BS, Kern CB, Wessels A. Origin and fate of cardiac mesenchyme. Dev Dyn. 2008;237:2804–2819. doi: 10.1002/dvdy.21725. [DOI] [PubMed] [Google Scholar]
- 27.Svensson EC, Huggins GS, Lin H, Clendenin C, Jiang F, Tufts R, et al. A syndrome of tricuspid atresia in mice with a targeted mutation of the gene encoding Fog-2. Nat Genet. 2000;25:353–356. doi: 10.1038/77146. [DOI] [PubMed] [Google Scholar]
- 28.Tevosian SG, Deconinck AE, Tanaka M, Schinke M, Litovsky SH, Izumo S, et al. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell. 2000;101:729–739. doi: 10.1016/s0092-8674(00)80885-5. [DOI] [PubMed] [Google Scholar]
- 29.Lu JR, McKinsey TA, Xu H, Wang DZ, Richardson JA, Olson EN. FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol Cell Biol. 1999;19:4495–4502. doi: 10.1128/mcb.19.6.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou B, Ma Q, Kong SW, Hu Y, Campbell PH, McGowan FX, et al. Fog2 is critical for cardiac function and maintenance of coronary vasculature in the adult mouse heart. J Clin Invest. 2009;119:1462–1476. doi: 10.1172/JCI38723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sassen S, Miska EA, Caldas C. MicroRNA: implications for cancer. Virchows Arch. 2008;452:1–10. doi: 10.1007/s00428-007-0532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 33.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 34.Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci U S A. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen PY, Meister G. microRNA-guided posttranscriptional gene regulation. Biol Chem. 2005;386:1205–1218. doi: 10.1515/BC.2005.139. [DOI] [PubMed] [Google Scholar]
- 36.Greytak S, Yelick PC, Huggins GS. Friend of GATA-2(FOG-2) regulates FOXM1 expression and cardiomyocyte proliferation. Circulation. 2010;122:A20281. (Abstract) [Google Scholar]