Abstract
The multidrug resistance 1 gene (MDR1) is an important candidate gene for influencing susceptibility to hepatocellular carcinoma (HCC). The objective of the present study was to evaluate the association of MDR1 polymorphisms with the risk of HCC in the Chinese Han population. A total of 353 HCC patients and 335 healthy subjects were enrolled in the study. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), created restriction site-PCR (CRS-PCR) and DNA sequencing methods were used to identify MDR1 gene polymorphisms. Two allelic variants (c.335T>C and c.3073A>C) were detected. The CC genotype of the c.335T>C polymorphism was associated with an increased risk of developing HCC compared to the TT genotype (OR = 2.161, 95%CI = 1.350-3.459, χ2 = 10.55, P = 0.0011). The risk of HCC was significantly higher for the CC genotype in the c.3073A>C polymorphism compared to the AA genotype in the studied populations (CC vs AA: OR = 2.575, 95%CI = 1.646-4.028, χ2 = 17.64, P < 0.0001). The C allele of the c.335T>C and c.3073A>C variants may contribute to the risk of HCC (C vs T of c.335T>C: OR = 1.512, 95%CI = 1.208-1.893, χ2 = 13.07, P = 0.0003, and C vs A of c.3073A>C: OR = 1.646, 95%CI = 1.322-2.049, χ2 = 20.03, P < 0.0001). The c.335T>C and c.3073A>C polymorphisms of the MDR1 gene were associated with the risk of occurrence of HCC in the Chinese Han population. Further investigations are needed to confirm these results in larger different populations.
Keywords: Hepatocellular carcinoma, Multidrug resistance 1 gene, Single nucleotide polymorphisms, Susceptibility, Risk factors
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common solid tumor and the third leading cause of cancer-related deaths worldwide 1-3. The estimated incidence of new HCC cases is approximately 500,000–1,000,000, causing 600,000 deaths globally each year 1. HCC shows significantly geographic variation, with a very high incidence in China corresponding to approximately 55% of annual new cases of HCC worldwide 1,4. HCC has been one of the most common causes of cancer-related deaths in China since the 1990s 5,6. Many environmental risk factors are associated with HCC, including chronic hepatitis B (HBV) or hepatitis C viral infections, exposure to dietary aflatoxin B1, cigarette smoking, alcohol consumption, diabetes mellitus, and conditions such as cirrhosis 7-10. Furthermore, it is generally accepted that genetic factors play important roles in the pathogenesis of HCC 11,12. The exact mechanism of hepatocarcinogenesis is still incompletely understood.
The human multidrug resistance 1 (MDR1) gene encodes a 170-kDa transmembrane protein, P-glycoprotein (Pgp), which is a membrane protein acting as an ATP-dependent exporter of xenobiotics from cells 13. This MDR1 protein anchors to the cell membrane and acts as an efflux transporter conferring resistance to a variety of natural cytotoxic drugs and potentially toxic xenobiotics 14-17. It has been reported that MDR1 is an important candidate gene influencing the risk for HCC. Previous studies have mainly focused on the expression of MDR1 (often referred to as Pgp) in HCC cell lines and HCC specimens, with heterogeneous and divergent results. It has been suggested that single nucleotide polymorphisms (SNPs) in the MDR1 gene may have an impact on the expression and function of Pgp, therefore influencing susceptibility to various diseases, including liver cancer 18-25. MDR1 is polymorphic, and at least 50 SNPs have been reported 17,23-30. Previous studies have reported the association of MDR1 gene polymorphisms with HCC risk factors 25,31,32. However, the association of the MDR1 gene c.335T>C and c.3073A>C SNPs with HCC has not been investigated. Thus, in the present study, we indentified these genetic polymorphisms in the MDR1 gene and evaluated their association with the risk of liver cancer in the Chinese population.
Subjects and Methods
Subjects
In this case-control study, 353 patients with HCC were enrolled from January 2008 to December 2011. HCC patients were diagnosed by doctors according to the standards established by the Chinese Society of Liver Cancer (CSLC). The control group consisted of 335 healthy subjects randomly selected from participants in health screening programs to exclude those with a medical history of surgery, cancer and other disease. All subjects were unrelated individuals of the Han nationality. Information was obtained regarding demographic data as well as related risk factors such as gender, age, smoking, drinking, hypertension, diabetes mellitus, serum a-FP levels, family history of HCC, and HBV serological markers [hepatitis-B surface antigen (HBs Ag), hepatitis-B surface antibody (HBs Ab), hepatitis-Be antigen (HBe Ag), antibody to hepatitis-Be antigen (anti-HBe Ab), and antibody to hepatitis-B core antigen (anti-HBc)] (Table 1). The current study was approved by the local Institutional Ethics Committee, and all subjects gave written informed consent to participate.
Table 1. Characteristics of the hepatocellular carcinoma (HCC) patients and controls.

Data are reported as number with percent in parentheses except for mean ± SD age. HBV = hepatitis B virus; HBs Ag = hepatitis B surface antigen; a-FP = alpha-fetoprotein. Patients and controls were compared by the chi-square (χ2) test. There were no statistically significant differences.
Primer design and polymerase chain reaction (PCR) amplification
Genomic DNA was extracted from peripheral venous blood using the standard phenol/chloroform extraction method. According to the DNA sequences (GenBank ID: NG_011513.1) and mRNA sequences (GenBank ID: NM_000927.4) of the MDR1 gene, specific PCR primers were designed using the Primer Premier 5.0 software (PREMIER Biosoft International, Canada). Primers, annealing temperature, region, fragment sizes, and selected restriction enzymes (MBI Fermentas, Germany) are shown in Table 2. PCR was carried out in a total volume of 20 µL containing 50 ng template DNA, 1X buffer (100 mM Tris-HCl, pH 8.3, 500 mM KCl, 0.25 µM primers, 2.0 mM MgCl2, 0.25 mM dNTPs, and 0.5 U Taq DNA polymerase (Promega, USA). The PCR protocol was 94°C for 5 min, followed by 35 cycles at 94°C for 30 s, annealing at the corresponding temperature (shown in Table 2) for 30 s and 72°C for 30 s, and a final extension at 72°C for 8 min. The PCR products were separated on 1.5% agarose gel and photographed under UV light.
Table 2. PCR, PCR-RFLP and CRS-PCR analysis used to genotype SNPs in the MDR1 gene.

PCR = polymerase chain reaction; PCR-RFLP = PCR-restriction fragment length polymorphism; CRS-PCR = created restriction site PCR; SNPs = single nucleotide polymorphisms. Underlined nucleotide indicates nucleotide mismatch enabling the use of the selected restriction enzymes for discriminating sequence variations.
MDR1 genotyping
c.335T>C was investigated using the created restriction site-PCR (CRS-PCR) method with one of the primers containing a nucleotide mismatch, which enables the use of restriction enzymes for discriminating sequence variations 33-35. c.3073A>C was detected by the PCR-restriction fragment length polymorphism (PCR-RFLP) method. Following the supplier's manual, 5-µL aliquots of PCR-amplified products were digested with 2 U restriction enzyme at 37°C for 10 h. The digested products were separated by 2.5% agarose gel electrophoresis and observed directly under UV light in order to determine the genotype of MDR1 polymorphisms. To confirm the genotype results obtained by CRS-PCR and PCR-RFLP, about 20% of PCR-amplified products were randomly selected for DNA sequencing (TaKaRa Biotechnology Co., Ltd., China).
Statistical analyses
All statistical analyses were performed using the Statistical Package for the Social Sciences software (SPSS, Windows version release 15.0; SPSS Inc., USA). The chi-square (χ2) test was applied to evaluate Hardy–Weinberg equilibrium in all individuals, as well as allele and genotype frequencies, and the general characteristics of the groups. Associations between MDR1 gene polymorphisms and HCC risk were estimated using odds ratios (OR) and 95% confidence intervals (95%CI) for the comparison of homozygotes (c.335T>C: CC vs TT, and c.3073A>C: CC vs AA), and heterozygotes (c.335T>C: TC vs TT, and c.3073A>C: AC vs AA), dominant model (c.335T>C: CC+TC vs TT, and c.3073A>C: CC+AC vs AA), recessive model (c.335T>C: CC vs TC+TT, and c.3073A>C: CC vs AC+AA) and allele contrast (c.335T>C: C vs T, and c.3073A>C: C vs A). For all statistical tests, P < 0.05 was defined as statistically significant.
Results
General characteristics of the subjects
A total of 688 subjects were recruited for this study, including 353 HCC patients and 335 controls. No significant differences between the patients and controls in terms of gender and age distribution suggested that matching of subjects based on these variables was adequate. Similarly, there were no significant differences in smoking status or drinking consumption, hypertension or diabetes mellitus between the patients and controls. The general characteristics of the populations are summarized in Table 1.
Detection and genotyping of MDR1 SNPs
In the present study, two allelic variants (c.335T>C and c.3073A>C) were investigated by the CRS-PCR and PCR-RFLP methods, respectively. Sequence analysis suggested that the c.335T>C polymorphism was caused by T to C mutations in the 5′-UTR of the human MDR1 gene. As was the case for the c.3073A>C variant, sequence analysis showed that this allelic variant was caused by A to C mutations in exon 22 of the human MDR1 gene. Furthermore, this polymorphism caused a leucine (Leu) to phenylalanine (Phe) amino acid replacement (p.Leu860Phe, Reference sequence GenBank IDs: NG_011513.1, NM_000927.4 and NP_000918.2). The PCR product of c.335T>C was digested with the NlaIII enzyme and divided into three genotypes: TT (208 bp), TC (208, 185, and 23 bp) and CC (185 and 23 bp; Table 2). The PCR product of c.3073A>C was digested with the MaeIII enzyme and divided into three genotypes: AA (163 and 86 bp), AC (249, 163, and 86 bp) and CC (249 bp; Table 2).
Allelic and genotypic frequencies
The chi-square (χ2) test for the c.335T>C and c.3073A>C variants in the studied subjects suggested that the polymorphic sites were in Hardy-Weinberg equilibrium (P > 0.05; Table 3). The allelic and genotypic frequencies of the c.335T>C and c.3073A>C polymorphisms are presented in Table 3. Allele T and allele A were the predominant alleles in the studied subjects in c.335T>C and c.3073A>C, respectively (Table 3). Regarding the c.335T>C variant, the allelic frequencies of the patients (T = 61.19%; C = 38.81%) were significantly different from those of controls (T = 70.45%; C = 29.55%; χ2 = 13.0743; P = 0.0003). Furthermore, the genotypic frequencies of the patients were not similar to those of the controls, with the differences being statistically significant (χ2 = 11.8639; P = 0.0027; Table 3). Similarly, for the c.3073A>C variant, the allelic frequencies of the patients (A = 55.24%; C = 44.76%) were significantly different from those of the controls (A = 67.01%; C = 32.99%; χ2 = 20.0272; P < 0.0001; Table 3). The genotypic frequencies of the patients were not similar to those of the controls, with the differences being statistically significant (χ2 = 18.4030; P = 0.0001; Table 3).
Table 3. Genotypic and allelic frequencies of c.335T>C and c.3073A>C polymorphisms in the studied subjects.
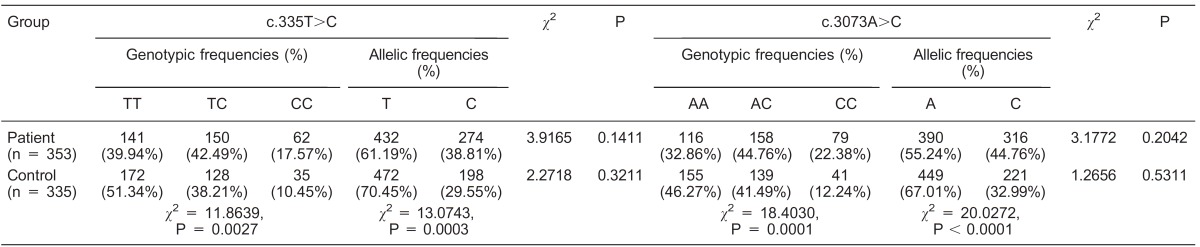
The chi-square (χ2) test was used to evaluate Hardy-Weinberg equilibrium in allele and genotype frequencies in patients and controls.
MDR1 polymorphisms and risk of HCC
Analysis of the association of genotypes/alleles from the c.335T>C and c.3073A>C SNPs with the risk of HCC suggested that there was a significantly increased risk of HCC (Table 4). Regarding the c.335T>C SNPs, a significantly increased risk of liver cancer was found in the homozygote comparison (CC vs TT: OR = 2.161, 95%CI = 1.350-3.459, χ2 = 10.55; P = 0.0011), heterozygote comparison (TC vs TT: OR = 1.430, 95%CI = 1.034-1.977, χ2 = 4.68; P = 0.0310), dominant model (CC+TC vs TT: OR = 1.587, 95%CI = 1.173-2.146, χ2 = 9.01; P = 0.0027), recessive model (CC vs TC+TT: OR = 1.826, 95%CI = 1.171-2.849, χ2 = 7.19; P = 0.0073), and allele contrast (C vs T: OR = 1.512, 95%CI = 1.208-1.893, χ2 = 13.07; P = 0.0003; Table 4). Similarly, there were significant differences in the c.3073A>C SNPs for homozygote comparison (CC vs AA: OR = 2.575, 95%CI = 1.646-4.028, χ2 = 17.64; P < 0.0001), heterozygote comparison (AC vs AA: OR = 1.519, 95%CI = 1.090-2.116, χ2 = 6.13; P = 0.0130), dominant model (CC+AC vs AA: OR = 1.759, 95%CI = 1.292-2.396, χ2 = 12.94; P = 0.0003), recessive model (CC vs AC+AA: OR = 2.067, 95%CI = 1.370-3.120, χ2 = 12.28; P = 0.0005), and allele contrast (C vs A: OR = 1.646, 95%CI = 1.322-2.049, χ2 = 20.03; P < 0.0001; Table 4).
Table 4. Association between hepatocellular carcinoma risk and MDR1 SNPs.

SNPs = single nucleotide polymorphisms. The association of MDR1 gene polymorphisms with hepatocellular carcinoma risk was estimated using odds ratios (OR) and 95% confidence intervals (95%CI) for the comparison of homozygotes, heterozygotes, dominant model, recessive model, and allele contrast by the chi-square (χ2) test.
Discussion
HCC is a common primary malignant tumor of the liver resulting from complex interactions between environmental and genetic factors. Genotypic variation has a key function in human phenotypic variability for cancer susceptibility. The present study provided novel information on the effects of two novel allelic variants (c.335T>C and c.3073A>C) of MDR1 on susceptibility to HCC. We observed a significant difference in allelic and genotypic frequencies between HCC patients and healthy controls (Table 3). Regarding the c.335T>C variants, the CC genotype was strongly associated with an increased risk of developing HCC compared to TT and TC/TT genotypes. The risk of HCC was significantly higher for the CC genotype in the c.3073A>C polymorphism compared to the AA genotype and AC/AA carriers. Thus, the C allele of both c.335T>C and c.3073A>C variants may contribute to the risk of HCC (C vs T of c.335T>C: OR = 1.512, 95%CI = 1.208-1.893; P = 0.0003 and C vs A of c.3073A>C: OR = 1.646, 95%CI = 1.322-2.049; P < 0.0001). The results of the present study suggest that the c.335T>C and c.3073A>C polymorphisms of the MDR1 gene are associated with the risk of occurrence of HCC in the Chinese Han population. Several previous studies have confirmed the relationship between MDR1 polymorphisms and risk factors for HCC 25,31,32. Wu et al. 25 analyzed the association between three polymorphisms (C1236T, G2677A/T, C3435T) of the MDR1 gene and the risk of recurrence after liver transplantation. The association between recurrence-free condition and being a 2677A carrier was significant (P = 0.019), but no significant association was observed with other polymorphisms 25. Wu et al. suggested that polymorphism of the MDR1 gene may be a valuable molecular marker for HCC recurrence after liver transplantation. Chen et al. 31 investigated the association between G2677T/A polymorphisms of the MDR1 gene and the risk of HCC 31 and suggested that 2677A may be an allele protecting against HCC, while 2677T may be a risk gene for HCC 31. Chen et al. 32 detected a correlation of two polymorphisms (C1236T and C3435T) of the MDR1 gene with the prognosis of HCC. The correlation between prognosis of HCC and the C3435T polymorphism was significant (P = 0.037), but no significant correlation was observed for the C1236T polymorphism (P = 0.762) 32.
These findings suggested that the C3435T could be a positive candidate molecular marker for the prognosis of HCC 32. To the best of our knowledge, this is the first report regarding the association of the MDR1 gene c.335T>C and c.3073A>C SNPs with the risk of HCC. The findings could provide new evidence for further analysis of the importance of the MDR1 polymorphism for the risk of HCC. Larger prospective studies will be needed to confirm these results in different populations.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–1917. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 3.Parikh S, Hyman D. Hepatocellular cancer: a guide for the internist. Am J Med. 2007;120:194–202. doi: 10.1016/j.amjmed.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 4.Schutte K, Bornschein J, Malfertheiner P. Hepatocellular carcinoma - epidemiological trends and risk factors. Dig Dis. 2009;27:80–92. doi: 10.1159/000218339. [DOI] [PubMed] [Google Scholar]
- 5.Chen JG, Zhang SW, Chen WQ. [Analysis of liver cancer mortality in the national retrospective sampling survey of death causes in China, 2. Zhonghua Yu Fang Yi Xue Za Zhi. 2010;44:383–389. [PubMed] [Google Scholar]
- 6.Zeng X, Liu S, Yu H, Ji L, Li L, Huang J, et al. DNA repair capacity, DNA-strand break repair gene polymorphisms, and the incidence of hepatocellular carcinoma in southwestern Guangxi of China. DNA Cell Biol. 2012;31:1384–1391. doi: 10.1089/dna.2012.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 8.Suriawinata A, Xu R. An update on the molecular genetics of hepatocellular carcinoma. Semin Liver Dis. 2004;24:77–88. doi: 10.1055/s-2004-860865. [DOI] [PubMed] [Google Scholar]
- 9.Bosch FX, Ribes J, Diaz M, Cleries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 10.Gomaa AI, Khan SA, Toledano MB, Waked I, Taylor-Robinson SD. Hepatocellular carcinoma: epidemiology, risk factors and pathogenesis. World J Gastroenterol. 2008;14:4300–4308. doi: 10.3748/wjg.14.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nault JC, Zucman-Rossi J. Genetics of hepatobiliary carcinogenesis. Semin Liver Dis. 2011;31:173–187. doi: 10.1055/s-0031-1276646. [DOI] [PubMed] [Google Scholar]
- 12.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 13.Fromm MF. Genetically determined differences in P-glycoprotein function: implications for disease risk. Toxicology. 2002;181-182:299–303. doi: 10.1016/S0300-483X(02)00297-4. [DOI] [PubMed] [Google Scholar]
- 14.Gervasini G, Carrillo Jose A, Garcia M, San Jose C, Cabanillas A, Benitez J. Adenosine triphosphate-binding cassette B1 (ABCB1) (multidrug resistance 1) G2677T/A gene polymorphism is associated with high risk of lung cancer. Cancer. 2006;107:2850–2857. doi: 10.1002/cncr.22332. [DOI] [PubMed] [Google Scholar]
- 15.Bodor M, Kelly EJ, Ho RJ. Characterization of the human MDR1 gene. AAPS J. 2005;7:E1–E5. doi: 10.1208/aapsj070101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borst P, Elferink RO. Mammalian ABC transporters in health and disease. Annu Rev Biochem. 2002;71:537–592. doi: 10.1146/annurev.biochem.71.102301.093055. [DOI] [PubMed] [Google Scholar]
- 17.Ambudkar SV, Kimchi-Sarfaty C, Sauna ZE, Gottesman MM. P-glycoprotein: from genomics to mechanism. Oncogene. 2003;22:7468–7485. doi: 10.1038/sj.onc.1206948. [DOI] [PubMed] [Google Scholar]
- 18.Jamroziak K, Mlynarski W, Balcerczak E, Mistygacz M, Trelinska J, Mirowski M, et al. Functional C3435T polymorphism of MDR1 gene: an impact on genetic susceptibility and clinical outcome of childhood acute lymphoblastic leukemia. Eur J Haematol. 2004;72:314–321. doi: 10.1111/j.1600-0609.2004.00228.x. [DOI] [PubMed] [Google Scholar]
- 19.Leonessa F, Clarke R. ATP binding cassette transporters and drug resistance in breast cancer. Endocr Relat Cancer. 2003;10:43–73. doi: 10.1677/erc.0.0100043. [DOI] [PubMed] [Google Scholar]
- 20.Kurzawski M, Drozdzik M, Suchy J, Kurzawski G, Bialecka M, Gornik W, et al. Polymorphism in the P-glycoprotein drug transporter MDR1 gene in colon cancer patients. Eur J Clin Pharmacol. 2005;61:389–394. doi: 10.1007/s00228-005-0926-5. [DOI] [PubMed] [Google Scholar]
- 21.Sohn JW, Lee SY, Lee SJ, Kim EJ, Cha SI, Kim CH, et al. MDR1 polymorphisms predict the response to etoposide-cisplatin combination chemotherapy in small cell lung cancer. Jpn J Clin Oncol. 2006;36:137–141. doi: 10.1093/jjco/hyi231. [DOI] [PubMed] [Google Scholar]
- 22.Vander Borght S, Komuta M, Libbrecht L, Katoonizadeh A, Aerts R, Dymarkowski S, et al. Expression of multidrug resistance-associated protein 1 in hepatocellular carcinoma is associated with a more aggressive tumour phenotype and may reflect a progenitor cell origin. Liver Int. 2008;28:1370–1380. doi: 10.1111/j.1478-3231.2008.01889.x. [DOI] [PubMed] [Google Scholar]
- 23.Taniguchi S, Mochida Y, Uchiumi T, Tahira T, Hayashi K, Takagi K, et al. Genetic polymorphism at the 5′ regulatory region of multidrug resistance 1 (MDR1) and its association with interindividual variation of expression level in the colon. Mol Cancer Ther. 2003;2:1351–1359. [PubMed] [Google Scholar]
- 24.Yu X, Xie H, Wei B, Zhang M, Wang W, Wu J, et al. Association of MDR1 gene SNPs and haplotypes with the tacrolimus dose requirements in Han Chinese liver transplant recipients. PLoS One. 2011;6:e25933. doi: 10.1371/journal.pone.0025933. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Wu L, Xu X, Shen J, Xie H, Yu S, Liang T, et al. MDR1 gene polymorphisms and risk of recurrence in patients with hepatocellular carcinoma after liver transplantation. J Surg Oncol. 2007;96:62–68. doi: 10.1002/jso.20774. [DOI] [PubMed] [Google Scholar]
- 26.Cavaco I, Gil JP, Gil-Berglund E, Ribeiro V. CYP3A4 and MDR1 alleles in a Portuguese population. Clin Chem Lab Med. 2003;41:1345–1350. doi: 10.1515/CCLM.2003.206. [DOI] [PubMed] [Google Scholar]
- 27.Chinn LW, Kroetz DL. ABCB1 pharmacogenetics: progress, pitfalls, and promise. Clin Pharmacol Ther. 2007;81:265–269. doi: 10.1038/sj.clpt.6100052. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmeyer S, Burk O, von Richter O, Arnold HP, Brockmoller J, Johne A, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo . Proc Natl Acad Sci U S A. 2000;97:3473–3478. doi: 10.1073/pnas.97.7.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaya P, Gunduz U, Arpaci F, Ural AU, Guran S. Identification of polymorphisms on the MDR1 gene among Turkish population and their effects on multidrug resistance in acute leukemia patients. Am J Hematol. 2005;80:26–34. doi: 10.1002/ajh.20427. [DOI] [PubMed] [Google Scholar]
- 30.Pechandova K, Buzkova H, Slanar O, Perlik F. Polymorphisms of the MDR1 gene in the Czech population. Folia Biol. 2006;52:184–189. [PubMed] [Google Scholar]
- 31.Chen YD, Yang F, Feng ST, Zhang YX. A case-control study on the association between genetic polymorphisms of MDR1 and hepatic cell cancer susceptibility. Chin Clin Oncol. 2009;14:1077–1081. [Google Scholar]
- 32.Chen XJ, Wang XG, Shen YJ, Zhou HK, Ni QF, Fei JG, et al. Correlation of MDR1 single nucleotide polymorphism with prognosis of hepatocellular carcinoma. J Chin Oncol. 2011;17:209–211. [Google Scholar]
- 33.Yuan ZR, Li JY, Li J, Zhang L, Gao X, Gao HJ, et al. Investigation on BRCA1 SNPs and its effects on mastitis in Chinese commercial cattle. Gene. 2012;505:190–194. doi: 10.1016/j.gene.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 34.Yuan ZR, Li J, Li JY, Gao X, Xu SZ. SNPs identification and its correlation analysis with milk somatic cell score in bovine MBL1 gene. Mol Biol Rep. 2013;40:7–12. doi: 10.1007/s11033-012-1934-z. [DOI] [PubMed] [Google Scholar]
- 35.Yuan ZR, Li JY, Li J, Gao X, Gao HJ, Xu SZ. Effects of DGAT1 gene on meat and carcass fatness quality in Chinese commercial cattle. Mol Biol Rep. 2013;40:1947–1954. doi: 10.1007/s11033-012-2251-2. [DOI] [PubMed] [Google Scholar]


