In nonsense-mediated mRNA decay (NMD), an exon-junction complex (EJC) located downstream from a termination codon acts as an enhancing signal, but is generally not required. We compared “EJC-enhanced” and “EJC-independent” modes of NMD with regard to their requirement for seven known NMD factors in human cells. This study suggests the existence of several mechanistically distinct branches of NMD and reveals a redundancy of the endo- and exonucleolytic mRNA degradation pathways in both modes of NMD.
Keywords: post-transcriptional gene regulation, mRNA turnover, mRNA surveillance, nonsense-mediated mRNA decay, exon-junction complex, endo- and exonucleolytic mRNA degradation; UPF1; UPF2; UPF3b; SMG1; SMG5; SMG6; SMG7
Abstract
Nonsense-mediated mRNA decay (NMD) is a eukaryotic post-transcriptional gene regulation mechanism that eliminates mRNAs with the termination codon (TC) located in an unfavorable environment for efficient translation termination. The best-studied NMD-targeted mRNAs contain premature termination codons (PTCs); however, NMD regulates even many physiological mRNAs. An exon-junction complex (EJC) located downstream from a TC acts as an NMD-enhancing signal, but is not generally required for NMD. Here, we compared these “EJC-enhanced” and “EJC-independent” modes of NMD with regard to their requirement for seven known NMD factors in human cells using two well-characterized NMD reporter genes (immunoglobulin μ and β-Globin) with or without an intron downstream from the PTC. We show that both NMD modes depend on UPF1 and SMG1, but detected transcript-specific differences with respect to the requirement for UPF2 and UPF3b, consistent with previously reported UPF2- and UPF3-independent branches of NMD. In addition and contrary to expectation, a higher sensitivity of EJC-independent NMD to reduced UPF2 and UPF3b concentrations was observed. Our data further revealed a redundancy of the endo- and exonucleolytic mRNA degradation pathways in both modes of NMD. Moreover, the relative contributions of both decay pathways differed between the reporters, with PTC-containing immunoglobulin μ transcripts being preferentially subjected to SMG6-mediated endonucleolytic cleavage, whereas β-Globin transcripts were predominantly degraded by the SMG5/SMG7-dependent pathway. Overall, the surprising heterogeneity observed with only two NMD reporter pairs suggests the existence of several mechanistically distinct branches of NMD in human cells.
INTRODUCTION
Nonsense-mediated mRNA decay (NMD) is a eukaryotic post-transcriptional mechanism to regulate gene expression. NMD affects the half-life of many physiological mRNAs and at the same time serves as a quality-control process that recognizes and degrades aberrant mRNAs with truncated open reading frames (for review, see Nicholson et al. 2010; Huang and Wilkinson 2012). Up to 10% of all mRNAs in Homo sapiens (Mendell et al. 2004; Wittmann et al. 2006; Chan et al. 2007; Huang et al. 2011; Yepiskoposyan et al. 2011; Tani et al. 2012), Saccharomyces cerevisiae (Lelivelt and Culbertson 1999; He et al. 2003; Guan et al. 2006; Johansson et al. 2007), Caenorhabditis elegans (Ramani et al. 2009), and Drosophila melanogaster (Rehwinkel et al. 2005) are regulated by NMD. The exact mechanism of substrate selection is not yet fully understood, but several mRNA features have been identified that can trigger NMD. These include a long 3′ untranslated region (UTR) that spatially separates the translation termination codon (TC) from the poly(A)-binding protein (PABP) (Amrani et al. 2004; Behm-Ansmant et al. 2007; Eberle et al. 2008; Ivanov et al. 2008; Silva et al. 2008; Singh et al. 2008), upstream open reading frames (uORFs), intron-containing 3′ UTRs (Thermann et al. 1998), an exon-junction complex (EJC) located downstream from the TC (Maquat 2004), and a premature termination codon (PTC), the classical and so far best examined feature. PTCs usually arise from genomic mutations or as a result of alternative splicing (Nicholson et al. 2010). PTC-containing (PTC+) mRNAs can be dangerous for the cell because they encode C-terminally truncated proteins with potentially detrimental functions.
Concerning the trans-acting factors involved in NMD, the three proteins UPF1, UPF2, and UPF3 (termed SMG2-4 in C. elegans) are conserved from yeast to humans and form a complex (He et al. 1997; Lykke-Andersen et al. 2000; Serin et al. 2001). UPF1 is a nucleic-acid-dependent ATPase and RNA helicase that binds with its N-terminal CH-rich domain to a natively unfolded C-terminal region of UPF2 (Clerici et al. 2009). In humans, UPF1 is a phosphoprotein, and both its phosphorylation–dephosphorylation cycle and its helicase activity are essential for NMD (Czaplinski et al. 1995; Weng et al. 1996; Bhattacharya et al. 2000; Ohnishi et al. 2003). With the last of its three MIF4G (middle portion of eIF4G) domains, UPF2 interacts with UPF3 (Kadlec et al. 2004) and thus represents the bridge between UPF1 and UPF3. Mammalian genomes encode two highly similar UPF3 genes, UPF3a and the X-chromosome-linked UPF3b (Lykke-Andersen et al. 2000), of which the UPF3b protein seems to be more effective in NMD than UPF3a (Kunz et al. 2006; Chan et al. 2009).
Additional NMD factors have been identified in metazoans. In mammals, these include SMG1, a phosphoinositide 3-kinase-related kinase (PIKK) that phosphorylates UPF1 (Page et al. 1999; Denning et al. 2001; Pal et al. 2001; Yamashita et al. 2001), and SMG5, SMG6, and SMG7, three proteins that contain a 14-3-3-like domain through which they interact with phosphorylated UPF1 (Okada-Katsuhata et al. 2012). SMG6 is an endonuclease that has been shown to cleave PTC-containing transcripts near the PTC in mammalian and D. melanogaster cells (Huntzinger et al. 2008; Eberle et al. 2009). SMG5 and SMG7 form a heterodimer (Ohnishi et al. 2003; Jonas et al. 2013), and the proline-rich C-terminus of SMG7 is sufficient to trigger degradation by a yet-unknown mechanism when tethered to a reporter mRNA (Unterholzner and Izaurralde 2004). A functional link between SMG5 and the proline-rich nuclear receptor coregulatory protein 2 (PNRC2) has recently been indicated to promote decapping (Cho et al. 2009, 2013). Furthermore, SMG8 and SMG9 were identified as subunits of the SMG1 complex in human cells that negatively regulate the kinase activity of SMG1 (Yamashita et al. 2009).
The emerging unified NMD model proposes a mechanistic and kinetic difference between proper translation termination and aberrant termination that results in NMD (for review, see Kervestin and Jacobson 2012). According to this model, NMD ensues when the ribosome at the TC fails to release properly from the mRNA, for example, due to the missing interaction of PABP with the eukaryotic release factor eRF3, which normally seems to trigger efficient translation termination on correctly positioned stop codons and which efficiently suppresses NMD when artificially brought into proximity of PTCs (Mangus et al. 2003; Amrani et al. 2004; Behm-Ansmant et al. 2007; Eberle et al. 2008; Ivanov et al. 2008; Silva et al. 2008; Singh et al. 2008). During aberrant translation termination at a PTC, UPF1 is thought to be recruited to the stalling ribosome by interaction with eRF3 (Czaplinski et al. 1998). In human cells, a ribosome-associated complex called SURF has been detected, consisting of the SMG1 complex (SMG1, SMG8, and SMG9), hypophosphorylated UPF1, eRF1, and eRF3 (Kashima et al. 2006). According to the proposed model, UPF1 and the SMG1 complex subsequently form a complex with UPF2 that in a classical NMD substrate is associated with an exon-junction complex (EJC; see below) positioned downstream from the PTC on the mRNA (Melero et al. 2012). This leads to the formation of the decay-inducing (DECID) complex (Kashima et al. 2006), in which the UPF1 helicase activity is derepressed by a structural rearrangement and the activated SMG1 kinase phosphorylates UPF1 at several serine/threonine–glutamine (S/TQ) motifs in the N- and C-terminal regions (Yamashita et al. 2001, 2009; Chakrabarti et al. 2011; Fiorini et al. 2012; Okada-Katsuhata et al. 2012). Phosphorylated UPF1, in turn, has been shown to interact with the decay-inducing factors SMG6, the heterodimer SMG5/SMG7, and PNRC2 (Cho et al. 2009; Okada-Katsuhata et al. 2012; Jonas et al. 2013). Whether the mRNA decay induced by SMG6-mediated endonucleolysis (Huntzinger et al. 2008; Eberle et al. 2009) and the deadenylation- and decapping-dependent exonucleolytic mRNA degradation (Chen and Shyu 2003; Lejeune et al. 2003; Couttet and Grange 2004) represent independent, redundant NMD pathways, whether they are functionally linked, or whether they act in a transcript-specific way is currently not clear (Muhlemann and Lykke-Andersen 2010; Cho et al. 2013).
It should be emphasized that many aspects of the above-outlined working model are based on studies with one or few specific NMD reporter mRNAs, and there is evidence suggesting that several different NMD routes acting on distinct subpopulations of NMD substrates might exist. For example, NMD pathways have been reported in human cells that can occur independently of UPF2 or independently of UPF3b (Chan et al. 2007; Gehring et al. 2009a). The determinants specifying a particular NMD substrate for one or the other pathway, however, remain to be elucidated.
In mammals, the presence of an EJC >30 nucleotides (nt) downstream from the TC functions as an important NMD enhancing factor (Stalder and Muhlemann 2008; Rebbapragada and Lykke-Andersen 2009). The EJC is a multimeric protein complex deposited by the splicing machinery 24 nt upstream of the spliced exon–exon boundary in metazoans (Le Hir et al. 2000; Sauliere et al. 2012; Singh et al. 2012). The EJC core consists of the four proteins eIF4AIII, Y14, MAGOH, and MLN51 (also known as Barentz) (Andersen et al. 2006; Bono et al. 2006), and additional factors associate with the EJC core in a dynamic way, including Pinin, SKAR, SRm160, RNPS1, Acinus, SAP18, UAP56, Aly/REF, and the two NMD factors UPF3b and SMG6 (Le Hir et al. 2000, 2001; Kashima et al. 2010; Bono and Gehring 2011; Murachelli et al. 2012). UPF3b binds the EJC core via its C-terminal low-complexity region (Gehring et al. 2003; Buchwald et al. 2010), and two conserved EJC-binding motifs (EBMs) were identified in the N-terminal part of SMG6 that interact with essentially the same surface on the EJC core as UPF3b (Kashima et al. 2010). EJCs located in the 5′ UTR and coding sequence of an mRNA are disassembled by the elongating ribosome by a process that involves the ribosome-bound protein PYM (Gehring et al. 2009b; Bono and Gehring 2011). After onset of translation, only EJCs positioned >30 nt downstream from the TC will therefore remain bound to the mRNA, and the vast majority of cellular mRNAs will be devoid of EJCs (Lejeune et al. 2002).
Initially, the presence of EJCs downstream from TCs was thought to be an essential signal to identify the TC as premature and elicit NMD (Maquat 2004), but it is meanwhile clear that mammalian NMD can also occur in the absence of an EJC downstream from the PTC (Zhang et al. 1998; Rajavel and Neufeld 2001; Delpy et al. 2004; LeBlanc and Beemon 2004; Buhler et al. 2006; Matsuda et al. 2007; Eberle et al. 2008; Singh et al. 2008). Since NMD is triggered upon aberrant translation termination, which can only occur after at least one ribosome has passed the entire coding sequence and thereby removed the EJCs, NMD-targeted mRNAs without introns downstream from the TC are most likely devoid of any bound EJCs, and we therefore refer to this NMD pathway as “EJC-independent NMD.” Noteworthy from an evolutionary perspective, such an EJC-independent mode of NMD has been also described in Arabidopsis thaliana, D. melanogaster, C. elegans, and Schizosaccharomyces pombe (which all have EJC factor homologs), as well as in S. cerevisiae (which appears to lack EJC factors altogether) (Gatfield et al. 2003; Longman et al. 2007; Kerenyi et al. 2008; Wen and Brogna 2010). In comparison to EJC-independent NMD, mRNAs containing an intron downstream from the PTC are generally more efficiently down-regulated by NMD (Buhler et al. 2006; Singh et al. 2008; see also Results). Hence, we refer to this mode of NMD as “EJC-enhanced NMD.”
Here, we examined in parallel EJC-independent and EJC-enhanced NMD using two different NMD reporter genes of which we compared relative mRNA levels of a PTC+ variant possessing a downstream intron with the identical construct lacking this last intron. As controls, PTC-free wild-type versions (wt) were also examined. As NMD reporters, we chose the immunoglobulin μ minigene system (mini-μ) and β-Globin constructs, for both of which EJC-independent NMD has been previously reported (Zhang et al. 1998; Buhler et al. 2006; Matsuda et al. 2007; Eberle et al. 2008). To discover potential differences between EJC-independent and EJC-enhanced NMD regarding the involvement of trans-acting factors, the effect of RNAi-mediated depletion of various NMD factors on both NMD modes was investigated. Our results confirmed that NMD in human cells can function independently of an EJC located downstream from the PTC and that the EJC acts as an NMD enhancer. We provide additional evidence for a previously suggested UPF2- and UPF3b-independent branch of the NMD pathway (Chan et al. 2007; Gehring et al. 2009a) and report a transcript-specific requirement for UPF2 and UPF3b in EJC-enhanced and EJC-independent NMD. Consistent with previous reports (Luke et al. 2007; Jonas et al. 2013), our data further indicate the existence of two at least partially redundant decay pathways in NMD of human cells—one involving SMG6-mediated endonucleolysis and the other SMG5/SMG7—and reveal mRNP-specific preferences for one or the other pathway.
RESULTS
PTC-containing mRNAs are decreased in their abundance independently of the presence or absence of an EJC downstream from the PTC
To compare the classical EJC-enhanced and the EJC-independent mode of NMD with regard to their requirements for NMD factors and degradation pathways, we tested two sets of NMD reporter constructs with or without an intron 3′ of the PTC under various NMD factor knockdown conditions. The mini-μ reporter construct, which is under the control of the human β-actin promoter, consists of six exons and a single point mutation generates a TAA stop codon in exon C3 in the PTC-containing (PTC+) variant (Fig. 1A). The intron between exons C3 and C4 was deleted in the mini-μ C3/C4 constructs, and in the mini-μ C3/H4 constructs, exon C4 was replaced by the same length of histone H4 sequence (Buhler et al. 2006). Mini-μ C3/H4 was generated to confirm that NMD observed with mini-μ C3/C4 was not due to a special “fail-safe sequence” in exon C4 that could compensate for the absence of the intron-mediated NMD-enhancing effect. The second set of NMD reporter constructs is based on a human–mouse hybrid β-Globin minigene consisting of three exons, and the PTC+ versions carry the nonsense codon TAG at amino acid position 39 in exon 2 (Fig. 1B; Zhang et al. 1998). The 5′ part including intron 2 originates from the human β-Globin gene, whereas exon 3 and the 3′ UTR stem from the mouse β-Globin gene. Deletion of intron 2 still led to a PTC-specific down-regulation of the mRNA to 13% in NIH 3T3 cells (Zhang et al. 1998), and a similar down-regulation was also observed in HeLa cells (Matsuda et al. 2007).
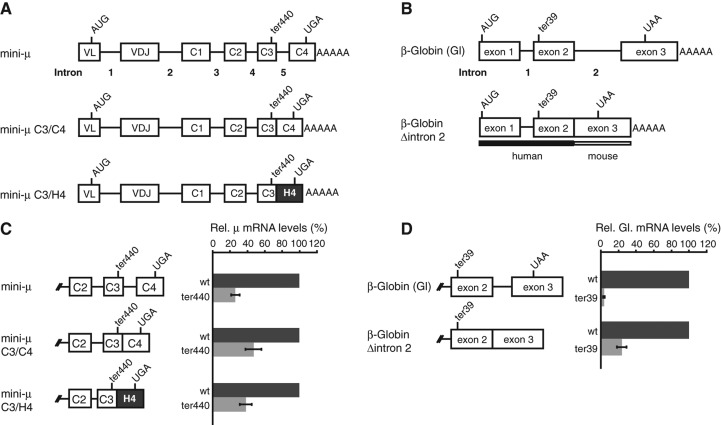
FIGURE 1.
Relative abundance of mini-μ and β-Globin NMD reporter transcripts with and without an intron downstream from the PTC. (A) Schematic representation of the mini-μ reporter constructs, each of which exists in the wild-type (wt) and the ter440 version, the latter contains a single point mutation in exon C3 generating a PTC at amino acid 440 (GAA → TAA). In the constructs mini-μ C3/C4 and C3/H4, intron 5 was deleted. Additionally, exon C4 was replaced with a sequence of identical length from the histone H4 gene in construct mini-μ C3/H4. (B) Schematic representation of the β-Globin (Gl) reporter constructs that either code for the wild-type ORF (wt) or harbor a single point mutation in exon 2 generating a PTC at amino acid 39 (CAG → TAG; ter39). The β-Globin construct is a hybrid between the human and mouse β-Globin (Zhang et al. 1998). In β-Globin Δintron2, intron 2 was deleted. Exons and introns are illustrated by boxes and lines, respectively, and the positions of the start (AUG) and termination (UGA or UAA) codons, as well as of the PTCs (ter440 or ter39), are indicated. (C) Relative mini-μ mRNA levels expressed in transiently transfected HeLa cells were measured 48 h post-transfection by reverse transcription followed by quantitative real-time PCR (RT-qPCR) and normalized to relative mRNA levels of cotransfected pGEM5 IRE β-Globin wt. Mean values and ±SEM of three or four independent experiments are shown. (D) Relative β-Globin mRNA levels expressed in transiently transfected HeLa cells were measured 48 h post-transfection by RT-qPCR and normalized to relative mRNA levels of pβ mini-μ wt IRE. Mean values and ±SEM of three independent experiments are shown.
We transiently expressed the mini-μ and β-Globin constructs in HeLa cells and determined the relative RNA levels 48 h post-transfection. Consistent with previous results indicating that downstream EJCs function as NMD enhancers (Buhler et al. 2006), the mRNA transcribed from the intron 5–containing mini-μ ter440 construct was 26% of its wt counterpart, whereas a deletion of the intron downstream from the PTC in C3/C4 and C3/H4 led to a weaker down-regulation of PTC+ mRNA to ∼47% and 38% of wt, respectively (Fig. 1C). Since the mini-μ C3/H4 ter440 mRNA levels are more reduced than the mini-μ C3/C4 ter440 levels, we used the mini-μ C3/H4 construct for most of the following experiments.
Also consistent with previous findings (Matsuda et al. 2007), the β-Globin ter39 mRNA level decreased strongly to 3.7% of wt, and removal of intron 2 in the β-Globin Δintron2 ter39 constructs led to a PTC-induced mRNA reduction of 24% of the corresponding wt (Fig. 1D). All human–mouse hybrid β-Globin constructs are spliced correctly, and a deletion of intron 2 has no influence on splicing of intron 1 (Supplemental Fig. 1). Correct splicing of the mini-μ constructs was previously confirmed (Buhler et al. 2006).
To test if a deletion of the 3′ most intron in the mini-μ constructs influences the effect of EJC factors on NMD, mini-μ and β-Globin reporter mRNA levels from constructs with or without the last intron were determined in cells in which we knocked down the EJC core factors eIF4AIII or Y14. As expected, knockdown of eIF4AIII and Y14 led to a strong stabilization of mini-μ ter440 mRNA (Supplemental Fig. 2A,B; see also Buhler et al. 2006). In contrast, eIF4AIII and Y14 knockdown had only a marginal yet detectable effect on the mRNA levels of mini-μ ter440 C3/C4 (Supplemental Fig. 2A,B; Buhler et al. 2006), indicating that in addition to the NMD-enhancing effect of an EJC positioned downstream from the PTC, EJCs could directly or indirectly affect NMD by other mechanisms (see Discussion). Compared with the marginal effects seen with mini-μ ter440 C3/C4, the β-Globin ter39 transcripts were significantly stabilized upon eIF4AIII or Y14 knockdown, irrespective of the presence of intron 2 (Supplemental Fig. 2C,D), confirming the previously published data (Matsuda et al. 2007).
FIGURE 2.
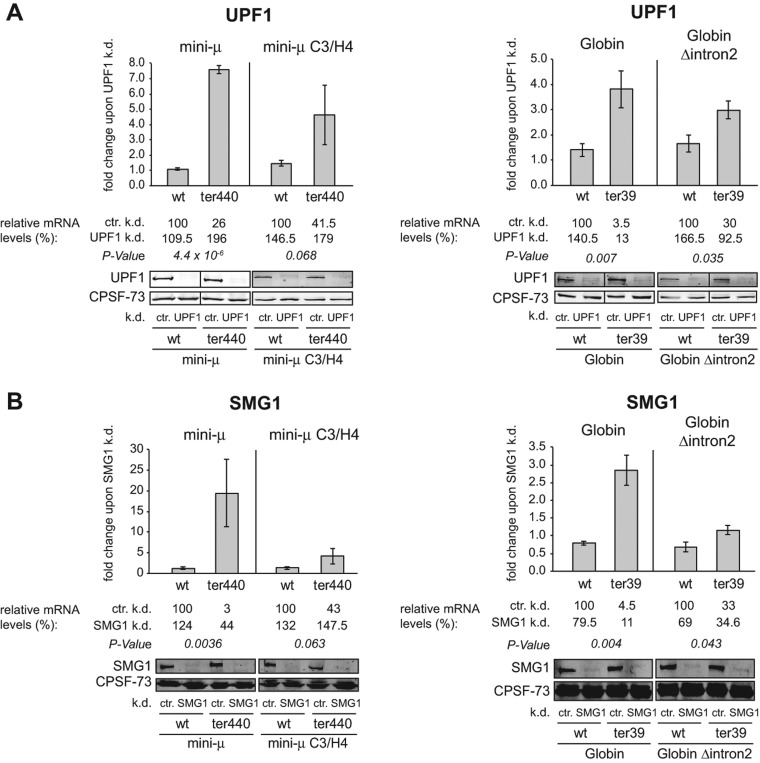
UPF1 and SMG1 are required for EJC-independent NMD. (A,B) Fold changes of relative mRNA levels of mini-μ and β-Globin constructs upon knockdown of UPF1 (A) or SMG1 (B) compared with a control knockdown (ctr.) in HeLa tTR-KRAB cells transiently transfected with the indicated reporter constructs are shown. Relative mRNA levels were determined 72 h post-transfection by RT-qPCR and normalized to the relative mRNA levels of cotransfected pGEM5 IRE β-Globin wt (for mini-μ) or pβ mini-μ wt IRE (for β-Globin). The relative mRNA levels are displayed below the diagrams, and the wt samples in the control knockdown were set as 100%. The efficiency of knockdowns was monitored by Western blotting. (k.d.) knockdown, (ctr.) control knockdown. (A) UPF1 or control knockdowns were induced by addition of DOX to HeLa tTR-KRAB-shUPF1 or HeLa tTR-KRAB-sh(scrambled) cells, except for mini-μ wt and mini-μ ter440, where the UPF1 and control knockdowns were induced by cotransfection of the respective pSUPERpuro plasmids. Mean values, fold changes, and ±SEM of three or four independent experiments are shown. P-values refer to the fold changes of PTC+ transcripts compared with the corresponding wt transcripts. (B) SMG1 or ctr. knockdowns were carried out in HeLa tTR-KRAB-shSMG1 or HeLa tTR-KRAB-sh(scrambled) cells as in A. Mean values, fold changes, and ±SEM of three independent experiments are shown. In all figures depicting data from knockdown experiments, the relative mRNA levels given below the histogram represent the mean of the corresponding relative mRNA levels in each independent experiment, whereas the “fold change” indicated in the histograms represents the mean of the corresponding fold changes in each independent experiment (for details, see Materials and Methods). Therefore, the depicted fold change in the histogram can deviate slightly from the ratio of the corresponding relative mRNA levels depicted below.
UPF1 is required for EJC-independent NMD
To facilitate RNAi-mediated knockdowns of individual NMD factors, we generated cell lines in which the expression of the respective shRNA can be induced by addition of doxycycline (DOX) to the cells for depleting each of the following NMD factors: UPF1, SMG1, UPF2, UPF3b, SMG5, SMG6, SMG7, and a control. The inducible knockdown system was established by integration of two lentiviral vectors. The first vector encodes a constitutively expressed tetracycline-repressor domain (tTR) fused to KRAB, a transcriptional repressor domain of human Kox1 that silences RNA polymerase II and III promoters within 3 kb from its DNA binding site by triggering heterochromatin formation, and a DsRed expression cassette for selection (Wiznerowicz and Trono 2003). The second lentiviral construct contains a tet operator (tetO) sequence to which the tTR-KRAB fusion protein binds in the absence of tetracycline, flanked by the shRNA expression cassette and a EGFP expression cassette for selection. Addition of DOX releases tTR-KRAB from the tetO, which leads to the production of the shRNA that subsequently eliminates the target mRNA.
First, we used this inducible knockdown system to test the requirement of UPF1 for the EJC-independent mode of NMD. The Western blots in Figure 2A show that the DOX-induced knockdown efficiently depleted UPF1 from the cells. Under UPF1 knockdown conditions, mRNA stabilization was lower with the Δintron constructs (mini-μ C3/H4 ter440 and β-Globin Δintron2 ter39) than with the corresponding constructs that still harbor the 3′-most intron (mini-μ ter440 and β-Globin ter39), consistent with the proposed NMD-enhancing function of EJCs located downstream from a PTC (Stalder and Muhlemann 2008; Rebbapragada and Lykke-Andersen 2009). Similar results were obtained with these reporter constructs when UPF1 was depleted by transiently transfected shRNA-expressing plasmids (Supplemental Fig. 3; Buhler et al. 2006) or transfection of chemically synthesized siRNA duplexes (Matsuda et al. 2007), confirming the requirement of UPF1 for both NMD modes and validating the use of our DOX-inducible knockdown cells.
FIGURE 3.
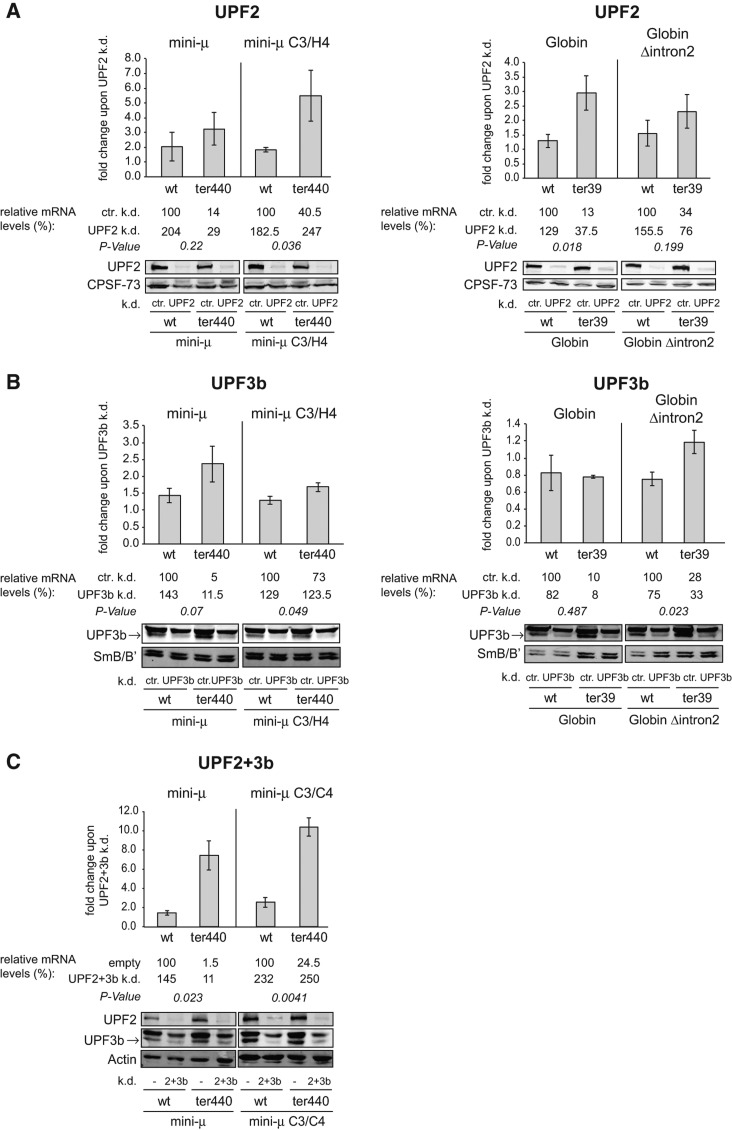
Transcript-specific roles for UPF2 and UPF3b and differential requirements in EJC-enhanced and EJC-independent NMD. (A) Fold changes of relative mini-μ and β-Globin mRNA levels upon knockdown of UPF2 in HeLa cells transiently transfected with reporter constructs are shown. Relative mini-μ and β-Globin mRNA levels were determined 72 h post-transfection by RT-qPCR and normalized to relative mRNA levels of cotransfected pNORM-7SL-AmdS. UPF2 or ctr. knockdowns were induced by addition of DOX to HeLa tTR-KRAB-shUPF2 or HeLa tTR-KRAB-sh(scrambled) cells. Mean values, fold changes, and ±SEM of three independent experiments are shown. The relative mRNA levels are displayed below the diagrams, and the wt samples in the control knockdown were set as 100%. The efficiency of the knockdowns was monitored by Western blotting. P-values were determined as in Figure 2A. (k.d.) knockdown; (ctr.) control knockdown. (B) As A, but with UPF3b knockdowns instead. (C) Fold changes of relative mini-μ mRNA levels upon knockdown of UPF2 and UPF3b in HeLa cells transiently transfected with reporter constructs are shown. Relative mini-μ mRNA levels were determined 96 h post-transfection by RT-qPCR and normalized to relative mRNA levels of cotransfected pmCMVrGPx1-TGC. Knockdowns with the mini-μ constructs were done by cotransfection of pSUPERpuro-shUPF2 and -shUPF3b plasmids or pSUPERpuro-empty as a control (empty/−) in HeLa cells. Data were analyzed as in A.
SMG1 depletion inhibits NMD of mini-μ constructs, but only weakly affects the β-Globin constructs
Given that UPF1 is a phosphoprotein and that its cycle of SMG1-mediated phosphorylation and PP2A-mediated dephosphorylation is essential for EJC-enhanced NMD in mammalian cells (Anders et al. 2003; Chiu et al. 2003; Ohnishi et al. 2003), we next asked if phosphorylation of UPF1 was also required for EJC-independent NMD. To this end, the kinase SMG1 was depleted using the DOX-inducible SMG1 knockdown cells, which resulted in a strong accumulation of mini-μ ter440 mRNAs (19.5-fold up-regulation compared with a mock knockdown), and NMD of mini-μ ter440 C3/H4 was completely inhibited (4.2-fold up-regulation to similar level as wt) (Fig. 2B). Compared with this, the 2.9-fold stabilization of β-Globin ter39 mRNA from 4.5% to 11% of wt was modest, and the relative mRNA level of β-Globin Δintron2 ter39 did not change under our SMG1 depletion conditions (Fig. 2B). Unexpectedly, the relative mRNA levels of both wt β-Globin constructs decreased to 79.5% and 69% of the control knockdown. Overall, it appears as if the β-Globin ter39 mRNAs, irrespective of whether they harbor an EJC downstream from the PTC or not, were less sensitive to reduced SMG1 concentrations than the mini-μ ter440 mRNAs.
UPF2 and UPF3b have transcript-specific roles, and they are differentially required in EJC-enhanced and EJC-independent NMD
Next, we examined the dependence of EJC-enhanced and EJC-independent NMD on UPF2 and UPF3b by expressing our mini-μ and β-Globin reporter constructs in DOX-inducible UPF2 or UPF3b knockdown cell lines (Fig. 3A,B). Depletion of UPF2 stabilized mini-μ ter440 mRNA 3.2-fold, which represents a modest NMD inhibition from 14% to 29% of the wt mRNA abundance, whereas the 5.5-fold increase of the EJC-independent mini-μ C3/H4 ter440 reporter transcript represents a complete NMD suppression (Fig. 3A). β-Globin ter39 mRNA was also significantly stabilized upon UPF2 depletion (threefold), and the β-Globin Δintron2 ter39 mRNA levels rose by 2.3-fold, which is not a statistically significant difference to the corresponding wt mRNA level, which also increased by 1.6-fold (P-value = 0.2). This result indicates differential sensitivities of different NMD-targeted transcripts for limiting UPF2 concentrations, consistent with previous studies reporting examples of UPF2 independent NMD (Gehring et al. 2005, 2009a; Ivanov et al. 2008). Furthermore, the data of the β-Globin constructs are comparable with a previous study that used siRNAs for the UPF2 knockdown (Matsuda et al. 2007).
UPF3b interacts with Y14/MAGOH and eIF4AIII and can therefore be regarded as an EJC component that physically links the EJC to UPF1 and UPF2 (Kim et al. 2001; Gehring et al. 2003; Chamieh et al. 2008; Buchwald et al. 2010). Knockdown of UPF3b provoked a moderate up-regulation of both mini-μ ter440 (2.4-fold) and of mini-μ ter440 C3/H4 mRNA (1.7-fold) compared with the mock knockdown (Fig. 3B). Contrary to our expectation, β-Globin ter39 mRNA was unaffected by the UPF3b knockdown, and the β-Globin Δintron2 ter39 levels were only slightly increased by 1.2-fold. This stabilization is statistically significant compared with the corresponding wt that decreased upon UPF3b depletion to 75% of the control knockdown (P-value = 0.023) (Fig. 3B). Overall, the UPF3b knockdowns affected all four NMD reporters only weakly if at all, which might indicate that UPF3a could functionally substitute for UPF3b when UPF3b concentrations are limiting, as has been previously shown (Chan et al. 2009). Alternatively, the residual UPF3b protein might still be sufficient to sustain NMD. Our results differ from the previously reported twofold stabilization of β-Globin ter39 mRNA and the unchanged levels of β-Globin Δintron2 ter39 upon UPF3b knockdown (Matsuda et al. 2007). The reason for these differences is unknown.
Finally, because of the reported UPF2 and UPF3b-independent NMD pathways (Gehring et al. 2005, 2009a; Chan et al. 2007), we also depleted UPF2 and UPF3b simultaneously by transiently transfecting shRNA-encoding pSUPERpuro plasmids in cells expressing the mini-μ C3/C4 reporters and compared the effects of the single factor knockdowns with those of the double knockdown (Fig. 3C). Compared with the single knockdowns, NMD of mini-μ ter440 mRNA was much more strongly suppressed in the double knockdown, and even a complete NMD inhibition was observed for mini-μ C3/C4 ter440 mRNA in the combined UPF2 and UPF3b knockdown. Thus, in both cases, UPF2 and UPF3b appear to be involved in the same pathway, but it seems that EJC-independent NMD is more strongly compromised upon reduced UPF2 and UPF3b levels.
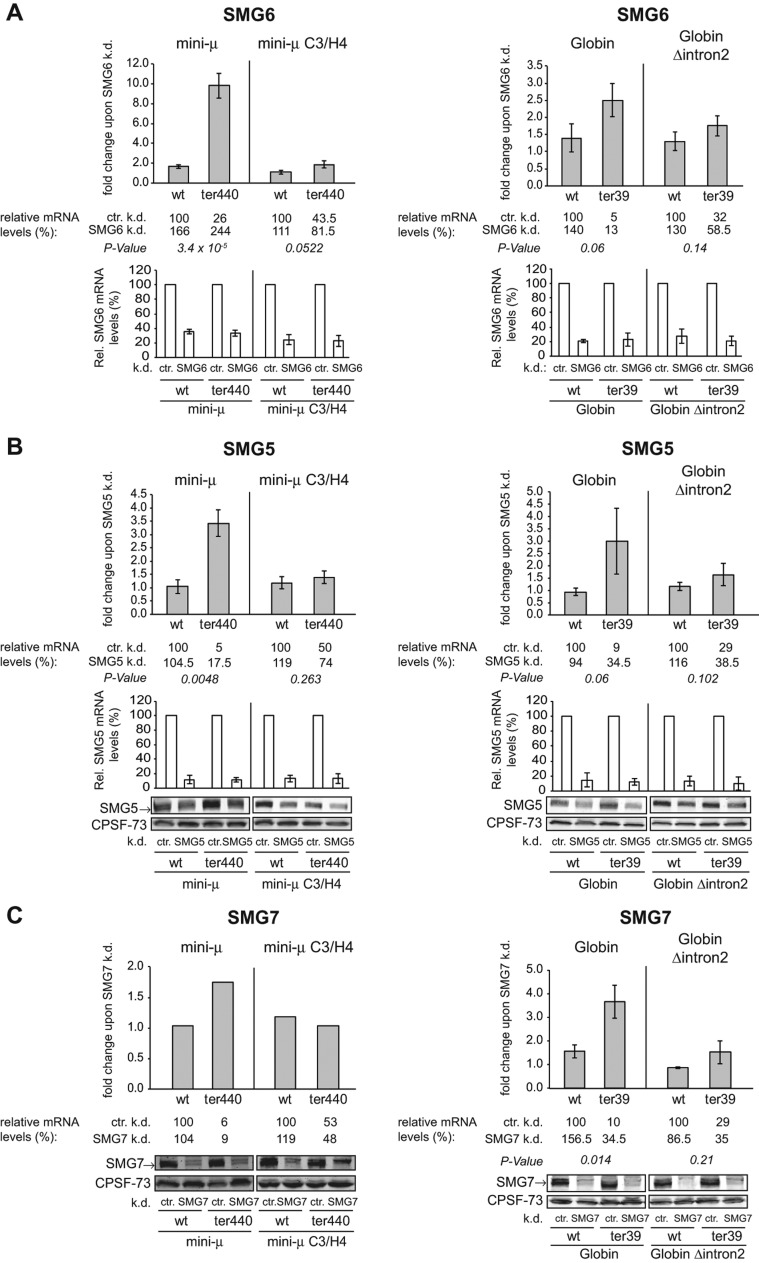
Single knockdowns of SMG6, SMG5, or SMG7 hardly affect EJC-independent NMD
In addition to the previously described NMD factors, which appear to be mainly involved in NMD substrate recognition, we tested the influence of SMG5, SMG6, and SMG7 on our reporter transcripts, three proteins that preferentially interact with phosphorylated UPF1 and directly or indirectly trigger the degradation of the NMD-committed mRNAs (Unterholzner and Izaurralde 2004; Fukuhara et al. 2005; Huntzinger et al. 2008; Eberle et al. 2009; Okada-Katsuhata et al. 2012; Cho et al. 2013). SMG6 has been characterized as an endonuclease that cleaves the target mRNA in classical NMD substrates harboring an EJC downstream from the PTC (Huntzinger et al. 2008; Eberle et al. 2009). Here, we wanted to find out if the EJC-independent NMD pathway also involves SMG6-mediated endonucleolysis. For these experiments, we induced SMG6 depletion by adding DOX to HeLa tTR-KRAB-shSMG6 cells. To assess the knockdown efficiency, SMG6 mRNA levels were measured by RT-qPCR due to the lack of a suitable SMG6 antibody at the time these experiments were performed. Four days after DOX addition, SMG6 mRNA was reproducibly reduced to 20%–35% of the levels in the control knockdowns (Fig. 4A), which is similar or even slightly better than the SMG6 mRNA reductions observed in Figure 5B that correlated with an efficient depletion of the SMG6 protein from the cells. Knockdown of SMG6 elevated the mini-μ ter440 mRNA 9.8-fold, which represents a complete inhibition of NMD. In contrast, under even slightly better knockdown conditions, EJC-independent NMD of mini-μ C3/H4 ter440 mRNA was only partially inhibited (1.9-fold up-regulation), suggesting that the EJC-enhanced mode of NMD might be more dependent on SMG6 than the EJC-independent mode. Partial suppression of NMD was also observed with both intron 2–containing and intron 2–lacking versions of β-Globin reporters. In the EJC-enhanced mode, the β-Globin ter39 mRNA increased 2.5-fold to 13% of wt and in the EJC-independent mode, a nonsignificant (P-value = 0.14) 1.8-fold increase to 60% of wt was detected (Fig. 4A). In XRN1-depleted cells, a 3′ fragment resulting from the SMG6-mediated endonucleolytic cleavage of three EJC-enhanced NMD reporters was previously detected by Northern blotting (Eberle et al. 2009). We therefore used the same approach to detect putative 3′ fragments originating from the EJC-independent NMD substrates mini-μ C3/H4 ter440 and β-Globin Δintron2 ter39 (Supplemental Fig. 4). A weak signal, barely above background, at the expected position of the 3′ decay intermediate could be detected for the β-Globin ter39 mRNA as well as for the β-Globin Δintron2 ter39 mRNA. Also for mini-μ ter440, a 3′ fragment could be reproducibly detected (Supplemental Fig. 4C), but the corresponding 3′ fragment was not detectable for PTC+ mini-μ C3/C4 (data not shown) or C3/H4 (Supplemental Fig. 4B). Based on the observed partial inhibition of NMD of mini-μ C3/H4 ter440 mRNA (Fig. 4A), it seems likely that SMG6-mediated endocleavage also contributes to NMD of this reporter mRNA and that our assay conditions were just not sensitive enough to detect the very unstable 3′ decay intermediate. However, it could also be that SMG6 plays an endocleavage-independent role in the EJC-independent NMD of mini-μ.
FIGURE 4.
Single knockdowns of SMG6, SMG5, or SMG7 hardly affect EJC-independent NMD. (A–C) Fold changes of relative mRNA levels of mini-μ or β-Globin constructs upon knockdown of SMG6 (A), SMG5 (B), or SMG7 (C) compared with a control knockdown (ctr.) in HeLa tTR-KRAB cells transiently transfected with the indicated reporter constructs are shown. Relative mini-μ or β-Globin mRNA levels were determined 72 h post-transfection by RT-qPCR. SMG6, SMG5, SMG7, or ctr. knockdowns were induced by addition of DOX to HeLa tTR-KRAB-shSMG6, -shSMG5, -shSMG7, or -sh(scrambled) cells, respectively. Mean values, fold changes, and ±SEM of two to four independent experiments are shown. The relative mRNA levels are displayed below the diagrams, and the wt samples in the control knockdown were set as 100%. The efficiency of the knockdowns was monitored by RT-qPCR (SMG6 [A], SMG5 [B]) and/or Western blotting (SMG5 [B], SMG7 [C]). P-values were determined as in Figure 2A. ±SEM and P-values were calculated when n ≥ 3. (k.d.) knockdown; (ctr.) control knockdown. (A) Relative mini-μ or β-Globin mRNA levels were normalized to relative mRNA levels of cotransfected pGEM5 IRE β-Globin wt (for mini-μ) or pβ mini-μ wt IRE (for β-Globin). (B) Relative mini-μ or β-Globin mRNA levels were normalized to relative mRNA levels of cotransfected pNORM-7SL-AmdS. (C) Relative mini-μ or β-Globin mRNA levels were normalized to relative mRNA levels of cotransfected pNORM-7SL-AmdS (for mini-μ) or pβ mini-μ wt IRE and pNORM-7SL-AmdS (for β-Globin).
FIGURE 5.
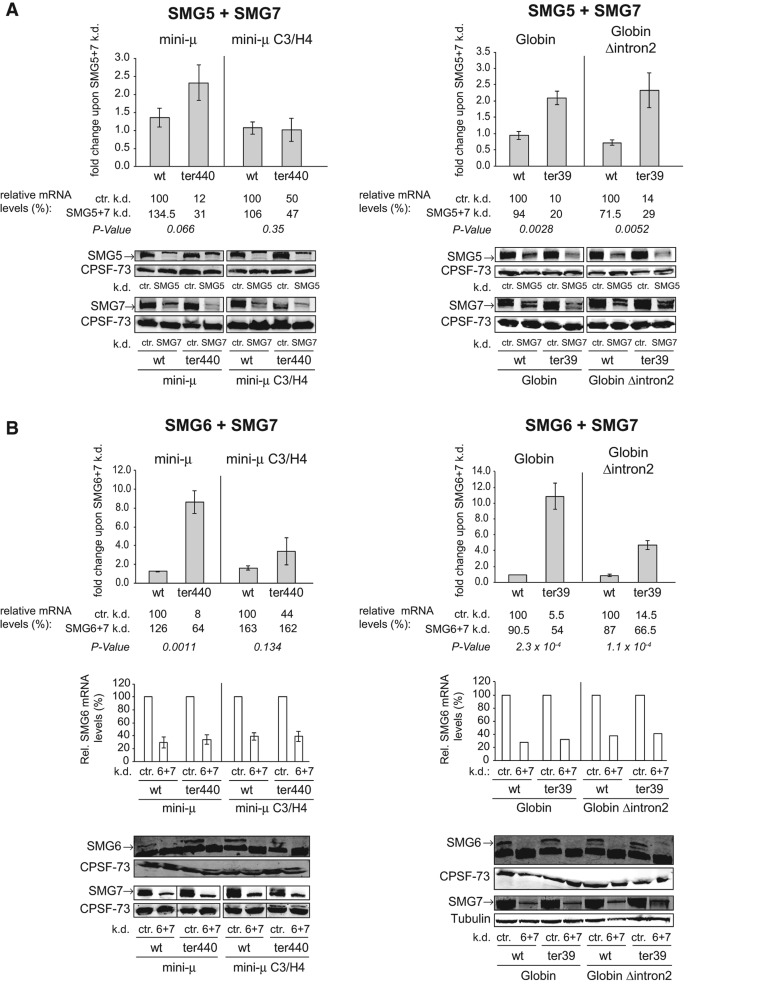
Codepletion of SMG6/SMG7 strongly inhibits EJC-enhanced and EJC-independent NMD, but a depletion of SMG5/SMG7 has much weaker effects. (A) Fold changes of relative mini-μ or β-Globin mRNA constructs upon a combined knockdown of SMG5 and SMG7 compared with a control knockdown (ctr.) in HeLa cells transiently transfected with the indicated reporter constructs are shown. Relative mini-μ or β-Globin mRNA levels were determined 96 h post-transfection by RT-qPCR and normalized to relative mRNA levels of cotransfected pNORM-7SL-AmdS. Knockdowns were performed by cotransfection into HeLa cells of pSUPERpuro-shSMG5 and -shSMG7 or pSUPERpuro-sh(scrambled) as a control. Mean values, fold changes, and ±SEM of three or four independent experiments are shown. The relative mRNA levels are displayed below the diagrams, and the wt samples in the control knockdown were set as 100%. The efficiency of the knockdowns was monitored by Western blotting. P-values were determined as in Figure 2A. (k.d.) knockdown; (ctr.) control knockdown. (B) As A, but with a combined knockdown of SMG6 and SMG7. For Western blotting, a different SMG7 antibody was used here from that in A and Figure 4C. Additionally, relative mRNA levels of SMG6 are shown.
Since it has been shown that SMG6 and SMG5/SMG7 can act redundantly to degrade NMD targets (Luke et al. 2007; Jonas et al. 2013), we first addressed whether the SMG5/SMG7 heterodimer plays a role in the degradation of EJC-independent NMD targets. Contrary to the results of the SMG6 knockdowns, single knockdowns of SMG5 or SMG7 generally suppressed EJC-enhanced NMD of β-Globin ter39 better than of mini-μ ter440 (Fig. 4B,C). The effect of these knockdowns on the two EJC-independent NMD targets was marginal and not significantly different from the effect on the corresponding wt transcripts. Noteworthy, we observed that knockdown of SMG5 led to only a moderate decrease of SMG5 protein levels, despite SMG5 mRNA levels being reduced to <15% (Fig. 4B), and efficient depletion of SMG5 was only observed in SMG5 and SMG7 double knockdowns (Fig. 5A), suggesting that SMG5 and SMG7 form together a stable and long-living complex. Nevertheless, this modest reduction of SMG5 protein led to a 3.4-fold increase of mini-μ ter440, but had virtually no effect on the PTC+ mRNA expressed from the mini-μ C3/H4 reporter gene (Fig. 4B). β-Globin ter39 was also up-regulated on average threefold, but this effect varied considerably between the three independently performed experiments, and overall it was not statistically significant (P-value = 0.06). As for the EJC-independent mini-μ construct, β-Globin Δintron2 ter39 mRNA was not significantly up-regulated under these SMG5 knockdown conditions.
The behavior of the NMD reporters in the DOX-inducible HeLa tTR-KRAB-shSMG7 cells was very similar to the SMG5 knockdown cells, even though the knockdown of SMG7 was much more effective compared with the SMG5 knockdown (Fig. 4C). The mRNA levels of mini-μ ter440 and β-Globin ter39 were stabilized 1.75-fold and 3.7-fold upon SMG7 depletion, respectively, whereas the mRNA levels of the PTC+ constructs lacking the 3′-most intron did not significantly change (Fig. 4C). The finding that individual depletion of none of these three SMGs significantly inhibited degradation of the EJC-independent NMD targets suggests a redundancy among SMG5, SMG6, and SMG7 in the EJC-independent pathway.
Codepletion of SMG6/SMG7 strongly inhibits EJC-enhanced and EJC-independent NMD, whereas depletion of the SMG5/SMG7 heterodimer has much weaker effects
Because of the indicated redundancy in the SMG5-, SMG6-, and SMG7-mediated mRNA degradation pathways (Luke et al. 2007; Jonas et al. 2013), we decided to perform double knockdowns. First, we tested the effect of an SMG5/SMG7 codepletion. SMG5 and SMG7 interact with each other (Ohnishi et al. 2003; Jonas et al. 2013) and are therefore expected to function together in NMD. On the other hand, SMG5 has been recently shown to be involved in PNRC2-mediated decapping of NMD-targeted mRNAs independently of SMG7 (Cho et al. 2013), whereas the C-terminal domain of SMG7 is sufficient to elicit decay through XRN1-dependent exonucleolysis when tethered to an mRNA (Unterholzner and Izaurralde 2004). For simultaneous knockdown of SMG5 and SMG7, HeLa cells were transiently transfected with a mix of two pSUPERpuro plasmids expressing shRNAs against SMG5 and SMG7 mRNAs, respectively, together with the indicated NMD reporter construct and an additional reporter gene for normalization. Overall, the results from the SMG5/SMG7 double knockdowns (Fig. 5A) resembled the results obtained with the separate knockdowns (Fig. 4B,C): Codepletion of SMG5 and SMG7 had no effect on the mRNA levels of mini-μ C3/H4 ter440 and stabilized mini-μ ter440 mRNA by 2.3-fold, indicating that EJC-independent degradation of mini-μ ter440 mRNA does not require the SMG5/SMG7 heterodimer in the presence of SMG6 (Fig. 5A). Both β-Globin ter39 reporter mRNAs were stabilized by about twofold, irrespective of whether or not they harbored an EJC downstream from the PTC, revealing a transcript-specific NMD factor dependency.
Finding that SMG5/SMG7 codepletion only partially suppressed NMD of the β-Globin constructs and the SMG6 knockdown also only marginally stabilized these transcripts (Fig. 4A) is consistent with the proposed hypothesis that SMG6-mediated endonucleolysis and the SMG5/SMG7-mediated exonucleolytic decay pathway might represent two independent NMD decay routes (Luke et al. 2007; Jonas et al. 2013). To test this hypothesis, we attempted to block both putative degradation routes by a double knockdown of SMG6 and SMG7 (Fig. 5B). The codepletion was achieved by expressing two shRNAs encoded on transiently transfected pSUPERpuro plasmids in HeLa cells, similar to the above-mentioned SMG5 and SMG7 double knockdown. Indeed, codepletion of SMG6 and SMG7 led to a strong stabilization of all NMD reporter transcripts. For the EJC-enhanced reporters, mini-μ ter440 mRNA levels were stabilized 8.6-fold, and β-Globin ter39 mRNA levels were almost 11-fold up-regulated. In the EJC-independent NMD mode, mini-μ C3/H4 ter440 mRNA levels were restored to wt mRNA levels in the SMG6/SMG7 double knockdown, and β-Globin Δintron2 ter39 mRNA showed a highly significant 4.7-fold up-regulation (Fig. 5B). In summary, these results indicate that the degradation of NMD-targeted mRNAs can either be elicited by SMG6 or by SMG5/SMG7, and that the inhibition of one decay pathway leads to the usage of the other one.
DISCUSSION
We report here a comparison between EJC-enhanced and EJC-independent NMD with regard to the trans-acting factor requirements of these two NMD modes, which we investigated by knocking down the seven NMD factors UPF1, UPF2, UPF3b, SMG1, SMG5, SMG6, and SMG7. For the majority of RNAi-mediated knockdowns, we used a tTR-KRAB inducible gene silencing system in HeLa cells, in which the expression of an shRNA is turned on by adding DOX to the culture medium (Wiznerowicz and Trono 2003). This inducible gene silencing system allows simple, homogeneous, efficient, and reproducible knockdowns even in large amounts of cells. β-Globin ter39 and mini-μ ter440 were chosen as NMD reporter genes because it was previously shown for both that mRNAs without splicing downstream from the PTC were a substrate for NMD (Zhang et al. 1998; Buhler et al. 2006; Matsuda et al. 2007; Eberle et al. 2008). The chimeric human–mouse β-Globin (Zhang et al. 1998) construct was used because deletion of intron 2 in the entirely human construct leads to a decreased cytoplasmic transcript accumulation and abolishes splicing of intron 1 (Collis et al. 1990; Antoniou et al. 1998; Damgaard et al. 2008), whereas the human–mouse hybrid β-Globin Δintron2 construct is spliced correctly (Supplemental Fig. 1). Zhang and colleagues proposed that a cis-acting “fail-safe sequence” in exon 3 of the human–mouse hybrid β-Globin construct compensates for the absence of intron 2 as the NMD-inducing signal (Zhang et al. 1998), but an alternative interpretation of their data is that the deletion constructs tested to map the putative “fail-safe sequence” resulted in shortened 3′ UTRs, which most likely caused the decreased NMD efficiency, as shown for the mini-μ construct (Buhler et al. 2006).
Relaxed correlation between intron removal and EJC assembly complicates interpretation of results
It was generally thought—and the assumption for this study—that removal of the 3′-most intron in the mini-μ and β-Globin reporter constructs prevents the assembly of an EJC at this position. Two very recent publications challenged this strict correlation between introns and EJCs by revealing that only ∼80% of spliced exon–exon junctions carry an EJC at the classical −24 nt position, and on the other hand, 40%–50% of the EJCs detected in these studies located to noncanonical positions (Sauliere et al. 2012; Singh et al. 2012). Thus, we do not know with absolute certainty whether our mini-μ and β-Globin reporter mRNAs containing the 3′-most intron indeed harbor an EJC downstream from the PTC. Likewise, we cannot exclude that the constructs mini-μ C3/C4, C3/H4, and β-Globin Δintron 2 would assemble noncanonical EJCs in their 3′ UTRs. However, the effects of eIF4AIII and Y14 knockdowns, two EJC core factors, on two PTC+ reporter mRNAs that differ only by the presence or absence of an intron downstream from the PTC indicate whether there is an EJC placed in the 3′ UTR. Therefore, we compared the effect of eIF4AIII and Y14 knockdowns on mini-μ ter440 and on mini-μ C3/C4 ter440 transcripts and found a strong requirement for both factors for mini-μ ter440 degradation (i.e., EJC-enhanced NMD), but only a weak dependence for EJC-independent NMD (Supplemental Fig. 2A,B; see also Buhler et al. 2006). The weak inhibition of EJC-independent NMD by eIF4AIII and Y14 knockdowns does not necessarily mean that EJC components play a direct role in this type of NMD. There is ample evidence that EJCs promote mRNA translation in mammalian cells (Wiegand et al. 2003; Nott et al. 2004; Gudikote et al. 2005; Ma et al. 2008), and EJC depletion could therefore indirectly inhibit NMD by reducing the translation efficiency. This could also explain the previously published and herein confirmed finding that NMD acting on the β-Globin Δintron 2 ter39 mRNA needs eIF4AIII and Y14 (Matsuda et al. 2007).
UPF1 and SMG1 are required for both EJC-enhanced and EJC-independent NMD
Our study confirmed evidence from previous publications that NMD can act independently of a splicing event downstream from the PTC (Zhang et al. 1998; Buhler et al. 2006; Matsuda et al. 2007; Eberle et al. 2008; Singh et al. 2008), but that an EJC located >30 nt 3′ from the TC acts as an important trans-acting enhancer of NMD (Stalder and Muhlemann 2008; Rebbapragada and Lykke-Andersen 2009). PTC+ reporter mRNA levels were reduced in HeLa cells independently of a spliced intron downstream from the PTC, but the levels of EJC-enhanced NMD reporter mRNAs were significantly more strongly decreased (Fig. 1C,D). The destabilization of both EJC-enhanced and EJC-independent PTC+ mRNAs was dependent on the key NMD factor UPF1 (Fig. 2A). SMG1-mediated UPF1 phosphorylation and subsequent dephosphorylation are required for NMD in mammalian cells and C. elegans (Ohnishi et al. 2003). Our data indicate that SMG1-mediated UPF1 phosphorylation is also required for EJC-independent NMD, since mini-μ mRNAs lacking the intron downstream from the PTC are stabilized upon SMG1 knockdown (Fig. 2B).
Transcript-specific sensitivity for UPF2 or UPF3b depletions
UPF1 phosphorylation is promoted by UPF2 and UPF3b (Page et al. 1999; Grimson et al. 2004; Kashima et al. 2006). While Kashima and colleagues reported that UPF1 phosphorylation requires both UPF2 and UPF3b together, cases in which NMD can function without UPF2 or without UPF3b have been documented (Gehring et al. 2005, 2009a; Chan et al. 2007; Tarpey et al. 2007; Ivanov et al. 2008; Saltzman et al. 2008). We detected such UPF2- or UPF3b-independent NMD activity also in the EJC-independent mode (Fig. 3A,B), but the relative dependency on UPF2 and UPF3b appeared to correlate more with the transcript type than with the presence or absence of a 3′-UTR intron. The determinants for the different transcript preferences are not yet understood. It remains further to be investigated how the large conformational movement of the UPF1 CH domain can occur in the absence of UPF2, a rearrangement that is necessary to activate UPF1’s helicase activity (Chamieh et al. 2008; Chakrabarti et al. 2011).
Interestingly, codepletion of UPF2 and UPF3b both had a synergistic effect on mini-μ ter440 and mini-μ C3/C4 ter440 compared with individual knockdowns of these factors, suggesting that UPF2 and UPF3b are involved in the same pathway. However, while NMD of the EJC-independent transcript was completely inhibited, only a partial inhibition was observed for the EJC-enhanced reporter (Fig. 3). This difference could be explained by a higher sensitivity of EJC-independent NMD to reduced UPF2 and UPF3b concentrations, because the two factors probably have to contact the RNA-bound UPF1 by diffusion. In contrast, UPF2 and UPF3b are assumed to be already bound to an NMD-enhancing EJC, which would increase their local concentration and allow NMD to remain functional in the EJC-enhanced mode even under strongly reduced cellular concentrations.
Transcript-specific sensitivity to SMG6 depletion
Whereas knockdown of SMG5 or SMG7 individually had only modest or no effects on the mini-μ and β-Globin NMD reporter transcripts, a SMG6 knockdown led to a strong inhibition of EJC-enhanced NMD on mini-μ, but affected β-Globin only weakly (Fig. 4), suggesting that SMG6 might overall contribute more to NMD of mini-μ than SMG5 and SMG7. Vice versa, β-Globin ter39 increased only 2.5-fold upon SMG6 depletion and slightly more upon SMG5 and SMG7 knockdowns (threefold and 3.7-fold, respectively), suggesting that its degradation might rely more on SMG5 and SMG7 than on SMG6. This differential, transcript-specific effect suggests that there might exist mechanistically different degradation routes in human NMD, as previously proposed (Muhlemann and Lykke-Andersen 2010).
Evidence for two redundant degradation pathways in NMD
There are several lines of evidence that SMG5 and SMG7 function together in the same decay pathway. They form a heterodimer by interaction of their 14-3-3-like domains and bind together to phosphorylated SQ motifs in the C terminus of UPF1 (Okada-Katsuhata et al. 2012; Jonas et al. 2013). In the SMG5/SMG7 dimer, the C-terminal domain of SMG7 is required for inducing exonucleolytic degradation of the target transcript through yet-to-be identified factors (Unterholzner and Izaurralde 2004). In contrast, SMG6 was recently shown to interact through its 14-3-3-like domain with phosphorylated T28 at the N-terminus of UPF1 (Okada-Katsuhata et al. 2012) and to endonucleolytically cleave mRNA in the vicinity of the PTC with its C-terminal PIN domain (Huntzinger et al. 2008; Eberle et al. 2009).
The first indication that SMG6-mediated endonucleolytic cleavage and SMG5/SMG7-induced exonucleolytic degradation might act partially redundantly on NMD substrate mRNAs came from Luke and colleagues, who showed that single knockdowns of SMG6 and SMG7 only modestly stabilized an NMD reporter transcript, whereas an SMG6/SMG7 double knockdown caused a dramatic stabilization of the β-Globin NMD reporter (Luke et al. 2007). This was recently confirmed by Jonas et al. (2013). We observed the same with our PTC+ mini-μ and β-Globin reporter genes, irrespective of whether they contained an intron in the 3′ UTR or not (Fig. 5B), corroborating that there exist two at least partially redundant PTC-triggered decay pathways in human cells: one being SMG6-dependent and the other being SMG7-dependent. Furthermore, consistent with SMG5 and SMG7 functioning in the same decay pathway, an SMG5/SMG7 double knockdown affected our NMD reporter genes similarly to the individual knockdowns of these two factors, with the exception of β-Globin Δintron2 ter39, which was not affected by the single knockdowns (Fig. 4B,C) but showed a modest 2.3-fold increase in the double knockdown (Fig. 5A, right panel). Based on tethering assays, PNRC2 has been very recently shown to promote decapping of PTC+ transcripts in an SMG5-dependent and SMG7-independent manner (Cho et al. 2013), suggesting the existence of even a third distinctive RNA degradation route in human NMD. The researchers claimed that the PNRC2/SMG5 pathway contributed more strongly to the overall NMD efficacy than the SMG5/SMG7 pathway, which could explain the stronger stabilization of mini-μ ter440 upon SMG5 knockdown compared with the SMG7 knockdown (Fig. 4B,C). However, both single knockdowns affected β-Globin ter39 similarly, arguing against a major contribution by PNRC2/SMG5 to β-Globin NMD. While we have not investigated PNRC2, our data collectively reveal a partial redundancy between the SMG6-mediated endonucleolytic and the SMG5/SMG7-dependent exonucleolytic mRNA degradation pathways in both EJC-enhanced and EJC-independent modes of NMD, with mini-μ preferring the SMG6 pathway and β-Globin preferring the SMG5/SMG7-dependent pathway.
Different routes to eliminate NMD substrates
In summary, the results presented here underscore the emerging view that what we traditionally used to call “NMD” in reality represents not just one clearly defined mechanistic pathway, but rather a network of several interconnected pathways to degrade mRNAs that terminate aberrantly. Human cells seem not only to possess distinct routes to activate UPF1, but activated UPF1 also can engage mechanistically different routes to degrade the targeted mRNA. It will be a future challenge to elucidate the cis-acting signals and trans-acting factors that specify the exact pathway for a given mRNA from the event of aberrant translation termination to its clearance from the cell.
MATERIALS AND METHODS
Plasmids
β-Globin constructs (norm, Ter39, norm Δintron2, and Ter39 Δintron2) in pmCMV-Gl were received from the Maquat laboratory (Zhang et al. 1998) and are called herein β-Globin wt/ter39/Δintron2 wt/Δintron2 ter39. They represent hybrids between the human (V00499) and mouse β-Globin sequences (J00413), whereas the mouse sequence starts with exon 3 and contains a silent point mutation of T → G in exon 3. The constructs are under the control of the mouse CMV promoter. pβ mini-μ wt neo SpeI was cut with SpeI and XbaI and ligated with a likewise cut insert from pTRE-μ wt SpeI giving pβ mini-μ wt uA1. pβ mini-μ ter440 uA1 was created using site-directed mutagenesis with primers ls48 and ls49 (sequences are listed in the Supplemental Material). The pβ mini-μ C3/C4 (wt and ter440) and pβ mini-μ C3/H4 (wt and ter440) constructs are described in Buhler et al. (2006). pSUPERpuro plasmids used in knockdown experiments of UPF1 (Supplemental Fig. 3), UPF2 + UPF3b, SMG5 + SMG7, SMG6 + SMG7, and a control (scrambled) were generated as described (Brummelkamp et al. 2002; Paillusson et al. 2005; Nicholson et al. 2012); the shRNA target sequences are listed in the Supplemental Material. pSUPERpuro-empty is described in Yepiskoposyan et al. (2011). For normalization of mRNA levels in transient transfections, pβ mini-μ wt IRE, pGEM5 IRE β-Globin wt, pNORM-7SL-AmdS, and pmCMVrGPx1-TGC were used. In pβ mini-μ wt IRE, an iron-responsive element (IRE) was cloned into the SalI site of the corresponding pβ mini-μ wt construct. pGEM5 IRE β-Globin wt is described in Thermann et al. (1998). For generating pNORM-7SL-AmdS, we cloned a construct consisting of the 7SL-RNA gene and an insertion of a fragment of the Kluyveromyces lactis acetamidase gene (AmdS) between the SalI and XbaI restriction sites into the pNORM backbone that is based on the pBS vector. The 7SL-AmdS normalizer construct is controlled by the 7SL-RNA promoter, which is recognized by RNA polymerase III. pmCMVrGPx1-TGC is described in Moriarty et al. (1997). The sequences of all ORFs were confirmed by sequencing.
Cell lines
All pSUPERpuro-shRNA-mediated knockdowns were done in HeLa cells. For single knockdowns of UPF1, UPF2, UPF3b, SMG1, SMG5, SMG6, SMG7, XRN1, and scrambled, the doxycycline-inducible tTR-KRAB knockdown system in HeLa cells was used (Wiznerowicz and Trono 2003). For this, cloning of lentiviral vectors and generation of HeLa tTR-KRAB-shRNA cells was done according to the published protocol. To generate the respective knockdown vectors, the pSUPERpuro plasmids were digested with BstXI and SalI to isolate the fragments containing the H1 promoter followed by the respective shRNA cassette. These fragments were then cloned into pLV-TH using the same restriction sites. The lentiviral particles were produced according to the published protocol (Wiznerowicz and Trono 2003). HeLa cells were transduced with pLV-tTR-KRAB-dsRed and sorted by FACS, followed by a second round of transduction with the pLV-TH-shRNA vectors. Post-transduction, all cell lines except from HeLa tTR-KRAB-shSMG5 were induced for 24–48 h with doxycycline to induce GFP expression and allow FACS sorting for dsRed/GFP-positive cells. After sorting, the cells were again cultured in doxycycline-free medium for expansion. The shRNA target sequences expressed in HeLa tTR-KRAB cells are listed in the Supplemental Material. To induce the expression of shRNAs and thereby RNAi-mediated knockdowns of the above-mentioned factors, doxycycline was added to the medium in a concentration of 10 µg/mL. HeLa cells were cultivated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS), penicillin (100 IU/mL), and streptomycin (100 μg/mL) (DMEM+/+). HeLa tTR-KRAB cells were kept in tetracycline-free DMEM+/+ medium. All cells were grown and kept in culture at 37°C and 5% CO2.
Cell experiments and RNAi-mediated knockdowns
To determine relative mRNA levels of the reporters in Figure 1C,D, 2 × 105 HeLa cells were seeded into six-well plates, and 24 h later, the cells were transiently transfected with 200 ng (pβ mini-μ) or 300 ng (pmCMV-Globin) of reporter plasmid and 100 ng of normalizer (pGEM5 IRE β-Globin wt or pβ mini-μ wt IRE) with DreamFect (OZ Biosciences). On the third day, the cells were split into a T 25-cm2 cell culture flask and were harvested 48 h after transfection.
For knockdowns mediated by transient transfection of pSUPERpuro plasmids, 2 × 105 HeLa cells were seeded into six-well plates. Twenty-four hours later, 300 ng of reporter construct, 100 ng of normalizer (pNORM-7SL-AmdS, pβ mini-μ wt IRE or pGEM5 IRE β-Globin wt, pmCMVrGPx1-TGC), and 400 ng (single knockdown) or 800 ng (double knockdown) of pSUPERpuro plasmid were transiently transfected into the cells with DreamFect (OZ Biosciences) according to the manufacturer’s guidelines. For Figure 3C and Supplemental Figure 3, 100 ng of reporter construct and 100 ng of pmCMVrGPx1-TGC normalizer were transfected. The cells were split into T 25-cm2 cell culture flasks 1 d later and selected with puromycin in a concentration of 1.5 µg/mL. Cells under SMG6/SMG7 double knockdown conditions were split 1 d later again into a T 75-cm2 flask. Twelve to 24 h before harvest, the cells were washed with PBS, and the puromycin-containing medium was exchanged with normal DMEM+/+ medium. Cells were harvested 4 d after transfection.
For knockdown experiments done with HeLa tTR-KRAB cells, 2 × 105 cells were seeded into six-well plates in DOX-containing medium. DOX was present in the medium until the day of harvest. Twenty-four hours after seeding, cells were transfected with 300 ng of reporter construct and 100 ng of normalizer (pNORM-7SL-AmdS, pβ mini-μ wt IRE or pGEM5 IRE β-Globin wt) and split 1 d later into a T 25-cm2 flask. The cells were harvested 3 d after transfection. For details, see Nicholson et al. (2012). Efficiencies of knockdowns were verified on the protein level by Western blotting and/or on the mRNA level by qPCR analysis.
Reverse transcription and quantitative real-time PCR (RT-qPCR)
Total RNA was extracted from the cells using guanidium thiocyanate:phenol:chloroform extraction (Nicholson et al. 2012) or the GenElute Mammalian Total RNA Miniprep Kit (Sigma-Aldrich). The RNA samples were digested with 15–20 units of RNase-free recombinant DNaseI (Roche) for 45 min at 37°C and afterward purified by phenol:chloroform:isoamyl alcohol extraction (25:24:1) and ethanol precipitation according to standard protocols. One microgram of RNA was reverse-transcribed with 300 ng of random hexamers, 0.4 mM dNTPs, 10 mM DTT, 1× AffinityScript RT Buffer, and 1 μL of AffinityScript Multiple Temperature Reverse Transcriptase (Agilent Technologies) according to the manufacturer’s protocol. Controls lacking reverse transcriptase were done to confirm the successful DNase digestion if 7SL-AmdS had to be detected. The cDNA was diluted to 8 ng/μL. Quantitative real-time PCR was done with 24 ng of cDNA and 1× Brilliant III Ultra-Fast QPCR Master Mix (Agilent Technologies) using TaqMan probes, which hybridized over exon–exon junctions. To detect 7SL-AmdS, 24 ng of cDNA and Brilliant III Ultra-Fast SYBR Green QPCR Master Mix (Agilent Technologies) were used in a SYBR Green assay. The sequences of the probes and primers are listed in the Supplemental Material. The samples were measured in duplicates using Rotor-Gene 6000 (Corbett) with the following conditions: 3 min at 95°C, 5 sec at 95°C, and 15 sec at 60°C in 40 cycles. Analysis was done according to Nicholson et al. (2012).
Statistics analysis
Fold changes of mRNA levels were determined for each independent experiment and averaged afterward. Mean values of relative mRNA levels were calculated from mRNA levels of the independent experiments. The ±SEM (standard error of the mean) was calculated with Excel (Microsoft Corporation), where n ≥ 3. P-values were determined using a one-tailed unpaired Student’s t-test, when n ≥ 3.
Western blotting
Whole cell extracts were electrophoresed on a 6%–15% SDS-PAGE. Proteins were transferred to Optitran BA-S 85 reinforced nitrocellulose membranes (Whatman GmbH) and probed with the rabbit polyclonal antibodies anti-hUPF1 (1:1000, RENT1 H-300, sc-48802, Santa Cruz Biotechnology or Lykke-Andersen et al. 2000), anti-hUPF2, anti-hUPF3b (both: 1:3000) (Lykke-Andersen et al. 2000), anti-hSMG5 (1:2000) (Franks et al. 2010), anti-hSMG6 (1:150, custom-made for our laboratory by Eurogentec; see de Turris et al. 2011), anti-hSMG7 (for Figs. 4C and 5A: 1:500 dilution of crude serum from a rabbit immunized with a bacterially expressed His-tagged polypeptide comprising the C-terminal amino acids 870–1137 of hSMG7; for Fig. 5B: 1:1000, A302–170A, Bethyl Laboratories), anti-CPSF-73 (1:5000; Jenny et al. 1996), anti-Actin (1:3000, Actin (20–33), A5060; Sigma-Aldrich), or with the mouse monoclonal antibodies anti-SMG1 (1:250, 6-RE13, sc-165563; Santa Cruz), anti-SmB/B′ (1:250 dilution of supernatant from the mouse hybridoma cell line Y12 that secretes an antibody against SmB/B′; Lerner et al. 1981), and anti-α-Tubulin (1:1000, B-7, sc-5286; Santa Cruz). As secondary antibodies, 1:10,000 diluted donkey-anti-rabbit or donkey-anti-mouse IRDye800CW (LI-COR Biosciences) was used. Fluorescent signals were detected using the Odyssey Infrared Imaging System (LI-COR Biosciences).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Jolanda Stettler, Maryam Syed, and Christoph Schweingruber for technical help; Simone C. Rufener for valuable comments on the manuscript; Lynne Maquat (Rochester University, New York), Andreas Kulozik (University of Heidelberg, Germany), and Torben Heick Jensen (Aarhus University, Denmark) for plasmids; Jens Lykke-Andersen (University of California, San Diego) and Silvia Barabino (University of Milano-Bicocca, Italy) for antibodies; and Didier Trono (EPFL, Switzerland) for providing the lentiviral system. This work was supported by grants to O.M. of the European Research Council (StG 207419), the Swiss National Science Foundation (31003A-127614 and 31003A-143717), and the canton of Bern.
REFERENCES
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 [DOI] [PubMed] [Google Scholar]
- Anders KR, Grimson A, Anderson P 2003. SMG-5, required for C. elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J 22: 641–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR 2006. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science 313: 1968–1972 [DOI] [PubMed] [Google Scholar]
- Antoniou M, Geraghty F, Hurst J, Grosveld F 1998. Efficient 3′-end formation of human β-globin mRNA in vivo requires sequences within the last intron but occurs independently of the splicing reaction. Nucleic Acids Res 26: 721–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E 2007. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J 26: 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW 2000. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6: 1226–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono F, Gehring NH 2011. Assembly, disassembly and recycling: The dynamics of exon junction complexes. RNA Biol 8: 24–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono F, Ebert J, Lorentzen E, Conti E 2006. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 126: 713–725 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553 [DOI] [PubMed] [Google Scholar]
- Buchwald G, Ebert J, Basquin C, Sauliere J, Jayachandran U, Bono F, Le Hir H, Conti E 2010. Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC–UPF3b complex. Proc Natl Acad Sci 107: 10050–10055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Steiner S, Mohn F, Paillusson A, Muhlemann O 2006. EJC-independent degradation of nonsense immunoglobulin-μ mRNA depends on 3′ UTR length. Nat Struct Mol Biol 13: 462–464 [DOI] [PubMed] [Google Scholar]
- Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Le Hir H, Conti E 2011. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell 41: 693–703 [DOI] [PubMed] [Google Scholar]
- Chamieh H, Ballut L, Bonneau F, Le Hir H 2008. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol 15: 85–93 [DOI] [PubMed] [Google Scholar]
- Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA II, Wilkinson MF 2007. An alternative branch of the nonsense-mediated decay pathway. EMBO J 26: 1820–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WK, Bhalla AD, Le Hir H, Nguyen LS, Huang L, Gecz J, Wilkinson MF 2009. A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat Struct Mol Biol 16: 747–753 [DOI] [PubMed] [Google Scholar]
- Chen CY, Shyu AB 2003. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol Cell Biol 23: 4805–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu SY, Serin G, Ohara O, Maquat LE 2003. Characterization of human Smg5/7a: A protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA 9: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kim KM, Kim YK 2009. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol Cell 33: 75–86 [DOI] [PubMed] [Google Scholar]
- Cho H, Han S, Choe J, Park SG, Choi SS, Kim YK 2013. SMG5–PNRC2 is functionally dominant compared with SMG5–SMG7 in mammalian nonsense–mediated mRNA decay. Nucleic Acids Res 41: 1319–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Mourao A, Gutsche I, Gehring NH, Hentze MW, Kulozik A, Kadlec J, Sattler M, Cusack S 2009. Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J 28: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collis P, Antoniou M, Grosveld F 1990. Definition of the minimal requirements within the human β-globin gene and the dominant control region for high level expression. EMBO J 9: 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couttet P, Grange T 2004. Premature termination codons enhance mRNA decapping in human cells. Nucleic Acids Res 32: 488–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Weng Y, Hagan KW, Peltz SW 1995. Purification and characterization of the Upf1 protein: A factor involved in translation and mRNA degradation. RNA 1: 610–623 [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev 12: 1665–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J 2008. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol Cell 29: 271–278 [DOI] [PubMed] [Google Scholar]
- de Turris V, Nicholson P, Orozco RZ, Singer RH, Muhlemann O 2011. Cotranscriptional effect of a premature termination codon revealed by live-cell imaging. RNA 17: 2094–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpy L, Sirac C, Magnoux E, Duchez S, Cogne M 2004. RNA surveillance down-regulates expression of nonfunctional κ alleles and detects premature termination within the last κ exon. Proc Natl Acad Sci 101: 7375–7380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning G, Jamieson L, Maquat LE, Thompson EA, Fields AP 2001. Cloning of a novel phosphatidylinositol kinase-related kinase: Characterization of the human SMG-1 RNA surveillance protein. J Biol Chem 276: 22709–22714 [DOI] [PubMed] [Google Scholar]
- Eberle AB, Stalder L, Mathys H, Orozco RZ, Muhlemann O 2008. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol 6: e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55 [DOI] [PubMed] [Google Scholar]
- Fiorini F, Boudvillain M, Le Hir H 2012. Tight intramolecular regulation of the human Upf1 helicase by its N- and C-terminal domains. Nucleic Acids Res 41: 2404–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Singh G, Lykke-Andersen J 2010. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell 143: 938–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E 2005. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 17: 537–547 [DOI] [PubMed] [Google Scholar]
- Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E 2003. Nonsense-mediated mRNA decay in Drosophila: At the intersection of the yeast and mammalian pathways. EMBO J 22: 3960–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE 2003. Y14 and hUpf3b form an NMD-activating complex. Mol Cell 11: 939–949 [DOI] [PubMed] [Google Scholar]
- Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE 2005. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell 20: 65–75 [DOI] [PubMed] [Google Scholar]
- Gehring NH, Lamprinaki S, Hentze MW, Kulozik AE 2009a. The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol 7: e1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW 2009b. Disassembly of exon junction complexes by PYM. Cell 137: 536–548 [DOI] [PubMed] [Google Scholar]
- Grimson A, O’Connor S, Newman CL, Anderson P 2004. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA decay in Caenorhabditis elegans. Mol Cell Biol 24: 7483–7490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q, Zheng W, Tang S, Liu X, Zinkel RA, Tsui KW, Yandell BS, Culbertson MR 2006. Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast. PLoS Genet 2: e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudikote JP, Imam JS, Garcia RF, Wilkinson MF 2005. RNA splicing promotes translation and RNA surveillance. Nat Struct Mol Biol 12: 801–809 [DOI] [PubMed] [Google Scholar]
- He F, Brown AH, Jacobson A 1997. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol 17: 1580–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell 12: 1439–1452 [DOI] [PubMed] [Google Scholar]
- Huang L, Wilkinson MF 2012. Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip Rev RNA 3: 807–828 [DOI] [PubMed] [Google Scholar]
- Huang L, Lou CH, Chan W, Shum EY, Shao A, Stone E, Karam R, Song HW, Wilkinson MF 2011. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol Cell 43: 950–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E 2008. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14: 2609–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE 2008. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27: 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Minvielle-Sebastia L, Preker PJ, Keller W 1996. Sequence similarity between the 73-kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science 274: 1514–1517 [DOI] [PubMed] [Google Scholar]
- Johansson MJ, He F, Spatrick P, Li C, Jacobson A 2007. Association of yeast Upf1p with direct substrates of the NMD pathway. Proc Natl Acad Sci 104: 20872–20877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S, Weichenrieder O, Izaurralde E 2013. An unusual arrangement of two 14-3-3-like domains in the SMG5–SMG7 heterodimer is required for efficient nonsense-mediated mRNA decay. Genes Dev 27: 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec J, Izaurralde E, Cusack S 2004. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol 11: 330–337 [DOI] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S 2006. Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Jonas S, Jayachandran U, Buchwald G, Conti E, Lupas AN, Izaurralde E 2010. SMG6 interacts with the exon junction complex via two conserved EJC-binding motifs (EBMs) required for nonsense-mediated mRNA decay. Genes Dev 24: 2440–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerenyi Z, Merai Z, Hiripi L, Benkovics A, Gyula P, Lacomme C, Barta E, Nagy F, Silhavy D 2008. Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J 27: 1585–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervestin S, Jacobson A 2012. NMD: A multifaceted response to premature translational termination. Nat Rev Mol Cell Biol 13: 700–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Kataoka N, Dreyfuss G 2001. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon–exon junction complex. Science 293: 1832–1836 [DOI] [PubMed] [Google Scholar]
- Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH 2006. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA 12: 1015–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ 2000. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J 19: 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ 2001. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 20: 4987–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc JJ, Beemon KL 2004. Unspliced Rous sarcoma virus genomic RNAs are translated and subjected to nonsense-mediated mRNA decay before packaging. J Virol 78: 5139–5146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Ishigaki Y, Li X, Maquat LE 2002. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: Dynamics of mRNP remodeling. EMBO J 21: 3536–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Li X, Maquat LE 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell 12: 675–687 [DOI] [PubMed] [Google Scholar]
- Lelivelt MJ, Culbertson MR 1999. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol 19: 6710–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA Jr, Steitz JA 1981. Monoclonal antibodies to nucleic acid-containing cellular constituents: Probes for molecular biology and autoimmune disease. Proc Natl Acad Sci 78: 2737–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman D, Plasterk RH, Johnstone IL, Caceres JF 2007. Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev 21: 1075–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Azzalin CM, Hug N, Deplazes A, Peter M, Lingner J 2007. Saccharomyces cerevisiae Ebs1p is a putative ortholog of human Smg7 and promotes nonsense-mediated mRNA decay. Nucleic Acids Res 35: 7688–7697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103: 1121–1131 [DOI] [PubMed] [Google Scholar]
- Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J 2008. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell 133: 303–313 [DOI] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A 2003. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE 2004. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5: 89–99 [DOI] [PubMed] [Google Scholar]
- Matsuda D, Hosoda N, Kim YK, Maquat LE 2007. Failsafe nonsense-mediated mRNA decay does not detectably target eIF4E-bound mRNA. Nat Struct Mol Biol 14: 974–979 [DOI] [PubMed] [Google Scholar]
- Melero R, Buchwald G, Castano R, Raabe M, Gil D, Lazaro M, Urlaub H, Conti E, Llorca O 2012. The cryo-EM structure of the UPF–EJC complex shows UPF1 poised toward the RNA 3′ end. Nat Struct Mol Biol 19: 498–505 [DOI] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC 2004. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 36: 1073–1078 [DOI] [PubMed] [Google Scholar]
- Moriarty PM, Reddy CC, Maquat LE 1997. The presence of an intron within the rat gene for selenium-dependent glutathione peroxidase 1 is not required to protect nuclear RNA from UGA-mediated decay. RNA 3: 1369–1373 [PMC free article] [PubMed] [Google Scholar]
- Muhlemann O, Lykke-Andersen J 2010. How and where are nonsense mRNAs degraded in mammalian cells? RNA Biol 7: 28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murachelli AG, Ebert J, Basquin C, Le Hir H, Conti E 2012. The structure of the ASAP core complex reveals the existence of a Pinin-containing PSAP complex. Nat Struct Mol Biol 19: 378–386 [DOI] [PubMed] [Google Scholar]
- Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O 2010. Nonsense-mediated mRNA decay in human cells: Mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci 67: 677–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson P, Joncourt R, Muhlemann O 2012. Analysis of nonsense-mediated mRNA decay in mammalian cells. Curr Protoc Cell Biol 55: p27.4.1–27.4.61 [DOI] [PubMed] [Google Scholar]
- Nott A, Le Hir H, Moore MJ 2004. Splicing enhances translation in mammalian cells: An additional function of the exon junction complex. Genes Dev 18: 210–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S 2003. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell 12: 1187–1200 [DOI] [PubMed] [Google Scholar]
- Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S 2012. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res 40: 1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page MF, Carr B, Anders KR, Grimson A, Anderson P 1999. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol 19: 5943–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillusson A, Hirschi N, Vallan C, Azzalin CM, Muhlemann O 2005. A GFP-based reporter system to monitor nonsense-mediated mRNA decay. Nucleic Acids Res 33: e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M, Ishigaki Y, Nagy E, Maquat LE 2001. Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA 7: 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajavel KS, Neufeld EF 2001. Nonsense-mediated decay of human HEXA mRNA. Mol Cell Biol 21: 5512–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani AK, Nelson AC, Kapranov P, Bell I, Gingeras TR, Fraser AG 2009. High resolution transcriptome maps for wild-type and nonsense-mediated decay-defective Caenorhabditis elegans. Genome Biol 10: R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbapragada I, Lykke-Andersen J 2009. Execution of nonsense-mediated mRNA decay: What defines a substrate? Curr Opin Cell Biol 21: 394–402 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E 2005. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA 11: 1530–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ 2008. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol 28: 4320–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauliere J, Murigneux V, Wang Z, Marquenet E, Barbosa I, Le Tonqueze O, Audic Y, Paillard L, Roest Crollius H, Le Hir H 2012. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat Struct Mol Biol 19: 1124–1131 [DOI] [PubMed] [Google Scholar]
- Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE 2001. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol Cell Biol 21: 209–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AL, Ribeiro P, Inacio A, Liebhaber SA, Romao L 2008. Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA 14: 563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Rebbapragada I, Lykke-Andersen J 2008. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol 6: e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ 2012. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 151: 750–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder L, Muhlemann O 2008. The meaning of nonsense. Trends Cell Biol 18: 315–321 [DOI] [PubMed] [Google Scholar]
- Tani H, Imamachi N, Salam KA, Mizutani R, Ijiri K, Irie T, Yada T, Suzuki Y, Akimitsu N 2012. Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol 9: 1370–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey PS, Lucy Raymond F, Nguyen LS, Rodriguez J, Hackett A, Vandeleur L, Smith R, Shoubridge C, Edkins S, Stevens C, et al. 2007. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet 39: 1127–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE 1998. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J 17: 3484–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E 2004. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell 16: 587–596 [DOI] [PubMed] [Google Scholar]
- Wen J, Brogna S 2010. Splicing-dependent NMD does not require the EJC in Schizosaccharomyces pombe. EMBO J 29: 1537–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW 1996. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol 16: 5477–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand HL, Lu S, Cullen BR 2003. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc Natl Acad Sci 100: 11327–11332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann J, Hol EM, Jack HM 2006. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol 26: 1272–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiznerowicz M, Trono D 2003. Conditional suppression of cellular genes: Lentivirus vector-mediated drug-inducible RNA interference. J Virol 77: 8957–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S 2001. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev 15: 2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, et al. 2009. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev 23: 1091–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yepiskoposyan H, Aeschimann F, Nilsson D, Okoniewski M, Muhlemann O 2011. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA 17: 2108–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sun X, Qian Y, Maquat LE 1998. Intron function in the nonsense-mediated decay of β-globin mRNA: Indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA 4: 801–815 [DOI] [PMC free article] [PubMed] [Google Scholar]