This paper describes an enzymatic analysis of RNA terminal phosphate cyclase. This enzyme was previously thought to only act on RNAs terminated with a 3′-phosphate, but the authors demonstrate that it can cyclize 2′-phosphates as well, albeit at a much slower rate. The results provide a plausible explanation for the organization of bacterial operons which contain both the cyclase and an RNA ligase.
Keywords: 2′,3′-cyclic phosphate ends; RNA repair
Abstract
RNA terminal phosphate cyclase catalyzes the ATP-dependent conversion of a 3′-phosphate RNA end to a 2′,3′-cyclic phosphate via covalent enzyme-(histidinyl-Nϵ)-AMP and RNA(3′)pp(5′)A intermediates. Here, we report that Escherichia coli RtcA (and its human homolog Rtc1) are capable of cyclizing a 2′-phosphate RNA end in high yield. The rate of 2′-phosphate cyclization by RtcA is five orders of magnitude slower than 3′-phosphate cyclization, notwithstanding that RtcA binds with similar affinity to RNA3′p and RNA2′p substrates. These findings expand the functional repertoire of RNA cyclase and suggest that phosphate geometry during adenylate transfer to RNA is a major factor in the kinetics of cyclization. RtcA is coregulated in an operon with an RNA ligase, RtcB, that splices RNA 5′-OH ends to either 3′-phosphate or 2′,3′-cyclic phosphate ends. Our results suggest that RtcA might serve an end healing function in an RNA repair pathway, by converting RNA 2′-phosphates, which cannot be spliced by RtcB, to 2′,3′-cyclic phosphates that can be sealed. The rtcBA operon is controlled by the σ54 coactivator RtcR encoded by an adjacent gene. This operon arrangement is conserved in diverse bacterial taxa, many of which have also incorporated the RNA-binding protein Ro (which is implicated in RNA quality control under stress conditions) as a coregulated component of the operon.
INTRODUCTION
RNA 2′,3′-cyclic phosphate ends play important roles in RNA metabolism, especially as intermediates in RNA repair by polynucleotide ligases (Popow et al. 2012; Remus and Shuman 2013). Escherichia coli RtcB exemplifies a new clade of RNA ligases that join 2′,3′-cyclic phosphate and 5′-OH RNA ends to yield a 3′-5′ phosphodiester splice junction (Englert et al. 2011; Popow et al. 2011; Tanaka and Shuman 2011; Tanaka et al. 2011a). RtcB is encoded in a two-gene operon in E. coli along with the RNA cyclase enzyme RtcA, which converts RNA 3′-phosphate ends to 2′,3′-cyclic phosphates (Fig. 1A; Genschik et al. 1998). The genetic organization of RtcA and RtcB prompted the suggestion that they provide the healing and sealing functions, respectively, in an RNA repair pathway (Tanaka and Shuman 2011). In this scenario, the healing refers to the restoration of ligatable 2′,3′-cyclic phosphate ends in the event that the inciting RNA damage directly generates RNA 3′-phosphates, or that the 2′,3′-cyclic phosphate products of RNA transesterification are further processed to a 3′-phosphate by a 2′,3′-cyclic phosphodiesterase. The fact that the rtcBA operon in E. coli is regulated by the σ54 coactivator RtcR (encoded by the rtcR gene, located immediately upstream of rtcBA and transcribed in the opposite orientation) suggested that the RNA repair functions are induced in response to cellular stress (Genschik et al. 1998), conceivably as a means to ameliorate the effects of bacterial ribotoxins that are turned on in stress situations.
FIGURE 1.
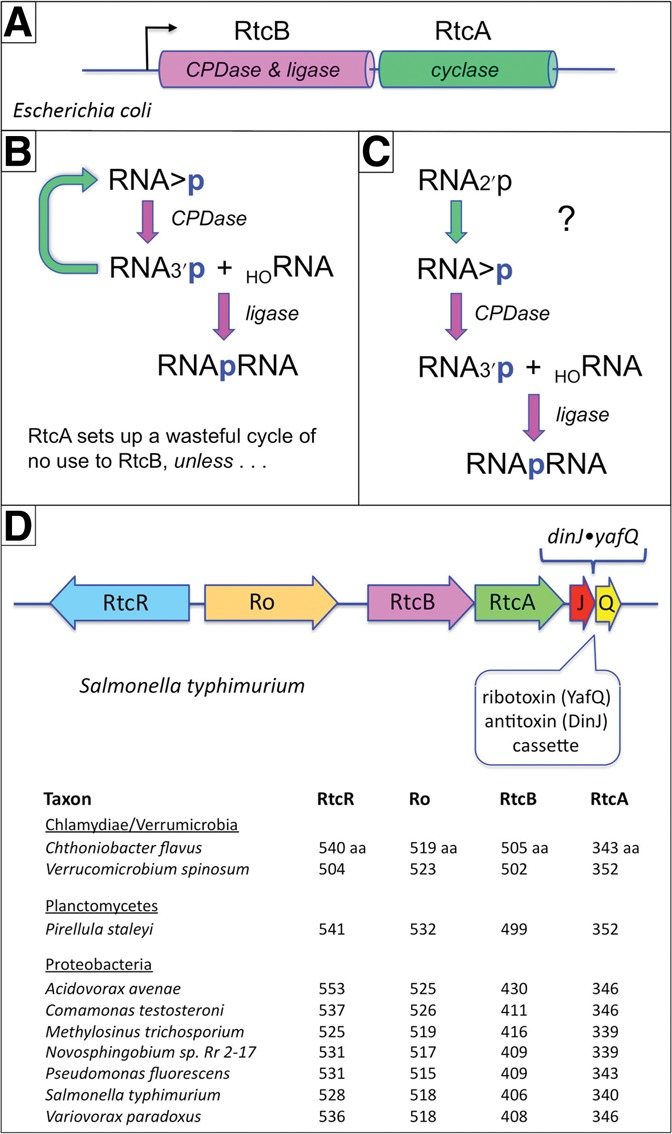
Bacterial rtcBA operons and pathway options for RtcA in RNA repair. (A) The RNA ligase RtcB is encoded in a two-gene operon in E. coli along with the RNA cyclase RtcA. (B) The cyclic phosphodiesterase activity of RtcB hydrolyzes a 2′,3′-cyclic phosphate to a 3′-phosphate. The ligase activity of RtcB then joins the RNA 3′-phosphate to a 5′-OH RNA end to yield a 3′-5′ phosphodiester splice junction. RtcA converts RNA 3′-phosphate ends to 2′,3′-cyclic phosphates, a reaction that appears wasteful from the perspective of RtcB. (C) Were RtcA able to cyclize RNA 2′-phosphate ends, which are not substrates for sealing by RtcB, it might serve a useful function in an RNA repair pathway, as shown. (D) Schematic diagram of the Salmonella typhimurium operon in which rtcB and rtcA are preceded by a co-oriented ORF encoding Ro and followed by a co-oriented toxin-antitoxin cassette encoding the mRNA ribotoxin YafQ and its antitoxin DinJ. The operon is regulated by RtcR, a σ54 coactivator encoded by an adjacent, oppositely oriented gene. Exemplary bacterial taxa that have an RtcR-regulated ro-RtcB-RtcA operon are listed; the sizes (aa) of the component proteins are shown.
The biochemical rationale for the two-component repair system was subsequently deflated by the discovery that RtcB is adept at joining 3′-phosphate/5′-OH ends or 2′,3′-cyclic phosphate/5′-OH ends (Tanaka et al. 2011b; Chakravarty et al. 2012). We now appreciate that RtcB executes a four-step pathway of end joining that requires GTP as an energy source and Mn2+ as a cofactor (Tanaka et al. 2011b; Chakravarty et al. 2012; Chakravarty and Shuman 2012). RtcB first reacts with GTP to form a covalent RtcB-(histidinyl337-N)-GMP intermediate. It then hydrolyzes the RNA 2′,3′-cyclic phosphate end to a 3′-phosphate and transfers guanylate from His337 to the RNA 3′-phosphate to form an RNA(3′)pp(5′)G intermediate. Finally, RtcB catalyzes the attack of an RNA 5′-OH on the RNA(3′)pp(5′)G end to form the splice junction and liberate GMP.
Given that RtcB has both cyclic phosphodiesterase and 3′-PO4/5′-OH ligase activities, the biochemical advantage to its coregulation with RtcA is not obvious. The combined action of the RtcA cyclase and RtcB cyclic phosphodiesterase seems to create a futile cycle (Fig. 1B) that is energetically wasteful, insofar as cyclization consumes ATP. RtcA generates 2′,3′-cyclic phosphate ends via three steps: reaction with ATP to form a covalent RtcA-(histidinyl309-Nϵ)-AMP intermediate; transfer of adenylate to the RNA 3′-phosphate to form an RNA(3′)pp(5′)A intermediate; and attack of the vicinal O2′ on the 3′-phosphorus to form a 2′,3′-cyclic phosphate product and release AMP (Filipowicz et al. 1985; Genschik et al. 1998; Billy et al. 1999; Tanaka et al. 2010). Whereas RtcB and RtcA are structurally unrelated proteins with no similarities in their active sites (Tanaka et al. 2010; Chakravarty et al. 2011; Englert et al. 2012), they do share two mechanistic features: (1) reliance on a histidine nucleophile for covalent nucleotidyl transfer; and (2) synthesis of an RNA 3′-phosphodiester via a covalently activated RNA(3′)pp(5′)N intermediate.
The functions of RNA cyclase in cellular RNA metabolism are unknown, and it remains unclear whether cyclization of 3′-phosphates is the only biochemical pathway in which RtcA enzymes participate. For example, E. coli RtcA catalyzes adenylylation of 5′-phosphate ends of DNA or RNA strands to form A(5′)pp(5′)DNA and A(5′)pp(5′)RNA products (Chakravarty and Shuman 2011). Biological scenarios in which the 5′ adenylyltransferase activity of RtcA might play a role include: (1) adenylylation of DNA or RNA 5′ ends as a means of protecting them against exonucleolytic decay, analogous to the function of the eukaryal mRNA cap; (2) installing an inverted 5′ adenylate as a “mark” that targets RNA or DNA molecules for downstream events, e.g., localization, translation, recombination, etc.; or (3) the generation of AppDNA or AppRNA ends that serve as substrates for ligation of AppDNA/RNA to 3′-OH termini by a separate repair enzyme (Chakravarty and Shuman 2011).
With respect to the connection of RtcA and RtcB, we considered the prospect that RNA cyclase might be capable of acting on RNA 2′-phosphate ends, which are not substrates for sealing by RtcB (Tanaka et al. 2011b). In this scenario, RtcA would serve an end healing function in an RNA repair pathway, by converting RNA 2′-phosphates to 2′,3′-cyclic phosphates that can then be sealed by RtcB (Fig. 1C). The goal of the present study was to determine whether RtcA can cyclize a 2′-phosphate end.
RESULTS AND DISCUSSION
RtcA cyclizes an RNA 2′-phosphate end
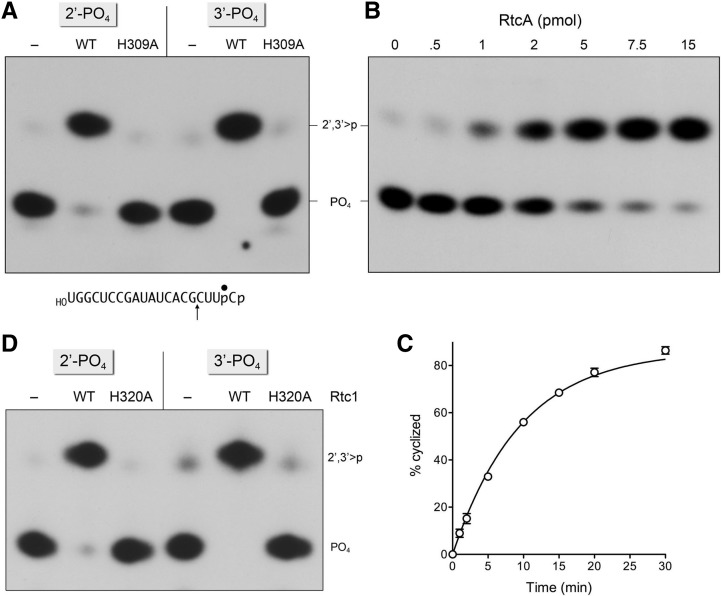
In the experiment in Figure 2A, RtcA was reacted with otherwise identical 20-mer RNAs with 3′-phosphate or 2′-phosphate ends and a single radiolabel between the 3′-terminal and penultimate nucleosides (Fig. 2A). The RNA3′p and RNA2′p substrates (20 nM) were incubated for 30 min at 37°C with 10 mM MgCl2, 100 µM ATP, and either 1.5 µM wild-type RtcA or the active site mutant H309A. To enable resolution of the reactants and products with different 3′ ends, the RNAs were digested with RNase T1 prior to analysis by denaturing PAGE (Fig. 2A). RNase T1 incised the substrates 3′ of the most distal guanosine to yield the 32P-labeled tetranucleotides HOCUUpC3′p or HOCUUpC2′p, which migrated identically during PAGE (Fig. 2A, lanes –). In the presence of RtcA, the RNA3′p and RNA2′p substrates were both converted into an RNA > p product that, after incision by RNase T1, yielded a 2′,3′-cyclic phosphate-terminated tetranucleotide, HOCUUpC > p, that migrated slower during PAGE than the phosphomonoester-terminated tetranucleotides (Fig. 2A). RtcA effected near-quantitative cyclization of the 2′-phosphate and 3′-phosphate ends. In contrast, the RtcA H309A mutant, which is unable to form the covalent RtcA–AMP intermediate, was unreactive with either RNA3′p or RNA2′p. These results establish that E. coli RtcA can indeed cyclize an RNA 2′-phosphate end in high yield.
FIGURE 2.
RtcA cyclizes a 2′-phosphate end. (A) Reaction mixtures (10 µL) containing 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 2 mM DTT, 100 µM ATP, 20 nM 20-mer RNA2′p (2′-PO4) or RNA3′p (3′-PO4) substrate (depicted at bottom with the 32P label denoted by •), and either no enzyme (lanes –) or 1.5 µM wild-type E. coli RtcA or mutant RtcA-H309A were incubated for 30 min at 37°C. The reactions were quenched with EDTA. The products were digested with RNase T1 and then analyzed by urea-PAGE. An autoradiogram of the gel is shown. The terminal phosphorylation states of the 32P-labeled RNase T1 fragments are indicated at right. (B) Reaction mixtures (10 µL) containing 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 2 mM DTT, 100 µM ATP, 20 nM (0.2 pmol) 20-mer RNA2′p (2′-PO4) substrate, and RtcA as specified were incubated for 30 min at 37°C. (C) A reaction mixture (70 µL) containing 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 2 mM DTT, 100 µM ATP, 20 nM 20-mer RNA2′p (2′-PO4), and 1.5 µM RtcA was incubated at 37°C. Aliquots (10 µL) were withdrawn at the times specified and quenched immediately with SDS and EDTA. After phenol-chloroform extraction, the mixtures were digested with RNase T1 and analyzed by PAGE. The extent of cyclization is plotted as a function of reaction time. Each datum is the average of three separate experiments ±SEM. A nonlinear regression curve fit of the data to a one-phase association (executed in Prism) is shown. The apparent rate constant for 2′-phosphate cyclization was 0.1 ± 0.003 min−1. (D) RNA 2′-phosphate cyclase activity of human Rtc1. Reaction mixtures (10 µL) containing 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 2 mM DTT, 100 µM ATP, 20 nM 20-mer RNA2′p (2′-PO4) or RNA3′p (3′-PO4) substrate, and either no enzyme (lanes –) or 1.5 µM wild-type human Rtc1 or mutant Rtc1-H320A were incubated for 30 min at 37°C. The reactions were quenched with EDTA. The products were digested with RNase T1 and then analyzed by urea-PAGE. An autoradiogram of the gel is shown.
We performed similar experiments with human RNA cyclase Rtc1 (Genschik et al. 1997; Tanaka and Shuman 2009) and found that it, too, converted the RNA3′p and RNA2′p substrates to RNA > p products in a reaction that depended on the His320 nucleophile that forms the covalent Rtc1–AMP intermediate (Fig. 2D).
Kinetics of 2′-phosphate and 3′-phosphate cyclization by RtcA
The extent of cyclization of the RNA2′p substrate during a 30-min reaction at 37°C was proportional to the concentration of input RtcA up to 0.5 µM RtcA; the reaction was effectively complete at 0.75 and 1.5 µM RtcA (Fig. 2B). The kinetics of 2′-phosphate cyclization by 1.5 µM RtcA are shown in Figure 2C. The data fit well to a one-phase association with an apparent rate constant of 0.1 ± 0.003 min−1.
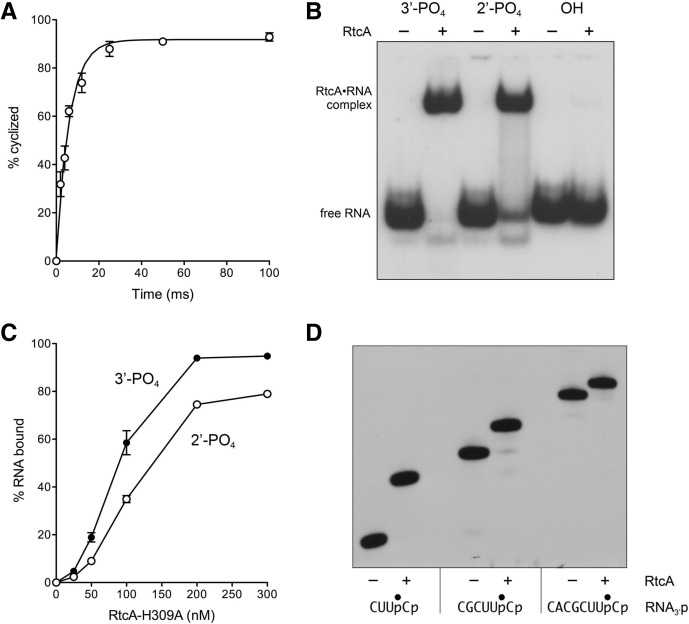
The rate of 3′-phosphate cyclization by RtcA under conditions of enzyme excess was too fast to measure manually. Therefore, we employed a rapid mix-quench apparatus to access the reaction on a millisecond time scale. Equal volumes of enzyme and substrate solutions were mixed at 22°C to achieve final concentrations of 20 nM RNA3′p and 200 nM RtcA. ATP and Mg2+ were omitted in order to ensure that cyclic product formation reflected a single-turnover reaction by preformed RtcA–AMP. We determined by enzyme titration in the absence of ATP that preformed RtcA–AMP comprised 25% of the RtcA preparation. Prior studies had established that, whereas RtcA requires a divalent cation to form the RtcA–AMP intermediate, preformed RtcA–AMP does not need a metal cofactor to execute 3′-phosphate cyclization (Chakravarty et al. 2011). The rapid mix reactions were quenched with SDS and EDTA. The SDS was removed by extraction with phenol-chloroform prior to treating the mixtures with RNase T1 and analyzing the products by denaturing PAGE. The kinetic profile showed that the RNA > p product accumulated steadily up to 25 msec and plateaued at 100 msec with an endpoint of 93% cyclization (Fig. 3A). Nonlinear regression fitting of the data to a one-phase association yielded an apparent rate constant of 163 ± 14 sec−1.
FIGURE 3.
Kinetics of 3′-phosphate cyclization, binding of RtcA to 3′-phosphate and 2′-phosphate RNAs, and minimized RNA substrate for cyclization. (A) Kinetics. Reaction mixtures containing 50 mM Tris-HCl (pH 7.4), 2 mM DTT, 20 nM 32P-labeled 20-mer RNA3′p, and 200 nM RtcA were incubated at 22°C. The extent of 3′-phosphate cyclization is plotted as a function of reaction time. Each datum is the average of three separate experiments ±SEM. A nonlinear regression curve fit of the data to a one-phase association (executed in Prism) is shown. (B) RNA binding. Reaction mixtures containing 50 mM Tris-HCl (pH 7.4), 1 mM DTT, 20 nM 32P-labeled 20-mer RNA3′p (3′-PO4) or RNA2′p (2′-PO4) or RNAOH (OH) substrate, and either no enzyme or 300 nM of RtcA-H309A (where indicated by +) were incubated for 20 min at 37°C. The mixtures were placed on ice, adjusted to 10% glycerol and 0.04% Triton-X100, and then analyzed by native PAGE. The 32P-labeled free RNAs and RtcA•RNA complexes were visualized by autoradiography. (C) Reaction mixtures containing 50 mM Tris-HCl (pH 7.4), 1 mM DTT, 20 nM of a 20-mer RNA3′p (3′-PO4) or RNA2′p (2′-PO4) substrate, and RtcA-H309A as indicated were incubated for 20 min at 37°C. The extents of RtcA•RNA complex formation are plotted as a function of input RtcA. Each datum is the average of three independent RtcA titration experiments (±SEM). (D) Minimized RNA substrate. Reaction mixtures (10 µL) containing 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 2 mM DTT, 100 µM ATP, 20 nM of 4-mer, 6-mer, or 8-mer RNA3′p substrates labeled with 32P at the penultimate phosphate, and either no enzyme (−) or 200 nM RtcA (+) were incubated for 30 min at 37°C. The reactions were quenched by adding an equal volume of 90% formamide and 50 mM EDTA, and then analyzed by urea-PAGE. An autoradiogram of the gel is shown. The nucleotide sequences of the RNA3′p strands are shown below the autoradiogram; the position of the 32P-label is denoted by •.
Binding of RtcA to 2′-phosphate and 3′-phosphate terminated RNAs
To interrogate the role of terminal phosphate position in RtcA binding to RNA, we exploited an electrophoretic mobility shift assay format that had been used previously to demonstrate that: (1) human RNA cyclase Rtc1 forms a discrete stable complex with 3′-phosphate-terminated RNA and (2) RNA3′p binding by Rtc1 is independent of enzyme adenylylation, insofar as the H320A mutant bound RNA as well as wild-type Rtc1 (Tanaka and Shuman 2009). Here, otherwise identical 20-mer RNAs (20 nM) with 3′-phosphate, 2′-phosphate, or 2′,3′-hydroxyl ends and a single radiolabel between the 3′-terminal and penultimate nucleosides were incubated with 300 nM RtcA-H309A in the absence of ATP or divalent cation. The mixtures were then analyzed by electrophoresis through a native 10% polyacrylamide gel. RtcA formed a discrete protein•RNA complex with the 2′-phosphate and 3′-phosphate RNAs that was well resolved from the free RNA (Fig. 3B). In contrast, there was no detectable binding of RtcA to the RNAOH ligand (Fig. 3B), thereby attesting to a stringent requirement for a terminal phosphate for RtcA to engage its substrate.
The dependence of RNAp binding as a function of RtcA concentration is shown in Figure 3C. Taking the slope of the binding curve in the linear range of RtcA dependence (25 to 200 nM) as a metric of RNA binding affinity (calculated by linear regression in Prism), we surmise that RtcA binds 2′-phosphate RNA 80% as well as it binds 3′-phosphate RNA.
Minimized substrate for cyclization
Prior studies had shown that, whereas 12-mer, 11-mer, or 10-mer RNA3′p substrates could be cyclized quantitatively by RtcA and Rtc1, these enzymes were unable to cyclize mononucleotide 5′,3′-bisphosphate (pNp) or dinucleotide-3′-phosphate (NpNp) substrates (Reinberg et al. 1985; Genschik et al. 1997, 1998; Tanaka and Shuman 2009; Tanaka et al. 2010). The minimal effective substrate for human RNA cyclase is a trinucleotide-3′-phosphate (Reinberg et al. 1985). Here, we found that E. coli RtcA could quantitatively cyclize 8-mer, 6-mer, and 4-mer RNA3′p substrates (Fig. 3D).
Cyclization of DNA with a single 3′-terminal ribonucleotide
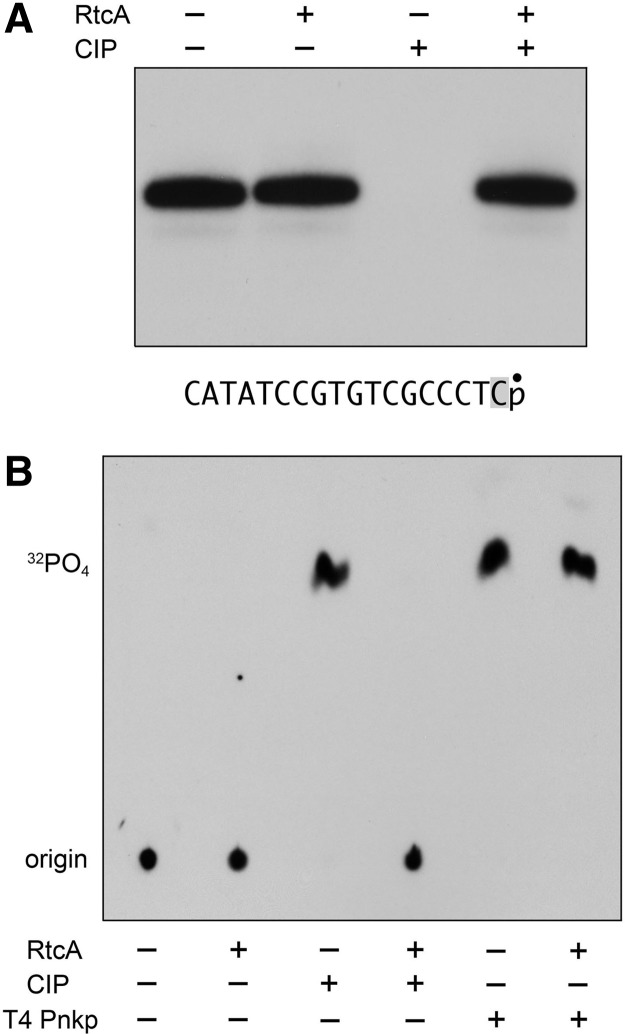
Although DNA 3′-phosphate ends obviously cannot be cyclized, Filipowicz et al. (1985) showed that prolonged incubation of a DNA3′p substrate with excess human RNA cyclase generated low amounts of DNA3′ppA. The observation that cold DNA3′p was 500- to 1000-fold less effective than an otherwise identical cold RNA3′p in inhibiting cyclization of a radiolabeled RNA3′p substrate by human or E. coli RNA cyclases (Genschik et al. 1997, 1998) signified that the feeble use of DNA3′p as an acceptor for adenylate transfer reflected, in large part, its weak interaction with the cyclase enzyme. Here, we queried the basis for discrimination of RNA vs. DNA by testing whether RtcA could cyclize an 18-mer substrate, D17R13′p, consisting of 17 deoxynucleotides and a single 3′-terminal ribonucleotide, with a 32P-label at the 3′-phosphate. The substrate was prepared by templated enzymatic ligation of a cold D17R1OH strand, 5′-d(CATATCCGTGTCGCCCT)(rC), to a 5′ 32P-labeled 10-mer DNA strand, 5′-pATTCCGATAG, followed by digestion of the isolated D17R1pD10 strand with RNase A to yield D17R13′p. The gel-purified D17R13′p oligonucleotide (shown in Fig. 4A) was reacted with RtcA, and the products were analyzed by PAGE, either directly or after treatment with calf intestine alkaline phosphatase (CIP). The radiolabel in the input D17R13′p strand was removed completely by CIP. The reaction with RtcA rendered the labeled strand resistant to CIP (Fig. 4A). The lack of a change in electrophoretic mobility of the oligonucleotide after reaction with RtcA is consistent with the CIP-resistant species being the cyclized product D17R1 > p, rather than D17R1ppA, which would migrate above the substrate during PAGE. Analysis of the reaction products by PEI-cellulose TLC showed that, whereas CIP treatment of the input D17R13′p oligonucleotide (which remained at the chromatographic origin) quantitatively liberated 32P inorganic phosphate, prior reaction with RtcA rendered the product resistant to hydrolysis by CIP (Fig. 4B). However, the 32P-label in both the substrate and the RtcA reaction product was sensitive to hydrolysis by T4 polynucleotide kinase-phosphatase (Pnkp) (Fig. 4B), which releases Pi from either 3′-phosphate or 2′,3′-cyclic phosphate termini (Das and Shuman 2013) but would not be able to liberate Pi from D17R1ppA. We surmise that a single terminal ribonucleoside suffices for productive cyclization by RtcA.
FIGURE 4.
Cyclization of DNA with a single 3′-terminal ribonucleotide. (A) Reaction mixtures (10 µL) containing 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 2 mM DTT, 100 µM ATP, 20 nM D17R13′p substrate (depicted at bottom with the 32P label denoted by • and the terminal ribonucleotide highlighted in gray), and either 400 nM RtcA (where indicated by +) or no added enzyme (−) were incubated for 30 min at 37°C. The mixtures were then treated with calf intestine phosphatase (CIP, where indicated by +) for 30 min at 37°C. The CIP-treated and mock-treated samples were mixed with an equal volume of formamide and EDTA, and then analyzed by electrophoresis through a 40-cm 20% polyacrylamide gel containing 7 M urea in 45 mM Tris borate, 1.2 mM EDTA. An autoradiogram of the gel is shown. (B) Reaction mixtures (10 µL) containing 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 2 mM DTT, 100 µM ATP, 0.2 µM D17R13′p substrate, and either 4 µM RtcA (where indicated by +) or no added enzyme (−) were incubated for 30 min at 37°C. The mixtures were then treated with either CIP or T4 polynucleotide kinase-phosphatase (Pnkp) for 30 min at 37°C. The treated and mock-treated samples were adjusted to 1 M formic acid and analyzed by PEI-cellulose TLC with 0.75 M LiCl as the mobile phase. An autoradiograph of the chromatogram is shown. The origin and the species corresponding to 32PO4 are indicated on the left.
Mechanistic implications for RNA cyclase
The experiments presented above establish that RtcA can cyclize an RNA 2′-phosphate with high efficiency, albeit slowly compared to its reaction at an RNA 3′-phosphate end, which is faster by a factor of 105. This huge rate disparity contrasts with the rather modest effect of terminal phosphate position on the binding of RtcA to its RNA substrate. We infer that RtcA discriminates kinetically between 3′- and 2′-phosphates at the level of adenylate transfer chemistry from RtcA–AMP to RNAp to form the RNAppA intermediate and/or the intramolecular reaction of RNAppA to form RNA > p and release AMP. The RNA adenylylation step of the RtcA pathway entails the attack of a terminal phosphate oxygen on the phosphorus atom of RtcA-(His309-Nϵ)–AMP to generate RNAppA and expel His309. The RNA adenylylation reaction is metal-independent and is thought to be catalyzed by essential basic residues that coordinate the RNA terminal phosphate (Tanaka and Shuman 2009; Chakravarty et al. 2011). A critical requirement for catalysis of RNA adenylylation is that the phosphate oxygen nucleophile be positioned apical to the histidine-Nϵ leaving group. We speculate that the extreme rate disparity between 3′- and 2′-phosphate cyclization reflects an unfavorable alignment of the 2′-phosphate with respect to the His309–AMP adduct. Phosphate position may also influence the subsequent cyclization step, in which the vicinal ribose hydroxyl is activated for attack on RNAppA, e.g., whereby the geometry of the ribose O3′ with respect to the AMP leaving group of RNA2′ppA is not auspicious for an in-line displacement. We favor the idea that a 2′-phosphate drastically slows the rate of the RNA adenylylation step, because we did not detect the accumulation of an adenylylated RNA species (migrating above the RNA > p product during PAGE) during reaction of a molar excess of RtcA with the RNA2′p substrate (data not shown). If the intramolecular cyclization of RNA2′ppA was rate-limiting, we would expect to see significant accumulation of an adenylated RNA intermediate. Even during the very rapid single-turnover reaction of RtcA with the RNA3′p substrate, we observed only very scant amounts of a putative RNAppA species, signifying that intramolecular cyclization is faster than RNAp adenylylation, whether at a 2′-phosphate or a 3′-phosphate end. It is worth noting that the rate of 2′-phosphate cyclization by RtcA (0.1 min−1) is similar to the rate of the 5′-phosphate adenylylation reaction (0.18 min−1) (Chakravarty and Shuman 2011). A full accounting of the terminal phosphate position effect on polynucleotide adenylylation and cyclization will hinge on obtaining new crystal structures of RtcA in complexes with 3′-phosphate, 2′-phosphate, and 5′-phosphate substrates. The delineation of a tetranucleotide as a minimized RtcA substrate (and the ability to replace all but the 3′-terminal nucleotides by deoxys) will aid that effort.
Is RtcA in a pathway with RtcB?
The precipitating question for this study—can RNA cyclases convert a 2′-phosphate to a 2′,3′-cyclic phosphate?—has been answered in the affirmative for the cyclases RtcA and Rtc1 from E. coli and humans, respectively. Thus, it is possible that RtcA serves a 2′-phosphate end healing function in an RNA repair pathway driven by RtcB. The rate of 2′-phosphate cyclization in vitro, though slow in comparison to 3′-phosphate cyclization, is not a deal-breaker, given the ability of RtcA to bind stably at an RNA 2′-phosphate end and to cyclize it in near-quantitative yield. Until we know more about the physiology of RtcA and RtcB (e.g., what stresses trigger their expression and what are their natural substrates for end-modification and repair?), we can only answer “maybe” to the issue of whether RtcA and RtcB are agents in a common biological pathway in E. coli. Nonetheless, for this scenario to be plausible, there must be some means of generating RNA 2′-phosphate ends in E. coli as substrates for RtcA. Bacterial ribotoxins are transesterifying endoribonucleases that yield 2′,3′-cyclic phosphate RNA breaks. It is noteworthy that E. coli and many other bacteria encode an RNA 2′,3′-cyclic phosphoesterase that hydrolyzes cyclic phosphates to yield an RNA 2′-phosphate product (Kanai et al. 2009).
Phylogenetics of rtcBA
Many bacterial taxa encode RtcA and RtcB homologs, though it is usually not the case that they are genetically linked on the respective bacterial chromosomes as observed for the E. coli rtcBA operon. RtcB and RtcA homologs are also found in many archaeal proteomes, but they are, to our inspection, never genetically clustered in an operon. Thus, the coregulation of RtcB and RtcA is the exception, not the rule. A list of 32 bacteria that do have an rtcBA gene cluster adjacent to an rtcR gene is compiled in Supplemental Table S1. These include diverse taxa from four different phyla, with a preponderance of examples from the several divisions of Proteobacteria. The ⇦rtcR•rtcB–rtcA⇨ operon organization seen in E. coli is maintained in most of the other taxa, i.e., the rtcR gene is upstream of rtcB and divergently transcribed, and rtcB precedes rtcA. There are notable exceptions. In Hahella chejuensis (a γ-proteobacterium), the gene arrangement is rtcR–rtcA–rtcB⇨, in which rtcR is upstream of rtcB but is transcribed in the same direction as rtcBA. In Pelobacter carbinolicus (a δ-proteobacterium), the arrangement is ⇦rtcR•rtcA–rtcB⇨, whereby the rtcBA order is reversed, and the rtcA ORF precedes rtcB. A conserved feature of the bacterial rtcBA operon is the presumptive coregulation of RtcB and RtcA production by the σ54 coactivator RtcR. In E. coli, rtcBA expression is undetectable under normal growth conditions but can be induced by expressing a deleted version of RtcR that lacks an N-terminal auto-inhibitory module (Genschik et al. 1998). Identifying the stresses and molecular signals that activate RtcR and induce rtcBA should provide instructive clues to the RNA transactions to which the repair enzymes are directed.
Genetic association of bacterial Ro protein with rtcBA
Many of the bacteria that have an rtcR•rtcBA cluster encode additional proteins within the putative operon, by interposing co-oriented ORFs, either upstream of rtcB or between rtcB and rtcA. The most prevalent variant of the operon encodes the RNA-binding protein Ro upstream of rtcB. The ⇦rtcR•ro–rtcB–rtcA⇨ operon organization applies to 10 bacterial taxa (Fig. 1D). The ∼60-kDa bacterial Ro protein, like its eukaryal Ro counterpart, is a toroidal ring composed of HEAT repeats plus a vWFA domain (Stein et al. 2005; Ramesh et al. 2007). Ro binds small RNAs, especially misfolded RNAs, and is thought to function in RNA quality control, including the recognition or processing of damaged RNAs (Chen et al. 2003). Single-strand RNA binds within the central cavity of the Ro ring; double-strand RNA binds on the outer surface of the ring (Stein et al. 2005; Fuchs et al. 2006). Wolin and colleagues have provided keen insights into the functions of bacterial Ro in Deinococcus radiodurans, where the Ro homolog (named Rsr) is induced during recovery from ultraviolet irradiation, associates with small noncoding RNAs, and contributes to the UV-resistance of Deinococcus (Chen et al. 2000). Their studies implicate Deinococcus Ro, together with exoribonucleases, in heat-stress-induced maturation of ribosomal RNA and, together with polynucleotide phosphorylase, in starvation-induced rRNA decay (Chen et al. 2007; Wurtmann and Wolin 2010). Indeed, Deincococcus Ro and polynucleotide phosphorylase interact physically to form an RNA-scaffolded degradation complex (Chen et al. 2013).
The prototypal Ro-containing ⇦rtcR•ro–rtcB–rtcA⇨ operon is found in Salmonella typhimurium, as illustrated in Fig. 1D. Consistent with the original E. coli findings, expression of the S. typhimurium Ro gene is undetectable under normal growth conditions but can be induced by production of a plasmid-encoded constitutively active N-terminal truncation variant of S. typhimurium RtcR (Chen et al. 2013). The genomic “gap” between the Ro and RtcB ORFs encodes two small noncoding RNAs that associate with S. typhimurium Ro (Chen et al. 2013). This noncoding gap is a conserved feature of other Ro-containing bacterial rtcBA operons (data not shown). We doubt it is mere coincidence that diverse bacterial taxa genetically cluster and coregulate an RNA quality control determinant (Ro) with two enzymes implicated in RNA end-modification (RtcA) and RNA repair (RtcB).
What is even more striking is that the two ORFs immediately downstream from the S. typhimurium rtcA gene comprise a dinJ yafQ antitoxin:ribotoxin cassette (Fig. 1D). YafQ is a site-specific endoribonuclease (an “mRNA interferase”) that associates with the ribosome, incises mRNA at in-frame AAA lysine codons followed by a purine, and thereby arrests translation (Prysak et al. 2009; Armalyte et al. 2012). We can speculate that the effects of stress-induced ribotoxic damage caused by YafQ are modulated by coexpressing clustered genes that influence the recognition, decay, and/or repair of the broken RNAs.
MATERIALS AND METHODS
RNA cyclase proteins
Wild-type E. coli RtcA and mutant H309A were produced as N-terminal His10-tagged fusion proteins and purified from soluble bacterial extracts by sequential Ni-agarose, anion-exchange, and gel-filtration chromatography steps as described (Chakravarty et al. 2011). Wild-type human Rtc1 and mutant H320A were purified from soluble bacterial extracts by sequential Ni-agarose chromatography, ammonium sulfate precipitation, and gel-filtration chromatography steps as described (Tanaka and Shuman 2009). Protein concentrations were determined by using the Bio-Rad dye reagent with bovine serum albumin as the standard.
RNA substrates
The 20-mer RNA3′p substrate was prepared by enzymatic addition of [5′-32P]pCp to a 19-mer RNA and then gel-purified (Das and Shuman 2013). The radiolabeled RNA3′p strand was treated with E. coli RtcA and ATP to generate a 2′,3′-cyclic phosphate derivative, RNA > p. A 2′-phosphate-terminated derivative, RNA2′p, was produced by reacting RNA > p with the cyclic phosphodiesterase domain of Arabidopsis thaliana tRNA ligase (Das and Shuman 2013). The conversion of RNA3′p to RNA > p and then to RNA2′p was verified by digestion of the RNAs with RNase T1 and analysis of the 32P-labeled terminal fragments by urea-PAGE. A 2′,3′-hydroxyl-terminated derivative, RNAOH, was produced by reacting RNA3′p with purified T4 polynucleotide kinase-phosphatase (Pnkp) for 30 min at 37°C in a reaction mixture (100 µL) containing 100 mM Tris acetate (pH 6.0), 10 mM MgCl2, 1 µM RNA3′p, and 50 µM T4 Pnkp. The RNAOH product was recovered by phenol-chloroform extraction and precipitation with ethanol.
Cyclization assay
Reaction mixtures containing 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 2 mM DTT, 20 nM 32P-labeled RNA2′p or RNA3′p substrate, and wild-type or mutant RtcA were incubated at 37°C. The reactions were quenched by adding either an equal volume of 100 mM EDTA or 100 mM EDTA plus 1% SDS. The samples were extracted with phenol-chloroform to remove the SDS. The RNAs in the aqueous phase were digested with RNase T1 (1000 U; Fermentas) for 30 min at 37°C. The samples were supplemented with 0.5 volume of 90% formamide, 0.01% bromophenol blue/xylene cyanol, 50 mM EDTA, and then analyzed by electrophoresis (at 50 W constant power) through a 40-cm 20% polyacrylamide gel containing 8 M urea in 45 mM Tris borate, 1.2 mM EDTA. The 32P-labeled tetranucleotides HOCUUpCp and HOCUUpC > p were visualized by autoradiography of the gel and, where specified, quantified by scanning the gel with a Fuji Film BAS-2500 imager.
Rapid kinetic analysis of 3′-phosphate cyclization
A Kintek RQF3 rapid chemical quench apparatus was used to assay the reaction of RNA3′p with a 10-fold molar excess of RtcA at 22°C in the absence of added ATP and Mg2+, in the range of 2 msec- to 100 msec-reaction times. The reaction mixtures (40 µL) contained 50 mM Tris-HCl (pH 7.4), 2 mM DTT, 20 nM 32P-labeled RNA3′p, and 200 nM RtcA. The rapid kinetic measurements were initiated by mixing two solutions (20 µL each). One solution contained 50 mM Tris-HCl (pH 7.4), 2 mM DTT, and 40 nM 32P-labeled RNA3′p. The other solution contained 50 mM Tris-HCl (pH 7.4), 2 mM DTT, and 400 nM RtcA. The reactions were quenched by rapid mixing with 1% SDS, 100 mM EDTA to a final volume of 150 µL. The mixtures were extracted with phenol-chloroform. The aqueous phase was removed and then digested with RNase T1. Aliquots of the samples were mixed with an equal volume of 90% formamide, 50 mM EDTA and analyzed by electrophoresis through a 20% polyacrylamide gel containing 7 M urea in 45 mM Tris borate, 1.2 mM EDTA. The 32P-labeled tetranucleotides HOCUUpC > p and HOCUUpC3′p were visualized by autoradiography and quantified by scanning the gel.
RNA binding assay
Reaction mixtures containing 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 2 mM DTT, 20 nM 32P-labeled RNA2′p or RNA3′p or RNAOH substrate, and RtcA-H309A were incubated for 20 min at 37°C. The mixtures were placed on ice and adjusted to 10% glycerol, 0.04% Triton-X 100 before analyzing them by electrophoresis (at 100 V constant voltage) through a 15-cm 10% polyacrylamide gel containing 23 mM Tris borate, 0.6 mM EDTA. The free radiolabeled 32P-labeled RNAs and the RtcA•RNA complexes were visualized by autoradiography and, where specified, quantified by scanning the gel.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Anupam Chakravarty and Barbara Remus for advice and assistance. This research was supported by NIH grant GM46330. S.S. is an American Cancer Society Research Professor.
REFERENCES
- Armalyte J, Jurenaite M, Beinoraviciute G, Teiserskas J, Suziedelience E 2012. Characterization of Escherichia coli dinJ-yafQ toxin-antitoxin system using insights from mutagenesis data. J Bacteriol 194: 1523–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billy E, Hess D, Hofsteenge J, Filipowicz W 1999. Characterization of the adenylation site in the RNA 3′-terminal phosphate cyclase from Escherichia coli. J Biol Chem 274: 34955–34960 [DOI] [PubMed] [Google Scholar]
- Chakravarty AK, Shuman S 2011. RNA 3′-phosphate cyclase (RtcA) catalyzes ligase-like adenylylation of DNA and RNA 5′-monophosphate ends. J Biol Chem 286: 4117–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty AK, Shuman S 2012. The sequential 2′,3′ cyclic phosphodiesterase and 3′-phosphate/5′-OH ligation steps of the RtcB RNA splicing pathway are GTP-dependent. Nucleic Acids Res 40: 8558–8567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty AK, Smith P, Shuman S 2011. Structures of RNA 3′-phosphate cyclase bound to ATP reveal the mechanism of nucleotidyl transfer and metal-assisted catalysis. Proc Natl Acad Sci 108: 21034–21039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty AK, Subbotin R, Chait BT, Shuman S 2012. RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc Natl Acad Sci 109: 6072–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Quinn AM, Wolin SL 2000. Ro ribonucleoproteins contribute to the resistance of Deinococcus radiodurans to ultraviolet irradiation. Genes Dev 14: 777–782 [PMC free article] [PubMed] [Google Scholar]
- Chen X, Smith JD, Shi H, Yang DD, Flavell RA, Wolin SL 2003. The Ro autoantigen binds misfolded U2 small nuclear RNAs and assists mammalian cell survival after UV irradiation. Curr Biol 13: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Chen X, Würtmann EJ, Van Batavia J, Zybailov B, Washburn MP, Wolin SL 2007. An ortholog of the Ro autoantigen functions in 23S rRNA maturation in D. radiodurans. Genes Dev 21: 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Taylor DW, Fowler CC, Galan JE, Wang HW, Wolin SL 2013. An RNA degradation machine sculpted by Ro autoantigen and noncoding RNA. Cell 153: 166–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das U, Shuman S 2013. Mechanism of RNA 2′,3′-cyclic phosphate end-healing by T4 polynucleotide kinase-phosphatase. Nucleic Acids Res 41: 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert M, Sheppard K, Aslanian A, Yates JR, Söll D 2011. Archaeal 3′-phosphate RNA splicing ligase characterization identified the missing component in tRNA maturation. Proc Natl Acad Sci 108: 1290–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert M, Xia S, Okada C, Nakamura A, Tanavde V, Yao M, Eom SH, Koningsberg WH, Söll D, Wang J 2012. Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3′-terminal phosphate and 5′-OH. Proc Natl Acad Sci 109: 15235–15240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Strugala K, Konarska M, Shatkin AJ 1985. Cyclization of RNA 3′-terminal phosphate by cyclase from HeLa cells proceeds via formation of N(3′)pp(5′)A activated intermediate. Proc Natl Acad Sci 82: 1316–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Stein AJ, Fu C, Reinisch KM, Wolin SL 2006. Structural and biochemical basis for misfolded RNA recognition by the Ro autoantigen. Nature Struct Mol Biol 13: 1002–1009 [DOI] [PubMed] [Google Scholar]
- Genschik P, Billy E, Swianiewicz M, Filipowicz W 1997. The human RNA 3′-terminal phosphate cyclase is a member of a new family of proteins conserved in Eukarya, Bacteria and Archaea. EMBO J 16: 2955–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P, Drabikowski K, Filipowicz W 1998. Characterization of the Escherichia coli RNA 3′-terminal phosphate cyclase and its σ54-regulated operon. J Biol Chem 273: 25516–25526 [DOI] [PubMed] [Google Scholar]
- Kanai A, Sato A, Fukuda Y, Okada K, Matsuda T, Sakamoto T, Muto Y, Yokoyama S, Kawai G, Tomita M 2009. Characterization of a heat-stable enzyme possessing GTP-dependent RNA ligase activity from a hyperthermophilic archaeon, Pyrococcus furiosus. RNA 15: 420–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Lürhmann R, Söll D, et al. 2011. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science 331: 760–764 [DOI] [PubMed] [Google Scholar]
- Popow J, Schleiffer A, Martinez J 2012. Diversity and roles of (t)RNA ligases. Cell Mol Life Sci 69: 2657–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prysak MH, Mozdzierz CJ, Cook AM, Zhu L, Zhang Y, Inouye M, Woychik NA 2009. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol Microbiol 71: 1071–1087 [DOI] [PubMed] [Google Scholar]
- Ramesh A, Savva CG, Holzenburg A, Sacchettini JC 2007. Crystal structure of Rsr, an ortholog of the antigenic Ro protein, links conformational flexibility to RNA binding activity. J Biol Chem 282: 14960–14967 [DOI] [PubMed] [Google Scholar]
- Reinberg D, Arenas J, Hurwitz J 1985. The enzymatic conversion of 3′-phosphate terminated RNA chains to 2′,3′-cyclic phosphate derivatives. J Biol Chem 260: 6088–6097 [PubMed] [Google Scholar]
- Remus BS, Shuman S 2013. A kinetic framework for tRNA ligase and enforcement of a 2′-phosphate requirement for ligation highlights the design logic of an RNA repair machine. RNA 19: 659–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein AJ, Fuchs G, Fu C, Wolin SL, Reinisch KM 2005. Structural insights into RNA quality control: The Ro autoantigen binds misfolded RNAs via its central cavity. Cell 121: 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Shuman S 2009. Structure-activity relationships in human RNA 3′-phosphate cyclase. RNA 15: 1865–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Shuman S 2011. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J Biol Chem 286: 7727–7731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Smith P, Shuman S 2010. Structure of the RNA 3′-phosphate cyclase–adenylate intermediate illuminates nucleotide specificity and covalent nucleotidyl transfer. Structure 18: 449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Meineke B, Shuman S 2011a. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J Biol Chem 286: 30253–30257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Chakravarty AK, Maughan B, Shuman S 2011b. Novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-hydroxyl ligation reactions. J Biol Chem 286: 43134–43143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtmann EJ, Wolin SL 2010. A role for a bacterial ortholog of the Ro autoantigen in starvation-induced rRNA degradation. Proc Natl Acad Sci 107: 4022–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]