Abstract
Bacterial cells have developed multiple strategies to communicate with their surrounding environment. The intracellular compartment is separated from the milieu by a relatively impermeable cell envelope through which small molecules can passively diffuse, while larger macromolecules, such as proteins, can be actively transported. In Gram-negative bacteria, the cell envelope is a double membrane, which houses several supramolecular protein complexes that facilitate the trafficking of molecules. For example, bacterial pathogens use these types of machines to deliver toxins into target eukaryotic host cells, thus subverting host cellular functions. Six different types of nanomachines, called Type I - Type VI secretion systems (T1SS - T6SS), can be readily identified by their composition and mode of action. A remarkable feature of these protein secretion systems is their similarity to systems with other biological functions, such as motility or the exchange of genetic material. The T6SS has provided a refreshing view on this concept since it shares similarity with the puncturing device of bacteriophages, which is used by these viruses to inject their DNA into bacterial target cells. In contrast, the bacterial T6SS transports toxins into other bacteria, engaging a ferocious competition for the colonization of their environment. Moreover, as with few other secretion systems, the T6SS is capable of injecting toxins into eukaryotic cells, which contributes to a successful infection. This highlights the multifunctional aspects of the T6SS, and our understanding of its mechanistic details is an intense field of investigation with significant implications for ecology, agriculture and medicine.
Protein secretion in Gram-negative bacteria
All cells are surrounded by an envelope whose prime function is to delineate the contour of the cell unit [1]. This envelope prevents the cellular content from leakage and protects it from disruption by damaging agents. However, cells cannot survive without establishing an active communication with the external environment. For example, waste generated by the cell metabolism needs to be released and nutrients essential for growth need to be taken up. Consequently, many molecular machines are in place within the cell envelope, which provide multiple paths for exchange of ions, small molecules or macromolecules, such as proteins. Bacteria are single-cell organisms and are classified as Gram-negative or Gram-positive depending on the structure of their cell envelope [1,2]. Both have a cytoplasmic membrane, but Gram-negative bacteria have an additional asymmetrical bilayer called the outer membrane. In this case, proteins targeted to the outside (exoproteins) have to pass two successive membrane hurdles. Over the course of evolution, these bacteria have developed sophisticated nanomachines, which are macromolecular complexes spanning both membranes and which guide the exoproteins from the inside to the outside [3,4]. These exoproteins can be toxins targeted to host cells in the case of bacterial pathogens but also enzymes whose role is to digest non-utilisable complex food sources into basics nutrients that can then be readily taken up into the cell.
Why so many different secretion systems?
To date, six different protein secretion systems have been characterized in detail [5]. The real difference between these systems is simply in the components that form the transport machinery. In some cases, one protein inserted into the outer membrane will be sufficient to give access to the external medium, while the rest of the information needed for targeting is contained in the exoprotein. This is the case for the T5SS whose archetype is the IgA protease from Neisseria meningitidis [6]. In contrast, others, such as the T3SS, have hugely complex structures composed of not less than 15-20 proteins, which assemble into a gigantic puzzle whose shape and localisation within the cell envelope is visible using cryo-electron microscopy (cryo-EM) [7,8], as first visualized with Salmonella typhimurium [9]. These complex secretion machines span both cytoplasmic and outer membranes and can be powered using ATP as fuel to expel proteins to the outside. Importantly, a single organism will not necessarily use only one type of secretion system. For example, Pseudomonas aeruginosa has them all [10] except for the T4SS, which was discovered in Agrobacterium tumefaciens [11,12].
The fundamental question is why does an organism put so many different systems in place to perform the same job? The answer might be two-fold; firstly, they might do different jobs and secondly, the organism did not plan which system to adopt in advance. Some secretion systems, such as the T1SS and T2SS, essentially release enzymes, such as proteases or lipases, into the surroundings and these enzymes can then chew up food for further assimilation by the cell. Note that in pathogens, these enzymes can cause severe damage to the tissues. Moreover, the exotoxin A of P. aeruginosa [13] is released in a T2SS-dependent manner and then taken up by retrograde transport into host target cells where it disrupts cellular functions [14]. In the case of the T3SS and T4SS, the delivery of toxins to target cells is more direct since it involves cell-to-cell contact via a specific structure of the secretion system that emerges at the surface of the bacterial cell. In the case of the T4SS, this emerging part resembles a conjugative pilus, which is used to exchange DNA between bacteria during the conjugation/mating process [15]. One may imagine that bacteria have evolved a system initially used to exchange DNA into a system that is used to inject toxins into eukaryotic cells. Another example of this evolution paradigm is the transformation of an apparatus used to assemble a flagellum at the cell surface (an appendage involved in bacterial motility) into an apparatus that assembles a needle-like structure better suited to contact and inject toxins into target cells [16]. Along these lines, the T2SS has adapted the contractile capability of Type IV pili to design a shorter piston-like structure, or pseudopilus, involved in propelling the T2SS substrate outside the cell [17,18].
The Type VI secretion system resembles the bacteriophage structure
Slight modification of already existing macromolecular complexes and the adaptation of their initial function to achieve a new goal is a common feature in the development of secretion systems. In all cases, the idea is to use an existing machine, providing access to the external medium, providing cell contact capacity or transferring macromolecules, such as DNA, thus turning it into a proper protein delivery system. The T6SS is the most recently discovered secretion type [19]. The initial observation described clusters of about 15 conserved genes of which two were encoding components similar to those found in the T4SS (IcmF and DotU). Because of this, the suspicion emerged that this cluster could be encoding a new secretion machine. However, the exact nature of the remaining components was unknown due to lack of similarity with proteins already in the databases. When the common ancestor is very ancient, the amino acid sequence similarity between evolved proteins is hardly identifiable. However, what is frequently conserved is the overall structure and fold. This is where the breakthrough came from, since two T6SS components (namely VgrG and Hcp) were finally recognized as homologous to the gp27/gp5 and gp19 bacteriophage proteins [20-22] (Figure 1).
Figure 1. The Type VI secretion system.
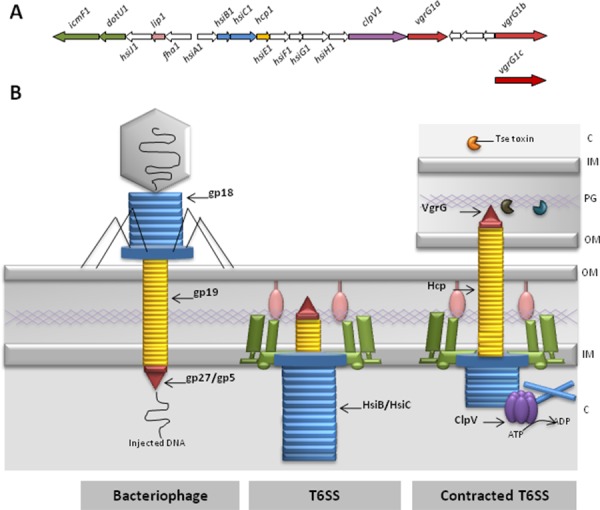
(A) The H1-type V1 secretion system (H1-T6SS) cluster from Pseudomonas aeruginosa. The nomenclature is as published previously [19]. The colour code used for the genes corresponds to the encoded proteins as shown in panel B.
(B) Protein secretion via the type V1 secretion system (T6SS) is compared with the bacteriophage DNA injection mechanism. On the left, a bacteriophage has landed on the surface of a bacterium. The DNA in the capsid is injected via the tail into the cytoplasm (C) of the bacterium. Highlighted is the tail tube (yellow), which is made with gp19, the tail sheath (blue) made with gp18 subunits and the puncturing device (red) made with gp27 and gp5. Next to the phage, the T6SS is represented and basically is an inverted phage tail. The colour code corresponds to the genes indicated in panel A and highlights the similarities with the phage. The HsiB/HsiC complex is the Pseudomonas aeruginosa T6SS sheath structure (also known as VipA/VipB or TssB/TssC) (see also Figure 2). TheT6SS tube is composed of Hcp (yellow) and the puncturing device is known as VgrG (red). The green and pink components are a representation of the IcmF/DotU/Lip complex, which connects the inner membrane (IM) and outer membrane (OM) through the peptidoglycan layer (PG). Upon contact with another bacterial cell (as shown on the right), the sheath contracts and pushes the puncturing device and tube through the cell envelope and punches the target bacterial cell. In the case of the P. aeruginosa H1-T6SS, three toxins are then injected (Tse1-3). Once the contraction has taken place, the sheath is disassembled by the AAA ATPase ClpV (purple) and the system is reset for a new round of toxin injection. For more details see main text.
The gp proteins are central components of the bacteriophage's tail, such as in the T4 phage [23] (Figure 1). Bacteriophages are bacterial viruses that inject their DNA into target bacterial cells. These bacteria are used as cell factories to replicate the phage DNA and to encode the structural components of the bacteriophage. The new virus particles are released by bursting the host bacterium and are then ready to infect new target cells. In brief, gp19 is the basic unit of the tail phage tube and displays structural homology with Hcp (Figure 1). The three-dimensional (3D) structure of the Hcp protein from P. aeruginosa was solved in 2006 [20], and Hcp forms a hexameric structure with a central channel of 40 Å. These hexamers can pile up on top of each other forming nanotubes resembling the gp19 tail phage tube [24,25]. The gp27 and gp5 proteins form a heterotrimeric complex (gp273/gp53) in which gp5 is made of a series of small beta strands which, once assembled into a trimer, form a sharp and rigid needle structure [26]. This needle is placed on the trimeric gp27 complex, which is sitting on top of the gp19 tail tube. Interestingly, the VgrG protein is made of two domains; the N-terminal domain resembling gp27, while the C-terminus has a gp5-like structure [21,27].
The function of the phage gp27/gp5 puncturing device is to create a hole in the outer membrane of the target cell and to allow injection of the phage DNA through the gp19 tube (Figure 1). This puncturing event also involves a tail sheath that surrounds the gp19 tube. This sheath structure is composed of more than a hundred copies of a protein called gp18, is contractile, and upon contraction pushes the tail tube and the puncturing device through the outer membrane and into the bacterial cell [28]. Electron microscopy studies have shown that two proteins of the T6SS, generically named VipA/VipB or TssB/TssC, form tubular structures, resembling cogwheels when viewed from the top [29,30] (Figure 2) and this structure cycles between extended and contracted conformations, further highlighting the conserved contractile mechanism [31,32]. These two proteins assemble to form a complex analogous to the gp18 structure [33].
Figure 2. The tail sheath-like structure of the T6SS.
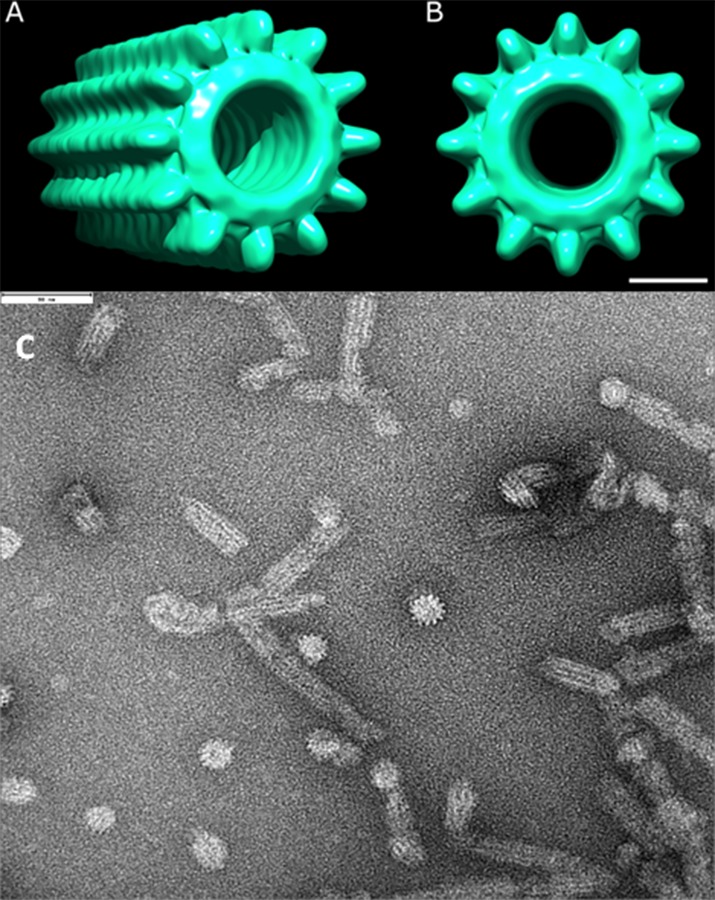
Model for the Pseudomonas aeruginosa HsiB/HsiC type V1 secretion system (T6SS) tail sheath as seen from the side (A) or from the top (B). The model is based on electron microscopy images as shown in (C) and also described elsewhere [30]. In panel C the scale shown at the top left corner is 90 nm.
The set of conserved T6SS components not only include TssB and TssC but also TssA, TssEFG and TssK. It is not yet known what the role of these proteins is in the T6SS, but since the homology between the phage tail and the T6SS is becoming obvious, the trail should lead us towards a more direct comparison with gp proteins that form the baseplate of the phage [34]. The baseplate is a complex that is central to the phage tail structure [35]. It contains proteins, such as gp48/gp54, which are essential to initiate elongation of the gp19 tail tube. Other proteins, such as gp6/gp25/gp53, form part of a wedge surrounding the puncturing device gp27/gp5. Finally, a subset of gp proteins act as adapters and short tail fibers, such as gp9 and gp12. The similarity between gp25 and TssE has now been demonstrated [36] and this trail is likely to take us towards the identification of further structural similarities between other T6SS components and some of the gp proteins named above. One should note here that it is unlikely that IcmF (TssM) and DotU (TssL) or the Lip (TssJ) components of the T6SS share any similarity with phage components. Instead, it has been shown that they are membrane bound, that they interact tightly with each other and are likely to form the docking area for the T6SS within the membrane, thus substituting for the docking role of the phage tail fibers [37,38].
In light of the previous observations, it is likely that the T6SS is a phage tail-like structure, with the major difference being that the phage punctures the cell from the outside to inject DNA, whereas the T6SS punctures the cell from the inside to secrete proteins (Figure 1).
The Type VI secretion system delivers bacterial toxins
The phrase “Tell me who you haunt and I will tell you who you are” comes from “Don Quixote” by Miguel de Cervantes and is now a French proverb. This quote introduces the concept that the function of a secretion system is primarily deduced from the type of proteins/effectors transported by the system. As mentioned previously, the T2SS from P. aeruginosa can have multiple functions, such as food acquisition via the secretion of lipases/proteases and cytotoxicity by the release of the ADP-ribosylating exotoxin A.
In the case of the T6SS, very few effectors were characterized until recently. The first effector to be identified was a VgrG protein from Vibrio cholerae (VgrG-1), which has a C-terminal extension in addition to the gp27/gp5 domain [21]. This C-terminal extension is an actin cross-linking domain (ACD) [39]. The ACD of VgrG-1, once injected into macrophages or amoeba, impairs the phagocytic activity of the cell by acting on the cytoskeleton and may cause cell death [40]. Another example where the secreted protein is a C-terminal extension of a VgrG protein is the VgrG1 from Aeromonas hydrophila [41]. In this case, the C-terminal extension is a VIP-2 domain with ADP ribosyltransferase activity. Injection of this domain into HeLa cells causes actin cytoskeleton disruption and apoptosis. Through these two examples, it has been suggested that the T6SS is a bacterial pathogen device whose primary function (as with the T3SS or T4SS) is to subvert signalling in eukaryotic cells and to take advantage of this process and survive in the infected host.
The reality is clearly different. In 2010, Hood and collaborators made the striking observation that one of the three T6SSs in P. aeruginosa delivers three toxins into other bacteria [42] (Figure 1). Two of these toxins, namely Tse1 and Tse3, are injected into the periplasm of Gram-negative bacteria [43]. There, they degrade the peptidoglycan, which provides shape and rigidity to the bacterial cell. As a result, the target bacterium rounds up and eventually bursts. Tse2's function is unknown but it is likely that it is injected into the cytosol, inhibiting further cell growth [42]. Each of the genes encoding the Tse toxins is organized in tandem with another gene coding for a specific immunity protein which, when expressed, protects the cells from the activity of the toxin. Since then, a whole family of T6SS-associated toxin/immunity couples have been reported [44], for example, in V. cholerae, Burkholderia species or Serratia marcescens [45-47].
This is a very exciting and important observation, which completely turned around the vision one had of the role of T6SS. The hypothesis now is that the T6SS is a key weapon not exclusively to attack a host but to compete with other bacteria living in the same host or environmental niche. Thus, it is proposed that the T6SS shapes the composition of a microbial population in one particular niche or within a mixed-species biofilm. It may also still be an important host colonization factor in the case of polymicrobial infections, such as in the lungs of cystic fibrosis patients.
Different bacteria use the T6SS in different ways. For example, P. aeruginosa uses the T6SS weapon against bacteria that show aggressive behaviour. The recent study by Basler and collaborators showed that when V. cholerae is mixed with P. aeruginosa, both bacteria produce active T6SS which results in the rounding up and lytic death of the Vibrio cells [48]. Interestingly, when the V. cholerae cells included in the mix are T6SS-defective, no P. aeruginosa-dependent T6SS killing is observed. This demonstrates that P. aeruginosa uses its T6SS in a tit-for-tat strategy and in this case, responds to any T6SS-dependent attack of Vibrio cells.
Is the puncturing device a multifunctional delivery system?
It is now clear that effectors from the T6SS can come at least in two flavours. One set of effectors is a C-terminal extension of the VgrG puncturing device and is mostly targeted to eukaryotic cells. The second set is made of “free” toxins involved in bacterial competition and killing. However, recent developments suggest that it might not be such a clear-cut case. Indeed, VgrG-3 from V. cholerae has a C-terminal extension, whose function is also to target the bacterial peptidoglycan [49,50]. Immediately downstream of the gene encoding VgrG-3, one can find a gene that encodes the cognate immunity protein, TsaB [49].
The question behind this observation is how are the “free” toxins secreted? One obvious suggestion is that although they are not covalently linked to a VgrG protein, they might potentially interact with the C-terminus of one such protein. A large number of VgrG proteins do not have any C-terminal extension and are termed canonical VgrG, as opposed to evolved VgrG (which have a C-terminal extension). It is thus a possibility that these canonical VgrGs may interact with “free” toxins via their C-terminus and will carry the effector into target cells as occurs with evolved VgrGs. In this case, it is possible that the VgrGs effectively have a dual function that is puncturing the cell envelope and transporting the effectors. Furthermore, it has also been proposed that proteins, such as members of the PAAR family, could bind to the tip of the VgrG proteins and be used as adaptors for additional T6SS toxins [51,52].
What are the next challenges?
Remarkably, 3D reconstruction based on cryo-EM images have provided a great deal of information about the assembly and structure of the bacteriophage tails. By contrast, structural data on the T6SS are rather scarce, and most of the knowledge is inferred from comparison with the phage. There are only a few images available, for example, the T6SS tail sheath-like structure [30,53] and the in situ cryo-EM images of a long tubule extending into the cytoplasm [54]. However, there is still a long way to go in following the bacteriophage trail and providing further structural comparisons.
This idea of the tail sheath contracting and pushing the puncturing device into target cells is appealing, but needs further demonstration. Importantly, there have been several reports describing that in the T6SS, an AAA ATPase (i.e. ClpV) is involved in recycling the tail-sheath components VipA/VipB after sheath contraction [32]. Sheath disassembly might then allow for a new round of assembly/contraction and injection of toxins into target cells. In the absence of ClpV, it is proposed that T6SS contraction happens only once and thus only one delivery of toxins occurs, resulting in poor T6SS activity [55]. Understanding the dynamic of the T6SS is an important challenge. Reports presenting live images of bacterial cells expressing GFP- or mCherry-tagged T6SS components are flourishing and could be the development needed to address this question [31,56,57]. In these images, bacteria can be seen battling against each other through flashes of fluorescence.
A great deal is to be expected from the discovery of new effectors and the greater the variety, the greater the functional potential of the T6SS. In the case of P. Aeruginosa, the genes encoding these effectors are not necessarily clustered with the T6SS genes, making their identification more complex. Recently, it has been suggested that a superfamily of phospholipases is a T6SS effector. The genes encoding these Type VI lipase effectors (tle) are not adjacent to complete T6SS gene clusters but adjacent to genes encoding orphan VgrG proteins [58]. Again, how the T6SS effectors are transported is still unclear, and whether the effectors are transported through the T6SS tube or are bound to VgrGs forming the T6SS tip awaits further demonstration.
Finally, it seems that the T6SS may play a role against both eukaryotic and prokaryotic cells. The role played by bacterial competition in pathogenesis and its importance (if any) is yet to be understood.
In conclusion, the T6SS has attracted the immediate attention of a number of microbiologists. Its unique structure and relationship with a bacterial virus will foster understanding of evolution mechanisms. Its increasingly relevant role in bacterial competition and its ability to control bacterial population makes it highly relevant to ecology, agriculture and medicine. Finally, the structural complexity and functional diversity of the T6SS is a vast playground for cutting edge research and understanding of basic principles in biology.
Acknowledgments
I would like to thank Agnes Sagfors for the design of the T6SS figure, Tillmann Pape for the electron microscopy images and modelling and Luke Allsopp for his critical review of the manuscript. The work on T6SS in Alain Filloux's laboratory is supported by the Medical Research Council Programme Grant MR/K001930/1 and the Wellcome Trust Grant WT091939.
Abbreviations
- Cryo-EM
cryo-electron microscopy
- P. aeruginosa
Pseudomonas aeruginosa
- T1SS
Type I secretion system
- T2SS
Type II secretion system
- T3SS
Type III secretion system
- T4SS
Type IV secretion system
- T5SS
Type V secretion system
- T6SS
Type VI secretion system
Disclosures
The author declares that he has no disclosures.
The electronic version of this article is the complete one and can be found at: http://f1000.com/prime/reports/b/5/52
References
- 1.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneewind O, Missiakas DM. Protein secretion and surface display in Gram-positive bacteria. Philos Trans R Soc Lond, B, Biol Sci. 2012;367:1123–39. doi: 10.1098/rstb.2011.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filloux A. Secretion signal and protein targeting in bacteria: a biological puzzle. J Bacteriol. 2010;192:3847–9. doi: 10.1128/JB.00565-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filloux A. Protein Secretion Systems in Pseudomonas aeruginosa: An Essay on Diversity, Evolution, and Function. Front Microbiol. 2011;2:155. doi: 10.3389/fmicb.2011.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desvaux M, Hébraud M, Talon R, Henderson IR. Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol. 2009;17:139–45. doi: 10.1016/j.tim.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Leo JC, Grin I, Linke D. Type V secretion: mechanism(s) of autotransport through the bacterial outer membrane. Philos Trans R Soc Lond, B, Biol Sci. 2012;367:1088–101. doi: 10.1098/rstb.2011.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornelis GR. The type III secretion injectisome, a complex nanomachine for intracellular ‘toxin’ delivery. Biol Chem. 2010;391:745–51. doi: 10.1515/bc.2010.079. [DOI] [PubMed] [Google Scholar]
- 8.Galán JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–73. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 9.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galán JE, Aizawa SI. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–5. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718180333
- 10.Bleves S, Viarre V, Salacha R, Michel GPF, Filloux A, Voulhoux R. Protein secretion systems in Pseudomonas aeruginosa: A wealth of pathogenic weapons. Int J Med Microbiol. 2010;300:534–43. doi: 10.1016/j.ijmm.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451–85. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waksman G, Fronzes R. Molecular architecture of bacterial type IV secretion systems. Trends Biochem Sci. 2010;35:691–8. doi: 10.1016/j.tibs.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Filloux A. The underlying mechanisms of type II protein secretion. Biochim Biophys Acta. 2004;1694:163–79. doi: 10.1016/j.bbamcr.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Spooner RA, Smith DC, Easton AJ, Roberts LM, Lord JM. Retrograde transport pathways utilised by viruses and protein toxins. Virol J. 2006;3:26. doi: 10.1186/1743-422X-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wallden K, Rivera-Calzada A, Waksman G. Type IV secretion systems: versatility and diversity in function. Cell Microbiol. 2010;12:1203–12. doi: 10.1111/j.1462-5822.2010.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pallen MJ, Gophna U. Bacterial flagella and Type III secretion: case studies in the evolution of complexity. Genome Dyn. 2007;3:30–47. doi: 10.1159/000107602. [DOI] [PubMed] [Google Scholar]
- 17.Durand E, Alphonse S, Brochier-Armanet C, Ball G, Douzi B, Filloux A, Bernard C, Voulhoux R. The assembly mode of the pseudopilus: a hallmark to distinguish a novel secretion system subtype. J Biol Chem. 2011;286:24407–16. doi: 10.1074/jbc.M111.234278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korotkov KV, Sandkvist M, Hol WGJ. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol. 2012;10:336–51. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filloux A, Hachani A, Bleves S. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology (Reading, Engl) 2008;154:1570–83. doi: 10.1099/mic.0.2008/016840-0. [DOI] [PubMed] [Google Scholar]
- 20.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordoñez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–30. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1002370
- 21.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci USA. 2007;104:15508–13. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1092890
- 22.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 2006;103:1528–33. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1030236
- 23.Leiman PG, Arisaka F, van Raaij MJ, Kostyuchenko VA, Aksyuk AA, Kanamaru S, Rossmann MG. Morphogenesis of the T4 tail and tail fibers. Virol J. 2010;7:355. doi: 10.1186/1743-422X-7-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ballister ER, Lai AH, Zuckermann RN, Cheng Y, Mougous JD. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc Natl Acad Sci USA. 2008;105:3733–8. doi: 10.1073/pnas.0712247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA. 2009;106:4154–9. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1158739
- 26.Kanamaru S, Leiman PG, Kostyuchenko VA, Chipman PR, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415:553–7. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1003946
- 27.Hachani A, Lossi NS, Hamilton A, Jones C, Bleves S, Albesa-Jové D, Filloux A. Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. J Biol Chem. 2011;286:12317–27. doi: 10.1074/jbc.M110.193045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leiman PG, Shneider MM. Contractile tail machines of bacteriophages. Adv Exp Med Biol. 2012;726:93–114. doi: 10.1007/978-1-4614-0980-9_5. [DOI] [PubMed] [Google Scholar]
- 29.Bönemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 2009;28:315–25. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1162646
- 30.Lossi NS, Manoli E, Förster A, Dajani R, Pape T, Freemont P, Filloux A. The HsiB1C1 (TssB-TssC) complex of the Pseudomonas aeruginosa type VI secretion system forms a bacteriophage tail sheathlike structure. J Biol Chem. 2013;288:7536–48. doi: 10.1074/jbc.M112.439273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basler M, Mekalanos JJ. Type 6 secretion dynamics within and between bacterial cells. Science. 2012;337:815. doi: 10.1126/science.1222901. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717953914
- 32.Kapitein N, Bönemann G, Pietrosiuk A, Seyffer F, Hausser I, Locker JK, Mogk A. ClpV recycles VipA/VipB tubules and prevents non-productive tubule formation to ensure efficient type VI protein secretion. Mol Microbiol. 2013;87:1013–28. doi: 10.1111/mmi.12147. [DOI] [PubMed] [Google Scholar]
- 33.Aksyuk AA, Leiman PG, Kurochkina LP, Shneider MM, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria. EMBO J. 2009;28:821–9. doi: 10.1038/emboj.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1159482
- 34.Yap ML, Mio K, Leiman PG, Kanamaru S, Arisaka F. The baseplate wedges of bacteriophage T4 spontaneously assemble into hubless baseplate-like structure in vitro. J Mol Biol. 2010;395:349–60. doi: 10.1016/j.jmb.2009.10.071. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718180334
- 35.Mesyanzhinov VV, Leiman PG, Kostyuchenko VA, Kurochkina LP, Miroshnikov KA, Sykilinda NN, Shneider MM. Molecular architecture of bacteriophage T4. Biochemistry Mosc. 2004;69:1190–202. doi: 10.1007/s10541-005-0064-9. [DOI] [PubMed] [Google Scholar]
- 36.Lossi NS, Dajani R, Freemont P, Filloux A. Structure-function analysis of HsiF, a gp25-like component of the type VI secretion system, in Pseudomonas aeruginosa. Microbiology (Reading, Engl) 2011;157:3292–305. doi: 10.1099/mic.0.051987-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aschtgen M, Thomas MS, Cascales E. Anchoring the type VI secretion system to the peptidoglycan: TssL, TagL, TagP… what else? Virulence. 2010;1:535–40. doi: 10.4161/viru.1.6.13732. [DOI] [PubMed] [Google Scholar]
- 38.Ma L, Lin J, Lai E. An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its walker a motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J Bacteriol. 2009;191:4316–29. doi: 10.1128/JB.00029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718180335
- 39.Ma AT, Mekalanos JJ. In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc Natl Acad Sci USA. 2010;107:4365–70. doi: 10.1073/pnas.0915156107. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718180336
- 40.Ma AT, McAuley S, Pukatzki S, Mekalanos JJ. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe. 2009;5:234–43. doi: 10.1016/j.chom.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1160123
- 41.Suarez G, Sierra JC, Erova TE, Sha J, Horneman AJ, Chopra AK. A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J Bacteriol. 2010;192:155–68. doi: 10.1128/JB.01260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718180337
- 42.Hood RD, Singh P, Hsu F, Güvener T, Carl MA, Trinidad RRS, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/5430960
- 43.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475:343–7. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/12268956
- 44.Russell AB, Singh P, Brittnacher M, Bui NK, Hood RD, Carl MA, Agnello DM, Schwarz S, Goodlett DR, Vollmer W, Mougous JD. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe. 2012;11:538–49. doi: 10.1016/j.chom.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718180338
- 45.Benz J, Sendlmeier C, Barends TRM, Meinhart A. Structural insights into the effector-immunity system Tse1/Tsi1 from Pseudomonas aeruginosa. PLoS ONE. 2012;7:e40453. doi: 10.1371/journal.pone.0040453. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718180339
- 46.Murdoch SL, Trunk K, English G, Fritsch MJ, Pourkarimi E, Coulthurst SJ. The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J Bacteriol. 2011;193:6057–69. doi: 10.1128/JB.05671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718180340
- 47.Schwarz S, West TE, Boyer F, Chiang W, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 2010;6:e1001068. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718180341
- 48.Basler M, Ho BT, Mekalanos JJ. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell. 2013;152:884–94. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718180343
- 49.Brooks TM, Unterweger D, Bachmann V, Kostiuk B, Pukatzki S. Lytic activity of the Vibrio cholerae type VI secretion toxin VgrG-3 is inhibited by the antitoxin TsaB. J Biol Chem. 2013;288:7618–25. doi: 10.1074/jbc.M112.436725. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717990025
- 50.Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc Natl Acad Sci USA. 2013;110:2623–8. doi: 10.1073/pnas.1222783110. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718180348
- 51.Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500:350–3. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718061906
- 52.Filloux A. Microbiology: a weapon for bacterial warfare. Nature. 2013;500:284–5. doi: 10.1038/nature12545. [DOI] [PubMed] [Google Scholar]
- 53.Bönemann G, Pietrosiuk A, Mogk A. Tubules and donuts: a type VI secretion story. Mol Microbiol. 2010;76:815–21. doi: 10.1111/j.1365-2958.2010.07171.x. [DOI] [PubMed] [Google Scholar]
- 54.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–6. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/14021965
- 55.Zheng J, Ho B, Mekalanos JJ. Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS ONE. 2011;6:e23876. doi: 10.1371/journal.pone.0023876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brunet YR, Espinosa L, Harchouni S, Mignot T, Cascales E. Imaging type VI secretion-mediated bacterial killing. Cell Rep. 2013;3:36–41. doi: 10.1016/j.celrep.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 57.LeRoux M, Leon JA de, Kuwada NJ, Russell AB, Pinto-Santini D, Hood RD, Agnello DM, Robertson SM, Wiggins PA, Mougous JD. Quantitative single-cell characterization of bacterial interactions reveals type VI secretion is a double-edged sword. Proc Natl Acad Sci USA. 2012;109:19804–9. doi: 10.1073/pnas.1213963109. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717970167
- 58.Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, Mougous JD. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature. 2013;496:508–12. doi: 10.1038/nature12074. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717997056


