Highlights
-
•
Rotavirus cores: biochemical and biophysical characterisation.
-
•
Packaging of single stranded RNA into rotavirus cores.
-
•
Competitive packaging assays.
-
•
Non-specificity of RNAs packaging into rotavirus cores.
-
•
Infectivity of subviral rotavirus particles.
Keywords: Rotavirus, Rotavirus cores, RNA packaging in vitro
Abstract
Rotavirus (RV) cores were released from double-layered particles (DLPs) by high concentrations of CaCl2, purified and ‘opened’ by treatment with EDTA or EGTA. Under appropriate in vitro conditions DLPs have been shown to have transcriptase and ‘open cores’ replicase activity. Furthermore, it has been demonstrated that transcriptase activity and infectivity of native cores can be restored by transcapsidation with VP6, VP7 and VP4. The missing link for particle reconstitution in vitro has been the manipulation of ‘open cores’ to become functionally active cores again. The experiments described here were undertaken with the aim of exploring packaging of RV RNAs into opened cores in vitro. Rotavirus cores were opened by approximately 200 μM EGTA, leading to the release of genomic dsRNA. Conversely, RV cores were found to be stable in the presence of minimum concentrations of Ca2+, Mg2+, spermidine3+ and cobalthexamine3+ of between 40 and 300 μM. Aggregates of purified cores were resolved in the presence of 0.3 mM deoxycholate (minimum concentration). Core shells opened with EGTA were reconstituted by the addition of di- or trivalent cations within 2 min of the opening procedure. Addition of purified, baculovirus recombinant-expressed VP6 to native and reconstituted cores led to the formation of DLPs or DLP-like particles, which upon transfection into MA104 cells were infectious. The rescued infectivity likely originated in part from unopened and in part from reconstituted cores. Radiolabelled RV (+) ssRNAs could be packaged into reconstituted cores and DLPs, as indicated by resistance to RNase I digestion. The packaging reaction was, however, not RV RNA sequence-specific, since unrelated ssRNAs, such as those transcribed from HIV-2 cDNAs, were also packaged. The kinetics of packaging of homologous and heterologous RNAs were similar, as evidenced by competitive packaging assays. None of the packaged in vitro engineered RNA segments has so far been rescued into infectious virus.
1. Introduction
Rotaviruses (RVs) are a major cause of acute gastroenteritis in infants and young children and impose a heavy disease burden worldwide resulting in substantial mortality (450,000–500,000 children of <5-year-old/annum), mainly in developing countries in sub-Saharan Africa, Asia and S America (Parashar et al., 2003, 2009; Tate et al., 2012). Two live, attenuated RV vaccines have been licensed in over 100 countries since 2006 and are in wide use (Desselberger et al., 2009; Yen et al., 2011) with so far very encouraging outcomes in relation to the reduction of hospitalisation for RV-associated acute gastroenteritis (AGE) and also of mortality (Yen et al., 2011; Patel et al., 2012; Soares-Weiser et al., 2012; Gastañaduy et al., 2013; Lee et al., 2013).
Rotavirus propagation is naturally initiated by an infectious virus particle (virion) adsorbing to a susceptible cell via specific cellular receptors, followed by the steps of a viral replication cycle: penetration, uncoating, transcription, translation, viral nucleic acid replication, assembly of viral components (morphogenesis), and release of virions by cell lysis. Rotavirus early morphogenesis and viral RNA replication take place in cytoplasmic inclusion bodies termed ‘viroplasms’ (Estes and Greenberg, 2013).
It has been possible to mimic and explore portions of the viral morphogenesis in vitro. Virions, consisting of triple–layered particles (TLPs), can be stripped first of their outer layer (containing VP7 and VP4) by mild EDTA treatment (Estes et al., 1979) to obtain double-layered particles (DLPs). From those the middle layer (consisting of VP6) can be removed in the presence of high concentrations of CaCl2 (Bican et al., 1982) to obtain ‘core’ particles. [Similar procedures have been used to prepare cores of LA virus (Naitow et al., 2001).] The core particles contain the 11 genomic segments of dsRNA, the RNA-dependent RNA polymerase (RdRp, VP1) and the capping enzyme (VP3) within a scaffold made of VP2. The structure of these components within the core has been studied in some detail (Prasad et al., 1996; Lawton et al., 1997; Pesavento et al., 2003; McClain et al., 2010; Settembre et al., 2011; Trask et al., 2012a,b; Estrozi et al., 2013), and structure–function relationships of the RdRp complexed with RNA, VP3, and VP2 have been explored (Lu et al., 2008; Ogden et al., 2012; Trask et al., 2012a,b). Isolated empty core-like particles have been obtained from VP2-expressing baculovirus recombinants, but found to be tightly cell-associated. Only the mild detergent deoxycholate (DOC) and SDS were able to solubilise VP2 (Labbé et al., 1991). At high concentrations of recombinant-expressed VP2, unusual forms (helix-like structures) were observed (Zeng et al., 1994).
DLPs and core particles are non-infectious in conventional cell culture infection assays. However, by successive addition (transcapsidation) of VP6, VP7 and VP4 to core particles in vitro, it was possible to partially reconstitute infectivity (Chen and Ramig, 1993a,b), and by careful quantitation of the in vitro transcapsidation system full infectivity could be restored (Trask and Dormitzer, 2006). Trans-capsidation of native cores with VP6 alone restores transcriptase activity (Kohli et al., 1993) which in turn can be inhibited by reacting the DLPs with monoclonal antibodies directed to VP6 (Kohli et al., 1994; Thouvenin et al., 2001). Upon transfection into susceptible cells DLPs are infectious (Bass et al., 1992; Chen and Ramig, 1993b). Native RV cores can be destabilised by dialysis against low salt buffer containing low concentrations of EDTA. Such preparations, termed ‘open cores’, accept externally added single-stranded (ss) RV RNA molecules of positive (+) polarity and of homologous or heterologous origin (i.e. from a different RV strain) as templates for RNA replication in vitro to yield dsRNA (Chen et al., 1994; Tortorici et al., 2003). Biochemically Zeng et al. (1996) defined a complex of VP1, VP2 and (+) ssRNA as the minimal replicase particle. The missing link for particle reconstitution completely in vitro is the ability to reconstitute cores out of their components in vitro to obtain functionally active structures.
Patton's group has shown that viral RNA replication occurred simultaneously with packaging of viral ssRNA into cores (Gallegos and Patton, 1989; Patton and Gallegos, 1990), and the detailed conditions of minus-strand RNA synthesis were worked out (Chen and Patton, 1998, 2000; Patton et al., 1996; Wentz et al., 1996; Patton et al., 1999; Tortorici et al., 2003). In addition, biochemical functions of open cores have been analysed (Chen et al., 1999; Patton and Chen, 1999).
Two RV nonstructural proteins, NSP2 and NSP5 (encoded by RV RNA segments 7,8, or 9 [depending on strain] and 11, respectively), are essential for the formation of viroplasms and thus for RV replication (Berois et al., 2003; Campagna et al., 2005; Eichwald et al., 2002, 2004; Fabbretti et al., 1999; Patton, 2001; Patton et al., 1997; Schuck et al., 2001; Silvestri et al., 2004; Taraporewala et al., 1999; Taraporewala and Patton, 2001; Torres-Vega et al., 2000; Vasquez-Del Carpio et al., 2004). The packaging of the 11 segments of RV RNA is very tightly controlled, but the mechanisms governing packaging are unknown. Primary replication complexes (consisting of one ss(+)RNA, VP1 and VP3, possibly attached to VP2) may specifically interact to form a native core, or ssRNAs may be pulled into preformed empty cores (concerted vs core-filling model, McDonald and Patton, 2011). For RVs, there are better arguments for the concerted than the core-filling packaging mechanism. When native cores are opened, the genomic dsRNA molecules are released (Chen et al., 1994). They cannot be repackaged, due to their intrinsic stiffness and rigidity as isolated molecules in solution (Kapahnke et al., 1986). Early attempts to introduce RV ss(+)RNA constructs into viroplasms in order to have them rescued into infectious progeny virus have failed (Gorziglia and Collins, 1992; Silvestri et al., 2004). Our long term aim is to find conditions permitting rescue of infectivity by reconstruction of RV particles in vitro from its components. Until now, only helper virus-dependent systems have succeeded in rescuing transfected or intracellularly produced RV RNA segments into infectious progeny (Komoto et al., 2006; Trask et al., 2010; Troupin et al., 2010).
The early steps of RV morphogenesis constitute a very complex and over large parts unexplored process that has so far resisted manipulations intended to rescue infectivity from engineered primary components. The work described here was carried out to further characterise rotavirus cores, to study conditions of their structural stability, and to explore mechanisms by which RV (+) ssRNA is packaged. It is shown that core particles opened in vitro can be reconstituted and that transcapsidation of such core structures is possible. In vitro conditions are described permitting the packaging of RV ssRNAs into opened cores. However, the packaging of ssRNAs is sequence-non-specific since HIV-2 RNAs transcribed in vitro from cDNA fragments are also packaged, and since specificity could not be demonstrated by competitive packaging assays. So far, rescue of infectivity from in vitro packaged RV particles has not succeeded.
2. Materials and methods
2.1. Viruses and cell culture
The bovine RV RF strain (G6 P6[1]) and the porcine RV OSU strain (G5P9[7]) were used. Rotavirus was propagated on confluent monolayers of MA104 cells or KJ cells (another embryonic monkey kidney cell line transduced with the gene encoding the V protein of SV5, as described for other cell lines by Young et al. (2003)) in the presence of 0.5 μg/ml of trypsin (Sigma type IX-S, cat. No. 0303) as previously described (Graham et al., 1987; Arnoldi et al., 2007). Infectivity titers were measured using standard procedures and recorded as TCID50/ml. Recombinant baculovirus was grown on Spodoptera frugiperda (Sf9) cells in suspension or as monolayers at 26 °C in Hinks medium supplemented with 10% fetal calf serum as described (Charpilienne et al., 2001; Cohen et al., 1989).
2.2. Virus purification and protein concentration
Rotavirus was purified from cell cultures infected at low m.o.i. after the appearance of complete CPE by ultracentrifugation, decafluoropentane (Vertrel XF, Dupont) extraction of the pellet suspension, and two subsequent equilibrium ultracentrifugation steps on CsCl gradients essentially as described by Patton et al. (2000) and Arnoldi et al., 2007. Two well separated gradient bands were obtained which contain TLPs (density 1.36 g/ml) and DLPs (density 1.38 g/ml). TLP and DLP suspensions were desalted by passing over G25 coarse Sephadex columns (0.7 cm3 column per 50 μl of virus in CsCl). The protein concentration of twice CsCl gradient-purified suspensions of TLPs and DLPs was determined by use of the Coomassie Protein Assay Reagent of Pierce (Perbio Science France) or by the Bradford assay (BioRad), following the manufacturers’ instructions. Serial twofold dilutions of a solution of 2 mg/ml of bovine serum albumin were used as standards. In addition, RV TLP and DLP preparations were analysed by micro spectrophotometry (Nanodrop 1000, Thermo Scientific) by which maxima of the spectra were seen at 260 nm wavelength. OD values at 260 nm wavelength were correlated with protein content measured in parallel, and the following relationship was established: [Protein in μg/ml] = OD260 × 68.3 (N = 41; r = 0.900; P < 0.001). The numbers of particles was estimated from the total MW of their protein components.
2.3. Preparation of viral mRNA from DLPs
The DLPs from CsCl gradient purified rotavirus were used to transcribe viral mRNA in vitro, either unlabelled or labeled with α[32P]-UTP, and then extracted with phenol/chloroform, as described by Patton et al. (2000) and Charpilienne et al., 2002.
2.4. Cloning of rotavirus cDNA and of rotavirus cDNA fusion constructs
Complementary DNA (cDNA) of dsRNA segments of RV strain OSU (G5P9[7]) was made and cloned into the pcDNA3 vector (Invitrogen) as described by Afrikanova et al. (1998) and Fabbretti et al. (1999).
2.5. In vitro transcription of cDNA constructs
Rotavirus cDNA clones were amplified with primers carrying the T7 polymerase promoter sequence (5′end) and suitable restriction endonuclease sites for defining the exact 3′end. Amplicons were synthesised by PCR using KOD polymerase (Novagen) according to the manufacturer's protocol. Thermocycler conditions were: 95 °C for 2 min, 40 cycles of 95 °C for 20 s, 63 °C for 30 s, 70 °C for 3 min, with a final extension at 70 °C for 10 min. Amplicons were purified using a Qiagen PCR column cleanup kit and digested in TAS buffer (30 mM Tris acetate, 65 mM KOAc, 10 mM MgOAc, 0.5 DTT, 0.5 mM BSA, 0.4 mM spermidine dihydrochloride, pH 7.9) with the appropriate restriction endonucleases (Promega) for 2 h at 37 °C prior to re-purification with a Qiagen Minelute kit according to the manufacturer's instructions.
In vitro transcription was carried out using the MEGAscript T7 kit (Ambion) according to the manufacturer's instructions. RNA was synthesised either unlabelled or was radiolabelled with 10 uCi of α-32P-UTP (NEN, specific radioactivity 3000 Ci/mM). For radiolabelling experiments, the concentration of unlabelled UTP used was one tenth of the concentration used for non-radioactive RNA synthesis. After transcription the reaction mixture was treated with DNase and then purified by proteinase K treatment, phenol-chloroform extraction and ethanol precipitation; the pellets were resuspended in small volumes of nuclease-free distilled water. Alternatively, RNA transcripts were purified by the Qiagen Minelute RNA purification kit. The products were checked for size and homogeneity by electrophoresis on 0.8% agarose gels in 20 mM MOPS-Tris buffer pH 7.7, followed by staining with ethidium bromide or autoradiography. Purified RNAs were quantitated by Nanodrop spectrophotometry.
Heterologous ssRNAs were derived from HIV-2 cDNA fragments as described below.
2.6. Preparation of virus cores and core-like particles
Viral cores were prepared from purified DLPs by treatment with 1.2 M CaCl2, followed by ultracentrifugation at 90,000 × g (Beckman TLA100.4) and 4 °C for 15 min and resuspension in water (Bican et al., 1982) or in 200 μM CaCl2
Alternatively, viral core-like particles (CLPs-VP1/VP2/VP3) were prepared from baculovirus recombinant-expressed DLP-like particles (DLPs-VP1/VP2/VP3/VP6) (Zeng et al., 1996) by similar treatment.
2.7. Purification of rotavirus protein VP6
Baculovirus recombinant expressed protein VP6 of the bovine RV RF strain was purified as described (Charpilienne et al., 2002). In brief, infected Sf9 cells were harvested at 3–5 days post infection, and the clarified supernatant was centrifuged in a Beckman 45 Ti rotor at 80,000 × g and 4 °C for 30 min. The pellet was resuspended in 50 mM MOPS (3-(N-morpholino)propanesulfonic acid) buffer, pH 6.0, adjusted to 0.3 M CaCl2 and centrifuged at 13,000 × g for 10 min. The supernatant containing purified VP6 was dialysed against water (nitrocellulose filter, 25 nm, Millipore) at room temperature for 10 min. Under these conditions, VP6 was obtained that had assembled to large polymers (tubules) (Lepault et al., 2001), and such preparations were used for reconstruction assays.
2.8. Reconstitution experiments with ‘opened cores’
Virus cores prepared from DLPs (see above) were ‘opened’ by treatment with 0.5–1 mM EGTA or EDTA, similar to the procedure of Chen et al (1994), but in a more controlled way. All reactions were in Hepes buffer, 10–50 mM, pH 7.4 or in 50 mM Tris–HCl buffer pH 7.4 if not otherwise stated. Reconstitution of core structures was explored by the addition of 2–5 mM MgCl2, CaCl2, spermidine (Invitrogen) or cobalthexamine chloride (Fluka) at time intervals over several minutes after the start of the EGTA treatment. Purified recombinant VP6 (see above) was then added to reconstitute DLP-like structures.
In order to define the conditions of stabilisation of cores as tightly as possible, some experiments were carried out using EGTA-buffered calcium calibration solutions with known concentrations of free Ca2+ and EGTA buffer with known amounts of Mg2+ ions added (Molecular Probes Inc., Eugene, OR 97402-9132, USA).
2.9. Packaging of RV and heterologous RNAs into ‘opened cores’
Rotavirus cores (60 ng, approximately 5 × 108 particles) were opened with 800 μM EGTA in the presence of RV mRNA or heterologous ss RNAs, alone or in mixtures (in competition experiments), and radiolabelled or unlabelled, for 2 min and then a packaging cocktail was added. The latter was derived from a packaging reaction used by Qiao et al. (1995) for packaging work with bacteriophage phi6 and contained the following components (in final concentrations): Tris–HCl, pH 7.4, 15 mM; MgCl2, 1 mM; MnCl2 300 μM; NaCl 8 mM; dithiothreitol, 1 mM; Na2EDTA, 80 μM; rNTPs, 250 μM each; RNase inhibitor, 1 unit [RNasin, N Engl Biolabs]; PEG 4000, 1–3%). The final concentration of EGTA present was 600 μM, radiolabelled viral RNAs were present at 10 ng/20 μl, and unlabelled viral RNAs at 30–150 ng/20 μl. Radiolabelled RNA associated with reconstituted cores which was resistant to treatment with RNase ONE was considered as packaged (Qiao et al., 1995). Cores treated in this way were transcapsidated with recombinant VP6 as described above. Reconstituted core structures or DLPs (produced by transcapsidation) were treated with 10 units of RNase I (Promega) at room temperature for 15 min, following the procedure of Qiao et al., 1995, and then analysed by agarose gel electrophoresis as described below.
In order to test for the specificity of packaging, the following RNAs were produced:
RV:
RNA segment 7 (encoding NSP2), transcribed in vitro from a T7 promoter containing plasmid, 1061 nt; RNA segment 11 (encoding NSP5), transcribed in vitro from a T7 promoter containing plasmid, 667 nt (Fabbretti et al., 1999)
HIV:
RNA fragment of HIV-2 ROD strain, nt pos 514-1618, transcribed in vitro from an RT-PCR amplicon in reverse orientation, 1104 nt; RNA fragment of HIV-2 ROD strain, nt pos 1-751, transcribed in vitro from RT-PCR amplicon in forward orientation, 751 nt.
RNAs were transcribed in vitro, either in the presence of unlabelled rNTPs, or in the presence of α-32P-UTP as described above.
2.10. Transfection assays with subviral particles
Transfections were carried out as described (Bass et al., 1992; Chen and Ramig, 1993b), using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
2.11. Electrophoresis
Using a slight modification of the procedure described by Gallegos and Patton (1989), samples were analysed on 0.6% non-denaturing agarose (Invitrogen) minigels using 10–20 mM MOPS buffer adjusted to pH 7.7 with 1 M Trizma base. Electrophoresis was at 90 V and 20–22 mA until the dye marker bromophenol blue had reached the bottom of the gel. Gels were then stained wet with 0.5 μg/ml of ethidium bromide dissolved in electrophoresis buffer, or dried and silver-stained, initially using the BioRad Silver Stain Plus kit following manufacturer's instructions (Charpilienne et al., 2002). Alternatively, the silver staining procedure as originally published by Gottlieb and Chavko (1987) was used. All gels were photographed and the images stored by computer (using Adobe Photoshop and Image Twain software).
Densitometry was carried out using the image J system (http://rsbweb.nih.gov/ij/).
2.12. Electron microscopy
Samples were applied to air-glow-discharged carbon-coated grids, blotted immediately with filter paper for 1–2 s, and negatively stained with 1% uranyl acetate solution for 2 × 5 s. The grids were examined in a Philips CM12 electron microscope operated at 80 keV (Erk et al., 2003).
2.13. Dynamic light scattering
The measurements were carried out at 16 °C with a DynaPro-99 P instrument equipped with a temperature-controlled microsampler (Protein Solutions). A sensitivity of 30% was used. The measurements were performed using 13 μl of particle suspension in water. The detergent deoxycholate (DOC) was added at various concentrations up to a final concentration of 450 μM. Two or three independent measurements of 20 acquisitions were carried out for each experiment. The data were analysed using the Dynamics 4.0 software with correlator flex-99. The refractive index and viscosity of water were applied. The percentage of mass distribution was plotted against different Rh(M) values, and the calculated mean hydrodynamic radius [Rh(M)] and the polydispersity index (PI, in percent) values are indicated.
2.14. Statistics
Slopes and intercepts of the kinetics of competitive packaging reactions were calculated by the method of linear regression. Differences in the slopes and intercepts were assessed by the ANCOVA test (http://ude1.edu/∼mcdonald/statancova.html).
3. Results
Titration of divalent and trivalent cation concentrations required to stabilise RV cores and action of chelating agents.
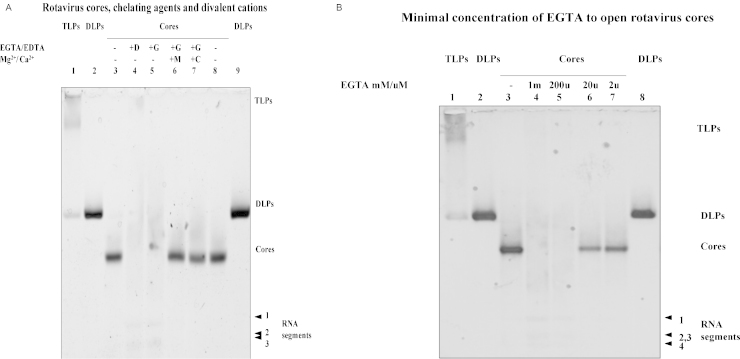
One of us (J.C.) had previously observed (2002, unpublished) that the core-disintegrating effect of chelating agents could be antagonised by simultaneous addition of divalent cations (Fig. 1A). Native rotavirus cores (intact controls in lanes 3 and 8) were opened by 1 mM EDTA (lane 4) or 1 mM EGTA (lane 5), but, as expected, this event was prevented by the simultaneous addition of 5 mM CaCl2 (lane 7) or 5 mM MgCl2 (lane 6). The same concentrations of MnCl2 or ZnCl2 did not have this effect (results not shown). Opening of the core liberates the genomic RV dsRNA segments (Fig. 1A, lanes 4, 5; Fig. 1B, lanes 4, 5). The minimal concentration of EGTA to open cores is approximately 200 μM (Fig. 1B). This was confirmed by re-titration with more narrowly spaced dilutions of EGTA (results not shown). Since EGTA chelation is more Ca2+ specific, we decided to use EGTA for all subsequent experiments.
Fig. 1.
(A) Effect of chelating agents EDTA/EGTA and of divalent cations (Mg2+, Ca2+) on native RV cores, Agarose gel 0.6%, electrophoresis with 10 mM MOPS/Tris buffer, pH 7.7, followed by silver staining. Lane 1: TLPs of bovine RV RF strain (slightly degraded to DLPs); lanes 2, 9: DLPs; lanes 3, 8: native RV cores; lane 4: cores + 1 mM EDTA; lane 5: cores + 1 mM EGTA; lane 6: RV cores + 1 mM EGTA + 5 mM MgCl2 (simultaneously); lane 7: RV cores + 1 mM EGTA + 5 mM CaCl2 (simultaneously). All reactions and reagents were in 20 mM Hepes buffer, pH 7.4. The large segments of genomic RNA (s1–s3) are seen in lanes 4 and 5. (B) Titration of EGTA concentration enabling opening of RV cores. Agarose gel 0.6%, electrophoresis with 10 mM MOPS/Tris buffer, pH 7.7, followed by silver staining. Lane 1: TLPs; lanes 2, 8: DLPs; lane 3: native RV cores; lanes 4–7: mixture of RV cores with EGTA at concentrations of 1 mM, 200 μM, 20 μM, 2 μM, respectively.
Using the same procedure and EGTA-buffered calibration solutions with defined concentrations of free Ca2+ and Mg2+ ions (Molecular Probes, Eugene, OR, USA), we determined the minimum free Ca2+ and Mg2+ concentrations stabilising the cores to be 50 μM and 300 μM, respectively (Table 1). Divalent cations could be replaced by trivalent cations, such as spermidine3+ and cobalthexamine3+, which were found to stabilise cores at minimal concentrations of 40–50 μM (Table 1).
Table 1.
Minimum concentrations of reagents to open and stabilise rotavirus cores.
| Reagent | Action | Minimum concentration |
|---|---|---|
| EGTA | Open cores in Ca2+ and Mg2+ free solution | 200 μM |
| Ca2+ | Stabilise cores in EGTA buffered solution | 50 μM |
| Mg2+ | Stabilise cores in EGTA buffered solution | 300 μM |
| Spermidine3+ | Stabilise cores in buffered solution | 50 μM |
| Cobalthexamine3+ | Stabilise cores in buffered solution | 40 μM |
3.1. Isolated rotavirus cores form aggregates
Isolated native cores have the tendency to form aggregates. Purified rotavirus DLPs and cores were studied by dynamic light scattering (DLS) to determine the mean particle size and characterise the homogeneity (polydispersity index, PI, in percent) of the suspensions. DLPs presented in a very monodisperse distribution (PI 9%) of particles of approximately 32 nm hydrodynamic radius and were used as a calibrator (Supplementary Figure S1A). Untreated purified cores were very polydisperse (PI 34%): the non-symmetrical form, the apparent median radius of 62 nm and the width of the peak reflected the presence of aggregates of heterogeneous size (Supplementary Figure S1B). Treatment of core suspensions with electrolytes (0.5 M NaCl or 5 mM MgCl2) or with buffers of different pH (5–9) did not result in major changes (results not shown). However, treatment with deoxycholate (DOC) at submillimolar concentrations (80–450 μM) produced a more homogeneous, and closer to monodisperse suspension (PI 20%) of particles of 25 nm radius (Supplementary Figure S1D): the transition of core aggregates to a virtually monodisperse suspension was achieved at approximately 300 μM of DOC (Supplementary Figure S1C). These observations strongly suggest that it were hydrophobic rather than electrostatic forces that produced the particle aggregation. In the native core suspension aggregates will contain an average of approximately 15 core particles/aggregate. The DLS measurements of the hydrodynamic radii of monodisperse rotavirus cores and DLPs are in excellent agreement with previously obtained cryo-EM data (Prasad et al., 1988; Yeager et al., 1990).
3.2. Opening of cores by EGTA can be blocked by subsequent addition of divalent cations
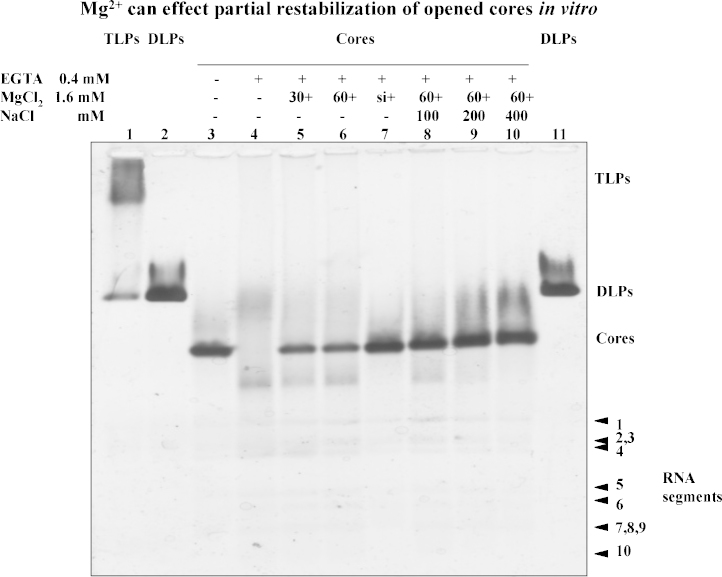
Blockage of core opening within short time periods is demonstrated in Fig. 2. The firm band of core particles (Fig. 2, lane 3) is replaced by a smear spanning an area from slightly above the core band to a band above RNA segment 1 (Fig. 2, lane 4). When a 4-molar excess of divalent cations (Mg2+) was added at 30–60 s after the start of a mild EGTA treatment (with 2× the minimal concentration required to open cores), the destabilisation of core particles was partially blocked (Fig. 2, lanes 5 and 6); this was in contrast to cores being protected upon simultaneous addition of the two reagents (Fig. 2, lane 7; Fig. 1A, lane 6). Recovery of stabilised core structures was not possible when divalent cations were added later than 2 min after the start of the EGTA treatment (results not shown). Partial recovery of cores (Fig. 2, lane 6) by addition of Mg2+ at 60 s after the start of EGTA treatment is improved in the presence of 100–400 mM NaCl (Fig. 2, lanes 8–10). Viral genomic dsRNA segments have to a large extent been released (Fig. 2, lanes 4–10), but probably not completely (see below).
Fig. 2.
Partial stabilisation of opened RV cores in vitro by the addition of Mg2+ ions. Agarose gel 0.6%, electrophoresis with 10 mM MOPS/Tris buffer, pH 7.7, followed by silver staining. Lane 1: TLPs (partially degraded to DLPs); lanes 2 and 11: DLPs; lane 3: native RV cores; lane 4: cores + 400 μM EGTA; lane 5: cores + 400 μM EGTA + 1.6 mM MgCl2 after 30 s; lane 6: cores + 400 μM EGTA + 1.6 mM MgCl2 after 60 s; lane 7: cores + [400 μM EGTA +1.6 mM MgCl2] used simultaneously; lanes 8–10: reactions as described for lane 6 in the presence of NaCl 100 mM (lane 8), 200 mM (lane 9), and 400 mM (lane 10). All reactions were in 10 mM Hepes buffer, pH 7.4.
In order to test the stability of reconstituted core structures, they were opened with 400 μM EGTA, treated with 1.6 mM CaCl2 after 60 s in the presence of 100 mM NaCl, followed by the addition of 1.6 mM EGTA after 30 min: now the cores could not be opened (results not shown).
Empty core-like particles (produced by treatment of DLP-like particles containing VP1, VP2, VP3 and VP6 with chaotropic agent) were not broken up by treatment with 10 mM EGTA, i.e. 50× the concentration of molecules sufficient to open native cores (Supplementary Figure S2), suggesting that fragmentation (‘opening’) of native cores by EGTA was due to chelation of divalent cations which neutralised the excess of negative charges of the enclosed genomic dsRNA segments.
Effect of the addition of recombinant VP6 to native cores and reconstituted core structures
3.3. VP6 was produced in and purified from VP6-expressing baculovirus recombinant-infected Sf9 insect cell cultures.
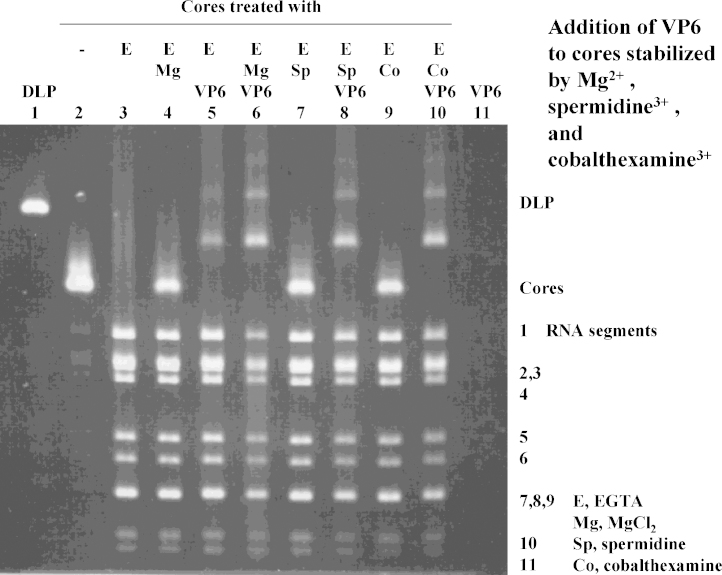
Purified VP6 was added to cores that had been opened by EGTA and then been treated with Mg2+ 60 s later. The results are shown in Fig. 3. Upon addition of VP6, cores treated with EGTA and then Mg2+ 60 s later (Fig. 3, lane 4), formed complexes (DLP-like structures) larger than cores (Fig. 3, line 6), migrating between cores (Fig. 3, lines 2, 4) and DLPs (Fig. 3, line 1) and some migrating slower than DLPs. [Fig. 3, lane 3 shows irreversible decomposition of core structures.]. Particles opened with EGTA and treated with VP6 directly (with no Mg2+ added; Fig. 3, lane 5) formed structures similar to those seen in lane 6 (reconstituted cores transcapsidated with VP6 in the presence of Mg2+), but DLP-like structures were less abundant. Very similar effects to those observed with Mg2+ were recorded when the polycations spermidine3+ (Fig. 3, lanes 7, 8) or cobalthexamine3+ (Fig. 3, lanes 9, 10) were used.
Fig. 3.
Formation of rotavirus DLPs and intermediates by addition of recombinant VP6 to cores opened by EGTA and restabilised by Mg2+, spermidine3+, and cobalthexamine3+. Agarose gel 0.8%, electrophoresis with 10 mM MOPS/Tris buffer, pH 7.7, followed by staining with ethidium bromide. Lane 1: Purified DLPs; lane 2: purified cores: lane 3: cores + 400 μM EGTA; lane 4: cores + 400 μM EGTA + 1.6 mM MgCl2 after 1 min; lane 5: cores + 400 μM EGTA + VP6; lane 6: cores + 400 μM EGTA + 1.6 mM MgCl2 after 1 min + VP6; lane 7: cores + 400 μM EGTA + 300 μM spermidine3+ after 1 min; lane 8: cores + 400 μM EGTA + 300 μM spermidine3+ after 1 min; lane 9: cores + 400 μM EGTA + 350 μM cobalthexamine3+ after 1 min; lane 10: cores + 400 μM EGTA + 350 μM cobalthexamine3+ after 1 min + VP6; lane 11: 400 μM EGTA + 1.6 mM MgCl2 + VP6.
3.4. Electron microscopy
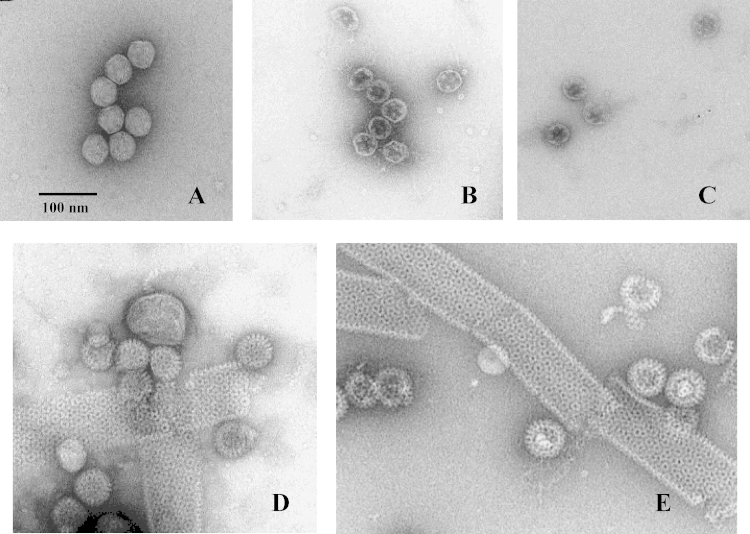
Purified cores (in 75 mM Tris–HCl buffer, pH 7.4) appear as regular hexa- to polygonal particles with a smooth surface. There were very few free dsRNA molecules seen (Fig. 4A). After treatment with 600 μM EGTA for 1 min (Fig. 4B) virtually all core particles have changed: the VP2 layer is marginalised, and the particles appear as empty shells (containing the contrast dye). Inside the VP2 layer little ‘knobs’ in 5-fold symmetry were observed that correspond to VP1/VP3 complexes (demonstrated by staining with VP1- and VP3-specific guinea pig antibodies followed by anti-guinea pig IgG immunogold-labelled secondary antibody; Desselberger, Lever and Crowther, unpublished results). Plenty of dsRNA molecules were seen outside of particles (Fig. 4B and Supplementary Figure S3), many of them with ‘knobs’ (VP1/VP3 complexes) attached to them. A time course of core treatment with EGTA demonstrated an increasing decomposition of their protein components after more than 2 min from the beginning of the treatment (results not shown). EGTA-treated core particles that were reacted after 1 min with 2.5 mM MgCl2 (Fig. 4C) appeared to be ‘stabilised’. Some cores seemed to acquire a rough surface, and some of them appeared to be filled with electron dense material. When purified native cores were reacted with baculovirus recombinant-expressed and purified VP6 (of bovine rotavirus RF strain), large numbers of VP6 tubules, many DLPs and many free VP6 trimers were seen (Fig. 4D). In some cases the cores were wrapped by VP6 originating from the tubules. Transcapsidation was a quick event and almost complete at 2 min after addition of VP6 (results not shown). When restabilised core structures (opened with 0.6 mM EGTA and treated with 5 mM MgCl2 1 min later) were reacted with VP6, many DLPs were seen as well, some appearing to be empty, some containing electron-dense material (Fig. 4E). The transcapsidation process was less effective, when the preparations of purified VP6 lacked tubules, suggesting that the VP6 concentration was too low (results not shown).
Fig. 4.
Electron microscopy of different rotavirus preparations. (A) Purified native cores; (B) Cores opened with 600 μM EGTA; C. Cores opened with 600 μM EGTA and treated with 2.4 mM MgCl2 after 1 min; (D) Purified native cores reacted with baculovirus recombinant-expressed and purified VP6; (E) Restabilised cores as shown in C, treated with purified VP6. The calibration bar indicates 100 nm [see Text for details]. A higher magnification of panel B is provided in Supplementary Figure S2.
3.5. Infectivity of native cores, reconstituted cores and in vitro reconstituted DLPs
Preparations of purified native cores, opened cores, reconstituted cores, and of DLPs reconstituted from both, native and reconstituted cores with baculovirus recombinant-expressed VP6 were transfected into confluent monolayers of MA104 cells. Great care was taken to avoid contamination. The results of three independent experiments are shown in Supplementary Table S1 (ST1). They demonstrate some infectivity of reconstituted cores after transcapsidation with VP6, with DLPs produced by encapsidation of native cores serving as positive controls (Chen and Ramig, 1993b).
To investigate whether infectivity of stabilised, VP6-transcapsidated RV cores was due to reclosing of opened cores or to the presence of residual unopened cores, cores were opened with EGTA and stabilised with Mg2+ after 30, 60, and 120 s at which time points recombinant VP6 was added; at that time points, aliquots of opened cores were also reacted with VP6 without prior treatment with Mg2+. Only in the cases where cores were stabilised with Mg2+ within one min was infectivity recovered (Supplementary Table S2, ST2). [This experiment was once reproduced with exactly the same result.] However, with these data alone, the rescue of infectivity shortly after opening and stabilisation of cores cannot be interpreted with certainty as rescue derived from stabilised and not from unopened cores. The issue was addressed further by testing in parallel the infectivity of DLPs reconstituted from EGTA-opened cores to which VP6 was added in the presence or absence of Mg2+, spermidine3+ or cobalthexamine3+. In three experiments it was shown that stabilisation of cores permitted the rescue of more infectivity by transcapsidation with VP6 (followed by transfection) than was possible after addition of VP6 alone (Table 2). This suggests that at least some of the infectivity rescued may have been due to transcapsidation of stabilised cores and not only the presence of possibly unopened cores.
Table 2.
Rescue of infectivity from rotavirus cores after various treatments in vitro followed by liposome-mediated transfection into KJ cells.
| Treatment of cores | Titration of yield (TCID50/ml) |
||
|---|---|---|---|
| Exp 1 | Exp 2 | Exp 3 | |
| None | Neg | Neg | Neg |
| EGTA | Neg | Neg | Neg |
| EGTA + Mg2+ | Neg | Neg | Neg |
| EGTA + VP6 | 100* | Neg | 2000 |
| EGTA + Mg2+ + VP6 | 1000* | 10,000 | 20,000 |
| EGTA + spermidine3+ | Neg | ND+ | Neg |
| EGTA + spermidine3+ + VP6 | 100 | ND | 20,000 |
| EGTA + cobalthexamine3+ | Neg | ND | Neg |
| EGTA + cobalthexamine3+ + VP6 | 1000 | ND | 5000 |
| None + Mg2+ + VP6 | ND | ND | 20,000 |
* Tested twice, + Not done.
3.6. Packaging of rotavirus RNA produced in vitro into opened and stabilised cores
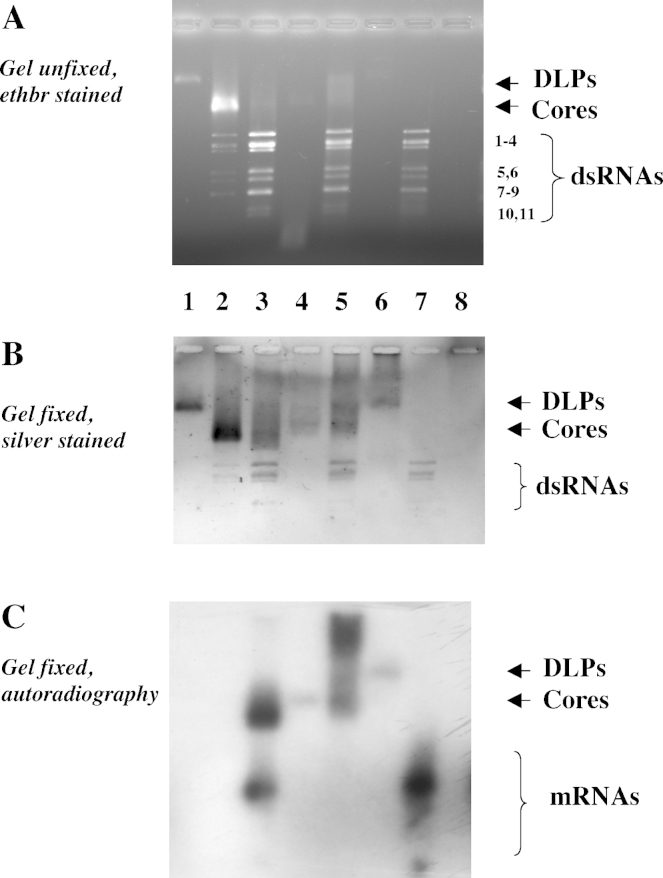
Based on these results, RV cores were opened with EGTA and reconstituted with divalent cations in the presence of small amounts (<10 ng) of 32P-radiolabelled, (+)ss RV RNA, produced in vitro either from DLPs (all 11 segments) or from T7 promoter-driven cDNAs contained in plasmids or amplicons (individual segments). To optimise packaging, a ‘packaging mixture’ was devised and optimised (see Section 2), which was derived from the solution used by L Mindich's group for packaging of bacteriophage phi6 ssRNA segments into cores in vitro (Qiao et al., 1995). After cation addition, cores were treated with RNase I and the reaction products analysed on non-denaturing agarose gels. It was shown (Fig. 5A–C) that opened RV cores could successfully package radiolabelled RV RNA (Fig. 5C, lane 3). The radiolabelled RNA was protected from RNase I which digests released genomic dsRNA and excess external ssRNA (Fig. 5C, lane 4). The difference in ssRNA binding to cores before and after RNase I treatment testifies to the high affinity of RV cores to ssRNA (Fig. 5C, lanes 3 and 4) and also to the fact that externally bound ssRNA is accessible to RNase I digestion. Moreover, the reconstituted core structures could be transcapsidated with VP6 and still contain the in vitro packaged ssRNA in an RNase I-protected form (Fig. 5C, lane 6). Experiments to show replication of ssRNA packaged into reconstituted core particles remained unsuccessful (results not shown). Due to small quantities of core particles which contained packaged ssRNAs, their density was not assessed. However, they comigrated with native cores during electrophoresis on non-denaturing gels.
Fig. 5.
In vitro packaging of 32P-labelled rotavirus mRNA (transcribed from DLPs) into particles: Protection from RNase I digestion. 0.8% agarose gel, MOPS-Tris buffer 10 mM, pH 7.7. (A) Gel unfixed, stained with ethidium bromide; (B) Gel fixed, silver stained; (C) Gel fixed, dried and autoradiographed. Lane 1: DLPs; lane 2: native RV cores; lane 3: cores + RV mRNA + EGTA + packaging mixture; lane 4: as lane 3 + RNase I; lane 5: cores + mRNA + EGTA + packaging mixture + VP6; lane 6: as lane 5 + RNase I; lane 7: mRNA [the dsRNA seen in panels A and B originates from the DLPs which have been pelleted by ultracentrifugation after the in vitro transcription reaction and may have been partially damaged]; lane 8: as lane 7 + RNase I. Cores amount used: 60 ng/20 μl, or approximately 5 × 108 particles.
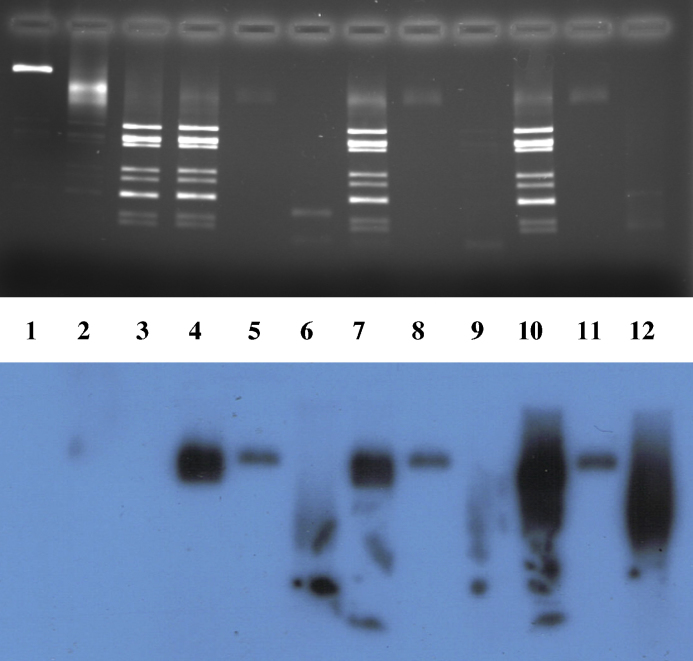
In order to explore the specificity of ssRNA packaging, opened RV cores were tested for their ability to package heterologous ssRNA. Transcripts of ssRNA (751–1104 bases in length), obtained from HIV-2 amplicons, were tested in parallel with segment-specific RV ssRNA transcripts (667–1061 bases in length). It was reproducibly found that the heterologous RNAs were packaged to an extent very similar to that of packaging of RV RNA (Fig. 6, lanes 7–9 and 10–12 in comparison with lanes 4–6). Various measures, e.g. heat pre-treatment of viral RNA (Supplementary Figure S4), variation of the ion concentrations of the packaging mixture (Supplementary Figure S5), or elevating the packaging reaction temperature to 37 °C (results not shown) did not succeed in increasing packaging specificity.
Fig. 6.
Non-specificity of packaging of ssRNA into rotavirus cores in vitro, Lane 1: DLPs; lane 2: native RV cores; lane 3: cores + EGTA; lane 4: cores + EGTA + 32P-labelled RV RNA 11 + packaging mixture; lane 5: as lane 4 + RNase I; lane 6: RV RNA11; lanes 7–9: treatment as in lanes 4–6, respectively, using 32P-labelled HIV-2 RNA751; lanes 10–12: treatment as in lanes 4–6, respectively, using 32P-labelled HIV-2 RNA1104.
3.7. Packaging competition assays
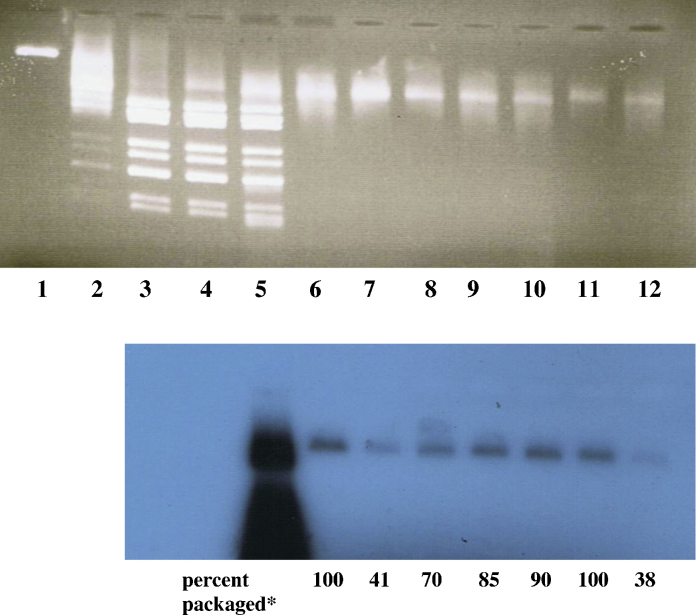
Based on these observations, the specificity of ssRNA packaging was explored further by competitive packaging assays in which constant amounts of 32P-labelled RV and HIV-2 RNAs competed for packaging with varying amounts of unlabelled, homologous or heterologous RNAs. Fig. 7 illustrates a typical experiment. The degree of decrease of packaged radiolabelled RNA by various amounts of unlabelled RNA was quantitated by the Image J program, taking the density of the not competed packaged radiolabelled material as 100%. The data of three experiments are shown in Table 3. From a number of such experiments the percentage decrease of packaging of radiolabelled ssRNA by competing unlabelled ssRNA was plotted against the amounts of unlabelled RNA in the reaction. The data are summarised in Supplementary Figure S6. The variance of experimental values around the straight lines [calculated by linear regression] was rather large (even in the homologous competition assays). The slopes and intercepts of homologous and heterologous competitive packaging reactions (Supplementary Figure S6; Table 4) were not significantly different by ANCOVA covariance analysis (P > 0.05), supporting the statement made above in assessing Fig. 6.
Fig. 7.
Inhibition of packaging of radiolabelled ssRNA by cold ssRNA (basis for competitive packaging assay), Lane 1: DLPs; lane 2: native RV cores; lane 3: cores + EGTA; lane 4: as lane 3 + packaging mixture; as lane 4 + radiolabelled RV RNA7; lane 6: as lane 5 + RNase I; lanes 7–10: as lane 6, with unlabelled RV RNA added before packaging: 100 ng (7), 50 ng(8), 25 ng (9), 13 ng (10); lane 11: cores + EGTA + radiolabelled HIV-2 1104 RNA + packaging mixture + RNase I; lane 12: as lane 11, with 100 ng of unlabelled HIV-2 1104 RNA added before packaging. * Determined by densitometric analysis of scanned autoradiograph using the Image J programme ‘Image J’ (http://rsbweb.nih.gov/ij/).
Table 3.
Competitive inhibition of packaging of radiolabelled RNA by unlabelled homologous or heterologous RNA.
| Exp/Track | Radiolabelled RNA | Unlabelled RNA (ng) | % packageda | |
|---|---|---|---|---|
| U73-3, | 6 | RV RNA 7 | – | 100 |
| 7 | “ | RV RNA 7 (105) | 31 | |
| 8 | “ | HIV 1100 RNA (125) | 65 | |
| U74-4, | 6 | RV RNA 7 | – | 100 |
| 7 | “ | RV RNA 7 (105) | 40 | |
| 9 | “ | HIV 1100 RNA (125) | 65 | |
| U77-2, | 6 | RV RNA 11 | – | 100 |
| 7 | “ | RV RNA11 (132) | 30 | |
| 8 | “ | “(66) | 63 | |
| 10 | “ | HIV 750 RNA (111) | 66 | |
| 11 | “ | “(56) | 76 | |
Determined by densitometric analysis of autoradiograph using program ‘Image J’.
Table 4.
Competition for packaging experiments: slopes and intercepts of linear regression functions in which the percent inhibition of packaging of radiolabelled RNA was plotted against the amounts of added unlabelled RNA.
| Radiolab. | Cold | Slope | Intercept | N | R | P | Slope | |
|---|---|---|---|---|---|---|---|---|
| RNA | RNA | ratio | ||||||
| RV | RV | y = 0.384x | + | 3.422 | 16 | 0.853 | <0.001 | |
| RV | HIV | y = 0.341x | + | 6.009 | 12 | 0.783 | <0.003 | 0.89 |
| HIV | HIV | y = 0.276x | + | 8.226 | 16 | 0.732 | <0.001 | |
| HIV | RV | y = 0.239x | + | 11.453 | 28 | 0.730 | <0.001 | 0.87 |
| RV hom | HIV hom | 0.72 | ||||||
| RV het | HIV het | 0.70 | ||||||
The slopes and intercepts were found not to differ significantly by the ANCOVA test (http://ude1.edu/∼mcdonald/statancova.html).
Despite extensive attempts at optimisation, the packaging assay was of low specificity, and the procedure was thus of insufficient quality to discriminate putative packaging signals on native RV RNAs or their mutants.
4. Discussion
Rotavirus core assembly in the infected cell is tightly controlled but not well understood. The inability of in vitro production of functional cores de novo is a major stumbling block in engineering viable RV particles from its components, whilst transcapsidation of native cores under appropriate conditions in vitro can achieve restoration of full infectivity (Trask and Dormitzer, 2006). Better knowledge of core assembly will help on the way towards developing a helper virus-free reverse genetics system for RVs (Trask et al., 2010). The availability of such a system would permit the detection of RNA packaging signals, e.g. as described for one bluetongue virus RNA segment (Matsuo and Roy, 2009).
The work presented here was carried out to define the conditions under which RV ‘opened cores’ are produced and to attempt reconstitution of ‘open cores’ in vitro with possible restoration of functions. The minimum concentration of EGTA sufficient to open cores was determined and, conversely, the minimum concentrations of di- and trivalent cations needed to stabilise cores. We observed that RV core preparations decompose within one week at 4 °C, but are much more stable when stored in the presence of 200 μM Ca2+ (results not shown). The requirements of divalent cations to stabilise TLPs (Ca2+; Estes et al., 1979) and VP6 assemblies (Zn2+; Erk et al., 2003) have been investigated previously. In the absence of divalent cations (mainly Ca2+) RV cores explode, liberating their genomic dsRNA. In their isolated form genomic RV dsRNAs have a minimal persistence length of 1125 Å, i.e. double the diameter of RV cores (Kapahnke et al., 1986). Divalent and trivalent cations were shown to reconstitute core structures but only within a very tight time frame after initiation of a treatment with EGTA. These data and the fact that baculovirus recombinant-expressed, empty core-like particles were not broken up by 50× the amount of EGTA sufficient to open native cores speak for the interaction of the cationic compounds with the genomic dsRNA inside native cores rather than for their interaction with proteins.
Rotavirus cores are difficult to work with since they have a tendency to aggregate.
Native RV cores purified from DLPs loose the monodispersity of the DLP preparation and form aggregates of 13–17 particles as described previously (Bican et al., 1982; Labbé et al., 1991; Zeng et al., 1994). Only DOC and SDS were reported to desaggregate the VP2 protein, the main scaffolding component of the cores (Labbé et al., 1991). Here we determined the minimum concentration of DOC needed to resolve core aggregates whilst maintaining their particular structure. We considered using DOC as desaggregating agent for core preparations, but as it acts as an emulsifier and mild detergent, we abstained from using it in transcapsidation experiments.
It was found that ‘opened cores’ could be reconstituted into core-like structures by the addition of divalent or trivalent cations (Ca2+, Mg2+, spermidine3+, cobalthexamine3+) to ‘opened core’-EGTA mixtures within the narrow time window of approximately 1–2 min as evidenced by EM and electrophoresis on non-denaturing gels. The question whether such reconstituted, ‘restored’ cores are functional or whether residual non-opened ones maintain function is important in the context. Some EM evidence and viral infectivity data argue at least in part for some function of ‘restored’ cores. Although, upon opening, cores release their genomic dsRNA quickly, upon treatment with cationic reagents within 1–2 min of opening electron-dense materials can be observed (also after transcapsidation with VP6) that is considered to contain components of the genome including the replication machinery.
Preparations of cores and ‘opened cores’ did not have residual infectivity on their own when tested in lipofectamine-mediated transfection assays (Bass et al., 1992; Chen and Ramig, 1993b). When baculovirus recombinant-expressed purified VP6 (Charpilienne et al., 2002) was added to native and ‘restored’ cores, infectivity was partially rescued, since cores transcapsidated with VP6 regain transcriptase activity (Kohli et al., 1993). From our experience, VP6 preparations which actively transcapsidated cores, always contained tubules. The formation of tubules depends on the concentration of VP6, on pH and ionic strength of the solution (Lepault et al., 2001). By non-denaturing gel analysis, transcapsidated cores were seen as a ladder-like set of particle subpopulations of different size. At present, their true composition is speculative. However, by EM VP6-encapsidated cores (native or ‘restored’) were clearly seen. The infectivity differences of preparations of ‘opened cores’ and ‘restored’ cores after VP6 transcapsidation in part signify the differences of infectivity of unopened and ‘restored’ cores, but the data were very variable. This may be due to the tendency of isolated native cores to form aggregates (Labbé et al., 1991; Zeng et al., 1994; this work).
We demonstrated that radiolabelled naked RV (+) ssRNAs can be packaged in vitro into opened and reconstituted cores where they are partially protected from RNase I treatment. [The great affinity of cores to ssRNA has been shown for bluetongue virus, belonging to another genus of the Reoviridae family (Loudon and Roy, 1992).] Preliminary experiments did not detect any infectivity of restored cores obtained after a packaging reaction and after transcapsidation with VP6 (results not shown). So far, no evidence has been obtained that RV RNA packaged in vitro is replicated.
In further experiments it was shown that the packaging of ssRNA into core-like structures is not virus-specific. The kinetics of competitive packaging of viral homotypic (RV) and heterotypic (HIV-2) RNAs confirmed a lack of specificity of RNA packaging, rendering this procedure unsuitable to test for packaging signals of wildtype or mutant RV RNAs in vitro.
Numerous attempts to introduce RV ssRNA into ‘opened cores’ followed by treatment with divalent or trivalent cations, transcapsidation with VP6 and infectivity testing in transfection assays have so far been unsuccessful to rescue infectious RV (U Desselberger, unpublished results). This identifies a main difficulty in attempting reconstruction of the early morphogenesis events of RV particles in vitro. Native isolated cores are relatively stable when suspended in the presence of divalent cations (200 μM Ca2+, unpublished results), but break apart in water within a few days. The decomposition process is very rapid as soon as divalent cations which neutralise the overall negative charge of the genomic dsRNA are removed, e.g. by chelating agents. Under the aspect of dsRNA flexibility, ‘one has to assume an intimate protein–RNA interaction or other forces in order to induce additional bending and packaging into the core’ (Kapahnke et al., 1986). Thus, it is futile to attempt repackaging of free RV dsRNA into cores. In natural infections it is the single stranded RNA segments of positive polarity that are packaged (Gallegos and Patton, 1989; Patton and Gallegos, 1990). Whilst the opening of cores appears to be reversible within narrow time limits, the loss of genomic dsRNA segments is irreversible.
A carefully balanced presence of NSP2 and NSP5 (Berois et al., 2003; Patton, 2001; Schuck et al., 2001; Taraporewala et al., 1999; Taraporewala and Patton, 2001) may be required to give such reconstitution experiments in vitro a chance of success. NSP2 has an NTPase activity (Taraporewala et al., 1999) and possibly acts as a molecular motor for packaging (Patton et al., 2003). NSP5 interacts strongly with VP2 (Berois et al., 2003) and is a possible modulator of the NSP2 function (Patton et al., 2003). Both NSP2 and NSP5 are needed for the formation of viroplasms during virus replication (Fabbretti et al., 1999), and there is evidence that RNA replication occurs in the viroplasm (Silvestri et al., 2004). Inhibition of NSP2 and NSP5 expression by siRNA or intrabodies prevents RNA replication and the production of infectious virus (Campagna et al., 2005; Silvestri et al., 2004; Vascotto et al., 2004). It is also possible that viral proteins VP1 and VP3 have to be added to in vitro packaging mixtures; this remains to be explored.
Furthermore, the recent findings that RV viroplasms interact with lipid droplets in the infected cell and that compounds disturbing the lipid droplet homoeostasis decrease the production of infectious RV progeny (Cheung et al., 2010) may explain the lack of success so far of in vitro reconstitution experiments for functional cores. It is likely that the in vitro conditions explored did not appropriately mimic the intracellular environment of RV early morphogenesis.
For the three-segmented dsRNA phages of the Cystoviridae family (phi6 and others) an in vitro packaging system has been devised (Olkkonen et al., 1990; Qiao et al., 1997; Mindich, 1999; Poranen et al., 2001; Poranen and Tuma, 2004): two viral enzymes, the packaging NTPase (P4) and the RdRp (P2), have been found to be essential for the nucleation step in addition to the major procapsid protein (P1). Once the single-stranded RNAs of positive polarity are packaged, they can be replicated, and by the addition of the nucleocapsid coat protein (P8) subviral particles be made that are infectious under specified conditions. However, the cystovirus packaging system is based on the pre-existence of an empty procapsid into which the ss(+)RNA segments are pulled in a particular order. For RV, the core-filling model of packaging is much less likely than the concerted packaging model (McDonald and Patton, 2011). A stunning example of a mammalian virus produced in a cell-free extract de novo with relatively simple means it that of poliovirus (Molla et al., 1991).
It is hoped that an in vitro system for the reconstitution of rotavirus particles from its components can be devised. Following unsuccessful early attempts (Gorziglia and Collins, 1992; Silvestri et al., 2004), recently helper virus-dependent reverse genetics (RG) systems have been established for RVs (Komoto et al., 2006; Troupin et al., 2010; Trask et al., 2010), but attempts so set up a helper virus-independent RG system for RVs have so far been elusive (Richards et al., 2013, and unpublished results of several other research groups). The availability of a helper virus-free system of reverse genetics (‘génétique inverse’) for RVs will have far reaching consequences for basic and applied research. In vitro reconstitution of infectious virus as recently achieved for bluetongue virus (Lourenco and Roy, 2011) will be a possibility to explore for RVs. Reconstitution of the RV core complex in vitro from its components (proteins and RNAs) to functional entities will be essential to resolve the sequence of events during early RV particle morphogenesis.
Acknowledgements
UD thanks INRA, Jouy-en-Josas/France, CNRS, Gif-sur-Yvette/France, and ICGEB, Trieste/Italy, for institutional support of his activities as a visiting research fellow. Thanks are due to A. Charpilienne, P. Vende, M. Campagna, C. Marchetti, M. Bestagno, W. Li and A. L’Hernault for support in the laboratory. Reagents were kindly supplied by MA McCrae, University of Warwick, UK, J Patton, NIAID, NIH, Bethesda, MD 20892, USA, and R. Randall, University of St. Andrews, UK. Discussions with D. Bamford, M.K. Estes, L. Mindich, J. Patton, B.V.V. Prasad and P. Tavarez were much appreciated.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplementary material related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.virusres.2013.09.034.
Contributor Information
Ulrich Desselberger, Email: ud207@medschl.cam.ac.uk.
James Richards, Email: james.richards@imperial.ac.uk.
Luba Tchertanov, Email: luba.tchertanov@lbpa.ens-cachan.fr.
Jean Lepault, Email: lepault@vms.cnrs-gif.fr.
Andrew Lever, Email: amll1@medschl.cam.ac.uk.
Oscar Burrone, Email: burrone@icgeb.org.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Afrikanova I., Fabbretti E., Miozzo M.C., Burrone O.R. Rotavirus NSP5 phosphorylation is up-regulated by interaction with NSP2. J. Gen. Virol. 1998;79:2679–2686. doi: 10.1099/0022-1317-79-11-2679. [DOI] [PubMed] [Google Scholar]
- Arnoldi F., Campagna M., Eichwald C., Desselberger U., Burrone O.R. Interaction of rotavirus polymerase VP1 with nonstructural protein NSP5 is stronger than that with NSP2. J. Virol. 2007;81:2128–2137. doi: 10.1128/JVI.01494-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass D.M., Baylor M.R., Chen C., Mackow E.M., Brémont M., Greenberg H.B. Liposome-mediated transfection of intact viral particles reveals that plasma membrane penetration determines permissivity of tissue culture cells to rotavirus. J. Clin. Invest. 1992;90:2313–2320. doi: 10.1172/JCI116119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berois M., Sapin C., Erk I., Poncet D., Cohen J. Rotavirus non-structural protein NSP5 interacts with the major core protein VP2. J. Virol. 2003;77:1757–1763. doi: 10.1128/JVI.77.3.1757-1763.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bican P., Cohen J., Charpilienne A., Scherrer R. Purification and characterization of bovine rotavirus cores. J. Virol. 1982;43:1113–1117. doi: 10.1128/jvi.43.3.1113-1117.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagna M., Eichwald C., Vascotto F., Burrone O.R. RNA interference of rotavirus segment 11 mRNA reveals the essential role of NSP5 in the virus replicative cycle. J. Gen. Virol. 2005;86:1483–1489. doi: 10.1099/vir.0.80598-0. [DOI] [PubMed] [Google Scholar]
- Charpilienne A., Nejmeddine M., Berois M., Parez N., Neumann E., Hewat E., Trugnan G., Cohen J. Individual rotavirus-like particles containing 120 molecules of fluorescent protein are visible in living cells. J. Biol. Chem. 2001;276:29361–29367. doi: 10.1074/jbc.M101935200. [DOI] [PubMed] [Google Scholar]
- Charpilienne A., Lepault J., Rey F., Cohen J. Identification of rotavirus VP6 residues located at the interphase with VP2 that are essential for capsid assembly and transcriptase activity. J. Virol. 2002;76:7822–7831. doi: 10.1128/JVI.76.15.7822-7831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Ramig R.F. Rescue of infectivity by in vitro transcapsidation of rotavirus single-shelled particles. Virology. 1993;192:422–429. doi: 10.1006/viro.1993.1057. [DOI] [PubMed] [Google Scholar]
- Chen D., Ramig R.F. Rescue of infectivity by sequential in vitro transcapsidation of rotavirus core particles with inner capsid and outer capsid proteins. Virology. 1993;194:743–751. doi: 10.1006/viro.1993.1315. [DOI] [PubMed] [Google Scholar]
- Chen D.Y., Patton J.T. Rotavirus RNA replication requires a single-stranded 3′ end for efficient minus-strand synthesis. J. Virol. 1998;72:7387–7396. doi: 10.1128/jvi.72.9.7387-7396.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D.Y., Patton J.T. De novo synthesis of minus-strand RNA by the rotavirus RNA polymerase in a cell-free system involves a novel mechanism of initiation. RNA. 2000;6:1455–1467. doi: 10.1017/s1355838200001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Zeng C.Q.Y., Wentz M.J., Gorziglia M., Estes M.K., Ramig R.F. Template-dependent, in vitro replication of rotavirus RNA. J. Virol. 1994;68:7030–7039. doi: 10.1128/jvi.68.11.7030-7039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D.Y., Luongo C.L., Nibert M.L., Patton J.T. Rotavirus open cores catalyze 5′-capping and methylation of exogenous RNA: evidence that VP3 is a methyltransferase. Virology. 1999;265:120–130. doi: 10.1006/viro.1999.0029. [DOI] [PubMed] [Google Scholar]
- Cheung W., Gill M., Esposito A., Kaminski C.F., Courousse N., Chwetzoff S., Trugnan G., Keshavan N., Lever A., Desselberger U. Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication. J. Virol. 2010;84:6782–6798. doi: 10.1128/JVI.01757-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J., Charpilienne A., Chilmonczyk S., Estes M.K. Nucleotide sequence of bovine rotavirus gene 1 and expression of the gene product in baculovirus. Virology. 1989;171:131–140. doi: 10.1016/0042-6822(89)90519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desselberger U., Manktelow E., Li W., Cheung W., Iturriza-Gómara M., Gray J. Rotaviruses and rotavirus vaccines. Br. Med. Bull. 2009;90:37–51. doi: 10.1093/bmb/ldp009. [DOI] [PubMed] [Google Scholar]
- Eichwald C., Vascotto F., Fabbretti E., Burrone O.R. Rotavirus NSP5: Mapping phosphorylation sites and kinase activation and viroplasm localization domains. J. Virol. 2002;76:3461–3470. doi: 10.1128/JVI.76.7.3461-3470.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichwald C., Rodriguez J.F., Burrone O.R. Characterization of rotavirus NSP2/NSP5 interactions and the dynamics of viroplasm formation. J. Gen. Virol. 2004;85:625–634. doi: 10.1099/vir.0.19611-0. [DOI] [PubMed] [Google Scholar]
- Erk I., Huet J.C., Duarte M., Duquerroy S., Rey F., Cohen J., Lepault J. A zincion controls assembly and stability of the major capsid protein of rotavirus. J. Virol. 2003;77:3595–3601. doi: 10.1128/JVI.77.6.3595-3601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes M.K., Greenberg H.B. Rotaviruses. In: Knipe D.M., Howley P.M., et al, editors. Fields’ Virology. 6th ed. Lippincott, Williams and Wilkins; Philadelphia: 2013. [Google Scholar]
- Estes M.K., Graham D.Y., Smith E.M., Gerba C.P. Rotavirus stability and inactivation. J. Gen. Virol. 1979;43:403–409. doi: 10.1099/0022-1317-43-2-403. [DOI] [PubMed] [Google Scholar]
- Estrozi L.F., Settembre E.C., Goret G., McClain B., Zhang X., Chen J.Z., Grigorieff N., Harrison S.C. Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles. J. Mol. Biol. 2013;425:124–132. doi: 10.1016/j.jmb.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbretti E., Afrikanova I., Vascotto F., Burrone O.E. Two nonstructural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structure in vivo. J. Gen. Virol. 1999;80:333–339. doi: 10.1099/0022-1317-80-2-333. [DOI] [PubMed] [Google Scholar]
- Gallegos C.O., Patton J.T. Characterization of rotavirus replication intermediates: a model for the assembly of single-shelled particles. Virology. 1989;172:616–627. doi: 10.1016/0042-6822(89)90204-3. [DOI] [PubMed] [Google Scholar]
- Gastañaduy P.A., Sánchez-Uribe E., Esparza-Aguilar M., Desai R., Parashar U.D., Patel M., Richardson V. Effect of rotavirus vaccine on diarrhea mortality in different socioeconomic regions of Mexico. Pediatrics. 2013;131:e1115–e1120. doi: 10.1542/peds.2012-2797. [DOI] [PubMed] [Google Scholar]
- Gorziglia M.I., Collins P.L. Intracellular amplification and expression of a synthetic analog of rotavirus genomic RNA bearing a foreign marker gene: mapping cis-acting nucleotides in the 3’-noncoding region. Proc. Natl. Acad. Sci. U.S.A. 1992;89:5784–5788. doi: 10.1073/pnas.89.13.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb M., Chavko M. Silver staining of native and denatured eukaryotic DNA in agarose gels. Anal. Biochem. 1987;165:33–37. doi: 10.1016/0003-2697(87)90197-7. [DOI] [PubMed] [Google Scholar]
- Graham A., Kudesia G., Allen A.M., Desselberger U. Reassortment of human rotavirus possessing genome rearrangements with bovine rotavirus: evidence for host cell selection. J. Gen.Virol. 1987;68:115–122. doi: 10.1099/0022-1317-68-1-115. [DOI] [PubMed] [Google Scholar]
- Kapahnke R., Rappold W., Desselberger U., Riesner D. The stiffness of dsRNA: hydrodynamic studies on fluorescence-labelled RNA segments of bovine rotavirus. Nucleic Acids Res. 1986;14:3215–3228. doi: 10.1093/nar/14.8.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli E., Pothier P., Tosser G., Cohen J., Sandino A.M., Spencer E. In vitro reconstitution of rotavirus transcriptional activity using viral cores and recombinant baculovirus expressed VP6. Arch. Virol. 1993;133:451–458. doi: 10.1007/BF01313782. [DOI] [PubMed] [Google Scholar]
- Kohli E., Pothier P., Tosser G., Cohen J., Sandino A.M., Spencer E. Inhibition of in vitro reconstitution of rotavirus transcriptionally active particles by anti-VP6 monoclonal antibodies. Arch. Virol. 1994;135:193–200. doi: 10.1007/BF01309778. [DOI] [PubMed] [Google Scholar]
- Komoto S., Sasaki J., Taniguchi K. Reverse genetics system for introduction of site-specific mutations into the double-stranded RNA genome of infectious rotavirus. Proc. Natl. Acad. Sci. U. S. A. 2006;103(12):4646–4651. doi: 10.1073/pnas.0509385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé M., Charpilienne A., Crawford S.E., Estes M.K., Cohen J. Expression of rotavirus VP2 produces empty corelike particles. J. Virol. 1991;65:2946–2952. doi: 10.1128/jvi.65.6.2946-2952.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton J.A., Zeng C.Q., Mukherjee S.K., Cohen J., Estes M.K., Prasad B.V. Three-dimensional analysis of recombinant rotavirus-like particles with intact and amino-terminal-deleted VP2: implications for the architecture of the VP2 capsid layer. J. Virol. 1997;71:7353–7360. doi: 10.1128/jvi.71.10.7353-7360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L.A., Franzel L., Atwel, l J., Datta S.D., Friberg I.K., Goldie S.J., Reef S.E., Schwalbe N., Simons E., Strebel P.M., Sweet S., Suraratdecha C., Tam Y., Vynnycky E., Walker N., Walker D.G., Hansen P.M. The estimated mortality impact of vaccinations forecast to be administered during 2011–2020 in 73 countries supported by the GAVI Alliance. Vaccine. 2013;31(Suppl. 2):B61–B72. doi: 10.1016/j.vaccine.2012.11.035. [DOI] [PubMed] [Google Scholar]
- Lepault J., Petitpas I., Erk I., Navaza J., Bigot D., Dona M., Vachette P., Cohen J., Rey F.A. Structural polymorphism of the major capsid protein of rotavirus. EMBO J. 2001;20:1498–1507. doi: 10.1093/emboj/20.7.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudon P.T., Roy P. Interaction of nucleic acids with core-like and subcore-like particles of bluetongue virus. Virology. 1992;191:231–236. doi: 10.1016/0042-6822(92)90184-q. [DOI] [PubMed] [Google Scholar]
- Lourenco S., Roy P. In vitro reconstitution of Bluetongue virus infectious cores. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13746–13751. doi: 10.1073/pnas.1108667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., McDonald S.M., Tortorici M.A., Tao Y.J., Vasquez-Del Carpio R., Nibert M.L., Patton J.T., Harrison S.C. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure. 2008;16:1678–1688. doi: 10.1016/j.str.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo E., Roy P. Bluetongue virus VP6 acts early in the replication cycle and can form the basis of chimeric virus formation. J. Virol. 2009;83:8842–8848. doi: 10.1128/JVI.00465-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain B., Settembre E., Temple B.R., Bellamy A.R., Harrison S.C. X-ray crystal structure of the rotavirus inner capsid particle at 3.8 Å resolution. J. Mol. Biol. 2010;397:587–599. doi: 10.1016/j.jmb.2010.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald S.M., Patton J.T. Assortment and packaging of the segmented rotavirus genome. Trends Microbiol. 2011;19:136–144. doi: 10.1016/j.tim.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindich L. Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage phi6. Microbiol. Mol. Biol. Rev. 1999;63:149–160. doi: 10.1128/mmbr.63.1.149-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla A., Paul A.V., Wimmer E. Cell-free, de novo synthesis of poliovirus. Science. 1991;254:1647–1651. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- Naitow H., Canady M.A., Lin T., Wickner R.B., Johnson J.E. Purification, crystallization, and preliminary X-ray analysis of L-A: a dsRNA yeast virus. J. Struct. Biol. 2001;135:1–7. doi: 10.1006/jsbi.2001.4371. [DOI] [PubMed] [Google Scholar]
- Ogden K.M., Ramanathan H.N., Patton J.T. Mutational analysis of residues involved in nucleotide and divalent cation stabilization in the rotavirus RNA-dependent RNA polymerase catalytic pocket. Virology. 2012;431:12–20. doi: 10.1016/j.virol.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olkkonen V.M., Gottlieb P., Strassman J., Qiao X., Bamford D.H., Mindich L. In vitro assembly of infectious nucleocapsids of bacteriophage phi6: formation of a recombinant double-stranded RNA virus. Proc. Natl. Acad. Sci. U.S.A. 1990;87:9173–9177. doi: 10.1073/pnas.87.23.9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar U.D., Hummelman E.G., Bresee J.S., Miller M.A., Glass R.I. Global illness and death caused by rotavirus disease in children. Emerg. Infect. Dis. 2003;9:565–572. doi: 10.3201/eid0905.020562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar U.D., Burton A., Lanata C., Boschi-Pinto C., Shibuya K., Steele D. Global mortality associated with rotavirus disease among children in 2004. J. Infect. Dis. 2009;200(Suppl. 1):S9–S15. doi: 10.1086/605025. [DOI] [PubMed] [Google Scholar]
- Patel M.M., Glass R., Desai R., Tate J.E., Parashar U.D. Fulfilling the promise of rotavirus vaccines: how far have we come since licensure? Lancet Infect Dis. 2012;12:561–570. doi: 10.1016/S1473-3099(12)70029-4. [DOI] [PubMed] [Google Scholar]
- Patton J.T. Rotavirus RNA replication and gene expression. Novartis Found. Symp. 2001;238:64–81. doi: 10.1002/0470846534.ch5. [DOI] [PubMed] [Google Scholar]
- Patton J.T., Chen D.Y. RNA-binding and capping activities of proteins in rotavirus open cores. J. Virol. 1990;73:1382–1390. doi: 10.1128/jvi.73.2.1382-1391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton J.T., Gallegos C.O. Rotavirus RNA replication: single-strand RNA extends from the replicase particle. J. Gen. Virol. 1990;71:1087–1094. doi: 10.1099/0022-1317-71-5-1087. [DOI] [PubMed] [Google Scholar]
- Patton J.T., Wentz M., Xiaobo J., Ramig R.F. Cis-acting signals that promote genome replication in rotavirus mRNA. J. Virol. 1996;70:3961–3971. doi: 10.1128/jvi.70.6.3961-3971.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton J.T., Jones M.T., Kalbach A.N., He Y.W., Xiaobo J. Rotavirus RNA polymerase requires the core shell protein to synthesize the double-stranded RNA genome. J. Virol. 1997;71:9618–9626. doi: 10.1128/jvi.71.12.9618-9626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton J.T., Chnaiderman J., Spencer E. Open reading frame in rotavirus mRNA specifically promotes synthesis of double-stranded RNA: template size also affects replication efficiency. Virology. 1999;264:167–180. doi: 10.1006/viro.1999.9989. [DOI] [PubMed] [Google Scholar]
- Patton J.T., Chizhikov V., Taraporewala Z., Chen D.Y. Virus replication. In: Gray J., Desselberger U., editors. Rotaviruses. Methods and Protocols. Humana Press; Totowa, New Jersey: 2000. pp. 33–66. [Google Scholar]
- Patton J.T., Kearney K., Taraporewala Z. Rotavirus genome replication: role of RNA-binding proteins. In: Desselberger U., Gray J., editors. Viral Gastroenteritis. Elsevier Science; Amsterdam: 2003. pp. 165–183. [Google Scholar]
- Pesavento J.B., Estes M.K., Prasad B.V.V. Structural organisation of the genome in rotavirus. In: Desselberger U., Gray J., editors. Viral Gastroenteritis. Elsevier Science B.V; Amsterdam: 2003. pp. 115–127. [Google Scholar]
- Poranen M.M., Tuma R. Self-assembly of double-stranded RNA bacteriophages. Virus Res. 2004;101:93–100. doi: 10.1016/j.virusres.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Poranen M.M., Paatero A.O., Tuma R., Bamford D.H. Self-assembly of a viral molecular machine from purified protein and RNA constituents. Mol. Cell. 2001;7:845–854. doi: 10.1016/s1097-2765(01)00228-3. [DOI] [PubMed] [Google Scholar]
- Prasad B.V., Wang G.J., Clerx J.P., Chiu W. Three-dimensional structure of rotavirus. J. Mol. Biol. 1988;199:269–275. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- Prasad B.V.V., Rothnagel R., Zeng C.Q.Y., Jakana J., Lawton J.A., Chiu W., Estes M.K. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature. 1996;382:471–473. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- Qiao X., Casini G., Qiao J., Mindich L. In vitro packaging of individual genomic segments of bacteriophage phi 6 RNA: serial dependence relationships. J. Virol. 1995;169:2926–2931. doi: 10.1128/jvi.69.5.2926-2931.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X., Qiao J., Mindich L. Stoichiometric packaging of the three genome segments of double-stranded RNA bacteriophage phi6. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4074–4079. doi: 10.1073/pnas.94.8.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J.E., Desselberger U., Lever A.M. Experimental pathways towards developing a rotavirus reverse genetics system: synthetic full length rotavirus ssRNAs are neither infectious nor translated in permissive cells. PLoS One. 2013;8(9):e74328. doi: 10.1371/journal.pone.0074328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P., Taraporewala Z., McPhie P., Patton J.T. Rotavirus nonstructural protein NSP2 self-assembles into octamers that undergo ligand-induced conformational changes. J. Biol. Chem. 2001;276:9679–9687. doi: 10.1074/jbc.M009398200. [DOI] [PubMed] [Google Scholar]
- Settembre E.C., Chen J.Z., Dormitzer P.R., Grigorieff N., Harrison S.C. Atomic model of an infectious rotavirus particle. EMBO J. 2011;30:408–416. doi: 10.1038/emboj.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri L.S., Taraporewala Z.F., Patton J.T. Rotavirus replication: Plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J. Virol. 2004;78:7763–7774. doi: 10.1128/JVI.78.14.7763-7774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares-Weiser K., Maclehose H., Bergman H., Ben-Aharon I., Nagpal S., Goldberg E., Pitan F., Cunliffe N. Vaccines for preventing rotavirus diarrhoea: vaccines in use. Cochrane Database Syst. Rev. 2012;11:CD008521. doi: 10.1002/14651858.CD008521.pub3. [DOI] [PubMed] [Google Scholar]
- Taraporewala Z., Patton J.T. Identification and characterization of the helix-destabilizing activity of the rotavirus nonstructural protein NSP2. J. Virol. 2001;75:4519–4527. doi: 10.1128/JVI.75.10.4519-4527.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraporewala Z., Chen D., Patton J.T. Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J. Virol. 1999;73:9934–9943. doi: 10.1128/jvi.73.12.9934-9943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J.E., Burton A.H., Boschi-Pinto C., Steele A.D., Duque J., Parashar U.D. WHO-coordinated Global Rotavirus Surveillance Network 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect. Dis. 2012;12(2):136–141. doi: 10.1016/S1473-3099(11)70253-5. [DOI] [PubMed] [Google Scholar]
- Thouvenin E., Schoehn G., Rey F., Petitpas I., Mathieu M., Vaney M.C., Cohen J., Kohli E., Pothier P., Hewat E. Antibody inhibition of the transcriptase activity of the rotavirus DLP: a structural view. J. Mol. Biol. 2001;307:161–172. doi: 10.1006/jmbi.2000.4479. [DOI] [PubMed] [Google Scholar]
- Torres-Vega M.A., Gonzalez R.A., Duarte M., Poncet D., Lopez S., Arias C.F. The C-terminal domain of rotavirus NSP5 is essential for its multimerization, hyperphosphorylation and interaction with NSP6. J. Gen. Virol. 2000;81:821–830. doi: 10.1099/0022-1317-81-3-821. [DOI] [PubMed] [Google Scholar]
- Tortorici M.A., Broering T.J., Nibert M.L., Patton J.T. Template recognition and formation of initiation complexes by the replicase of a segmented double-stranded RNA virus. J. Biol. Chem. 2003;278:32673–32682. doi: 10.1074/jbc.M305358200. [DOI] [PubMed] [Google Scholar]
- Trask S.D., Dormitzer P.R. Assembly of highly infectious rotavirus particles recoated with recombinant outer capsid proteins. J. Virol. 2006;80:11293–11304. doi: 10.1128/JVI.01346-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask S.D., Taraporewala Z.F., Boehme K.W., Dermody T.S., Patton J.T. Dual selection mechanisms drive efficient single-gene reverse genetics for rotavirus. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18652–18657. doi: 10.1073/pnas.1011948107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask S.D., Ogden K.M., Patton J.T. Interactions among capsid proteins orchestrate rotavirus particle functions. Curr. Opin. Virol. 2012;2:373–379. doi: 10.1016/j.coviro.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask S.D., McDonald S.M., Patton J.T. Structural insights into the coupling of virion assembly and rotavirus replication. Nat. Rev. Microbiol. 2012;10:165–177. doi: 10.1038/nrmicro2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troupin C., Dehée A., Schnuriger A., Vende P., Poncet D., Garbarg-Chenon A. Rearranged genomic RNA segments offer a new approach to the reverse genetics of rotaviruses. J. Virol. 2010;84:6711–6719. doi: 10.1128/JVI.00547-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vascotto F., Campagna M., Visintin M., Cattaneo A., Burrone O.R. Effects of intrabodies specific for rotavirus NSP5 during the virus replicative cycle. J. Gen. Virol. 2004;85:3285–3290. doi: 10.1099/vir.0.80075-0. [DOI] [PubMed] [Google Scholar]
- Vasquez-Del Carpio R., Gonzalez-Nilo F.D., Jayaram H., Spencer E., Prasad B.V.V., Patton J.T., Taraporewala Z.F. Role of the histidine triad-like motif in nucleotide hydrolysis by the rotavirus RNA-packaging protein NSP2. J. Biol. Chem. 2004;279:10624–10630. doi: 10.1074/jbc.M311563200. [DOI] [PubMed] [Google Scholar]
- Wentz M.J., Patton J.T., Ramig R.F. The 3′-terminal consensus sequence of rotavirus mRNA is the minimal promoter of negative-strand RNA synthesis. J. Virol. 1996;70:7833–7841. doi: 10.1128/jvi.70.11.7833-7841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager M., Dryden K.A., Olson N.H., Greenberg H.B., Baker T.S. Three-dimensional structure of rhesus rotavirus by cryoelectron microscopy and image reconstruction. J. Cell Biol. 1990;110:2133–2144. doi: 10.1083/jcb.110.6.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen C., Tate J.E., Patel M.M., Cortese M.M., Lopman B., Fleming J., Lewis K., Jiang B., Gentsch J., Steele D., Parashar U.D. Rotavirus vaccines: update on global impact and future priorities. Hum. Vaccin. 2011;7:1282–1290. doi: 10.4161/hv.7.12.18321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D.F., Andrejeva L., Livingstone A., Goodbourn S., Lamb R.A., Collins P.L., Elliott R.M., Randall R.E. Virus replication in engineered human cells that do not respond to interferons. J. Virol. 2003;77:2174–2181. doi: 10.1128/JVI.77.3.2174-2181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C.Q., Labbe M., Cohen J., Prasad B.V., Chen D., Ramig R.F., Estes M.K. Characterization of rotavirus VP2 particles. Virology. 1994;201:55–65. doi: 10.1006/viro.1994.1265. [DOI] [PubMed] [Google Scholar]
- Zeng C.Q., Wentz M.J., Cohen J., Estes M.K., Ramig R.F. Characterization and replicase activity of double-layered and single-layered rotavirus-like particles expressed from baculovirus recombinants. J. Virol. 1996;70:2736–2742. doi: 10.1128/jvi.70.5.2736-2742.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.