Abstract
Human epidermal growth factor receptor 2 (HER2) has been evaluated in breast cancer patients to identify those most likely to benefit from herceptin-targeted therapy. HER2 amplification, detected in 20-30% of invasive breast tumors, is associated with reduced survival and metastasis. The most frequently used technique for evaluating HER2 protein status as a routine procedure is immunohistochemistry (IHC). HER2 copy number alterations have also been evaluated by fluorescence in situ hybridization (FISH) in moderate immunoexpression (IHC 2+) cases. An alternative procedure to evaluate gene amplification is chromogenic in situ hybridization (CISH), which has some advantages over FISH, including the correlation between HER2 status and morphological features. Other methodologies have also been used, such as silver-enhanced in situ hybridization (SISH) and quantitative real-time RT-PCR, to determine the number of HER2 gene copies and expression, respectively. Here we will present a short and comprehensive review of the current advances concerning HER2 evaluation in human breast cancer.
Keywords: CISH, Breast cancer, HER2, Immunohistochemistry, FISH
Introduction
The v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma-derived oncogene homolog (avian), ERBB2/HER2 (other aliases: CD340, HER-2, HER-2/neu, MLN 19, NEU, NGL, TKR1) plays a role in the pathogenesis of a significant number of human cancers. This membrane receptor protein of the growth factor receptor gene family presents tyrosine kinase activity and is associated with cell growth, survival and differentiation. In human breast cancer, HER2 overexpression is reported in 20-30% of breast carcinomas 1-5 , mostly due to HER2 gene gains or amplification. HER2 overexpression is associated with constitutive activation of different pathways, in particular the PI3K and ERK pathways, leading to a significant increase in cell proliferation 6.
For breast cancer management, discrimination of HER2 status is crucial for determining therapy and prognosis, since HER2 alterations are associated with a shorter disease-free period, overall survival and resistance to tamoxifen antiestrogen therapy and other chemotherapy regimens, regardless of the nodal or hormone receptor status 4,5,7,8. Moreover, patients with HER2-positive breast cancers can benefit from Trastuzumab (Herceptin™, Genentech, Inc., USA), while patients presenting HER2 amplification without Ch17CEP duplication have apparently limited benefit from the addition of the anthracycline 9,10.
As a result of the importance of HER2 assessment in clinical practice, several methods have been described to evaluate its status. Currently, three types of assays, already approved by the FDA (U.S. Food and Drug Administration), have been described for HER2 evaluation in formalin-fixed paraffin-embedded samples. HER2 protein expression can be determined by immunohistochemistry (IHC), while copy number alterations can be determined by fluorescence in situ hybridization (FISH) or chromogenic in situ hybridization (CISH).
IHC has been the most commonly used assay for determining HER2 status. It is easy to perform and of relatively low cost. However, wide variation in sensitivity and specificity has been reported among commercially available antibodies 11. Its scoring is highly applicable to cases presenting negative (0 or 1+) or positive (3+) expression; however, tumors showing moderate protein expression (2+) are considered equivocal results and must be evaluated by other methods, such as FISH analysis 12.
In situ hybridization techniques are able to determine gene copy number using labeled DNA probes complementary to the target genomic sequences. For FISH, archival paraffin-embedded samples are pretreated to remove cytoplasmic and nuclear proteins, which can be a barrier to probe penetration, and the target DNA is denatured. Fluorescent-labeled probes are added to the tissue section to hybridize to HER2 gene sequences, whose signals are viewed with a fluorescence microscope. Tissue morphology and gene amplification are primarily disconnected, and although the nuclei can be identified by fluorescent DNA counterstain, such as DAPI (4′-6-diamidino-2-phenylindole), this does not always allow sufficient histopathological evaluation. Hematoxylin-and-eosin-stained sections from the same block are viewed in conjunction to enable morphologic analysis. The advantage of FISH testing is that the quantitative interpretation of the results is relatively straightforward and concordance rates among observers are higher than for IHC in some studies (for reviews, see Refs. 13,14).
More recently, CISH has emerged as a potential alternative to FISH for confirming ambiguous IHC results 15. CISH is a combination of in situ hybridization with antibodies or avidin conjugated with enzymes, such as alkaline phosphatase and peroxidase, to develop a chromogenic reaction similar to IHC staining. The principle of FISH is the hybridization of a fluorochrome-labeled DNA (probe) with a complementary target DNA sequence. A fluorescent counterstain is applied and the use of a fluorescent microscope with appropriate filters is necessary. Compared to FISH, CISH is much easier for pathologists to use for the analysis of gene amplification simultaneously with detailed morphologic features of tumors. Moreover, CISH signals do not diminish over time and can provide useful archives in the laboratories 9,16,17. This method has several advantages compared to FISH analysis, such as cost, the use of a light microscope, permanent staining, and available tissue morphology. Moreover, pathologists are more familiar with IHC labeling than with the FISH signal 18.
In addition to the CISH methodology, another bright-field in situ hybridization (BRISH) technique is the automated silver-enhanced in situ hybridization (SISH) developed by Ventana Medical System (Tucson, USA). SISH offers the advantages of a bright-field FISH test coupled with automation for HER2 amplification 17. It improves the efficiency and consistency of BRISH, reducing the risk of error. Probes for the HER2 gene and chromosome 17 are labeled with dinitrophenol. After DNA denaturation with enzyme digestion, goat anti-rabbit antibody conjugated to horseradish peroxidase is used as a chromogenic enzyme. Sequential addition of silver acetate as the source of ionic silver, hydroquinone, and hydrogen peroxide is used to yield a metallic silver precipitate at the probe site, which is visualized as a black dot. The slides are counterstained with hematoxylin for examination by light microscopy 17,19,20. A stable and discrete chromogenic reaction product is achieved allowing quantification of centromeric chromosome 17 and HER2 probe signals on the same slide by conventional bright-field light microscopy 19,21.
Methods based on the polymerase chain reaction (PCR) are also being increasingly applied to evaluate gene expression, in particular, quantitative real-time reverse transcription PCR (qRT-PCR), based on TaqMan methodology 11. This technique has successfully evaluated mRNAs expressed in mixed cell populations and specific mRNAs, especially those present in low copy numbers in a small number of cells or in small quantities of tissue. However, qRT-PCR suffers from the same drawback as other PCR-based methods. Besides isolating the tumor cell population within the tissue under evaluation, other technical aspects must be considered, including template quality, operator variability, and subjectivity in data analysis and reporting 14,22. Limitations related to tumor heterogeneity can be eliminated by the use of laser microdissection, although this seems to be impractical for routine diagnosis 14.
Due to the potential and application of these techniques, the aim of the current review is to summarize and compare available HER2 clinical routine findings by CISH methodology with IHC, FISH, SISH, and qRT-PCR results in breast cancer tissues.
Chromogenic in situ hybridization
HER2 amplification can be assessed by CISH in archival paraffin-embedded samples. This method is based on peroxidase- or alkaline phosphatase-labeled reporter antibodies that are detected using an enzymatic reaction 18. CISH was first used for HER2 screening by Tanner et al. 18, who demonstrated amplification by enzymatic detection as an additional method to be combined with IHC in breast cancer tumors. The main advantage of CISH is the use of chromogens instead of fluorochromes for signal identification, which can be viewed with a standard bright-field microscope.
The commercial system for CISH detection, known as the SPoT-Light HER2 CISH kit (Zymed Laboratories Inc., USA), recommends the following classification for HER2 status based on gene copy number in the nuclei: a) non-amplified, tumor cells with 2 to 5 brown intranuclear spots per nucleus; b) low-level amplification, when 6 to 10 signals per nucleus were detected in more than 50% of tumor cells or when a small coalescing signal cluster was identified; c) high-level amplification, defined as more than 10 copies per nucleus or when copy clusters were observed in more than 50% of cancer cells. Non-amplified tumors can be grouped as disomy, when 1 or 2 copies of the gene are present, or polysomy, when 3 to 5 copies were detected per nucleus in more than 50% of cancer cells.
CISH in comparison with other methodologies
Using the PubMed database (http://www.ncbi.nlm.nih.gov/pubmed, accessed on August 24, 2011), 112 studies were found using the key words: “breast cancer”, “HER2” and “CISH”. Of these, 67 presented adequate data to enable comparisons between CISH and other methods, comprising more than 7000 cases of breast cancer. The concordance rate between CISH and IHC was achieved when: 1) cases scored as negative (0 or 1+) or positive (3+) by IHC were non-amplified and amplified, respectively, by CISH; 2) cases scored as 2+ by IHC were considered to be amplified by CISH. CISH and FISH technique agreement was achieved when: 1) cases were considered to be amplified by both methods independent of the status of low and high amplification level; 2) cases were not amplified by either methodology. Table 1 shows the comparison between CISH and IHC and between CISH and FISH. The concordance varied from 52 to 100% and 82 to 100% when CISH results were compared to IHC and to FISH, respectively.
Table 1. Comparative analysis of chromogenic in situ hybridization (CISH), immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) results for breast cancer samples.
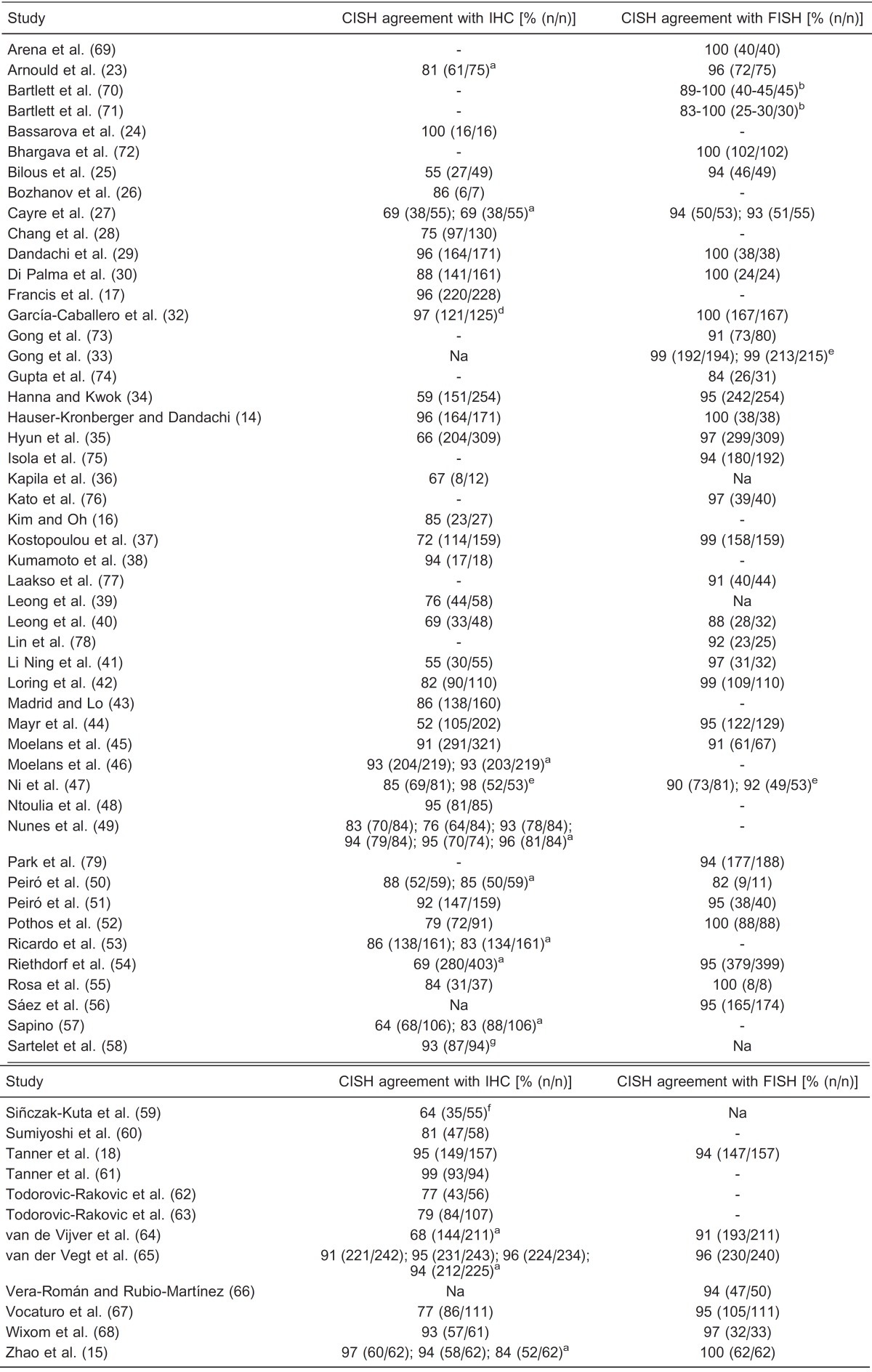
Na = data not available to calculate the percentage; (-) = method not performed. aDifferent antibodies were used. bData from different laboratories. dNo IHC 2+ cases. eTwo different groups of samples or different sites from the same tumor. fNo IHC 0-1+ cases. gCISH was performed on cytology samples.
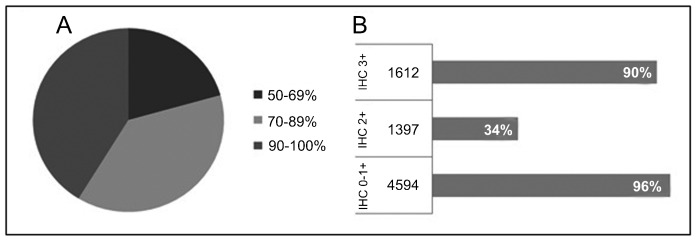
CISH compared to IHC. Forty-seven studies compared CISH to IHC (Table 1) and 41% showed more than 90% of agreement (Figure 1A). Studies presenting lower concordance rates can be explained by the high number of 2+ cases evaluated by IHC, in which amplification by CISH was not detected. A subset of these cases could involve chromosome 17 polysomy, which is frequently associated with IHC 2+ tumors 37,80.
Figure 1. Forty-seven studies (14-18,26-30,32,34-55,57-65,67,68) comparing CISH to IHC methodology for breast cancer samples. A, Agreement between the two methodologies ranging from50 to 100%. B, Percent agreement based on IHC subgroups (0-1+, 2+ and 3+) compared to amplified and non-amplified HER2 status by CISH. CISH = chromogenic in situ hybridization; IHC = immunohistochemistry.
While evaluating IHC 2+ tumors at seven different centers, Di Palma et al. 31 observed a variation concerning amplified or non-amplified cases. Three of 12 IHC 2+ cases presented discrepant results by CISH among the centers. For IHC 0-1+ or 3+ cases, 100% agreement was achieved by CISH, indicating that it is a highly reproducible method.
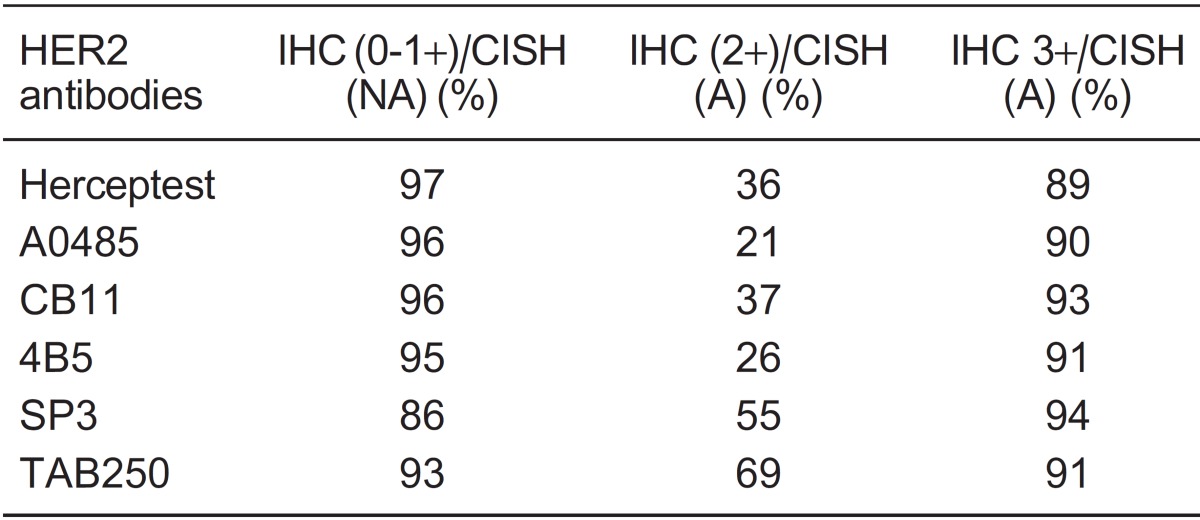
IHC 2+ cases. Cases scored as 2+ by IHC showed amplification using CISH in 34% of 1397 tumors, ranging from 0 to 100% (Figure 1B). Sixteen studies detected amplification in more than 60% of IHC 2+ tumors. Six different antibodies were used for HER2 immunostaining evaluation, two of them polyclonal (Herceptest and A0485) and four monoclonal (CB11, 4B5, SP3, and TAB250). Herceptest was provided by Dako, while the other antibodies were supplied by different manufacturers. The studies were grouped according to the antibody used to discriminate IHC 2+ status. The percentage of amplification by CISH ranged from 21 to 69% of tumors (Table 2). A higher amplification rate by CISH was detected in studies that used monoclonal antibodies (CB11, SP3 and TAB250), compared to studies that used polyclonal antibodies (A0485 and Herceptest). These results agree with two studies that had used different antibodies for the same sample 15,49. Nunes et al. 49 observed that amplification was detected in 8 and 38% of cases in the IHC 2+ subgroup by polyclonal antibodies (A0485 and Herceptest, respectively) and in 80, 91 and 92% by monoclonal antibodies (NCL-CB11, CM-CB11 and 4D5, respectively). Zhao et al. 15 also reported lower agreement between IHC 2+ and amplified tumors using polyclonal antibodies (A0485, 11%) than monoclonal antibodies (57 and 80% for CB11 and TAB250, respectively).
Table 2. Concordance between immunohistochemistry (IHC) and chromogenic in situ hybridization (CISH) according to the antibodies tested for HER2 staining.
According to Kostopoulou et al. 37, IHC 2+ tumors without gene amplification could indicate that at least some of these cases were either “overstained” or “over-read” in IHC analysis. The authors performed an immunohistochemical reevaluation of IHC 2+ cases and observed a decrease in, but not elimination of, this particular subgroup. Tumor heterogeneity is another plausible explanation for these findings. Kostopoulou et al. 37 reported that approximately 3% of their cases showed heterogeneous immunostaining and hybridization signals. Small areas from the same tumor were observed showing IHC 3+ and amplification next to areas showing IHC 2+ or 0 and non-amplification. The same intratumoral heterogeneity was described by Rosa et al. 55 using CISH, but not in IHC 2+ cases. The authors observed a pool of mixed cells presenting no amplification (2 or 3-5 copies) and high-level amplification in the same area. Hybridization heterogeneity was also observed by FISH in IHC 2+ tumors; the tumors presented low-level amplification or a mosaic mixture of high-level amplified and non-amplified cells 81. The major question is whether cells showing different levels of amplification make any difference or whether a threshold (or its value) percentage of amplified tumor cells is required to define non-amplified and amplified tumors 55.
IHC 0-1+ and 3+ cases. The distribution into subgroups according to IHC status is shown in Figure 1B. Of the 4594 cases scored as 0/1+ by IHC, 96% were confirmed as non-amplified, while 90% of 1612 cases classified as 3+ by IHC showed amplification by CISH (Figure 1B).
The concordance rate between IHC and CISH results varied according to the antibodies used for IHC (Table 2). Correlation between CISH and IHC for almost all antibodies was above 93 and 91% for 0/1+ and 3+ immunostaining, respectively. Breast tumors IHC 0/1+ evaluated by the SP3 antibody were non-amplified in 86% of the samples, while Herceptest 3+ was associated with amplification in 89% of the cases. Higher agreement values were observed for SP3 and Herceptest antibodies in the IHC 3+ and 0/1+ groups, respectively. Three studies 49,53,55 used the SP3 antibody in a total of 282 cases. Two of these used tissue microarray and observed a correlation between CISH and IHC results of 84 and 93% 53 and 93 and 100% 49 for 0/1+ and 3+ immunostaining, respectively. Rosa et al. 55 showed agreement in 85 and 100% of cases presenting IHC 0/1+ and 3+, respectively. Putative explanations for SP3 false-negative cases are the loss of the extracellular domain due to HER2 cleavage and the shedding of the SP3 binding site, despite gene amplification by CISH 53 or gene overexpression by qRT-PCR 55.
Herceptest 3+ was detected in 552 cases from 16 studies 14,17,27,29,30,34,38,42,45,46,49,59,60,62-64, in which 60 samples were not amplified by CISH. Nine studies evaluated the same cases by FISH 14,27,29,30,34,42,45,59,64. Among these, 7 presented adequate data that permitted comparison. Of the 23 discordant cases between CISH and IHC, 100% were also non-amplified by FISH, confirming the CISH results. The discrepancy between CISH and IHC 3+ analyzed by Herceptest may be due to false-positive IHC signals, probably due to overstaining 54. Confirmatory evaluation in IHC 3+ cases by CISH or FISH will ensure that patients receive the most appropriate therapeutic approach, avoiding expensive and cardiotoxic treatment for cases presenting IHC 3+ and non-amplified tumors.
CISH compared to FISH. Forty-two studies presenting 4460 breast cancer cases compared CISH to FISH analysis (Table 1). The overall agreement was 96%. Three reports (7%) revealed <90% concordance between the two methodologies 40,50,74. Peiró et al. 50 performed CISH and FISH on 11 paraffin-embedded samples and observed two IHC 2+ cases amplified by FISH, but not by CISH. The authors suggested that CISH could have a lower sensitivity compared to FISH and recommended that IHC 2+ cases without amplification by CISH analysis should be later submitted to the FISH methodology. Gupta et al. 74 observed that most (3/4) low-level amplification cases showed chromosome 17 polysomy by FISH, explaining the presence of more than 5 copies per nucleus. CISH was repeated in one case that presented a weak signal and was non-amplified. The false-positive case of amplification was caused by overinterpretation of the signal in the presence of background, which in this case was due to an endogenous peroxidase.
An agreement ranging from 90 to 95% was observed for 48% of studies, while 45% presented ≥96% concordance. These findings were expected, since CISH has been confirmed by many investigations as a robust and viable test to assess HER2 status 14,25,41,54,55. Moreover, CISH has some relevant advantages over FISH: 1) the analysis is faster; 2) interpretation is performed using equipment that already exists in routine histopathology laboratories, such as a standard light microscope; 3) it permits simultaneous evaluation of copy number alterations, tumor cell and surrounding tissue morphology on the same slide; 4) morphology is easier to analyze, particularly for distinguishing invasive cancer cells and in situ components; 5) permanent staining is produced allowing the samples to be archived indefinitely; 6) CISH is also easier to interpret for pathologists who are not familiar with fluorescence 9,15,18,23,25.
In FISH analysis, tissue morphology and gene amplification are primarily independent because tumor cells for copy number evaluation are based on nuclear DAPI or propidium iodide staining, which does not always permit adequate histopathological evaluation of the cells 14. FISH slides must be stored at a temperature of 4°C or lower and are subject to quenching of the fluorescent signal, whereas CISH-stained slides can be stored in standard slide files and the reaction product is permanent 15.
When grouping the 42 studies, disagreement between CISH and FISH results was observed in 194 cases (4%). Data were available for 132 of these, with 67 being amplified by FISH and not by CISH and the inverse occurring in 65. Explanations for this disagreement are differences in sample preparation 34, scoring system 25,35, tumor heterogeneity 44,75, material thickness 23 or the absence of a specific probe for the chromosome 17 centromere, which would be able to distinguish amplification from chromosomal polysomy by CISH analysis 27,73. A limitation of determining chromosomal polysomy is the time lost to retest the chromosome 17 centromere probe by CISH on a serial section. With the advent of dual-color CISH (dc-CISH), information regarding polysomy and gene amplification has been obtained in a single assay using probes for HER2 and the chromosome 17 centromere and has shown very strong agreement with FISH results 32,44,71,76,77. However, even using dc-CISH, disagreement with FISH results has been observed for high-level amplification cases, because the signal appears as a typical peroxidase-positive ‘cluster', in which the number of gene copies cannot be counted 32,76.
CISH compared to SISH. In a study of 230 breast cancer cases, Francis et al. 17 found a concordance rate of 96% for CISH and SISH using HER2 single-probe analysis and 95% for CISH and SISH with single- and dual-probe analysis, respectively. Similar results were observed by Park et al. 20 in a study in which 96% of 257 cases presented concordant data between CISH and SISH. According to the authors, the advantage of using SISH is the shorter time (6 h) needed to perform the procedure and automation of the method compared to CISH and FISH.
CISH compared to qRT-PCR. Rosa et al. 55 observed a correlation between transcript expression analysis and CISH in 90% of cases. Two discordant samples were amplified by CISH and non-overexpressed by qRT-PCR. This was probably caused by a difference in sample material; paraffin-embedded and fresh tumor tissue for CISH and qRT-PCR, respectively, or by the presence of normal cells. Kostopoulou et al. 37 reported mRNA overexpression in all cases amplified by CISH. In the IHC 2+ group, cases presenting or not polysomy showed similar mean mRNA values, while IHC 2+ cases without amplification or polysomy had mRNA expression values closer to normal samples. High concordance between gene expression and amplification status has also been observed in other studies using FISH 11,37,55.
Conclusion
Since the FDA (September 1998) approval of the use of the monoclonal antibody Tratuzumab for breast cancer treatment in HER2-positive cases, significant improvements in the overall prognosis for patients have been reported. Different methods have been developed to evaluate HER2 copy number alterations, including FISH, CISH, SISH, and qRT-PCR. CISH has been compared to non-routine and routine diagnostic methods, such as IHC, and has been used as an efficient alternative method to FISH for HER2 gene status elucidation.
Acknowledgments
Research supported by FAPESP and CNPq.
References
- 1.Coussens L, Yang-Feng TL, Liao YC, Chen E, Gray A, McGrath J, et al. Tyrosine kinase receptor with extensive homology to EGF receptor shares chromosomal location with neu oncogene. Science. 1985;230:1132–1139. doi: 10.1126/science.2999974. [DOI] [PubMed] [Google Scholar]
- 2.King CR, Kraus MH, Aaronson SA. Amplification of a novel v-erbB-related gene in a human mammary carcinoma. Science. 1985;229:974–976. doi: 10.1126/science.2992089. [DOI] [PubMed] [Google Scholar]
- 3.Ross JS, Fletcher JA. HER-2/neu (c-erb-B2) gene and protein in breast cancer. Am J Clin Pathol. 1999;112:S53–S67. [PubMed] [Google Scholar]
- 4.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 5.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 6.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 7.Carlomagno C, Perrone F, Gallo C, De Laurentiis M, Lauria R, Morabito A, et al. c-erb B2 overexpression decreases the benefit of adjuvant tamoxifen in early-stage breast cancer without axillary lymph node metastases. J Clin Oncol. 1996;14:2702–2708. doi: 10.1200/JCO.1996.14.10.2702. [DOI] [PubMed] [Google Scholar]
- 8.Tetu B, Brisson J, Plante V, Bernard P. p53 and c-erbB-2 as markers of resistance to adjuvant chemotherapy in breast cancer. Mod Pathol. 1998;11:823–830. [PubMed] [Google Scholar]
- 9.Lambros MB, Natrajan R, Reis-Filho JS. Chromogenic and fluorescent in situ hybridization in breast cancer. Hum Pathol. 2007;38:1105–1122. doi: 10.1016/j.humpath.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 10.Bartlett JM, Munro AF, Dunn JA, McConkey C, Jordan S, Twelves CJ, et al. Predictive markers of anthracycline benefit: a prospectively planned analysis of the UK National Epirubicin Adjuvant Trial (NEAT/BR9601) Lancet Oncol. 2010;11:266–274. doi: 10.1016/S1470-2045(10)70006-1. [DOI] [PubMed] [Google Scholar]
- 11.Vanden Bempt I, Vanhentenrijk V, Drijkoningen M, Wlodarska I, Vandenberghe P, De Wolf-Peeters C. Real-time reverse transcription-PCR and fluorescence in situ hybridization are complementary to understand the mechanisms involved in HER-2/neu overexpression in human breast carcinomas. Histopathology. 2005;46:431–441. doi: 10.1111/j.1365-2559.2005.02112.x. [DOI] [PubMed] [Google Scholar]
- 12.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 13.Hicks DG, Tubbs RR. Assessment of the HER2 status in breast cancer by fluorescence in situ hybridization: a technical review with interpretive guidelines. Hum Pathol. 2005;36:250–261. doi: 10.1016/j.humpath.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 14.Hauser-Kronberger C, Dandachi N. Comparison of chromogenic in situ hybridization with other methodologies for HER2 status assessment in breast cancer. J Mol Histol. 2004;35:647–653. doi: 10.1007/s10735-004-2186-6. [DOI] [PubMed] [Google Scholar]
- 15.Zhao J, Wu R, Au A, Marquez A, Yu Y, Shi Z. Determination of HER2 gene amplification by chromogenic in situ hybridization (CISH) in archival breast carcinoma. Mod Pathol. 2002;15:657–665. doi: 10.1038/modpathol.3880582. [DOI] [PubMed] [Google Scholar]
- 16.Kim GY, Oh YL. Chromogenic in situ hybridization analysis of HER-2/neu status in cytological samples of breast carcinoma. Cytopathology. 2004;15:315–320. doi: 10.1111/j.1365-2303.2004.00214.x. [DOI] [PubMed] [Google Scholar]
- 17.Francis GD, Jones MA, Beadle GF, Stein SR. Bright-field in situ hybridization for HER2 gene amplification in breast cancer using tissue microarrays: correlation between chromogenic (CISH) and automated silver-enhanced (SISH) methods with patient outcome. Diagn Mol Pathol. 2009;18:88–95. doi: 10.1097/PDM.0b013e31816f6374. [DOI] [PubMed] [Google Scholar]
- 18.Tanner M, Gancberg D, Di Leo A, Larsimont D, Rouas G, Piccart MJ, et al. Chromogenic in situ hybridization: a practical alternative for fluorescence in situ hybridization to detect HER-2/neu oncogene amplification in archival breast cancer samples. Am J Pathol. 2000;157:1467–1472. doi: 10.1016/S0002-9440(10)64785-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dietel M, Ellis IO, Hofler H, Kreipe H, Moch H, Dankof A, et al. Comparison of automated silver enhanced in situ hybridisation (SISH) and fluorescence ISH (FISH) for the validation of HER2 gene status in breast carcinoma according to the guidelines of the American Society of Clinical Oncology and the College of American Pathologists. Virchows Arch. 2007;451:19–25. doi: 10.1007/s00428-007-0424-5. [DOI] [PubMed] [Google Scholar]
- 20.Park K, Han S, Kim JY, Kim HJ, Kwon JE, Gwak G. Silver-enhanced in situ hybridization as an alternative to fluorescence in situ hybridization for assaying HER2 amplification in clinical breast cancer. J Breast Cancer. 2011;14:276–282. doi: 10.4048/jbc.2011.14.4.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cell Markers and Cytogenetics Committees College of American Pathologists. Clinical laboratory assays for HER-2/neu amplification and overexpression: quality assurance, standardization, and proficiency testing. Arch Pathol Lab Med. 2002;126:803–808. doi: 10.5858/2002-126-0803-CLAFHN. [DOI] [PubMed] [Google Scholar]
- 22.Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech. 2004;15:155–166. [PMC free article] [PubMed] [Google Scholar]
- 23.Arnould L, Denoux Y, MacGrogan G, Penault-Llorca F, Fiche M, Treilleux I, et al. Agreement between chromogenic in situ hybridisation (CISH) and FISH in the determination of HER2 status in breast cancer. Br J Cancer. 2003;88:1587–1591. doi: 10.1038/sj.bjc.6600943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bassarova AV, Nesland JM, Sedloev T, Lilleby W, Hristova SL, Trifonov DY, et al. Simultaneous bilateral breast carcinomas: a category with frequent coexpression of HER-2 and ER-alpha, high Ki-67 and bcl-2, and low p53. Int J Surg Pathol. 2005;13:239–246. doi: 10.1177/106689690501300302. [DOI] [PubMed] [Google Scholar]
- 25.Bilous M, Morey A, Armes J, Cummings M, Francis G. Chromogenic in situ hybridisation testing for HER2 gene amplification in breast cancer produces highly reproducible results concordant with fluorescence in situ hybridisation and immunohistochemistry. Pathology. 2006;38:120–124. doi: 10.1080/00313020600561518. [DOI] [PubMed] [Google Scholar]
- 26.Bozhanov SS, Angelova SG, Krasteva ME, Markov TL, Christova SL, Gavrilov IG, et al. Alterations in p53, BRCA1, ATM, PIK3CA, and HER2 genes and their effect in modifying clinicopathological characteristics and overall survival of Bulgarian patients with breast cancer. J Cancer Res Clin Oncol. 2010;136:1657–1669. doi: 10.1007/s00432-010-0824-9. [DOI] [PubMed] [Google Scholar]
- 27.Cayre A, Mishellany F, Lagarde N, Penault-Llorca F. Comparison of different commercial kits for HER2 testing in breast cancer: looking for the accurate cutoff for amplification. Breast Cancer Res. 2007;9:R64. doi: 10.1186/bcr1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang E, Lee A, Lee E, Lee H, Shin O, Oh S, et al. HER-2/neu oncogene amplification by chromogenic in situ hybridization in 130 breast cancers using tissue microarray and clinical follow-up studies. J Korean Med Sci. 2004;19:390–396. doi: 10.3346/jkms.2004.19.3.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dandachi N, Dietze O, Hauser-Kronberger C. Chromogenic in situ hybridization: a novel approach to a practical and sensitive method for the detection of HER2 oncogene in archival human breast carcinoma. Lab Invest. 2002;82:1007–1014. doi: 10.1097/01.lab.0000024360.48464.a4. [DOI] [PubMed] [Google Scholar]
- 30.Di Palma S, Collins N, Faulkes C, Ping B, Ferns G, Haagsma B, et al. Chromogenic in situ hybridisation (CISH) should be an accepted method in the routine diagnostic evaluation of HER2 status in breast cancer. J Clin Pathol. 2007;60:1067–1068. doi: 10.1136/jcp.2006.043356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Palma S, Collins N, Bilous M, Sapino A, Mottolese M, Kapranos N, et al. A quality assurance exercise to evaluate the accuracy and reproducibility of chromogenic in situ hybridisation for HER2 analysis in breast cancer. J Clin Pathol. 2008;61:757–760. doi: 10.1136/jcp.2007.053850. [DOI] [PubMed] [Google Scholar]
- 32.García-Caballero T, Grabau D, Green AR, Gregory J, Schad A, Kohlwes E, et al. Determination of HER2 amplification in primary breast cancer using dual-colour chromogenic in situ hybridization is comparable to fluorescence in situ hybridization: a European multicentre study involving 168 specimens. Histopathology. 2010;56:472–480. doi: 10.1111/j.1365-2559.2010.03503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong Y, Sweet W, Duh YJ, Greenfield L, Tarco E, Trivedi S, et al. Performance of chromogenic in situ hybridization on testing HER2 Status in breast carcinomas with chromosome 17 polysomy and equivocal (2+) herceptest results: a study of two institutions using the conventional and new ASCO/CAP scoring criteria. Am J Clin Pathol. 2009;132:228–236. doi: 10.1309/AJCP4M2VUZCLDALN. [DOI] [PubMed] [Google Scholar]
- 34.Hanna WM, Kwok K. Chromogenic in-situ hybridization: a viable alternative to fluorescence in-situ hybridization in the HER2 testing algorithm. Mod Pathol. 2006;19:481–487. doi: 10.1038/modpathol.3800555. [DOI] [PubMed] [Google Scholar]
- 35.Hyun CL, Lee HE, Kim KS, Kim SW, Kim JH, Choe G, et al. The effect of chromosome 17 polysomy on HER-2/neu status in breast cancer. J Clin Pathol. 2008;61:317–321. doi: 10.1136/jcp.2007.050336. [DOI] [PubMed] [Google Scholar]
- 36.Kapila K, Al-Awadhi S, Francis I. Her-2 neu (Cerb-B2) expression in fine needle aspiration samples of breast carcinoma: A pilot study comparing FISH, CISH and immunocytochemistry. J Cytol. 2011;28:54–56. doi: 10.4103/0970-9371.80731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kostopoulou E, Vageli D, Kaisaridou D, Nakou M, Netsika M, Vladica N, et al. Comparative evaluation of non-informative HER-2 immunoreactions (2+) in breast carcinomas with FISH, CISH and QRT-PCR. Breast. 2007;16:615–624. doi: 10.1016/j.breast.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 38.Kumamoto H, Sasano H, Taniguchi T, Suzuki T, Moriya T, Ichinohasama R. Chromogenic in situ hybridization analysis of HER-2/neu status in breast carcinoma: application in screening of patients for trastuzumab (Herceptin) therapy. Pathol Int. 2001;51:579–584. doi: 10.1046/j.1440-1827.2001.01255.x. [DOI] [PubMed] [Google Scholar]
- 39.Leong AS, Formby M, Haffajee Z, Clarke M, Morey A. Refinement of immunohistologic parameters for Her2/neu scoring validation by FISH and CISH. Appl Immunohistochem Mol Morphol. 2006;14:384–389. doi: 10.1097/01.pai.0000210415.53493.d4. [DOI] [PubMed] [Google Scholar]
- 40.Leong AS, Haffajee Z, Clarke M. Microwave enhancement of CISH for HER2 oncogene. Appl Immunohistochem Mol Morphol. 2007;15:88–93. doi: 10.1097/01.pai.0000209864.35828.13. [DOI] [PubMed] [Google Scholar]
- 41.Li Ning TE, Ronchetti R, Torres-Cabala C, Merino MJ. Role of chromogenic in situ hybridization (CISH) in the evaluation of HER2 status in breast carcinoma: comparison with immunohistochemistry and FISH. Int J Surg Pathol. 2005;13:343–351. doi: 10.1177/106689690501300406. [DOI] [PubMed] [Google Scholar]
- 42.Loring P, Cummins R, O'Grady A, Kay EW. HER2 positivity in breast carcinoma: a comparison of chromogenic in situ hybridization with fluorescence in situ hybridization in tissue microarrays, with targeted evaluation of intratumoral heterogeneity by in situ hybridization. Appl Immunohistochem Mol Morphol. 2005;13:194–200. doi: 10.1097/01.pai.0000132189.01233.6d. [DOI] [PubMed] [Google Scholar]
- 43.Madrid MA, Lo RW. Chromogenic in situ hybridization (CISH): a novel alternative in screening archival breast cancer tissue samples for HER-2/neu status. Breast Cancer Res. 2004;6:R593–R600. doi: 10.1186/bcr915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayr D, Heim S, Weyrauch K, Zeindl-Eberhart E, Kunz A, Engel J, et al. Chromogenic in situ hybridization for Her-2/neu-oncogene in breast cancer: comparison of a new dual-colour chromogenic in situ hybridization with immunohistochemistry and fluorescence in situ hybridization. Histopathology. 2009;55:716–723. doi: 10.1111/j.1365-2559.2009.03427.x. [DOI] [PubMed] [Google Scholar]
- 45.Moelans CB, de Weger RA, van Blokland MT, Ezendam C, Elshof S, Tilanus MG, et al. HER-2/neu amplification testing in breast cancer by multiplex ligation-dependent probe amplification in comparison with immunohistochemistry and in situ hybridization. Cell Oncol. 2009;31:1–10. doi: 10.3233/CLO-2009-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moelans CB, Kibbelaar RE, van den Heuvel MC, Castigliego D, de Weger RA, van Diest PJ. Validation of a fully automated HER2 staining kit in breast cancer. Cell Oncol. 2010;32:149–155. doi: 10.3233/CLO-2010-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ni R, Mulligan AM, Have C, O'Malley FP. PGDS, a novel technique combining chromogenic in situ hybridization and immunohistochemistry for the assessment of ErbB2 (HER2/neu) status in breast cancer. Appl Immunohistochem Mol Morphol. 2007;15:316–324. doi: 10.1097/01.pai.0000213138.01536.2e. [DOI] [PubMed] [Google Scholar]
- 48.Ntoulia M, Kaklamanis L, Valavanis C, Kafousi M, Stathopoulos E, Arapantoni P, et al. HER-2 DNA quantification of paraffin-embedded breast carcinomas with LightCycler real-time PCR in comparison to immunohistochemistry and chromogenic in situ hybridization. Clin Biochem. 2006;39:942–946. doi: 10.1016/j.clinbiochem.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 49.Nunes CB, Rocha RM, Reis-Filho JS, Lambros MB, Rocha GF, Sanches FS, et al. Comparative analysis of six different antibodies against Her2 including the novel rabbit monoclonal antibody (SP3) and chromogenic in situ hybridisation in breast carcinomas. J Clin Pathol. 2008;61:934–938. doi: 10.1136/jcp.2007.053892. [DOI] [PubMed] [Google Scholar]
- 50.Peiró G, Mayr D, Hillemanns P, Lohrs U, Diebold J. Analysis of HER-2/neu amplification in endometrial carcinoma by chromogenic in situ hybridization. Correlation with fluorescence in situ hybridization, HER-2/neu, p53 and Ki-67 protein expression, and outcome. Mod Pathol. 2004;17:227–287. doi: 10.1038/modpathol.3800006. [DOI] [PubMed] [Google Scholar]
- 51.Peiró G, Aranda FI, Adrover E, Niveiro M, Alenda C, Paya A, et al. Analysis of HER2 by chromogenic in situ hybridization and immunohistochemistry in lymph node-negative breast carcinoma: Prognostic relevance. Hum Pathol. 2007;38:26–34. doi: 10.1016/j.humpath.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 52.Pothos A, Plastira K, Plastiras A, Vlachodimitropoulos D, Goutas N, Angelopoulou R. Comparison of chromogenic in situ hybridisation with fluorescence in situ hybridisation and immunohistochemistry for the assessment of her-2/neu oncogene in archival material of breast carcinoma. Acta Histochem Cytochem. 2008;41:59–64. doi: 10.1267/ahc.07029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ricardo SA, Milanezi F, Carvalho ST, Leitão DR, Schmitt FC. HER2 evaluation using the novel rabbit monoclonal antibody SP3 and CISH in tissue microarrays of invasive breast carcinomas. J Clin Pathol. 2007;60:1001–1005. doi: 10.1136/jcp.2006.040287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riethdorf S, Hoegel B, John B, Ott G, Fritz P, Thon S, et al. Prospective multi-centre study to validate chromogenic in situ hybridisation for the assessment of HER2 gene amplification in specimens from adjuvant and metastatic breast cancer patients. J Cancer Res Clin Oncol. 2011;137:261–269. doi: 10.1007/s00432-010-0881-0. [DOI] [PubMed] [Google Scholar]
- 55.Rosa FE, Silveira SM, Silveira CG, Bergamo NA, Neto FA, Domingues MA, et al. Quantitative real-time RT-PCR and chromogenic in situ hybridization: precise methods to detect HER-2 status in breast carcinoma. BMC Cancer. 2009;9:90. doi: 10.1186/1471-2407-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saéz A, Andreu FJ, Segui MA, Bare ML, Fernandez S, Dinares C, et al. HER-2 gene amplification by chromogenic in situ hybridisation (CISH) compared with fluorescence in situ hybridisation (FISH) in breast cancer - A study of two hundred cases. Breast. 2006;15:519–527. doi: 10.1016/j.breast.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 57.Sapino A. [Diagnostic process in breast carcinoma: role of core biopsy and needle aspiration of axillary lymph nodes] Pathologica. 2003;95:258–259. [PubMed] [Google Scholar]
- 58.Sartelet H, Lagonotte E, Lorenzato M, Duval I, Lechki C, Rigaud C, et al. Comparison of liquid based cytology and histology for the evaluation of HER-2 status using immunostaining and CISH in breast carcinoma. J Clin Pathol. 2005;58:864–871. doi: 10.1136/jcp.2004.024224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siñczak-Kuta A, Tomaszewska R, Rudnicka-Sosin L, Okon K, Stachura J. Evaluation of HER2/neu gene amplification in patients with invasive breast carcinoma. Comparison of in situ hybridization methods. Pol J Pathol. 2007;58:41–50. [PubMed] [Google Scholar]
- 60.Sumiyoshi K, Shibayama Y, Akashi S, Nohara T, Iwamoto M, Kobayashi T, et al. Detection of human epidermal growth factor receptor 2 protein and gene in fine needle aspiration cytology specimens and tissue sections from invasive breast cancer: can cytology specimens take the place of tissue sections? Oncol Rep. 2006;15:803–808. [PubMed] [Google Scholar]
- 61.Tanner M, Jarvinen P, Isola J. Amplification of HER-2/neu and topoisomerase IIalpha in primary and metastatic breast cancer. Cancer Res. 2001;61:5345–5348. [PubMed] [Google Scholar]
- 62.Todorovic-Rakovic N, Jovanovic D, Neskovic-Konstantinovic Z, Nikolic-Vukosavljevic D. Comparison between immunohistochemistry and chromogenic in situ hybridization in assessing HER-2 status in breast cancer. Pathol Int. 2005;55:318–323. doi: 10.1111/j.1440-1827.2005.01831.x. [DOI] [PubMed] [Google Scholar]
- 63.Todorovic-Rakovic N, Jovanovic D, Neskovic-Konstantinovic Z, Nikolic-Vukosavljevic D. Prognostic value of HER2 gene amplification detected by chromogenic in situ hybridization (CISH) in metastatic breast cancer. Exp Mol Pathol. 2007;82:262–268. doi: 10.1016/j.yexmp.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 64.van de Vijver M, Bilous M, Hanna W, Hofmann M, Kristel P, Penault-Llorca F, et al. Chromogenic in situ hybridisation for the assessment of HER2 status in breast cancer: an international validation ring study. Breast Cancer Res. 2007;9:R68. doi: 10.1186/bcr1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van der Vegt B, de Bock GH, Bart J, Zwartjes NG, Wesseling J. Validation of the 4B5 rabbit monoclonal antibody in determining Her2/neu status in breast cancer. Mod Pathol. 2009;22:879–886. doi: 10.1038/modpathol.2009.37. [DOI] [PubMed] [Google Scholar]
- 66.Vera-Roman JM, Rubio-Martinez LA. Comparative assays for the HER-2/neu oncogene status in breast cancer. Arch Pathol Lab Med. 2004;128:627–633. doi: 10.5858/2004-128-627-CAFTNO. [DOI] [PubMed] [Google Scholar]
- 67.Vocaturo A, Novelli F, Benevolo M, Piperno G, Marandino F, Cianciulli AM, et al. Chromogenic in situ hybridization to detect HER-2/neu gene amplification in histological and ThinPrep-processed breast cancer fine-needle aspirates: a sensitive and practical method in the trastuzumab era. Oncologist. 2006;11:878–886. doi: 10.1634/theoncologist.11-8-878. [DOI] [PubMed] [Google Scholar]
- 68.Wixom CR, Albers EA, Weidner N. Her2 amplification: correlation of chromogenic in situ hybridization with immunohistochemistry and fluorescence in situ hybridization. Appl Immunohistochem Mol Morphol. 2004;12:248–251. doi: 10.1097/00129039-200409000-00011. [DOI] [PubMed] [Google Scholar]
- 69.Arena V, Pennacchia I, Vecchio FM, Carbone A. “CISH the FISH” for HER2: our laboratory experience. Am J Clin Pathol. 2010;134:347–348. doi: 10.1309/AJCPVH1IGEG8CPXB. [DOI] [PubMed] [Google Scholar]
- 70.Bartlett JM, Campbell FM, Ibrahim M, Wencyk P, Ellis I, Kay E, et al. Chromogenic in situ hybridization: a multicenter study comparing silver in situ hybridization with FISH. Am J Clin Pathol. 2009;132:514–520. doi: 10.1309/AJCPXY3MJ6GSRCYP. [DOI] [PubMed] [Google Scholar]
- 71.Bartlett JM, Campbell FM, Ibrahim M, O'Grady A, Kay E, Faulkes C, et al. A UK NEQAS ISH multicenter ring study using the Ventana HER2 dual-color ISH assay. Am J Clin Pathol. 2011;135:157–162. doi: 10.1309/AJCPVPRKK1ENEDGQ. [DOI] [PubMed] [Google Scholar]
- 72.Bhargava R, Lal P, Chen B. Chromogenic in situ hybridization for the detection of HER-2/neu gene amplification in breast cancer with an emphasis on tumors with borderline and low-level amplification: does it measure up to fluorescence in situ hybridization? Am J Clin Pathol. 2005;123:237–243. doi: 10.1309/C4PEBGB9LN830TVL. [DOI] [PubMed] [Google Scholar]
- 73.Gong Y, Gilcrease M, Sneige N. Reliability of chromogenic in situ hybridization for detecting HER-2 gene status in breast cancer: comparison with fluorescence ihybridization and assessment of interobserver reproducibility. Mod Pathol. 2005;18:1015–1021. doi: 10.1038/modpathol.3800432. [DOI] [PubMed] [Google Scholar]
- 74.Gupta D, Middleton LP, Whitaker MJ, Abrams J. Comparison of fluorescence and chromogenic in situ hybridization for detection of HER-2/neu oncogene in breast cancer. Am J Clin Pathol. 2003;119:381–387. doi: 10.1309/P40P2EAD42PUKDMG. [DOI] [PubMed] [Google Scholar]
- 75.Isola J, Tanner M, Forsyth A, Cooke TG, Watters AD, Bartlett JM. Interlaboratory comparison of HER-2 oncogene amplification as detected by chromogenic and fluorescence in situ hybridization. Clin Cancer Res. 2004;10:4793–4798. doi: 10.1158/1078-0432.CCR-0428-03. [DOI] [PubMed] [Google Scholar]
- 76.Kato N, Itoh H, Serizawa A, Hatanaka Y, Umemura S, Osamura RY. Evaluation of HER2 gene amplification in invasive breast cancer using a dual-color chromogenic in situ hybridization (dual CISH) Pathol Int. 2010;60:510–515. doi: 10.1111/j.1440-1827.2010.02553.x. [DOI] [PubMed] [Google Scholar]
- 77.Laakso M, Tanner M, Isola J. Dual-colour chromogenic in situ hybridization for testing of HER-2 oncogene amplification in archival breast tumours. J Pathol. 2006;210:3–9. doi: 10.1002/path.2022. [DOI] [PubMed] [Google Scholar]
- 78.Lin F, Shen T, Prichard JW. Detection of Her-2/neu oncogene in breast carcinoma by chromogenic in situ hybridization in cytologic specimens. Diagn Cytopathol. 2005;33:376–380. doi: 10.1002/dc.20401. [DOI] [PubMed] [Google Scholar]
- 79.Park K, Kim J, Lim S, Han S. Topoisomerase II-alpha (topoII) and HER2 amplification in breast cancers and response to preoperative doxorubicin chemotherapy. Eur J Cancer. 2003;39:631–634. doi: 10.1016/S0959-8049(02)00745-1. [DOI] [PubMed] [Google Scholar]
- 80.McCormick SR, Lillemoe TJ, Beneke J, Schrauth J, Reinartz J. HER2 assessment by immunohistochemical analysis and fluorescence in situ hybridization: comparison of HercepTest and PathVysion commercial assays. Am J Clin Pathol. 2002;117:935–943. doi: 10.1309/3643-F955-7Q6B-EWWL. [DOI] [PubMed] [Google Scholar]
- 81.Lewis JT, Ketterling RP, Halling KC, Reynolds C, Jenkins RB, Visscher DW. Analysis of intratumoral heterogeneity and amplification status in breast carcinomas with equivocal (2+) HER-2 immunostaining. Am J Clin Pathol. 2005;124:273–281. doi: 10.1309/J9VXABUGKC4Y07DL. [DOI] [PubMed] [Google Scholar]