Abstract
Biomarkers are becoming increasingly more important in clinical decision-making, as well as basic science. Diagnosing myocardial infarction (MI) is largely driven by detecting cardiac-specific proteins in patients' serum or plasma as an indicator of myocardial injury. Having recently shown that cardiac myosin binding protein-C (cMyBP-C) is detectable in the serum after MI, we have proposed it as a potential biomarker for MI. Biomarkers are typically detected by traditional sandwich enzyme-linked immunosorbent assays. However, this technique requires a large sample volume, has a small dynamic range, and can measure only one protein at a time.
Here we show a multiplex immunoassay in which three cardiac proteins can be measured simultaneously with high sensitivity. Measuring cMyBP-C in uniplex or together with creatine kinase MB and cardiac troponin I showed comparable sensitivity. This technique uses the Meso Scale Discovery (MSD) method of multiplexing in a 96-well plate combined with electrochemiluminescence for detection. While only small sample volumes are required, high sensitivity and a large dynamic range are achieved. Using this technique, we measured cMyBP-C, creatine kinase MB, and cardiac troponin I levels in serum samples from 16 subjects with MI and compared the results with 16 control subjects. We were able to detect all three markers in these samples and found all three biomarkers to be increased after MI. This technique is, therefore, suitable for the sensitive detection of cardiac biomarkers in serum samples.
Keywords: Molecular Biology, Issue 78, Cellular Biology, Biochemistry, Genetics, Biomedical Engineering, Medicine, Cardiology, Heart Diseases, Myocardial Ischemia, Myocardial Infarction, Cardiovascular Diseases, cardiovascular disease, immunoassay, cardiac myosin binding protein-C, cardiac troponin I, creatine kinase MB, electrochemiluminescence, multiplex biomarkers, ELISA, assay
Introduction
The measurement of low amounts of protein in complex biological samples, such as serum, is of growing clinical importance for patient management, as well as basic science. For instance, an increase in serum levels of cardiac biomarkers, such as troponin I, in a clinical setting is consistent with acute myocardial infarction (MI)1. To detect proteins in serum samples, standard enzyme-linked immunosorbent assay (ELISA) is the most often used technique as it has high sensitivity and allows for absolute quantification of the analyte. However, traditional ELISAs require a relatively large amount of sample (typically 100 μl), have high background signal in some biological fluids, and are restricted to the measurement of only one analyte per ELISA2.
Recently, a new immunoassay technique was introduced that circumvents many of these drawbacks. This modified assay, developed by MSD, uses electrochemiluminescence (ECL) for signal detection, which allows for a very low background and an increased sensitivity, enabling the use of small sample volumes. Electrochemiluminescence is based on the reporter molecule ruthenium (II) trisbipyridal, which is attached to the detection antibodies. This reporter molecule emits light at 620 nm upon the electrical stimulation of the bottom of the 96-well plate which has carbon electrodes integrated in them3. Also, by using spot coating, multiple capture antibodies can be coated into one well (up to 10 on a 96-well plate), allowing for simultaneous quantification of different proteins in a single sample4. This technique has recently been used to measure proinflammatory cytokine profiles in serum5,6. The multiplex plates from MSD compare favorably to other multiplex assay platforms7.
Using MSD as the primary assay platform, we further developed a custom made 3-plex plate that can simultaneously quantify the level of cardiac myosin binding protein-C (cMyBP-C), creatine-kinase MB (CK-MB), and cardiac troponin I (cTnI), and the results were compared with monoplex detection of cMyBP-C. CK-MB and cTnI are well-established biomarkers for MI. However, increases of these biomarkers can be caused by pathologies other than MI, e.g. myocarditis or renal failure8. This argues for the addition of additional biomarkers to increase the specificity of MI diagnosis. We have recently shown that cMyBP-C is also a potential biomarker for MI9. cMyBP-C is a thick filament associated protein that is expressed in the heart,10-12 but not in skeletal or smooth muscles. Thus, the increased level of cMyBP-C in the circulatory system is a specific indicator of cardiac damage13.
In this study, we compared uniplex detection of cMyBP-C with the use of a custom 3-plex assay to measure serum levels of cMyBP-C, CK-MB, and cTnI in serum of patients with MI. In the future, this signature technique might be used to diagnose MI in patients presenting with chest pain in the emergency room.
The institution review board (IRB) of the Loyola University Chicago approved the study for use of deidentified human samples and the use of the immunoassay (LU# 20392).
Protocol
1. Uniplex cMyBP-C Assay
The day before the experiment, coat the 96-well MSD bare standard plate with capture antibody. For cMyBP-C, use a 30 μl volume of mouse monoclonal anti-cMyBP-C antibody (gelatin free) at a concentration of 5 μg/ml diluted in phosphate buffered saline (PBS).
Since the well is hydrophobic, pipette the solution into the bottom corner of the well; then tap the sides of the plate to spread the solution over the entire well.
The plate is covered with a plate sealer and incubated O/N at 4 °C without shaking.
Remove the capture antibody solution by tapping the solution out over a sink and then on a stack of paper towels. Non-specific binding to the plate is blocked by adding 150 μl of a 5% (w/v) blocker A (high-grade BSA) solution in PBS to each well.
Seal the plate and incubate for 1 hr at RT while shaking at 700 rpm.
During the blocking step, prepare the standards and samples. Make the standard series by diluting recombinant cMyBP-C protein fragment (amino acids 1 - 271) to a starting concentration of 2,000 ng/ml in 1% (w/v) blocker A/PBS. Then serially dilute by a factor of 5 in 1% (w/v) blocker A/PBS. Total of 7 standards + 1 blank (1% (w/v) blocker A/PBS alone) were used.
Remove the blocking solution and wash the plate 3x with 150 μl 0.05% (v/v) Tween-20/PBS. Each time, remove the wash solution by inverting the plate above a sink. After the third wash step, vigorously flick the plate over a sink and pat the plate vigorously on a layer of paper towels until it is completely dry. This is a crucial step, as incubation volumes are small, and any remaining wash solution will significantly dilute the next incubation.
Pipette 25 μl of standards and samples into the wells. Seal the plate and incubate at RT, while shaking at 700 rpm for 1 hr.
Prepare the detection antibody solution at 1 μg/ml in 1% blocker A/PBS. A custom made cMyBP-C antibody (epitope amino acids 2 - 14) with MSD SULFO-TAG labeling serves as the detection antibody. Kits are available for SULFO-TAG labeling, making it a relatively simple procedure to label any antibody.
Repeat the wash step as described in step 1.4.
Add 25 μl of detection antibody solution to each well, seal the plate, and incubate at RT for 1 hr on a plate shaker set at 700 rpm.
During the incubation period, run MSD's demo-plate (plate with LED lights) to ensure proper function of the Sector Imager and software (refer Reagent section). Also, dilute the read-buffer, which is provided by MSD, by adding 5 ml of 4x read-buffer to 15 ml distilled water.
Repeat the wash step as described in step 1.4.
Add 150 μl of read-buffer to each well by using a multichannel pipette. Be sure to avoid air bubbles (reverse pipetting is a suitable method). After addition of the read-buffer, immediately scan the plate in the Sector Imager.
2. 3-plex cMyBP-C, CK-MB, and cTnI Assay
A 96-well 3-plex plate and diluent solutions are allowed to equilibrate to RT before use. This plate is pre-coated with capture antibodies for cMyBP-C, CK-MB, and cTnI by MSD.
Add 25 μl of 1% (w/v) blocker A/PBS to each well. Tap the sides of the plate to make sure that the entire well is covered by solution.
Seal the plate with a plate sealer, place the plate on a plate shaker, and shake at 700 rpm for 30 min.
In the meantime, a dilution series of individual calibrators is prepared. Because each well has three capture antibodies three calibrators need to be mixed and serially diluted before use. Dilute recombinant cMyBP-C, CK-MB (MSD), and cTnI (MSD) together in 1% (w/v) blocker A/PBS to create one sample with 2,000 ng/ml cMyBP-C, 100 ng/ml CK-MB, and 25 ng/ml cTnI (sample A).
A final volume of at least 100 μl is prepared for each standard. To complete the standard series, serially dilute the samples are by a factor of 5. Use 25 μl of sample A in 100 μl 1% (w/v) blocker A/PBS to create sample B. Then use 25 μl of sample B in 100 μl 1% (w/v) blocker A/PBS to create sample C, and so forth. Human serum samples from 16 subjects with MI and 16 control subjects were used to measure cMyBP-C, CK-MB, and cTnI levels. These samples were diluted 2x in 1% (w/v) blocker A/PBS.
Add 25 μl of the standards and diluted samples to the 25 μl already present in the wells of the 3-plex plate. It is important to also include a blank (diluent only). To establish the technique, samples are loaded in technical triplicates. Otherwise, duplicates are sufficient.
Seal the plate again (plate sealer can be reused) and incubate on a plate shaker (700 rpm) at RT for 2 hr.
Just before the incubation is finished, the detection antibody solution is prepared. The diluent used is 1% (w/v) blocker A in PBS. All three SULFO-tagged detection antibodies are diluted together into a final concentration of 1 μg/ml each. Detection antibodies are typically provided at a 50x stock concentration.
For a full 96-well plate, 2,700 μl of total diluted solution is needed (with pipetting error included). Add 54 μl of each detection antibody to 2,538 μl 1% (w/v) blocker A/PBS.
After 2 hr, remove solutions by vigorously flicking the plate above a sink. Wash the wells 3x with 150 μl of 0.05% Tween-20 in PBS. Add 25 μl of detection antibody solution to each well using a multichannel pipette, and the sides of the plate are tapped to ensure that the entire well is covered by solution.
Seal the plate and incubate for 1 hr while shaking at 700 rpm.
During the incubation period, run MSD's demo-plate (plate with LED lights) to ensure proper function of the Sector Imager and software. Also dilute the read-buffer, which is provided by MSD, by adding 5 ml of 4x read-buffer to 15 ml distilled water.
Repeat the wash step described in step 2.8.
Add 150 μl of read-buffer to each well using a multichannel pipette. Avoiding air bubbles is critical (reverse pipetting is a suitable method). After addition of the read-buffer, immediately scan the plate in the Sector Imager.
Representative Results
The basic principles and workflow of the 3-plex assay are shown in Figure 1, and the overall workflow is described in Table 1. Uniplex assays work along the same principle except that the entire bottom well is coated with one capture antibody. Signal detection is done by ECL in which an electrical signal is applied to the bottom of the well. This, in turn, initiates a local production of light through a chemical reaction of the read-buffer with the SULFO-TAG label on the detection antibody. This leads to the low background and high sensitivity of the assay. In Figure 2, the standard curve of cMyBP-C in the uniplex assay is compared to that of cMyBP-C in the 3-plex assay. Both assays show a very high sensitivity and a high dynamic range. Detection levels and quantification levels are shown in Table 2 and are comparable in both uniplex and 3-plex assays. Detection levels for cTnI and CK-MB are also shown in Figure 3 and Table 2. Inter-operator variability was determined by measuring identical samples (n = 2, 3 technical replicates) by two different operators and variability was found to be low (CV 8.5%). Cross reactivity of detection antibodies with the other two calibrators was studied by incubating individual standard curves of all three calibrators with single detection antibodies (Figure 4). Only low amount of cross-reactivity was seen between CK-MB calibrator and both cTnI and cMyBP-C detection antibodies while no cross-reactivity was observed between the other calibrators and detection antibodies.
As proof of the applicability of the assay for the detection of cardiac injury, serum levels of 16 subjects with MI were compared with a control group (n = 16). Using the 3-plex plate, serum levels of cMyBP-C, CK-MB, and cTnI were determined. All three biomarkers were increased in the MI subjects (see Figure 5) from 3.7-fold (cMyBP-C) to 12.2-fold (CK-MB), although variation in serum levels was the lowest in the cMyBP-C group.
| Process | Uniplex assay | 3-plex assay | |
| Experiment day 0 | Coat plate with capture antibody | Overnight | N/A |
| Experiment day 1 | Block the plate | 1 hr | 30 min |
| Prepare samples and standard series | |||
| Incubate with samples and standards | 1 hr | 2 hr | |
| Incubate with detection antibody | 1 hr | 1 hr | |
| Add detection reagent and read plate |
Table 1. Workflow overview of uniplex and multiplex assays.
| LLOD (ng/ml) | LLOQ (ng/ml) | ULOQ (ng/ml) | |
| cMyBP-C (uniplex) | 0.126 ± 0.007 | 0.567 ± 0.073 | 400 ng/ml |
| cMyBP-C (3plex) | 0.057 ± 0.022 | 0.469 ± 0.171 | 400 ng/ml |
| CK-MB (3-plex) | 0.038 ± 0.014 | 0.587 ± 0.213 | 100 ng/ml |
| cTnI (3plex) | 0.033 ± 0.011 | 0.147 ± 0.053 | 25 ng/ml |
Table 2. Detection limits of uniplex and 3-plex calibrators. LLOD, lower limit of detection, is defined as calculated concentration of the signal of the blank + 3 times standard deviation of the blank samples. LLOQ, lower limit of quantification, is defined as a value higher than LLOD, with a coefficient of variance (reciprocal of signal to noise ratio) lower than 20%, and a recovery between 80 - 120%. ULOQ, upper limit of quantification, is defined as the highest analyzed concentration that can be measured with a coefficient of variance lower than 20% and a recovery between 80 - 120%.
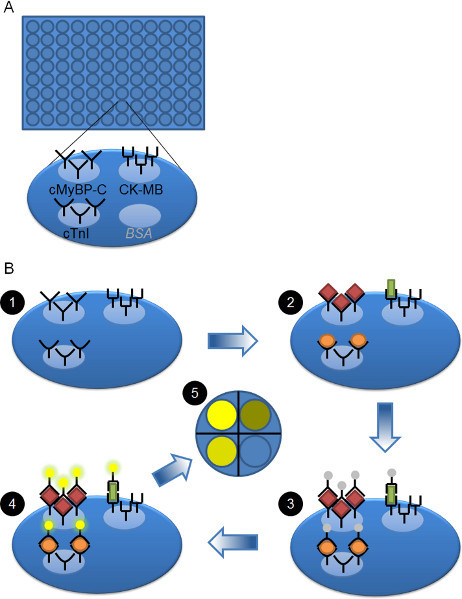 Figure 1. Schematic overview of plate and workflow.A) 96 well plate is shown. Each well is coated with three different capture antibodies, i.e. against cMyBP-C, CK-MB and cTnI. The fourth available spot is not used and is coated with BSA. B) Workflow of the 3-plex ELISA. The pre-coated plate (step 1) is blocked and then serum samples or calibrators are added to each well and allowed to bind (step 2). cMyBP-C (red diamonds), CK-MB (green rectangle), and cTnI (orange oval) bind to their respective capture antibodies. After washing away the unbound proteins, the SULFO-TAG-labeled detection antibodies are added (step 3). They will bind to the cMyBP-C, CK-MB, or cTnI proteins that are bound to their capture antibody. After washing away unbound detection antibody, read buffer is added and the plate is analyzed. An electrical signal to the bottom of the plate initiates a local chemical reaction of the SULFO-TAG with the read buffer, leading to the production of light (step 4). The amount of light is directly proportional to the amount of SULFO-TAG labeled detection antibody bound. The CCD imager records the light and can distinguish the different spots within each well and therefore the signal coming from each analyte (step 5).
Figure 1. Schematic overview of plate and workflow.A) 96 well plate is shown. Each well is coated with three different capture antibodies, i.e. against cMyBP-C, CK-MB and cTnI. The fourth available spot is not used and is coated with BSA. B) Workflow of the 3-plex ELISA. The pre-coated plate (step 1) is blocked and then serum samples or calibrators are added to each well and allowed to bind (step 2). cMyBP-C (red diamonds), CK-MB (green rectangle), and cTnI (orange oval) bind to their respective capture antibodies. After washing away the unbound proteins, the SULFO-TAG-labeled detection antibodies are added (step 3). They will bind to the cMyBP-C, CK-MB, or cTnI proteins that are bound to their capture antibody. After washing away unbound detection antibody, read buffer is added and the plate is analyzed. An electrical signal to the bottom of the plate initiates a local chemical reaction of the SULFO-TAG with the read buffer, leading to the production of light (step 4). The amount of light is directly proportional to the amount of SULFO-TAG labeled detection antibody bound. The CCD imager records the light and can distinguish the different spots within each well and therefore the signal coming from each analyte (step 5).
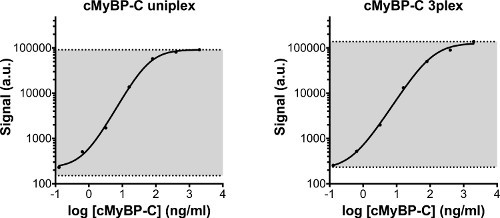 Figure 2. Standard curve of cMyBP-C calibrator measured in uniplex or in 3-plex. The same standard series was measured in a uniplex assay, and on the 3-plex plate. The standard series started at 2,000 ng/ml and was serially diluted 5x with the lowest concentration being 0.128 ng/ml. In addition, diluent was used as a blank sample. The detection area of the assay is displayed in grey. The lower line indicates the lower limit of detection (LLOD), which is defined as the calculated concentration of the blank signal + 3 times the standard deviation of the blank values. The upper limit of detection is defined as the highest concentration of the standard series that can reliably be measured. Both uniplex and 3-plex cMyBP-C standard curves are comparable. Detection limits are displayed in Table 2. a.u: arbitrary units.
Figure 2. Standard curve of cMyBP-C calibrator measured in uniplex or in 3-plex. The same standard series was measured in a uniplex assay, and on the 3-plex plate. The standard series started at 2,000 ng/ml and was serially diluted 5x with the lowest concentration being 0.128 ng/ml. In addition, diluent was used as a blank sample. The detection area of the assay is displayed in grey. The lower line indicates the lower limit of detection (LLOD), which is defined as the calculated concentration of the blank signal + 3 times the standard deviation of the blank values. The upper limit of detection is defined as the highest concentration of the standard series that can reliably be measured. Both uniplex and 3-plex cMyBP-C standard curves are comparable. Detection limits are displayed in Table 2. a.u: arbitrary units.
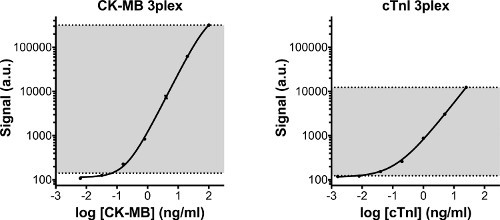 Figure 3. Standard curves of CK-MB and cTnI. Standard curves of CK-MB and cTnI, which were measured simultaneously with cMyBP-C on the 3-plex plate. Both calibrators show high sensitivity and a large dynamic range. In gray the detection area of the assay is displayed. The lower line indicates the lower limit of detection (LLOD), which is defined as the calculated concentration of the blank signal + 3 times the standard deviation of the blank values. The upper limit of detection is defined as the highest concentration of the standard series that can reliably be measured. Detection limits are displayed in Table 2. a.u: arbitrary units.
Figure 3. Standard curves of CK-MB and cTnI. Standard curves of CK-MB and cTnI, which were measured simultaneously with cMyBP-C on the 3-plex plate. Both calibrators show high sensitivity and a large dynamic range. In gray the detection area of the assay is displayed. The lower line indicates the lower limit of detection (LLOD), which is defined as the calculated concentration of the blank signal + 3 times the standard deviation of the blank values. The upper limit of detection is defined as the highest concentration of the standard series that can reliably be measured. Detection limits are displayed in Table 2. a.u: arbitrary units.
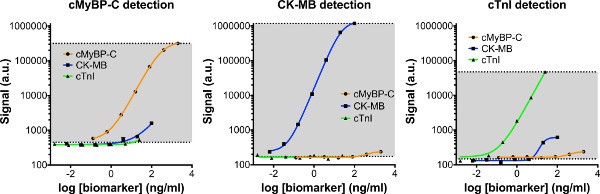 Figure 4. Cross-reactivity of 3-plex detection antibodies. Separate standard curves of cMyBP-C, CK-MB, and cTnI were prepared and incubated on the 3-plex plate. Each standard curve was than probed with individual detection antibodies, to assess the amount of cross-reactivity between individual detection antibodies (e.g. cMyBP-C) and the other calibrators (e.g. CK-MB and cTnI). Cross-reactivity was low in the 3-plex assay. Click here to view larger figure.
Figure 4. Cross-reactivity of 3-plex detection antibodies. Separate standard curves of cMyBP-C, CK-MB, and cTnI were prepared and incubated on the 3-plex plate. Each standard curve was than probed with individual detection antibodies, to assess the amount of cross-reactivity between individual detection antibodies (e.g. cMyBP-C) and the other calibrators (e.g. CK-MB and cTnI). Cross-reactivity was low in the 3-plex assay. Click here to view larger figure.
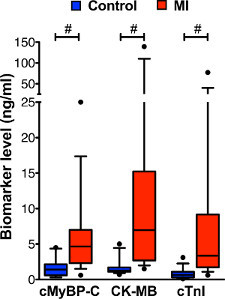 Figure 5. Biomarker levels in serum of MI and control subjects measured with 3-plex assay. Levels of cMyBP-C, CK-MB, and cTnI were measured simultaneously by 3-plex assay in serum samples of 16 control subjects and 16 subjects with MI. Data is displayed by box and whisker plots (whiskers represent 10th and 90th percentile). Increased levels of all three biomarkers were observed in the serum of subjects with MI compared with the control group. Because the data did not show a normal distribution (tested by D'Agostino Pearson Omnibus), the non-parametric Mann-Whitney statistical test was used to test for differences between control and MI groups. #
P < 0.001
Figure 5. Biomarker levels in serum of MI and control subjects measured with 3-plex assay. Levels of cMyBP-C, CK-MB, and cTnI were measured simultaneously by 3-plex assay in serum samples of 16 control subjects and 16 subjects with MI. Data is displayed by box and whisker plots (whiskers represent 10th and 90th percentile). Increased levels of all three biomarkers were observed in the serum of subjects with MI compared with the control group. Because the data did not show a normal distribution (tested by D'Agostino Pearson Omnibus), the non-parametric Mann-Whitney statistical test was used to test for differences between control and MI groups. #
P < 0.001
Discussion
In this study, we show the applicability of a multiplex immunoassay for the detection of multiple cardiac biomarkers in serum from patients. This technique has numerous advantages over traditional ELISA. First, ECL in the assay kit is used instead of colorimetric detection. In ECL, an electrical signal stimulates the local production of the measured property (light), thus uncoupling the stimulation event from the signal, which reduces background14. This allows for high sensitivity, as can be seen in Table 2.
Multiplexing the assay does not result in a loss of sensitivity for the detection of cMyBP-C, as can be seen in Figure 2 and Table 2. cMyBP-C measured in uniplex showed a sensitivity and detection range comparable to cMyBP-C measured simultaneously with CK-MB and cTnI13. Simultaneous measurements substantially reduce the volume of sample needed for measurements, which can be a critical factor when working with rare biological samples, as well as reducing time spent running multiple uniplex assays.
As with any immunoassay, the assay is highly dependent on the quality of the antibodies used. The affinity, specificity, and avidity of the capture and detection antibodies is the main determinant of the sensitivity of the assay15. Different antibody pairs should be tried to see which combination of capture and detection antibodies gives the highest sensitivity and dynamic range. When using multiplexing techniques, the capture antibodies need to be spot-coated on the well. This means that a low volume, highly concentrated solution is spotted on part of the well (see Figure 1) in contrast to solution coating used in the uniplex assay or in conventional ELISA. It should be assessed if the capture antibody performs as well with spot coating as it does with solution coating. Cross-reactivity of antibodies or calibrators with another antibody pair can also give rise to false-positive results2,15.
For this study, a custom made multiplex plate was designed in cooperation with MSD. Although a range of pre-coated multiplex plates are commercially available, this particular innovation required optimization of the assay because a novel (cMyBP-C) target was multiplexed with previously optimized catalog assays (CK-MB and cTnI). As such, an obvious drawback of this method is the added requirement for a specialized plate reader and plates containing electrodes to detect the ECL signal. Although running one multiplex assay versus multiple uniplex assays will reduce overall costs, a plate reader must be initially purchased2.
Using the 3-plex immunoassay the levels of biomarker release of cMyBP-C, CK-MB, and cTnI was measured in subjects with MI and compared to a control group. The fold change of increase over the control group differed between the three biomarkers (see Figure 4), as did the variation in the MI group. Variation in biomarker levels is expected in a collection of MI patients, as timing of blood sampling, infarct size, as well as a host of other factors likely contribute to the release of cardiac proteins and its subsequent degradation in the circulation. Whether the level of the individual biomarkers correlate with clinical parameters, such as infarct size, warrants further study.
Translating these findings directly to use in the emergency room to detect MI requires a number of challenges to be overcome. Prime among them is the time the protocol takes to produce results, which even with pre-coated plates takes 3 ½ hr. As with the troponin assays used today, the assay should be optimized for quick detection, enabling rapid diagnosis.
In conclusion, we have developed a signature assay in which two known cardiac biomarkers and one potential cardiac biomarker, cMyBP-C, were demonstrated to simultaneously detect the presence of myocardial injury in patients with MI by quantitation of cardiac protein serum titers.
Disclosures
A full patent application is pending (Application Serial No. 13/464,466, Pub. No. US 2012/0282618 A1 and Date: 05/04/12) to determine the risk factors associated with cMyBP-C degradation and release into human body fluid and quantitate the level of cMyBP-C in human body fluid.
Acknowledgments
This study was funded by National Institutes of Health grants R01HL105826 and K02HL114749 (Dr. Sadayappan), and American Heart Association Midwest Fellowships; 11PRE7240022 (Mr. Barefield) and 13POST14720024 (Dr. Govindan). The authors gratefully acknowledge the assistance of Dr. Jimmy Page, Jill Clampit and Dr. John Hudson, MSD, for their excellent technical and literature support in developing the assay.
References
- Thygesen K, et al. Third universal definition of myocardial infarction. Eur. Heart J. 2012;33:2551–2567. doi: 10.1093/eurheartj/ehs184. [DOI] [PubMed] [Google Scholar]
- Leng SX, et al. ELISA and multiplex technologies for cytokine measurement in inflammation and aging research. J. Gerontol. A. Biol. Sci. Med. Sci. 2008;63:879–884. doi: 10.1093/gerona/63.8.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson I, et al. Competitive electrochemiluminescence wash and no-wash immunoassays for detection of serum antibodies to smooth Brucella strains. Clin. Vaccine Immunol. 2009;16:765–771. doi: 10.1128/CVI.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsmore SF. Multiplexed protein measurement: technologies and applications of protein and antibody arrays. Nat. Rev. Drug. Discov. 2006;5:310–320. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altan ZM, et al. A long-acting tumor necrosis factor alpha-binding protein demonstrates activity in both in vitro and in vivo models of endometriosis. J. Pharmacol. Exp. Ther. 2010;334:460–466. doi: 10.1124/jpet.110.166488. [DOI] [PubMed] [Google Scholar]
- Patel KD, et al. High sensitivity cytokine detection in acute coronary syndrome reveals up-regulation of interferon gamma and interleukin-10 post myocardial infarction. Clin. Immunol. 2009;133:251–256. doi: 10.1016/j.clim.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Fu Q, Zhu J, Van Eyk JE. Comparison of multiplex immunoassay platforms. Clin. Chem. 2010;56:314–318. doi: 10.1373/clinchem.2009.135087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan VS, Jarolim P. How to interpret elevated cardiac troponin levels. Circulation. 2011;124:2350–2354. doi: 10.1161/CIRCULATIONAHA.111.023697. [DOI] [PubMed] [Google Scholar]
- Govindan S, et al. Cardiac myosin binding protein-C is a potential diagnostic biomarker for myocardial infarction. J. Mol. Cell Cardiol. 2012;52:154–164. doi: 10.1016/j.yjmcc.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barefield D, Sadayappan S. Phosphorylation and function of cardiac myosin binding protein-C in health and disease. J. Mol. Cell Cardiol. 2010;48:866–875. doi: 10.1016/j.yjmcc.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuster DW, et al. Cardiac myosin binding protein C phosphorylation in cardiac disease. J. Muscle Res. Cell Motil. 2012;33:43–52. doi: 10.1007/s10974-011-9280-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadayappan S, de Tombe PP. Cardiac myosin binding protein-C: redefining its structure and function. Biophys. Rev. 2012;4:93–106. doi: 10.1007/s12551-012-0067-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadayappan S. Cardiac myosin binding protein-C: a potential early-stage, cardiac-specific biomarker of ischemia-reperfusion injury. Biomark Med. 2012;6:69–72. doi: 10.2217/bmm.11.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichorova RN, et al. Biological and technical variables affecting immunoassay recovery of cytokines from human serum and simulated vaginal fluid: a multicenter study. Anal. Chem. 2008;80:4741–4751. doi: 10.1021/ac702628q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malekzadeh A, et al. Challenges in multi-plex and mono-plex platforms for the discovery of inflammatory profiles in neurodegenerative diseases. Methods. 2012;56:508–513. doi: 10.1016/j.ymeth.2012.03.017. [DOI] [PubMed] [Google Scholar]


