PIWI proteins contain symmetric dimethylarginines (sDMAs) that are targeted by the TUDOR protein family. This study investigated the roles of PIWI sDMAs and TUDOR proteins in the biogenesis of PIWI-interacting RNAs (piRNAs) in Bombyx mori-derived BmN4 cells, and identified BmPAPI as a piRNA biogenesis factor. BmPAPI interacted with PIWI proteins via sDMAs at the surface of the mitochondrial outer membrane. BmPAPI depletion resulted in 3′-end extensions of mature piRNAs, suggesting the existence of a piRNA 3′-terminal processing machinery at the mitochondrial surface.
Keywords: PIWI, piRNA, arginine methylation, TUDOR, BmPAPI, mitochondria
Abstract
PIWI proteins and their associated PIWI-interacting RNAs (piRNAs) protect genome integrity by silencing transposons in animal germlines. The molecular mechanisms and components responsible for piRNA biogenesis remain elusive. PIWI proteins contain conserved symmetrical dimethylarginines (sDMAs) that are specifically targeted by TUDOR domain-containing proteins. Here we report that the sDMAs of PIWI proteins play crucial roles in PIWI localization and piRNA biogenesis in Bombyx mori-derived BmN4 cells, which harbor fully functional piRNA biogenesis machinery. Moreover, RNAi screenings for Bombyx genes encoding TUDOR domain-containing proteins identified BmPAPI, a Bombyx homolog of Drosophila PAPI, as a factor modulating the length of mature piRNAs. BmPAPI specifically recognized sDMAs and interacted with PIWI proteins at the surface of the mitochondrial outer membrane. BmPAPI depletion resulted in 3′-terminal extensions of mature piRNAs without affecting the piRNA quantity. These results reveal the BmPAPI-involved piRNA precursor processing mechanism on mitochondrial outer membrane scaffolds.
INTRODUCTION
In animal gonads, PIWI proteins and their bound PIWI-interacting RNAs (piRNAs) silence transposons and other targets to maintain genome integrity (Malone and Hannon 2009; Senti and Brennecke 2010; Siomi et al. 2011). This genome defense system is highly conserved across a wide range of species, and mutations in PIWI or other piRNA biogenesis factors lead to overexpression of transposon, eventually resulting in sterility (Malone and Hannon 2009; Senti and Brennecke 2010; Siomi et al. 2011). The molecular mechanisms and components involved in piRNA biogenesis remain elusive (Ishizu et al. 2012). piRNAs are 24–31 nucleotides (nt) in length and show a highly complex mix of sequences with immense diversity (Malone and Hannon 2009; Senti and Brennecke 2010; Siomi et al. 2011; Ishizu et al. 2012). At their 3′-ends, piRNAs are 2′-O-methylated by HEN1 methyltransferase (Horwich et al. 2007; Kirino and Mourelatos 2007; Ohara et al. 2007; Saito et al. 2007). piRNAs can be subclassified as primary piRNAs and secondary piRNAs, which are generated by a distinct biogenesis machinery. Primary piRNAs are thought to be generated from long single-stranded RNA transcripts derived from defined genomic regions called piRNA clusters (Brennecke et al. 2007). Primary piRNAs are subsequently subjected to an amplification loop system, known as the Ping-pong cycle, to produce abundant quantities of secondary piRNAs (Brennecke et al. 2007; Gunawardane et al. 2007).
A unique structural feature of PIWI proteins is the presence of arginine methylation. Arginine methylation is an important post-translational modification that may occur as symmetrical dimethylarginines (sDMAs), asymmetrical dimethylarginines (aDMAs), or monomethylarginines (Krause et al. 2007; Bedford and Clarke 2009; Blackwell and Ceman 2012). sDMAs occur in “sDMA motifs” composed of multiple arginine-glycine (RG) and arginine-alanine (RA) repeats in various proteins, and modulate diverse cellular processes. PIWI proteins from various species commonly contain the sDMA motif typically at their N terminus, and evolutionarily conserved presence of PIWI sDMAs has been confirmed in mice, Drosophila, zebrafish, and Xenopus (Chen et al. 2009; Kirino et al. 2009; Nishida et al. 2009; Reuter et al. 2009; Vagin et al. 2009; Huang et al. 2011b). The PIWI sDMAs are mediated by PRMT5 methyltransferase, and their functional importance has been demonstrated by the studies on Drosophila null mutants of PRMT5, in which sDMAs are absent in any of the three Drosophila PIWI proteins, leading to transposon activation and defects in germline development (Gonsalvez et al. 2006; Anne et al. 2007; Kirino et al. 2009). The identification of PIWI sDMAs suggested a possible interactive network of PIWI proteins with proteins containing TUDOR domain that constitutes the TUDOR “royal family” of protein domains (Maurer-Stroh et al. 2003). TUDOR domain specifically recognizes and binds methylated lysines or arginines within target substrates to mediate protein–protein interactions (Chen et al. 2011; Pek et al. 2012). A subset of TUDOR domain-containing proteins specifically bind sDMAs, and several members have been strongly implicated as factors binding PIWI sDMAs and/or contributing to piRNA biogenesis (Siomi et al. 2010; Chen et al. 2011; Pek et al. 2012). Those TUDOR proteins, such as Spindle-E, TUDOR, Krimper, Tejas, Vreteno, and Kumo in Drosophila and TDRD1-9 in mice, play crucial roles in development of germline (Siomi et al. 2010; Chen et al. 2011; Pek et al. 2012) and possess an additional conserved structural element flanking the canonical TUDOR core domain, termed the extended TUDOR domain (Liu et al. 2010a,b; Chen et al. 2011; Handler et al. 2011).
Although PIWI sDMAs and their interacting TUDOR proteins are emerging as key molecular factors in the piRNA pathway, their mechanistic contributions to piRNA biogenesis remain poorly understood. In this study, we utilized Bombyx mori ovary-derived BmN4 cell system to analyze the function of PIWI sDMAs and piRNA biogenesis-related TUDOR proteins. BmN4 is a uniformly cultured germ cell system that expresses the Bombyx PIWI proteins, SIWI, and BmAGO3, and that synthesizes their bound piRNAs via fully functional primary and secondary piRNA biogenesis mechanisms (Kawaoka et al. 2009, 2011, 2013). Using BmN4 cells, we found that PIWI proteins harbor sDMAs that critically function in PIWI localization and piRNA biogenesis. RNAi screenings for Bombyx genes encoding TUDOR proteins identified BmPAPI, a Bombyx homolog of Drosophila PAPI, as the factor influencing piRNA length. Mitochondrial outer surface was the place where BmPAPI interacts with PIWI proteins, suggesting the presence of a BmPAPI-involved piRNA precursor processing mechanism on mitochondrial outer membrane scaffolds.
RESULTS
sDMAs are required for PIWI localization to perinuclear nuage-like granules in BmN4
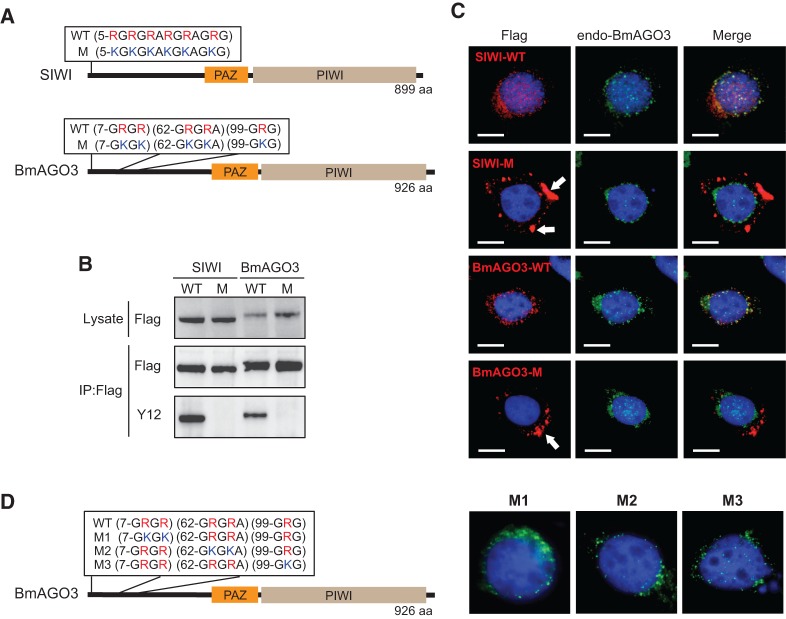
To elucidate the role of sDMAs in PIWI proteins, we generated stable BmN4 cell lines expressing FLAG-tagged wild-type SIWI or BmAGO3, or sDMA-lacking mutant SIWI or BmAGO3 whose sDMA motifs had been disrupted by substituting arginines with lysines (Fig. 1A). We purified the expressed PIWI proteins by FLAG-immunoprecipitation, and analyzed their sDMAs by Western blots using a Y12 antibody whose major epitope was sDMA (Lerner et al. 1981; Brahms et al. 2000). Since Y12 reacted with wild-type SIWI and BmAGO3 but not with their mutants (Fig. 1B), we confirmed that, in BmN4 cells, both SIWI and BmAGO3 contained sDMAs within their sDMA motifs and that mutations in these motifs abolished sDMAs. The identical expression levels of wild-type and mutant PIWI proteins (Fig. 1B) suggest no effect of sDMAs on PIWI stability in BmN4 cells.
FIGURE 1.
Arginine methylation is involved in PIWI localization. (A) Wild-type (WT) and mutant (M) PIWI protein sequences showing the arginines in sDMA motifs are substituted by lysines in the mutants. (B) Total protein lysates or anti-FLAG immunoprecipitates from BmN4 stable cell lines expressing wild-type or mutant PIWI were probed on Western blots with anti-FLAG or Y12 antibody (anti-sDMA). (C) Immunofluorescence staining of wild-type or mutant PIWI (red, left), endogenous BmAGO3 (green, middle), and DNA (blue) in BmN4 stable cell lines. A merged image of the three channels is shown on the right. Cytoplasmic aggregates of mutant PIWI are shown by arrows. Scale bar, 10 μm. (D) Sequences of wild-type (WT) BmAGO3 and its mutants (M1, M2, and M3), and immunostaining of the mutants (green) and DNA (blue) in BmN4 cells. M1–M3 mutants are localized in perinuclear granules.
PIWI proteins accumulate in amorphous, ribonucleoprotein-rich, perinuclear cytoplasmic granules that are termed nuage in Drosophila and intermitochondrial cement or chromatoid body in mice (Brennecke et al. 2007; Chuma et al. 2009). In BmN4 cells, SIWI was co-localized with BmAGO3 in perinuclear nuage-like granules (Fig. 1C; Supplemental Fig. S1). In contrast, mutant SIWI and BmAGO3 were commonly delocalized from granules and formed large aggregates in the cytoplasm, indicating that sDMAs are crucial in localizing PIWI proteins to the granules. Mutants for any one of the three sDMA motifs in BmAGO3 exhibited normal localization (Fig. 1D). Therefore, the abundance of sDMAs, and not their specific positions, is integral to normal BmAGO3 localization to granules. Xiol and colleagues reported that a similar BmAGO3 mutant construct with six arginine residue mutations diffused uniformly in the cytoplasm but did not form aggregates (Xiol et al. 2012). This difference may result from different numbers and positions of mutation sites.
Absence of BmAGO3 sDMAs increases length of bound piRNAs
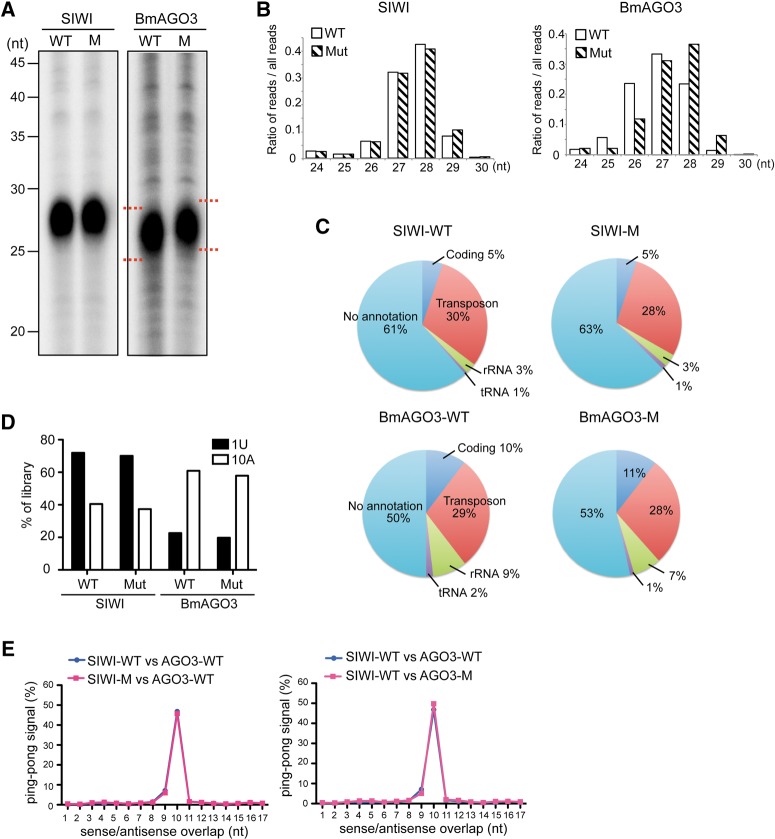
We subsequently investigated the function of PIWI sDMAs in piRNA biogenesis. We isolated and analyzed piRNAs bound to wild-type or mutant PIWI proteins and found that similar quantities of piRNAs remained bound to the mutant proteins compared with wild-type proteins (Fig. 2A). However, we observed a slight upper-shift of the band from mutant BmAGO3-bound piRNAs compared with those bound to wild type, suggesting that the lack of BmAGO3 sDMAs resulted in increased lengths of bound piRNAs. To confirm this observation, we gel-purified the piRNAs and performed deep sequencings, yielding approximately 9.5, 10, 8.5, and 8.5 million reads that aligned to the B. mori genome for piRNAs bound to wild-type SIWI, mutant SIWI, wild-type BmAGO3, and mutant BmAGO3, respectively. As shown in Fig. 2B, read lengths of the wild-type and mutant SIWI-bound piRNAs showed similar unimodal distribution patterns. Conversely, the read distribution of BmAGO3-bound piRNAs clearly revealed that the mutant-bound piRNAs were slightly longer than the wild-type-bound piRNAs, with average read lengths of 27.31 nt and 26.84 nt for mutant- and wild-type-bound piRNAs, respectively. The observation of the increased length of mutant BmAGO3-bound piRNAs is similar to the previous report (Xiol et al. 2012) and implies important roles of BmAGO3 sDMAs regarding piRNA processing and maturation. piRNA annotations and sequence distributions were not appreciably affected by the absence of sDMAs (Fig. 2C,D). Moreover, in both wild-type and mutant, we detected identical patterns of Ping-pong signals: sense-antisense piRNA pairs overlapping by 10 nt at their 5′-ends that were characteristic of a Ping-pong amplification cycle in secondary piRNA biogenesis (Fig. 2E; Brennecke et al. 2007; Kawaoka et al. 2009).
FIGURE 2.
Absence of BmAGO3 arginine methylations increases length of bound piRNAs. (A) piRNAs bound to wild-type or mutant PIWI were 5′-end-radiolabeled and analyzed by denaturing urea-PAGE. (B) Comparison of piRNA read-length distributions between wild-type and mutant SIWI (left graph) and BmAGO3 (right graph). (C) Pie charts summarizing the annotations of wild-type and mutant SIWI- and BmAGO3- bound piRNAs. (D) The preference for a first position U (1U) or a tenth position A (10A) in the piRNAs from wild-type and mutant SIWI and BmAGO3. (E) The distances between 5′-ends of SIWI- and BmAGO3-bound piRNAs across opposite genomic strands were plotted (Ping-pong analysis). Approximately 50% of reads conformed to a separation preference of 10 nt (Ping-pong signal).
BmPAPI is a TUDOR domain-containing protein affecting piRNA length
Since TUDOR domains specifically recognize sDMAs, we reasoned that length alterations in mutant BmAGO3-bound piRNAs could be attributed to the absence of interactions between BmAGO3 and TUDOR domain-containing proteins. We therefore decided to systematically analyze TUDOR proteins in BmN4 cells for their involvement in piRNA biogenesis. We retrieved 15 TUDOR protein sequences from B. mori genome that were homologous to Drosophila TUDOR proteins (Handler et al. 2011) by using the tblastn algorithm in SilkBase (http://silkbase.ab.a.u-tokyo.ac.jp/cgi-bin/index.cgi) (Table 1). To identify the TUDOR protein that influences piRNA biogenesis, the expression of each individual protein was silenced in BmN4 cells by RNAi using in vitro-synthesized double-stranded RNAs (dsRNAs; 400–600 bp). The efficiency of gene silencing and its influence in the piRNA pathway were at first confirmed by PIWI knockdowns. The RNAi targeting SIWI caused severe reduction in SIWI mRNA, protein, and also in piRNA-1, which specifically binds to SIWI, while microRNA (miRNA) pathway was not affected (Fig. 3A,B; Supplemental Fig. S2). On the other hand, BmAGO3 knockdown decreased the levels of BmAGO3 mRNA, protein, and piRNA-2, which specifically interacts with BmAGO3. A slight decrease in piRNA-2 following SIWI knockdown resulted from the concomitant decrease in BmAGO3 protein induced by SIWI knockdown.
TABLE 1.
Bombyx mori genes encoding TUDOR domain-containing proteins retrieved from Silkworm genome database
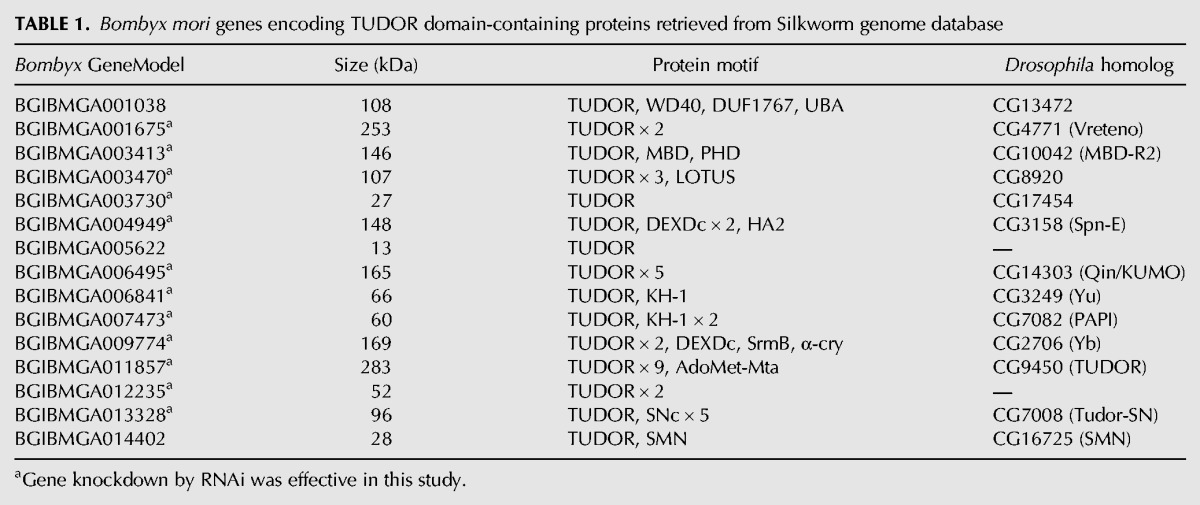
FIGURE 3.
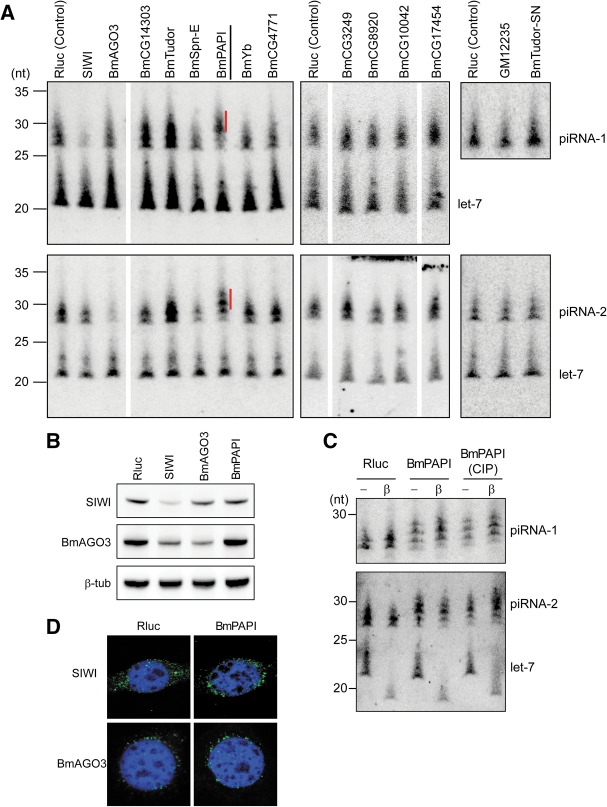
RNAi knockdowns of genes encoding TUDOR domain-containing proteins identified BmPAPI as a factor affecting piRNA length. (A) The indicated genes encoding TUDOR domain-containing proteins, PIWI (positive control), and Renilla luciferase (Rluc, negative control) were silenced by RNAi in BmN4 cells. Total RNAs from the cells were subjected to Northern blots against piRNA-1 (SIWI-bound), piRNA-2 (BmAGO3-bound), and let-7 miRNA (control). Quantifications of mRNA to confirm knockdown efficiencies are shown in Supplemental Figure S2. piRNAs that were extended as a result of BmPAPI knockdown are indicated with red lines. (B) Total cell lysates from BmN4 cells treated with dsRNAs targeting Rluc (control), SIWI, BmAGO3, and BmPAPI were subjected to Western blots with the indicated antibodies. (C) Total RNAs from Rluc- (control) and BmPAPI-silenced BmN4 cells were treated with NaIO4, subjected to β-elimination, and detected by Northern blots. Total RNA treated with phosphatase before the NaIO4 reaction was also analyzed. Let-7 was shortened by β-elimination due to its hydroxyl 3′ terminus, whereas piRNAs were not affected due to 2′-O-methylation at their 3′ termini. (D) Immunofluorescence staining of endogenous SIWI and BmAGO3 (green) and DNA (blue) in Rluc- or BmPAPI-silenced BmN4 cells.
Our RNAi experiments successfully reduced the mRNA levels of 12 Bombyx TUDOR proteins out of the 15 proteins that we retrieved (Table 1; Supplemental Fig. S2), and identified three TUDOR proteins as factors apparently responsible for piRNA biogenesis (Fig. 3A). Since we analyzed only two piRNAs, we cannot exclude the possibility that additional TUDOR proteins influence the biogenesis of other piRNA species. The Bombyx GeneModels of the three proteins are BGIBMGA004949, BGIBMGA007473, and BGIBMGA011857, which are homologous to Spindle-E (Spn-E), PAPI, and TUDOR in Drosophila, respectively. We therefore named BmSpn-E, BmPAPI, and BmTUDOR. BmSpn-E depletion resulted in decreased levels of both SIWI- and BmAGO3-bound piRNAs, whereas BmTUDOR depletion increased the levels of the piRNAs. These results are in agreement with previous reports on Drosophila ovaries which implicated Spn-E and TUDOR in piRNA biogenesis, and showed that piRNA levels were severely decreased in spn-E mutant flies and that tudor mutant contained increased piRNA levels (Lim and Kai 2007; Malone et al. 2009; Nishida et al. 2009). Strikingly, as observed in sDMA-lacking BmAGO3-bound piRNAs, the BmPAPI depletion did not affect quantity but increased the lengths of both SIWI- and BmAGO3-bound piRNAs (Fig. 3A). The 3′ termini of the extended piRNAs were not eliminated by periodate oxidation followed by β-elimination, and phosphatase treatment did not alter the piRNA band positions (Fig. 3C). These results indicate that the extended piRNAs in BmPAPI-depleted cells retained 2′-O-methylation at their 3′ termini. We also confirmed normal localization of SIWI and BmAGO3 in BmPAPI-depleted cells (Fig. 3D). These results strongly suggest that BmPAPI functions in piRNA biogenesis in a way that does not disturb HEN1-mediated piRNA methylation or PIWI protein localization.
BmPAPI specifically binds to sDMAs and interacts with SIWI and BmAGO3
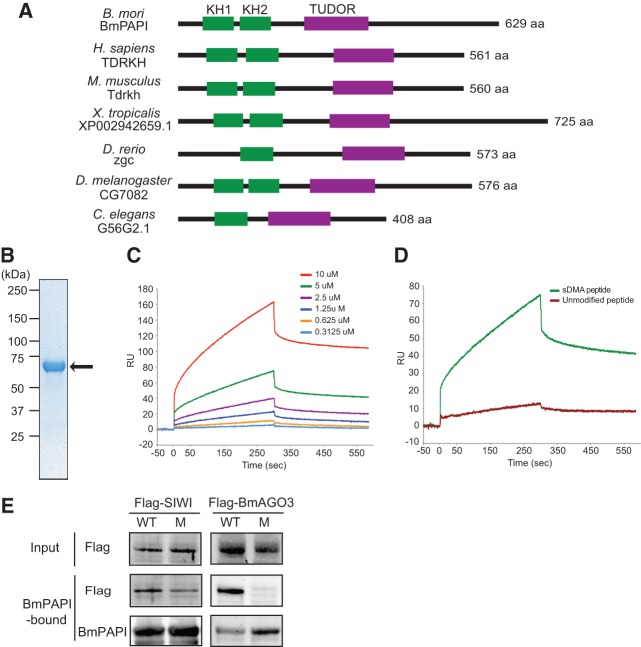
The BmPAPI gene is localized on scaf2886 of chromosome 3 in the B. mori genome. We isolated a full-length BmPAPI cDNA, which confirmed that the BmPAPI gene is ∼15 kb in length consisting of 13 exons, and encodes a 629-amino acid, 69-kDa protein (Supplemental Fig. S3A–C). Using BmPAPI as a query, we retrieved orthologs from human, mouse, Xenopus, zebrafish, Drosophila, and Caenorhabditis elegans genomes, which share two evolutionarily conserved domains: hnRNP K homology (KH) and TUDOR (Fig. 4A; Supplemental Fig. S3D). KH domains, known as nucleic acid recognition motifs, are tandemly arranged as KH1 and KH2 in many PAPI homologs. Zebrafish and C. elegans homologs lack the GXXG loop required for nucleotide recognition in their KH1 domain and thus contain only KH2 (Supplemental Fig. S3E). PAPI homologs also contain the extended TUDOR domain, which includes the core TUDOR domain sequence and additional conserved extensions (Supplemental Fig. S3F) as observed in other TUDOR proteins associated with the piRNA pathway (Liu et al. 2010a,b; Chen et al. 2011; Handler et al. 2011).
FIGURE 4.
BmPAPI specifically recognizes sDMAs and interacts with PIWI proteins. (A) Domain organizations of BmPAPI and its homologs, showing that the KH and TUDOR domains are evolutionarily conserved. (B) Coomassie-stained SDS-PAGE gel of the purified His-tagged ΔN-BmPAPI recombinant protein (indicated by the arrow). (C) Binding curves of ΔN-BmPAPI to an sDMA-containing peptide (MNLPPNPVIA[sDMA]G[sDMA]G[sDMA]G[sDMA]KPN) measured with an SPR binding assay. Curves represent measurements with an increasing concentration of the protein used from bottom to top as indicated. The vertical and horizontal axes show the response unit (RU) and the time scale (seconds). (D) Binding curves of ΔN-BmPAPI (5 μM) to the sDMA-containing peptide and an unmodified peptide (MNLPPNPVIARGRGRGRKPN). (E) Pull-down experiments to analyze the association of BmPAPI with PIWI proteins. His-BmPAPI immobilized on Ni-Dynabeads was incubated with the lysates from BmN4 stable cells expressing wild-type (WT) or mutant (M) PIWI proteins, and after extensive washing the eluates were probed with anti-FLAG antibody (for PIWI detection) and anti-His antibody (for BmPAPI detection).
To examine the biochemical properties of BmPAPI, we produced His-tagged BmPAPI recombinant protein in Escherichia coli. Although the full-length protein was insoluble, we were able to purify N-terminal deleted BmPAPI (ΔN-BmPAPI: amino acids 48–629) (Fig. 4B), which does not contain the transmembrane region predicted by the SOSUI program (Hirokawa et al. 1998). A surface plasmon resonance (SPR) binding assay revealed that ΔN-BmPAPI bound to an sDMA-containing peptide with a Kd of 27 ± 2 μM (Fig. 4C). However, ΔN-BmPAPI failed to bind unmodified peptide (Fig. 4D), suggesting that the TUDOR domain of BmPAPI specifically recognizes and interacts with sDMAs. To further examine the interaction between BmPAPI and PIWI proteins, we performed pull-down experiments. We immobilized the recombinant ΔN-BmPAPI protein on Ni-beads and subsequently incubated the beads with lysates from the BmN4 stable cell lines expressing wild-type or mutant PIWI proteins. After extensive washing, the eluates were subjected to Western blots to detect the bound PIWI proteins. As shown in Figure 4E, both wild-type and mutant SIWI were clearly detected in the BmPAPI-bound fraction. Although a slight decrease in mutant SIWI suggests a contribution of SIWI sDMAs to the BmPAPI association, the BmPAPI–SIWI interaction is not solely dependent on SIWI sDMAs. In contrast, only wild-type BmAGO3, and not mutant BmAGO3, was detected in the BmPAPI-bound fraction (Fig. 4E), suggesting that a direct association of the BmPAPI TUDOR domain with the BmAGO3 sDMA is essential for the BmPAPI–BmAGO3 interaction.
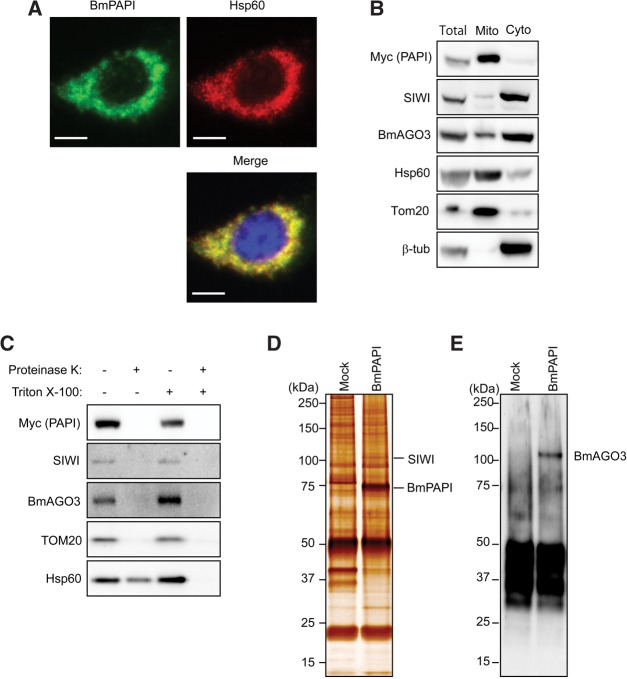
BmPAPI is localized on the outer surface of mitochondria
TargetP (Emanuelsson et al. 2000) and Psort II (Nakai and Horton 1999) prediction programs identified a putative mitochondrial localization signal that encompassed the transmembrane helix domain of the BmPAPI N terminus. This feature is conserved in the N-terminal regions of orthologs from other organisms such as humans, mice, Xenopus, zebrafish, and Drosophila. Indeed, we observed that stably expressed Myc-tagged BmPAPI co-localized with the mitochondrial protein Hsp60 in BmN4 cells (Fig. 5A), indicating mitochondrial localization of BmPAPI. In addition, we separated the mitochondrial fraction from cytoplasmic fraction and confirmed the presence of BmPAPI in the mitochondrial fraction by Western blots (Fig. 5B). Furthermore, SIWI and BmAGO3 were detected in the mitochondrial fraction, although the majority of the proteins were present in the cytoplasmic fraction, which is the presumed site of perinuclear granules (Fig. 5B). To further characterize the mitochondrial localization of BmPAPI, the mitochondrial fraction was treated with protease in the presence or absence of detergent, which disrupts mitochondrial membrane structure. Without detergent, the protease still was able to digest BmPAPI, SIWI, and BmAGO3 (Fig. 5C), suggesting that these proteins are commonly present on the outer surface of mitochondria. BmPAPI immunoprecipitates from the mitochondrial fraction contained a ∼100-kDa protein that was identified as SIWI by mass spectrometry (Fig. 5D). By Western blot, we also detected BmAGO3 in the immunoprecipitates (Fig. 5E). These results suggest that BmPAPI interacts with PIWI proteins on the outer surface of mitochondria and functions in piRNA biogenesis.
FIGURE 5.
BmPAPI is localized to mitochondrial outer membrane. (A) Immunofluorescence staining of stably expressed Myc-tagged BmPAPI (green, left) and Hsp60 (red, right) in BmN4 stable cell lines. Panels with DNA staining in blue were merged (lower panel) to show the overlap of the two fluorescence patterns. Scale bar, 10 μm. (B) The total protein lysate (Total), mitochondrial fraction (Mito), and cytoplasmic fraction (Cyto) were subjected to Western blots with the indicated antibodies. Hsp60 and Tom20 were used as markers for mitochondria, and β-tubulin was used as a cytoplasmic marker. (C) The mitochondrial fraction was treated with proteinase K in the presence or absence of Triton X-100. Hsp60 is localized in the intermembrane space and thus is digested only in the presence of detergent. Tom20 protrudes from the outer membrane and therefore is digested even without detergent treatment. Anti-Myc immunoprecipitates from the mitochondrial fraction of BmN4 cells (Mock) or Myc-tagged BmPAPI-expressing stable cells were developed by SDS-PAGE and subjected to silver-staining (D) or Western blot with anti-BmAGO3 (E).
BmPAPI depletion results in 3′-terminal extensions of mature piRNAs
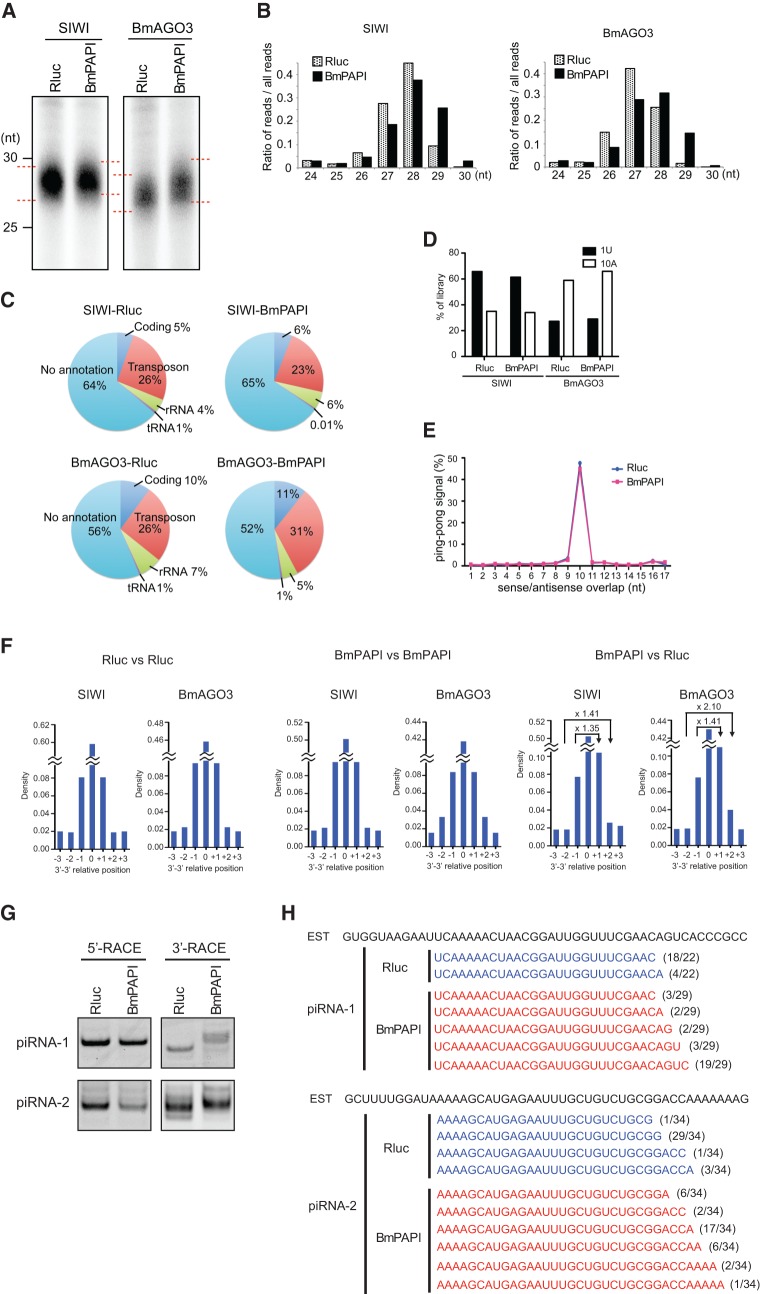
To examine the contribution of BmPAPI to piRNA biogenesis, we isolated and analyzed piRNAs from control (Rluc-depleted) cells and BmPAPI-depleted cells. Consistent with the analyses of individual piRNAs (Fig. 3A), total piRNAs bound to SIWI and BmAGO3 exhibited length extensions but no quantitative changes in BmPAPI-depleted cells (Fig. 6A). To examine the profiles of the extended piRNAs more comprehensively, we purified the piRNAs and performed deep sequencings, which yielded approximately 47.7, 33.7, 37.7, and 33.2 million reads for control SIWI, PAPI-depleted SIWI, control BmAGO3, and PAPI-depleted BmAGO3, respectively, that aligned to the B. mori genome. Consistent with gel mobility results (Fig. 6A), the size distributions of the reads clearly showed extended lengths of both SIWI- and BmAGO3-bound piRNAs from BmPAPI-depleted cells (Fig. 6B). BmPAPI depletion caused no remarkable changes in the annotations of SIWI- and BmAGO3-bound piRNA populations (Fig. 6C), and the majority of the piRNAs (97.7% and 97.9% of SIWI- and BmAGO3-bound piRNAs, respectively) derived from BmPAPI-depleted cells overlapped with those from control cells. In addition, upon BmPAPI depletion, we detected no appreciable changes in the features of 5′ terminus of primary piRNA biogenesis, a strong enrichment for U at their first position, and in the features of secondary piRNA biogenesis, a strong enrichment for A at their tenth position and Ping-pong signal (Fig. 6D,E). Given these results, we reasoned that the length extension occurs in 3′ terminus of mature piRNAs which could be caused by immature 3′-terminal processing.
FIGURE 6.
Characterization of piRNAs from BmPAPI-depleted cells. (A) BmN4 stable cell lines expressing FLAG-SIWI or FLAG-BmAGO3 were treated with dsRNAs targeting Rluc (control) or BmPAPI. SIWI- and BmAGO3-bound piRNAs were isolated from FLAG-tagged immunoprecipitates of the respective cell lysates. The piRNAs were 5′-end-labeled and analyzed by denaturing urea-PAGE. (B) Comparison of read-length distributions between the piRNAs from Rluc- (control) and BmPAPI-depleted cells. (C) Pie charts summarizing the annotations of SIWI- and BmAGO3-bound piRNAs from Rluc- or BmPAPI-depleted cells. (D) The preference for a first-position U (1U) or a tenth-position A (10A) in the piRNAs. (E) The Ping-pong analysis resulted in ∼50% of reads conforming to a separation preference of 10 nt (Ping-pong signal) in both Rluc- (control) and BmPAPI-depleted cells. (F) Relative terminal position analyses of piRNAs from BmPAPI-depleted cells compared with those from Rluc-depleted (control) cells. The relative position was defined as the 3′ genomic position of the piRNAs from BmPAPI-depleted cells minus the position of those from control cells. Fold changes between positive and negative positions were depicted in the panels. The analyses between the same samples were shown on the left (Rluc vs. Rluc) and middle (BmPAPI vs. BmPAPI) panels as negative controls that showed no density change between positive and negative positions. (G) 5′- and 3′-RACE of piRNA-1 and piRNA-2 in Rluc- (control) and BmPAPI-depleted cells. The PCR products of 3′-RACE in BmPAPI-depleted cells were longer than those in the control cells. (H) The populations of piRNA-1 and piRNA-2 clarified by 3′-RACE in Rluc- (control) and BmPAPI-depleted cells. piRNA-1 and piRNA-2 were aligned to rbmnc26i22 and bmnc17g01, the EST sequences from SilkBase, respectively. The numbers of each sequenced clone per total clones are indicated on the right.
We accordingly performed a relative terminal position analysis that calculated the distribution of the 3′-end relative positions for all pairwise combinations of overlapping piRNAs between the control and BmPAPI-depleted cells. As a control, running the analysis on a sample against itself (Rluc vs. Rluc and BmPAPI vs. BmPAPI) should result in a symmetrical distribution with the same density for positive and negative relative positions (e.g., +1 and +1, +2 and +2) (Fig. 6F). However, in the analyses on BmPAPI vs. Rluc, the distribution of the 3′-end relative positions was positively skewed, showing higher densities at +1 and +2 than those at −1 and −2. These results indicate that the 3′-ends of BmPAPI-depleted piRNAs occurred downstream from the 3′-ends of control piRNAs. Thus, on BmPAPI depletion, the piRNAs included 3′-end extensions. To confirm the 3′-end extensions in individual piRNAs, we performed RACE analyses of piRNA-1 and piRNA-2 in control and BmPAPI-depleted cells. These analyses revealed that 3′-RACE in BmPAPI-depleted cells produced longer products than those in control cells, while 5′-RACE products showed no difference in length (Fig. 6G). We cloned the 3′-RACE products and confirmed the 3′-end extensions of piRNA-1 and piRNA-2 in BmPAPI-depleted cells (Fig. 6H). These results strongly suggest that BmPAPI is involved in 3′-terminal processing machineries of piRNAs. In support of this inference, the BmN4 cell pellet with in vitro exonucleolytic piRNA 3′-end formation activity (Kawaoka et al. 2011) contained abundant BmPAPI along with other mitochondrial factors (Supplemental Fig. S4).
DISCUSSION
Since the discovery of PIWI sDMAs, TUDOR domain-containing proteins have attracted increasing attention as PIWI-binding factors involved in piRNA biogenesis. While studies in mice, Drosophila, and zebrafish have identified crucial roles for several TUDOR proteins in the piRNA pathway, detailed molecular functions of individual TUDOR proteins still remain ambiguous. The availability of efficient and convenient plasmid transfection and RNAi gene silencing techniques make BmN4 an ideal system to explore the function of TUDOR proteins, and we identified BmPAPI, BmSpn-E, and BmTUDOR as the Bombyx TUDOR proteins responsible for piRNA biogenesis. We were able to recapitulate in BmN4 cells the piRNA behaviors described previously in spn-E and tudor mutant flies (Lim and Kai 2007; Malone et al. 2009; Nishida et al. 2009): BmSpn-E or BmTUDOR depletion by RNAi resulted in severe decreases or increases in piRNA levels, respectively.
BmPAPI localizes and interacts with PIWI proteins at the outer membrane of mitochondria, suggesting that BmPAPI-involved piRNA biogenesis occurs at the mitochondrial surface. A link between the piRNA biogenesis and mitochondria has been implied by the earlier studies on Zucchini (Zuc), a member of the phospholipase D (PLD) superfamily in Drosophila. Zuc localizes to mitochondria (Saito et al. 2010; Huang et al. 2011a) and is essential for piRNA production in Drosophila ovaries (Pane et al. 2007; Haase et al. 2010; Olivieri et al. 2010; Saito et al. 2010). MitoPLD, the mammalian homolog of Zuc, is also a mitochondrial protein that plays crucial roles in piRNA biogenesis (Huang et al. 2011a; Watanabe et al. 2011). Zuc and MitoPLD possess an in vitro endonucleolytic activity toward single-stranded RNA to produce RNA fragment with 5′-monophosphate, a hallmark of mature piRNAs, and the activity is required for piRNA production in the cells (Ipsaro et al. 2012; Nishimasu et al. 2012). The possibility raised from these studies is that Zuc/MitoPLD cleaves piRNA precursors to generate 5′-ends of mature piRNAs at the mitochondrial surface. For the formation of piRNA 3′-ends, in vitro experiments using BmN4 cell lysates suggest that an unknown Trimmer exonucleolyticaly trims precursor piRNAs that are already loaded into PIWI proteins, followed by HEN1 methylation (Kawaoka et al. 2011). Since trimming-active BmN4 cell pellets contained various mitochondrial factors including BmPAPI, the formation of 3′-ends also might occur at the mitochondrial surface. Since BmPAPI depletion resulted in 3′-terminal extensions of mature piRNAs, we speculate that BmPAPI acts as a scaffold, interacting with PIWI proteins and other factors including unidentified Trimmer to stabilize piRNA 3′-end processing complexes and modulating the cleavage sites on the mitochondrial outer membrane (Fig. 7). An in vitro trimming assay using BmN4 cell lysate containing different levels of BmPAPI will be necessary to further corroborate the model.
FIGURE 7.
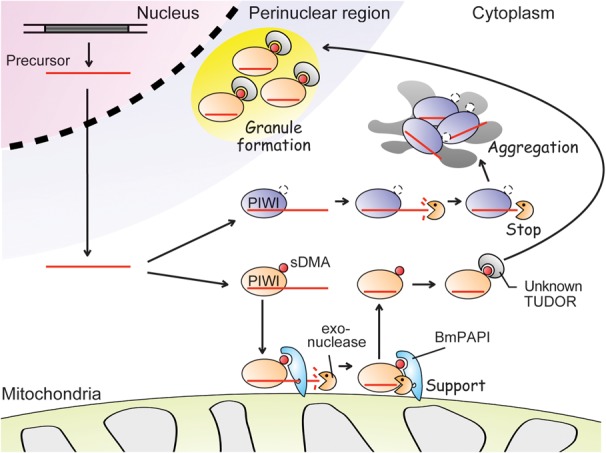
A proposed model for BmPAPI-involved piRNA biogenesis. The transcripts from piRNA clusters are shortened into piRNA precursors. The cleaved precursors are then loaded onto PIWI proteins. The unidentified exonuclease trims the precursor to form mature 3′-ends. BmPAPI interacts directly with PIWI proteins and supports the trimming activity. The sDMAs of PIWI proteins complexed with mature piRNAs are recognized by an unknown TUDOR domain-containing protein and are transferred to perinuclear granules. sDMA-lacking mutant PIWI proteins are not transferred, resulting in their aggregation in the cytoplasm.
Two tandemly arranged KH domains exist in BmPAPI. Since KH domains are nucleotide-binding elements (Valverde et al. 2008; Teplova et al. 2011), BmPAPI KH domains might be required to interact with piRNA precursors. It will be interesting to examine whether BmPAPI interacts with potential piRNA precursors and to elucidate the sequences. The TUDOR domain would be the source of the sDMA-specific binding capacity of BmPAPI. Interestingly, BmPAPI recognizes sDMAs of BmAGO3 for the BmPAPI–BmAGO3 interaction, whereas SIWI sDMAs are dispensable to the BmPAPI–SIWI interaction. Corresponding observations were made in PIWI mutant experiments showing that BmAGO3 mutant lacking sDMAs contained extended piRNAs presumably due to the lack of interaction with BmPAPI. Conversely, BmPAPI is likely to be able to associate with the SIWI mutant lacking sDMAs, thereby maintaining the lengths of bound piRNAs.
BmPAPI involvement in the PIWI/piRNA pathway is consistent with the result of earlier studies in Drosophila and mice. In Drosophila ovaries, PAPI interacts with PIWI proteins, particularly AGO3 (Liu et al. 2011). The absence of PAPI results in delocalization of AGO3 in nuage and transposon activation, implicating PAPI as a functional component in the piRNA pathway for transposon silencing (Liu et al. 2011). Proteomic surveys of mouse PIWI complexes have identified TDRKH/TDRD2, a mouse PAPI homolog, as a binding partner of mouse PIWI proteins (Chen et al. 2009; Vagin et al. 2009). During our manuscript preparation, Saxe and colleagues reported a functional characterization of mouse TDRKH, which plays crucial roles in spermatogenesis (Saxe et al. 2013). TDRKH localizes in mitochondria and interacts with MIWI and MIWI2 via sDMAs. TDRKH mutant mice showed 3′-end extension of piRNAs, suggesting its role in 3′-end formation of piRNAs. Therefore, mitochondrial localization, interaction with PIWI proteins, and functional involvement of piRNA 3′-end biogenesis are evolutionarily conserved characteristics of BmPAPI. Mouse TDRKH is expressed not only in testis but also in other tissues such as the brain and spinal cord (MitoCarta; http://www.broadinstitute.org/pubs/MitoCarta/) (Saxe et al. 2013), suggesting the involvement of PAPI in additional mitochondrial pathways.
Although BmPAPI depletion did not affect SIWI or BmAGO3 localization to perinuclear granules, mutant PIWI proteins lacking sDMAs formed large aggregates in the cytoplasm. Therefore, PIWI sDMAs are required not only for BmPAPI-involved piRNA biogenesis on mitochondria, but also for PIWI translocation from the mitochondria to perinuclear granules. This latter function could be mediated by other TUDOR domain-containing proteins (Fig. 7). It will be interesting to screen for TUDOR proteins that affect PIWI localization to further uncover PIWI–TUDOR interacting networks and to decode their functions in the piRNA pathway.
MATERIALS AND METHODS
BmN4 cell culture and generation of stable cell lines
BmN4 cells were cultured at 27°C in Insect-Xpress medium (Lonza). Plasmid constructs (pIZ/V5-His) encoding FLAG-tagged SIWI and FLAG-tagged BmAGO3 (Kawaoka et al. 2009) were a gift from S. Kawaoka and S. Katsuma. Mutant SIWI and BmAGO3 expression constructs in which arginines in sDMA motifs had been replaced with lysines were generated by site-directed mutagenesis (QuikChange XL, Stratagene) using the primers indicated in Supplemental Table S1. To generate the BmPAPI expression construct, a complete sequence of BmPAPI mRNA was first identified and confirmed by RACE as described below. The BmPAPI coding sequence with a C-terminal Myc-tag sequence then was amplified from BmN4 total RNA by RT-PCR using the following primers: forward (5′-TTTAAGCTTATGTCATTGAACACAAAATTAGCTTTGCCCATTGCTTTGGGCTTGTCCCTC-3′) and reverse (5′-AAAGCGGCCGCTCACAGATCCTCTTCAGAGATGAGTTTCTGCTCCTTTTCAAAAGCGGAC-3′). The PCR product was cloned into a pIZ/V5-His vector (Life Technologies) that contained a Zeocin-resistance gene for selection. The expression constructs were transfected into BmN4 cells using Escort transfection reagent (Sigma), and 500 μg/mL (final concentration) of Zeocin was added to the medium 3 d after transfection.
Determination of full-length cDNA sequence of BmPAPI by RACE
From SilkBase (http://silkbase.ab.a.u-tokyo.ac.jp/cgi-bin/index.cgi), we retrieved the BGIBMGA007473 GeneModel sequence as a significant homolog to Drosophila PAPI; this gene was named BmPAPI. The 5′- and 3′-UTR sequences of BmPAPI mRNA were isolated from BmN4 cells by RACE analysis using a FirstChoice RLM-RACE kit (Life Technologies) with the following primers: 5′-CGCTTCGACTGCTCTATCTCC-3′ (for 5′-RACE) and 5′-ATCTATCGTATCCCGACCCTTC-3′ (for 3′-RACE). We identified two sequence variants (162 nt and 102 nt) for the 5′ UTR, and a 252-nt sequence plus polyA was detected for 3′ UTR. We also identified the coding sequence of BmPAPI mRNA by RT-PCR using primers designed in 5′ UTR (5′-AATCGAGTAAAACGGCTCAACAGAA-3′) and 3′ UTR (5′-TTTCTGTACTCGCTTCCAATACG-3′), enabling determination of the full BmPAPI cDNA sequence. The length of the identified sequence (1890 nt) was longer than that of the BGIBMGA007473 GeneModel sequence (1638 nt), indicating an insertion of 84 amino acids (Supplemental Fig. S3A,B). The structure of the BmPAPI gene was determined by comparing the identified BmPAPI sequence with the B. mori genome sequence obtained from SilkDB (http://www.silkdb.org/silkdb/). The BmPAPI gene consisted of 13 exons and was localized to scaf2886 on chromosome 3 of the B. mori genome (Supplemental Fig. S3C).
Production of anti-SIWI antibody
Anti-SIWI antibody was prepared by immunizing rabbits with a synthetic peptide (HPDPMELYSDRKTDC; Genscript). Sera were affinity-purified over a column containing immobilized peptide, resulting in “S213” anti-SIWI antibody.
Western blot and immunoprecipitation
Western blot and immunoprecipitation were performed as described previously (Kirino et al. 2009). Cell lysates were prepared from BmN4 cells in a lysis buffer containing 20 mM Tris-HCl pH 7.4, 200 mM NaCl, 2.5 mM MgCl2, 0.5% NP-40, 0.1% Triton X-100 and complete protease inhibitor (Roche Diagnostics). For Western blots, the following antibodies were used: S213 anti-SIWI, anti-BmAGO3 (a gift from S. Kawaoka and S. Katsuma) (Kawaoka et al. 2009), Y12 (a gift from G. Dreyfuss), anti-Flag (Sigma), anti-Myc (Cell Signaling), anti-His (Cell Signaling), anti-Tom20 (Santa Cruz Biotechnology), anti-Hsp60 (Cell Signaling), and anti-β-tubulin (Developmental Studies Hybridoma Bank). Anti-FLAG M2 affinity gel (Sigma) was used for immunoprecipitation of FLAG-tagged proteins, whereas Dynabeads Protein A (Life Technologies) incubated with anti-Myc was used for Myc-immunoprecipitation. Mass-spec protein identification in BmPAPI-immunoprecipitates was performed by Taplin Mass Spectrometry Facility (Harvard Medical School), and the two peptides from SIWI, “VLPETISILR (125–134)” and “EGTIAPTSYNVIEDTTGLNPDR (824–845),” were detected from ∼100-kDa protein.
Immunofluorescence and confocal microscopy
For mitochondrial staining, BmN4 cells were incubated with 300 nM MitoTracker Red (Life Technologies) for 30 min at 27°C. Immunofluorescence staining was performed as previously described (Nakaya et al. 2008) using S213 anti-SIWI (diluted 1:2000), anti-BmAGO3 (1:5000), anti-FLAG (1:1000), anti-Myc (1:2000), and anti-Hsp60 (1:500) as primary antibodies. Alexa Fluor 488 goat anti-rabbit IgG and Alexa Fluor 555 goat anti-mouse IgG (Life Technologies) were used as secondary antibodies. After DNA counterstaining with ProLong Gold Antifade Reagent with DAPI (Life Technologies), images for Figures 1C,D and 5A were acquired using an Eclipse Ti-U microscope with NIS-Elements BR 3.2 software (Nikon). Additional images for Figure 3D and Supplemental Figure S1B were acquired using Leica SP5 confocal microscopy.
RNA isolation, labeling, and β-elimination
RNA isolation, labeling and β-elimination were performed as described previously (Kirino and Mourelatos 2007). Briefly, total RNA was isolated from BmN4 cells or immunoprecipitates using Trizol reagent (Life Technologies). For radiolabeling, isolated RNAs were treated with calf intestinal phosphokinase (CIP; New England Biolabs) and were subjected to 5′-end labeling using [γ-32P] ATP (American Radiolabeled Chemicals) and T4 polynucleotide kinase (New England Biolabs). Labeled RNAs were resolved by 15% PAGE containing 7 M urea and were visualized by storage phosphor autoradiography using a PharosFX Plus Molecular Imager and Quantity One software (Bio-Rad). For β-elimination reaction, RNAs were incubated with 10 mM NaIO4 at 0°C for 40 min in the dark. Subsequently, 1 M rhamnose (1/10 volume) was added to quench unreacted NaIO4 and incubated at 0°C for 30 min. β-elimination was then performed by adding equal volume of 2 M Lys-HCl (pH 8.5) followed by incubation at 45°C for 90 min. The treated RNAs were collected by ethanol precipitation.
Northern blot
Bm–piRNA-1 (5′-UCAAAAACUAACGGAUUGGUUUCGAAC-3′), Bm–piRNA-2 (5′-AAAAGCAUGAGAAUUUGCUGUCUGCGG-3′), and Bm–let-7 (5′-UGAGGUAGUAGGUUGUAUAGUA-3′) were analyzed by Northern blots. Total RNA was resolved by 15% PAGE containing 7 M urea, transferred to Hybond N+ membranes (GE Healthcare), and hybridized to 5′-end-labeled antisense probes detecting Bm–piRNA-1 (5′-GTTCGAAACCAATCCGTTAGTTTTTGA-3′), Bm–let-7 (5′-TACTATACAACCTACTACCTCA-3′) (Kawaoka et al. 2009), and Bm–piRNA-2 (5′-CCGCAGACAGCAAATTCTCATGCTTTT-3′).
RNAi knockdown of target gene expression in BmN4 cells
Templates for the in vitro synthesis of dsRNA were amplified via nested RT-PCR using the primers indicated in Supplemental Table S2. The first PCR was performed using a cDNA template produced by oligo-dT-based reverse transcription from BmN4 total RNA. The second PCR was designed to attach T7 promoter sequence (5′-TAATACGACTCACTATAGGG-3′) to both strands of the amplified DNA. The resultant products were used as templates in a T7 in vitro transcription system (MEGAscript T7, Ambion), followed by purification with MEGAclear (Ambion). dsRNA (5 μg) was transfected into 5 × 106 BmN4 cells using 4D-Nucleofector (Lonza), and cells were harvested four days after transfection. The amounts of targeted mRNAs were analyzed by Real-Time PCR as previously described (Kirino et al. 2009) using the primers indicated in Supplemental Table S3.
Expression and purification of recombinant BmPAPI protein
Nucleotides 142–1887 (amino acids 48–629) of the BmPAPI ORF were amplified by PCR using the following primers: forward (5′-AAACATATGACAATTGAAATTCATGTTC-3′) and reverse (5′-TTTGCGGCCGCCTTTTCAAAAGCGGACTTAC-3′). The product was cloned into pET-21a (Novagen) to attach a C-terminal His-tag sequence and was expressed in E. coli BL21-Codon-Plus (Stratagene) at 16°C for 16 h in the presence of 100 μM IPTG. The expressed protein was purified using a Ni-affinity column (linear gradient of 40–240 mM imidazole; Hi-Trap Chelating HP, GE Healthcare), and was further purified using Resource Q (linear gradient of 0–350 mM NaCl; GE Healthcare).
Surface plasmon resonance (SPR) binding assay
SPR binding experiments were performed using a SensiQ dual channel semi-automated SPR instrument (Pioneer) at a sensor temperature of 16°C with running buffer composed of 20 mM Tris-HCl pH 8.0, 220 mM NaCl, and 0.005% Tween-20. A biotinylated unmodified peptide (Bio-MNLPPNPVIARGRGRGRKPN) and an sDMA-containing peptide (Bio-MNLPPNPVIA[sDMA]G[sDMA]G[sDMA]G[sDMA]KPN), which we described previously (Kirino et al. 2010), were immobilized on two flow cells of an avidin-coupled sensor chip (BioCap, SensiQ). For immobilization, 150 μL of a 10 μg/mL peptide solution in running buffer was injected onto the sensor surface at 470 or 430 response units (RUs). The second flow cell of the chip was used for a blank control. BmPAPI protein was passed through flow cells of peptide-coupled chips at various concentrations and at a flow rate of 20 μL/min. The association and disassociation time were 100 sec and 420 sec, respectively. Kinetic constants were calculated from the sensor grams using Qdat software (SensiQ).
Pull-down binding assay
BmN4 cell lysates were prepared in a binding buffer containing 20 mM Tris-HCl pH 7.4, 100 mM NaCl, 2.5 mM MgCl2, 0.5% NP-40, 0.1% Triton X-100, and complete protease inhibitor cocktail (Roche Diagnostics). Recombinant BmPAPI protein immobilized on Dynabeads His-Tag Isolation and Pull-down (Life Technologies) was incubated with the lysate for 20 min at 4°C and was washed extensively with the binding buffer. Bound proteins were then eluted into an elution buffer containing 50 mM sodium-phosphate pH 8.0, 300 mM imidazole, 140 mM NaCl, and 0.02% Tween-20. Western blots of the proteins were subsequently performed.
Preparation of mitochondrial fraction and piRNA precursor-trimming active fraction
Mitochondrial fractions were prepared from BmN4 cells as described previously (Tomitsuka et al. 2003). Approximately 1 × 108 BmN4 cells were suspended with 5 mL sucrose gradient buffer composed of 0.25 M sucrose, 20 mM HEPES-KOH pH 7.5, and 3 mM EDTA, and homogenized by 7 mL daunce tissue grinder (Wheaton) on ice. The homogenate was made up to 10 mL and centrifuged at 600g for 15 min twice to pellet cell debris and nuclei. The supernatant was further centrifuged at 15,000g for 15 min and the pellet was resuspended with 5 mL buffer, followed by centrifugation again at 15,000g for 15 min. The pellet (mitochondrial fraction) and the supernatant (cytosolic fraction) were analyzed by Western blots. For immunoprecipitation, the mitochondrial fraction was resuspended with a lysis buffer and homogenized by dauncing. For protease treatment, the freshly isolated mitochondrial fraction was resuspended in 100 µL of sucrose gradient buffer and incubated with Proteinase K (200 µg/mL, Roche Diagnostics) in the presence or absence of 1% Triton X-100 at 4°C for 30 min. Digestion was terminated using phenylmethylsulfonylfluoride (2 mM, Sigma). The BmN4 cell lysate possessing piRNA precursor-trimming activity was prepared as described previously (Kawaoka et al. 2011). Briefly, BmN4 cells were suspended in equal volume of a buffer containing 30 mM HEPES-KOH pH 7.4, 100 mM KOAc, 2 mM Mg(OAc)2, 5 mM DTT, and complete protease inhibitor cocktail (Roche Diagnostics), and homogenized by dauncing. The homogenate was centrifuged at 1000g for 20 min at 4°C. The pellet (trimming active fraction) and supernatant (trimming inactive fraction) were analyzed by Western blots.
Next-generation sequencing of piRNAs and bioinformatics
piRNAs purified from PIWI immunoprecipitates were 5′-end-labeled and gel-purified. Directional ligation of adaptors and cDNA generation were performed using a Truseq Small RNA Sample Prep kit (Illumina). Deep sequencing was performed on a Genome Analyzer IIx (Illumina). Reads were trimmed at their 3′-ends to remove nucleotides which were identified with reduced quality due to Illumina Analyzer bias to identify bases with lower accuracy toward the 3′-ends of the reads. Trimming was performed as described in the BWA alignment software documentation (Li and Durbin 2009). The 3′-end ligated adaptor (RL3) was removed from the sequences using cutadapt (Martin 2011) with an acceptable error rate of 0.25 for the adaptor alignment on the reads. Reads that were <15 nt after adaptor removal were excluded from further analyses. Remaining reads were aligned to the B. mori (bm0) genome (downloaded from SilkDB; http://www.silkdb.org/silkdb/) using the BWA alignment program (Li and Durbin 2009) with default program parameters allowing for 0.04 fraction of missing alignments given a 2% uniform base error rate. To refine the library and avoid randomly aligned reads, all aligned reads <20 nt which aligned to more than one position on the genome were also excluded from further analyses. In relative terminal position analysis, we calculated the distribution of the 3′–3′-end relative positions for all pairwise combinations of overlapping piRNAs between two given samples. For each piRNA in the first sample, we identified all overlapping piRNAs from the second sample. We then calculated the 3′–3′-end relative positions between each piRNA from the first sample and all its overlapping ones from the second sample. The number of piRNA pairs at each specific relative position was normalized by dividing by the total number of piRNA pairs. Using these normalized counts, a density plot was created as described in Figure 6F.
RACE analyses of piRNA-1 and piRNA-2
For 5′-RACE, 500 ng of total RNA was ligated to 20 pmol RNA adaptor (5′-GUUCAGAGUUCUACAGUCCGACGAUC-3′) using T4 RNA ligase (Fisher Scientific) and subjected to RT-PCR using forward (5′-GTTCAGAGTTCTACAGTCCGACGATC-3′) and reverse (5′-GCCGTTCGAAACCAATCCGTTAG-3′ for piRNA-1; 5′-CGACCGCAGACAGCAAATTCTC-3′ for piRNA-2) primers by One Step SYBR Ex Taq qRT-PCR Kit (Takara). For 3′-RACE, total RNA was first incubated with 10 mM NaIO4 at 0°C for 40 min in the dark to disrupt the 3′-ends of RNAs other than the 3′-end-protected RNAs such as 2′-O-methylated piRNAs. After CIP treatment, 200 ng of the total treated RNA was ligated to 20 pmol RNA adaptor (5′-phos-UGGAAUUCUCGGGUGCCAAGG-dideoxycytidine-3′) using T4 RNA ligase (Fisher Scientific) and subjected to RT-PCR using forward (5′-GCCGTCAAAAACTAACGGATTGG-3′ for piRNA-1; 5′-CGGCAAAAGCATGAGAATTTGCTG-3′ for piRNA-2) and reverse (5′-GCCTTGGCACCCGAGAATTCCA-3′) primers with One Step SYBR Ex Taq qRT-PCR Kit. The PCR products from 3′-RACE were cloned with a StrataClone PCR cloning kit (Agilent Technologies).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Shinpei Kawaoka and Susumu Katsuma for providing BmN4 cells, PIWI antibodies and constructs, and for their technical advice; Gideon Dreyfuss for Y12 antibody; Takuma Suematsu for technical advice; Anastasios Vourekas for discussion; Mark Greene for encouragement and support; and the members of our laboratories for helpful discussions. Illumina next-generation sequencing was performed at the Genomics Core of Cedars-Sinai Medical Center, and we are grateful to Vincent Funari. This work was supported by the Cedars-Sinai Medical Center Research Fund, Martz Translational Breast Cancer Career Development Grant, and a Grant for Basic Science Research Project from The Sumitomo Foundation to Yohei K.; and NIH grant (GM072777) to Z.M.
REFERENCES
- Anne J, Ollo R, Ephrussi A, Mechler BM 2007. Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development 134: 137–146 [DOI] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG 2009. Protein arginine methylation in mammals: Who, what, and why. Mol Cell 33: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell E, Ceman S 2012. Arginine methylation of RNA-binding proteins regulates cell function and differentiation. Mol Reprod Dev 79: 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahms H, Raymackers J, Union A, de Keyser F, Meheus L, Luhrmann R 2000. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J Biol Chem 275: 17122–17129 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Chen C, Jin J, James DA, Adams-Cioaba MA, Park JG, Guo Y, Tenaglia E, Xu C, Gish G, Min J, et al. 2009. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc Natl Acad Sci 106: 20336–20341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Nott TJ, Jin J, Pawson T 2011. Deciphering arginine methylation: Tudor tells the tale. Nat Rev Mol Cell Biol 12: 629–642 [DOI] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Tanaka T, Nakatsuji N 2009. Ultrastructural characterization of spermatogenesis and its evolutionary conservation in the germline: Germinal granules in mammals. Mol Cell Endocrinol 306: 17–23 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Gonsalvez GB, Rajendra TK, Tian L, Matera AG 2006. The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr Biol 16: 1077–1089 [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, Pappin DJ, Chen C, Gordon A, Hannon GJ 2010. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev 24: 2499–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Olivieri D, Novatchkova M, Gruber FS, Meixner K, Mechtler K, Stark A, Sachidanandam R, Brennecke J 2011. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J 30: 3977–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa T, Boon-Chieng S, Mitaku S 1998. SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics 14: 378–379 [DOI] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD 2007. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 17: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Huang H, Gao Q, Peng X, Choi SY, Sarma K, Ren H, Morris AJ, Frohman MA 2011a. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell 20: 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HY, Houwing S, Kaaij LJ, Meppelink A, Redl S, Gauci S, Vos H, Draper BW, Moens CB, Burgering BM, et al. 2011b. Tdrd1 acts as a molecular scaffold for Piwi proteins and piRNA targets in zebrafish. EMBO J 30: 3298–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ 2012. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 491: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizu H, Siomi H, Siomi MC 2012. Biology of PIWI-interacting RNAs: New insights into biogenesis and function inside and outside of germlines. Genes Dev 26: 2361–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka S, Hayashi N, Suzuki Y, Abe H, Sugano S, Tomari Y, Shimada T, Katsuma S 2009. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA 15: 1258–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka S, Izumi N, Katsuma S, Tomari Y 2011. 3′ end formation of PIWI-interacting RNAs in vitro. Mol Cell 43: 1015–1022 [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Hara K, Shoji K, Kobayashi M, Shimada T, Sugano S, Tomari Y, Suzuki Y, Katsuma S 2013. The comprehensive epigenome map of piRNA clusters. Nucleic Acids Res 41: 1581–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z 2007. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol 14: 347–348 [DOI] [PubMed] [Google Scholar]
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z 2009. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol 11: 652–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Vourekas A, Sayed N, de Lima Alves F, Thomson T, Lasko P, Rappsilber J, Jongens TA, Mourelatos Z 2010. Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA 16: 70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause CD, Yang ZH, Kim YS, Lee JH, Cook JR, Pestka S 2007. Protein arginine methyltransferases: Evolution and assessment of their pharmacological and therapeutic potential. Pharmacol Ther 113: 50–87 [DOI] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA Jr, Steitz JA 1981. Monoclonal antibodies to nucleic acid-containing cellular constituents: Probes for molecular biology and autoimmune disease. Proc Natl Acad Sci 78: 2737–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Kai T 2007. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci 104: 6714–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang JY, Huang Y, Li Z, Gong W, Lehmann R, Xu RM 2010a. Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev 24: 1876–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Chen C, Guo Y, Lam R, Bian C, Xu C, Zhao DY, Jin J, MacKenzie F, Pawson T, et al. 2010b. Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc Natl Acad Sci 107: 18398–18403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Qi H, Wang J, Lin H 2011. PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development 138: 1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ 2009. Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M 2011. Cutadapt removes adaptor sequences from high-throughput sequencing reads. EMBnet J 17: 10–12 [Google Scholar]
- Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP 2003. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci 28: 69–74 [DOI] [PubMed] [Google Scholar]
- Nakai K, Horton P 1999. PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci 24: 34–36 [DOI] [PubMed] [Google Scholar]
- Nakaya T, Kawai T, Suzuki T 2008. Regulation of FE65 nuclear translocation and function by amyloid β-protein precursor in osmotically stressed cells. J Biol Chem 283: 19119–19131 [DOI] [PubMed] [Google Scholar]
- Nishida KM, Okada TN, Kawamura T, Mituyama T, Kawamura Y, Inagaki S, Huang H, Chen D, Kodama T, Siomi H, et al. 2009. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J 28: 3820–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, et al. 2012. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 491: 284–287 [DOI] [PubMed] [Google Scholar]
- Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T 2007. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol 14: 349–350 [DOI] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J 2010. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J 29: 3301–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schupbach T 2007. Zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell 12: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pek JW, Anand A, Kai T 2012. Tudor domain proteins in development. Development 139: 2255–2266 [DOI] [PubMed] [Google Scholar]
- Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS 2009. Loss of the Mili-interacting Tudor domain–containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol 16: 639–646 [DOI] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC 2007. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev 21: 1603–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC 2010. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev 24: 2493–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxe JP, Chen M, Zhao H, Lin H 2013. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J 32: 1869–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti KA, Brennecke J 2010. The piRNA pathway: A fly's perspective on the guardian of the genome. Trends Genet 26: 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Mannen T, Siomi H 2010. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev 24: 636–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA 2011. PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258 [DOI] [PubMed] [Google Scholar]
- Teplova M, Malinina L, Darnell JC, Song J, Lu M, Abagyan R, Musunuru K, Teplov A, Burley SK, Darnell RB, et al. 2011. Protein-RNA and protein-protein recognition by dual KH1/2 domains of the neuronal splicing factor Nova-1. Structure 19: 930–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomitsuka E, Goto Y, Taniwaki M, Kita K 2003. Direct evidence for expression of type II flavoprotein subunit in human complex II (succinate-ubiquinone reductase). Biochem Biophys Res Commun 311: 774–779 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA 2009. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev 23: 1749–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde R, Edwards L, Regan L 2008. Structure and function of KH domains. FEBS J 275: 2712–2726 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, et al. 2011. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell 20: 364–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiol J, Cora E, Koglgruber R, Chuma S, Subramanian S, Hosokawa M, Reuter M, Yang Z, Berninger P, Palencia A, et al. 2012. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol Cell 47: 970–979 [DOI] [PubMed] [Google Scholar]