Abstract
Emergence of avian influenza viruses with high lethality to humans, such as the currently circulating highly pathogenic A(H5N1) (emerged in 1996) and A(H7N9) cause serious concern for the global economic and public health sectors. Understanding the spatial and temporal interface between wild and domestic populations, from which these viruses emerge, is fundamental to taking action. This information, however, is rarely considered in influenza risk models, partly due to a lack of data. We aim to identify areas of high transmission risk between domestic poultry and wild waterfowl in China, the epicenter of both viruses. Two levels of models were developed: one that predicts hotspots of novel virus emergence between domestic and wild birds, and one that incorporates H5N1 risk factors, for which input data exists. Models were produced at 1 and 30 km spatial resolution, and two temporal seasons. Patterns of risk varied between seasons with higher risk in the northeast, central-east, and western regions of China during spring and summer, and in the central and southeastern regions during winter. Monte-Carlo uncertainty analyses indicated varying levels of model confidence, with lowest errors in the densely populated regions of eastern and southern China. Applications and limitations of the models are discussed within.
Keywords: H5N1, avian influenza, spatial modeling, uncertainty, China, waterfowl, poultry, Monte-Carlo
Introduction
Emerging infectious diseases in wildlife are a growing concern to human health. More than 75% of known emerging pathogens are zoonotic, or transmissible from animal to humans (1, 2). An increased global demand for meat products has caused rapid intensification of the domestic livestock industry (3), and improvements to transportation and market chains has brought humans and their agricultural systems closer together (4–6). Coupled with climate change and landscape fragmentation (7–9), incidence of emerging zoonoses is likely to continue to rise, with particular threat in developing regions such as China, India, and parts of Southeast Asia (7–10).
Two currently circulating avian influenza viruses, highly pathogenic A(H5N1) and low pathogenic A(H7N9) (hereafter H5N1 and H7N9) are of particular concern due to their high case-fatality rates (approximately 60 and 30% currently), and economic impact to the livestock industry and public health system (11). H5N1 first emerged in domestic geese in southern China in 1996 (12), and has since infected 60 countries across Asia, Africa, and Europe killing 374 people. It continues to persist with year 2013 reports of animal infection in Bangladesh, Bhutan, Cambodia, China, India, and Nepal (13) and human cases in Bangladesh, Cambodia, China, Egypt, Vietnam (11). H7N9 was first reported in a man from China who began showing symptoms in mid-February 2013. In the following 12 weeks, the number of human cases rapidly rose to 132 [as of 7 June 2013, (11)], all within China. At the time of this writing, human to human transmission is rare in both viruses, although concern exists that genetic mutation or reassortment could cause a human pandemic (14–16).
Wild birds, generally waterfowl and shorebirds (Orders Anseriformes and Charadriiformes), are the natural reservoir for low pathogenic avian influenza viruses (LPAIV) (17). LPAIV can mutate into lethal form, which commonly occurs upon entry into a high density host population, such as a poultry farm (18). This appears to have been the case with H5N1 (2), however, H7N9 remains an LPAIV (as defined by pathogenicity to chickens), despite having bridged the species gap and causing death in humans. Although many questions remain unanswered in these early months of H7N9 occurrence – such as which domestic or wild species are the reservoir for this virus – there is a growing body of knowledge from our H5N1 experience that might be applied toward understanding H7N9. In particular, understanding where wild and domestic birds have opportunity to interact on the landscape will be useful in identifying areas where disease transmission may be more likely to occur. These regions would become focal areas for surveillance and prevention.
A recent review of H5N1 risk models (19) noted that few studies have explicitly incorporated wild birds in transmission risk models, in part because obtaining adequate inputs for these populations is difficult. Our aim is to model regions where domestic and wild birds co-occur on the landscape, thereby presenting an opportunity for disease transfer. We take an iterative approach to understanding the spatial relationships between wild and domestic birds across the breeding and wintering seasons by building high spatial resolution (1 km) deterministic models based on wild and domestic bird co-occurrence, and subsequently by developing a model that incorporates H5N1 risk factors. The first set of models (co-occurrence) has broad utility for predicting areas for emergence of novel viruses, and potentially toward understanding H7N9. The second group of models provides insight on transmission potential at the domestic and wild bird interface for H5N1.
Materials and Methods
Study area
Transmission risk models were developed for China based on the importance of this region for H5N1 and H7N9, and the potential for emergence of novel viruses. China’s anthropogenic and natural landscapes differ greatly across the country, allowing for varying levels of disease risk, both spatially and temporally.
In addition to developing nationwide models for China, we focused on two areas of interest regarding transmission at the wild and domestic bird interface: Poyang Lake (PYL) Region in southeastern China and Qinghai Lake (QHL) in northwestern China. PYL, located in along the Yangtze River basin, is a complex wetland system that supports 8.8 million people, 14 million ducks, and 100,000 wintering migratory waterbirds, including 90% of the global population of endangered Siberian Cranes (Grus leucogeranus) (20). The majority of the human population at PYL lives in village settings, well-integrated within the agricultural landscape. Rice-cropping and free range duck farming are prevalent, and the demand for “healthy” wild meat has led to the rise of farmed wild waterfowl [Chinese spotbill (Anas poecilorhyncha), mallard (Anas platyrhynchos), northern pintail (Anas acuta), etc.], increasing the potential for wild and domestic populations to exchange virus (21, Cappelle, in review). QHL in contrast, is a remote arid region on the high-elevation Qinghai-Tibet Plateau with few poultry or free-ranging duck farms. Surprisingly, H5N1 outbreaks are common to both regions, and investigating the response using our transmission risk models to these very different regions is of particular interest.
Model design and inputs
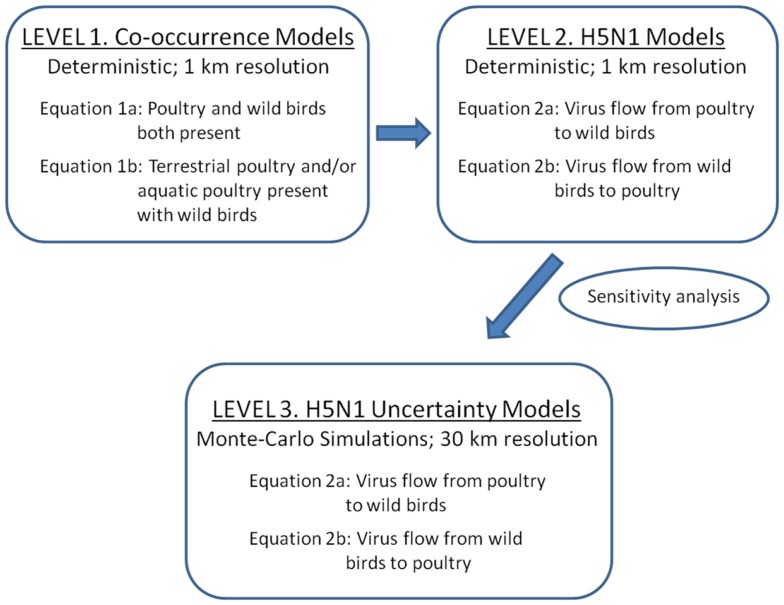
We developed three levels of models to predict risk for disease transmission between poultry and wild waterfowl in China (Figure 1). As a first step (Level 1), we developed simple overlay gridded maps that predict where poultry and wild waterfowl are likely to co-occur on the landscape at 1 km resolution. Poultry is defined as aquatic poultry (domestic ducks and geese) and terrestrial poultry (chickens). Wild waterfowl is defined as wild duck and goose species, and does not include farmed wild birds. The Level 1 models show us where domestic and wild birds are likely to be present within close proximity (<1 km) to allow for potential disease transfer via direct transmission or the environment.
Figure 1.
Three levels of spatial models implemented for assessing H5N1 transmission risk between wild and domestic birds in China. Level 1 and 2 deterministic models were developed to refine the transmission equations (1 km resolution). Level 1 models are co-occurrence models that predict where wild and domestic birds may come in contact. Level 2 models incorporate uni-directional equations for H5N1 transmission risk between poultry and wild birds. Level 3 models incorporate uncertainty using Monte-Carlo simulations at 30 km resolution.
Level 2 and 3 models incorporate H5N1 parameters. The models include four basic components that relate to classic compartmental models in epidemiology (22): infected and susceptible populations and their viral shedding and exposure rates (see equations below). These models use a hybrid density-dependent and environmental transmission approach, whereby direct transmission is defined by bird to bird transmission (within poultry or waterfowl) as well as the fecal to oral route facilitated by transmission through the land and water environment (23, 24). We first use a deterministic approach to develop and refine the model equations (Level 2), then apply Monte-Carlo simulations to incorporate estimates of uncertainty around the parameter inputs (Level 3).
Nine parameters were derived for the risk models (Table 1), including spatial and non-spatial parameters: terrestrial poultry density, aquatic poultry density, effective waterfowl population, contaminant containment scalar, incoming biosecurity scalar, terrestrial poultry viral shedding rate, aquatic poultry shedding rate, effective waterfowl population shedding rate, and viral uptake rate. Spatial inputs include poultry and waterfowl densities and biosecurity scalars. Non-spatial inputs, meaning parameters for which the current state of knowledge does not allow for differentiation across the spatial extent of our model, include viral shedding and uptake rates. These parameters values, which were taken from the literature (Table 1), were applied uniformly across each grid cell in the model.
Table 1.
Parameters of 1 km resolution transmission risk equations including the range of values, approach for sensitivity analyses, and reference for each.
| Parameter | Description | Value range for 1 km (mean, SD) | Value range for 30 km | Notes (Reference) |
|---|---|---|---|---|
| Pte | Terrestrial poultry density | 0–9418 (379.4, 745.7) chickens/km2 | 0–5871 chickens/km2 | Chicken densities for China (25 ) |
| Paq | Aquatic poultry density | 0–2796 (86.2, 164.7) ducks and geese/km2 | 0–2796 Ducks and geese/km2 | Duck and goose densities for China (25) |
| WEF | Effective waterfowl population | WEFbr: 0–0.32 (0.01, 0.04) | Wprbr: 0–0.29 | Distributions from (Prosser, in review) |
| Breeding season: WEFbr | WEFwi: 0–0.39 (0.006, 0.025) watefowl/km2 | Wprw i 0 to 0.39 | Population estimates from (43 , 44 ) | |
| Wintering season: WEFwi | Prevalence rates from (45–49) | |||
| Cte | Contaminant containment, terrestrial poultry (biosecure threshold Pte = 5000 birds/km2) | Biosecure = 0.75 and 0.25; non-biosecure = 1 (unitless) | Biosecure = 0.5; non-biosecure = 1 | Biosecure threshold of 5000 chickens per km2. Reduction of population by 0.25 or 0.75 given biosecure designation |
| Bte | Biosecurity, terrestrial poultry thresholds: Pte ≤ 50: Pbackyard = Pte × 1.0 50 < Pte ≤ 1000, Pbackyard: Pte × 0.5, Pte > 1000, Pbackyard = 1000 | Pte × Bte = 0–1000 poultry/km2 | Pte × Bte = 0–1000 poultry/km2 | Tri-part equation: At densities ≤ 50, 100% of population is backyard poultry From 50 to 1000, half are backyard poultry At greater than 1000, backyard poultry is limited to 1000 |
| Vte | Viral shedding rate, terrestrial poultry | 101.4 and 109.8 EID50 | 100, 109.8, 106.8 EID50 | Viral shedding rates per individual per day from (50–52) |
| Vaq | Viral shedding rate, aquatic poultry | 101 and 105.7 EID50 | 0, 106.5, 102.98 EID50 | Viral shedding rates per individual per day from (53–57) |
| Vwf | Viral shedding rate, wild waterfowl | 102.5 and 106.5 EID50 | 102.5, 106.5, 104.77 EID50 | Viral shedding rates per individual per day from (58) |
| U | Viral uptake = consumption rate of virus in the environment/minimum load for infection | 10−15/(104.7–101.8) EID50 | 1.58e−17, 1.99e−20, 1.99e−20 ∑50 | Consumption rate of virus in environment 10-15 (37); minimum viral load of 104.7 EID50 (and 101.8 EID50) to initiate infection with low pathogenic AIV (23, 59, 60) |
We modeled terrestrial (Pte) and aquatic poultry (Paq) populations by disaggregating census data and applying regression models for China’s top three poultry species: chickens, ducks, and geese (25, Prosser, in review). Poultry densities were modeled at 1 km resolution across the extent of China with units of birds per km2. For waterfowl, because the number of species and individual susceptibility to H5N1 is diverse (26), we aimed to derive an “effective waterfowl population” for each 1 km grid cell. Here we first modeled population estimates for each species by taking the total population for a given species and distributing it evenly across the extent of its range (Prosser, in review). We then applied prevalence rates [from the literature, see Table 1; (Prosser, in review)] to estimate the proportion of the population that may be (a) shedding virus if disease is present, or (b) susceptible to infection if disease is present in domestic species. To get the total effective waterfowl population (WEF), we then summed the resulting numbers of wild birds for each grid cell. Units for WEF are birds per km2. This process was conducted separately for the breeding and wintering seasons (WEFbr and WEFwi) as population distributions vary greatly due to the migratory patterns of waterfowl (Prosser, in review).
Two biosecurity scalars were developed to reduce the effective poultry populations: a contaminant containment parameter (Cte) that moderates virus flow from poultry farms to the environment, and a biosecurity parameter (Bte) that moderates exposure of poultry to virus [Table 1; (Prosser, in review)]. The parameters were derived based on the assumption that more biosecure farms control the flow of potential pathogens from the farm to the environment (Cte term), and protect themselves from incoming pathogens from the environment and other farms (Bte term); for example by cleansing vehicles before entering the farm, housing animals in structures secure from wild species, etc. Due to the difference in farming systems for chickens versus aquatic poultry [housed versus free range (27)], the biosecurity scalars were applied to the terrestrial poultry populations only. Cte was given a value of 1 for cells with less than 5,000 terrestrial poulty/km2 (i.e., no change in effective population) and 0.5 for cells with higher concentrations of birds (i.e., reducing the effective population by one half). Bte uses a tri-part scalar to reduce the effective population of chickens based on an estimate of the proportion of birds in backyard farms versus larger chicken houses [details in (28, Prosser, in review)]. The assumption made here is that if virus is transferred through the environment, only the free-ranging “backyard” poultry would be exposed to virus coming from wild birds. However, from our field studies, we also understand that movement of poultry feces and usage of water from different sources is complex and provides opportunity for virus transfer even to housed poultry. Therefore, we also ran the models using the entire poultry population as the effective population size (removing the Bte term from the model).
Virus shedding rates for terrestrial poultry (Vte), aquatic poultry (Vaq), and waterfowl (Vwf) were taken from the literature (Table 1). Units are EID50 (amount of virus that causes infection in 50% of embryos) per individual per day. The virus uptake term (U), was taken from the literature (Table 1), and includes the consumption rate of virus in the environment (per day per bird) divided by the minimum load to infection (in units of EID50). The U term acts as a contact rate modifier for the number of susceptible birds in the population.
Transmission risk equations
Level 1 models
We developed two Level 1 models, both of which are based on presence or absence of bird groups and not effective population numbers. The first model generated a simple binary risk map with the assumptions that (a) transmission risk is bi-directional (equal probability) between poultry and waterfowl and (b) transmission would only occur if both poultry and waterfowl were predicted to be “present”:
| (1) |
where P01 and W01 are the predicted presence of poultry and waterfowl, respectively. Parameter values are 1 or 0 (present or absent). A T01A value of 0 indicates no transmission risk (i.e., no co-occurrence) between domestic and wild species because both are not present within a cell. A transmission risk value of 1 indicates the potential for virus transmission between wild and domestic populations. The second Level 1 model measures the co-occurrence of waterfowl with one or both types of poultry. Transmission potential of these varying combinations may have implications for surveillance and control, which is why we included this iteration of the models. Pt01 is predicted presence of terrestrial poultry, and Pa01 is predicted presence of aquatic poultry. Parameter values are 1 or 0 (present or absent), and model output is 2, 1, or 0 represented in decreasing threat of transmission risk for the three output values (both types of poultry are present along with waterfowl; one type of poultry is present along with waterfowl; or either poultry or waterfowl are not present in the grid cell):
| (2) |
Level 2 and 3 models
Level 2 and 3 models are based on where poultry and waterfowl are found together on the landscape, but also include effective population size, H5N1 shedding rates, and virus uptake for each group. We developed uni-directional equations for transmission potential from poultry to waterfowl versus waterfowl to poultry (Eqs 3 and 4) due to differences in farming structure and movement of virus through the environment (see above). The equations include compartments (grouped by brackets below) for the amount of virus entering the environment from infected birds (effective populations of infected birds times shedding rates) and amount of virus being taken up by susceptible individuals (effective susceptible populations times the uptake rate):
| (3) |
| (4) |
where Pte and Paq represent densities of terrestrial and aquatic poultry; Cte and Bte as biosecurity scalars; Vte, Vaq, and Vwf as viral shedding rates for terrestrial poultry, aquatic poultry, and waterfowl; WEF as the effective waterfowl population; and U as the virus uptake rate. Model output is a measure of the transmission risk between poultry and waterfowl in units of predicted number of cases. Due to limits in the state of knowledge of shedding and uptake rates, we do not presume to use the model outputs as direct measures of the risk of transmission within a grid cell at this time; alternatively, we expressed the output in relative format by grouping the choropleth legend into 0 risk and 4 quantiles representing low, medium, high, and highest risk (29, 30). The Level 3 models used Monte-Carlo simulations to incorporate uncertainty around model inputs and to map estimates of error on a spatial basis (31, 32). Uncertainty for the poultry variables, Pte and Paq, was described using a normal distribution. This was determined by fitting a random sample of poultry estimates across 25 bootstrapped layers for 100 spatial locations [fitdistrplus package, R, (33, 34)]. We used best estimates and minimum – maximum limits within triangular and truncated normal distributions for the remaining variables [mc2d package, R, (34, 35)] to perform the triangular and truncated normal distributions. Each simulation was run for 10,000 iterations to ensure model convergence. We calculated the coefficient of variation (CV = standard deviation divided by the mean) to estimate uncertainty of model predictions using a bootstrap procedure for the poultry models (25) and Monte-Carlo analysis for the waterfowl indices (Prosser, in review).
Spatial and temporal scale of analysis
The Level 1 and 2 deterministic equations were modeled at 1 km resolution in a geographic framework using ArcGIS 10.0 (ESRI, Redlands California) and Python (www.python.org). The Level 3 models, which incorporated the Monte-Carlo-based uncertainty (31, 32), were run at the coarser resolution of 30 km, approximately the average county size for China. Models were run separately for two temporal seasons relating the annual chronology of wild waterfowl to transmission risk: the breeding season (spring and summer months, generally April to July) and the wintering season (November to March).
Results
Risk models
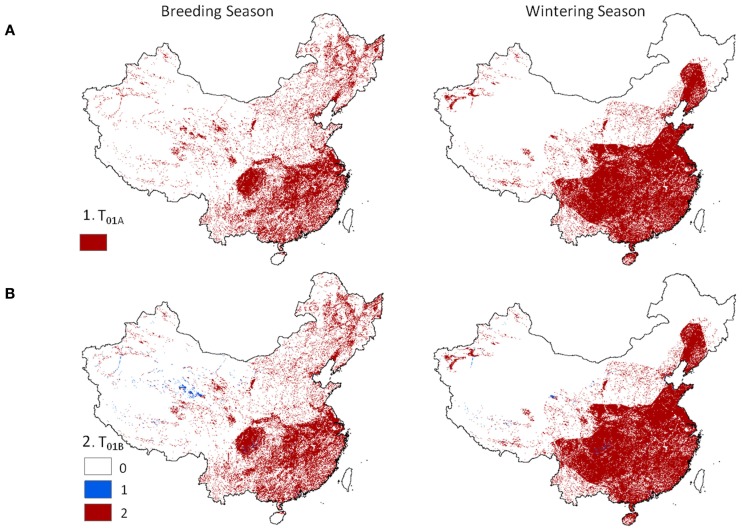
Level 1 models, identifying locations where poultry and wild waterfowl co-occur showed distinct patterns across China (Figure 2). Dense concentrations of positive risk grid cells were present across much of southeastern China during both seasons. Northeastern and western China showed more localized patterns of transmission risk, and wider extent for the breeding season than the wintering season. The spatial pattern was similar when considering the areas of co-occurrence of poultry with wild birds (Eq. 1) and presence of one or both poultry groups in combination with wild waterfowl (Eq. 2). Localized regions existed in the west where only one poultry type (usually chickens) was present in combination with wild waterfowl (Figure 2, lower panel).
Figure 2.
Hotspot regions of potential disease transmission between domestic and wild birds in China. Models are 1 km resolution co-occurrence for China’s domestic poultry and wild Anatidae waterfowl: (A) domestic poultry and wild Anatidae are predicted present (Eq. 1), and (B) terrestrial and aquatic poultry are predicted present in combination with wild Anatidae (red) versus presence of one poultry group (blue) with wild Anatidae (Eq. 2).
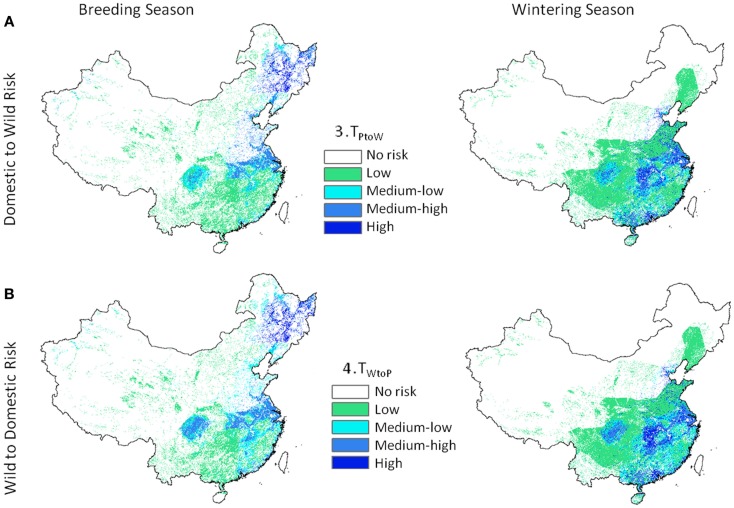
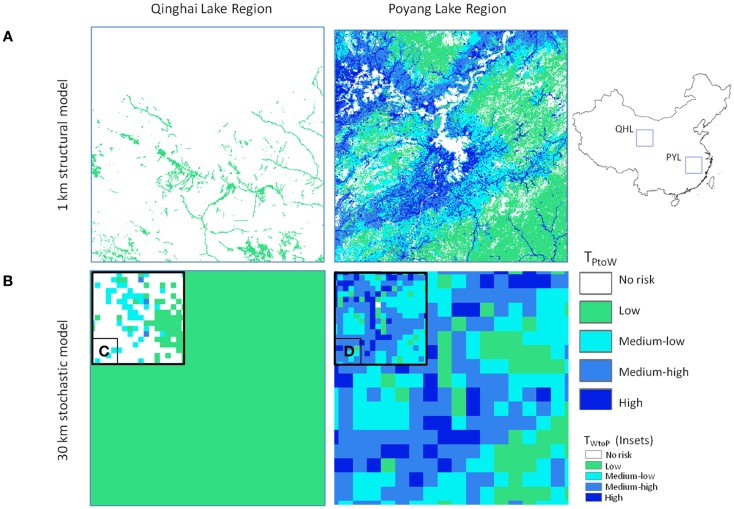
Transmission risk for the 1 km deterministic poultry to wild equations (Eq. 3, TP to W) ranged from 0 to 164.8 e−9 with units of predicted number of cases per day. Wild to poultry deterministic transmission risk was lower (0 to 5.19 e−11). Ranges for the 30 km Monte-Carlo models were 0 to 67.0 e−9 and 0 to 16.3 e−12 for Eqs 3 and 4, respectively. The mean level of risk was greater for the poultry to waterfowl models (Eq. 3) by approximately two orders of magnitude (Table 2). Spatial patterns of disease risk were similar across the broad scale of China (Figure 3). Both the 1 km deterministic and 30 km Monte-Carlo models showed distinct spatial patterns between seasons (Figures 3 and 4, respectively). During the breeding season, highest levels of risk (in both directions) were localized patches in northeastern China and along the Yangtze River plain of south-central China. For the wintering season, higher levels of risk were confined to southern and eastern China, particularly along the major river basins. Winter models had higher means than breeding season models by 46% Eq. (3) and 53% Eq. (4). Focused model predictions for the PYL and QHL sub-regions showed contrasting results between seasons (Figure 5). PYL had moderate to high transmission risk in both the winter and summer seasons whereas QHL showed low risk during the winter season and moderate risk during the summer breeding season.
Table 2.
Differences in mean values across all cells for two modeling approaches (Level 2 and 3) and four transmission scenarios.
| Model | Eq. 3 breeding season | Eq. 3 wintering season | Eq. 4 breeding season | Eq. 4 wintering season |
|---|---|---|---|---|
| Level 2 (deterministic) | 3.82E−10 | 7.13E−10 | 1.48E−13 | 3.13E−13 |
| Level 3 (Monte-Carlo) | 1.18E−09 | 1.66E−09 | 6.03E−13 | 8.39E−13 |
| Coefficient of variation | 144 | 147 | 219 | 223 |
Units of Level 2 and 3 model output are predicted number of cases per day.
Figure 3.
Highly pathogenic H5N1 transmission risk between domestic poultry and wild Anatidae waterfowl at 1 km resolution for China. Level 2 models include H5N1-specific transmission factors and are uni-directional with (A) representing transmission risk from domestic to wild birds (Eq. 3), and (B) from wild birds to domestic (Eq. 4).
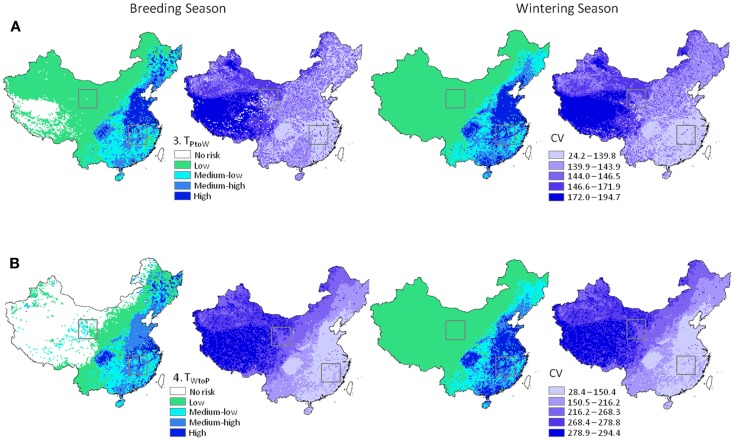
Figure 4.
H5N1 transmission risk between wild and domestic birds in China and associated uncertainty predictions at 30 km resolution. Risk maps represented as mean and CV (left and right in each pair of maps, respectively). (A) Top panel represents transmission risk from poultry to wild waterfowl; (B) bottom panel represents transmission risk from wild waterfowl to poultry. Maps are symbolized using quantiles. Black boxes correspond to the Qinghai Lake and Poyang Lake Regions outlined in Figure 5.
Figure 5.
Comparison of model outputs for Qinghai Lake (QHL) and Poyang Lake (PYL) sub-regions for (A) 1 km deterministic and (B) 30 km Monte-Carlo model outputs using Eq. 3 (poultry to wild transmission risk) for the winter season. Insets (C,D) show comparisons for the breeding season Eq. 4 (wild to poultry transmission).
Patterns in uncertainty of the Level 3 Monte-Carlo simulation models were similar across seasons and uni-directional equations. We carefully investigated mean input and output for the models between the Level 2 deterministic and Level 3 Monte-Carlo simulations (36). Means were nearly identical for the 1 and 30 km models, indicating a lack of bias in the resampling process (Table 3); we also noted higher means for the input parameters that were modeled using the triangular distribution of the Monte-Carlo models (Table 3, Section B). These models initially used a global (fixed) minimum and maximum value for all cells which is less computationally intensive but has the effect of increasing the mean values. We then reran the models using individual minimum and maximum values for each cell which reduced the mean values to match the 30 km deterministic models; however, to avoid negative values for input parameters such as waterfowl abundance and prevalence, we truncated the triangular distributions to fit within each parameter’s input range (Table 1). Truncating the distributions increased means for each parameter (Table 3, Section B); however, the values were closer to the 30 km deterministic models than the models using a global min/max, and thus used in the final models reported. The most uncertain areas of prediction were located in the western part of the country, and the least uncertain areas were located in the south and east (Figure 4). Models encompassing virus flow from wild birds to poultry (Eq. 4) had CVs that were 40% higher than models describing virus flow from poultry to wild birds (Eq. 3).
Table 3.
Comparisons of mean model outputs for 1 km deterministic, 30 km deterministic, and 30 km Monte-Carlo models of H5N1 transmission risk between wild and domestic birds in China.
| (A) Parameter | 1 km Deterministic | 30 km Deterministic | 30 km Monte-Carlo | |
|---|---|---|---|---|
| TRUNCATED NORMAL DISTRIBUTIONS | ||||
| Pte (chickens/km2) | 379 | 378 | 379 | |
| Paq (ducks and geese/km2) | 86 | 86 | 86 | |
| (B) Parameter | 1 km Deterministic | 30 km Deterministic | 30 km Monte-Carlo global min/max | 30 km Monte-Carlo individual min/max truncateda |
| TRIANGULAR DISTRIBUTIONS | ||||
| WEFbr (waterfowl/km2) | 0.01 | 0.01 | 0.13 | 0.03 |
| WEFwi (waterfowl/km2) | 0.006 | 0.006 | 0.099 | 0.037 |
| Pte × Bte (chickens/km2) | 183 | 184 | 395 | 227 |
| Cte (poultry/km2) | 1.00 | 1.00 | 0.83 | 0.83 |
aThe 30 km Monte-Carlo individual min/max truncated values were used in the final models, and were closer to the 30 km deterministic means than the models that used a global min/max value for the distributions. (A) Notes values for parameters using truncated normal distributions for the Monte-Carlo simulations. (B) Notes values for parameters using triangular distributions for the Monte-Carlo simulations.
Discussion
Model summary and interpretation
The objective of this study was to lay the foundation for a systematic modeling approach to investigate and predict spatial and temporal patterns of disease transmission risk between poultry and wild waterfowl populations in China. We explicitly took a multi-level approach toward modeling transmission risk between wild and domestic waterfowl in China. The Level 1 deterministic models demonstrated, at a fine resolution, predicted locations of co-occurrence of wild and domestic waterfowl distributions. High risk hotspots during the wintering season were observed in the southern and eastern lowland regions of China (Figure 2). These areas have concentrated poultry populations, particularly free-grazing ducks in association with rice farming, and are important wintering areas for many migratory waterfowl species. Hotspot regions of risk during the breeding season were observed in the northeastern and central-eastern China and had a greater geographic extent but more localized pattern in comparison to the winter risk models. The difference in pattern can be explained in part by waterfowl that tend to breed in the north and high-elevation western regions – areas where wetland habitat is distributed in a patchier, more localized pattern than the extensive lowland wetlands and rice paddies of the southeast (Prosser, in review).
Level 2 and 3 models, although unrealistically simplistic for predicting absolute risk of transmission, takes a first attempt at incorporating H5N1-specific parameters and population numbers for the different bird groups. Virus shedding and uptake rates were included as constants in the model. As our knowledge increases, these constants can be replaced with inputs parameterized to reflect geographic heterogeneity. In particular, the numerator of the uptake term seems unrealistically low (10−15). This term was taken directly from a traditional compartmental epidemiological model (37) which is designed to run iterative steps through time. This term has a large affect on the output value of our model, which is represented by a single risk value (in time). Improvements to this term would improve the overall output of our model. In light of these issues, we have used the model to highlight regions of H5N1 transmission risk in relation to other locations.
The effect of the addition of seasonal bird population size was apparent in the model results for QHL and PYL focal areas (Figure 5). Hundreds of thousands of migratory waterfowl return to the PYL region in the winter and reside amongst some of the highest poultry densities in the country. In the QHL region, waterfowl migrate away from the cold and arid plateau for the winter months and risk is lower year-round due to the low poultry densities in the area. Risk for QHL changes during the breeding season (Figure 5C) with the return of tens of thousands of nesting waterfowl to the region. The differences in risk could not be predicted without explicitly incorporating the ecology of the wild bird populations, which is one of the strengths of our approach.
Although waterfowl species were considered as a composite, the contribution of significant species was still evident in certain areas. A concentrated section of risk was observed in northeastern China wintering models (Figures 2 and 3, right panel) which followed the distribution of a single species, the greater white-fronted goose (Anser albifrons). The effect of this species was most noticeable in the Level 1 risk models (Figure 2) because input values from the poultry and wild bird populations were given equal weight. The pattern was present in Level 2 models to a lesser degree (Figure 3) since prevalence rates for the greater white-fronted goose were low in comparison to other species (2.2%).
The virus shedding and uptake rates spanned four to ten orders of magnitude. Due to the large range in values, and that we extracted them directly from the literature, we chose to keep these rates fixed in the Monte-Carlo models so we could more clearly assess the effects of our modeled input parameters (wild bird and poultry distributions, and biosecurity and contaminant containment parameters). One term of particular interest was the biosecurity term (Pte × Bte), which had a substantial effect on the model results. Until data are available to generate a better estimate, we developed two sets of models have been developed for use (28).
Level 3 models showed a similar pattern across China to the deterministic models, confirming that the number of simulations was sufficient for the mean values to converge toward results of the deterministic models (32). This allowed us to identify areas of greatest variability. The lowest predicted errors were in the southeast (Figure 4), which can be explained in part by the high poultry densities in this region (25). The highest predicted errors were in the western regions where waterfowl populations are more localized and sparsely distributed.
Utility and limits of the models
These models should be considered as a starting point to refine predictions of risk for H5N1 outbreaks. In contrast to existing temporal dynamic models of H5N1 transmission (23, 24, 37), our models focus on transmission risk at the interface of wild and domestic species and illustrate that even simple incorporation of transmission parameters modify the risk map. These are the first spatially based models to incorporate waterfowl distributions from population data. Our approach complements existing dynamic epidemiological models as well as studies which have mapped statistical relationships between outbreak events and environmental or anthropogenic risk factors without having complete information on an entire region or populations (19, 38–42). Our spatial characterization of the susceptible populations and the stepwise examination of the effects of adding model complexity should form a basis for more sophisticated refinement of transmission parameters as these are recognized in other studies.
With growing interest in predicting the risk of future transmission of H5N1 or other emerging strains such as H7N9, all three levels of model have utility. Level 1 models identify areas where transmission can occur due to co-occurrence of wild and domestic species. Level 2 models incorporate population size, prevalence, transmission, and biosecurity parameters. The model structure allows inputs to be changed and new maps to be created. Level 3 models show that the coarser county scale is appropriate for informing surveillance and prevention measures, and is more realistic for assessment. Identification of locations of greater uncertainty in the predictions also helps inform policy decision-making. Applications for the findings of this study may include use by health experts and wildlife officials who are interested in using the poultry to wild risk models (Figure 4A) to identify regions where wild migratory birds are at higher risk of exposure to new and evolving virus strains from poultry. Poultry farmers and health officials may use the wild to poultry risk models to identify areas where farming practices or vaccination programs should be enhanced to protect poultry from exposure to wild birds. As the models take a combined density-dependent and environmental transmission approach, the results may also help target environmental surveillance programs.
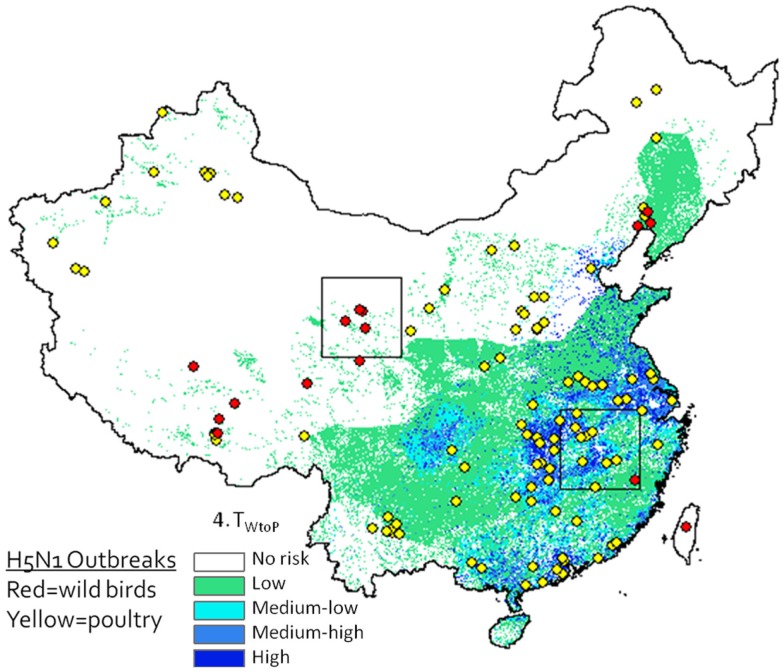
A desirable step would be to validate the model predictions using avian surveillance and outbreak data. A strong match exists between outbreak locations and our predicted risk areas (Figure 6), but this may be misleading as our model is intended to predict risk at the interface between poultry and waterfowl, and it is unknown whether these poultry outbreaks were caused by transmission from wild birds (versus poultry) and whether wild bird outbreaks were caused by poultry (versus wild birds). Validation would require geographic and temporal data on infections in wild and domestic birds including information on the type of host that caused the infection. Such detailed surveillance data do not yet exist, and deriving the infecting population from the virus isolates is difficult – even the use of phylogenetic analyses may not definitively answer this question as intermediary transmissions may occur between outbreak events. The spatial and temporal relationships between the wild and domestic waterfowl distributions in our risk models do identify relationships that might guide future targeted, more intensive sampling, and surveillance studies.
Figure 6.
H5N1 outbreak data (2003–2009) plotted against Level 2 deterministic 1 km resolution H5N1 transmission risk models (Eq. 4). Wild bird cases shown by red circles, poultry by yellow circles. Although spatial associations between outbreak data and risk map appear to be high, since the model is intended to predict risk at the interface between poultry and waterfowl, this type of presentation may be misleading as it is not known whether yellow dots represent poultry cases caused by transmission from wild birds (versus poultry) and whether red dots represent wild bird cases caused by poultry (versus wild birds).
Through a structured approach to predicting transmission risk between domestic poultry and wild waterfowl in China, we were able to separate the spatial relationships between poultry and waterfowl from the disease-specific factors to better understand the contributions of each to transmission risk. We explicitly incorporated uncertainty measures with our risk predictions and conducted sensitivity analyses to understand the effects of uncertainty on the model outputs. It is the first analysis of its kind and one of the few that focuses specifically on interactions between the wild and domestic bird populations, providing a unique contribution to our growing knowledge on the topic of wild birds and avian influenza transmission.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by funding from the United States Geological Survey Victims and Vectors Program. We thank Ruth DeFries of Columbia University for expertise in remote sensing, and Lisa Vormwald and two anonymous reviewers for strengthening earlier versions of this manuscript. The use of trade, product, or firm names in this publication is for descriptive purposes only and does not imply endorsement by the U.S. Government.
References
- 1.Taylor LH, Latham SM, Woolhouse MEJ. Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci (2001) 356(1411):983–9 10.1098/rstb.2001.0888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander DJ. An overview of the epidemiology of avian influenza. Vaccine (2007) 25(30):5637–44 10.1016/j.vaccine.2006.10.051 [DOI] [PubMed] [Google Scholar]
- 3.Delgado C. Rising consumption of meat and milk in developing countries has created a new food revolution. J Nutr (2003) 133(11):3907S–10 [DOI] [PubMed] [Google Scholar]
- 4.Daszak P, Cunningham AA, Hyatt AD. Wildlife ecology – emerging infectious diseases of wildlife – threats to biodiversity and human health. Science (2000) 287(5452):443–9 10.1126/science.287.5452.443 [DOI] [PubMed] [Google Scholar]
- 5.Brown C. Emerging zoonoses and pathogens of public health significance – an overview. Rev Sci Tech (2004) 23(2):435–42 [DOI] [PubMed] [Google Scholar]
- 6.Morse SS. Factors in the emergence of infectious diseases. Emerg Infect Dis (1995) 1(1):7–15 10.3201/eid0101.950102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kovats RS, Campbell-Lendrum DH, McMichel AJ, Woodward A, Cox JSH. Early effects of climate change: do they include changes in vector-borne disease? Philos Trans R Soc Lond B Biol Sci (2001) 356(1411):1057–68 10.1098/rstb.2001.0894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McMichael AJ. Global climate change: will it affect vector-borne infectious diseases? Intern Med J (2003) 33(12):552U–4 10.1111/j.1445-5994.2003.00492.x [DOI] [PubMed] [Google Scholar]
- 9.McMichael AJ, Woodruff RE, Hales S. Climate change and human health: present and future risks. Lancet (2006) 367(9513):859–69 10.1016/S0140-6736(06)68079-3 [DOI] [PubMed] [Google Scholar]
- 10.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, et al. Global trends in emerging infectious diseases. Nature (2008) 451(7181):990U–4 10.1038/nature06536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Health Organization Summary and assessment as of 07 June 2013. In: Monthly Risk Assessment Summary: Influenza at the Human-Animal Interface. (2013). Available from: http://www.who.int/influenza/human_animal_interface/Influenza_Summary_IRA_HA_interface_26Apr13.pdf
- 12.Xu XY, Subbarao K, Cox NJ, Guo YJ. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology (1999) 261(1):15–9 10.1006/viro.1999.9820 [DOI] [PubMed] [Google Scholar]
- 13.OIE Update on Highly Pathogenic Avian Influenza in Animals: Type H5 and H7. (2013). Available from: http://www.oie.int/animal-health-in-the-world/update-on-avian-influenza/2013/
- 14.Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature (2012) 486(7403):420–8 10.1038/nature10831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell CA, Fonville JM, Brown AEX, Burke DF, Smith DL, James SL, et al. The potential for respiratory droplet-transmissible A/H5N1 influenza virus to evolve in a mammalian host. Science (2012) 336(6088):1541–7 10.1126/science.1222526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu H, Wang D, Kelvin DJ, Li L, Zheng Z, Yoon SW, et al. Infectivity, transmission, and pathology of human H7N9 influenza in ferrets and pigs. Science (2013) 341(6142):183–6 10.1126/science.1239844 [DOI] [PubMed] [Google Scholar]
- 17.Alexander DJ. A review of avian influenza in different bird species. Vet Microbiol (2000) 74(1–2):3–13 10.1016/S0378-1135(00)00160-7 [DOI] [PubMed] [Google Scholar]
- 18.Webby RJ, Webster RG. Emergence of influenza A viruses. Philos Trans R Soc Lond B Biol Sci (2001) 356(1416):1817–28 10.1098/rstb.2001.0997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert M, Pfeiffer DU. Risk factor modelling of the spatio-temporal patterns of highly pathogenic avian influenza (HPAIV) H5N1: a review. Spat Spatiotemporal Epidemiol (2012) 3(3):173–83 10.1016/j.sste [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takekawa JY, Prosser DJ, Newman SH, Muzaffar SB, Hill NJ, Yan B, et al. Victims and vectors: highly pathogenic H5N1 and the ecology of water birds. Avian Biol Res (2010) 3:51–73 10.3184/175815510X12737339356701 [DOI] [Google Scholar]
- 21.FAO H5N1 HPAI in different species. Understanding Avian Influenza. (Chap. 3). Animal Production and Health Manual. Rome: FAO; (2007). p. 1–15. [Google Scholar]
- 22.Bailey NT. Infectious Diseases of Humans: Dynamics and Control. 2nd ed Oxford: Hafner Press, MacMillan Publishing Co. (1975). [Google Scholar]
- 23.Roche B, Lebarbenchon C, Gauthier-Clerc M, Chang CM, Thomas F, Renaud F, et al. Water-borne transmission drives avian influenza dynamics in wild birds: the case of the 2005-2006 epidemics in the Camargue area. Infect Genet Evol (2009) 9(5):800–5 10.1016/j.meegid.2009.04.009 [DOI] [PubMed] [Google Scholar]
- 24.Rohani P, Breban R, Stallknecht DE, Drake JM. Environmental transmission of low pathogenicity avian influenza viruses and its implications for pathogen invasion. Proc Natl Acad Sci U S A (2009) 106(25):10365–9 10.1073/pnas.0809026106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prosser DJ, Wu J, Ellis EC, Gale F, Van Boeckel TP, Wint W, et al. Modelling the distribution of chickens, ducks, and geese in China. Agric Ecosyst Environ (2011) 141:381–9 10.1016/j.agee.2011.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark L, Hall J. Avian influenza in wild birds: status as reservoirs, and risks to humans and agriculture. Ornithol Monogr (2006) 2006(60):3–29 10.2307/40166825 [DOI] [Google Scholar]
- 27.FAO Biosecurity for highly pathogenic avian influenza. Animal Production and Health Manual No. 165. Rome: FAO; (2008). 80 p [Google Scholar]
- 28.Prosser DJ. Wild Birds and Emerging Diseases: Modeling Avian Influenza Transmission Risk Between Domestic and Wild Birds in China [Doctoral Dissertation]. College Park (MD): University of Maryland; (2012). [Google Scholar]
- 29.Brewer CA. Basic mapping principles for visualizing cancer data using geographic information systems (GIS). Am J Prev Med (2006) 30(2):S25–36 10.1016/j.amepre.2005.09.007 [DOI] [PubMed] [Google Scholar]
- 30.Brewer CA, Pickle L. Evaluation of methods for classifying epidemiological data on choropleth maps in series. Ann Assoc Am Geogr (2002) 92(4):662–81 10.1111/1467-8306.00310 [DOI] [Google Scholar]
- 31.Morgan MG, Henrion M. Uncertainty: A Guide to Dealing with Uncertainty in Quantitative Risk and Policy Analysis. New York, NY: Cambridge University Press; (1990). 332 p. [Google Scholar]
- 32.Kroese DP, Taimre T, Botev ZI. Handbook of Monte Carlo Methods. Hoboken, NJ: John Wiley & Sons, Inc; (2010). [Google Scholar]
- 33.Delignette-Muller ML, Pouillot R, Denis JB, Dutang C. Use of the Package Fitdistrplus to Specify a Distribution from Non-Censored or Censored Data. (2011) [cited 2012 Jun]. Available from: http://cran.r-project.org/web/packages/fitdistrplus/index.html [Google Scholar]
- 34.R Core Development Team R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; (2012). Available from: http://www.R-project.org/ [Google Scholar]
- 35.Pouillot R, Delingnette-Muller ML, Kelly DL, Denis JB. The mc2d Package. (2010). Available from: http//riskassessment.r-forge.r-project.org/ [Google Scholar]
- 36.Burmaster DE, Anderson PD. Principles of good practice for the use of Monte Carlo techniques in human health and ecological risk assessments. Risk Anal (1994) 14(4):477–81 10.1111/j.1539-6924.1994.tb00265.x [DOI] [PubMed] [Google Scholar]
- 37.Liu R, Duvvuri VRSK, Wu J. Spread pattern formation of H5N1 avian influenza and its implications for control strategies. Math Model Nat Phenom (2008) 3(7):161–79 10.1051/mmnp:2008048 [DOI] [Google Scholar]
- 38.Fang LQ, de VlasSJ, Liang S, Looman CW, Gong P, Xu B, et al. Environmental factors contributing to the spread of H5N1 avian influenza in mainland china. PLoS ONE (2008) 3(5):e2268. 10.1371/journal.pone.0002268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilbert M, Xiao X, Pfeiffer DU, Epprecht M, Boles S, Czarnecki C, et al. Mapping H5N1 highly pathogenic avian influenza risk in Southeast Asia. Proc Natl Acad Sci U S A (2008) 105(12):4769–74 10.1073/pnas.0710581105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin V, Pfeiffer DU, Zhou X, Xiao X, Prosser DJ, Guo F, et al. Spatial distribution and risk factors of highly pathogenic avian influenza (HPAI) H5N1 in China. PLoS Pathog (2011) 7(3):e1001308. 10.1371/journal.ppat.1001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfeiffer DU, Minh PQ, Martin V, Epprecht M, Otte MJ. An analysis of the spatial and temporal patterns of highly pathogenic avian influenza occurrence in Vietnam using national surveillance data. Vet J (2007) 174(2):302–9 10.1016/j.tvjl.2007.05.010 [DOI] [PubMed] [Google Scholar]
- 42.Tiensin T, Ahmed SS, Rojanasthien S, Songserm T, Ratanakorn P, Chaichoun K, et al. Ecologic risk factor investigation of clusters of avian influenza A (H5N1) virus infection in Thailand. J Infect Dis (2009) 199(12):1735–43 10.1086/599207 [DOI] [PubMed] [Google Scholar]
- 43.Cao L, Barter M, Lei G. New Anatidae population estimates for eastern China: implications for current flyway estimates. Biol Conserv (2008) 141(9):2301–9 10.1016/j.biocon.2008.06.022 [DOI] [Google Scholar]
- 44.Delany S, Scott D. Waterbird Population Estimates: Fourth Edition. 4th ed Wageningen: Wetlands International; (2006). 239 p [Google Scholar]
- 45.Gaidet N, Dodman T, Caron A, Balança G, Desvaux S, Goutard F, et al. Influenza surveillance in wild birds in Eastern Europe, the Middle East, and Africa: preliminary results from an ongoing FAO-led survey. J Wildl Dis (2007) 43(3):S22–8 [Google Scholar]
- 46.Hesterberg U, Harris K, Stroud D, Guberti V, Busani L, Pittman M, et al. Avian influenza surveillance in wild birds in the European Union in 2006. Influenza Other Respi Viruses (2009) 3(1):1–14 10.1111/j.1750-2659.2008.00058.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kou Z, Li Y, Yin Z, Guo S, Wang M, Gao X, et al. The survey of H5N1 flu virus in wild birds in 14 provinces of China from 2004 to 2007. PLoS ONE (2009) 4(9):e6926. 10.1371/journal.pone.0006926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Munster VJ, Baas C, Lexmond P, Waldenström J, Wallensten A, Fransson T, et al. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog (2007) 3(5):e61. 10.1371/journal.ppat.0030061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olsen B, Munster VJ, Wallensten A, Waldenström J, Osterhaus AD, Fouchier RA. Global patterns of influenza a virus in wild birds. Science (2006) 312(5772):384–8 10.1126/science.1122438 [DOI] [PubMed] [Google Scholar]
- 50.Jeong OM, Kim MC, Kim MJ, Kang HM, Kim HR, Kim YJ, et al. Experimental infection of chickens, ducks and quails with the highly pathogenic H5N1 avian influenza virus. J Vet Sci (2009) 10(1):53–60 10.4142/jvs.2009.10.1.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shortridge KF, Zhou NN, Guan Y, Gao P, Ito T, Kawaoka Y, et al. Characterization of avian H5N1 influenza viruses from poultry in Hong Kong. Virology (1998) 252(2):331–42 10.1006/viro.1998.9488 [DOI] [PubMed] [Google Scholar]
- 52.Yu Z, Song Y, Zhou H, Xu X, Hu Q, Wu H, et al. Avian influenza (H5N1) virus in waterfowl and chickens, Central China. Emerg Infect Dis (2007) 13(5):772–5 10.3201/eid1305.061209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen H, Deng G, Li Z, Tian G, Li Y, Jiao P, et al. The evolution of H5N1 influenza viruses in ducks in southern China. Proc Natl Acad Sci U S A (2004) 101(28):10452–7 10.1073/pnas.0403212101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perkins LE, Swayne DE. Pathogenicity of a Hong Kong-origin H5N1 highly pathogenic avian influenza virus for emus, geese, ducks, and pigeons. Avian Dis (2002) 46(1):53–63 10.1637/0005-2086(2002)046[0053:POAHKO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 55.Phuong DQ, Dung NT, Jørgensen PH, Handberg KJ, Vinh NT, Christensen JP. Susceptibility of Muscovy (Cairina moschata) and mallard ducks (Anas platyrhynchos) to experimental infections by different genotypes of H5N1 avian influenza viruses. Vet Microbiol (2011) 148(2–4):168–74 10.1016/j.vetmic.2010.09.007 [DOI] [PubMed] [Google Scholar]
- 56.Sturm-Ramirez KM, Ellis T, Bousfield B, Bissett L, Dyrting K, Rehg JE, et al. Reemerging H5N1 influenza viruses in Hong Kong in 2002 are highly pathogenic to ducks. J Virol (2004) 78(9):4892–901 10.1128/JVI.78.9.4892-4901.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sturm-Ramirez KM, Hulse-Post DJ, Govorkova EA, Humberd J, Seiler P, Puthavathana P, et al. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J Virol (2005) 79(17):11269–79 10.1128/JVI.79.17.11269-11279.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brown JD, Stallknecht DE, Swayne DE. Experimental infection of swans and geese with highly pathogenic avian influenza virus (H5N1) of Asian lineage. Emerg Infect Dis (2008) 14(1):136–42 10.3201/eid1401.070740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ito T, Okazaki K, Kawaoka Y, Takada A, Webster RG, Kida H. Perpetuation of influenza-A viruses in Alaskan waterfowl reservoirs. Arch Virol (1995) 140(7):1163–72 10.1007/BF01322743 [DOI] [PubMed] [Google Scholar]
- 60.Lu H, Castro AE. Evaluation of the infectivity, length of infection, and immune response of a low-pathogenicity H7N2 avian influenza virus in specific-pathogen-free chickens. Avian Dis (2004) 48(2):263–70 10.1637/7064 [DOI] [PubMed] [Google Scholar]