Abstract
We produced three highly purified recombinant antigens rLipL32, rLipL41, and rLigA-Rep (leptospiral immunoglobulin-like A repeat region) for the detection of Leptospira-specific antibodies in an enzyme-linked immunosorbent assay (ELISA). The performance of these recombinant antigens was evaluated using 121 human sera. Among them, 63 sera were microscopic agglutination test (MAT)-confirmed positive sera from febrile patients in Peru, 22 sera were indigenous MAT-negative febrile patient sera, and 36 sera were from patients with other febrile diseases from Southeast Asia, where leptospirosis is also endemic. Combining the results of immunoglobulin M (IgM) and IgG detection from these three antigens, the overall sensitivity is close to 90% based on the MAT. These results suggest that an ELISA using multiple recombinant antigens may be used as an alternative method for the detection of Leptospira-specific antibodies.
Introduction
Leptospirosis is caused by spirochaetes of the genus Leptospira. It is considered to be the most widespread bacterial zoonotic disease in the world and recognized as an emerging infectious disease.1,2 More than 1.7 million cases of severe leptospirosis are reported each year, with case mortality rate about 10%.3 It is most common in developing countries, particularly in the Caribbean, Latin America, the Indian subcontinent, Southeast Asia, and Oceania, although locally acquired cases in industrialized countries are well described.1,4–13 In recent years, a new trend in human leptospirosis outbreaks related to outdoor recreational activities or natural disasters has been observed.14–22 Symptoms of leptospirosis are non-specific and may be easily confused with other febrile illnesses, such as dengue or malaria, which require different treatment regimens. Therefore, timely diagnosis is essential, because antibiotic therapy provides the greatest benefit when initiated early in the course of illness.23,24
Currently, the microscopic agglutination test (MAT) is the standard serological method for the diagnosis of leptospirosis, in which whole cell antigens representing different serogroups of leptospirosis are mixed with serum samples and examined by dark-field microscopy for agglutination.25 It is technically complex and time consuming, because the method requires many serovars of Leptospira to be cultured.26 Culturing Leptospira to obtain the whole cell antigen is particularly labor intensive and requires special precautions to prevent infection of laboratory staff. Because MAT relies on the detection of antibodies to leptospiral antigens, it is limited by the low sensitivity when acute serum samples were tested.27 Commercial assays based on Leptospira whole cell antigen are available in rapid formats amenable for point-of-care use. However, field evaluations of these assays suggested low sensitivities (39–72%) during the early phase of infection.28–36 Recently, the whole genome sequences of several serovars of Leptospira were reported.37–40 Hundreds of genes encoding surface exposed lipoproteins and outer membrane proteins have now been identified as candidates for the development of rapid diagnostics of leptospirosis. Among them, LipL32, LipL41, and leptospiral immunoglobulin-like A (LigA) proteins may be excellent target antigens for such a rapid diagnostic test. All three proteins are present only in pathogenic Leptospira species and have been shown to be exposed on the Leptospira cell surface.41–44 LipL32 and LipL41 are also the most abundant cell surface proteins, and all three proteins are highly conserved across a broad range of pathogenic leptospiral serovars.42,45,46 Lastly, these three antigens have been shown to be immunogenic, and they have shown promise as target antigens for serologic diagnosis.47,48
In this study, we cloned the genes of LipL32, LipL41, and part of the repeat region in LigA antigens into Escherichia coli expression vectors. The expressed recombinant proteins were then purified by affinity chromatography on nickel column. A panel of 85 human sera from Peru (63 MAT positive and 22 MAT negative sera) was used to evaluate the potential of these recombinant antigens to be used as diagnostic reagents in enzyme-linked immunosorbent assays (ELISAs).
Materials and Methods
Bacterial strains and vectors.
The genomic DNA of L. interrogans serovar Copenhageni strain Fiocruz L1-130 (ATCC, Manassas, VA) was used as the template for cloning of all recombinant proteins. Escherichia coli Top10 (Life Technologies, Grand Island, NY) was used for general cloning. The cloned genes were inserted into pET28a (EMD Millipore, Billerica, MA) for the expression of recombinant proteins in E. coli BL21 (DE3) (Life Technologies, Grand Island, NY) under the control of phage T7 lac promoter.49
Recombinant antigen preparation.
Cloning of the gene coding for LipL32, LipL41, and the repeat region of LigA proteins into the expression vector pET28a.
A primer pair (LipL32f [5′-GGTGGTCATATGGGTCTGCCAAGCCTAAAAAGC-3′] and LipL32r [5′-CCGCTCGAGCTTAGTCGCGTCAGAAGCAGC-3′]) was designed by using the nucleotide sequence of the open reading frame for LipL32 from strain L1-130 (GenBank accession no. AF245281.1). The coding region of full length protein minus predicted signal peptide for LipL32 (amino acids 25–272) was amplified by polymerase chain reaction (PCR) using genomic DNA isolated from L. interrogans strain L1-130 as the template. The primer pair for LipL41 (LipL41f [5′-GGTGGTCATATGGCTACAGTCGATGTAGAATATCC-3′] and LipL41r [5′-CCGCTCGAGCTTTGCGTTGCTTTCATCAACG-3′]) was designed for the coding region of full length protein minus predicted signal peptide for LipL41 (amino acids 22–355), and primer pair for LigA (LigAf [5′-AAGAATCATATGGCAGCCTTAGTTTCTATTTCTGT-3′] and LigAr [5′-CGCCTCGAGAATATCCGTATTAGAGGAATTCCA-3′]) was designed for the coding region of amino acids 312–630. Each PCR product was digested with NdeI and XhoI and ligated into the expression vector pET28a. The resulting plasmids contained a sequence coding His tag at both N and C termini of LipL32, LipL41, and the repeat region of LigA. Top10 competent cells were transformed with the ligation mixture, and colonies were screened for the presence of inserts with the correct size. The final sequences were confirmed by DNA sequencing of the resulting plasmid.
Expression and purification of the recombinant LipL32, LipL41, and LigA proteins.
E. coli BL21 (DE3) was transformed with plasmids carrying the LipL32, LipL41, or LigA insert. The recombinant E. coli colony with high expression levels of the desired protein was cultured overnight in Overnight Express Medium TB (EMD Millipore, Billerica, MA) in the presence of kanamycin (50 mg/L) at 37°C with shaking at 200 rpm. Cell pellets from 500 mL cultures were resuspended in 20 mL buffer A of 20 mM Tris·HCl, pH 8.0, and 0.5 M NaCl after centrifugation. Cells were ruptured by sonication (Ultrasonic Liquid Processor Model VirSonic 475; VIRTIS Company, Gardiner, NY) five times at setting 3 for 10 seconds each time, with cooling on ice for 1 minute between each sonication. Cell extract was centrifuged at 10,000 × g for 30 minutes at 4°C in a Thermo centrifuge (model IEC MultiRF; Thermo Scientific, Waltham, MA). The recombinant LipL32 (rLipL32) and recombinant LigA (rLigA-Rep) were expressed in soluble form, but recombinant LipL41 (rLipL41) was expressed as an inclusion body. For the purification of rLipL32 or rLigA-Rep, the cell lysate supernatant was applied onto a 3 mL nickel column (Ni-NTA) equilibrated with 20 mM Tris, pH 8.0, 0.5 M NaCl, and 10 mM imidazole (Hisbind buffer). The column was washed extensively with 30 mL Hisbind buffer. The recombinant protein was eluted from the column in a step gradient of 3 mL of 25, 50, 100, 200, 400, and 600 mM imidazole in Hisbind buffer. Fractions of 3 mL were collected and analyzed by sodium dodecyl sulfate - polyacrylamide gel electrophoresis (SDS-PAGE) for purity. Peak fractions were pooled and stored at −20°C until use. For the purification of rLipL41, the pellets of inclusion body were resuspended in Hisbind buffer containing 8 M urea by vortexing, placed on a shaker at room temperature for an additional 10 minutes, and centrifuged for 30 minutes at 10,000 × g. The supernatant was applied onto a 3 mL nickel column (Ni-NTA) equilibrated with the same buffer containing 8 M urea. The column was washed extensively with 30 mL Hisbind buffer containing 8 M urea. The rLipL41 was eluted from the column as described above for rLipL32 or rLigA-Rep in the presence of 8 M urea. Peak fractions were pooled for refolding.
Refolding of rLipL41 protein.
Refolding of rLipL41 protein in 8 M urea in Hisbind buffer was achieved by sequential dialysis with decreasing concentration of urea in buffer A containing 1 mM (ethylenedinitrilo) tetraacetic acid (EDTA) and 1 mM dithiothreitol (DTT) and finally, buffer A containing 1 mM EDTA and 1 mM DTT without urea. Before dialysis, the protein concentration of the eluates from the nickel column was adjusted to less than 1 mg/mL and dialyzed against 10 volumes of 6 M urea in buffer A for 60 minutes at 4°C. The same procedure was repeated with 4, 2, and 1 M urea in buffer A. The final dialysis was in 10 volumes of the eluates of buffer A without urea with one initial change of buffer at 60 minutes and finally, overnight at 4°C. The refolded rLipL41 was stored at −20°C.
ELISA.
ELISA was used in the detection of immunoglobulin M (IgM) and IgG antibodies against rLipL32, rLipL41, and rLigA-Rep. Different amounts (0.15, 0.3, 0.45, and 0.6 μg/well) of each antigen were used to coat the ELISA plate to determine the optimum amount for coating. The optimal amount was determined to be 0.3 μg/well, because 0.45 and 0.6 μg did not increase the signal. Microtiter plates (96 well) were coated for 40 hours at 4°C with recombinant antigen diluted in phosphate buffered saline (PBS) and blocked with 10% skim milk in PBS for 1 hour. Patient sera diluted 1:100 in PBS with 5% skim milk were then added to the plate, incubated for 1 hour at room temperature, and washed three times for 10 minutes each with 0.1% Tween-20 in PBS. Peroxidase-conjugated rabbit anti-human IgG (Santa Cruz Biotechnology, Dallas, TX) at 1:4,000-dilution or anti-human IgM (Dako, Carpinteria, CA) at 1:1,000 dilution was added. After 1 hour of incubation at room temperature, the plates were washed as previously described before the addition of the 2,2′-azinobis (3-ethylbenzthiazoline-6-sulfonate) (ABTS) substrate (Kirkegaard & Perry, Gaithersburg, MD). Optical density at 405 nm (OD405) was measured after 30 minutes of incubation at room temperature in a plate reader (Molecular Devices, Sunnyvale, CA).
Human sera.
In total, 85 sera collected from individual febrile patients (63 MAT positive sera with titers greater than 100 against different Leptospira serovars [Bataviae, Bratislava, Icterohaemorrhagiae, and Varillal] and 22 MAT negative sera) from the Iquitos area of Peru were received from Naval Medical Research Unit 6, Lima, Peru. The 22 negative sera were used as local negative controls to calculate cutoff values for the ELISA. The sample collection date after onset of fever is listed in Table 1; 36 archived sera from patients residing in the leptospirosis-endemic area of Southeast Asia with other febrile illness (9 patients with scrub typhus, 9 patients with murine typhus, 9 patients with spotted fever-type rickettsioses, and 9 patients with Q fever) were used as other control specimens.
Table 1.
Summary of the sample collection date after onset of fever
| Group | Leptospirosis | Local control |
|---|---|---|
| Number of sera | 63 | 22 |
| Number of sera with day after onset of fever | 60 | 20 |
| Range (days) | 0–44 | 0–71 |
| Median (days) | 16 | 5 |
| Number of sera between 0 and 7 days | 22 | 14 |
| Number of sera between 8 and 20 days | 17 | 2 |
| Number of sera > 21 days | 21 | 4 |
Ethics.
The study was approved by the Naval Medical Research Center Institutional Review Board (Case Number PJT61 and Protocol NMRCD.2000.0006) in compliance with all applicable federal regulations governing the protection of human subjects. Informed consent was obtained from all study participants.
Results
Production of rLipL32, rLipL41, and rLigA-Rep.
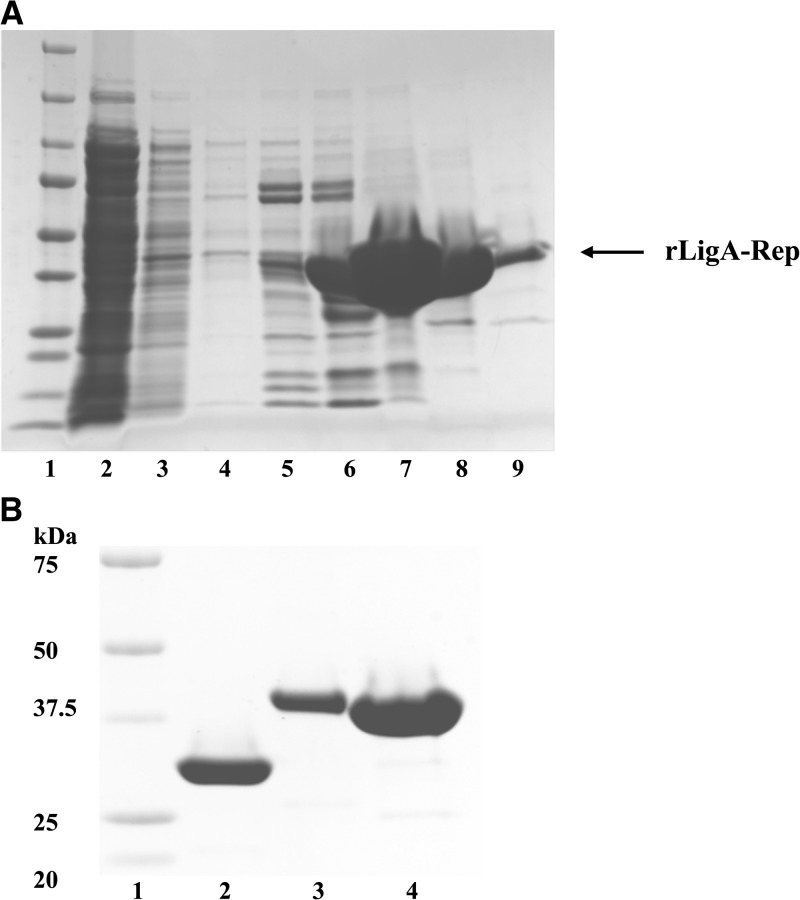
LipL32 and LipL41 are highly conserved immunodominant cell surface proteins among pathogenic leptospiral serovars. The Lig protein family has three members, LigA, -B, and -C, all with high molecular mass.44,47,50 These Lig proteins have 12–13 tandem bacterial Ig-like repeat domains. The first six domains from the amino terminus are highly conserved among different strains.46 The coding region of amino acids 312–630 of LigA consists of repeat domains three, four, five, and part of six. It is 98.5% homologous to the same domains from LigB within the same strain. The percentage of DNA sequence identity of LigB among different strains of leptospira ranges from 67.9% to 97.1%.46 The rLipL32 and rLigA-Rep were purified under the native condition, and the rLipL41 was purified under the denatured condition. Fractions collected during different steps of the purification procedure were analyzed by SDS-PAGE (Figure 1A). The rLipL32, rLipL41, and rLigA-Rep were highly purified using nickel column (Figure 1B).
Figure 1.
Purification and characterization of rLipL32, rLipL41, and rLigA-Rep. SDS-PAGE profile for the purification of rLigA-Rep as an example for recombinant protein expressed in BL21 (DE3). The fractions of the nickel column affinity chromatography were analyzed by SDS-PAGE. Lane 1, molecular mass marker; lane 2, starting material before loaded onto the column; lane 3, fraction of wash; lanes 4–9, fractions of the eluate with 25, 50, 100, 200, 400, and 600 mM imidazole, respectively. (B) Purified recombinant proteins. Lane 1, molecular mass marker; lane 2, rLipL32; lane 3, rLipL41; lane 4, rLigA-Rep.
IgM and IgG ELISA results for patient sera.
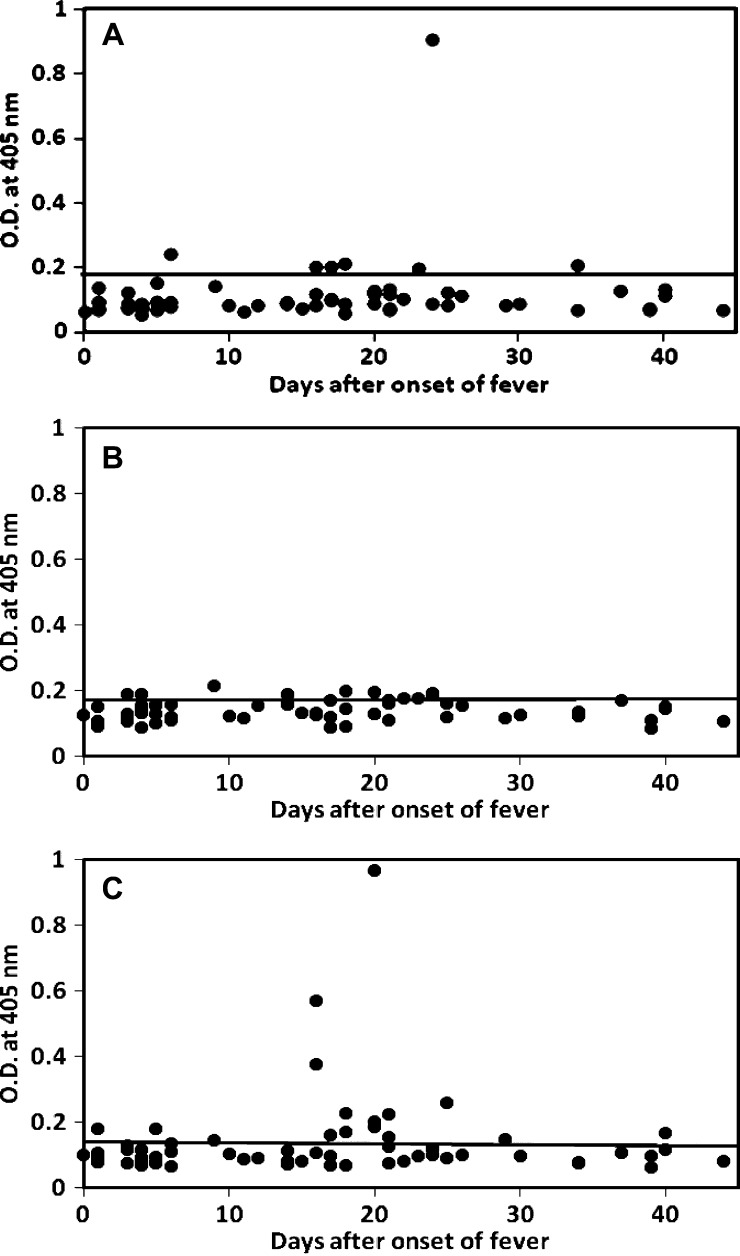
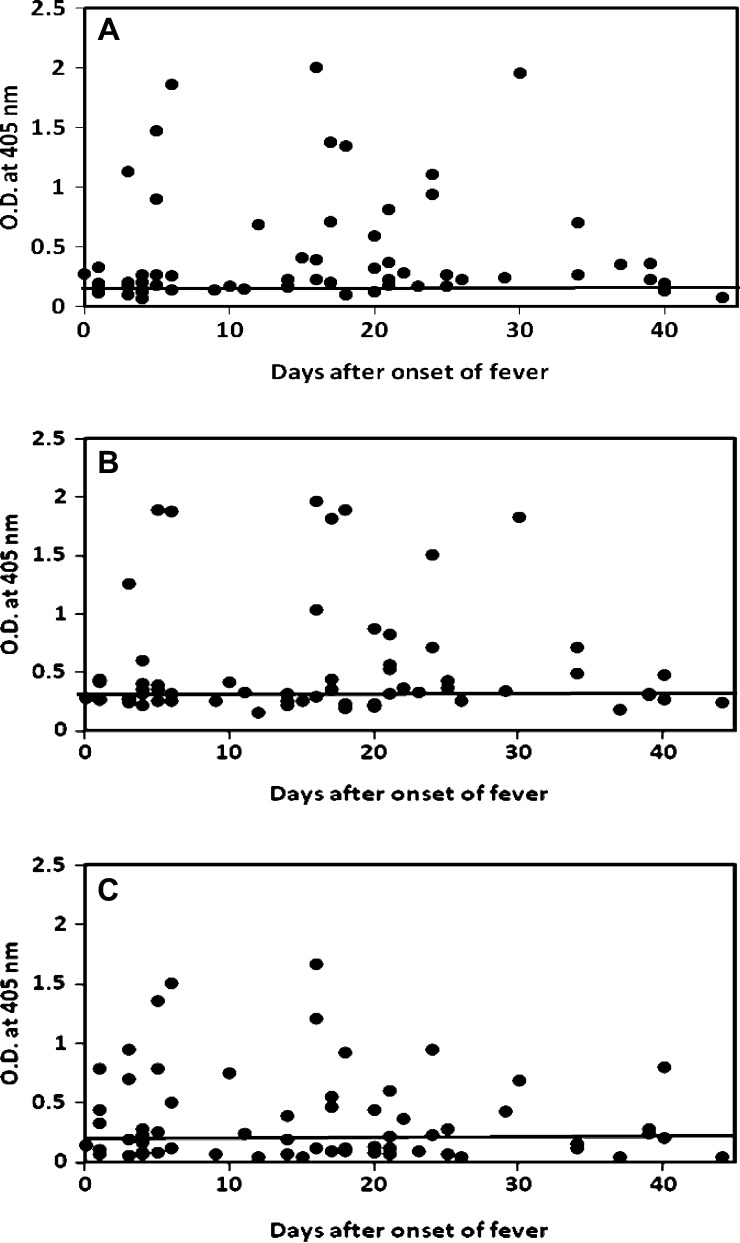
The purified rLipL32 and rLigA-Rep and refolded rLipL41 were used as antigens to detect the presence of IgM and IgG specific for Leptospira in an ELISA. The IgM ELISA results for 63 leptospirosis serum samples against rLipL32, rLipL41, and rLigA-Rep are shown in Figure 2A –C, respectively, and the IgG ELISA results for 63 leptospirosis serum samples against rLipL32, rLipL41, and rLigA-Rep are shown in Figure 3A –C, respectively. Table 2 lists the results of patient samples that had specific IgM and IgG antibodies against each recombinant antigen. There were 7 (11%) and 39 (62%) samples that showed detectable IgM and IgG against rLipL32, respectively; 8 (13%) and 33 (52%) samples that showed detectable IgM and IgG against rLipL41, respectively; and 15 (24%) and 33 (52%) samples that showed detectable IgM and IgG against rLigA-Rep, respectively. Patient samples that had specific IgM and IgG against any one of these antigens were 24 (38%) and 50 (79%), respectively.
Figure 2.
IgM ELISA results for 63 MAT-positive patient samples using recombinant proteins. Specific IgM against (A) rLipL32, (B) rLipL41, and (C) rLigA-Rep were tested. The x axis indicates the number of days after the onset of illness, and the y axis indicates the OD405. For three samples missing the days after onset of fever information, day 16 (medium day) was used to plot. The cutoff values were 0.189 for rLipL32, 0.180 for rLipL41, and 0.142 for rLigA-Rep (mean of 22 negative controls plus 2.3 SDs for 95% confidence level).
Figure 3.
IgG ELISA results for 63 MAT-positive patient samples using recombinant proteins. Specific IgG against (A) rLipL32, (B) rLipL41, and (C) rLigA-Rep were tested. The x axis indicates the number of days after the onset of illness, and the y axis indicates the OD405. For three samples missing the days after onset of fever information, day 16 (medium day) was used to plot. The cutoff values were 0.199 for rLipL32, 0.330 for rLipL41, and 0.207 for rLigA-Rep (mean of 22 negative controls plus 2.3 SDs for 95% confidence level).
Table 2.
Summary of ELISA results for IgM and IgG responses against rLipL32, rLipL41, and rLigA-Rep in patient sera
| Group | No. of sera | No. (%) positive against | |||||||
|---|---|---|---|---|---|---|---|---|---|
| IgM | IgG | ||||||||
| L32* | L4* | Lig* | Com† | L32* | L41* | Lig* | Com† | ||
| Leptospirosis | 63 | 7 (11) | 8 (13) | 15 (24) | 24 (38) | 39 (62) | 33 (52) | 33 (52) | 50 (79) |
| Local control | 22 | 1 (5) | 0 (0) | 1 (5) | 2 (9) | 1 (5) | 0 (0) | 1 (5) | 2 (9) |
| Other control‡ | 36 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
L32, L41, and Lig represent rLipL32, rLipL41, and rLigA-Rep, respectively.
Sera that have antibody levels above cutoff values against at least one antigen.
Sera from patients with other febrile illness (nine patients with scrub typhus, nine patients with murine typhus, nine patients with spotted fever-type rickettsioses, and nine patients with Q fever).
Combining IgM and IgG positive specimens, the sensitivity of the ELISA using one single antigen ranged from 62% to 65% (Table 3) (rLipL32, 65%; rLipL41, 62%; rLigA-Rep, 63%). The samples that had specific antibodies (IgG or IgM) from different combinations of either rLipL32 or rLipL41, rLipL32 or rLigA-Rep, rLipL41 or rLigA-Rep, and rLipL32, or rLipL41, or rLigA-Rep were 53 (84%), 52 (83%), 45 (71%), and 57 (90%), respectively (Table 3). Of 63 MAT positive sera, 60 sera have the known collection date after onset of fever, and of 22 local controls, 20 controls have that information (Table 1). There is a wide range of collection date for the MAT negative local controls, but more than one-half of them were collected in the first 7 days after onset of fever (Table 1). The MAT-positive samples were collected evenly throughout the 44 day period. The IgM responses were only detected in five (23%) samples among those samples collected in the first 7 days after onset of fever. The percentage increased to 53% among the samples collected between days 8 and 20 and dropped back to 43% for the samples collected between 21 and 44 days after onset of fever (Table 4). The IgG responses were detected in 68%, 71%, and 95% samples among those samples collected during days 0–7, 8–20, and 21–44 after onset of fever, respectively (Figures 2 and 3 and Table 4).
Table 3.
Analysis of different combinations of ELISA data using rLipL32, rLipL41, and rLigA-Rep as the antigen (have detectable IgM or IgG)
| Group | No. of sera | No. (%) positive against | ||||||
|---|---|---|---|---|---|---|---|---|
| L32* | L41* | Lig* | L32 + L41* | L32 + Lig* | L41 + Lig* | L32 + L41 + Lig* | ||
| Leptospirosis | 63 | 41 (65) | 39 (62) | 40 (63) | 53 (84) | 52 (83) | 45 (71) | 57 (90) |
| Local control | 22 | 2 (9) | 0 (0) | 2 (9) | 2 (9) | 4 (18) | 2 (9) | 4 (18) |
L32, L41, and Lig represent rLipL32, rLipL41, and rLigA-Rep, respectively.
Table 4.
Antibody responses against rLipL32, rLipL41, and rLigA-Rep according to days after onset of fever in leptospirosis patient sera (only 60 of 63 leptospirosis patient sera have the information for days after onset of fever)
| Range after onset of fever (days) | No. of sera | No. (%) positive against | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IgM | IgG | M + G‡ | ||||||||
| L32* | L4* | Lig* | Com† | L32* | L41* | Lig* | Com† | |||
| 0–7 | 22 | 1 (5) | 2 (9) | 2 (9) | 5 (23) | 12 (55) | 10 (45) | 12 (55) | 15 (68) | 18 (82) |
| 8–20 | 17 | 2 (12) | 4 (24) | 7 (41) | 9 (53) | 9 (53) | 7 (41) | 7 (41) | 12 (71) | 16 (94) |
| 21–44 | 21 | 4 (19) | 2 (10) | 5 (24) | 9 (43) | 15 (71) | 14 (67) | 12 (57) | 20 (95) | 20 (95) |
L32, L41, and Lig represent rLipL32, rLipL41, and rLigA-Rep, respectively.
Sera that have antibody levels above cutoff values against at least one antigen.
Have detectable IgM or IgG.
In 63 MAT-confirmed patient sera used in this study, the sera with the highest agglutinating titers against the serovars Bataviae, Bratislava, Icterohaemorrhagiae, and Varillal were 10, 29, 12, and 12, respectively (Table 5). Among 10 patient sera with the highest serovar Bataviae titers, more samples had IgG antibodies to rLipL32 than rLipL41 or rLigA-Rep. Among 29 patient sera with the highest serovar Bratislava titers, the rLigA-Rep IgG ELISA had the highest number of positive followed by rLipL32 and rLipL41. Among 12 patient sera with the highest serovar Icterohaemorrhagiae titers, 75% of sera had IgG antibodies to rLipL32 or rLigA-Rep, and 58% of sera had IgG antibodies to rLipL41 above the cutoff value. Among 12 patient sera with the highest serovar Varillal titers, 67% of sera had IgG antibodies to rLipL32 or rLipL41, and 33% of sera had IgG antibodies to rLigA-Rep above cutoff value.
Table 5.
Comparison of the IgG responses against rLipL32, rLipL41, and rLigA-Rep in 63 leptospirosis patient sera with primary infecting serovar by ELISA
| Serovar* | No. of sera | No. (%) positive against | |||
|---|---|---|---|---|---|
| rLipL32 | rLipL41 | rLigA-Rep | Com† | ||
| Bataviae | 10 | 7 (70) | 4 (40) | 4 (40) | 8 (80) |
| Bratislava | 29 | 15 (52) | 14 (48) | 16 (55) | 22 (76) |
| Icterohaemorrhagiae | 12 | 9 (75) | 7 (58) | 9 (75) | 10 (83) |
| Varillal | 12 | 8 (67) | 8 (67) | 4 (33) | 10 (83) |
If the sample reacts with more than one serovar by MAT, the serovar with the highest agglutinating titers is listed.
Sera that have IgG against at least one specific antigen.
Discussion
The diagnosis of leptospirosis relies mainly on serological methods. The current serologic gold standard, MAT, is complex and time consuming. The performance of MAT requires knowledge of the prevalent Leptospira strains in a particular region and the maintenance of a large panel of leptospiral cultures.51 The agglutination of Leptospira is thought to be mediated by lipopolysaccharide (LPS)-specific total antibodies, including both IgM and IgG.52 The LPS varies among different serovars with low cross-reactivity; therefore, a large number of different serovars need to be cultured and maintained for agglutination tests, such as MAT. Leptospiral proteins with a high degree of sequence homology among various serovars have a potential advantage compared with serovar-specific LPS antigens. LipL32 and LigA each exhibits 99% amino acid sequence homology across a broad range of pathogenic Leptospira species, whereas a 95% homology has been shown for LipL41.42,46 Recombinant proteins are much easier to prepare and standardize, which lead to better batch to batch consistency compared with leptospiral LPS preparation using culture based methods.
In this study, we produced three recombinant immunogenic antigens, rLipL32, rLipL41, and rLigA-Rep, to explore the feasibility of using them for the detection of Leptospira-specific antibodies in ELISA. All three proteins showed similar overall sensitivity (62–65%) (Table 3) and high specificity individually (greater than 90%) (Table 3). Not every serum had detectable IgM or IgG against these recombinant antigens. Among 63 positive sera, 4, 4, and 11 only had IgM against rLipL32, rLipL41, and rLigA-Rep, respectively, and 10, 3, and 2 only had IgG against rLipL32, rLipL41, and rLigA-Rep, respectively. Of 50 IgG-positive samples, 17 of them were also IgM positive. There were 8, 2, and 1 patient sera that had specific antibodies against only rLipL32, rLipL41, and rLigA-Rep, respectively.
Guerreiro and others52 previously showed, using IgG immunoblotting with whole cell antigens, that LipL32 was recognized by 37% and 84% of acute and convalescent phase sera, respectively, and that LipL41 was recognized by 21% and 36% of acute and convalescent phase sera, respectively. Similar findings were also reported by Flannery and others53 using recombinant antigens LipL32 and LipL41 in IgG ELISA.53 The recombinant LipL32 was recognized by 56% and 94% of acute and convalescent phase sera, and LipL41 was recognized by 24% and 44% of acute and convalescent phase sera. In both studies, the majority of the patient samples had the highest agglutinating titers to reference strains of the serovar Icterohaemorrhagiae. Table 4 showed that rLipL32 was recognized by 55% and 71% of days 0–7 and 21–44 sera, respectively, and that rLipL41 was recognized by 45% and 67% of days 0–7 and 21–44 sera, respectively. Our data indicated that, even with highly conserved proteins (such as LipL32 and LipL41), one antigen was unable to detect antibodies in all sera from different serovar infections. Our recombinant proteins have been derived from a strain of L. interrogans. Therefore, the test results were reasonably good, because the majority of this panel consists of infections with serovars that belong to L. interrogans. However, data from 15 Thai samples showed that these three recombinant antigens could also detect antibodies in samples with the highest agglutinating titers against other serovars, such as Cynopteri (L. kirschneri), Panama (L. kirschneri), and Sejroe (L. borgpetersenii) (Chen H-W and others, unpublished results). This result strongly suggests the broadness of the assay using the combination of conserved antigens.
All three antigens showed robust IgG and weak IgM interactions to the sera of infected patients. They could be the results of previous exposures to Leptospira infections, which is quite common in a highly endemic area. Several studies have documented IgG immunoblot reactivity to leptospiral proteins during acute-phase illness in the absence of specific IgM.52–55 The predominant humoral responses during acute-phase infection were thought to be IgM antibodies directed primarily against carbohydrate epitopes.56,57 These findings suggest that early host immune response to Leptospira infection is characterized by both IgM and IgG specific for different moieties, similar to processes observed in Borrelia and Treponema infections.58–60 As shown in Table 4, more samples had detectable IgG than IgM in all three stages. There was no indication of early detection of IgM in the samples that we evaluated. The percentage of samples that had detectable IgG against recombinant antigens increased from early (68%; days 0–7) to late stage (95%; days 21–44). The purpose of the original study for febrile patients was to determine the seroprevalence of different diseases, and therefore, the outcomes from these leptospirosis patients were not followed.
The combination of results from rLipL32 and rLipL41 only gave a sensitivity of 84% and a specificity of 91% (Table 3). The combination of ELISA results from rLipL32, rLipL41, and rLigA-Rep showed an increase in the assay sensitivity to 90%, whereas the specificity dropped to 82% in the local control (Table 3). The low specificity is the cumulative non-specific interactions of rLipL32 and rLigA-Rep. No false positive was found among the local control against rLipL41 in ELISA. Both rLipL32 and rLigA-Rep had two false positives among the local control. To further improve the performance of the ELISA, we plan to include additional immunogens (such as LipL21 and OmpL1) to increase the assay's sensitivity. The cross-reactive epitopes, which cause false positives, may be identified by pepscan and subsequently truncated from LipL32 or LigA to improve the assay's specificity. This study is a pilot study to show the potential of using multiple recombinant antigens for the detection of Leptospira-specific antibodies. In addition, a larger number of samples with the information of the collection date after onset of fever and paired acute and convalescent phase samples will be needed for future study.
ACKNOWLEDGMENTS
We are grateful to all the participants in the study and all the medical and administrative staff who assisted in the collection of patient samples.
Disclaimer: The views expressed in this article are the views of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, the Department of Defense, or the US Government. E.S.H., C.G., E.C., E.H., R.M., D.H.T., T.J.K., and W.-M.C. are employees of the US Government. This work was prepared as part of their official duties. Title 17 U.S.C. §105 provides that “Copyright protection under this title is not available for any work of the United States Government.”
Footnotes
Financial support: This research was supported by the Naval Medical Research Center (research work unit 6000.RAD1.J.A0310) and the US Department of Defense Global Emerging Infections Systems Research Program (work unit number: 847705.82000.25GB.B0016).
Authors' addresses: Hua-Wei Chen, Zhiwen Zhang, Eric Hall, Tadeusz J. Kochel, and Wei-Mei Ching, Naval Medical Research Center, Silver Spring, MD, E-mails: Hua-Wei.Chen@med.navy.mil, Zhiwen.Zhang@med.navy.mil, Eric.Hall@med.navy.mil, Tad.Kochel@med.navy.mil, and WeiMei.Ching@med.navy.mil. Eric S. Halsey, Carolina Guevara, Enrique Canal, and Drake H. Tilley, Naval Medical Research Unit No. 6, Lima, Peru, E-mails: Eric.Halsey@med.navy.mil, Carolina.Guevara@med.navy.mil, Enrique.Canal@med.navy.mil, and Drake.Tilley@med.navy.mil. Ryan Maves, Naval Medical Center San Diego, San Diego, CA, E-mail: Ryan.Maves@med.navy.mil.
References
- 1.Pappas G, Papadimitriou P, Siozopoulou V, Christou L, Akritidis N. The globalization of leptospirosis: worldwide incidence trends. Int J Infect Dis. 2008;12:351–357. doi: 10.1016/j.ijid.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartskeerl RA. Molecular Detection of Human Bacterial Pathogens. Boca Raton, FL: CRC Press Taylor and Francis Group; 2012. Leptospira; pp. 1169–1188. [Google Scholar]
- 4.Cruz LS, Vargas R, Lopes AA. Leptospirosis: a worldwide resurgent zoonosis and important cause of acute renal failure and death in developing nations. Ethn Dis. 2009;19:37–41. [PubMed] [Google Scholar]
- 5.Sethi S, Sharma N, Kakkar N, Taneja J, Chatterjee SS, Banga SS, Sharma M. Increasing trends of leptospirosis in northern India: a clinico-epidemiological study. PLoS Negl Trop Dis. 2010;4:579. doi: 10.1371/journal.pntd.0000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Victoriano AF, Smythe LD, Gloriani-Barzaga N, Cavinta LL, Kasai T, Limpakarnjanarat K, Ong BL, Gongal G, Hall J, Coulombe CA, Yanagihara Y, Yoshida S, Adler B. Leptospirosis in the Asia Pacific region. BMC Infect Dis. 2009;9:147. doi: 10.1186/1471-2334-9-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim VK. Leptospirosis: a re-emerging infection. Malays J Pathol. 2011;33:1–5. [PubMed] [Google Scholar]
- 8.Reller ME, Bodinayake C, Nagahawatte A, Devasiri V, Kodikara-Arachichi W, Strouse JJ, Flom JE, Dumler JS, Woods CW. Leptospirosis as frequent cause of acute febrile illness in southern Sri Lanka. Emerg Infect Dis. 2011;17:1678–1684. doi: 10.3201/eid1709.100915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourhy P, Collet L, Lernout T, Zinini F, Hartskeerl RA, van der Linden H, Thiberge JM, Diancourt L, Brisse S, Giry C, Pettinelli F, Picardeau M. Human leptospira isolates circulating in Mayotte (Indian Ocean) have unique serological and molecular features. J Clin Microbiol. 2012;50:307–311. doi: 10.1128/JCM.05931-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fonzar UJ, Langoni H. Geographic analysis on the occurrence of human and canine leptospirosis in the city of Maringá, state of Paraná, Brazil. Rev Soc Bras Med Trop. 2012;45:100–105. doi: 10.1590/s0037-86822012000100019. [DOI] [PubMed] [Google Scholar]
- 11.Lau CL, Dobson AJ, Smythe LD, Fearnley EJ, Skelly C, Clements AC, Craig SB, Fuimaono SD, Weinstein P. Leptospirosis in American Samoa 2010: epidemiology, environmental drivers, and the management of emergence. Am J Trop Med Hyg. 2012;86:309–319. doi: 10.4269/ajtmh.2012.11-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C, Wang H, Yan J. Leptospirosis prevalence in Chinese populations in the last two decades. Microbes Infect. 2012;14:317–323. doi: 10.1016/j.micinf.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Vinetz JM, Glass GE, Flexner CE, Mueller P, Kaslow DC. Sporadic urban leptospirosis. Ann Intern Med. 1996;125:794–798. doi: 10.7326/0003-4819-125-10-199611150-00002. [DOI] [PubMed] [Google Scholar]
- 14.Sejvar J, Bancroft E, Winthrop K, Bettinger J, Bajani M, Bragg S, Kaiser R, Marano N, Popovic T, Tappero J, Ashford D, Mascola L, Vugia D, Perkins B, Rosenstein N. Leptospirosis in “Eco-Challenge” atheletes, Malaysian Borneo, 2000. Emerg Infect Dis. 2003;9:702–707. doi: 10.3201/eid0906.020751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan J, Borstein SL, Karpati AM, Bruce M, Bolin CA, Austin CC, Woods CW, Lingappa J, Langkop C, Davis B, Graham DR, Proctor M, Ashford DA, Bajani M, Bragg SL, Shutt K, Perkins BA, Tappero JW. Outbreak of leptospirosis among triathlon participants and community residents in Springfield, Illinois, 1998. Clin Infect Dis. 2002;34:1593–1599. doi: 10.1086/340615. [DOI] [PubMed] [Google Scholar]
- 16.Centers for Disease Control and Prevention Outbreak of leptospirosis among white-water rafters—Costa Rica, 1996. MMWR Morb Mortal Wkly Rep. 1997;46:577–579. [PubMed] [Google Scholar]
- 17.Boland M, Sayers G, Cileman T, Bergin C, Sheehan N, Creamer E, O'Connell M, Jones L, Zochowski W. A cluster of leptospirosis cases in canoeists following a competition on the River Liffey. Epidemiol Infect. 2004;132:195–200. doi: 10.1017/s0950268803001596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radl C, Müller M, Revilla-Fernandez S, Karner-Zuser S, de Martin A, Schauer U, Karner F, Stanek G, Balcke P, Hallas A, Frank H, Fürnschlief A, Erhart F, Allerberger F. Outbreak of leptospirosis among triathlon participants in Langau, Austria, 2010. Wien Klin Wochenschr. 2011;123:751–755. doi: 10.1007/s00508-011-0100-2. [DOI] [PubMed] [Google Scholar]
- 19.Kouadio IK, Aljunid S, Kamigaki T, Hammad K, Oshitani H. Infectious diseases following natural disasters: prevention and control measures. Expert Rev Anti Infect Ther. 2012;10:95–104. doi: 10.1586/eri.11.155. [DOI] [PubMed] [Google Scholar]
- 20.Amilasan AS, Ujiie M, Suzuki M, Salva E, Belo MC, Koizumi N, Yoshimatsu K, Schmidt WP, Marte S, Dimaano EM, Villarama JB, Ariyoshi K. Outbreak of leptospirosis after flood, the Philippines, 2009. Emerg Infect Dis. 2012;18:91–94. doi: 10.3201/eid1801.101892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin CY, Chiu NC, Lee CM. Leptospirosis after typhoon. Am J Trop Med Hyg. 2012;86:187–188. doi: 10.4269/ajtmh.2012.11-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith JK, Young MM, Wilson KL, Craig SB. Leptospirosis following a major flood in Central Queensland, Australia. Epidemiol Infect. 2012;25:1–6. doi: 10.1017/S0950268812001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faine SB, Adler B, Bolin C, Perolat P. Leptospira and Leptospirosis. 2nd ed. Melbourne, Australia: MediSci; 1999. [Google Scholar]
- 24.WHO . Human Leptospirosis: Guidance for Diagnosis, Surveillance and Control. Malta: World Health Organization; 2003. [Google Scholar]
- 25.Levett PN. Manual of Clinical Microbiology. 9th ed. Washington, DC: American Society for Microbiology Press; 2007. Leptoapira; pp. 963–970. [Google Scholar]
- 26.Faine S. Guidelines for the Control of Leptospirosis. Geneva: World Health Organization; 1982. [Google Scholar]
- 27.Cumberland PC, Everard COR, Levett PN. Assessment of the efficacy of the IgM enzyme-linked immunosorbent assay (ELISA) and microscopic agglutination test (MAT) in the diagnosis of acute leptospirosis. Am J Trop Med Hyg. 1999;61:731–734. doi: 10.4269/ajtmh.1999.61.731. [DOI] [PubMed] [Google Scholar]
- 28.Smits HL, Eapen CK, Sugathan S, Kuriakose M, Gasem MH, Yersin C, Sasaki D, Pujianto B, Vestering M, Abdoel TH, Gussenhoven GC. Lateral-flow assay for rapid serodiagnosis of human leptospirosis. Clin Diagn Lab Immunol. 2001;8:166–169. doi: 10.1128/CDLI.8.1.166-169.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Effler PV, Bogard AK, Domen HY, Katz AR, Higa HY, Sasaki DM. Evaluation of eight rapid screening tests for acute leptospirosis in Hawaii. J Clin Microbiol. 2002;40:1464–1469. doi: 10.1128/JCM.40.4.1464-1469.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levett PN, Branch SL. Evaluation of two enzyme-linked immunosorbent assay methods for detection of immunoglobulin M antibodies in acute leptospirosis. Am J Trop Med Hyg. 2002;66:745–748. doi: 10.4269/ajtmh.2002.66.745. [DOI] [PubMed] [Google Scholar]
- 31.Vijayachari P, Sugunan AP, Sehgal SC. Evaluation of Lepto Dri Dot as a rapid test for the diagnosis of leptospirosis. Epidemiol Infect. 2002;129:617–621. doi: 10.1017/s0950268802007537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bajani MD, Ashford DA, Bragg SL, Woods CW, Aye T, Spiegel RA, Plikaytis BD, Perkins BA, Phelan M, Levett PN, Weyant RS. Evaluation of four commercially available rapid serologic tests for diagnosis of leptospirosis. J Clin Microbiol. 2003;41:803–809. doi: 10.1128/JCM.41.2.803-809.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sehgal SC, Vijayachari P, Sugunan AP, Umapathi T. Field application of Lepto lateral flow for rapid diagnosis of leptospirosis. J Med Microbiol. 2003;52:897–901. doi: 10.1099/jmm.0.05064-0. [DOI] [PubMed] [Google Scholar]
- 34.McBride AJ, Athanazio DA, Reis MG, Ko AI. Leptospirosis. Curr Opin Infect Dis. 2005;18:376–386. doi: 10.1097/01.qco.0000178824.05715.2c. [DOI] [PubMed] [Google Scholar]
- 35.Blacksell SD, Smythe L, Phetsouvanh R, Dohnt M, Hartskeerl R, Symonds M, Slack A, Vongsouvath M, Davong V, Lattana O, Phongmany S, Keoluangkot V, White NJ, Day NP, Newton PN. Limited diagnostic capacity of two commercial assays for the detection of leptospira immunoglobulin M antibodies in Laos. Clin Vaccine Immunol. 2006;3:1166–1169. doi: 10.1128/CVI.00219-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desakorn V, Wuthiekanun V, Thanachartwet V, Sahassananda D, Chierakul W, Apiwattanaporn A, Day NP, Limmathurotsakul D, Peacock SJ. Accuracy of a commercial IgM ELISA for the diagnosis of human leptospirosis in Thailand. Am J Trop Med Hyg. 2012;86:524–527. doi: 10.4269/ajtmh.2012.11-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren SX, Fu G, Jiang XG, Zeng R, Miao YG, Xu H, Zhang YX, Xiong H, Lu G, Lu LF, Jiang HQ, Jia J, Tu YF, Jiang JX, Gu WY, Zhang YQ, Cai Z, Sheng HH, Yin HF, Zhang Y, Zhu GF, Wan M, Huang HL, Qian Z, Wang SY, Ma W, Yao ZJ, Shen Y, Qiang BQ, Xia QC, Guo XK, Danchin A, Saint Girons I, Somerville RL, Wen YM, Shi MH, Chen Z, Xu JG, Zhao GP. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature. 2003;422:888–893. doi: 10.1038/nature01597. [DOI] [PubMed] [Google Scholar]
- 38.Nascimento AL, Ko AI, Martins EA, Monteiro-Vitorello CB, Ho PL, Haake DA, Verjovski-Almeida S, Hartskeerl RA, Marques MV, Oliveira MC, Menck CF, Leite LC, Carrer H, Coutinho LL, Degrave WM, Dellagostin OA, El-Dorry H, Ferro ES, Ferro MI, Furlan LR, Gamberini M, Giglioti EA, Góes-Neto A, Goldman GH, Goldman MH, Harakava R, Jerônimo SM, Junqueira-de-Azevedo IL, Kimura ET, Kuramae EE, Lemos EG, Lemos MV, Marino CL, Nunes LR, de Oliveira RC, Pereira GG, Reis MS, Schriefer A, Siqueira WJ, Sommer P, Tsai SM, Simpson AJ, Ferro JA, Camargo LE, Kitajima JP, Setubal JC, Van Sluys MA. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J Bacteriol. 2004;186:2164–2172. doi: 10.1128/JB.186.7.2164-2172.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bulach DM, Zuerner RL, Wilson P, Seemann T, McGrath A, Cullen PA, Davis J, Johnson M, Kuczek E, Alt DP, Peterson-Burch B, Coppel RL, Rood JI, Davies JK, Adler B. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc Natl Acad Sci USA. 2006;103:14560–14565. doi: 10.1073/pnas.0603979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Picardeau M, Bulach DM, Bouchier C, Zuerner RL, Zidane N, Wilson PJ, Creno S, Kuczek ES, Bommezzadri S, Davis JC, McGrath A, Johnson MJ, Boursaux-Eude C, Seemann T, Rouy Z, Coppel RL, Rood JI, Lajus A, Davies JK, Médigue C, Adler B. Genome sequence of the saprophyte leptospira biflexa provided insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS One. 2008;3:e1607. doi: 10.1371/journal.pone.0001607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, Levett PN, Gilman RH, Willig MR, Gotuzzo E, Vinetz JM. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis. 2003;3:757–771. doi: 10.1016/s1473-3099(03)00830-2. [DOI] [PubMed] [Google Scholar]
- 42.Haake DA, Chao G, Zuerner RL, Barnett JK, Barnett D, Mazel M, Matsunaga J, Levett PN, Bolin CA. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect Immun. 2000;68:2276–2285. doi: 10.1128/iai.68.4.2276-2285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shang ES, Summers TA, Haake DA. Molecular cloning and sequence analysis of the gene coding LipL41, a surface-exposed lipoprotein of pathogenic Leptospira species. Infect Immun. 1996;64:2322–2330. doi: 10.1128/iai.64.6.2322-2330.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsunaga J, Barocchi MA, Croda J, Young TA, Sanchez Y, Siqueira I, Bolin CA, Reis MG, Riley LW, Haake DA, Ko AI. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol Microbiol. 2003;49:929–945. doi: 10.1046/j.1365-2958.2003.03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cullen PA, Xu X, Matsunaga J, Sanchez Y, Ko AI, Haake DA, Adler B. Surfaceome of Leptospira spp. Infect Immun. 2005;73:4853–4863. doi: 10.1128/IAI.73.8.4853-4863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mcbride AJ, Cerqueira GM, Suchard MA, Moreira AN, Zuerner RL, Reis MG, Haake DA, Ko AI, Dellagostin OA. Genetic diversity of the leptospiral immunoglobulin-like (Lig) genes in pathogenic Leptospira spp. Infect Genet Evol. 2009;9:196–205. doi: 10.1016/j.meegid.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palaniappan RU, Chang YF, Jusuf SS, Artiushin S, Timoney JF, McDonough SP, Barr SC, Divers TJ, Simpson KW, McDonough PL, Mohammed HO. Cloning and molecular characterization of an immunogenic LigA protein of Leptospira interrogans. Infect Immun. 2002;70:5924–5930. doi: 10.1128/IAI.70.11.5924-5930.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doungchawee G, Kositanont U, Niwetpathomwat A, Inwisai T, Sagarasaeranee P, Haake DA. Early diadnosis of leptospirosis by immunoglobulin M immunoblot testing. Clin Vaccine Immunol. 2008;15:492–498. doi: 10.1128/CVI.00152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 50.Koizumi N, Watanabe H. Leptospiral immunoglobulin-like proteins elicit protective immunity. Vaccine. 2004;22:1545–1552. doi: 10.1016/j.vaccine.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 51.Chappel RJ, Goris M, Palmer MF, Hartskeerl RA. Impact of proficiency testing on results of the microscopic agglutination test for diagnosis of leptospirosis. J Clin Microbiol. 2004;42:5484–5488. doi: 10.1128/JCM.42.12.5484-5488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guerreiro H, Croda J, Flannery B, Mazel M, Reis MG, Levett PN, Ko AI, Haake DA. Leptospiral proteins recognized during the humoral immune response to leptospirosis in humans. Infect Immun. 2001;69:4958–4968. doi: 10.1128/IAI.69.8.4958-4968.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flannery B, Costa D, Carvalho FP, Guerreiro H, Matsunaga J, DaSilva ED, Ferreira AGP, Riley LW, Reis MG, Haake DA, Ko AI. Evaluation of recombinant Leptospira antigen-based enzyme-linked immunosorbent assays for the serodiagnosis of leptospirosis. J Clin Microbiol. 2001;39:3303–3310. doi: 10.1128/JCM.39.9.3303-3310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chapman AJ, Adler B, Faine S. Antigens recognized by the human immune response to infection with Leptospira interrogans serovar hardjo. J Med Microbiol. 1988;25:269–278. doi: 10.1099/00222615-25-4-269. [DOI] [PubMed] [Google Scholar]
- 55.Chapman AJ, Everard CO, Faine S, Adler B. Antigens recognized by human immune response to severe leptospirosis in Barbados. Epidemiol Infect. 1991;107:143–155. doi: 10.1017/s0950268800048779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adler B, Murphy AM, Locarnini SA, Faine S. Detection of specific antileptospiral immunoglobulins M and G in human serum by solid-phase enzyme-linked immunosorbent assay. J Clin Microbiol. 1980;11:452–457. doi: 10.1128/jcm.11.5.452-457.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Terpstra WJ, Ligthart GS, Schoone GJ. ELISA for the detection of specific IgM and IgG in human leptospirosis. J Gen Microbiol. 1985;131:377–385. doi: 10.1099/00221287-131-2-377. [DOI] [PubMed] [Google Scholar]
- 58.Engstrom SM, Shoop E, Johnson RC. Immunoblot interpretation criteria for serodiagnosis of early lyme disease. J Clin Microbiol. 1995;55:419–427. doi: 10.1128/jcm.33.2.419-427.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Magnarelli LA, Ijdo JW, Padula SJ, Flavelli RA, Fikrig E. Serologic diagnosis of Lyme borreliosis by using enzyme-linked immunosorbent assays with recombinant antigens. J Clin Microbiol. 2000;38:1735–1739. doi: 10.1128/jcm.38.5.1735-1739.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmidt BL, Edjlalipour M, Luger A. Comparative evaluation of nine different enzyme-linked immunosorbent assays for determination of antibodies against Treponema pallidum in patients with primary syphilis. J Clin Microbiol. 2000;38:1279–1282. doi: 10.1128/jcm.38.3.1279-1282.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]